Abstract
Purpose
Late genitourinary (GU) toxicity after radiation therapy limits the quality of life of prostate cancer survivors; however, efforts to explain GU toxicity using patient and dose information have remained unsuccessful. We identified patients with a greater congenital GU toxicity risk by identifying and integrating patterns in genome-wide single nucleotide polymorphisms (SNPs).
Methods and Materials
We applied a preconditioned random forest regression method for predicting risk from the genome-wide data to combine the effects of multiple SNPs and overcome the statistical power limitations of single-SNP analysis. We studied a cohort of 324 prostate cancer patients who were self-assessed for 4 urinary symptoms at 2 years after radiation therapy using the International Prostate Symptom Score.
Results
The predictive accuracy of the method varied across the symptoms. Only for the weak stream endpoint did it achieve a significant area under the curve of 0.70 (95% confidence interval 0.54–0.86; P = .01) on hold-out validation data that outperformed competing methods. Gene ontology analysis highlighted key biological processes, such as neurogenesis and ion transport, from the genes known to be important for urinary tract functions.
Conclusions
We applied machine learning methods and bioinformatics tools to genome-wide data to predict and explain GU toxicity. Our approach enabled the design of a more powerful predictive model and the determination of plausible bio-markers and biological processes associated with GU toxicity.
Introduction
Prostate cancer is 1 of the most common malignancies for American men, with ~161,000 new cases diagnosed annually (1). Radiation therapy (RT), the use of ionizing radiation to induce tumor cell death, is an important treatment option for prostate cancer. However, RT for prostate cancer can lead to late genitourinary (GU) toxicity, negatively affecting patients’ quality of life after therapy (2). Patients exhibit various symptoms classified as lower urinary tract syndrome (LUTS). LUTS has 3 broad categories: symptoms that deteriorate bladder emptying (voiding or obstructive), storage of urine in the bladder (storage or irritative), and symptoms experienced after urination (after micturition) (3).
Efforts have been made to establish an association between the incidence of RT-induced GU toxicity and the amount of RT dose spill to organs in the urinary tract such as the bladder neck, trigone, and urethra (4–7). However, the benefits of using dosimetric quantities to predict this endpoint remain unproved, partially owing to the variability in bladder shape causing discrepancies between the planned and delivered dose (8). This has motivated investigations of the genetic variations, mainly single nucleotide polymorphisms (SNPs), which modify inherent normal tissue sensitivity to radiation (9, 10). Genome-wide association studies (GWASs) using single-SNP association tests have identified loci tagged by risk SNPs for urinary endpoints (11–13). However, the single-SNP association methods used by those GWAS analyses faced difficulties in replication (14) owing to the large number of hypotheses being tested simultaneously and inherently small effect size of an individual SNP, limiting the statistical power (15). Machine learning-based multivariate modeling is an alternative approach that considers many important SNPs simultaneously and combines the small effects of the SNPs to achieve greater predictive power by aggregating the effect sizes of the predictors (16). Random forest (RF) is a multivariate method that has been widely applied in several GWASs but not in the setting of RT-induced toxicity (16–20). The following characteristics make RF an attractive method for GWAS: (1) it performs well in high-dimensional problems in which the number of predictors is high relative to the sample size (21, 22); (2) it provides the relative importance of predictors that can be used to highlight genes or biological processes for possible associations with the phenotype (16, 18); and (3) properties of random processes, such as bootstrap sampling and random feature subset selection, help to reduce model variance due to aggregation of trees with low correlation (21). To enhance the robustness of models to noise in a prediction target, a “preconditioning step” was introduced before RF training, which has been shown to identify patients with greater RT toxicity risk (16).
The present study was initiated to address the imminent clinical need for better explanation and prediction of RT-induced GU toxicity. The primary goal was to predict a congenital GU toxicity risk by using genome-wide SNP predictors. To this end, we used a machine learning method (preconditioned RF regression [PRFR] reported by Oh et al [16]) to build a reliable predictive model and a bioinformatics method to identify the biological correlates associated with RT-induced GU toxicity.
