Abstract
Next-generation sequencing (NGS) studies are becoming routinely used for the detection of novel and clinically actionable DNA variants at a pangenomic scale. Such analyses are now used in the clinical practice to enable precision medicine. Formalin-fixed paraffin-embedded (FFPE) tissues are still one of the most abundant source of cancer clinical specimen, unfortunately this method of preparation is known to degrade DNA and therefore compromise subsequent analysis. Some studies have reported that variant detection can be performed on FFPE samples sequenced with NGS techniques, but few or none have done an in-depth coverage analysis and compared the influence of different state-of-the-art FFPE DNA extraction kits on the quality of the variant calling. Here, we generated 42 human whole-exome sequencing data sets from fresh-frozen (FF) and FFPE samples. These samples include normal and tumor tissues from two different organs (liver and colon), that we extracted with three different FFPE extraction kits (QIAamp DNA FFPE Tissue kit and GeneRead DNA FFPE kit from Qiagen, Maxwell™ RSC DNA FFPE Kit from Promega). We determined the rate of concordance of called variants between matched FF and FFPE samples on all common variants (representing at least 86% of the total number of variants for SNVs). The concordance rate is very high between all matched FF / FFPE pairs, with equivalent values for the three kits we analyzed. On the other hand, when looking at the difference between the total number of variants in FF and FFPE, we find a significant variation for the three different FFPE DNA extraction kits. Coverage analysis shows that FFPE samples have less good indicators than FF samples, yet the coverage quality remains above accepted thresholds. We detect limited but statistically significant variations in coverage indicator values between the three FFPE extraction kits. Globally, the GeneRead and QIAamp kits have better variant calling and coverage indicators than the Maxwell kit on the samples used in this study, although this kit performs better on some indicators and has advantages in terms of practical usage. Taken together, our results confirm the potential of FFPE samples analysis for clinical genomic studies, but also indicate that the choice of a FFPE DNA extraction kit should be done with careful testing and analysis beforehand in order to maximize the accuracy of the results.
Introduction
Next-generation sequencing (NGS) approaches have proven to be a cost-effective and relevant method for the identification of novel and clinically actionable variants across many genes in a single test [1–7]. NGS is nowadays commonly used in clinical molecular diagnostic for the detection of germline and somatic variants [4–6, 8–10]. NGS can be used to detect the full range of DNA variations, i.e. single-nucleotide variants (SNVs), insertions/deletions (INDELs), translocations and copy-number changes. The main advantage of NGS over traditional techniques, such as Sanger sequencing, is the greater level of multiplexing of genes that NGS can offer, along with the ability to detect mutations across an entire gene as opposed to PCR based methods that focus on specific single nucleotide variants.
Using NGS approaches for routine clinical testing implicates testing for the various type of samples that may be used in the laboratory and that should be generated with minimally invasive techniques for the patient. Most molecular tests are performed on fresh (and/or frozen) tissues (e.g. blood samples or biopsies), mainly because this minimizes the risk of DNA degradation in the sample. However, in most clinical molecular pathology settings, FF tissues are rare, due to the complexities of the logistic chain for the preparation, collection and storage of such samples. Instead, FFPE is the method of choice (and sometimes the gold standard method) for clinicians. FFPE specimen are much easier to prepare and to store, but it is well established that formalin fixation results in DNA damage. Formaldehyde reacts with DNA and proteins, resulting in DNA-DNA, DNA-RNA, and DNA-protein molecules that are covalently linked by methylene bridges. Formaldehyde is also known to induce oxidation and deamination reactions and the formation of cyclic bases derivatives. These chemical modifications have the potential to alter molecular testing through inhibition of enzymatic reparation of DNA or direct changes at single base or sequence levels. Furthermore, crosslinks lead to DNA fragmentation that render sequencing and analysis even more complicated [11–16].
The dramatic decrease in NGS related costs associated with the demonstrated capability of detection of clinically actionable targets and affordable large-scale computational power have triggered the creation of large projects (such as the precision medicine initiative [17]) aiming at bringing the benefits of large scale genomic analyses to patients suffering from a variety of diseases. In this context, there is a strong interest in analyses based on FFPE samples, due to their large abundance in clinical biobanks.
A number of studies have already established that NGS can be performed with DNA from FFPE samples. For instance, in one of the earliest attempts, Schweiger and colleagues showed that short reads sequencing could be applied to FFPE samples to detect genomic variations and copy number alterations in normal and tumor breast tissue, in spite of using low coverage [18]. A more recent study examined hybridization-capture of twenty-seven cancer-related genes with NGS on paired FF and FFPE samples, detecting a high degree of concordance and agreement between them [19]. Several studies have assessed the performance of various forms of DNA sequencing (whole-exome, whole-genome, targeted exon sequencing), and even tried RNA sequencing on FFPE samples [20–23]. Those studies have found a high degree of concordance between FF and FFPE samples and concluded that NGS-based analysis of FFPE samples could be used in both prospective and retrospective studies with the possibility to uncover clinically important genes. It is also worth noticing that some groups specifically tackled the question of the effect of FFPE storage time on the quality of the results. For instance, Hegegaard and colleagues found a high concordance of variants in FF and FFPE samples stored for fewer than three years [20]. Carrick et al. [23] analyzed a set of samples stored from 3 to 32 years and reported that 90% of the samples could provide data with sufficient quantity and quality for data mining regardless of storage time, although specimens stored for longer periods of time had significantly lower coverage of the target regions and lower average read depth. At last there is at least one study that used FFPE material for causal variants discovery: Bagnall and colleagues found significant variants in two genes associated to sudden death syndrome by performing exome sequencing on FFPE samples [24].
However, few studies analyzed in details the sequencing coverage for matched FF/FFPE samples and studied the impact of recent state-of-the-art DNA extraction kits that are compatible with systems allowing an automation of sample preparation. For instance, Janecka and colleagues [25] compared eight different commercial kits for preparing DNA from FFPE samples, but only analyzed the quality of the DNA obtained and did not perform variant analysis. More recently, Bonfiglio et al. [26] compared two solutions-based exome capture technologies by comparing coverage and variant detection, but did not address the problem of different extraction kits. Astolfi et al. [27] performed whole-exome sequencing on four gastrointestinal stromal tumor samples either extracted from FF or FFPE, and analyzed the quality of the DNA extracted and the variants called on the samples. They concluded on the feasibility of WES based analysis of FFPE samples compared to FF, but they did not test the influence of FFPE DNA extraction kits on the results. Heydt and colleagues [28] evaluated five automated DNA extraction systems and five DNA quantification systems on FFPE samples, focusing mainly the analysis on DNA quality related parameters, but they did not include in-depth coverage analysis and whole-exome sequencing variant calling analysis.
High-quality DNA extraction with automated sample preparation is of course an important point to consider, that may facilitate the implementation of routine NGS based analysis of FFPE samples for large scale, high-throughput clinical precision medicine projects in the near future. In this study, we performed whole-exome deep sequencing of 42 human FF and FFPE samples, including tumor and normal samples extracted from liver and colon tissues. DNA was extracted from FFPE samples using three different commercial kits, namely the QIAamp DNA FFPE Tissue kit (QIAGEN), the GeneRead DNA FFPE kit (QIAGEN) and the Maxwell RSC DNA FFPE kit (Promega). We selected those kits as robust and well-established methods for DNA extraction from FFPE samples and also for their ability to be included in automated systems for sample preparation. Extraction of FF samples was performed with either a QIAamp DNA Micro Kit or Maxwell™ RSC Blood DNA kit. After sequencing, we did a detailed sequencing coverage analysis for all samples, and then finally performed variants analysis. At each step, we compared FF and FFPE conditions for matched pairs of samples, and checked the effects of the three different extraction kits on FFPE samples. FFPE artifacts and tumor-specific variants annotation were also analyzed.
