Abstract
Next-generation DNA sequencing technologies have led to a massive accumulation of genomic and transcriptomic data from patients and healthy individuals. The major challenge ahead is to understand the functional significance of the elements of the human genome and transcriptome, and implications for diagnosis and treatment. Genetic screens in mammalian cells are a powerful approach to systematically elucidate gene function in health and disease states. In particular, recently developed CRISPR/Cas9-based screening approaches have enormous potential to uncover mechanisms and therapeutic strategies for human diseases. The focus of this review is the use of CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) for genetic screens in mammalian cells. We introduce the underlying technology and present different types of CRISPRi/a screens, including those based on cell survival/proliferation, sensitivity to drugs or toxins, fluorescent reporters, and single-cell transcriptomes. Combinatorial screens, in which large numbers of gene pairs are targeted to construct genetic interaction maps, reveal pathway relationships and protein complexes. We compare and contrast CRISPRi and CRISPRa with alternative technologies, including RNA interference (RNAi) and CRISPR nuclease-based screens. Finally, we highlight challenges and opportunities ahead.
Main text
Genetic screens are powerful tools to uncover molecular players in a biological process of interest, and to assign functions to the elements of a genome. While model organisms such as budding yeast have been used extensively for cell-based genetics for decades (due to the often-invoked “awesome power of yeast genetics”), mammalian cells are relatively late arrivals. Until recently, RNA interference (RNAi) and expression of open reading frames (ORFs) from DNA vectors were the standard tools for loss-of-function and gain-of-function genetic screens, respectively, in mammalian cells (Fig. 1). Remarkably, a rapidly expanding set of genetic tools based on clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology has been developed over the last 5 years only (Fig. 1), and is already rivaling and, in many cases, outperforming pre-CRISPR tools. These recent developments are paving the way for a new era, defined by the “awesome power of mammalian cell genetics”.
Figure 1. Technologies to perturb gene function in mammalian cells for pooled genetic screens.
Loss-of-function technologies include: RNA interference (RNAi); CRISPR nuclease (CRISPRn), in which Cas9-mediated DNA cleavage directed to the coding region of a gene by a single guide RNA (sgRNA) results in error-prone repair by cellular non-homologous end joining pathways, thereby disrupting gene function (in particular when frame shifts are introduced); and CRISPR interference (CRISPRi), in which catalytically dead Cas9 (dCas9) fused to a transcriptional repressor domain (e.g., KRAB) is recruited to the transcription start site (TSS) of an endogenous gene, as specified by an sgRNA, to repress transcription.
Gain-of-function technologies include: Overexpression of Open Reading Frames (ORFs) as transgenes; and CRISPR activation (CRISPRa), in which transcriptional activators are recruited via sgRNAs and dCas9 to TSSs of endogenous genes to induce their overexpression. To achieve high levels of overexpression with a single sgRNA, CRISPRa methods recruit more than one transcriptional activator to a given TSS. Multiple activator domains are either directly fused to dCas9 (e.g., VP64, p65 and RTA in the VPR system18), recruited to a protein scaffold fused to dCas9 (e.g. VP64 fused to superfolder GFP (sfGFP) and an antibody single-chain variable fragment (scFv) targeting a GCN4 epitope, which are recruited to a tandem array of 10 copies of the GCN4 epitope in the SunCas system19, 20), or recruited to an RNA scaffold fused to the sgRNA (e.g. p65 and HSF1 transcriptional activation domains fused to MS2 coat protein (MCP), dimers of which are recruited to MS2 RNA hairpins in the Synergistic Activation Mediator (SAM) system21.
CRISPRi and CRISPRa: precision control of gene expression
The CRISPR system is a bacterial adaptive immune system1, the centerpiece of which is a programmable DNA endonuclease that can be reconstituted from only two components: the protein Cas9 from Streptococcus pyogenes (or analogous proteins from other species) and a single guide RNA (sgRNA) that directs the nuclease activity to complementary sequences in the substrate DNA2. CRISPR/Cas9 can be used for genome editing in mammalian cells3–6, by introducing defined deletions or by altering genomic sequences as directed by a homology repair (HR) template for cellular HR repair of CRISPR-mediated DNA cuts. In the absence of a HR template, DNA cuts introduced by CRISPR/Cas9 are subject to non-homologous end joining (NHEJ) repair. NHEJ repair is error-prone and commonly results in short deletions. When introduced within a protein-coding ORF, such deletions can lead to frame shifts that result in a loss of function of the encoded protein. Several genetic screening platforms for mammalian cells have been implemented based on this approach7–9, which we will refer to as CRISPR nuclease (CRISPRn) (Fig. 1).