Methods and Materials
Clinical data
Under compliance of the institutional review board, a cohort of 368 prostate cancer patients were enrolled at the Mount Sinai Hospital. The clinical variables and outcomes were collected prospectively. The patients underwent brachytherapy with or without external beam RT with curative intent. The patients were followed up for GU symptoms using the patient-reported International Prostate Symptom Score (IPSS) (23). The IPSS questionnaire consists of 6 grades (0, no symptoms, to 5, most severe) for the following 7 symptoms: incomplete emptying, frequency, intermittency, urgency, weak stream, straining, and nocturia. We studied 324 patients with ≥1 available IPSS assessment during the 2 years ± 6 months after RT. For each of the 7 IPSS symptom endpoints, the patients were further stratified by the baseline performance (good, defined as a score 0 or 1 for individual symptoms vs other), and patients with good baseline status were analyzed. A toxicity event for a symptom was defined as a maximum score increase of ≥3 from baseline to 2 years ± 6 months. Of the 7 endpoints, 4 had event rates that were >10% and were subject to analysis. These included urinary frequency, urge, nocturia, and weak stream (Table 1). The patients who met the selection criteria for each symptom were randomly split into training (two-thirds) and hold-out validation (one-third) data sets with matching toxicity event rates. This configuration of disjoint training and validation sets is able to produce a single prediction model and thus can be readily validated externally or interpreted using previous biological knowledge.
Table 1.
Overall association strengths between SNPs and 4 genitourinary endpoints and predictive performance of multi-SNP PRFR models
| Symptom category | Symptom name | Samples (train/test; n) | Event rate | Inflation factor | SNPs with P < .001 (n) | PRFR performance | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 5-CV | Using 100% training | |||||||
|
|
|
|||||||
| AUC (95% CI) | AUC (95% CI) | P value | ||||||
| Irritative (storage) | Frequency | 119/60 | 0.23 | 0.97 | 539 | 0.60 (0.58–0.63) | 0.64 (0.49–0.80) | .06 |
| Urgency | 161/81 | 0.16 | 0.98 | 758 | 0.53 (0.5–0.58) | 0.53 (0.35–0.72) | .38 | |
| Nocturia | 111/56 | 0.17 | 1.03 | 977 | 0.54 (0.51–0.56) | 0.55 (0.35–0.73) | .33 | |
| Obstructive (voiding) | Intermittency | 164/82 | 0.10 | |||||
| Weak stream | 149/75 | 0.18 | 0.98 | 823 | 0.67 (0.64–0.70) | 0.70 (0.54–0.86) | .01 | |
| Straining | 196/98 | 0.05 | ||||||
| After micturition | Incomplete emptying | 168/84 | 0.10 | |||||
Abbreviations: 5-CV = 5-fold cross-validation; AUC = area under curve; CI = confidence interval; PRFR = preconditioned random forest regression; SNPs = single nucleotide polymorphisms.
Genotype data and population structure
The patients were genotyped for 606,563 germline SNPs using the Affymetrix, version 6.0, array (Affymetrix, Santa Clara, CA). Preprocessing of the SNP has been described in Appendix E1 (available online at www.redjournal.org). The effect of population structure on the SNP association strengths was assessed using 2 methods: (1) the inflation factor was measured in a genome-wide association test for each symptom endpoint; and (2) a principal component analysis on the genotype data was performed using the software EIGENSOFT (24), where the ethnic diversity in the cohort was represented by 3 significant (false-discovery rate-adjusted P < .05) principal axes (Fig. E1; available online at www.redjournal.org). Ethnicity-based toxicity prediction was performed in a logistic model using the 3 principal components as predictors.
PRFR model training and validation
Before PRFR modeling, the univariate association strength in the training data was measured for each of the predictors, including those SNPs that had passed the quality control, and the available clinical variables. The associations for the following 14 clinical covariates were investigated (11): tumor stage, androgen deprivation therapy (ADT), RT type, smoking status, hypertension, diabetes, Gleason score, α-blockers, age, initial prostate-specific antigen score, total biologically effective dose to a tumor, prostate volume, baseline IPSS, and self-identified ethnicity. In addition, for those who received ADT, the ADT duration and interval (between the end of ADT and the start of RT) were tested. The χ2 test was used for SNPs and categorical clinical variables; continuous clinical variables were evaluated using logistic regression. Based on the resulting P values, the number of variables for modeling was reduced. Because of the much larger number of hypotheses in genotype data, a more stringent P value cutoff of .001 was applied to the SNPs, instead of the conventional cutoff of P ≤ .05 used for clinical variables. The predictors with P values less than the respective threshold were used as predictors for the PRFR model.
