Abstract
Age-related macular degeneration (AMD) is a progressive blinding disease and represents the leading cause of visual impairment in the aging population. AMD affects central vision which impairs one’s ability to drive, read and recognize faces. There is no cure for this disease and current treatment modalities for the exudative form of the disease require repeated intravitreal injections which may be painful, are incompletely efficacious, and represent a significant treatment burden for both the patient and physician. As such, AMD represents a significant and important clinical problem.
It is anticipated that in three years’ time, 196 million individuals will be affected with AMD. Over 250 billion dollars per year are spent on care for AMD patients in the US. Over half of the heritability is explained by two major loci, thus AMD is considered the most well genetically defined of the complex disorders. A recent GWAS on 43,566 subjects identified novel loci and pathways associated with AMD risk, which has provided an excellent platform for additional functional studies. Genetic variants have been investigated, particularly with respect to anti-VEGF treatment, however to date, no pharmacogenomic associations have been consistently identified across these studies. It may be that if the goal of personalized medicine is to be realized and biomarkers are to have predictive value for determining the magnitude of risk for AMD at the genetic level, one will need to examine the relationships between these pathways across disease state and relative to modifiable risk factors such as hypertension, smoking, body mass index, and hypercholesterolemia. Further studies investigating protective alleles in populations with low AMD prevalence may lead to this goal.
Introduction
Age-related macular degeneration (AMD) is a progressive blinding disease and is the leading cause of visual impairment in the aging population, especially in those over 55 years of age, for which there remains no cures. In 3 years’ time, an anticipated 196 million individuals will be affected with AMD (1). As of 2013, 11 million new cases were being diagnosed per year in the United States alone (2), this is more than double the number of Alzheimer disease cases diagnosed per year (3). The yearly cost for direct health care due to AMD in the U.S. is $255 billion, which is almost half the direct cost for all vision loss ($513 billion), making AMD the leading cause of visual disability in the developed world and the third globally (4). AMD compromises one’s central, fine vision, which impairs the ability to drive, recognize faces and read. As we age, changes take place in the retina, and include small hard drusen, which are considered part of normal aging and 95% of the population over the age of 43 has such changes (5). Drusen are lipoproteinaceous deposits and acellular debris. When vision becomes compromised due to these changes becoming pathophysiological accompanied by hypo and/or hyper pigmentation of the Retinal Pigment Epithelium (RPE) cells of the retina,), enlargement or increase in confluence to the drusen, the term age-related macular degeneration is used. This is considered the early/intermediate stages of AMD. In the more advanced stages of AMD, the advanced dry form is termed geographic atrophy (death of cells) and the wet or exudative form is termed neovascular AMD (this is the growth of abnormal blood vessels from beneath the retina). At present, most therapies are directed toward treatment of neovascular AMD, target established abnormal blood vessel growth through antibody-based inhibition of VEGF, and demonstrate a range of efficacy. For a small subset of patients, anti-VEGF treatment results in stable to improved visual acuity without the need for ongoing treatment. However, the majority of patients require indefinite treatment, do not regain vision, or demonstrate progression of disease despite therapies. For in depth reviews on the clinical description of AMD, please see Yonekawa (6), Miller (7), Rufai (8), Sacconi (9), and Gemenetz (10). With 22 million individuals expected to be diagnosed with AMD by 2050 (4), unless the disease can be prevented, delayed, or treated effectively, affected individuals will continue to suffer and health care costs will rise exponentially.
Risk Factors
Men and women can both be affected with AMD, though some reports show that woman are 1.3 times at greater risk for AMD (11). Age and a positive family history of AMD are the two strongest risk factors for AMD. Twin studies demonstrating greater concordance in monozygotic (37%) than dizygotic (19%) twins% (12) as well as studies demonstrating clustering of AMD in families, a hallmark of a disease with complex inheritance, were the first to underscore the genetic basis for AMD (13,14). It has been shown that an individual with a sibling or a parent with AMD is 12-27 times more susceptible than someone from the general population to develop AMD (15).