This review will focus on screening approaches that do not rely on genome editing, but instead on reversible control of gene expression. Nuclease-dead mutants of Cas9 (dCas9) were found to maintain sgRNA-directed binding of specific DNA sequences, which could block transcription of these genes in bacteria10, a process termed CRISPR interference (CRISPRi). CRISPRi based on dCas9 can also represses transcription in mammalian cells, but it is more effective when transcriptional repressor domains are fused to dCas911 (Fig. 1). In particular, fusions of dCas9 to a Krüppel-associated box (KRAB12) domain, which promotes heterochromatin13, were found to potentiate CRISPRi in human cells11.
dCas9 can also be used to activate gene expression, in an approach termed CRISPR activation (CRISPRa). Early implementations of CRISPRa used simple fusions of dCas9 to an activator domain, most commonly VP64, but only achieved modest activation of the targeted gene with single sgRNAs11, 14–17. While this limitation can be overcome by simultaneously using several sgRNAs targeting the same gene, pooled genetic screens generally require effective perturbation of the targeted gene with a single sgRNA. Therefore, CRISPRa constructs recruiting more than one activator domain with a single sgRNA were developed, using one of three strategies (Fig. 1): First, as direct fusions to dCas9, exemplified by the VPR approach18 (tripartite fusion of VP64 and the activation domains of the p65 subunit of NFκB and Epstein-Barr virus R transactivator, Rta). Second, via a protein scaffold, exemplied by the SunTag19, 20 (VP64 fused to superfolder GFP (sfGFP) and an antibody single-chain variable fragment (scFv) targeting a GCN4 epitope, which are recruited to a tandem array of 10 copies of the GCN4 epitope). Third, via an RNA scaffold, exemplified by the SAM approach21 (Synergistic Activation Mediator, in which p65 and HSF1 transcriptional activation domains fused to MS2 coat protein (MCP), dimers of which are recruited to MS2 RNA hairpins).
In combination, CRISPRi and CRISPRa can control transcript levels of endogenous genes over several orders of magnitude19. Thus, they can be used to define the precise relationship between a phenotype of interest and the levels of a given gene product in an otherwise isogenic background. An important application in the context of chemical biology is the measurement of the sensitivity to a compound as a function of the expression level of the putative target of the compound – which can help to establish that the compound does indeed act through the intended target22.
Pooled genetic screens: discovery for biology and medicine
To identify protein-coding genes or other genetic elements that function in a biological process of interest, a genetic screen can be carried out in which large numbers of genes are targeted and those with the strongest phenotypes are selected for validation and mechanistic follow-up experiments. The first genome-scale screens in mammalian cells based on CRISPRn, CRISPRi and CRISPRa were all implemented as pooled screens (Table 1). In a pooled CRISPR-based screen, libraries of sgRNA expression constructs are stably integrated into the genomes of mammalian cells via lentiviral transduction, using conditions in which each cell typically integrates only one sgRNA construct (Fig. 2). A number of different strategies can then be used to determine a phenotype of interest caused by sgRNA-directed gain-of-function or loss-of-function.
Table 1.
Published genome-scale CRISPRi and CRISPRa screens in mammalian cells
| Screening mode | Cell type | Targeted genes | Phenotype | Reference |
|---|---|---|---|---|
| CRISPRi | K562 leukemia cells | Protein-coding | Growth | 19, 24 |
| K562 leukemia cells | Protein-coding | Sensitivity to bacterial toxin | 19 | |
| K562 leukemia, HeLa cervical cancer, U87 glioblastoma, MCF7 and MDA-MB-231 mammary adenocarcinoma, HEK293T and human induced pluripotent stem cells | lncRNAs | Growth | 26 | |
| K562 leukemia cells | Protein-coding | Activation of the unfolded protein response (FACS-based) | 30 | |
| HT29 colorectal cancer and MIAPACA2 pancreatic cancer cells | Protein-coding | Growth | 51 | |
| CRISPRa | K562 leukemia cells | Protein-coding | Growth | 19, 24 |
| K562 leukemia cells | Protein-coding | Sensitivity to bacterial toxin | 19 | |
| A375 melanoma cells | Protein-coding | Resistance to BRAF inhibitor | 21 | |
| A375 melanoma cells | lncRNAs | Resistance to BRAF inhibitor | 87 |
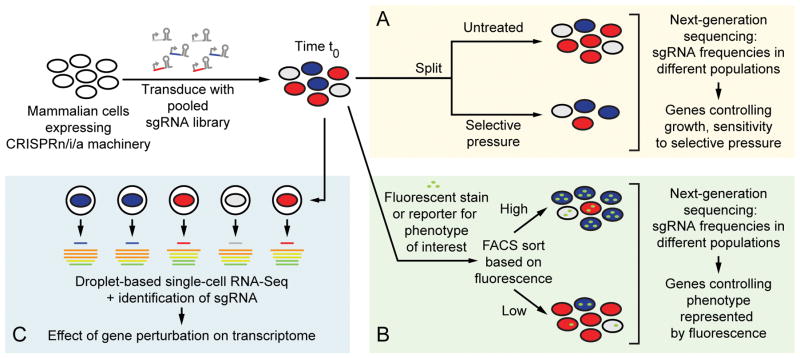
Figure 2. Strategies for pooled genetic screens using CRISPRi or CRISPRa.
