Abstract
Ultrasound (US) imaging is the most commonly performed cross-sectional diagnostic imaging modality in the practice of medicine. It is low-cost, non-ionizing, portable, and capable of real-time image acquisition and display. US is a rapidly evolving technology with significant challenges and opportunities. Challenges include high inter- and intra-operator variability and limited image quality control. Tremendous opportunities have arisen in the last decade as a result of exponential growth in available computational power coupled with progressive miniaturization of US devices. As US devices become smaller, enhanced computational capability can contribute significantly to decreasing variability through advanced image processing. In this paper, we review leading machine learning (ML) approaches and research directions in US, with an emphasis on recent ML advances. We also present our outlook on future opportunities for ML techniques to further improve clinical workflow and US-based disease diagnosis and characterization.
Keywords: Deep Learning, Elastography, Machine Learning, Medical Ultrasound, Sonography
I. Introduction
Ultrasound (US) is one of the core diagnostic imaging modalities, and is routinely used as the first line of medical imaging for evaluation of internal body structures, including solid organ parenchyma, blood vessels, the musculoskeletal system, and the fetus. US has become a ubiquitous diagnostic imaging tool owing to several major advantages over other medical imaging methods such as computed tomography (CT) and magnetic resonance imaging (MRI). These key advantages include real-time imaging, no use of ionizing radiation, and better cost effectiveness than CT and MRI in many situations. In addition, US is portable, requires no shielding, and utilizes conventional electrical power sources and is therefore well suited to point-of-care applications, especially in under-resourced settings. As the field progresses, US, especially when combined with other technologies, has the potential to be an in-home biosensor, providing ambulatory, long duration, and non-intrusive monitoring with real-time biofeedback.
US also presents unique challenges, including operator dependence, noise, artifacts, limited field of view, difficulty in imaging structures behind bone and air, and variability across different manufacturers’ US systems. Dependence on operator skill is particularly limiting. Many healthcare providers who are not imaging specialists do not use US at the point of care owing to a lack of skill in acquiring and interpreting images. For those that do, high inter- and intra-operator variability remains a significant challenge in clinical decision making. As a result of high inter-operator variability, US-derived tumor measurements are not accepted in most cancer drug trials, and US is therefore generally not used clinically for serial oncologic imaging. Automated US image analysis promises to play a crucial role in addressing some of these challenges.
Recent surveys of ML for medical imaging, such as [1], [2], [3], [4] primarily focus on CT, MRI, and microscopy. In this review, we focus on the use of machine learning (ML) in US. The objective of this paper is to review how recent advances in ML have helped accelerate US image analysis adoption by modeling complicated multidimensional data relationships that answer diagnosis and disease severity classification questions. We have two goals: (1) to highlight contributions that utilize ML advances to solve current challenges in medical US, (2) to discuss future opportunities that will utilize ML techniques to further improve clinical workflow and US-based disease diagnosis and characterization. Our survey is nonexclusive, as we mainly focus on work within the past five years, where ML, particularly deep learning (DL), has started to have a major impact. We also emphasize solutions at the system level, which is an important aspect, due to the unique characteristics of the US image generation workflow. Fig. 1 shows that US image processing involves more than simply a classification step, but additionally includes preprocessing and various types of analysis depending on several possible applications.
Fig. 1.