With 50% or more of the heritability of AMD already explained by two major loci harboring coding and non-coding variation at chromosomes 1q (CFH) and 10q (ARMS2/HTRA1), AMD is considered the most well genetically defined complex disorders (16–24). This is further emphasized by the largest genome wide association study to date for AMD conducted by the International age-related macular degeneration genomics consortium (IAMDGC) that was sponsored by the NEI/NIH/CIDR and comprised of 27 study partners from throughout the world (24). In the IAMDGC study of 43,566 subjects, mostly of European ancestry comprised of 17,832 controls, 16,144 advanced AMD cases and 6,657 with intermediate AMD, 52 independently associated variants spanning 34 loci were identified at genome-wide significant association (P < 5.0×10−8) (24). Though the custom chip that was interrogated had common variants, it also contained protein coding and rare variants (frequency of 0.1%). After burden testing, only rare variants in the previously associated AMD genes, CFH, CFI, and TIMP3 reached genome wide significance. However, this highlighted the importance of CFI and TIMP3 to AMD pathophysiology (25–27). All these data are available to the research community through dbGAP (https://www.ncbi.nlm.nih.gov/gap/ddb/).
Clinical utility of SNP findings: what has been done and where do we go from here?
The clinical utility and translational significance of disease associated SNPs varies by pathology. Recent work has increased our understanding of disease pathways and hence the potential clinical significance of genetic variation in AMD. In the case of the recent GWAS experiment (24), genes were found to function not only in known AMD pathways but highlighted the importance of additional pathways as well, including complement activation, collagen synthesis, lipid metabolism/cholesterol transport, receptor-mediated endocytosis, endodermal cell differentiation, and extracellular matrix organization. Of importance was that ten variants located in seven extracellular matrix genes (COL15A1, COL8A1, MMP9, PCOLCE, MMP19, CTRB1-CTRB2 and ITGA7) were associated with only advanced AMD (neovascular AMD and geographic atrophic AMD) and not intermediate AMD, suggesting that extracellular remodeling pathway(s) of the retina/RPE are activated in the development of the more advanced stages of AMD. Pathway analysis may also indicate that there may be upstream regulators/modulators for the gene/variant of interest that are important as therapeutic targets. This will further allow for greater insight into the relationship between genetic variation and disease mechanism as well as identify novel therapeutic targets. However, appropriate in vivo and in vitro models are needed to assess these relationships.
Greater understanding of multifactorial AMD risk is also advancing the future possibility of a personalized approach to AMD screening and treatment. The goal of personalized medicine is to predict individual disease risk based upon analysis of his or her environment and genetic variation. Moreover, this information is hypothesized to aid in identification of individually significant therapeutic targets and to predict a patient’s response to therapy. The use of genetic testing to predict progression to advanced AMD in the clinical setting has not been proven to improve outcomes and is not recommend for routine, general use by the American Academy of Ophthalmology (28,29). However, it has been demonstrated that adding genotype information, particularly from the CFH and ARMS2/HTRA1 loci, to phenotypic features from clinical examination, increases the precision of predicting risk of advanced AMD (30). Genotype has also been examined with respect to treatment response with inconsistent results. For example, some studies have suggested that the beneficial effect of the Age Related Eye Disease Study (AREDS) vitamin supplements (antioxidants with zinc) in reducing progression to advanced AMD was influenced by genotype (31,32). However, other analyses did not validate this finding (28,33). As stated previously, most therapeutics for AMD are directed towards the advanced neovascular form of AMD. Several studies have investigated the relationship of genetic variants, particularly in CFH, ARMS2/HTRA1, and the VEGF pathway, and a patient's response to anti-VEGF treatment. However to date, no pharmacogenomic associations have been consistently identified for anti-VEGF therapies across these studies. An initial GWAS of treatment response to anti-vegf therapy for neovascular AMD led to the identification of a variant in the olfactory receptor gene, OR52B4 (42). However, additional studies will need to be performed to replicate such results.
As stated earlier, AMD is etiologically complex, meaning it has many epidemiologic and genetic factors that influence susceptibility to risk. While some of these epidemiological risk factors are modifiable, and include body-mass index (BMI), smoking cigarettes and blood lipid and cholesterol levels, other factors are not, and include genotype at a given risk locus, sex, ethnicity, and age (for reviews please see Klein (43) and DeAngelis and Pennington (44)). Further insight into AMD mechanism and risk mitigation can be gained by examining similarly complex disease states that demonstrate overlapping pathophysiology with AMD including Alzheimer disease and cardiovascular disease (44–60). It may be that if biomarkers are to have predictive value for determining the magnitude of risk for AMD at the genetic level, one will need to examine the relationships between these pathways across disease state and relative to modifiable risk factors such as hypertension, smoking, body mass index, and hypercholesterolemia. One would hypothesize that if similar pathways, for example, lipid metabolism/cholesterol biosynthesis, are involved in the pathophysiology of AMD, then proteins that are intrinsic to these pathways would function differently between patients with and without AMD. It is the investigation of those differences that would lead to personalized, targeted treatment pathways as depicted in Figure 1.