Materials and methods
FF and FFPE samples
We ordered matched FFPE and FF samples from company AMS BIOTECHNOLOGY EUROPE Ltd. We included tissues from two different organs (liver and colon) in this study. For each tissue, all samples come from the same individual and include both normal and tumor samples from fresh-frozen and formalin-fixed, paraffin-embedded tissues. All the samples were frozen or formalin-fixed in 2013 and 2015. As we processed the samples in 2016, we therefore have a storage time of maximum three years for FF and FFPE. In total we analyzed 42 samples, of which 26 FFPE (10 processed with the Qiagen GeneRead DNA FFPE kit, 8 processed with the Promega Maxwell RSC DNA FFPE kit and 8 processed with the Qiagen QIAamp DNA FFPE Tissue kit) and 16 FF (8 processed with the Maxwell RSC Blood DNA kit, 8 processed with the QIAamp DNA Micro kit). For the analysis, we build a list of sample pairs, associating FF and FFPE samples according to the tissue type and extraction kits. As we have less FF samples than FFPE, the FF samples were repeatedly paired to the FFPE samples extracted with different kits. Finally we have a list of 26 sample pairs, of which 10 FF/QIAamp samples paired with 10 FFPE/GeneRead samples, 8 FF/Maxwell samples paired with 8 FFPE/Maxwell samples and 8 FF/QIAamp samples paired with 8 FFPE/QIAamp samples (see S1 and S2 Tables for the details).
Purification and quality control of DNA from FF and FFPE samples
All the DNA extractions were made from 10 μm thick sample slices. For FFPE samples, we used the QIAamp DNA FFPE Tissue kit (QIAGEN), the GeneRead DNA FFPE kit (QIAGEN) and the Maxwell RSC DNA FFPE kit (Promega). For the FF samples we used the QIAamp DNA Micro Kit (QIAGEN) or the Maxwell™ RSC Blood DNA Kit (Promega). All samples were extracted after an overnight proteinaseK digestion step at 65°C. All Maxwell™ extractions were performed according to the manufacturer’s protocols, on a Maxwell RSC device from Promega. QIAGEN extractions were also performed according to the manufacturer’s instructions, including systematically a RNAse treatment and an optimized elution step. After extraction, all DNA samples were quantified in fluorescence and in duplicate, using Quant-iT™ dsDNA Assay Kits (Invitrogen). The quality of the DNA for all samples has been assessed by loading an aliquot of ∼ 20 ng on a TapeStation 4200 from Agilent to determine the DNA Integrity Number (DIN).
Sequencing of DNA exome libraries and analysis
Exomes were captured using the Agilent Sureselect All Exons Human V5 kit (Agilent Technologies, santa Clara, CA, USA) according to manufacturer’s instructions, with an input of 200 ng. Final libraries were sequenced on a HiSeq2000 with 100 bp paired-end reads (samples were pooled by three on each lane).
Bioinformatics analysis
The reads were mapped to the human genome (GRCh37) using BWA 0.7.12 [29]. Picard Tools 2.6.0 was used to flag duplicate reads and we applied the GATK for indel realignment, base quality score recalibration, SNPs and indels calling using the Haplotype Caller algorithm across all samples simultaneously according to the GATK Best Practices recommendations [30]. After calling we filtered the vcf files for variants having a coverage ≥ 13 and a mapping quality ≥ 43 (as done by Munchel and colleagues in their study [22]). For the coverage analysis, we used SAMtools 1.3.1 [31], BEDtools 2.21.0 [32] and a custom Python script to generate all the statistics. For the analysis of variants, we used Picard Tools to select variants on their quality and discriminate between SNPs and indels. A custom script was also used to compare the variants between FF and FFPE samples. For the somatic analysis, we followed a protocol described in [22]. Briefly, we selected SNVs for colon and liver tumors that were not present in normal samples. This selection was done for FF and FFPE conditions separately, then we counted how many of those tumor-specific variants were in common between the FF and FFPE conditions. All the variants were further annotated with snpEff [33], the COSMIC catalog of somatic mutations in cancer [34] and the KEGG database of biological pathways [35]. The VAF (Variant Allele Frequency) values were calculated as the ratio between the depth of the alternative allele (AD) and the total depth (DP). The AD and DP values were extracted from the VCF files. For the average gene coverage, we used the tool sambamba [36] to calculate the read coverage for all the position of a given gene from the alignments files (option “depth base”).
Results
DNA quality analysis
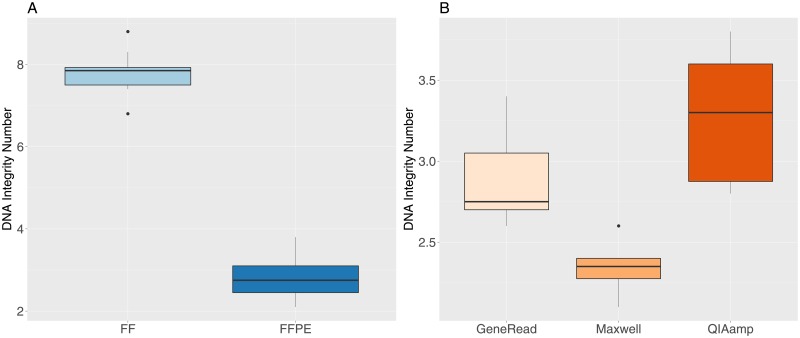
We checked the DNA quality for all samples by measuring the DNA Integrity Number (DIN, the equivalent of the RNA Integrity Number [37] for DNA). Unsurprisingly, the DIN values are much lower in FFPE compared to FF samples (Fig 1A), and the difference is highly significant (t-test t = 34.9, df = 33.9, p-value = 2.2e-16). Lower FFPE DIN values indicate more fragmented DNA and a lower molecular weight for those samples. Furthermore, we find a significant difference for the DIN values between the three extraction methods for the FFPE samples (Fig 1B, one-way anova, F-value = 19.7, df = 2, p-value = 1.03e-5). Samples treated with the Maxwell kit have the lowest values, followed by GeneRead samples and finally the QIAamp samples.
Fig 1. DNA Integrity Number (DIN) values for FF and FFPE samples.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
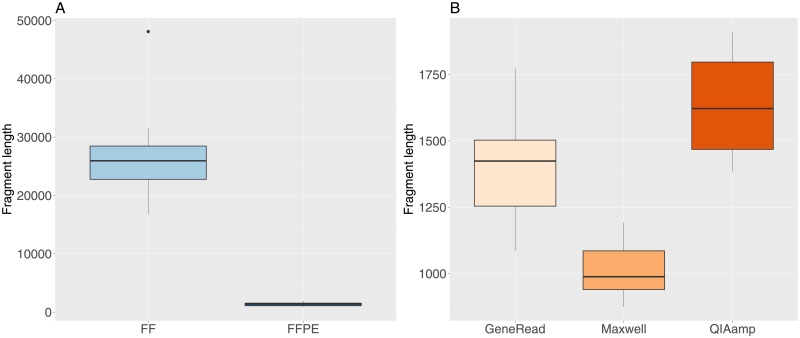
The DNA fragment length was obtained from the readouts of the DIN analysis (length in base pairs of the main peak). The median fragment length is very significantly shorter in FFPE (1368 bp) compared to FF samples (25946 bp, Fig 2A, t-test t = 14.3, df = 15.036, p-value = 3.7e-10). There are also significant differences between the median fragment length for the three different extraction methods (Fig 1B, with increasing fragment length for the Maxwell (median value 988 bp), GeneRead (1424 bp) and QIAamp (1622 bp) kits (one-way anova, F-value = 24.15, df = 2, p-value = 2.2e-6).
