Abstract
There is still no consensus on the true origin of fatal yellowing, one of the most important diseases affecting oil palm (Elaeis guineensis Jacq.) plantations. This study involved two-dimensional liquid chromatography coupled with tandem mass spectrometry (2D-UPLC-MSE) analyses to identify changes in protein profiles of oil palms affected by FY disease. Oil palm roots were sampled from two growing areas. Differential accumulation of proteins was assessed by comparing plants with and without symptoms and between plants at different stages of FY development. Most of the proteins identified with differential accumulation were those related to stress response and energy metabolism. The latter proteins include the enzymes alcohol dehydrogenase and aldehyde dehydrogenase, related to alcohol fermentation, which were identified in plants with and without symptoms. The presence of these enzymes suggests an anaerobic condition before or during FY. Transketolase, isoflavone reductase, cinnamyl alcohol dehydrogenase, caffeic acid 3-O-methyltransferase, S-adenosylmethionine synthase, aldehyde dehydrogenase and ferritin, among others, were identified as potential marker proteins and could be used to guide selection of FY-tolerant oil palm genotypes or to understand the source of this anomaly. When comparing different stages of FY, we observed high accumulation of alcohol dehydrogenase and other abiotic stress related-proteins at all disease stages. On the other hand, biological stress-related proteins were more accumulated at later stages of the disease. These results suggest that changes in abiotic factors can trigger FY development, creating conditions for the establishment of opportunistic pathogens.
1. Introduction
The main oil source of plant origin in the world is the oil palm (Elaeis guineensis Jacq.). This species has large economic and social importance in producing countries. The fruit of this palm species contains palm oil and palm kernel oil, used in processed foods, pharmaceuticals and cosmetics, as well as for sustainable energy generation [1, 2].
The first reports of FY date to the 1980s, and its etiology remains unknown. A good deal of research has been done to understand FY’s cause, considering biotic factors [3, 4, 5, 6, 7, 8, 9], but there is still no consensus on the true origin of this anomaly. The current trend is to focus on abiotic factors, mainly associated with water balance distribution [10, 11, 12, 13], as well as the limitation of drainage, soil nutrition and oil palm root system [14, 15].
Due to inadequate soil management, growing areas suffer from soil compaction, reducing drainage and physical impedance, associated with long flooded periods (up to six months). In these conditions, it is impossible for the plants to maintain their regular metabolic activities, because in waterlogged conditions the root system cannot properly metabolize energy and suffers from fermentation [16, 17]. Consequently, anaerobic metabolism triggers an increase in glycolysis, increasing gene transcription of enzymes related to ethanol fermentation. In addition to these alterations, carbohydrate metabolism produces more substrates for fermentation [17]. Concomitantly, the activity of the antioxidant system increases, and in the final stages, opportunistic pathogens attack the roots [18].
In this context, it is very important to identify alterations at the molecular level in plants with FY versus healthy ones. This can shed light on the tolerance mechanism associated with this problem. In this respect, proteomic techniques enable obtaining a protein profile with precision and sensitivity with the help of mass spectrometry and bioinformatics tools. These techniques have been used to analyze plant responses to different environmental conditions, including soil flooding [19, 20, 21]. In a recent work, Vargas et al. (2016) [22] established a protocol for analysis of metabolites in oil palm leaves, which can contribute to the identification of biochemical markers for FY. In addition, techniques in proteomics should help improve the knowledge about metabolic changes related to FY tolerance or development.
Our hypothesis is that the abiotic factors can favor the start of FY and this problem can be aggravated by biological agents during its development. The objective of this study was to obtain the proteome differential of plants with and without apparent FY symptoms, to identify proteins related with the tolerance, start and/or development of FY in oil palms.
2. Material and methods
2.1. Plant material
Oil palm roots were sampled in field conditions of two areas in August 2016 (after a period of higher rainfall, when the incidence of FY in the field is greater), in a sandy yellow dystrophic latosol in the municipality of Mojú, Pará state, in northern Brazil (1°26’S and 48°26’W, 21 m above sea level). One area belongs to the company Marborges Agroindústria S.A. (area I) and another to the company Biopalma (area II). Sampling was carried out at Marborges S.A and Biopalma S.A farms with their logistic support, safety instructions and authorizations. No specific permission were required for these locations/activities and the study did not involve endangered or protected species. The region has tropical climate with mean annual temperature of 25°C and average rainfall 2,319 mm, mainly distributed from January to August. The plants of area I are progenies of Deli x Lamé of planting dated of 2000. Plants of area II are progenies of Deli x Nigeria and the planting date of 2010. The plants were cultivated in full sunlight with spacing of 9.0 x 9.0 m. The standard crop management was performed in relation to soil nutrition and control of pathogens and insects. Irrigation was not necessary due to the abundant rainfall during the entire crop development. Asymptomatic plants and plants with symptoms in the initial, intermediate and late stages of FY symptoms were collected according to the classification proposed by Souza et al. (2000) [23].
The roots were collected 1 m from a stipe basis in a hole with and 50 x 50 x 20 cm length, width and depth, respectively. After washing with water, the roots were kept in liquid nitrogen and transported to the laboratory of Instituto Tecnológico Vale.
For comparisons between plant proteomes with and without FY symptoms, roots from fifteen plants were pooled in order to obtain three biological replicates for each condition, each replicate consisting of roots from five plants. For the proteomic analyzes between the different stages of FY development, roots from five plants were pooled to obtain one sample for each stage. Three analytical replicates (LC-MS runs) were obtained from each sample.
2.2. Protein extraction and quantification
Proteins were isolated following the SDS (sodium dodecyl sulfate)/phenol protocol proposed by Wang (2006) [24] with some modifications (S1 Table). The protein concentration of each sample was measured on the Qubit 2.0 fluorometer (Invitrogen, Thermo Fisher Scientific), using Qubit protein assay kit according to the manufacturer's protocol.
2.3. Protein digestion
For protein digestion, 50 μg of proteins from each sample were treated with 5 mM of dithiothreitol (DTT) for 25 minutes at 56°C and then with 14 mM of iodoacetamide (IAA) for 30 minutes at room temperature. Then residual quenching of the IAA was performed by adding 5 mM of DTT for 15 minutes at room temperature. After 1/5 (v/v) dilution of the samples, ammonium bicarbonate (50 mM) was added to CaCl2 (1 mM) for all the samples, followed by addition of 20 ng/μL of trypsin (Trypsin Gold, Promega, WI, USA). The samples were left for digestion for 16 hours at 37°C. The enzymatic reaction was stopped by adding 0.4% trifluoroacetic acid (TFA).
2.4. Protein desalting
Samples were desalted using a C18 Sep-Pack column (Oasis) for solid-phase extraction. The column was conditioned with 3 mL of 100% acetonitrile (ACN); equilibrated with 1 mL of 50% ACN 50%/0.1% formic acid and then 0.1% TFA (3 mL). The samples were loaded into the column and washed with 3 mL of 0.1% TFA; equilibrated with 0.1% formic acid (1 mL). The samples were then eluted with, in order, 50% ACN/0.1% formic acid (2 mL) and 80% ACN/0.1% formic acid (1 mL), followed by drying in a vacuum concentrator and resuspension using 50 μL of ammonium formate 10 mM, before UPLC-MS injection.
2.5. 2D-UPLC- mass spectrometric analysis
An aliquot containing (4.5 μg of each sample was loaded for separation into an Nano Acquity UPLC® System (Waters Corp.) equipped with 2D online dilution technology. The first chromatographic dimension of the peptide fraction was ascertained under basic (pH = 10) conditions in a BEH C18 300 Å, 5 μM 300 um x 50 mm reverse phase column (XBridgeTM, Waters Corp.). This was performed at a flow rate of 2 μL/min. Eluent A was aqueous 20 mM FA (pH = 10) and eluent B was neat ACN. All samples were analyzed using a five-step fractionation method. The fractions were eluted from the first dimension using a composition of 10.8, 14.0, 16.7, 20.4 or 65% of eluent B, respectively.
The fractionation process was programmed to start immediately after completion of sample loading (20 min at 10 μL/min with 3% B). Each first dimension elution step was performed with 20 min run time using a flow rate of 2 μL/min. Eluent peptide was mixed online with 10 μL/min of 0.1% TFA solution (1:10 dilution) before being trapped in the trapping column (100 μm x 100 mm), packed with 1.7 μm 100 Å silica-based C18 (Symmetry, Waters Corp, Milford, MA).