Figure 1.
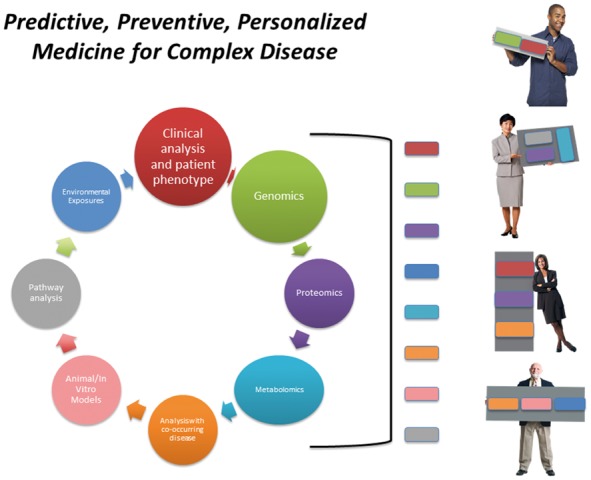
Predictive, preventive, personalized medicine. Model for approaching the integrative analysis of patient phenotype in the context of individual ‘omic’, environmental and experimental data. The significance of each of these factors for an individual patient may vary as demonstrated.
In a recent study by Burgess and Davey Smith (61), Mendelian randomization was employed to assess known lipid gene associations with AMD risk on 33,536 individuals utilizing coronary artery disease as a positive control and Alzheimer disease as a negative control. Specifically in this instance, a Mendelian randomization approach was utilized to measure 185 variants that included gene proxies for lipid therapy, spanning 157 genes to examine the effect of HDL cholesterol, LDL cholesterol and triglycerides on AMD risk. They found that variants in the CETP gene (also identified in the GWAS by Fritsche et al. (24)), were associated with AMD risk, and that inhibiting CETP to increase HDL-cholesterol levels may increase AMD risk. Thus, they concluded that the mechanism for the known HDL-cholesterol/AMD association (62) is through the modulation of CETP. This clever study provides a foundation to test a feasible in vivo or in vitro model of AMD for potential AMD therapy.
Currently, both in vivo and in vitro models for the study of AMD are limiting. There have been some successes at recapitulating aspects of the AMD phenotype in various mouse models of AMD (63–69). Though the mouse has a retina and RPE, humans are the only species with a macula, the 2mm region in the retina, where AMD manifests. Additionally, it is unclear if AMD is a localized disease (occurring only in the affected eye tissues), a systemic disease, or if there are aspects of the disease that are a combination of both. One area of intense effort to further the field, particularly in the area of multi-omic approaches for AMD, is the collection and assessment of fresh donor eye tissue. We and others are actively engaged in creating well phenotyped repositories with short duration post mortem time (6 h or less) (70). Such tissues can then be used for studies including epigenetics, RNASeq, microRNAs, metabolomics, and proteomics. Results can then be tied back to the donors to assay biomarkers in the serum or plasma. Results from these studies can then be widely shared with the scientific research community.
Protective alleles and their role in AMD
Much interest has been given to the role of protective alleles in complex disease (for review please see Harper (71)) and that it has been hypothesized that protective alleles may prove to be a better therapeutic target than alleles that increase disease risk.
Prevalence of AMD varies greatly by ethnicity, with whites of European descent having the greatest disease burden. A recent study by Wong et al. (1) calculated pooled prevalence of ethnically diverse population-based studies of AMD (mapped to an age range of 45–85 years) and confirmed that the prevalence was greatest among those of European descent at 12.3% - 30% with increasing age. Disease burden, although slightly less, is still significant amongst Hispanics (10.4%), Africans (7.5%) and Asians (7.4%) (1). Still others have estimated the disease burden within the United States as lower with non-Hispanic White Europeans having the highest at almost 7.3% and African-Americans at 2.4% (72). Regardless, it is clear that the prevalence of AMD varies by ethnicity and racial group and therefore the role of genetic variants, environmental exposures, and their interplay in AMD susceptibility will likely as well.