Fig 2. DNA fragment length values for FF and FFPE samples.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
Coverage analysis
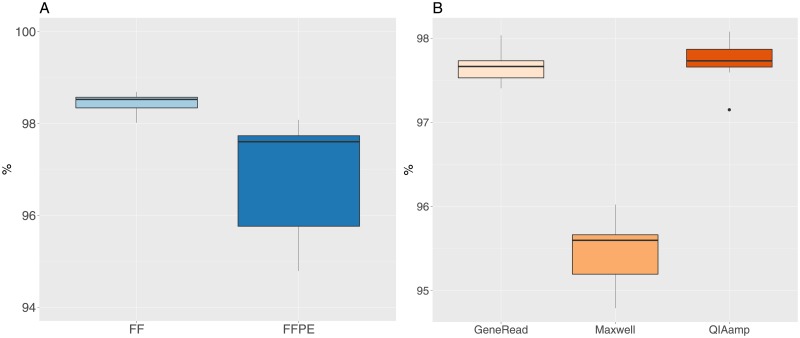
Initial analysis of the mapped reads revealed an unequal number of reads between the samples (median 123M, IQR 27M). In order to be able to compare the samples, we re-sampled all the files to 80M reads. One sample was eliminated from the study, having a value of 73M reads, which is below our usual quality threshold. Re-sampled files were mapped to the human genome, with a median percentage of reads mapped of 99.90% (minimum value 99.73, maximum value 99.96). The median percentage of reads mapping outside the target regions (as defined by the exome capture kit) is 20.3% (min. 18.1%, max. 23.2%), which is within the values that are usually accepted for human whole-exome analysis. The percentage of positions having a coverage greater than or equal to 30X is very high for both FF and FFPE samples (Fig 3A, mean values of 98.4 and 97% respectively), but the difference between the two groups is significant (t-test t = 6.4, df = 28.49, p-value = 4.8e-7). For FFPE samples, there is a significant difference between the three extraction methods (one-way anova F = 165, df = 2, p-value = 2.3e-14), with the Maxwell kit having less coverage than the other two methods (Fig 3B).
Fig 3. Percentage of positions having a coverage greater then or equal to 30X.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
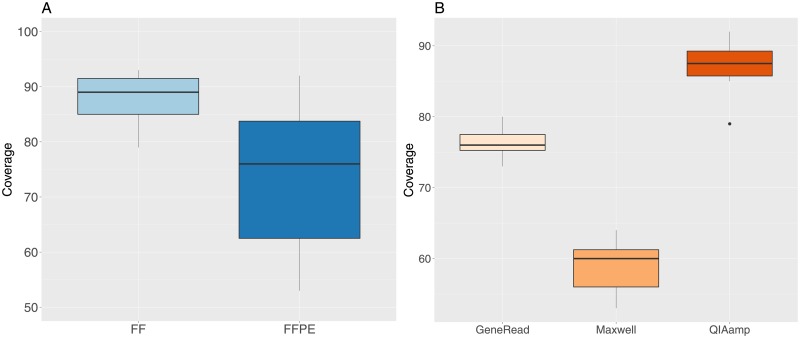
The mean median coverage value for all samples is 80X (Fig 4A), but there is a significant difference (t-test t = 5.03, df = 36.8, p-value = 1.3e-05) between FF (mean value of 87.6X) and FFPE (mean value of 74.3X) samples. The median coverage is also significantly different between the the three extraction methods (one-way anova F = 149.7, df = 2, p-value = 6.5e-14), with increasing values for the Maxwell, GeneRead and QIAamp methods (Fig 4B).
Fig 4. Median coverage values for FF and FFPE samples.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
The percentage of duplicated reads is an important indicator of the sequencing quality for further analysis. We have a median value of 10% for all samples (Fig 5A), but the values are significantly different for FF (median value 7.9%) and FFPE (median value 12.2%, t-test t = -3.06, df = 37.7, p-value = 0.004). For FFPE values only, there is a significant difference between the three extraction methods (Fig 5B, one-way anova F = 21, df = 2, p-value = 6.4e-6). The lowest level is observed for the Maxwell method, then follows the QIAamp method and finally the GeneRead method who has the highest level of duplicated reads.
Fig 5. Percentage of duplicated reads for FF and FFPE samples.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
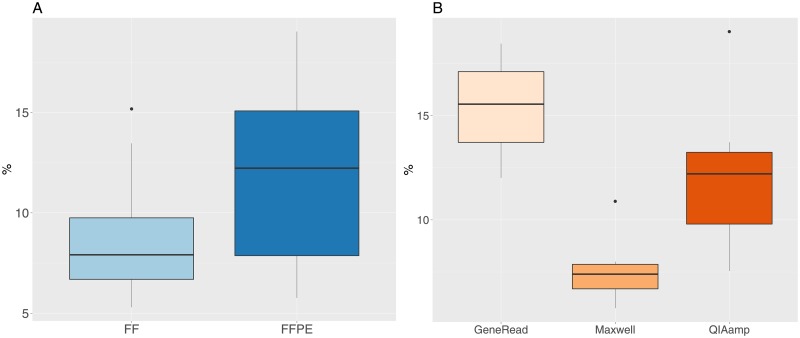
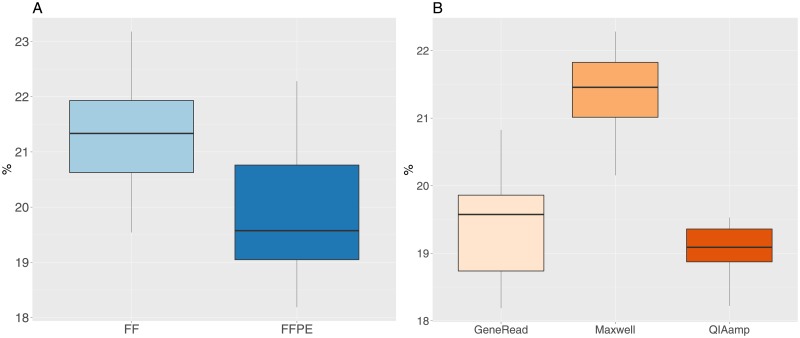
The percentage of reads mapping outside the target regions is slightly lower in FFPE (median value 19.6) compared to FF samples (median value 21.3, t-test t = 4.1, df = 35.1, p-value = 0.0002, Fig 6A). For the FFPE values, we observe a significant difference between the three extraction methods (Fig 6B, one-way anova F = 27, df = 2, p-value = 9.2e-7). The Maxwell kit has the highest median value (21.4) for the percentage of reads mapping outside the target regions, while the QIAamp and GeneRead have lower values (19.1 and 19.6 respectively). However, the percentage values for the three kits are quite close.
Fig 6. Percentage of reads mapping outside target regions.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
Variant analysis and FFPE artifacts
To analyze the potential sequencing artifacts and biases induced by the FFPE treatment, we used 25 matched pairs of both normal and tumor samples of liver and colon tissue. After whole exome sequencing, we characterized all pairs by calling both single nucleotide (SNVs) and small insertion-deletion (INDELs) events, and filtering out low quality calls (see Methods). In order to assess the reliability of base calls from FFPE samples, we determined the common positions between the FF and FFPE lists of variants, and then we used calls from the FF samples as the reference and classified FFPE base calls as concordant if they are the same or discordant if they are not the same. For each pair we can calculate a concordance rate as the number of concordant bases divided by the number of common positions.