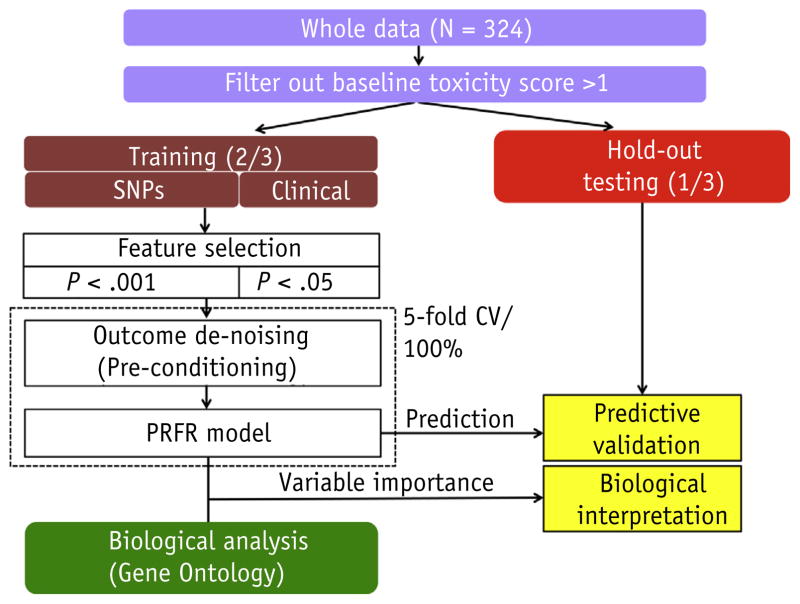
Modeling of the PRFR consisted of a preconditioning step (25) to create continuous surrogate outcomes from the original binary outcomes, followed by RF regression using the surrogate outcomes as a prediction target. More specifically, preconditioned outcomes were produced using logistic regression, coupled with the principal components, computed using a set of important SNPs such that the correlation between the original outcomes and the preconditioned outcomes is maximized, thereby refining the original outcomes and providing a more informative input for further statistical learning. Next, the preconditioned outcomes were used in RF modeling. More details on the implementation of PRFR are described in the supplemental material of the study by Oh et al (16). Validation of the PRFR model was always performed independently of the model training in the use of the hold-out data set (Fig. 1). Training of PRFR was completed in 2 stages for different purposes:
Fig. 1.
Flowchart describing our modeling pipeline. Abbreviations: CV = cross validation; PRFR = preconditioned random forest regression; SNPs = single nucleotide polymorphisms.
Fivefold cross-validation (CV): CV was used for testing stability and comparing PRFR against other baseline models. Unlike conventional CV, validation occurred in the hold-out data instead of the left-out fold. CV was repeated 100 times with randomized fold configuration. Permutation-based variable importance measures (VIMs) for the SNPs in the PRFR model were obtained by taking an average of 5 × 100 VIM results. The 500 models resulting from the 5 × 100 iterations were tested in the hold-out validation data set. The predictive performance of the PRFR model in the validation set, measured by an area under the curve (AUC), was compared against 5 competing models: (1) RF without preconditioning; (2) least absolute shrinkage and selection operator; (3) preconditioned least absolute shrinkage and selection operator; (4) the PRFR model retrained with a fewer number of SNPs, first the top 50% and then the top 75% of SNPs based on the VIMs; and (5) ethnicity-based model (see method 2).
Model building on the whole training data: the PRFR model was built using all the samples in the training set and was tested in the hold-out validation set to obtain the performance of the “finalized” model. Owing to the randomness in RF learning, the training was repeated 500 times, and the resultant predictions were averaged. The significance of the prediction was measured using Mason and Graham’s test (26), with a null hypothesis that the obtained AUC would not be >0.5. The statistical analyses were performed with the R language using the packages glm (27), ranger (28), and GenABEL (29). The described pipeline for our modeling approach is summarized in Figure 1.
Analysis of biological plausibility of the PRFR model
The PRFR model was investigated for its relevance to GU toxicity based on the functional annotations for the SNPs in the model. First, a proportion of SNPs among the top 50%, 75%, and 100% quartiles of the highest VIMs, which produced the greatest validation AUC, were extracted. The list of genes near the SNPs within 20,000 base pairs were found using annotations from the Genome Reference Consortium GRCh37. The biological relevance of the resultant candidate genes was studied in 2 ways: (1) gene ontology (GO) enrichment analysis was performed to discover the GO terms for the biological processes that were significantly enriched with the gene list (Appendix E2; available online at www.redjournal.org); and (2) to discover from the candidate genes a subset of interacting proteins with similar functions, MetaCore (Thompson Reuters, New York, NY) was used to search a manually curated protein–protein interaction database to discover a cluster of proteins connected to each other. Next, a systematic literature search on the proteins in the largest cluster for their relevance to LUTS was performed. Articles were searched in July 2017 from PubMed using a search scheme with the keywords shown in Fig. E2 (available online at www.redjournal.org).
Results
Univariate associations of predictors
The association P values for the 14 covariates with respect to the 4 GU symptoms are shown in Table E1 (available online at www.redjournal.org). No significant association between the covariates and any GU endpoints was found after Bonferroni’s correction. Analysis of the treatment type resulted in a P value of .03 for nocturia (odds ratio 0.23, 95% confidence interval [CI] 0.04–0.89) and thus was included in the PRFR model. However, its inclusion did not significantly improve the performance.