The Population Architecture Using Genomics and Epidemiology (PAGE) study demonstrated that AMD risk appears to differ with respect to lipid metabolism and cholesterol-related genes in Mexican Americans, Asian Americans, African Americans and non-Hispanic White Europeans when all types of AMD were examined (73). In fact, none of the major risk variants for AMD i.e. ARMS2 or CFH were significant in the non-white European populations after correction for multiple testing (73). Similarly, Cheng et al. (74) found a different variant in a known CETP lipid gene (previously associated with AMD risk in whites of European descent) as well as novel AMD lipid/cholesterol genes associated with AMD risk in East Asians. Moreover, the CETP risk variant was found to interact with high serum HDL levels in individuals of Japanese ancestry and Chinese from Singapore (74). While these examples illustrate the differences in genetic variation and allele frequency at important loci they do not explain why certain ethnicities may have a slightly lower disease prevalence. It may be that protective alleles may be more important in different populations. For example, in a study of individuals from Timor-Leste, it was found that AMD occurs at a very low frequency (< 1%) (75). When examining AMD-disease associated SNPs, those with AMD in this cohort did not have risk alleles at the two most associated AMD loci, CFH Y402H (rs1061170) and ARMS2/HTRA1 A69S (rs10490924). However, the Timor population as a whole had increased frequency of protective alleles at CFH (19), CFHR2 (76), and C2 (77) underscoring the importance of those alleles. As others have shown, for other complex disorders protective alleles may be as important, if not more important in than risk alleles (71). Studying other populations where AMD prevalence is low in the context of environmental exposures along with genetic variants will require well-powered studies to move forward for the goal of personalized medicine to be fully realized.
Conflict of Interest statement. None declared.
Funding
This research was supported by the Macular Degeneration Foundation; CARL MARSHALL REEVES & MILDRED ALMEN REEVES FOUNDATION, INC.; National Institutes of Health (EY014800), and an Unrestricted Grant from Research to Prevent Blindness, Inc., New York, NY, to the Department of Ophthalmology & Visual Sciences, University of Utah. Primary Children’s Health Foundation Early Career Development Award; The Eunice Kennedy Shriver National Institute of Child Health and Human Development and the Office of Research on Women’s Health of the National Institute of Health under Award Number K12HD085852.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.-Y., Wong T.Y. (2014) Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health, 2, e106–e116. [DOI] [PubMed] [Google Scholar]
- 2. Rein D.B., Wittenborn J.S., Zhang X., Honeycutt A.A., Lesesne S.B., Saaddine J. (2009) Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatment. Arch. Ophthalmol., 127, 533–540. [DOI] [PubMed] [Google Scholar]
- 3. Aging N.I. (2011) Alzheimer’s disease fact sheet. National Institute on Aging. https://www.nia.nih.gov/alzheimers/publication/alzheimers-disease-fact-sheet; date last accessed June 6, 2017. [Google Scholar]
- 4. Age-Related Macular Degeneration: Facts & Figures (2015) BrightFocus Foundation.http://www.brightfocus.org/macular/article/age-related-macular-facts-figures; date last accessed June 6, 2017.
- 5. Klein R., Klein B.E., Linton K.L. (1992) Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology, 99, 933–943. [DOI] [PubMed] [Google Scholar]
- 6. Yonekawa Y., Miller J.W., Kim I.K. (2015) Age-related macular degeneration: advances in management and diagnosis. J. Clin. Med., 4, 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller J.W. (2013) Age-related macular degeneration revisited–piecing the puzzle: the LXIX Edward Jackson memorial lecture. Am. J. Ophthalmol., 155, 1–35.e13. [DOI] [PubMed] [Google Scholar]