The results for SNVs and INDELs are shown in Tables 1 and 2. The average number of variants is significantly lower in FFPE for both SNVs and INDELs (mean values of 40938 SNV variants for FF, 39748 for FPPE, t-test t = 4.2, df = 25.1, p-value = 0.0003, mean values of 3680 INDELs for FF, 3370 for FFPE, t-test t = 5.2, df = 26.7, p-value = 1.7e-05). The number of common positions for filtered base calls represent minimum 86% of the total number of positions for SNVs, and a minimum of 67% for INDELs. The percentage of concordance for SNVs on common positions is minimum 99.98%, and 98.44% for INDELs.
Table 1. Single nucleotide variants (SNVs) analysis between FF and FFPE sample pairs.
NFF: Number of SNV in FF samples, NFFPE: number of SNV in FFPE samples, NPos: number of common positions between FF and FFPE samples, Nco: number of concordant positions, Ndi: number of discordant positions, P: concordance rate (Nco/NPos * 100).
| Pair | NFF | NFFPE | NPos | Nco | Ndi | P |
|---|---|---|---|---|---|---|
| 1 | 41008 | 40165 | 39778 | 39772 | 6 | 99.98 |
| 2 | 40902 | 40157 | 39691 | 39685 | 6 | 99.98 |
| 3 | 40950 | 40361 | 39913 | 39906 | 7 | 99.98 |
| 4 | 41078 | 40011 | 39643 | 39639 | 4 | 99.99 |
| 5 | 40705 | 40295 | 39355 | 39351 | 4 | 99.99 |
| 6 | 40999 | 40134 | 39570 | 39567 | 3 | 99.99 |
| 7 | 40571 | 40481 | 39634 | 39629 | 5 | 99.99 |
| 8 | 40568 | 40623 | 39568 | 39564 | 4 | 99.99 |
| 9 | 41008 | 40716 | 40075 | 40068 | 7 | 99.98 |
| 10 | 40902 | 40764 | 40048 | 40044 | 4 | 99.99 |
| 11 | 40902 | 40712 | 40004 | 40001 | 3 | 99.99 |
| 12 | 40950 | 40712 | 40094 | 40089 | 5 | 99.99 |
| 13 | 41078 | 40788 | 40102 | 40098 | 4 | 99.99 |
| 14 | 41078 | 40779 | 40116 | 40110 | 6 | 99.99 |
| 15 | 40705 | 40794 | 39631 | 39626 | 5 | 99.99 |
| 16 | 40999 | 40852 | 40046 | 40042 | 4 | 99.99 |
| 17 | 40571 | 40586 | 39473 | 39468 | 5 | 99.99 |
| 18 | 40568 | 41009 | 39848 | 39844 | 4 | 99.99 |
| 19 | 41239 | 37396 | 36851 | 36846 | 5 | 99.99 |
| 20 | 41201 | 37834 | 37239 | 37236 | 3 | 99.99 |
| 21 | 41115 | 37721 | 37185 | 37179 | 6 | 99.98 |
| 22 | 41189 | 36215 | 35751 | 35745 | 6 | 99.98 |
| 23 | 40963 | 38097 | 37588 | 37582 | 6 | 99.98 |
| 24 | 41074 | 38132 | 37577 | 37572 | 5 | 99.99 |
| 25 | 41133 | 38369 | 37860 | 37858 | 2 | 99.99 |
Table 2. Insertion-deletion events (INDELs) analysis between FF and FFPE sample pairs.
NFF: Number of INDELs in FF samples, NFFPE: number of INDELs in FFPE samples, NPos: number of common positions between FF and FFPE samples, Nco: number of concordant positions, Ndi: number of discordant positions, P: percentage of concordance Nco/NPos.
| Pair | NFF | NFFPE | NPos | Nco | Ndi | P |
|---|---|---|---|---|---|---|
| 1 | 3609 | 3402 | 3176 | 3140 | 36 | 98.87 |
| 2 | 3612 | 3369 | 3169 | 3122 | 47 | 98.52 |
| 3 | 3610 | 3435 | 3193 | 3154 | 39 | 98.78 |
| 4 | 3624 | 3428 | 3168 | 3122 | 46 | 98.55 |
| 5 | 3762 | 3445 | 3178 | 3136 | 42 | 98.68 |
| 6 | 3770 | 3446 | 3212 | 3170 | 42 | 98.69 |
| 7 | 3749 | 3587 | 3315 | 3276 | 39 | 98.82 |
| 8 | 3712 | 3589 | 3239 | 3203 | 36 | 98.89 |
| 9 | 3609 | 3568 | 3258 | 3210 | 48 | 98.53 |
| 10 | 3612 | 3589 | 3267 | 3221 | 46 | 98.59 |
| 11 | 3612 | 3538 | 3240 | 3196 | 44 | 98.64 |
| 12 | 3610 | 3588 | 3267 | 3216 | 51 | 98.44 |
| 13 | 3624 | 3565 | 3251 | 3206 | 45 | 98.62 |
| 14 | 3624 | 3567 | 3271 | 3235 | 36 | 98.90 |
| 15 | 3762 | 3611 | 3293 | 3245 | 48 | 98.54 |
| 16 | 3770 | 3614 | 3336 | 3292 | 44 | 98.68 |
| 17 | 3749 | 3629 | 3311 | 3268 | 43 | 98.70 |
| 18 | 3712 | 3711 | 3346 | 3310 | 36 | 98.92 |
| 19 | 3723 | 2931 | 2740 | 2703 | 37 | 98.65 |
| 20 | 3770 | 2999 | 2826 | 2797 | 29 | 98.97 |
| 21 | 3763 | 2957 | 2791 | 2759 | 32 | 98.85 |
| 22 | 3732 | 2736 | 2593 | 2570 | 23 | 99.11 |
| 23 | 3600 | 2952 | 2779 | 2756 | 23 | 99.17 |
| 24 | 3635 | 2972 | 2788 | 2752 | 36 | 98.71 |
| 25 | 3651 | 3036 | 2839 | 2810 | 29 | 98.98 |
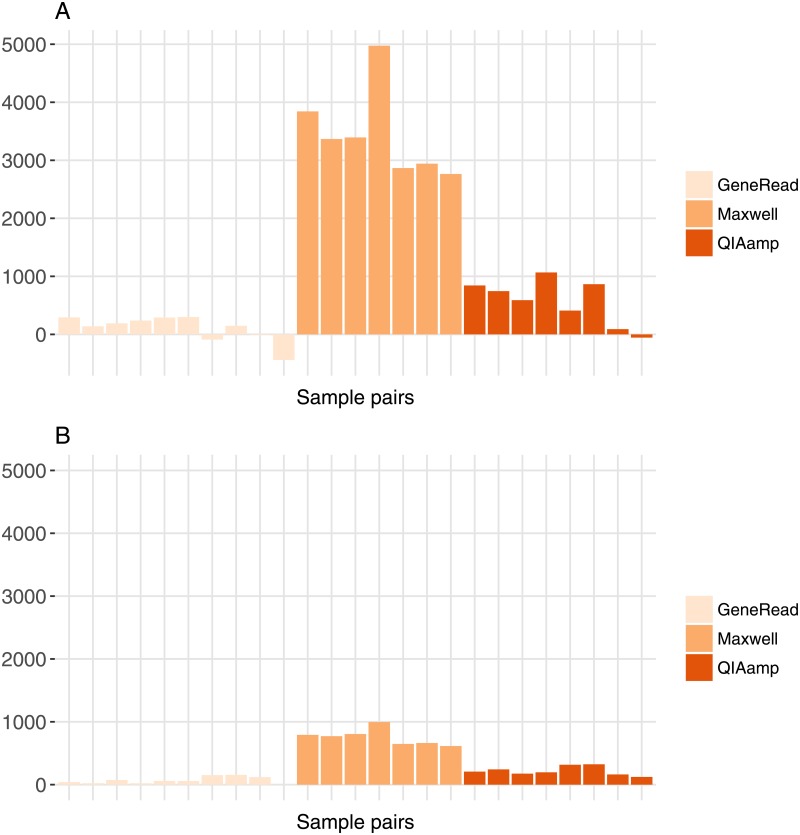
To evaluate the impact of the different extraction kits, we calculated the difference in number of variants for each pair of matched FF/FFPE samples for SNVs and INDELs (Fig 7A and 7B). There is a significant variation in difference values for the three extraction kits for both SNVs (one-way anova, F-Value = 108.6, df = 2, p-value = 3.9e-12) and INDELs (one-way anova, F-Value = 135.5, df = 2, p-value = 4.3e-13). We can see on the graph that difference values are low for GeneRead and QIAamp, while values for Maxwell are much higher, especially for SNVs. The GeneRead kit has the lowest median value, which might be due to the fact that this kit is designed to minimize the number of artifacts induced in FFPE samples (by enzymatic correction). While the difference values for SNVs represent on average 1% of the total number of variants for GeneRead and QIAamp, they increase to around 10% of the total number of variants for the Maxwell kit (Fig 7A). The profile of the variations is similar for INDELs (Fig 7B).