The strength of the genome-wide associations varied across the 4 GU endpoints (Table 1; Fig. E3; available online at www.redjournal.org). Nocturia returned the highest number of SNPs (n =977) with P <.001, and frequency returned the lowest (n = 539). No notable inflation was detected for any of the GU endpoints. The inflation factors ranged from 0.97 to 1.03 (Fig. E3; available online at www.redjournal.org).
Performance of PRFR models in predicting GU outcomes
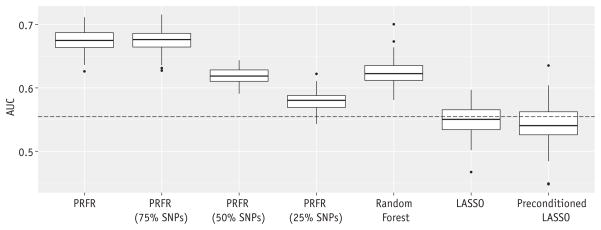
Similar to the variability of the genome-wide association strengths, the performance of the PRFR models also varied across symptoms (Table 1). A weak stream was predicted using the PRFR model, with the highest classification performance (fivefold CV AUC of 0.67, 95% CI 0.64–0.70; AUC for the model built using the whole training data of 0.70, 95% CI 0.54–0.86). Also, only for this endpoint, weak stream, did the AUC using the whole training data reach statistical significance (P = .01). For weak stream, the PRFR approach significantly outperformed the RF and linear models (P < .001); reducing the number of SNPs based on the VIMs decreased the AUC when the top 50%, but not the top 75%, of SNPs were used (Fig. 2). Ethnicity-based prediction recorded an AUC of 0.55 for this endpoint. Given the absence of the ethnic pattern, it is unlikely that the prediction of the PRFR model was driven by ethnicity-specific SNPs.
Fig. 2.
Hold-out validation area under the curve (AUC) and its fluctuation over 5-fold cross-validation for the weak stream endpoint as predicted by 7 models. Dashed line indicates AUC for ethnicity-based prediction. Abbreviations: LASSO =least absolute shrinkage and selection operator; PRFR = preconditioned random forest regression; SNPs = single nucleotide polymorphisms.
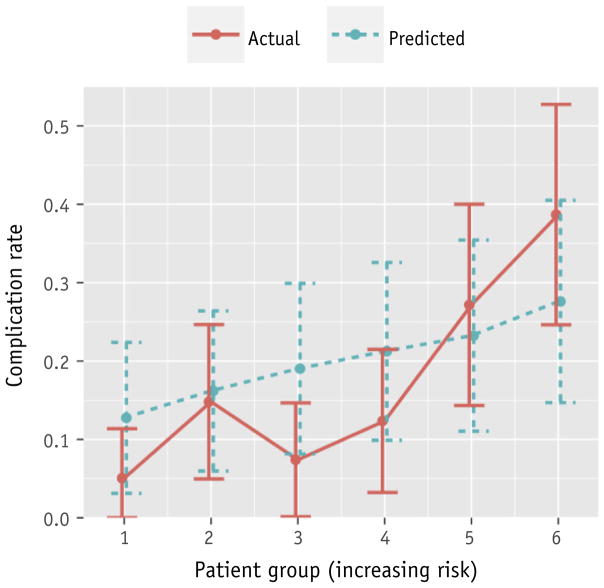
The ability of the PRFR model to identify those patients with a greater risk of a weak stream was examined further. A risk stratification plot was generated, in which the data from 75 patients in the hold-out validation set were sorted by increasing predicted risk and divided into 6 equal-size bins. Next, the actual rate of the toxicity was calculated for each bin. Figure 3 shows the degree of discrepancy between the actual and predicted risks at each of the 6 risk bins. The Hosmer-Lemeshow test on the stratified risk scores resulted in a P value of .49, indicating good agreement between the predicted and observed outcomes.
Fig. 3.
Risk stratification plot for a weak stream endpoint. Error bars represent 1 standard error.