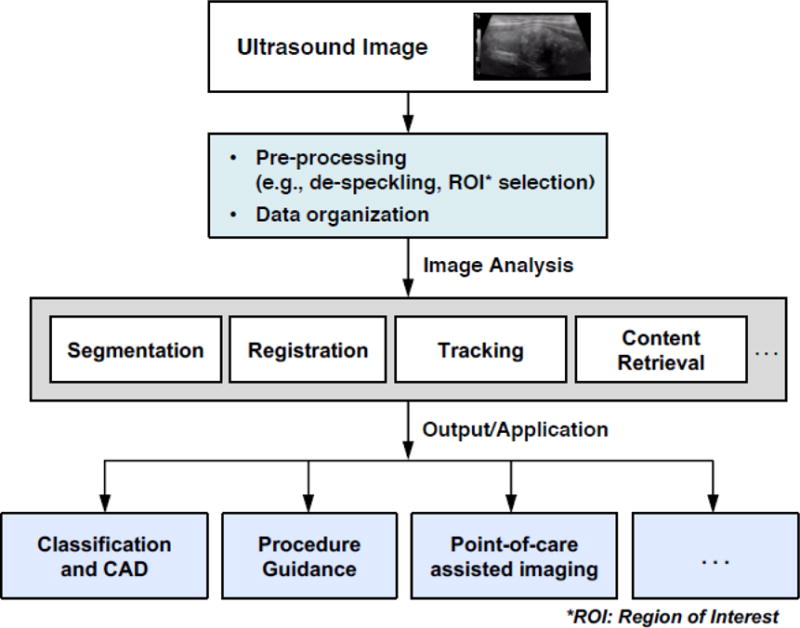
Overview of Ultrasound processing system workflow.
This article is divided into four sections: (i) an overview of basic principles of US, (ii) an overview of ML, (iii) ML for US, and (iv) summary and outlook.
II. Overview of US Imaging
2.1 US Imaging
Medical US images are formed by using an US probe to transmit mechanical wave pulses into tissue. Sound echoes are generated at boundaries where different tissues exhibit acoustic impedance differences. These echoes are recorded and displayed as an anatomic image, which may contain characteristic artifacts including signal dropout, attenuation, speckle, and shadows. Image quality is highly dependent on multiple factors, including force exerted on the US transducer, transducer location, and orientation.
Using various signal-to-image reconstruction approaches, several different types of images can be formed using US equipment. The most well known and routinely used clinically is a B-mode image, which displays the acoustic impedance of a two-dimensional cross-section of tissue. Other types of US imaging display blood flow (Doppler imaging and contrast-enhanced US), motion of tissue over time (M-mode), the anatomy of a three-dimensional region (3D US), and tissue stiffness (elastography).
2.2 US Elastography
US elastography is a relatively new imaging technique of which there are two main types in current clinical use: 1) strain elastography, where image data is compared before and after application of external compression force to detect tissue deformation, and 2) shear wave elastography (SWE), which uses acoustic energy to move tissue, generating shear waves that extend laterally in tissue. These shear waves can be tracked to compute shear wave velocity, which is algebraically related to tissue stiffness measured as the tissue Young’s modulus.
Tissue stiffness is a useful biomarker for pathologic processes, including fibrosis and inflammation, leading to several additional clinical applications for medical US. A diagnostic imaging gap recently addressed by US elastography is the evaluation of chronic liver disease [5]–[8]. US elastography liver stiffness measurements have been shown to be a promising liver fibrosis staging biomarker, and as a result highly relevant to chronic liver disease risk stratification. These technologies have the potential to replace liver biopsy as the diagnostic standard of care for key biologic variables in chronic liver disease. SWE has also been used to assess breast lesions [9]–[11], thyroid nodules [12]–[15], musculoskeletal conditions [16]–[20], and prostate cancer [21]–[23].
Fig. 2 is an example of a SWE image (the colored pixels) overlaid on top of a B-mode US image, acquired for liver fibrosis staging. Tissue stiffness measurements are obtained by placing a region of interest (ROI) inside the SWE image box. Similar to B-mode US, elastography also suffers from inter- and intra-observer variability [8]. This represents an area of opportunity for ML-based automated image analysis improvement. We will discuss this in detail in Section 4.4.
Fig. 2.