Fig 7. Difference in number of variants between FF and FFPE samples for all matched FF/FFPE pairs.
A: SNVs. B: INDELs.
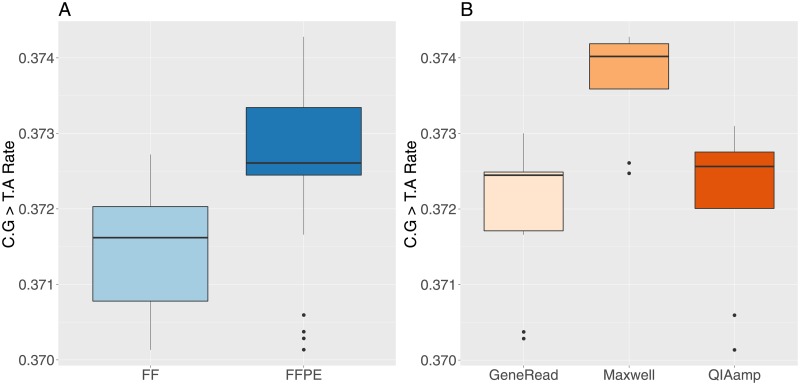
FFPE DNA has been shown to have artifacts created by formalin fixation and sample preparation that trigger enhanced cytosine deamination [38–40]. These artifacts show up as non-reproducible C > T or G > A (C.G > T.A) substitutions. We therefore analyzed combined C > T, G > A substitutions in SNV variants for all samples. As expected, the number of substitutions is higher in FFPE samples (Fig 8A, t-test t = -3.8, df = 37.9, p-value = 0.0005) and the values are slightly higher in Maxwell treated samples, followed by QIAamp and GeneRead (Fig 8B, one-way anova, F-value = 9.5, df = 2, p-value = 0.009). The GeneRead kit has the lowest values for the C > T, G > A substitution rates, which is most likely due to the artifact correction capabilities included in this kit (by enzymatic activity). Although the difference is statistically significant, it is worth noticing that the increase in rate value between FF and FFPE, and between the extraction methods remains small in absolute value. As shown on Fig 8, the median value difference between FFPE and FF C.G > T.A rates is equal to 0.0098 (≈ 1%, the median value differences between Maxwell and GeneRead for FFPE data only is equal to 0.0015, the median value differences between Maxwell and GeneRead for FFPE data only is equal to 0.00011, i.e. ≈ 0.01%).
Fig 8. Rate of SNVs C.G > T.A substitution in FF and FFPE samples.
A: FF and FFPE samples. B: FFPE samples grouped by extraction method.
Since we had matched normal and tumor tissues, we performed somatic mutation analysis to evaluate the potential impact and interest of the SNVs called. Upon subtraction of variants present in matched normal samples, we found 394 putative somatic variants in liver FF samples, 333 in liver FFPE samples, 436 in colon FF samples and 458 in colon FFPE samples (S3 Table). We observe that a total of 165 tumor-specifc SNVs for colon show overlap between FF and FFPE (representing 38% and 36% of tumor-specific FF and FFPE SNVs respectively, see S3 Table) and that 53 tumor-specific SNVs for liver do show overlap between FF and FFPE (representing 13% and 16% of tumor-specific FF and FFPE SNVs respectively, see S3 Table). A selection of the annotated variants is shown in Table 3, and the complete list of annotated variants for colon and liver tissues is available as S4 Table. A number of variants overlap with the COSMIC database and are found in well-established tumor-related biological pathways. For instance, the KRAS, PTEN and APC genes are well-established tumor drivers in many different tumor types, including colon cancer [2]. We selected the top 5 mutated genes in liver (TERT, CTNNB1, TP53, ALB, ARID1A, [41]) and top 4 mutated genes in colon (APC, TP53, SYNE1, PIK3CA, [2]) cancer according to two recent studies and analyzed the mean coverage in FF, FFPE and for the three FFPE extraction kits for all colon and liver tumor samples (S5 Table). The average coverage values for all genes is greater than 30X for all genes in FF and FFPE samples, excepted for PIK3CA in FFPE samples. Interestingly, average coverage values are lower in FFPE compared to FF samples in many cases, and the Maxwell kit has lower average cover than the other kits, although there are exceptions to this tendency (e.g. TP53 for colon and liver samples, TERT and ARID1A for liver samples).
Table 3. Selection of annotated tumor-specific variants found in common between FF and FFPE colon samples.
Chr: chromosome number. Position: position of the variant on the chromosome. Ns: number of samples in which the variant was found. COSMIC ID: COSMIC database identification code. Gene Symbol: HGNC gene symbol. Pathways: selection of KEGG or REACTOME pathways in which the gene is involved. FFV: mean variant allelic frequency (%) for FF samples. FFD: mean read depth for the position for FF samples (coverage). FEV: mean variant allelic frequency (%) for FFPE samples. FED: mean read depth for the position for FFPE samples (coverage).