Interpretation of biological plausibility of the PRFR model
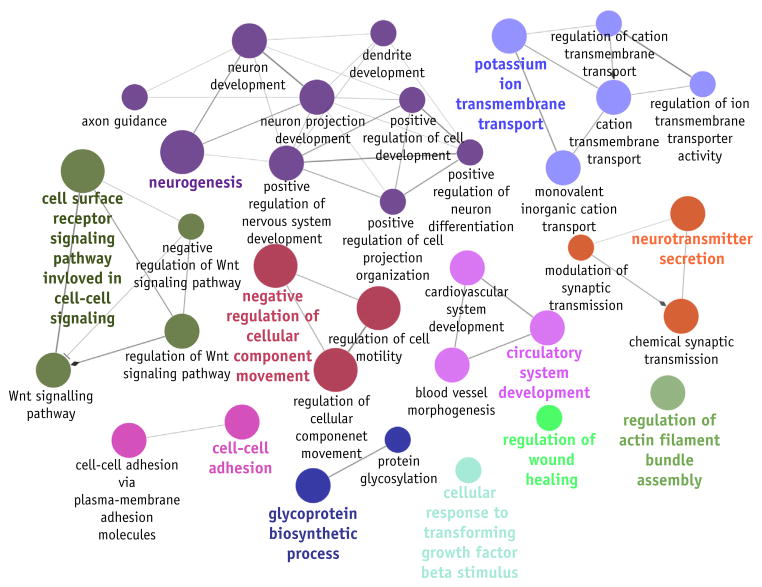
Owing to the statistical significance of its hold-out AUC value, the weak stream PRFR model was interpreted for its biological relevance. The top 75% of SNPs based on the VIMs (the smallest set of SNPs at the optimal AUC as shown in Fig. 2) resulted in 241 genes. From these genes, 34 significantly enriched GO biological process terms and 11 functional groups were identified (Fig. 4; Appendix E2; available online at www.redjournal.org). The GO biological process with the lowest P value was “negative regulation of cellular component movement” (GO ID, GO:0051271; P = 1.1 × 10−6). The largest functional group, containing 9 GO terms, was related to neurogenesis, with a group P value of 6.4 × 10−6. The second largest group, with 5 GO terms, was associated with ion transport.
Fig. 4.
Overrepresented gene ontology (GO) terms in the genes associated with the weak stream endpoint. A node size indicates a degree of significance of a GO term. Eleven functional groups are shown in different colors. A list of individual terms and their significance are provided in Appendix E2 (available online at www.redjournal.org).
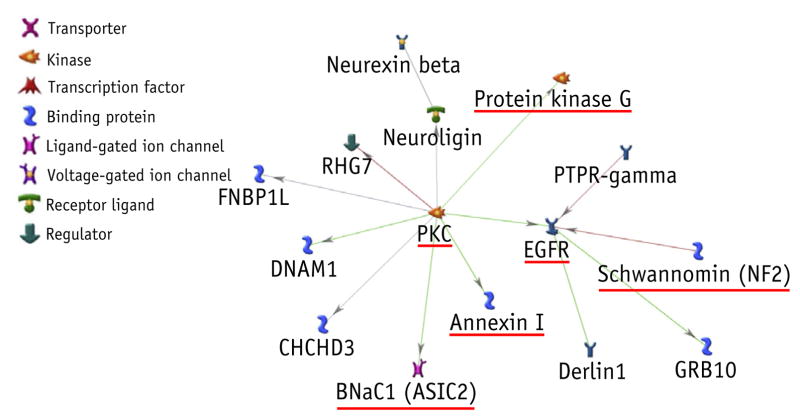
MetaCore detected a cluster of 15 proteins that were connected to each other with previously known direct protein–protein interactions (Fig. 5). From the systematic literature search, we found that 7 proteins in the cluster—protein kinase C (PKC), annexin I, protein kinase G, epidermal growth factor receptor (EGFR), schwannomin, acid-sensing ion channel 2, and neurexin—have been previously proven to be associated with LUTS (Table E2; available online at www.redjournal.org). These proteins, with the exception of neurexin, were interconnected, forming a subcluster within the 15-protein cluster.
Fig. 5.
A cluster of gene products in the weak stream model. Underscored nodes are the proteins that have been shown to be relevant to lower urinary tract syndrome in the systematic literature search (Table E2; available online at www.redjournal.org). Green, red, and gray connections indicate activation, inhibition, and unspecified, respectively. Abbreviations: EGFR = epidermal growth factor receptor; PKC protein kinase C. (A color version of this figure is available at www.redjournal.org.)
Discussion
A clinically actionable prediction model for RT-induced late GU toxicity has been lacking. In particular, patient-specific genetic variation has been largely overlooked in the context of predictive modeling of the toxicity, with the exception of a study by De Langhe et al (10), which used 343 SNPs that were chosen based on relevance to the cellular response to ionizing radiation as predictors. In contrast, in the present study, the entire genome was agnostically searched for SNPs that could be informative of GU toxicity without previous knowledge of particular genes or biological processes. Those SNPs were then integrated into a prediction model. This task poses a challenge to validation owing to the high number of variables to be considered and resultant risk of overfitting. Nevertheless, our machine learning approach significantly predicted a weak stream endpoint in hold-out validation. Further post hoc analysis of the model based on GO enrichment analysis and a literature survey showed the plausibility of the biology highlighted by the model.