The mobile phase for the second chromatographic dimension (low pH RS) was 0.1% FA in water (immobile phase A) and 0.1% FA in ACN (mobile phase B). The second dimension column was 100 μm x 10 mm C18 packed with changed surface hybrid (CSH) 1.8 mm particles (Acquity UPLC M-Class CSH C18, Waters Corp., Milford, MA). The flow rate for the second dimension separation was 400 nL.min-1, while the column was maintained at 55°C. A 40-minute gradient from 3 to 40% B was used to separate peptides in the second separation dimension. The column was then washed using 90% B for 1 minute and equilibrated with 3% B for 7 minutes before returning to the next of fractionation.
Mass spectra were obtained with a Synapt G2-S spectrometer equipped with standard electrospray ionization (ESI) source (Waters). For all measurements, the mass spectrometer was operated in positive ion resolution mode. Mass spectra were acquired in continuum mode over an m/z range of 50–1200, using a capillary voltage of 2.6 KV, source temperature of 100°C, source offset voltage of 100 V, cone gas flow of 50 L/h and cone voltage of 40 V. The spectral acquisition time at each energy setting was 0.5 seconds. A solution of 0.2 μM Glu1-fibrinopeptide (785.8427 Da) was used as a lock-mass solution, delivered at a flow rate of 0.5 μL/min using an auxiliary pump of the liquid chromatography system. The lock-mass was sampled every 30 sec using 0.1 second scans over the same mass range.
2.4. Experimental design and data analysis
We compared asymptomatic and symptomatic roots of plants collected in two different areas. Datasets were analyzed separately. A comparison was also made between proteomes obtained from roots in the initial, intermediate and advanced stages (stages 1, 5 and 9, respectively) in order to identify differential accumulation throughout the progression of FY symptoms. This last experiment was performed only with the proteomes of roots of plants sampled in the first area.
The peptic identification list was generated by the Protein Lynx Global Server (PLGS) 3.0.2 (Waters Corp, Milford, MA, USA) using a combination of exact mass and MSE fragment data. Processed spectra were then searched against a custom protein database compiled from Elaeis guineensis Jacq. at the website of the National Center for Biotechnology Information (NCBI:, 04/2016). Management and validation of mass spectrometry data were performed using the Scaffold Q+ (Scaffold version 4.5.1, Proteome Software Inc., Portland, OR). Protein identification was only accepted if the peptide identification probability was greater than 90% and proteins greater 95% accordingly to the peptideprophet and proteinprophet algorithms [25]. Differentially expressed proteins were determined by applying a permutation test with significance level grater than 95% (p < 0.05). Statistical significance (P-values) for quantitative measurements are available in S2A, S3A and S4A Tables. With few exceptions, in this study we used the cutoff criterion of more or less abundant proteins of log2 fold change ≥ 1 for more abundant proteins and log2 fold change ≤ -1 for less abundant proteins. In addition, we highlighted proteins identified in at least two replicates. Functional annotation of proteins was performed with Blast2GO version 4.0 (Biobam). The heatmap with proteins involved in stress response and energy metabolism was calculated by the R statistical software, through the utilization of the heatmap.2 function available in the gplots R package.
3. Results
3.1. Oil palm root protein profile from FY occurrence areas
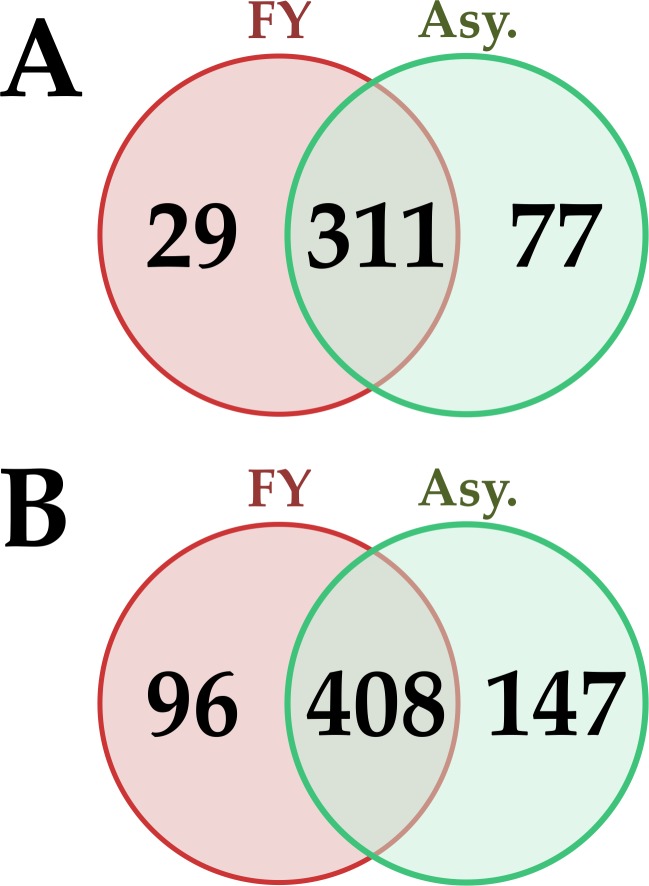
All told, 417 and 651 proteins were identified and quantified in roots of oil palms sampled from areas I and II, respectively. The set of proteins presented some distinctions between the plants of the two areas. All the proteins identified in this study are detailed in S2, S3 and S4 Tables. Proteomic data distribution is displayed in the Venn diagram (Fig 1).
Fig 1. Venn diagram of proteins identified in oil palm roots of plants with symptoms (FY.), asymptomatic (Asy.), and in both conditions (intersection).
(A) Proteins from plants sampled in area I. (B) Proteins from plant sampled from area II.
Comparing plant protein profiles with and without FY symptoms, 127 proteins were up-regulated and 162 were down-regulated in plants with FY symptoms. In plants of area II, 179 and 239 proteins were up- and down-regulated, respectively.
Among the most differentially accumulated proteins present in the current dataset are those involved in the production of energy and proteins related to different mechanisms of stress response.
3.2. Proteins related to biotic and abiotic stresses
In plants of area I, several proteins related to stress responses were differentially abundant (Table 1).
Table 1. Differentially abundant proteins directly or indirectly related to stress response in oil palm roots of area I with log2 fold change ≥ 1 or ≤ -1.