Mammalian cells expressing CRISPRi or CRISPRa machinery are transduced by pooled lentiviral libraries for expression of single guide RNAs (sgRNAs) with low multiplicity of infection, resulting in stable integration of mostly a single sgRNA expression construct per cell. (A) The effect of sgRNAs on cell viability and growth or sensitivity to a selective pressure can be quantified by determining the frequencies of cells expressing each sgRNAs in populations at the t0 timepoint and after culture in the absence or presence of selective pressure. (B) The effect of sgRNAs on a phenotype monitored by a fluorescent reporter or antibody can be quantified by next-generation sequencing of populations separated based on phenotype by fluorescence-activated cell sorting (FACS). (C) CRISPR-based genetic screens can be coupled to droplet-based single-cell RNA sequencing to obtain high-dimensional transcriptomic phenotypes for each genetic perturbation.
The most straightforward phenotype for a pooled screen is proliferation and survival – in other words, cellular fitness (Fig. 2A). Fitness (relative to wild type) can be quantified for each sgRNA by comparing the frequencies of cells expressing sgRNAs at an initial time point (t0) to those at a later time point23. These frequencies are determined isolating genomic DNA from the different cell populations, PCR-amplifying the locus expressing the sgRNA, and subjecting the PCR product to next-generation sequencing. CRISPRi and CRISPRa screens for growth phenotypes have yielded complementary insights19, 24. CRISPRi phenotypes were found for a large number “gold-standard”25 essential genes (including many housekeeping genes such as ribosomal subunits and genes involved in DNA replication). Essential genes that are unique to specific cell types are of particular interest in cancer research, where cancer-specific vulnerabilities are potential therapeutic targets. Intriguingly, a growth-based CRISPRi screen targeting long non-coding RNAs (lncRNAs) identified cell-type specific essential lncRNAs26. CRISPRa revealed a smaller set of genes whose overexpression was detrimental to growth, which was highly enriched in tumor suppressors and developmental transcription factors19.
A variation of the growth-based screen is the sensitization/resistance screen (Fig. 2A), in which the extent to which sgRNAs affect the sensitivity to a selective pressure (such as treatment with a drug-like compound or toxin) is quantified by comparing the frequencies of sgRNA constructs between treated and untreated cell populations that were cultured in parallel23. The selective pressure can be thought of as a tool to query a specific area of cell biology. For example, CRISPRi/a screens for sensitization to bacterial toxins revealed cellular trafficking pathways highjacked by the toxin, as well as the metabolic pathway involved in biosynthesis of the toxin’s cell surface receptor19. For some hit genes, CRISPRi loss-of-function and CRISPRa gain-of-function had opposite phenotypes. In other cases, CRISPRi and CRISPRa yielded complementary information19. For example, genes not expressed in the cell type used for the screen cannot have a CRISPRi loss-of-function phenotype, but may have a CRISPRa gain-of-function phenotype that sheds light on the biological process of interest. Vice versa, overexpression of a single subunit of a multi-subunit protein complex may not have a gain-of-function phenotype, whereas knockdown of individual subunit may disrupt the function of the complex and result in a loss-of-function phenotype.
Screens investigating genetic modifiers of cellular sensitivity to a drug or drug-like compound are of particular interest to chemical biologists. For drug candidates found in phenotypic screens, it is often not trivial to identify the molecular target of the compound, and its mechanism of action. As we have recently reviewed elsewhere, genetic screens are a powerful approach to identifying the relevant molecular targets of novel compounds27. Beyond the identification of the direct target of a drug, such screens can also identify other cellular factors that mediate resistance to the compound, or synergistic targets for combination therapy. Examples from our own work include the identification of 19S proteasomal subunit levels as a biomarker predictive of multiple myeloma patient response to the 20S proteasome inhibitor carfilzomib28, and the identification of PI3Kδ inhibitors and dexamethasone as a synergistic combination therapy in B-cell precursor acute lymphoblastic leukemia29. CRISPRa in particular has enormous potential for the elucidation of drug resistance mechanisms in cancer cells, which are thought to arise frequently from gain-of-function events. A CRISPRa screen in BRAF(V600E) melanoma cells for resistance to the BRAF inhibitor PLX-4720 recapitulated previously known resistance mechanisms, such as EGFR and ERK pathway activation, but also revealed novel resistance mechanisms involving G protein-coupled receptors21. Importantly, this CRISPRa gain-of-function screen for PLX-4720 resistance yielded complementary information to a parallel CRISPRn loss-of-function screen8.