| Chr | Position | Ns | COSMIC ID | Gene Symbol | Pathways | FFV | FFD | FEV | FED |
|---|---|---|---|---|---|---|---|---|---|
| chr1 | 228109626 | 10 | COSM4389123 | WNT9A | WNT signaling pathway | 24 | 208 | 22 | 428 |
| Basal cell carcinoma | |||||||||
| Melanogenesis | |||||||||
| Pathways in cancer | |||||||||
| Hedgehog signaling pathway | |||||||||
| chr2 | 231867435 | 10 | COSM5499569 | SPATA3 | 21 | 116 | 26 | 145 | |
| chr2 | 238990381 | 9 | COSM5471498 | UBE2F-SCLY | Selenoaminoacid metabolism | 23 | 153 | 22 | 226 |
| COSM5471497 | SCLY | ||||||||
| chr3 | 98188932 | 10 | COSM262658 | OR5K1 | Olfactory transduction | 22 | 166 | 19 | 85 |
| chr5 | 112175594 | 9 | COSM19705 | APC | WNT signaling pathway | 21 | 142 | 22 | 73 |
| CTC-554D6.1 | Endometrial cancer | ||||||||
| Pathways in cancer | |||||||||
| Colorectal cancer | |||||||||
| Basal cell carcinoma | |||||||||
| chr5 | 140432413 | 7 | COSM125200 | PCDHB1 | Signaling by Rho GTPases | 24 | 79 | 25 | 54 |
| chr6 | 36982783 | 10 | COSM3076428 | FGD2 | 22 | 130 | 18 | 211 | |
| chr10 | 50315817 | 10 | COSM259693 | VSTM4 | 22 | 180 | 21 | 286 | |
| COSM259692 | |||||||||
| chr10 | 89692910 | 10 | COSM5032 | PTEN | Pathways in cancer | 31 | 153 | 31 | 81 |
| Tight junction | |||||||||
| Prostate cancer | |||||||||
| PI signaling system | |||||||||
| Melanoma, Glioma | |||||||||
| P53 signaling pathway | |||||||||
| chr11 | 64083221 | 3 | COSM300694 | TRMT112 | Peroxisome | 15 | 60 | 10 | 120 |
| ESRRA | Nuclear receptor transcription | ||||||||
| PRDX5 | Generic transcription | ||||||||
| chr11 | 76796018 | 3 | COSM4592217 | CAPN5 | 25 | 88 | 27 | 178 | |
| chr12 | 25398284 | 9 | COSM1140133 | KRAS | Pathways in cancer | 19 | 88 | 20 | 61 |
| COSM49168 | Prostate cancer | ||||||||
| COSM520 | Endometrial cancer | ||||||||
| Acute myeloid leukemia | |||||||||
| Non small cell lung cancer | |||||||||
| Glioma, Thyroid cancer | |||||||||
| Colorectal cancer | |||||||||
| ERBB signaling pathway | |||||||||
| VEGF signaling pathway | |||||||||
| chr13 | 111109670 | 3 | COL4A2-AS2 | ECM receptor interaction | 48 | 169 | 48 | 212 | |
| COL4A2 | Pathways in cancer | ||||||||
| Small cell lung cancer | |||||||||
| Focal adhesion | |||||||||
| chr20 | 62076690 | 9 | COSM2932051 | KCNQ2, | Developmental biology | 23 | 112 | 19 | 190 |
| COSM2932050 | RP11-358D14.2 | Potassium channels | |||||||
| COSM2932052 | RP11-358D14.2 | Neuronal system | |||||||
| COSM4100400 | |||||||||
| chrX | 37028002 | 10 | COSM1319445 | FAM47C | 46 | 74 | 45 | 95 | |
| COSM1319446 |
Discussion
In this study, we compared DNA samples from FF and FFPE commercial tissue samples, using whole-exome high-throughput sequencing. We systematically analyzed coverage indicators and performed variant analysis between FF and FFPE samples, and we also analyzed the influence of three different FFPE DNA extraction kits by comparing matched pairs of FF and FFPE samples.
Our results show that the quality of the DNA extracts, as measured by the DNA integrity number (DIN), is significantly lower in FFPE samples, which indicates more degraded DNA molecules. This result is expected, since it is a well-established fact that the FFPE process contributes to fragmentation, cross-linking and chemical modifications of FFPE derived nucleic acids [11, 12]. Within the FFPE samples, we observe a limited but nevertheless statistically significant difference in DIN values between the three kits, with the Maxwell kit having the lowest median DIN value. The DNA fragment length in the libraries is dramatically lower in FFPE samples compared to FF (almost 20 times smaller) and we also found a significant but much less important difference in fragment length values for the three extraction kits, with the Maxwell kit having the smallest fragment length.
The coverage analysis shows a significant difference between FF and FFPE samples for multiple indicators, with a lower percentage of positions having a coverage greater than or equal to 30X, a lower median coverage value, a higher percentage of duplicated reads and a slightly lower percentage of reads mapping outside the target regions in FFPE samples. Regarding the three FFPE extraction kits we observe moderated but significant differences for coverage indicators. The GeneRead and QIAamp are quite similar in terms of all the coverage statistics we have monitored, while the Maxwell kit has lower values for median coverage and percentage of positions having a coverage greater than or equal to 30x and a higher percentage of reads mapping outside the target regions, but on the other hand has a lower percentage of duplicated reads. Note that some metrics can be difficult to interpret together. For instance, the percentage of duplicated reads is higher for sampled treated with the Generead kit compared to Maxwell, but at the same time the coverage is higher for GeneRead samples compared to Maxwell.
As could be expected, these results indicate a lower coverage quality in FFPE samples. We find also a significant effect of the extraction kit protocols we have tried on the coverage quality metrics. However, it is worth noting that the minimum value for the percentage of positions having a coverage of 30X or more for all FFPE samples is 94%, which is well above our usual quality threshold of at least 80% applied for exome sequencing studies (see for instance [4–7]). The same applies for the percentage of duplicated reads, where the maximum values for all FFPE samples is 19% while our quality threshold is at most 25%. On the other hand, for the median coverage indicator, there are a few samples below our threshold of at least 60X, but still 86% of the samples are above this criteria. Taken together, these results show that the FFPE samples have lower coverage quality and that we can detect rather small but significant differences between the three extraction methods we have analyzed, but the resulting sequences are above usual quality standards for whole-exome sequencing.
For the variant analysis, our results detect a significant but small decrease in the number of SNVs or INDELs called in the FFPE compared to the FF sample pairs. However, the number of common variants between FF and FFPE pairs is very high in all cases (minimum 86% of the total number of SNVs), and the percentage of concordance for the common positions is also very high (99.98% mininum for SNVs). These results show that variant calling results in FFPE samples are highly similar to FF samples. Consistent with this global result, our analysis of tumor-specific variants found in both FF and FFPE samples shows that a large number of them are potentially impacting various genes and biological pathways relevant to cancer. Our results are concordant with previous studies related to whole-exome sequencing and other forms of high-throughput sequencing for FFPE samples [18–20, 22, 23]. Although we found a high percentage of concordance for variants detection between FF and FFPE samples, we detected limited but significant variations in the total number of variants difference between the three FFPE extraction kits, with a higher decrease in variants for FFPE samples extracted with the Maxwell extraction kit that can reach up to 10% of the total number of variants for SNVs.
The tumor-specific variants analysis found several candidates that overlap with knownn and well-established COSMIC variants, for genes that are found in canonical tumor-related biological pathways. For instance, genes such as KRAS, PTEN and APC are well-known tumor drivers in several types of cancer. The number of tumor-specific variants we found in FF and FFPE samples, as well as the proportion of those variants that are common between FF and FFPE are similar to values found in other exome based studies [22]. The relative low values of the common tumor-specific variants between FF and FFPE samples illustrate the feasability as well as the challenges associated with the analysis of concordant somatic mutation calls between FF and FFPE samples.