| Protein name | MWa (kDa) | Accession | Log2 FCb |
|---|---|---|---|
| Hypersensitive-induced response protein 1 | 31 | gi|743755084 | FYc |
| Nucleoside diphosphate kinase B-like | 17 | gi|743827402 | FY |
| Nucleoside diphosphate kinase B | 16 | gi|743778585 | FY |
| Glucan endo-1,3-beta-glucosidase-like | 37 | gi|743785400 | 6.51 |
| Patellin-3-like | 54 | gi|743816880 | 5.77* |
| Patellin-3-like | 61 | gi|743764635 | 4.35 |
| Hypersensitive-induced response protein 1 X2 | 31 | gi|743798950 | 2.32 |
| 22.0 kDa class IV heat shock protein-like | 22 | gi|743765011 | 2.22 |
| Acidic endochitinase-like | 31 | gi|743796702 | 2.14 |
| Hydroxyacylglutathione hydrolase cytoplasmic | 29 | gi|743891582 | 1.92 |
| Apyrase 2 | 50 | gi|743789264 | 1.91 |
| 14-3-3-like protein D isoform X1 | 30 | gi|743757050 | 1.69 |
| Chaperone protein ClpB1 | 101 | gi|743756256 | 1.57 |
| Guanine nucleotide-binding protein subunit beta-like protein A | 36 | gi|743794305 | 1.51 |
| Formamidase C869.04 | 50 | gi|743761007 | 1.42 |
| Peroxiredoxin | 17 | gi|192910922 | 1.29 |
| Leucine aminopeptidase 2, chloroplastic-like | 56 | gi|743857317 | 1.29 |
| Glycine-rich RNA-binding protein | 16 | gi|648174145 | 1.22 |
| Beta-1,3-glucanase | 36 | gi|192910884 | 1.2 |
| Flavonoid 3',5'-methyltransferase-like | 27 | gi|743813658 | 1.18 |
| Caffeoyl-CoA O-methyltransferase-like isoform X1 | 22 | gi|743813662 | 1.14 |
| 18.1 kDa class I heat shock protein | 18 | gi|743810653 | 1.14 |
| Uncharacterized protein phloem protein 2-like A4-like | 20 | gi|743855845 | 1.12 |
| Guanine nucleotide-binding protein subunit beta-like protein A | 35 | gi|743772066 | -1.25 |
| Thaumatin-like protein 1b | 25 | gi|743826113 | -1.26 |
| Oryzain alpha chain-like | 51 | gi|743805669 | -1.27 |
| Lipoxygenase homology domain-containing protein 1-like | 19 | gi|743778359 | -1.31 |
| Superoxide dismutase [Cu-Zn], chloroplastic | 23 | gi|743852970 | -1.38 |
| Pathogenesis-related protein | 18 | gi|192910872 | -1.44 |
| Glucan endo-1,3-beta-glucosidase-like | 36 | gi|743875101 | -1.53 |
| Annexin D1-like | 36 | gi|743849454 | -1.75* |
| Glutathione S-transferase | 24 | gi|448872672 | -1.85 |
| Universal stress protein A-like protein | 24 | gi|743845102 | -2.05 |
| Membrane steroid-binding protein 2 | 29 | gi|743776234 | -2.22 |
| Aspartic proteinase oryzasin-1-like | 59 | gi|743794899 | -2.5 |
| Osmotin-like protein | 28 | gi|743775988 | -3.26 |
| Aspartic protease in guard cell 1-like | 48 | gi|743766057 | Asyd. |
| Profilin 2 | 14 | gi|192910850 | Asy. |
| Pathogenesis-related protein PRB1-2-like | 23 | gi|743761748 | Asy. |
| Fumarilacetoacetase | 47 | gi|743767417 | Asy. |
| 20 kDa chaperonin, chloroplastic-like | 27 | gi|743774176 | Asy. |
| Peptidyl-prolyl cis-trans isomerase FKBP12 isoform X1 | 12 | gi|743849924 | Asy. |
| Universal stress protein A-like protein | 18 | gi|743773844 | Asy.* |
| Mannose/glucose-specific lectin-like isoform X2 | 21 | gi|743759608 | Asy.* |
| Pathogenesis-related protein PR-4-like | 15 | gi|743774487 | Asy.* |
| Subtilisin-like protease | 81 | gi|743774266 | Asy.* |
| Ferritin-4, chloroplastic-like | 29 | gi|743873486 | Asy.* |
| L-ascorbate oxidase homolog | 61 | gi|743793209 | Asy. |
| Peroxidase 17-like | 39 | gi|743840871 | Asy. |
| L-ascorbate peroxidase 6, chloroplastic | 39 | gi|743816733 | Asy. |
| Peroxidase 12-like, partial | 22 | gi|743763659 | Asy. |
| Superoxide dismutase [Cu-Zn] | 15 | gi|743845883 | Asy. |
| L-ascorbate peroxidase, cytosolic-like | 28 | gi|743787774 | Asy.* |
In plants of this area, proteins well-known for their importance in plant defense response such as glucan endo-1,3-beta-glucosidase, acidic endochitinase, apyrase 2, flavonoid 3', 5'-methyltransferase, patellins and caffeoyl-CoA O-methyltransferase were up-regulated in plants with FY symptoms. Among proteins with reduced accumulation were thaumatin, lipoxygenases, some pathogenesis-related proteins, annexins, subtilisin and osmotin (Table 1). On the other hand, most of the proteins involved in the response mechanisms of oil palm roots of area I were more accumulated or identified only in asymptomatic plants. Still regarding asymptomatic plants, among the specific proteins identified from the antioxidant system in oil palm roots sampled from area I were L-ascorbate peroxidase, and superoxide dismutase [Cu-Zn].
In plants sampled from area II, we can highlight two proteins (LOC105031936 and LOC105038753) that had similarity (blastp) with sieve element occlusion (accessions gi|743864273 and gi|743765428, NCBI). These proteins were exclusively found in plants with FY symptoms (Table 2). Most of the proteins which were found only in plants with symptoms or that were up-regulated in this condition are related to defense against fungal pathogens. On the other hand, stress-related proteins involved in response against biotic and abiotic stress, such as S-adenosylmethionine synthase, transketolase and isoflavone reductases, among others, and those involved in the antioxidant system, were identified only in asymptomatic plants.
Table 2. Differentially abundant proteins directly or indirectly related to the stress response in roots of plants of area II with log2 fold change ≥ 1 or ≤ -1.
| Protein name | MWa (kDa) | Accession | Log2 FCb |
|---|---|---|---|
| L-ascorbate peroxidase 4 | 31 | gi|743779328 | FYc |
| Nucleoredoxin 1–1 | 69 | gi|743840630 | FY |
| Glutathione S-transferase | 25 | gi|743792918 | FY |
| Cationic peroxidase 1-like | 34 | gi|743782272 | FY |
| Glutathione S-transferase F11-like | 25 | gi|743889616 | FY |
| Cationic peroxidase SPC4-like | 24 | gi|743817481 | FY |
| Mavicyanin-like | 18 | gi|743859187 | FY |
| Allene oxide cyclase 1, chloroplastic-like | 27 | gi|743756470 | FY |
| Alpha carbonic anhydrase 7-like | 33 | gi|743864471 | FY |
| Chemocyanin-like | 13 | gi|743800853 | FY |
| Alpha-mannosidase | 116 | gi|743813312 | FY |
| Ubiquitin-like isoform X2 | 14 | gi|743857302 | FY |
| Serine protease EDA2 isoform X2 | 50 | gi|743777910 | FY |
| Peroxidase 12-like, partial | 22 | gi|743763659 | FY* |
| Manganese superoxide dismutase | 27 | gi|406870049 | FY |
| Monodehydroascorbate reductase,chloroplastic X2 | 55 | gi|743854818 | FY* |
| Germin-like protein 5–1 | 31 | gi|743855137 | FY* |
| Pathogenesis-related protein 1-like | 17 | gi|743844417 | FY* |
| Mannose/glucose-specific lectin-like X1 | 31 | gi|743759606 | FY* |
| Subtilisin-like protease SBT3.5 | 73 | gi|743829002 | FY* |
| Beta-1,3-glucanase | 36 | gi|192910882 | FY* |
| Protein LOC105031936 | 81 | gi|743864273 | FY* |
| Protein LOC105038753 | 81 | gi|743765428 | FY* |
| Aspartic proteinase oryzasin-1-like | 59 | gi|743794899 | 5.81 |
| Peroxiredoxin | 17 | gi|192910922 | 4.82 |
| Putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 | 17 | gi|743877681 | 4.72 |
| Osmotin-like protein | 28 | gi|743775988 | 4.15 |
| Caffeoyl-CoA O-methyltransferase-like | 38 | gi|743813686 | 3.57 |
| Pathogenesis-related protein PRB1-2-like | 24 | gi|743761746 | 3.5 |
| L-ascorbate peroxidase, cytosolic-like | 28 | gi|743787774 | 3.44 |
| Oil palm profilin-like allergen PF2 | 14 | gi|34223519 | 3.38 |
| Chaperone protein ClpB1 | 101 | gi|743756256 | 3.35* |
| Glycine-rich RNA-binding protein | 16 | gi|648174145 | 3.19 |
| Hypersensitive-induced response protein 1 X1 | 34 | gi|743798946 | 2.8 |
| Leucine-rich repeat extensin-like protein 2 | 70 | gi|743772323 | 2.52 |
| Universal stress protein A-like protein | 24 | gi|743845102 | 2.16 |
| GTP-binding nuclear protein Ran1B-like | 25 | gi|743769294 | 2.10 |
| Pathogenesis-related protein PRB1-2-like | 23 | gi|743761748 | 2.07 |