- 8. Rufai S.R., Almuhtaseb H., Paul R.M., Stuart B.L., Kendrick T., Lee H., Lotery A.J. (2017) A systematic review to assess the ‘treat-and-extend’ dosing regimen for neovascular age-related macular degeneration using ranibizumab. Eye (Lond), 10.1038/eye.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sacconi R., Corbelli E., Querques L., Bandello F., Querques G. (2017) A review of current and future management of geographic atrophy. Ophthalmol. Ther., 6, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gemenetzi M., Patel P.J. (2017) A systematic review of the treat and extend treatment regimen with anti-VEGF agents for neovascular age-related macular degeneration. Ophthalmol. Ther., 6, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudnicka A.R., Kapetanakis V.V., Jarrar Z., Wathern A.K., Wormald R., Fletcher A.E., Cook D.G., Owen C.G. (2015) Incidence of late-stage age-related macular degeneration in American Whites: systematic review and meta-analysis. Am. J. Ophthalmol., 160, 85–93.e3. [DOI] [PubMed] [Google Scholar]
- 12. Hammond C.J., Webster A.R., Snieder H., Bird A.C., Gilbert C.E., Spector T.D. (2002) Genetic influence on early age-related maculopathy: a twin study. Ophthalmology, 109, 730–736. [DOI] [PubMed] [Google Scholar]
- 13. Silvestri G., Johnston P.B., Hughes A.E. (1994) Is genetic predisposition an important risk factor in age-related macular degeneration?. Eye (Lond), 8 (Pt 5), 564–568. [DOI] [PubMed] [Google Scholar]
- 14. Seddon J.M., Ajani U.A., Mitchell B.D. (1997) Familial aggregation of age-related maculopathy. Am. J. Ophthalmol., 123, 199–206. [DOI] [PubMed] [Google Scholar]
- 15. Shahid H., Khan J.C., Cipriani V., Sepp T., Matharu B.K., Bunce C., Harding S.P., Clayton D.G., Moore A.T., Yates J.R.W.. et al. (2012) Age-related macular degeneration: the importance of family history as a risk factor. Br. J. Ophthalmol., 96, 427–431. [DOI] [PubMed] [Google Scholar]
- 16. Klein R.J., Zeiss C., Chew E.Y., Tsai J.-Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T.. et al. (2005) Complement factor H polymorphism in age-related macular degeneration. Science, 308, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards A.O., Ritter R. 3rd, Abel K.J., Manning A., Panhuysen C., Farrer L.A. (2005) Complement factor H polymorphism and age-related macular degeneration. Science, 308, 421–424. [DOI] [PubMed] [Google Scholar]
- 18. Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R.. et al. (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science, 308, 419–421. [DOI] [PubMed] [Google Scholar]
- 19. Hageman G.S., Anderson D.H., Johnson L.V., Hancox L.S., Taiber A.J., Hardisty L.I., Hageman J.L., Stockman H.A., Borchardt J.D., Gehrs K.M.. et al. (2005) A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A, 102, 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rivera A., Fisher S.A., Fritsche L.G., Keilhauer C.N., Lichtner P., Meitinger T., Weber B.H.F. (2005) Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet., 14, 3227–3236. [DOI] [PubMed] [Google Scholar]
- 21. Dewan A., Liu M., Hartman S., Zhang S.S.-M., Liu D.T.L., Zhao C., Tam P.O.S., Chan W.M., Lam D.S.C., Snyder M.. et al. (2006) HTRA1 promoter polymorphism in wet age-related macular degeneration. Science, 314, 989–992. [DOI] [PubMed] [Google Scholar]
- 22. Yang Z., Camp N.J., Sun H., Tong Z., Gibbs D., Cameron D.J., Chen H., Zhao Y., Pearson E., Li X.. et al. (2006) A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science, 314, 992–993. [DOI] [PubMed] [Google Scholar]
- 23. Deangelis M.M., Ji F., Adams S., Morrison M.A., Harring A.J., Sweeney M.O., Capone A. Jr, Miller J.W., Dryja T.P., Ott J.. et al. (2008) Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology, 115, 1209–1215.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fritsche L.G., Igl W., Bailey J.N.C., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M.. et al. (2016) A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet., 48, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J., Ohno-Matsui K., Yoshida T., Kojima A., Shimada N., Nakahama K., Safranova O., Iwata N., Saido T.C., Mochizuki M.. et al. (2008) Altered function of factor I caused by amyloid beta: implication for pathogenesis of age-related macular degeneration from Drusen. J. Immunol., 181, 712–720. [DOI] [PubMed] [Google Scholar]