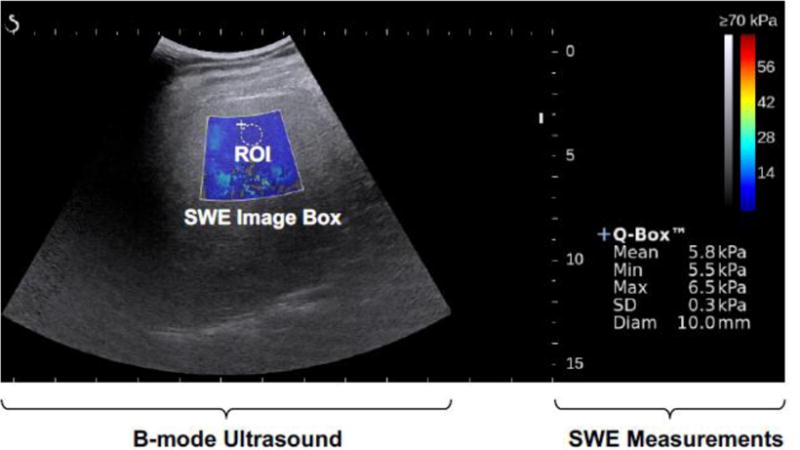
Example SWE color map overlaid on a B-mode ultrasound image.
2.3 Contrast-Enhanced US (CEUS)
Contrast-enhanced US utilizes gas-filled microbubbles for dynamic evaluation of microvasculature and macrovasculature. At present, US contrast agents are exclusively intravascular blood pool agents. Differentiation between benign and malignant focal liver lesions is an application of particular clinical interest [24]–[26]. The late phase of contrast enhancement allows for real-time characterization of washout, a critical feature in the differentiation of benign liver lesions (e.g. hemangioma, focal nodular hyperplasia, adenoma, regenerative nodule) and malignant liver lesions (e.g. hepatocellular carcinoma, cholangiocarcinoma, metastasis). The DEGUM study – a multicenter German study that analyzed 1328 focal liver lesions – reported a 90.3% accuracy of CEUS for focal liver lesions with a 95.8% sensitivity, 83.1% specificity, 95.4% positive predictive value, and 95.9% negative predictive value for distinguishing benign and malignant liver lesions [27]. Other areas of clinical interest include evaluation of focal renal lesions [28], thyroid nodules[29], splenic lesions [30] and prostate cancer [31]. CEUS limitations include operator dependence, motion sensitivity, and the need for a good acoustic window. Advanced US image processing offers potential opportunities to augment CEUS by mitigating these limitations.
III. Overview of Machine Learning (ML)
ML is an interdisciplinary field that aims to construct algorithms that can learn from and make predictions on data [32], [33]. It is part of the broad field of artificial intelligence and overlaps with pattern recognition. Substantial progress has been made in applying ML to natural language processing (NLP), computer vision (e.g., image and text search, face recognition), video surveillance, financial data analysis and many other domains. Recent progress in deep learning, a form of ML, has been dramatic, resulting in significant performance advances in international competitions and wide commercial adoption. The application of ML to diverse areas of computing is gaining popularity rapidly, not only because of more powerful hardware, but also because of the increasing availability of free and open source software, which enable ML to be readily implemented. The purpose of this section is to introduce ML approaches and capabilities to US researchers and clinicians. Historical reviews of the field and its relationship with pattern recognition can be found elsewhere [34]–[36]. The following essential concepts are introduced at a level appropriate for understanding this review: supervised and unsupervised learning, learning based on handcrafted features, deep learning, testing and performance metrics.
3.1 Supervised vs. Unsupervised Learning
Most ML applications for US involve supervised learning, in which a classifier is trained on a database of US images labeled with desired classification outputs. For example, a classifier could be trained to output a value of 1 for input images of malignant tumors and a value of 0 for benign tumors. Once a classifier is trained, it can be used to classify previously unseen test images.
Unsupervised learning involves finding clusters or similarities in data, with no labels provided. This can be useful for applications such as content-based retrieval, or to determine features that can distinguish different classes of data.