While growth and sensitization/resistance screens can be used to probe many areas of biology, not all biological questions can be translated into a growth or survival phenotype. An alternative approach is pooled screening based on fluorescence-activated cell sorting (FACS). A cell population transduced with an sgRNA library is subjected to FACS, and subpopulations are isolated based on a fluorescent signal (Fig. 2B). The fluorescent signal can be generated by a chemical probe or a genetically encoded reporter for a cellular process of interest, or by a fluorescently labeled antibody detecting a molecular species of interest. Of course, cells can also be separated based on combinations of fluorescent signals in different channels. As for growth-based screens, the frequencies of cells expressing each sgRNA are quantified by next-generation sequencing, and a quantitative phenotype is calculated for each sgRNA based on its frequency ratio between the populations, relative to non-targeting negative-control sgRNAs. A recently published FACS-based CRISPRi screen revealed loss-of-function events that activated the Ire1 branch of the unfolded protein response30. In our own previous work, we used a FACS approach to identify cellular factors controlling the exit from latency of HIV31 and to identify the molecular target of ISRIB, a drug-like inhibitor of the integrated stress response32.
Both growth- and FACS-based strategies for pooled genetic screens separate cells based on “low-dimensional” phenotypes: along the dimension of growth/survival rate, or along one or few dimensions of fluorescence parameters, respectively. An in-depth characterization of the phenotype caused by perturbation of hit genes found in a primary screen requires a series of secondary assays. An exciting recent innovation are pooled genetic screens coupling perturbation of target genes using CRISPRi30 or CRISPRn33–35 to droplet-based single-cell RNA sequencing (Fig. 2C). Thus, a very “high-dimensional” phenotype is measured for each sgRNA, generating a wealth of data and enabling clustering of hit genes based on the similarity of the transcriptomic consequence of their perturbation. At the same time, the single-cell resolution of such screens also enables the dissection of stochastic and other non-genetic determinants of cellular states, such as cell cycle phases30.
Genetic interaction maps reveal cellular pathways
Following the successful completion of a genetic screen, the interpretation of the observed hit genes, and the prioritization of genes for mechanistic follow-up studies can be a major challenge. A general approach to revealing functional connections and pathway relationships between hit genes is the quantification of genetic interactions between them. Genetic interactions are quantitatively defined as deviation of the observed phenotype of a combinatorial gene perturbation from the expected phenotype based on the observed phenotypes of the individual perturbations. We have discussed elsewhere the nontrivial question of how to define the expected phenotype23. A genetic interaction usually indicates a functional relationship between the two genes. If the combinatorial phenotype of two genes is less severe than expected, the genetic interaction is called “buffering” or epistatic, and can indicate that the two genes act in a linear pathway or encode subunits of protein complex (Fig. 3A). Conversely, if the combinatorial phenotype of two genes is more severe than expected, the genetic interaction is called “synergistic” – or, in extreme cases, “synthetic lethal”, and the genes may be acting in parallel, partially redundant pathways that can compensate for each others’ loss of function (Fig. 3A).
Figure 3. Genetic interactions reveal functional relationships between genes.
(A, B) Genetic interactions are defined as the deviation of the observed phenotype of a combinatorial gene perturbation from the expected phenotype based on the observed phenotypes of the individual perturbations. They indicate a functional relationship between the two genes, as visualized by the pathway motifs. X, Y, Z: genes. P: observed phenotype.
(C) Systematic genetic interaction maps reveal functional clusters of genes based on the similarity of their genetic interaction patterns, as exemplified by our previously published map for ricin resistance40. Examples for different types of interactions are highlighted: members of protein complexes (the TRAPP complex and others), paralogues (RAB1A and RAB1B), and regulatory pathways (the small GTPase ARF1 and its nucleotide exchange factor GBF1).
While a buffering genetic interaction can point to a pathway relationship between genes, it does generally not reveal which of the genes acts upstream and which downstream in the pathway. However, if two genes have opposite phenotypes, and the combinatorial phenotype matches that of one of the two genes (i.e., it masks the effect of the other gene), that gene can be hypothesized to act downstream in the pathway (Fig. 3B). It is important to stress that genetic interactions can only create a hypothesis for the nature of functional relationships between genes. Mechanistic experiments are required to test this hypothesis.
Systematic quantification of pairwise genetic interactions in a large set of genes, first implemented in yeast36–39, results in a genetic interaction map (Fig. 3C). Beyond detecting individual gene pairs with strong genetic interactions, genetic interaction maps also enable clustering of genes based on the similarity of their genetic interaction patterns with all other genes, which reveals functionally related groups of genes and can uncover the function of previously uncharacterized genes. Pooled genetic screens with pairwise genetic perturbations can yield high-quality genetic interaction maps in mammalian cells, using an experimental and computational approach we first developed based on RNAi23, 40. More recently, similar approaches have been implemented based on CRISPRi41 and CRISPRn42–45. Importantly, CRISPR-based approaches can in principle enable simultaneous gain-of-function and loss-of-function perturbations in the same cell, either by co-expressing different types of sgRNA with RNA scaffolds for transcriptional modulators (as described for CRISPRn + CRISPRa46 and CRISPRi + CRISPRa47), or by using orthogonal Cas9 proteins48. Combinations of gain-of-function and loss-of-function perturbations in genetic interaction maps should greatly enhance our ability to identify directional pathway relationships: since unidirectional perturbation of genes acting in a pathway (such as a biosynthetic pathway or a signaling pathway) often will result in individual phenotypes of the same “sign”, the upstream or downstream position of genes in the pathway can generally not be deduced from their genetic interactions (Fig. 3A). However, CRISPRi and CRISPRa perturbation of the same gene often results in phenotypes of the opposite “sign”19, and thus CRISPRi targeting of one gene in a pathway and CRISPRa targeting of another gene in the pathway can generate opposing phenotypes. As discussed previously, genetic interactions between perturbations of opposing phenotypes can reveal the upstream and downstream positions of the perturbed genes in the pathway (Fig. 3B).