- 26. Fagerness J.A., Maller J.B., Neale B.M., Reynolds R.C., Daly M.J., Seddon J.M. (2009) Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet., 17, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen W., Stambolian D., Edwards A.O., Branham K.E., Othman M., Jakobsdottir J., Tosakulwong N., Pericak-Vance M.A., Campochiaro P.A., Klein M.L.. et al. (2010) Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A, 107, 7401–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chew E.Y. Klein M.L. Clemons T.E. Agrón E. and Abecasis G.R. (2015) Genetic testing in persons with agerelated macular degeneration and the use of the AREDS supplements: to test or not to test.. Ophthalmology, 122, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chew E.Y. (2017) Nutrition, genes, and age-related macular degeneration: what have we learned from the trials?. Ophthalmologica, 10.1159/000473865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perlee L.T., Bansal A.T., Gehrs K., Heier J.S., Csaky K., Allikmets R., Oeth P., Paladino T., Farkas D.H., Rawlings P.L.. et al. (2013) Inclusion of genotype with fundus phenotype improves accuracy of predicting choroidal neovascularization and geographic atrophy. Ophthalmology, 120, 1880–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Awh C.C., Hawken S., Zanke B.W. (2015) Treatment response to antioxidants and zinc based on CFH and ARMS2 genetic risk allele number in the Age-Related Eye Disease Study. Ophthalmology, 122, 162–169. [DOI] [PubMed] [Google Scholar]
- 32. Awh C.C., Zanke B.W., Re: Chew. et al. (2015) No clinically significant association between CFH and ARMS2 genotypes and response to nutritional supplements: AREDS report number 38 (Ophthalmology 2014;121:2173-80). Ophthalmology, 122, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chew E.Y., Klein M.L., Clemons T.E., Agrón E., Ratnapriya R., Edwards A.O., Fritsche L.G., Swaroop A., Abecasis G.R. and Age-Related Eye Disease Study Research Group (2014) No clinically significant association between CFH and ARMS2 genotypes and response to nutritional supplements: AREDS report number 38. Ophthalmology, 121, 2173–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchihashi T., Mori K., Horie-Inoue K., Gehlbach P.L., Kabasawa S., Takita H., Ueyama K., Okazaki Y., Inoue S., Awata T.. et al. (2011) Complement factor H and high-temperature requirement A-1 genotypes and treatment response of age-related macular degeneration. Ophthalmology, 118, 93–100. [DOI] [PubMed] [Google Scholar]
- 35. McKibbin M., Ali M., Bansal S., Baxter P.D., West K., Williams G., Cassidy F., Inglehearn C.F. (2012) CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br. J. Ophthalmol., 96, 208–212. [DOI] [PubMed] [Google Scholar]
- 36. Finger R.P., Wickremasinghe S.S., Baird P.N., Guymer R.H. (2014) Predictors of anti-VEGF treatment response in neovascular age-related macular degeneration. Surv. Ophthalmol., 59, 1–18. [DOI] [PubMed] [Google Scholar]
- 37. Hagstrom S.A., Ying G., Pauer G.J.T., Sturgill-Short G.M., Huang J., Maguire M.G., Martin D.F. and Comparison of Age-Related Macular Degeneration Treatments Trials (CATT) Research Group (2014) VEGFA and VEGFR2 gene polymorphisms and response to anti-vascular endothelial growth factor therapy: comparison of age-related macular degeneration treatments trials (CATT). JAMA Ophthalmol., 132, 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hermann M.M., van Asten F., Muether P.S., Smailhodzic D., Lichtner P., Hoyng C.B., Kirchhof B., Grefkes C., den Hollander A.I., Fauser S. (2014) Polymorphisms in vascular endothelial growth factor receptor 2 are associated with better response rates to ranibizumab treatment in age-related macular degeneration. Ophthalmology, 121, 905–910. [DOI] [PubMed] [Google Scholar]
- 39. Cruz-Gonzalez F., Cabrillo-Estévez L., López-Valverde G., Cieza-Borrella C., Hernández-Galilea E., González-Sarmiento R. (2014) Predictive value of VEGF A and VEGFR2 polymorphisms in the response to intravitreal ranibizumab treatment for wet AMD. Graefes Arch. Clin. Exp. Ophthalmol., 252, 469–475. [DOI] [PubMed] [Google Scholar]
- 40. Chen G., Tzekov R., Li W., Jiang F., Mao S., Tong Y. (2015) Pharmacogenetics of complement factor H Y402H polymorphism and treatment of neovascular AMD with anti-VEGF agents: a meta-analysis. Sci. Rep., 5, 14517.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kepez Yildiz B., Ozdek S., Ergun M.A., Ergun S., Yaylacioglu Tuncay F., Elbeg S. (2016) CFH Y402H and VEGF Polymorphisms and anti-VEGF treatment response in exudative age-related macular degeneration. Ophthalmic Res., 56, 132–138. [DOI] [PubMed] [Google Scholar]
- 42. Riaz M., Lorés-Motta L., Richardson A.J., Lu Y., Montgomery G., Omar A., Koenekoop R.K., Chen J., Muether P., Altay L.. et al. (2016) GWAS study using DNA pooling strategy identifies association of variant rs4910623 in OR52B4 gene with anti-VEGF treatment response in age-related macular degeneration. Sci. Rep., 6, 37924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klein R. (2007) Overview of progress in the epidemiology of age-related macular degeneration. Ophthalmic Epidemiol., 14, 184–187. [DOI] [PubMed] [Google Scholar]