Currently, no conclusive models derived from dose and/or patient characteristics are available for GU toxicity. Yahya et al (5) presented multivariate prediction models for late GU symptoms (dysuria, hematuria, incontinence, and frequency) and identified the presence of baseline GU symptoms as the most important predictor, which has been observed in other studies (30). This variable was controlled by analyzing only those patients who had minimal baseline symptoms. Given the absence of significant correlation between the baseline IPSS and the endpoints (Table E1; available online at www.redjournal.org), it is unlikely that the proposed SNP-based bio-markers are confounded by the baseline condition. Thor et al (4) predicted different symptom categories of GU toxicity using dose–volume parameters. The AUC values in a validation data set ranged from 0.51 to 0.64. Compared with these data, the results from our PRFR method indicate that genomic profiling might be able to complement the dose-only models in projecting GU toxicity risks. However, accurate dosimetric data were not available for testing this hypothesis.
The predictive accuracy of our PRFR method varied across the symptoms studied. Two obstructive symptoms with an event rate of <10% did not qualify for further analysis. The variable symptom rates and predictive performance suggest that the irritative and obstructive symptoms should be studied separately rather than as an aggregation, which was already demonstrated by Yahya et al (31). Further studies on using radiation dose distribution could help elucidate the difference between the 2 categories because the symptoms might originate from different urinary tract structures and be affected by different biological mechanisms (3).
Two major machine learning approaches in GWASs are RF and regression type models such as linear or logistic regression. Several studies have reported that RF tends to improve the predictive power compared with other machine learning methods, especially in the problems with a larger number of features than samples (32), likely because of the random process during RF modeling (22). Moreover, the preconditioning idea coupled with RF improved the predictive power significantly compared with conventional RF.
The key biological processes and gene products that were discovered from the PRFR weak stream model aligned well with the previously known etiology of LUTS, which is not necessarily radiation-induced. In particular, the GO enrichment analysis revealed neurogenesis as the most prominent biological process. The lower urinary tract is innervated by various peripheral nerves, including pelvic, hypogastric, and pudendal nerves, which control essential urinary functions such as the sensation of bladder filling, phasic contraction of smooth muscles in the bladder, and contraction of sphincters for urination (33). Studies have reported nerve damage after RT for prostate cancer with a possible association to erectile dysfunction (34, 35), which has not been studied to a large extent for GU toxicity. The inability to recover from damage to the nervous system caused by interventions such as RT could explain part of the toxicity risk. The second most influential biological process in the analysis pertained to ion transport. This can be related to the detrusor smooth muscle, a muscular wall of the bladder. This muscular machinery is also important for urination, and K+ ion channels are vital for detrusor smooth muscle contraction and relaxation (36).
Among the 241 genes in the PRFR weak stream model, a 15-protein cluster, of which 7 proteins were previously associated with urinary disorder, was discovered. Six of these proteins formed a subnetwork, with PKC acting as a network hub (connected to protein kinase G, EGFR, acid-sensing ion channel 2, and annexin I). PKC is expressed in bladder smooth muscle cells (37), and its activation has been shown to increase smooth muscle contractile forces (38), which could explain its association with the weak stream. It has also been identified by Oh et al (16) in the protein networks for RT-induced erectile dysfunction and rectal bleeding. Another notable protein in the network is EGFR, a protein known for stimulation of cell proliferation and movement. It has been implicated in bladder wall smooth cell proliferation resulting from a sustained stretch of the bladder wall (39), which is a likely consequence of an obstructive symptom such as weak stream.
One of the limitations of the present study was the use of only heritable genetic factors. Moreover, as previously mentioned, the heterogeneity of RT dose distributions should be considered to investigate the dose effect on the toxicities and facilitate external validation of the model. It seems reasonable to assume that predictive power could be significantly increased if the actual dosimetric drivers of damage were better known on a patient-by-patient basis. This might require better imaging of each fraction to accumulate a usefully accurate dose map of the delivered dose to the bladder. The present analysis likely benefited from using data from a single clinic with a consistent treatment philosophy, thereby reducing interpatient dosimetric variability compared with multicenter cohorts. Another limitation was the endpoint definition at a single follow-up point of 2 years after RT, which was determined based on a study by Kerns et al (11), in which the peak in the overall GU symptom score occurred 1 to 2 years after RT. Further research on the earlier onset of GU toxicity could highlight the difference in genetic components and biological processes between acute and late complications.