Comparison of technologies for pooled genetic screens
For researchers planning to embark on a genetic screen, an important question is which of the available screening approaches (Fig. 1) they should use. There are currently only few empirical comparisons between the different loss-of-function technologies, and it is important to keep in mind that CRISPR-based approaches are still being optimized, while RNAi is a relatively mature technology. An early comparison between next-generation RNAi based on our high-complexity shRNA library and our first-generation CRISPRi library found that they performed comparably in a pooled screen for toxin resistance49. However, the second-generation CRISPRi library would likely outperform RNAi, since it is vastly improved over the first-generation version24, most importantly by incorporating empirical information for transcription start sites and optimizing the position of the sgRNA with respect to the transcription start site, and thereby to the nucleosome-free region just downstream. Similarly, a direct comparison of CRISPRi, CRISPRn and RNAi in screens for essential genes reported the best performance for CRISPRn50; however, the study did not utilize state-of-the art CRISPRi reagents. A meta-analysis of CRISPRn and CRISPRi screens revealed that our second-generation CRISPRi library performed comparably with the best CRISPRn libraries in detecting “gold-standard” essential genes24. As CRISPRn and CRISPRi technologies mature, results delivered by the various screening approaches will show which technology, if any, consistently performs the best. Most likely, CRISPRi and CRISPRn will yield complementary results, since each method has different caveats and sources of false-positive results51. Furthermore, some genes may only have a phenotype on complete knockout, whereas the biological function of essential genes may only be revealed by partial knockdown52, 53.
Importantly, there are several differences between available technologies based on first principles, and each approach can have advantages and disadvantages depending on the intended application, as described below.
A. Loss-of-function technologies (summarized in Table 2)
Table 2.
Comparison of loss-of-function approaches
| CRISPRi | CRISPRn | RNAi | |
|---|---|---|---|
| Number of components | 2 | 2 | 1 |
| Outcome in individual cells | Deterministic | Stochastic | Deterministic |
| Loss of function | Knockdown | Complete KO possible | Knockdown |
| Specificity | Very high; but caveat of bidirectional promoters | Some off-targets | Many off-targets |
| Non-specific toxicity | Not measurable | Due to DNA damage, especially when targeting amplified regions | With high siRNA/shRNA levels |
| Reversibility | Yes | No | Yes |
| Targeting long noncoding RNAs | Yes | Requires paired sgRNAs | No |
| Differential targeting of splice isoforms | No | Generally no | Yes |
| Targeting cis-regulatory DNA elements | Yes | Yes | No |
In the pre-CRISPR era, the approach of choice for loss-of-function screens in mammalian cells was RNAi. (We will not discuss insertional mutagenesis approaches, since these are practically limited to use in haploid cell lines54.) Unlike CRISPR-based approaches, RNAi utilizes endogenous protein machinery in mammalian cells, and therefore delivery of a single component (siRNAs or shRNAs) is sufficient to trigger gene knockdown. On the one hand, this can be an advantageous for biological systems in which delivery of Cas9 constructs is inefficient. On the other hand, competition of exogenous siRNAs/shRNAs with endogenous substrates of the RNAi machinery can lead to non-specific toxicity55. The major drawback of RNAi is the pervasiveness of off-target effects, which can lead to false-positive results in genetic screens56, 57. While the problem of false-positives can be overcome by using ultracomplex shRNA libraries23, 40, 49 or by systematically comparing large numbers of parallel genetic screens58, the scale of such experiments can be prohibitive. Also, the off-target effects of any given siRNA/shRNA still encumber follow-up studies for individual hit genes from a screen.
CRISPRi lacks the notorious off-target effects of RNAi and enables highly specific knockdown of target genes11, 19. The exquisite specificity of CRISPRi is likely due to the narrow region around transcription start sites in which CRISPRi is active19. One caveat of CRISPRi is that targeting of bidirectional promoters can knock down both adjacent genes51. As many as 13% of human genes have transcription start sites within 1kb of each other51. However, given that CRISPRi in mammalian cells is most effective when sgRNAs target regions just downstream of transcription start sites19, 24, careful analysis of the relative phenotypes obtained from several sgRNAs targeting different sites along bidirectional promoters should help to pinpoint which of the flanking genes is more likely to be responsible for an observed phenotype.