- 44. Pennington K.L. and DeAngelis M.M. (2016) Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye and Vision.,3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohno-Matsui K. (2011) Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog. Retin. Eye Res., 30, 217–238. [DOI] [PubMed] [Google Scholar]
- 46. Logue M.W., Schu M., Vardarajan B.N., Farrell J., Lunetta K.L., Jun G., Baldwin C.T., Deangelis M.M., Farrer L.A. (2014) Search for age-related macular degeneration risk variants in Alzheimer disease genes and pathways. Neurobiol. Aging, 35, 1510.e7–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keenan T.D.L., Goldacre R., Goldacre M.J. (2014) Associations between age-related macular degeneration, Alzheimer disease, and dementia: record linkage study of hospital admissions. JAMA Ophthalmol., 132, 63–68. [DOI] [PubMed] [Google Scholar]
- 48. Stefanova N.A., Kozhevnikova O.S., Vitovtov A.O., Maksimova K.Y., Logvinov S.V., Rudnitskaya E.A., Korbolina E.E., Muraleva N.A., Kolosova N.G. (2014) Senescence-accelerated OXYS rats: a model of age-related cognitive decline with relevance to abnormalities in Alzheimer disease. Cell Cycle, 13, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williams M.A., Silvestri V., Craig D., Passmore A.P., Silvestri G. (2014) The prevalence of age-related macular degeneration in Alzheimer’s disease. J. Alzheimers Dis., 42, 909–914. [DOI] [PubMed] [Google Scholar]
- 50. Butler J.M., Sharif U., Ali M., McKibbin M., Thompson J.P., Gale R., Yang Y.C., Inglehearn C., Paraoan L. (2015) A missense variant in CST3 exerts a recessive effect on susceptibility to age-related macular degeneration resembling its association with Alzheimer’s disease. Hum. Genet., 134, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Romano G.L., Platania C.B.M., Drago F., Salomone S., Ragusa M., Barbagallo C., Di Pietro C., Purrello M., Reibaldi M., Avitabile T.. et al. (2017) Retinal and Circulating miRNAs in Age-Related Macular Degeneration: An In vivo Animal and Human Study. Front Pharmacol., 8, 168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu J., Uchino M., Sastry S.M., Schaumberg D.A. (2014) Age-related macular degeneration and the incidence of cardiovascular disease: a systematic review and meta-analysis. PLoS One, 9, e89600.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feehan M., Hartman J., Durante R., Morrison M.A., Miller J.W., Kim I.K., DeAngelis M.M. (2011) Identifying subtypes of patients with neovascular age-related macular degeneration by genotypic and cardiovascular risk characteristics. BMC Med. Genet., 12, 83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan J.S.L., Wang J.J., Liew G., Rochtchina E., Mitchell P. (2008) Age-related macular degeneration and mortality from cardiovascular disease or stroke. Br. J. Ophthalmol., 92, 509–512. [DOI] [PubMed] [Google Scholar]
- 55. Fernandez A.B., Wong T.Y., Klein R., Collins D., Burke G., Cotch M.F., Klein B., Sadeghi M.M., Chen J. (2012) Age-related macular degeneration and incident cardiovascular disease: the Multi-Ethnic Study of atherosclerosis. Ophthalmology, 119, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hogg R.E., Woodside J.V., Gilchrist S.E.C.M., Graydon R., Fletcher A.E., Chan W., Knox A., Cartmill B., Chakravarthy U. (2008) Cardiovascular disease and hypertension are strong risk factors for choroidal neovascularization. Ophthalmology, 115, 1046–1052.e2. [DOI] [PubMed] [Google Scholar]
- 57. Hyman L., Schachat A.P., He Q., Leske M.C.. Age-Related Macular Degeneration Risk Factors Study Group (2000) Hypertension, cardiovascular disease, and age-related macular degeneration. Arch. Ophthalmol., 118, 351–358. [DOI] [PubMed] [Google Scholar]
- 58. Grassmann F., Kiel C., Zimmermann M.E., Gorski M., Grassmann V., Stark K. International AMD Genomics Consortium (IAMDGC) Heid I.M., Weber B.H.F. (2017) Genetic pleiotropy between age-related macular degeneration and 16 complex diseases and traits. Genome Med., 9, 29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Delcourt C., Michel F., Colvez A., Lacroux A., Delage M., Vernet M.H. and POLA Study Group (2001) Associations of cardiovascular disease and its risk factors with age-related macular degeneration: the POLA study. Ophthalmic Epidemiol., 8, 237–249. [DOI] [PubMed] [Google Scholar]
- 60. Cheung C.M.G., Wong T.Y. (2014) Is age-related macular degeneration a manifestation of systemic disease? New prospects for early intervention and treatment. J. Intern. Med., 276, 140–153. [DOI] [PubMed] [Google Scholar]