| Patellin-3-like | 61 | gi|743764635 | 1.97 |
| Aspartic proteinase in guard cell 1-like | 48 | gi|743766057 | 1.86 |
| 17.4 kDa class III heat shock protein | 18 | gi|743774135 | 1.80 |
| Germin-like protein 5–1 | 24 | gi|743762743 | 1.79 |
| Aldo-keto reductase 2 | 38 | gi|743814040 | 1.46 |
| Formate dehydrogenase, mitochondrial | 41 | gi|743838587 | 1.41 |
| Peroxidase 3-like | 35 | gi|743820630 | 1.29 |
| Remorin-like | 21 | gi|743866636 | 1.19 |
| 26S proteasome non-ATPase regulatory subunit 2 homolog A-like | 98 | gi|743776123 | 1.12 |
| Profilin-2-like | 14 | gi|743795378 | -1.01 |
| Protein IN2-1 homolog B-like | 28 | gi|743892338 | -1.13 |
| Protein DJ-1 homolog D-like | 41 | gi|743834058 | -1.22 |
| Universal stress protein A-like protein | 19 | gi|743784546 | -1.28 |
| Heat shock protein 81-1-like | 80 | gi|743807690 | -1.44 |
| Enoyl-[acyl-carrier-protein] reductase [NADH] 1, chloroplastic | 40 | gi|743808818 | -1.59 |
| Serine carboxypeptidase-like | 59 | gi|743780890 | -1.62 |
| Skin secretory protein xP2-like | 17 | gi|743886010 | -1.68 |
| Subtilisin-like protease | 81 | gi|743774266 | -1.78 |
| Subtilisin-like protease | 82 | gi|743778980 | -1.85 |
| Peroxidase 4-like | 35 | gi|743818796 | -2.03 |
| Hypersensitive-induced response protein 1 | 31 | gi|743755088 | -2.01 |
| Guanine nucleotide-binding protein subunit beta-like protein A | 36 | gi|743794305 | -2.19 |
| Cationic peroxidase SPC4-like | 38 | gi|743817476 | -2.19 |
| Superoxide dismutase [Cu-Zn], chloroplastic | 23 | gi|743852970 | -2.7 |
| Proteasome subunit alpha type-7 | 27 | gi|743855249 | -2.74 |
| Aminopeptidase M1-like | 101 | gi|743769768 | -2.78 |
| Chaperonin CPN60-2, mitochondrial-like | 61 | gi|743826168 | -3.54 |
| Uncharacterized protein YDL057W isoform X1 | 32 | gi|743797589 | -3.58 |
| Apyrase 2 | 50 | gi|743789264 | -5.25 |
| Uncharacterized protein LOC105034060 | 28 | gi|743875660 | -6.16 |
| Isoflavone reductase-like protein isoform X1 | 34 | gi|743871605 | Asyd* |
| Isoflavone reductase-like protein | 28 | gi|743871643 | Asy.* |
| Oryzain alpha chain-like | 51 | gi|743822079 | Asy. |
| BAG family molecular chaperone regulator 7 | 48 | gi|743872681 | Asy. |
| Tuliposide A-converting enzyme 2, chloroplastic-like | 35 | gi|743860456 | Asy.* |
| Caffeic acid 3-O-methyltransferase-like | 40 | gi|743832206 | Asy.* |
| Cinnamyl alcohol dehydrogenase 2-like | 39 | gi|743861182 | Asy.* |
| Annexin D2-like | 36 | gi|743801141 | Asy.* |
| Annexin D2-like isoform X2 | 36 | gi|743773114 | Asy.* |
| Subtilisin-like protease SDD1 | 82 | gi|743874008 | Asy.* |
| Subtilisin-like protease SDD1 | 82 | gi|743874008 | Asy.* |
| Beta-galactosidase 15 isoform X1 | 99 | gi|743813552 | Asy. |
| S-adenosylmethionine synthase | 43 | gi|743783184 | Asy.* |
| Profilin-1-like | 14 | gi|743799588 | Asy. |
| 16.9 kDa class I heat shock protein 2-like | 18 | gi|743772279 | Asy.* |
| Linoleate 9S-lipoxygenase 5 | 99 | gi|743830998 | Asy.* |
| Allene oxide synthase 2-like | 54 | gi|743767001 | Asy.* |
| Nudix hydrolase 3 isoform X1 | 81 | gi|743767540 | Asy. |
| UDP-glucuronic acid decarboxylase 6 isoform X1 | 38 | gi|743874409 | Asy.* |
| Dihydroxy-acid dehydratase, chloroplastic | 67 | gi|743852824 | Asy. |
| Membrane steroid-binding protein 2 | 29 | gi|743776234 | Asy.* |
| Leucine-rich repeat extensin-like protein 5 | 71 | gi|743802043 | Asy. |
| Cysteine synthase | 34 | gi|743774724 | Asy.* |
| Syntaxin-71-like isoform X2 | 30 | gi|743795463 | Asy. |
| Protein phosphatase 2C 62 isoform X1 | 31 | gi|743758875 | Asy. |
| 17.3 kDa class II heat shock protein-like | 17 | gi|743799089 | Asy.* |
| Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase-like isoform X2 | 44 | gi|743763413 | Asy. |
| Proteasome subunit alpha type-4 | 27 | gi|743873727 | Asy. |
| Transketolase, chloroplastic | 81 | gi|743854750 | Asy.* |
| Glutathione S-transferase omega-like 2 | 39 | gi|743771606 | Asy. |
| Peroxidase 15-like | 35 | gi|743839458 | Asy. |
| Thioredoxin reductase NTRB | 39 | gi|743821878 | Asy. |
| Glutathione S-transferase 3 | 24 | gi|743844790 | Asy. |
| Peroxidase 72-like | 36 | gi|743838248 | Asy. |
| Thioredoxin H1 | 13 | gi|743759544 | Asy.* |
| Uncharacterized protein LOC105042730 | 59 | gi|743775881 | Asy. |
| Peroxidase 3-like | 36 | gi|743768213 | Asy.* |
3.3. Energy metabolism
Overall, proteins related to energy metabolism presented greater accumulation in asymptomatic oil palm roots of the two sampling areas. Most of these proteins are involved in carbohydrate metabolism, mainly in glycolysis (Tables 3 and 4). Noteworthy, enzymes involved in energy metabolism under anaerobic conditions were also identified.
Table 3. Differentially abundant proteins related to energy production in roots of plants of area I with log2 fold change ≥ 1 or ≤ -1.
| Protein name | MWa (kDa) | Accession | Log2 FCb |
|---|---|---|---|
| Pyruvate kinase, cytosolic isozyme | 41 | gi|743816338 | 1.52 |
| Glyceraldehyde-3-phosphate dehydrogenase GAPCP1, chloroplastic-like | 57 | gi|743855918 | 1.51 |
| Fructose-bisphosphate aldolase 1, chloroplastic-like | 42 | gi|743800712 | -1.17 |
| Pyruvate kinase, cytosolic isozyme | 46 | gi|743852444 | -2.32 |
| D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like | 65 | gi|743809510 | Asyc. |
| 2,3-bisphosphoglycerate-independent phosphoglycerate mutase-like | 61 | gi|743843487 | Asy. |
| 2-isopropylmalate synthase A-like | 68 | gi|743821259 | Asy.* |
| Aldehyde dehydrogenase family 2 member B7, mitochondrial-like | 62 | gi|743767790 | Asy.* |
Table 4. Differentially abundant proteins related to energy production in roots of plants of area II with log2 fold change ≥ 1 or ≤ -1.
| Protein name | MWa (kDa) | Accession | Log2 FCb |
|---|---|---|---|
| Pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial-like | 68 | gi|743880374 | FYc* |
| Succinate-semialdehyde dehydrogenase, mitochondrial isoform X2 | 54 | gi|743768016 | FY |
| Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial isoform X1 | 57 | gi|743759731 | FY |
| V-type proton ATPase catalytic subunit A | 68 | gi|743809830 | 3.32 |
| Uncharacterized oxidoreductase At4g09670-like | 40 | gi|743773279 | 1.34 |
| Pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial-like isoform X2 | 46 | gi|743811207 | 1.22 |
| Aldehyde dehydrogenase family 2 member B7, mitochondrial-like | 62 | gi|743767790 | -1.16 |
| Isocitrate dehydrogenase [NADP] | 47 | gi|743755796 | -1.95 |
| Bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 | 57 | gi|743876340 | -2.25 |
| Dihydrolipoyl dehydrogenase, mitochondrial-like | 57 | gi|743855576 | -2.26* |
| 2-isopropylmalate synthase A-like | 55 | gi|743821259 | -2.74 |
| Pyruvate kinase, cytosolic isozyme | 68 | gi|743775291 | -4.2 |
| UDP-sugar pyrophosphorylase | 55 | gi|743804515 | Asy.d |
| 6-phosphogluconate dehydrogenase, decarboxylating 1-like | 54 | gi|743826796 | Asy. |
| Cytochrome b5-like | 15 | gi|743765463 | Asy. |
| V-type proton ATPase subunit B 2-like isoform X2 | 54 | gi|743797544 | Asy.* |
| V-type proton ATPase subunit G-like | 12 | gi|743757417 | Asy. |
| Uncharacterized protein LOC105037637 | 45 | gi|743892552 | Asy.* |
Still regarding proteins involved in the anaerobic metabolism, a sequence (gi|743767790) of the enzyme aldehyde dehydrogenase was identified in plant samples from the two growing areas with greater accumulation in asymptomatic plants, as shown in Tables 3 and 4. Moreover, alcohol dehydrogenases were also detected in the proteomes of plants from both areas, with high intensity in all analyzed samples (S2 and S3 Tables).