Off-target effects of CRISPRn are an area of active investigation due to their relevance for therapeutic gene-editing applications. Several studies suggest that CRISPRn off-target effects may be more pervasive and harder to predict than initially thought59, 60, but efforts are underway to develop CRISPRn systems with reduced off-targets. An additional source of false-positive results in CRISPRn screens is the non-specific toxicity due to DNA damage that is observed with sgRNAs targeting amplified DNA sequences61, 62, which does not occur with CRISPRi24, 51.
There is a fundamental difference in the mode of loss-of-function between RNAi and CRISPRi on the one hand, and CRISPRn on the other hand: While RNAi and CRISPRi knock down gene expression deterministically to a certain level in all cells, CRISPRn introduces DNA breaks that are then subject to stochastic, error-prone DNA repair. In some cells, CRISPRn will result in short deletions that cause a frame-shift, leading to a complete loss-of-function of the targeted gene. In other cells, the repair outcome can be a short in-frame deletion, which may not significantly affect the function of the targeted gene. This stochastic nature of CRISPRn can lead to false-negative phenotypes of sgRNAs in pooled screens, but it can also be used to map functionally relevant domains of proteins63. Stochastic outcomes of CRISPRn are especially a concern when several genes are simultaneously targeted in the same cell for the generation of genetic interaction maps, since the fraction of cells in which the two targeted genes are both biallelically inactivated may be relatively small43. Unlike CRISPRn, CRISPRi and RNAi are reversible, thus enabling the time-resolved investigation of gene function during biological processes. While complete knockout of genes using CRISPRn may achieve stronger phenotypes than CRISPRi-based knockdown51, partial knockdown can enable the investigation of cellular functions of essential genes, and may more accurately predict the outcome of pharmacological inhibition of the gene product.
The differences in the mechanisms underlying RNAi, CRISPRi and CRISPRn have implications for the types of genetic elements that can be targeted by these approaches. RNAi targets mature cytosolic RNAs for degradation, and is therefore uniquely suited to selectively target different splice isoforms derived from the same gene. CRISPRi blocks the initiation of transcription, enabling the targeting of not only protein-coding, but also nuclear long non-coding RNAs (lncRNAs) with a single sgRNA19, 26, whereas RNAi is not effective in the nucleus64. Individual CRISPRn sgRNAs introduce short deletions unlikely to disrupt lncRNA function; however, paired-sgRNA libraries can be used with CRISPRn to remove entire lncRNA loci65. Since CRISPRn and CRISPRi target genomic DNA, both can be used to probe the function of cis-regulatory DNA elements such as enhancers66–68 and super-enhancers69.
B. Gain-of-function approaches (summarized in Table 3)
Table 3.
Comparison of gain-of-function approaches
| CRISPRa | ORF/transgene expression | |
|---|---|---|
| Number of components | ≥ 2 | 1 |
| Extent of overexpression | Can achieve high overexpression by multiplexing sgRNAs or activator domains | Can achieve high overexpression |
| Specificity | Very high; but caveat of bidirectional promoters | Very high |
| Limitations for long ORFs | No | Yes |
| Differential overexpression of splice isoforms | No | Yes |
| Expression of variants not matching host cell genotype | No | Yes |
In the pre-CRISPR era, gain-of-function screens in mammalian cells were enabled by genome-scale lentiviral ORF expression libraries70. The construction and validation of such libraries was a painstaking effort, and the comparative ease of CRISPRa sgRNA library construction is a clear advantage. Furthermore, the broad distribution of ORFs sizes poses a challenge for pooled propagation and screening; ORF expression screens targeting different genes are therefore generally implemented as arrayed screens71, 72. Lentiviral packaging efficiency decreases with increasing vector length73, thus complicating the expression of long ORFs. Conversely, all elements of a CRISPRa sgRNA library have the same length, regardless of the lengths of the targeted genes, and are suitable for pooled screens19, 21.
An important difference between CRISPRa screens and ORF expression screens is that the former induce the transcription of endogenous genes and recapitulate the expression of different splice isoforms, whereas the latter express specific isoforms, but can be used to express mutant versions of proteins that differ from the genotype of the host cell. Therefore, ORF expression screens can elucidate the cellular phenotypes of large numbers of point mutations in a given protein of interest71, 74, 75.
CRISPRa has been shown to activate targeted genes with high specificity15, 19, 21, 76. However, the caveat of bidirectional promoters, described for CRISPRi above, likely applies to CRISPRa as well.
The choice of CRISPRa versus ORF expression will hinge on the biological question of interest: gain-of-function screens for endogenous genes are much easier to implement with CRISPRa, whereas screens for the impact of different mutations will require ORF expression.
As shown in Figure 1, different CRISPRa systems are currently in use. A recent study76 directly compared a large number of published CRISPRa systems and found the VPR18, SunTag19, 20 and SAM21 approaches to perform the best in HEK293T cells. Across a number of different cell lines and target genes, none of the three approaches consistently outperformed the others. Importantly, the rules for optimal sgRNA target sites appeared to be the same across the different CRISPRa systems76.