We found a significant but limited increased rate of C.G > T.A substitutions in FFPE samples (≈ 1%) compared to FF, and also a significant but very limited C.G > T.A substitution rate variations between the three FFPE extraction methods (maximum ≈ 0.01% between the GeneRead and Maxwell kits). C.G > T.A changes artifacts are caused by a chemical reaction called cytosine deamination. This change can happen spontaneously in vivo and is corrected by intracellular enzymes uracil DNA glycosylase (UDG) and 5-methylcytosine DNA glycosylase (with the latter repairing specifically changes occurring at CpG dinucleotides sites) [42, 43]. Formalin fixation has been reported to play an important deamination role [19, 39, 44, 45], but other factors such as UV irradiation, pH, hypoxia and heat can also have an effect [46–48]. In order to reduce deamination linked artifacts in FFPE, researchers have tried to measure the benefits of adding a pre-treatment step with UDG, which seems to be effective, with a more important effect on older specimens [39, 40, 44]. In fact, the GeneRead kit includes UDG precisely in order to repair this type of artifact. On our FFPE samples, the GeneRead kit has a significant but very limited effect on on the C.G > T.A substitution rate compared to the other kits, which might be due to the fact that the samples used in this study are only maximum two years old, whereas UDG effects has been shown to be more efficient in older specimens.
To summarize our results on the three FFPE DNA extraction kits we have analyzed in this study, we compiled all the coverage and variant calling indicators in Table 4. These indicators are of course important, but we also included indicators related to the usage of the extraction kits at the bench, more especially how easy or practical they are in the perspective of using them in projects where large numbers of samples will be processed. Although such indicators clearly do not directly affect the quality of the results, they might be important secondary indicators of high practical importance when choosing an extraction kit for a large-scale project. We also included in the table qualitative rankings (from one to three stars) indicating how the three kits performed relative to each other for all the indicators with the samples included in this study. Globally, Table 4 shows that the three kits performed very well regarding the concordance of variant calling between FF and FFPE sample pairs. However, when looking at the various indicators of Table 4, we can see that the Qiagen kits (GeneRead, QIAamp) perform better on several indicators compared to the Maxwell™ kit on the samples used in this study, but it is also worth noticing that this kit has several technical and practical advantages, such as a cassette system that is easy to use and a large number of samples that can be processed per run. Few studies have analyzed the effect of DNA extraction systems and kits. Heydt et al. [28] included a Maxwell 16 DNA extraction system (an older version of the machine we used in this study) in their analysis of five DNA extraction systems on FFPE samples, and concluded that this system and associated extraction kit was giving better results in terms of DNA concentration. However, a previous study focusing on the same Maxwell 16 system found a higher DNA concentration using another extraction kit [49]. Those two studies used a different Maxwell automated extraction system and associated FFPE extraction kit, which may explain the different results.
Table 4. Coverage, variant calling and technical quality indicators for the three FFPE DNA extraction kits.
Values are given for quantitative indicators. The number of stars between brackets (one, two or three) indicate the relative ranking of a given kit for the indicator and for the samples we have analyzed in this study. Technical indicators (qualitative and quantitative) describe how easy or practical it is to use the kits, especially with the aim of analyzing large number of samples, based on our experience in this study.
| GeneRead | Maxwell | QIAamp | |
|---|---|---|---|
| Coverage and variants indicators | |||
| DNA Integrity Number median value | 2.35 (**) | 1.75 (*) | 3.3 (***) |
| DNA fragment length in bp | 1424 (**) | 988 (*) | 1622 (***) |
| Median percentage of positions with coverage ≥ 30X 1 | 97.6 (***) | 95.5 (*) | 97.7 (***) |
| Median coverage values 1 | 76X (**) | 60X (*) | 87.5X (***) |
| Percentage of duplicated reads 1 | 15.5 (*) | 7.4 (***) | 12.2 (*) |
| Percentage of reads mapping outside target regions1 | 19.6 (**) | 21.4 (*) | 19.1 (**) |
| C.T > G.A conversion rate in SNV calls | 0.372 | 0.374 | 0.372 |
| Median value for the variant difference FF/FFPE 2 | 214 (***) | 3367 (*) | 667 (**) |
| Median value for the percentage of concordance FF/FFPE 3 | 99.9 (***) | 99.9 (***) | 99.9 (***) |
| Technical indicators for the extraction process | |||
| Purification technique easiness 4 | (*) | (***) | (*) |
| Possibility of elution step optimization 5 | (**) | (**) | |
| Output tube format and transfer 6 | (***) | (*) | (***) |
| Max number of samples per run | 12 (**) | 16 (***) | 12 (**) |
| Input material quantity (nb of 10 μm tissue slices / extraction) 7 | 1 (*) | 1 to 16 (***) | 1 to 8 (**) |
1 The values are calculated on bam files normalized to 80M reads.
2 The value corresponds to the absolute value of the difference between the number of variants in FF versus FFPE sample pairs.
3 The percentage of concordance is determined on the common variants positions between FF and FFPE sample pairs.
4 Purification for the Maxwell kit is based on magnetic beads and cassettes, GeneRead and QIAamp kits use columns which are more time consuming to manipulate.
5 The elution volume is fixed for the Maxwell kit, leaving no room for optimization, while with the QIAGEN kits, it is possible to optimize the final concentration by playing with the elution volume and/or to warm up the elution buffer to increase efficiency.
6 For the Maxwell kit, the format of the output tube (0.5 ml) is not practical, and there are magnetic beads residues in the tubes, necessitating a transfer in a more adapted tube for further processing. These problems are not present for the QIAGEN kits.
7 The higher the number of tissue slices per extraction, the more flexible it is to obtain enough material for sequencing.
In summary, our results demonstrate that high-throughput whole-exome analysis of variants on FFPE samples have high quality of coverage and a very high percentage of variants concordance when compared to paired FF samples. Limited but significant variations in coverage and variant calling indicators can be detected for the three different FFPE DNA extraction kits on the samples included in this study. The values of all the indicators are above usually accepted thresholds, but the differences suggest that the selection of a kit for a large scale project (e.g. precision medicine) should be done with care and thorough testing beforehand.
Supporting information
(PDF)
(PDF)
(PDF)
(XLSX)
(PDF)
Acknowledgments
We would like to thank Florence Jobard, Caroline Horgues, Jeanne-Antide Perrier, Emmanuel Menard, David Derbala, Johan Tassin for excellent technical advice and assistance. Stéphane Meslage and Ghislain Septier were very helpful for the whole-exome analysis pipeline and for the calculation of the whole-exome coverage statistics. This work has been developed and supported by the LABEX (laboratory of excellence program investment for the future) GENMED (Medical Genomics).