3.4. Protein profiling of different stages of FY symptoms
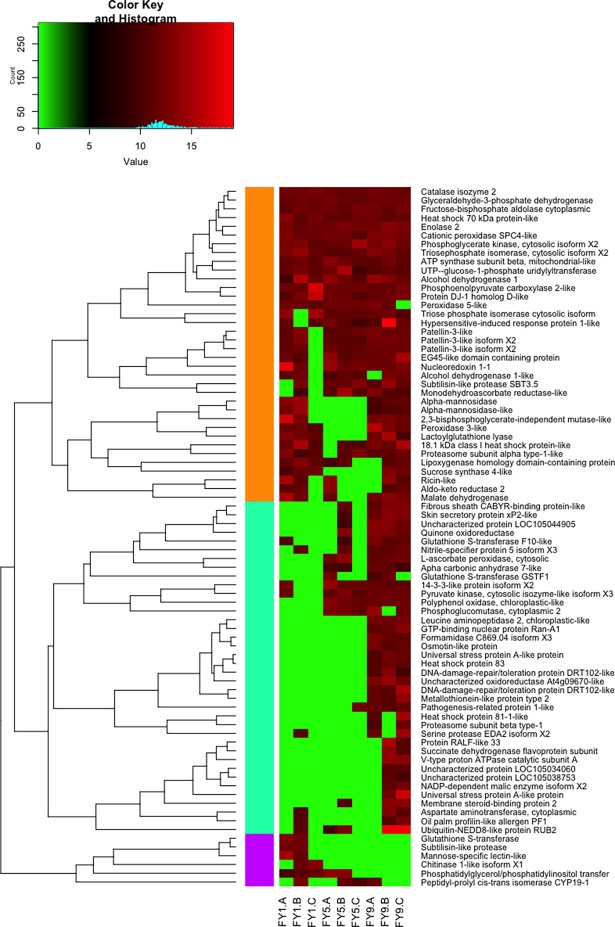
Root proteomes of three different stages of plants afflicted with FY revealed 367 proteins, which were grouped in 197 clusters. As expected, the majority of the detected proteins are related to stress, defense and energy metabolism, including processes such as transport, signalization and oxi-reduction.
A hierarchical grouping based on protein accumulation profiles involved in response to stress and energy metabolism at different stages of symptom severity revealed the formation of three groups indicated by orange, green and purple bars, respectively (Fig 2). The first group (orange bar) consisted mainly of proteins that showed high accumulation in the three stages of severity of FY symptoms. Noteworthy are those involved in the antioxidant system and the metabolism of carbohydrates. In addition, sequences of alcohol dehydrogenase were identified with high intensity from the onset of symptoms, and were grouped together with other proteins involved in energy production. The second group (green bar) included proteins with high accumulation, mainly in the most advanced stage of FY severity (FY9). In this group, we observed proteins related to biotic stress response and ROS homeostasis. The third group (purple bar) consisted of a small number of proteins identified only in the initial (FY1) or intermediate (FY5) stages. This group consisted of only six proteins, with the majority being related to the response to biotic and abiotic stresses.
Fig 2. Hierarchical grouping of differentially expressed proteins related to stress response and energy metabolism in oil palm roots at three stages of severity of FY symptoms.
Three analytical replicates were analyzed for each stage: initial (FY1.A, FY1.B and FY1.C), intermediate (FY5.A, FY5.B and FY5.C) and advanced (FY9.A, FY9.B and FY9.C).
4. Discussion
4.1. Stress-related proteins and their possible relationships with FY
The lower accumulation of stress-responsive proteins in plants with fatal yellowing symptoms and the identification of some of these proteins only in asymptomatic plants suggest that the two cultivated fields may be in areas susceptible to FY development, but asymptomatic plants still exhibited greater resistance to a stress, which could be related to FY development. In this context, proteins identified in this study may be fundamental to the mechanism of resistance to FY.
Among them we identified proteins involved in phenylpropanoid and lignin metabolism to be down-regulated in plants with FY symptoms. Transketolase and isoflavone reductase are involved in phenylpropanoid synthesis, which are important to stress response [26, 27]. Henkes et al. (2001) [26] verified that partial inhibition in transketolase activity resulted in decreased production of phenylpropanoid metabolism products like lignin, as well as chlorophyll and carotene, in tobacco. Isoflavone reductase is essential for the response to various biotic and abiotic stresses [27], by participating in the synthesis of phytoalexins that help increase tolerance in plants submitted to different environmental stresses. The decrease in phenylpropanoid biosynthesis can be considered one of the determining factors for aggravation of FY symptoms. Down-regulation of those enzymes, as found in this study, can be determinant of the FY development in symptomatic plants.
Concerning lignin metabolism, the enzymes cinnamyl alcohol dehydrogenase and caffeic acid 3-O-methyltransferase were also identified only in asymptomatic plants. These enzymes participate in the biosynthetic pathway of lignin. Caffeic acid 3-O-methyltransferase belongs to a family of methyltransferases that are dependent on S-adenosylmethionine. One sequence related to S-adenosylmethionine synthase was also identified only in asymptomatic plants. This enzyme also plays a crucial role in methionine biosynthetic metabolism, besides protein biosynthesis. The up-regulation of S-adenosylmethionine synthase in plants has been related to biotic and abiotic stress responses, like those caused by herbivore insects and salt stress [28].
Lignin is the second most abundant biopolymer in plants after cellulose and it is essential for the structural rigidity, and defense mechanism of plants, acting as a physical barrier to pathogen attack [29]. Thus, the identification of caffeic acid 3-O-methyltransferase, S-adenosylmethionine synthase and cinnamyl alcohol dehydrogenase only in roots of asymptomatic plants suggests that root lignification is a fundamental process against aggravation of FY symptoms, promoting greater resistance against the appearance of necroses already seen in roots of plants with FY [14] and dissemination of opportunistic pathogens in roots of weakened plants.
Other proteins involved in the response to pathogen attacks are the sieve occlusion elements (SEO), which act in phloem tubes for immediate sealing after injuries. They have also been found to be efficient in avoiding photoassimilate loss and pathogen diffusion by the phloem [30]. Froelich et al. (2011) [31] performed studies with these this protein class in A. thaliana and reported its positive effects on defense against pathogens, in addition to not showing any obstruction of sieve. On the other hand, there is a possibility that agglomerate formation may obstruct the sieve translocation [30], which impairs the mass fluxes inside the plant. Recently, Srivastava et al. (2016) [32] also observed that the expression of these proteins increases the tolerance to salinity stress in tobacco. In this way, this class of proteins does not only act in response to biotic stress. However, more research is necessary to understand the enhanced accumulation of these proteins in FY conditions in oil palms.
Considering plants with symptoms at different stages of FY (initial, intermediate and advanced), proteins related to biotic stresses were more abundant in roots with more advanced symptoms (Fig 2), which is consistent with the hypothesis of increased infection by opportunistic pathogens. Therefore, even though a wide variety of stress response related proteins were identified in asymptomatic plants, this does not imply that the development of FY symptoms is initiated by a biotic agent. However, the hypothesis that FY has its initial symptoms occasioned by a biotic agent should not be ruled out.
4.2. Proteins associated with antioxidation and detoxification
FY is largely an unknown syndrome or pathology in oil palms andscarce physiological evidences are available to understand this species’ behavior in environments subject to flooding or waterlogged soil. The FY symptoms have been associated with soil compaction, resulting in lower hydraulic conductivity and porosity. This fact has already been observed in clayey soils containing oil palm crops with high FY incidence [33, 6, 34].
In waterlogged soils, the absorption of nutrients like Ca and P is impaired, and some elements like Fe and Mn can be reduced to more soluble forms and reach toxic levels to the plant [35, 11, 14, 36, 37]. Concomitantly, these effects can trigger oxidative stress [35, 38]. This effect can be enhanced when flooding and post flooding are combined with high irradiance and temperature, as it regularly occurs in the Amazon Region.
Proteins related to the antioxidant system had greater accumulation in asymptomatic plants in the two oil palm growing areas (Tables 1 and 2). The formation of reactive oxygen species is related to the regulation of signaling pathways and initial responses to various environmental stresses, including excessive accumulation of essential elements. Concomitantly, the antioxidant system acts for O2 and H2O2 elimination in different subcellular compartments [39, 40, 20]. These mechanisms have been shown to be fundamental for the resistance to oxidative stress such as that caused by soil flooding, based on previous studies carried out with proteomic analyses in crops submitted to this stress [19, 20, 21].