- 61. Burgess S., Davey Smith G. (2017) Mendelian randomization implicates high-density lipoprotein cholesterol-associated mechanisms in etiology of age-related macular degeneration. Ophthalmology, 10.1016/j.ophtha.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Leeuwen R., Klaver C.C.W., Vingerling J.R., Hofman A., van Duijn C.M., Stricker B.H.C., de Jong P.T.V.M. (2004) Cholesterol and age-related macular degeneration: is there a link?. Am. J. Ophthalmol., 137, 750–752. [DOI] [PubMed] [Google Scholar]
- 63. Ding J.-D., Johnson L.V., Herrmann R., Farsiu S., Smith S.G., Groelle M., Mace B.E., Sullivan P., Jamison J.A., Kelly U.. et al. (2011) Anti-amyloid therapy protects against retinal pigmented epithelium damage and vision loss in a model of age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A, 108, E279–E287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang L., Kondo N., Cano M., Ebrahimi K., Yoshida T., Barnett B.P., Biswal S., Handa J.T. (2014) Nrf2 signaling modulates cigarette smoke-induced complement activation in retinal pigmented epithelial cells. Free Radic. Biol. Med., 70, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hasegawa E., Sweigard H., Husain D., Olivares A.M., Chang B., Smith K.E., Birsner A.E., D’Amato R.J., Michaud N.A., Han Y.. et al. (2014) Characterization of a spontaneous retinal neovascular mouse model. PLoS One, 9, e106507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Veleri S., Lazar C.H., Chang B., Sieving P.A., Banin E., Swaroop A. (2015) Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis. Model Mech., 8, 109–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ding J.-D., Kelly U., Groelle M., Christenbury J.G., Zhang W., Bowes Rickman C. (2014) The role of complement dysregulation in AMD mouse models. Adv. Exp. Med. Biol., 801, 213–219. [DOI] [PubMed] [Google Scholar]
- 68. Ramkumar H.L., Zhang J., Chan C.-C. (2010) Retinal ultrastructure of murine models of dry age-related macular degeneration (AMD). Prog. Retin. Eye Res., 29, 169–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Obert E., Strauss R., Brandon C., Grek C., Ghatnekar G., Gourdie R., Rohrer B. (2017) Targeting the tight junction protein, zonula occludens-1, with the connexin43 mimetic peptide, αCT1, reduces VEGF-dependent RPE pathophysiology. J. Mol. Med., 95, 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morgan D.J., DeAngelis M.M. (2015) Differential gene expression in age-related macular degeneration. Cold Spring Harb. Perspect. Med., 5, a017210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Harper A.R., Nayee S., Topol E.J. (2015) Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet., 16, 689–701. [DOI] [PubMed] [Google Scholar]
- 72. Klein R., Chou C.-F., Klein B.E.K., Zhang X., Meuer S.M., Saaddine J.B. (2011) Prevalence of age-related macular degeneration in the US population. Arch. Ophthalmol., 129, 75–80. [DOI] [PubMed] [Google Scholar]
- 73. Restrepo N.A., Spencer K.M., Goodloe R., Garrett T.A., Heiss G., Bůžková P., Jorgensen N., Jensen R.A., Matise T.C., Hindorff L.A.. et al. (2014) Genetic determinants of age-related macular degeneration in diverse populations from the PAGE Study. Invest Ophthalmol. Vis. Sci., 10.1167/iovs.14-14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cheng C.-Y., Yamashiro K., Jia Chen L., Ahn J., Huang L., Huang L., Cheung C.M.G., Miyake M., Cackett P.D., Yeo I.Y.. et al. (2015) New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat. Commun., 6, 6063.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morrison M.A., Magalhaes T.R., Ramke J., Smith S.E., Ennis S., Simpson C.L., Portas L., Murgia F., Ahn J., Dardenne C.. et al. (2015) Ancestry of the Timorese: age-related macular degeneration associated genotype and allele sharing among human populations from throughout the world. Front. Genet., 6, 238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang H., Morrison M.A., Dewan A., Adams S., Andreoli M., Huynh N., Regan M., Brown A., Miller J.W., Kim I.K.. et al. (2008) The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med. Genet., 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gold B., Merriam J.E., Zernant J., Hancox L.S., Taiber A.J., Gehrs K., Cramer K., Neel J., Bergeron J., Barile G.R.. et al. (2006) Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet., 38, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]