Conclusions
Genome-wide SNP data were used to predict the incidence of 4 GU toxicity symptoms after RT. PRFR was used to combine the effects of hundreds of SNPs, and its predictive performance was compared with those of other multivariate strategies. Only 1 of the endpoints (weak stream) resulted in a statistically significant prediction model (P = .01), which was confirmed on the hold-out validation data. Although the performance varied across the symptoms, these results suggest that PRFR is an effective approach for risk stratification using genome-wide data. By ranking the importance of SNPs in PRFR and applying bioinformatics tools, the biological processes and proteins implicated in radiation injury were identified. Many of the same genes had previously been identified in the reported data as related to urinary tract function. On further validation, the predictive model could help design personalized RT for prostate cancer and discover novel biomarkers.
Supplementary Material
Summary.
Genitourinary toxicity after radiation therapy limits the quality of life of prostate cancer survivors. We identified patients with a greater risk of genitourinary toxicity by discovering and integrating genome-wide risk signatures using machine learning methods. We applied preconditioned random forest regression to predict 4 urinary symptoms after radiation therapy. For weak stream, the method achieved an area under the curve of 0.7 on a hold-out validation data set. Gene ontology analysis identified key biological processes, including neurogenesis and ion transport.
Acknowledgments
This research was funded in part through National Institutes of Health/ National Cancer Institute Cancer Center Support grant P30 CA008748 and the Breast Cancer Research Foundation.
Footnotes
Conflict of interest: none.
Supplementary material for this article can be found at www.redjournal.org.
References
- 1.American Cancer Society. [Accessed February 13, 2018];Key Statistics for Prostate Cancer. 2017 Available at: www.cancer.org/cancer/prostate-cancer/about/key-statistics.html.
- 2.Fransson P. Patient-reported lower urinary tract symptoms, urinary incontinence, and quality of life after external beam radiotherapy for localized prostate cancer—15 Years’ follow-up. A comparison with age-matched controls. Acta Oncol. 2008;47:852–861. doi: 10.1080/02841860701654325. [DOI] [PubMed] [Google Scholar]
- 3.Abrams P, Cardozo L, Fall M, et al. The standardisation of terminology in lower urinary tract function: Report from the standardisation sub-committee of the international continence society. Urology. 2003;61:37–49. doi: 10.1016/s0090-4295(02)02243-4. [DOI] [PubMed] [Google Scholar]
- 4.Thor M, Olsson C, Oh JH, et al. Urinary bladder dose-response relationships for patient-reported genitourinary morbidity domains following prostate cancer radiotherapy. Radiother Oncol. 2016;119:117–122. doi: 10.1016/j.radonc.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yahya N, Ebert MA, Bulsara M, et al. Dosimetry, clinical factors and medication intake influencing urinary symptoms after prostate radiotherapy: An analysis of data from the radar prostate radiotherapy trial. Radiother Oncol. 2015;116:112–118. doi: 10.1016/j.radonc.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan AN, Yorke ED, Marks LB, et al. Radiation dose-volume effects of the urinary bladder. Int J Radiat Oncol Biol Phys. 2010;76:S116–122. doi: 10.1016/j.ijrobp.2009.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghadjar P, Zelefsky MJ, Spratt DE, et al. Impact of dose to the bladder trigone on long-term urinary function after high-dose intensity modulated radiation therapy for localized prostate cancer. Int J Radiat Oncol Biol Phys. 2014;88:339–344. doi: 10.1016/j.ijrobp.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen ES, Muren LP, Sorensen TS, et al. Bladder dose accumulation based on a biomechanical deformable image registration algorithm in volumetric modulated arc therapy for prostate cancer. Phys Med Biol. 2012;57:7089–7100. doi: 10.1088/0031-9155/57/21/7089. [DOI] [PubMed] [Google Scholar]
- 9.Rosenstein BS, West CM, Bentzen SM, et al. Radiogenomics: Radiobiology enters the era of big data and team science. Int J Radiat Oncol Biol Phys. 2014;89:709–713. doi: 10.1016/j.ijrobp.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Langhe S, De Meerleer G, De Ruyck K, et al. Integrated models for the prediction of late genitourinary complaints after high-dose intensity modulated radiotherapy for prostate cancer: Making informed decisions. Radiother Oncol. 2014;112:95–99. doi: 10.1016/j.radonc.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Kerns SL, Stone NN, Stock RG, et al. A 2-stage genome-wide association study to identify single nucleotide polymorphisms associated with development of urinary symptoms after radiotherapy for prostate cancer. J Urol. 2013;190:102–108. doi: 10.1016/j.juro.2013.01.096. [DOI] [PubMed] [Google Scholar]