Still regarding protection against oxidative stress, ferritin is an important protein for plants’ detoxification. In plants, ferritins are present in the plastids and are involved in Fe transport and storage due to their high affinity and capacity to accommodate this element [41]. The increase in ferritin expression also occurs in response to oxidative stress, since the excess of Fe generates reactive oxygen species through the Haber-Weiss reaction [42, 43]. Increased levels of free Fe in cell is toxic, and the ferritin acts as a detoxification protein [36, 38, 44]. In this study, iron concentrations in the plants were not verified. However, it is common to observe an increase in the concentration of iron in plants submitted to hypoxic environments such as flooded soils [45, 43, 46].
Proteomic [47] and gene expression [48] assays reported accumulation of ferritin in plants submitted to flooding, suggesting that this protein plays a significant role in the defense of plants against oxidative stress in this condition.
Ferritin was identified only in asymptomatic plants of area I. Knowing the significant role of ferritin in iron homeostasis in plant cells, its detoxification could be an essential process in oil palm resistance to root structural weakness and greater tolerance to FY development. This enzyme together with other components of the antioxidant system can be considered as an important factor to the tolerance of FY in response to flooded or waterlogged soils.
Asymptomatic plants showed greater accumulation of proteins related to biotic and abiotic stress compared with plants showing symptoms. The synergic action of stress related-proteins in asymptomatic plants can enable higher tolerance to FY development in these genotypes.
Asymptomatic plants may comprise genotypes with adaptations that allow greater tolerance to different environmental stresses, biotic or abiotic. This is due to a more effective response, including expression of genes for the synthesis of proteins involved in mechanisms related to plant tolerance to stresses such as flooding, insect and pathogen defense, and response to oxidative stress that may occur in a variety of situations. However, few studies have been conducted in oil palm to identify Elaeis guineensis varieties with different genotypes or with different degrees of tolerance to FY. In a study of genetic characterization, Costa et al. (2014) [49] compared RAPD (Random Amplification polymorphic DNA) with a population of 51 oil palm plants also from Mojú, Pará State, Brazil, where 24 plants had the symptoms of FY and 27 were apparently healthy. The authors did not observe significant differences to genetically discriminate within groups of affected plants and healthy ones, not being able to attribute a genetic cause to FY.
Scientific advances have allowed the use of genome-wide molecular markers for the identification of genetic alterations, allowing the discrimination of genotypes (cultivars or varieties) of the same species with tolerance-related characteristics to several environmental stresses [50, 51, 52, 53, 54, 55]. Studies with this scope have given subsidies to plant breeding programs [56, 57], but genetic variability in oil palm individuals with and without FY symptoms still deserve further investigation.
4.3. Proteins involved in energy and fermentative metabolism
Components of the primary metabolism are involved in defense in plants [58]. Energy production is fundamental for the expression of genes for the biosynthesis of proteins involved in stress response mechanisms [59]. A positive regulation of transcripts involved in increased energy metabolism has been observed in response to biotic and abiotic stress [60]. However, in contrast to the high levels of energy metabolism proteins since the initial stage of the FY, proteins related to defense mechanisms were more abundant only in the advanced stages of the symptoms.
Additionally, we also identified high intensity of alcohol dehydrogenase in plant roots with and without the symptoms of the FY in the oil palms in both growing areas, and in the proteomes at all three stages of FY severity. Alcohol dehydrogenases are involved in alcoholic fermentation, where they catalyze the conversion of pyruvate to ethanol. During oxygen deficiency, there is a decrease in energy production through oxidative phosphorylation, and the fermentative metabolism promotes energy compensation through the recycling of NAD+ to the glycolytic pathway [61, 62]. Therefore, alcohol dehydrogenase plays a fundamental role in the maintenance of energy metabolism under anaerobic conditions.
The increase of proteins of the glycolytic pathway and involved in the anaerobic respiration related to the alcohol fermentation process has been identified as a key response of plants to hypoxia in flooded soils [19, 63]. In our results, aldehyde dehydrogenase was more accumulated in asymptomatic plants. Under anaerobic conditions, amounts of acetaldehyde are produced which can cause toxicity to the plant. At this time, the aldehyde dehydrogenase converts the alcetaldehyde to acetate. Thus, this enzyme has been related to higher plant survival under anaerobic conditions [64, 65, 66, 67, 68].
Overexpression of the ADH1 gene had no effect on flood tolerance in Arabidopsis, but its levels of expression under anaerobic conditions were found to be critical for plant survival under anaerobic conditions [62]. Moreover, Bertolde et al. (2014) [40] showed that in flooded Theobroma cacao tolerant genotype the ADH gene was overexpressed in comparison to the susceptible genotype. Thus ADH levels in oil palms with and without FY symptoms may not be a critical point for tolerance to the development of symptoms, but an indicator that these plants have been exposed to anaerobiosis.
Furthermore, there may be temporal differences in ADH expression between tolerant plants and more susceptible to hypoxia or anoxia conditions, as well as between different plant species, which would influence response mechanisms for anaerobic survival [69]. Therefore, it is important to emphasize that in this study, plants were analyzed under field conditions and it is not possible to infer the exact moment when plants were submitted to such stress. Thus, the difference between ADH expression “response-time” within plants with and without FY symptoms could also be considered as a factor influencing FY development.
The adaptation to hypoxia or anoxia includes metabolic alterations such as the synthesis of anaerobic stress proteins (ANPs), which includes enzymes of glycolysis, ethanol fermentation and carbohydrate metabolism [16, 70]. In addition to the enzymes already mentioned, other enzymes essential to carbohydrate metabolism were identified, mostly down-regulated in plants with symptoms, as can be seen in Tables 3 and 4. These enzymes were also found in plant roots at different stages of development of FY symptoms, as shown in Fig 2.
The relationship between the higher incidence of FY, increase in rainfall, soil flooding index [10, 12, 13] high levels of glycolytic pathway enzymes and the presence of aldehyde dehydrogenase, alcohol dehydrogenase and many antioxidant enzymes, suggests that the plants from the two areas of oil palm cultivation may have been submitted to a recent or constant state of hypoxia in the roots.
The higher accumulation of proteins involved in energy production, including aldehyde dehydrogenase, in asymptomatic plants, may also be associated with a more efficient response of those genotypes against FY development, through more efficient use of energy under anaerobic conditions.
5. Conclusions
This was the first study aiming to describe protein alterations associated with FY in oil palm roots. Protein analysis proved to be a powerful tool to shed light on the molecular mechanisms related to the tolerance and development of FY disease. Putative protein markers that could to guide selection of FY-tolerant oil palm genotypes were also identified.
Proteins related to anaerobic metabolism were found in all sampled plants, whether symptomatic or not, suggesting a recent or constant condition of hypoxia in their respective environments. Comparing different stages of FY symptoms’ severity, the higher intensity of alcohol dehydrogenase and energy related-proteins since the onset of symptoms contrasted with the increase of biotic stress related proteins in later stages of the syndrome. Our finding suggests that changes in abiotic factors may precede the occurrence of FY, paving the way for opportunistic pathogens.
Supporting information
(PDF)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
(DOC)
Acknowledgments
We would like to thank the companies Marborges and Biopalma, the “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior” (CAPES), the “Universidade Federal Rural da Amazônia” and the “Programa de Pós-Graduação em Biotecnologia aplicada à Agropecuária”.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Instituto Tecnológico Vale. The funder provided support in the form of salaries for authors RBSV, GO and ROA, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Trindade DR, Poltronieri LS, Furlan Jr. Abordagem sobre o estado atual das pesquisas para a identificação do agente causal do amarelecimento fatal do dendezeiro In: Poltronieri LS, Trindade DR, Santos IP, editors. Pragas e doenças de cultivos amazônicos. Embrapa Amazônia Oriental; 2005. pp.439–450. [Google Scholar]
- 2.Mekhilef S, Siga S, Saidur R. A review on palm oil biodiesel as a source of renewable fuel. Vol. 15, Renewable and Sustainable Energy Reviews. 2011. p. 1937–49. https://doi.org/10.1016/j.rser.2010.12.012 [Google Scholar]
- 3.Celestino Filho P, Louise C, Lucchini F. (1993). ESTUDOS DE TRANSMISSÃO DO AMARELECIMENTO FATAL DO DENDEZEIRO (Elaeis guinensis, Jacq) COM INSETOS SUSPEITOS. Anais do 14° Congresso brasileiro de entomologia. 1993;(January):p.194. [Google Scholar]
- 4.Silva HM, Celestino filho P, Trindade DR, Conceição HEO, Mulle AA, Alves RM. ESTADO ATUAL DOS CONHECIMENTOS SOBRE A DOENÇA AMARELECIMENTO FATAL (AF) DO DENDEZEIRO (Elaeis guinensis Jacq) NO ESTADO DO PARÁ. Workshop sobre a cultura de dendê. 1996: p.87-98.