Onwards and Upwards with CRISPRi and CRISPRa
The strong performance of CRISPRi and CRISPRa in genetic screens is remarkable, given how recently these technologies were developed. Over the next years, improvements in sgRNA design and dCas9 constructs for CRISPRi and CRISPRa may further enhance these approaches and overcome current limitations (Tables 2, 3). In parallel, variations of the CRISPRi/a approach in which dCas9 recruits functional domains for targeted mutagenesis77–79 or epigenome editing by modifying DNA80, 81 or histones82–84 will enable additional ways of probing genome function in pooled screens. While pooled CRISPRi and CRISPRa screens have so far mostly been carried out in cancer cell lines, applications in induced pluripotent stem cells (iPSCs) are feasible26, 85, and pooled screens in human iPSC-derived cell types have enormous potential to uncover cellular mechanisms and therapeutic targets in a wide range of diseases, such as neurodegenerative diseases86.
CRISPR-based technologies are key tools for precision medicine and biology. The recent revolution in sequencing technology has enabled us to “read” genomes and transcriptomes, enabling us to catalogue variation correlating with different biological phenotypes and disease states. CRISPRn and CRISPRi/a enable us to “write” genomes and transcriptomes, respectively. This capability is paving the way for highly specific manipulation of biological systems for both research purposes (the functional dissection of causal relationships) and therapeutic purposes (the correction of disease states).
Acknowledgments
We thank members of the Kampmann lab and the UCSF CRISPR Developers community for discussions. Research in the Kampmann lab is supported by NIH/NIGMS New Innovator Award DP2 GM119139, NIH/NINDS grant NIH/NCI grant U54 NS100717 (Tau Center Without Walls), K99/R00 CA181494, Allen Distinguished Investigator Award (Paul G. Allen Family Foundation), a Stand Up to Cancer/American Association for Cancer Research Innovative Research Grant, the Paul F. Glenn Center for Aging Research, a Calico/QB3 Longevity Award and the Tau Consortium. MK is a Chan Zuckerberg Biohub investigator.
Keywords
- CRISPR
clustered regularly interspaced short palindromic repeats, an adaptive immune system in bacteria, which involves cleavage of DNA sequences by the programmable nuclease Cas9 (or analogous proteins)
- dCas9
nuclease-dead Cas9 protein, which can be used to target functional protein domains to genomic loci specified by an sgRNA
- sgRNA
single guide RNA, a synthetic RNA construct that links a CRISPR RNA and a transactivating CRISPR RNA (tracrRNA) and is able of directing the sequence specificity of Cas9
- CRISPRi
CRISPR interference, transcriptional repression of endogenous genes by dCas9 alone, or dCas9 fused to transcriptional repressor domains
- CRISPRa
CRISPR activation, transcriptional overexpression of endogenous genes by dCas9-mediated recruitment of transcriptional activator domains
- CRISPRn
CRISPR nuclease, an approach to loss-of-function screens in which gene function is disrupted by error-prone non-homologous end joining repair following Cas9-mediated DNA cleavage
- Genetic interaction
Deviation of the observed phenotype of a combinatorial gene perturbation from the expected phenotype based on the observed phenotypes of the individual perturbations, which usually indicates a functional relationship between the two genes
- Genetic interaction map
Systematic quantification of all pairwise genetic interactions for a large set of genes, which can reveal functionally related genes based on the similarity of their genetic interaction patterns.
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 4.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koike-Yusa H, Li Y, Tan EP, del Velasco-Herrera MC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 8.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., 3rd Kruppel-associated boxes are potent transcriptional repression domains. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, EPRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2317–2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5 doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kampmann M. Elucidating drug targets and mechanisms of action by genetic screens in mammalian cells. Chemical communications. 2017 doi: 10.1039/c7cc02349a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta-Alvear D, Cho MY, Wild T, Buchholz TJ, Lerner AG, Simakova O, Hahn J, Korde N, Landgren O, Maric I, Choudhary C, Walter P, Weissman JS, Kampmann M. Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits. eLife. 2015;4:e08153. doi: 10.7554/eLife.08153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruth KA, Fang M, Shelton DN, Abu-Halawa O, Mahling R, Yang H, Weissman JS, Loh ML, Muschen M, Tasian SK, Bassik MC, Kampmann M, Pufall MA. Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. 2017;129:3000–3008. doi: 10.1182/blood-2017-02-766204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882e1821. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besnard E, Hakre S, Kampmann M, Lim HW, Hosmane NN, Martin A, Bassik MC, Verschueren E, Battivelli E, Chan J, Svensson JP, Gramatica A, Conrad RJ, Ott M, Greene WC, Krogan NJ, Siliciano RF, Weissman JS, Verdin E. The mTOR Complex Controls HIV Latency. Cell Host Microbe. 2016;20:785–797. doi: 10.1016/j.chom.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, Sokabe M, Mendez AS, Newton BW, Tang EL, Verschueren E, Johnson JR, Krogan NJ, Fraser CS, Weissman JS, Renslo AR, Walter P. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866e1817. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 35.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 37.