Data Availability
Whole-exome sequencing data has been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) which is hosted at the EBI, under accession number EGAS00001002631.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1. Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. Journal of Clinical Oncology. 2013;31(15):1806–1814. doi: 10.1200/JCO.2012.46.8934 [DOI] [PubMed] [Google Scholar]
- 2. consortium TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discovery. 2012;2(3):214–226. doi: 10.1158/2159-8290.CD-12-0012 [DOI] [PubMed] [Google Scholar]
- 4. Nicolas G, Wallon D, Charbonnier C, Quenez O, Rousseau S, Richard AC, et al. Screening of dementia genes by whole-exome sequencing in early-onset Alzheimer disease: input and lessons. European Journal of Human Genetics. 2016;24(5):710–716. doi: 10.1038/ejhg.2015.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerino M, Gorokhova S, Laforet P, Ben Yaou R, Salort-Campana E, Pouget J, et al. Genetic Characterization of a French Cohort of GNE-mutation negative inclusion body myopathy patients with exome sequencing. Muscle & Nerve. 2017; p. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 6. Bauché S, OeRegan S, Azuma Y, Laffargue F, McMacken G, Sternberg D, et al. Impaired Presynaptic High-Affinity Choline Transporter Causes a Congenital Myasthenic Syndrome with Episodic Apnea. The American Journal of Human Genetics. 2016;99(3):753–761. doi: 10.1016/j.ajhg.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruel AL, Franco B, Duffourd Y, Thevenon J, Jego L, Lopez E, et al. Fifteen years of research on oral–facial–digital syndromes: from 1 to 16 causal genes. Journal of Medical Genetics. 2017; p. jmedgenet–2016. doi: 10.1136/jmedgenet-2016-104436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. New England Journal of Medicine. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. New England Journal of Medicine. 2012;366(12):1090–1098. doi: 10.1056/NEJMoa1106968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feldman MY. Reactions of Nucleic Acids and NucleoDroteins with Formaldehyde. Progress in Nucleic Acid Research and Molecular Biology. 1973;13:1–49. doi: 10.1016/S0079-6603(08)60099-9 [DOI] [PubMed] [Google Scholar]
- 12. Auerbach C, Moutschen-Dahmen M, Moutschen J. Genetic and cytogenetical effects of formaldehyde and related compounds. Mutation Research/Reviews in Genetic Toxicology. 1977;39(3-4):317–361. doi: 10.1016/0165-1110(77)90011-2 [DOI] [PubMed] [Google Scholar]
- 13. Bresters D, Schipper M, Reesink H, Boeser-Nunnink B, Cuypers H. The duration of fixation influences the yield of HCV cDNA-PCR products from formalin-fixed, paraffin-embedded liver tissue. Journal of Virological Methods. 1994;48(2-3):267–272. doi: 10.1016/0166-0934(94)90125-2 [DOI] [PubMed] [Google Scholar]
- 14. Karlsen F, Kalantari M, Chitemerere M, Johansson B, Hagmar B. Modifications of human and viral deoxyribonucleic acid by formaldehyde fixation. Laboratory Investigation. 1994;71(4):604–611. [PubMed] [Google Scholar]
- 15. Inadome Y, Noguchi M. Selection of higher molecular weight genomic DNA for molecular diagnosis from formalin-fixed material. Diagnostic Molecular Pathology. 2003;12(4):231–236. doi: 10.1097/00019606-200312000-00007 [DOI] [PubMed] [Google Scholar]
- 16. Farragher SM, Tanney A, Kennedy RD, Harkin DP. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochemistry and Cell Biology. 2008;130(3):435–445. doi: 10.1007/s00418-008-0479-7 [DOI] [PubMed] [Google Scholar]
- 17. Collins FS, Varmus H. A new initiative on precision medicine. New England Journal of Medicine. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number-and mutation-analysis. PloS One. 2009;4(5):e5548 doi: 10.1371/journal.pone.0005548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spencer DH, Sehn JK, Abel HJ, Watson MA, Pfeifer JD, Duncavage EJ. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. The Journal of molecular diagnostics. 2013;15(5):623–633. doi: 10.1016/j.jmoldx.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hedegaard J, Thorsen K, Lund MK, Hein AMK, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PloS One. 2014;9(5):e98187 doi: 10.1371/journal.pone.0098187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of FFPE tumor samples to guide precision cancer medicine. Nature Medicine. 2014;20(6):682 doi: 10.1038/nm.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munchel S, Hoang Y, Zhao Y, Cottrell J, Klotzle B, Godwin AK, et al. Targeted or whole genome sequencing of formalin fixed tissue samples: potential applications in cancer genomics. Oncotarget. 2015;6(28):25943–61. doi: 10.18632/oncotarget.4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carrick DM, Mehaffey MG, Sachs MC, Altekruse S, Camalier C, Chuaqui R, et al. Robustness of next generation sequencing on older formalin-fixed paraffin-embedded tissue. PloS One. 2015;10(7):e0127353 doi: 10.1371/journal.pone.0127353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagnall RD, Ingles J, Yeates L, Berkovic SF, Semsarian C. Exome sequencing-based molecular autopsy of formalin-fixed paraffin-embedded tissue after sudden death. Genetics in Medicine. 2017; p. advance online publication. doi: 10.1038/gim.2017.15 [DOI] [PubMed] [Google Scholar]
- 25. Janecka A, Adamczyk A, Gasińska A. Comparison of eight commercially available kits for DNA extraction from formalin-fixed paraffin-embedded tissues. Analytical biochemistry. 2015;476:8–10. doi: 10.1016/j.ab.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 26. Bonfiglio S, Vanni I, Rossella V, Truini A, Lazarevic D, Dal Bello MG, et al. Performance comparison of two commercial human whole-exome capture systems on formalin-fixed paraffin-embedded lung adenocarcinoma samples. BMC Cancer. 2016;16(1):692 doi: 10.1186/s12885-016-2720-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Astolfi A, Urbini M, Indio V, Nannini M, Genovese CG, Santini D, et al. Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST). BMC Genomics. 2015;16(1):892 doi: 10.1186/s12864-015-1982-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heydt C, Fassunke J, Künstlinger H, Ihle MA, König K, Heukamp LC, et al. Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics. PloS One. 2014;9(8):e104566 doi: 10.1371/journal.pone.0104566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Research. 2016; p. gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–2034. doi: 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 2006;7(1):3 doi: 10.1186/1471-2199-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hofreiter M, Jaenicke V, Serre D, Haeseler Av, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Research. 2001;29(23):4793–4799. doi: 10.1093/nar/29.23.4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil-DNA glycosylase. Oncotarget. 2012;3(5):546–558. doi: 10.18632/oncotarget.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Do H, Wong SQ, Li J, Dobrovic A. Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clinical Chemistry. 2013;59(9):1376–1383. doi: 10.1373/clinchem.2012.202390 [DOI] [PubMed] [Google Scholar]
- 41. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nature Genetics. 2015;47(5):505–511. doi: 10.1038/ng.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annual Review of Genetics. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA-occurrence, consequences and repair. Oncogene. 2002;21(58):8935 doi: 10.1038/sj.onc.1205996 [DOI] [PubMed] [Google Scholar]
- 44. Wong SQ, Li J, Tan AY, Vedururu R, Pang JMB, Do H, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Medical Genomics. 2014;7(1):23 doi: 10.1186/1755-8794-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clinical Chemistry. 2015;61(1):64–71. doi: 10.1373/clinchem.2014.223040 [DOI] [PubMed] [Google Scholar]
- 46. Ehrlich M, Norris KF, Wang RY, Kuo KC, Gehrke CW. DNA cytosine methylation and heat-induced deamination. Bioscience Reports. 1986;6(4):387–393. doi: 10.1007/BF01116426 [DOI] [PubMed] [Google Scholar]
- 47. Barak Y, Cohen-Fix O, Livneh Z. Deamination of Cytosine-containing Pyrimidine Photodimers in UV-irradiated DNA SIGNIFICANCE FOR UV LIGHT MUTAGENESIS. Journal of Biological Chemistry. 1995;270(41):24174–24179. doi: 10.1074/jbc.270.41.24174 [DOI] [PubMed] [Google Scholar]
- 48. Lee CH, Wu CL, Shiau AL. Hypoxia-induced cytosine deaminase gene expression for cancer therapy. Human Gene Therapy. 2007;18(1):27–38. doi: 10.1089/hum.2005.239 [DOI] [PubMed] [Google Scholar]
- 49. Khokhar SK, Mitui M, Leos NK, Rogers BB, Park JY. Evaluation of Maxwell® 16 for automated DNA extraction from whole blood and formalin-fixed paraffin embedded (FFPE) tissue. Clinical Chemistry and Laboratory Medicine (CCLM). 2012;50(2):267–272. doi: 10.1515/cclm.2011.763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(XLSX)
(PDF)
Data Availability Statement
Whole-exome sequencing data has been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) which is hosted at the EBI, under accession number EGAS00001002631.