- 12.Kerns SL, Dorling L, Fachal L, et al. Meta-analysis of genome wide association studies identifies genetic markers of late toxicity following radiotherapy for prostate cancer. EBioMedicine. 2016;10:150–163. doi: 10.1016/j.ebiom.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnett GC, Thompson D, Fachal L, et al. A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol. 2014;111:178–185. doi: 10.1016/j.radonc.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Barnett GC, Coles CE, Elliott RM, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: A prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 15.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 16.Oh JH, Kerns S, Ostrer H, et al. Computational methods using genome-wide association studies to predict radiotherapy complications and to identify correlative molecular processes. Scientific reports. 2017;7:43381. doi: 10.1038/srep43381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein BA, Hubbard AE, Cutler A, et al. An application of random forests to a genome-wide association dataset: Methodological considerations & new findings. BMC Genet. 2010;11:49. doi: 10.1186/1471-2156-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunetta KL, Hayward LB, Segal J, et al. Screening large-scale association study data: Exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TT, Huang J, Wu Q, et al. Genome-wide association data classification and snps selection using two-stage quality-based random forests. BMC Genomics. 2015;16(Suppl 2):S5. doi: 10.1186/1471-2164-16-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan J, Stegle O, Beyer A. A random forest approach to capture genetic effects in the presence of population structure. Nat Commun. 2015;6:7432. doi: 10.1038/ncomms8432. [DOI] [PubMed] [Google Scholar]
- 21.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 22.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry MJ, Fowler FJ, Jr, O’Leary MP, et al. Measuring disease-specific health status in men with benign prostatic hyperplasia: Measurement committee of the American Urological Association. Med Care. 1995;33:AS145–AS155. [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Paul D, Bair E, Hastie T, et al. “Preconditioning” for feature selection and regression in high-dimensional problems. Ann Stat. 2008;36:1595–1618. [Google Scholar]
- 26.Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: Statistical significance and interpretation. Quart J Roy Meteor Soc. 2002;128:2145–2166. [Google Scholar]
- 27.Simon N, Friedman J, Hastie T, et al. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright MN, Ziegler A. Ranger: A fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77:1–17. [Google Scholar]
- 29.Aulchenko YS, Ripke S, Isaacs A, et al. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 30.Ghadjar P, Jackson A, Spratt DE, et al. Patterns and predictors of amelioration of genitourinary toxicity after high-dose intensity-modulated radiation therapy for localized prostate cancer: Implications for defining postradiotherapy urinary toxicity. Eur Urol. 2013;64:931–938. doi: 10.1016/j.eururo.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yahya N, Ebert MA, Bulsara M, et al. Urinary symptoms following external beam radiotherapy of the prostate: Dose-symptom correlates with multiple-event and event-count models. Radiother Oncol. 2015;117:277–282. doi: 10.1016/j.radonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Tibshirani RJ. The LASSO problem and uniqueness. Electron J Statist. 2013;7:1456–1490. [Google Scholar]
- 33.Fowler CJ, Griffiths D, de Groat WC. The neural control of micturition. Nat Rev Neurosci. 2008;9:453–466. doi: 10.1038/nrn2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiBiase SJ, Wallner K, Tralins K, et al. Brachytherapy radiation doses to the neurovascular bundles. Int J Radiat Oncol Biol Phys. 2000;46:1301–1307. doi: 10.1016/s0360-3016(99)00551-9. [DOI] [PubMed] [Google Scholar]
- 35.Nolan MW, Marolf AJ, Ehrhart EJ, et al. Pudendal nerve and internal pudendal artery damage may contribute to radiation-induced erectile dysfunction. Int J Radiat Oncol Biol Phys. 2015;91:796–806. doi: 10.1016/j.ijrobp.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Petkov GV. Central role of the BK channel in urinary bladder smooth muscle physiology and pathophysiology. Am J Physiol Regul Integr Comp Physiol. 2014;307:R571–584. doi: 10.1152/ajpregu.00142.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hypolite JA, Malykhina AP. Regulation of urinary bladder function by protein kinase C in physiology and pathophysiology. BMC Urol. 2015;15:110. doi: 10.1186/s12894-015-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hristov KL, Smith AC, Parajuli SP, et al. Large-conductance voltage-and Ca2+-activated K+ channel regulation by protein kinase C in guinea pig urinary bladder smooth muscle. Am J Physiol Cell Physiol. 2014;306:C460–C470. doi: 10.1152/ajpcell.00325.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estrada CR, Adam RM, Eaton SH, et al. Inhibition of EGFR signaling abrogates smooth muscle proliferation resulting from sustained distension of the urinary bladder. Lab Invest. 2006;86:1293–1302. doi: 10.1038/labinvest.3700483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.