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 38.Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Molecular cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 39.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 40.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, Hyman AA, Leproust EM, McManus MT, Weissman JS. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du D, Roguev A, Gordon DE, Chen M, Chen SH, Shales M, Shen JP, Ideker T, Mali P, Qi LS, Krogan NJ. Genetic interaction mapping in mammalian cells using CRISPR interference. Nat Methods. 2017;14:577–580. doi: 10.1038/nmeth.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, Sanchez KS, Thomas A, Kuo CC, Du D, Roguev A, Lewis NE, Chang AN, Kreisberg JF, Krogan N, Qi L, Ideker T, Mali P. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong AS, Choi GC, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, Shalek AK, Fraenkel E, Lu TK. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbluh J, Mercer J, Shrestha Y, Oliver R, Tamayo P, Doench JG, Tirosh I, Piccioni F, Hartenian E, Horn H, Fagbami L, Root DE, Jaffe J, Lage K, Boehm JS, Hahn WC. Genetic and Proteomic Interrogation of Lower Confidence Candidate Genes Reveals Signaling Networks in beta-Catenin-Active Cancers. Cell Syst. 2016;3:302–316e304. doi: 10.1016/j.cels.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kampmann M, Horlbeck MA, Chen Y, Tsai JC, Bassik MC, Gilbert LA, Villalta JE, Kwon SC, Chang H, Kim VN, Weissman JS. Next-generation libraries for robust RNA interference-based genome-wide screens. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3384–E3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 51.Rosenbluh J, Xu H, Harrington W, Gill S, Wang X, Vazquez F, Root DE, Tsherniak A, Hahn WC. Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat Commun. 2017;8:15403. doi: 10.1038/ncomms15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, Gilbert LA, Li A, Mateo R, Smith M, Glenn JS, Carette JE, Khosla C, Bassik MC. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nature chemical biology. 2016;12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy T, Kampmann M. CRISPulator: a discrete simulation tool for pooled genetic screens. BMC Bioinformatics. 2017;18:347. doi: 10.1186/s12859-017-1759-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, Brummelkamp TR. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 55.Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaelin WG., Jr Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC. Defining a Cancer Dependency Map. Cell. 2017;170:564–576e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 2017;14:547–548. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, Kost-Alimova M, Gill S, Xu H, Ali LD, Jiang G, Pantel S, Lee Y, Goodale A, Cherniack AD, Oh C, Kryukov G, Cowley GS, Garraway LA, Stegmaier K, Roberts CW, Golub TR, Meyerson M, Root DE, Tsherniak A, Hahn WC. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer discovery. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, DeWeck A, Abramowski D, Wan J, Shirley MD, Neshat SY, Rakiec D, de Beaumont R, Weber O, Kauffmann A, McDonald ER, 3rd, Keen N, Hofmann F, Sellers WR, Schmelzle T, Stegmeier F, Schlabach MR. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer discovery. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 63.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS, Wei W. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nature genetics. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, Johannessen CM, Silver SJ, Nguyen C, Murray RR, Hieronymus H, Balcha D, Fan C, Lin C, Ghamsari L, Vidal M, Hahn WC, Hill DE, Root DE. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, Bagul M, Kamburov A, Imielinski M, Hogstrom L, Zhu C, Yang X, Pantel S, Sakai R, Watson J, Kaplan N, Campbell JD, Singh S, Root DE, Narayan R, Natoli T, Lahr DL, Tirosh I, Tamayo P, Getz G, Wong B, Doench J, Subramanian A, Golub TR, Meyerson M, Boehm JS. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer cell. 2016;30:214–228. doi: 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 74.Brenan L, Andreev A, Cohen O, Pantel S, Kamburov A, Cacchiarelli D, Persky NS, Zhu C, Bagul M, Goetz EM, Burgin AB, Garraway LA, Getz G, Mikkelsen TS, Piccioni F, Root DE, Johannessen CM. Phenotypic Characterization of a Comprehensive Set of MAPK1/ERK2 Missense Mutants. Cell Rep. 2016;17:1171–1183. doi: 10.1016/j.celrep.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majithia AR, Tsuda B, Agostini M, Gnanapradeepan K, Rice R, Peloso G, Patel KA, Zhang X, Broekema MF, Patterson N, Duby M, Sharpe T, Kalkhoven E, Rosen ED, Barroso I, Ellard S, Consortium UKMD, Kathiresan S, O’Rahilly S, Consortium UKCL, Chatterjee K, Florez JC, Mikkelsen T, Savage DB, Altshuler D Myocardial Infarction Genetics C. Prospective functional classification of all possible missense variants in PPARG. Nature genetics. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N, Collins JJ, Church G. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 80.Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7:46545–46556. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic acids research. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwon DY, Zhao YT, Lamonica JM, Zhou Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat Commun. 2017;8:15315. doi: 10.1038/ncomms15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So PL, Conklin BR. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kampmann M. A CRISPR Approach to Neurodegenerative Diseases. Trends in molecular medicine. 2017;23:483–485. doi: 10.1016/j.molmed.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY, Yoon CH, Boehm JS, Lander ES, Zhang F. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]