- 5.Sarria G a, Torres G a, Aya H, Ariza J, Rodriguez J, Velez D, et al. Phytophthora sp. es el responsable de las lesiones iniciales de la Pudrición del cogollo (PC) de la Palma de aceite en Colombia. Palmas. 2008;29:31–41. [Google Scholar]
- 6.Martínez G, Sarria G, Torres G, Varón F, Drenth A, Guest D. Nuevos hallazgos sobre la Pudrición del cogollo de la palma de aceite en Colombia: biología, detección y estrategias de manejo. Palmas. 2014;35(1):11–7. [Google Scholar]
- 7.Sarria GA, Martinez G, Varon F, Drenth A, Guest DI. Histopathological studies of the process of Phytophthora palmivora infection in oil palm. Eur J Plant Pathol. 2016;145(1):39–51. https://doi.org/10.1007/s10658-015-0810-9 [Google Scholar]
- 8.Torres G a, Sarria G a, Varon F, Coffey MD, Elliot ML, Martinez G. First Report of Bud Rot Caused by Phytophthora palmivora on African Oil Palm in Colombia. Plant Dis. 2010;94(9):1163 https://doi.org/10.1094/PDIS-94-9-1163A [DOI] [PubMed] [Google Scholar]
- 9.Torres GA, Sarria GA, Martinez G, Varon F, Drenth A, Guest DI. Bud Rot Caused by Phytophthora palmivora: A Destructive Emerging Disease of Oil Palm. Phytopathology [Internet]. 2016;(Turner 1981):PHYTO-09-15-024. Available from: http://apsjournals.apsnet.org/doi/10.1094/PHYTO-09-15-0243-RVW [DOI] [PubMed] [Google Scholar]
- 10.Acosta A, Munevar F. Bud Rot in Oil Palm Plantations: Link to Soil Physical Properties and Nutrient Status. Better Crop Int. 2003;17(2):22–5. [Google Scholar]
- 11.Munévar M. F.; Acosta G. A.; Gómez C. P. Factores edáficos asociados con la Pudrición del cogollo de la palma de aceite en Colombia. Palmas. 2001; 22(2). 9–19. [Google Scholar]
- 12.Venturieri A, Fernandes WR, Boari ADJ. RELAÇÃO ENTRE OCORRÊNCIA DO AMARELECIMENTO FATAL DO DENDEZEIRO (Elaeis guineensis Jacq.) E VARIÁVEIS AMBIENTAIS NO ESTADO DO PARÁ. An XIV Simp Bras Sensoriamento Remoto. 2009;(Abril):523–30. [Google Scholar]
- 13.Martínez LG. Pudrición del cogollo, marchitez sorpresiva, anillo rojo y marchitez letal en la palma de aceite en América. Palmas. 2010; 31: 43–53. [Google Scholar]
- 14.Albertazzi H. Burgarelli J. & Chinchilla C. Onset of spear rot symptoms in oil and prior (and contemporary) events. ASD Oil Palm Papers. 2005; 28: 21–41. [Google Scholar]
- 15.Boari AJ. O desafio das pesquisas com a etiologia do Amarelecimento Fatal (AF) da cultura da palma de óleo. In: Ramalho Filho A, Motta PEF, Freitas PL, Teixeira WG. Zoneamento Agroecológico, Produção E Manejo Para a Cultura Da Palma de Óleo Na Amazônia. Rio de Janeiro. Embrapa solos. 2010. pp153–165. [Google Scholar]
- 16.Drew MMC. Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol [Internet]. 1997;48(1):223–50. https://doi.org/10.1146/annurev.arplant.48.1.223 [DOI] [PubMed] [Google Scholar]
- 17.Voesenek LACJ, Bailey-Serres J. Flood adaptive traits and processes: An overview. Vol. 206, New Phytologist. 2015. p. 57–73. https://doi.org/10.1111/nph.13209 [DOI] [PubMed] [Google Scholar]
- 18.Bostock RM, Pye MF, Roubtsova T V. Predisposition in Plant Disease: Exploiting the Nexus in Abiotic and Biotic Stress Perception and Response. Annu Rev Phytopathol [Internet]. 2014;52(1):517–49. https://doi.org/10.1146/annurev-phyto-081211-172902 [DOI] [PubMed] [Google Scholar]
- 19.Alam I, Lee DG, Kim KH, Park CH, Sharmin SA, Lee H, et al. Proteome analysis of soybean roots under waterlogging stress at an early vegetative stage. J Biosci. 2010;35(1):49–62. https://doi.org/10.1007/s12038-010-0007-5 [DOI] [PubMed] [Google Scholar]
- 20.Yu F, Han X, Geng C, Zhao Y, Zhang Z, Qiu F. Comparative proteomic analysis revealing the complex network associated with waterlogging stress in maize (Zea mays L.) seedling root cells. Proteomics. 2015;15(1):135–47. https://doi.org/10.1002/pmic.201400156 [DOI] [PubMed] [Google Scholar]
- 21.Campos NA, Alves JD, De Souza KRD, Porto BN, Magalhães MM, Da Silva GJ, et al. Evolution of aerenchyma formation in a maize breeding program. Plant Omics. 2016;9(1):19–25. [Google Scholar]
- 22.Vargas LHG, Neto JCR, de Aquino Ribeiro JA, Ricci-Silva ME, Souza MT, Rodrigues CM, et al. Metabolomics analysis of oil palm (Elaeis guineensis) leaf: evaluation of sample preparation steps using UHPLC–MS/MS. Metabolomics. 2016;12(10). https://doi.org/10.1007/s11306-016-1100-z [Google Scholar]
- 23.Souza RLR, Veiga AS, Ramos EJA. Denpasa: Amarelecimento fatal em dendezeiro: identificação prática. 2000. 27p. [Google Scholar]
- 24.Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27(13):2782–6. doi: 10.1002/elps.200500722 [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. https://doi.org/10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 26.Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. A Small Decrease of Plastid Transketolase Activity in Antisense Tobacco Transformants Has Dramatic Effects on Photosynthesis and Phenylpropanoid Metabolism. Plant Cell. 2001;13(March):535–51. https://doi.org/10.1105/tpc.13.3.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Q, Li N, Dong L, Zhang D, Fan S, Jiang L, et al. Overexpression of Soybean Isoflavone Reductase (GmIFR) Enhances Resistance to Phytophthora sojae in Soybean. Front Plant Sci [Internet]. 2015;6 https://doi.org/10.3389/fpls.2015.01024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan R, Wang H, Wang Y, Yu D. Proteomic analysis of soybean defense response induced by cotton worm (prodenia litura, fabricius) feeding. Proteome Sci [Internet]. 2012;10(1):16 https://doi.org/10.1186/1477-5956-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neutelings G. Lignin variability in plant cell walls: Contribution of new models. Vol. 181, Plant Science. 2011. p. 379–86. https://doi.org/10.1016/j.plantsci.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 30.Ernst AM, Jekat SB, Zielonka S, Muller B, Neumann U, Ruping B, et al. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc Natl Acad Sci [Internet]. 2012;109(28):E1980–9. https://doi.org/10.1073/pnas.1202999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Froelich DR, Mullendore DL, Jensen KH, Ross-Elliott TJ, Anstead JA, Thompson GA, et al. Phloem Ultrastructure and Pressure Flow: Sieve-Element-Occlusion-Related Agglomerations Do Not Affect Translocation. Plant Cell [Internet]. 2011;23(12):4428–45. https://doi.org/10.1105/tpc.111.093179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava VK, Raikwar S, Tuteja R, Tuteja N. Ectopic expression of phloem motor protein pea forisome PsSEO-F1 enhances salinity stress tolerance in tobacco. Plant Cell Rep. 2016;35(5):1021–41. https://doi.org/10.1007/s00299-016-1935-9 [DOI] [PubMed] [Google Scholar]
- 33.Silveira RI, Veiga AS, Ramos EJA, Parente JR. Evolução da sintomatologia do amarelecimento fatal a adubações com omissão de macro e micronutrientes. Denpasa. 2000. 35p. [Google Scholar]
- 34.Cristancho JA, Alfonso OA, Molina DL. Revisión de literatura sobre el papel del suelo y la nutrición de plantas en la Pudrición del cogollo de la palm a de aceite The Role of Soil and Plant Nutrition on Oil Palm Bud Rot Disease: A Review. 2012;33. [Google Scholar]
- 35.Bernardes MSR. Relatório de visita à plantações de Paricatuba, na Denpasa, visando à identificação das causas do AF. In: Pesquisa sobre amarelecimento fatal em dendezeiro. Denpasa. 2001. [Google Scholar]
- 36.Villón M. (2007). Drenaje. 1st ed. Editorial tecnológica de Costa Rica; 2007. [Google Scholar]
- 37.Laing D. Defeciência temporária de cálcio como causa primária do amarelecimento fatal do dendezeiro. Informações Agronômicas. 2012: 137. [Google Scholar]
- 38.Briat JF, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, et al. New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Vol. 105, Annals of Botany. 2010. p. 811–22. https://doi.org/10.1093/aob/mcp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hossain Z, Nouri MZ, Komatsu S. Plant cell organelle proteomics in response to abiotic stress. Vol. 11, Journal of Proteome Research. 2012. p. 37–48. https://doi.org/10.1021/pr200863r [DOI] [PubMed] [Google Scholar]
- 40.Bertolde FZ, Almeida AAF, Pirovani CP. Analysis of gene expression and proteomic profiles of clonal genotypes from Theobroma cacao subjected to soil flooding. PLoS One. 2014;9(10). https://doi.org/10.1371/journal.pone.0108705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. Vol. 22, EMBO Journal. 2003. p. 1959–68. doi: 10.1093/emboj/cdg215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.HENDRY GAF, BROCKLEBANK KJ. IRON‐INDUCED OXYGEN RADICAL METABOLISM IN WATERLOGGED PLANTS. New Phytol. 1985;101(1):199–206. doi: 10.1111/j.1469-8137.1985.tb02826.x [DOI] [PubMed] [Google Scholar]
- 43.Bottcher A, Mazzafera P. Ferritin as part of the antioxidative machinery in plants under stress. In: Ferritin: Functions, Biosynthesis and Regulation. 2012. p. 65–81. [Google Scholar]
- 44.Llanos R de, Martínez-Garay CA, Fita-Torró J, Romero AM, Martínez-Pastor MT, Puig S. Soybean ferritin expression in Saccharomyces cerevisiae modulates iron accumulation and resistance to elevated iron concentrations. Appl Environ Microbiol. 2016;82(10):3052–60. https://doi.org/10.1128/AEM.00305-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majerus V, Bertin P, Lutts S. Abscisic acid and oxidative stress implications in overall ferritin synthesis by African rice (oryza glaberrima steud.) seedlings exposed to short term iron toxicity. Plant Soil. 2009;324(1):253–65. [Google Scholar]
- 46.Morales-Olmedo M, Ortiz M, Selles G. Effects of transient soil waterlogging and its importance for rootstock selection. Chil J Agric Res. 2015;75(August):45–56. [Google Scholar]
- 47.Kamal AHM, Rashid H, Sakata K, Komatsu S. Gel-free quantitative proteomic approach to identify cotyledon proteins in soybean under flooding stress. J Proteomics. 2015;112:1–13. doi: 10.1016/j.jprot.2014.08.014 [DOI] [PubMed] [Google Scholar]
- 48.Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010;51(1):21–37. doi: 10.1093/pcp/pcp163 [DOI] [PubMed] [Google Scholar]
- 49.Costa MR, Boari AJ, Fortes ACR, Nascimento SV. Estimativa da variabilidade genéica do dendezeiro (Elaeis guineensis Jacq.) por marcadores RAPD em área de ocorrência da doença amarelecimento fatal. Revista Sodebras. 2014; 9(101):40–43. [Google Scholar]
- 50.1. Metwali EMR, Carle R, Schweiggert RM, Kadasa NM, Almaghrabi OA. Genetic diversity analysis based on molecular marker and quantitative traits of the response of different tomato (Lycopersicon esculentum Mill.) cultivars to drought stress. Arch Biol Sci. 2016;68(2).:427–38. doi: 10.2298/ABS150629126M [Google Scholar]
- 51.Freeg HA, Anis GB, Abo-Shousha AA, El-Banna AN, El-Sabagh A. Genetic Diversity Among Some Rice Genotypes with Different Drought Tolerance based on SSR Markers. Cercet Agron Mold [Internet]. 2016;49(3). doi: 10.1515/cerce-2016-0024 [Google Scholar]
- 52.Pradhan SK, Barik SR, Sahoo A, Mohapatra S, Nayak DK, Mahender A, et al. Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice. PLoS One. 2016;11(8). doi: 10.1371/journal.pone.0160027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh D, Pal M, Singh CK, Taunk J, Jain P, Chaturvedi AK, et al. Molecular scanning and morpho-physiological dissection of component mechanism in Lens species in response to aluminium stress. PLoS One. 2016;11(7). doi: 10.1371/journal.pone.0160073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nadeem MZ, Nawaz MN, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment. 2017: 1314–3530 doi: 10.1080/13102818.2017.1400401 [Google Scholar]
- 55.Wang J, Li R, Mao X and Jing R. Functional Analysis and Marker Development of TaCRT-D Gene in Common Wheat (Triticum aestivum L.). Front. Plant Sci. 2017; 8:1557 doi: 10.3389/fpls.2017.01557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang GL. Molecular marker-assisted breeding: A plant breeder’s review. Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools. 2016;1:431–72. doi: 10.1007/978-3-319-22521-0_15 [Google Scholar]
- 57.Govindaraj M, Vetriventhan M, Srinivasan M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet Res Int [Internet]. 2015;2015:1–14. doi: 10.1155/2015/431487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci [Internet]. 2014;5 https://doi.org/10.3389/fpls.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, et al. Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem. 2002;277(12):10555–61. doi: 10.1074/jbc.M104863200 [DOI] [PubMed] [Google Scholar]
- 60.Less H, Angelovici R, Tzin V, Galili G. Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell [Internet]. 2011;23(4):1264–71. https://doi.org/10.1105/tpc.110.082867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy RA, Rumpho ME, Fox TC. Anaerobic Metabolism in Plants. Plant Physiol. 1992;100:1–6. doi: 10.1104/pp.100.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ismond KP. Enhanced Low Oxygen Survival in Arabidopsis through Increased Metabolic Flux in the Fermentative Pathway. PLANT Physiol [Internet]. 2003;132(3):1292–302. doi: 10.1104/pp.103.022244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nanjo Y, Skultety L, Ashraf Y, Komatsu S. Comparative proteomic analysis of early-stage soybean seedlings responses to flooding by using gel and gel-free techniques. J Proteome Res. 2010;9(8):3989–4002. https://doi.org/10.1021/pr100179f [DOI] [PubMed] [Google Scholar]
- 64.Ismail AM, Ella ES, Vergara G V., Mackill DJ. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann Bot. 2009;103(2):197–209. https://doi.org/10.3389/fpls.2013.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakazono M, Tsuji H, Li YH, Saisho D, Arimura S, Tsutsumi N, et al. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 2000;124(2):587–98. doi: 10.1104/pp.124.2.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuji H, Meguro N, Suzuki Y, Tsutsumi N, Hirai A, Nakazono M. Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice. FEBS Lett. 2003;546(2–3):369–73. doi: 10.1016/S0014-5793(03)00631-8 [DOI] [PubMed] [Google Scholar]
- 67.Meguro N, Tsuji H, Tsutsumi N, Nakazono M, Hirai A. Involvement of aldehyde dehydrogenase in alleviation of post-anoxic injury in rice. In: Abiotic Stress Tolerance in Plants. 2006. p. 111–9. doi: 10.1007/1-4020-4389-9_7 [Google Scholar]
- 68.Miro B, Ismail AM. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front Plant Sci [Internet]. 2013;269(1). doi: 10.3389/fpls.2013.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haque ME, Kawaguchiand K, Komatsu S. Analysis of Proteins in Aerenchymatous Seminal Roots of Wheat Grown in Hypoxic Soils under Waterlogged Conditions. Protein Pept Lett. 2011;18(9):912–24. doi: 10.2174/092986611796011455 [DOI] [PubMed] [Google Scholar]
- 70.Subbaiah CC, Sachs MM. Molecular and cellular adaptations of maize to flooding stress. Vol. 91, Annals of Botany. 2003. p. 119–27. https://doi.org/10.1093/aob/mcf210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
A) Quantitative samples view. B) Protein report. C) Quantitative peptide report. D) Quantitative spectrum report.
(XLS)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.