A type of learning that falls between supervised and unsupervised learning is termed weakly supervised learning [37]. A significant challenge in building up large US image databases has been the time involved for expert annotation to support supervised learning. Annotation effort can be simplified by reducing the detail of information provided by the expert. For example, an image containing a tumor can be labeled as such, without having to annotate the precise location or boundaries. The ability to train a classifier with this type of less detailed information is termed weakly supervised learning. These and other types of learning, such as reinforcement learning, are described in detail elsewhere [38].
3.2 Learning Based on Handcrafted Features
Traditionally, ML has involved computing handcrafted features that are believed to be able to distinguish between classes of data. These features are then used to train and test a classifier. For US, common types of features are morphologic, e.g., lesion area or perimeter, or textural [39], based on information in the frequency domain [40], or parameter fitting. Often a large number of candidate features are computed and then a feature selection algorithm is applied to select the best features or a dimensionality reduction algorithm [41] is applied to combine the features into a smaller composite set.
A classifier is then trained to form a feature mapping to compute desired outputs. It is important to constrain the classifier so that it does not overfit to the training data, because overfitting results in model errors that do not generalize beyond the training set to new data. The need to avoid overfitting is one of the main reasons feature selection or extraction algorithms are applied before training a classifier. Avoiding overfitting is a special concern for US research, which has thus far involved relatively small databases. Over the years, many supervised learning classification algorithms have been developed and many have been applied to US for handcrafted features. The most common approaches applied in the surveyed papers are random forests [42], support vector machines [43], [44], and multilayer feedforward networks [45]–[47], also known as artificial neural networks.
3.3 Deep Learning (DL)
The effort and domain expertise involved in handcrafting features has led researchers to seek algorithms that can learn features automatically from data. DL is a particularly powerful tool for extracting nonlinear features from data. This is particularly promising in US, where predictable acoustic patterns are typically neither obvious nor easily hand-engineered. Fig. 3 illustrates high-level differences between conventional ML and DL. The fast adoption of DL has been enabled by faster algorithms, more capable Graphics Processing Unit (GPU)-based computing, and large data sets.
Fig. 3.
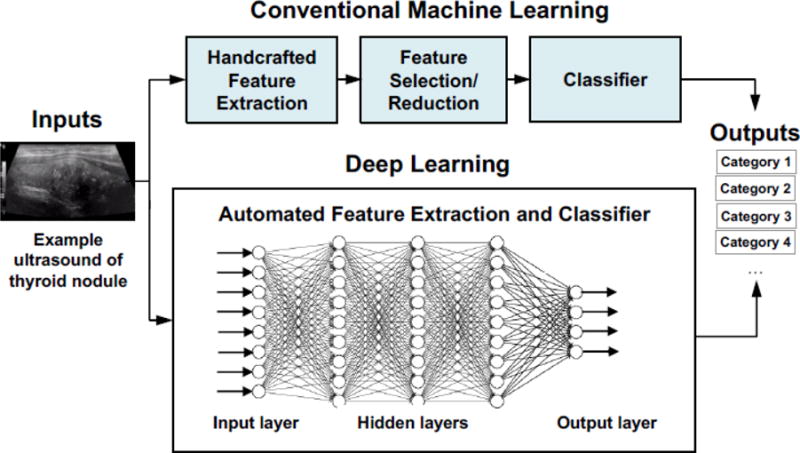
Conventional machine learning vs. deep learning.
DL extends multilayer feedforward networks from the two layers of weights used in the past to multiple layers. Fig. 4 is an example of a generic supervised DL pipeline that includes both the learning phase and the deployment phase. In the learning phase, labeled samples (e.g. labeled US thyroid nodule images) are randomly divided into training/test sets or training/validation/test sets. The training data is used for finding the weights for each of the layers. During the process, features are discovered automatically and a model is learned. The validation set is used for optimizing the network parameters. The test data is used for estimating the performance of the learned network. This model estimation and selection technique is called cross-validation [48]. During the deployment phase, the machine applies the model learned to make a prediction on a new, unlabeled input (e.g., an unlabeled US thyroid nodule image that the machine has not seen before).
Fig. 4.
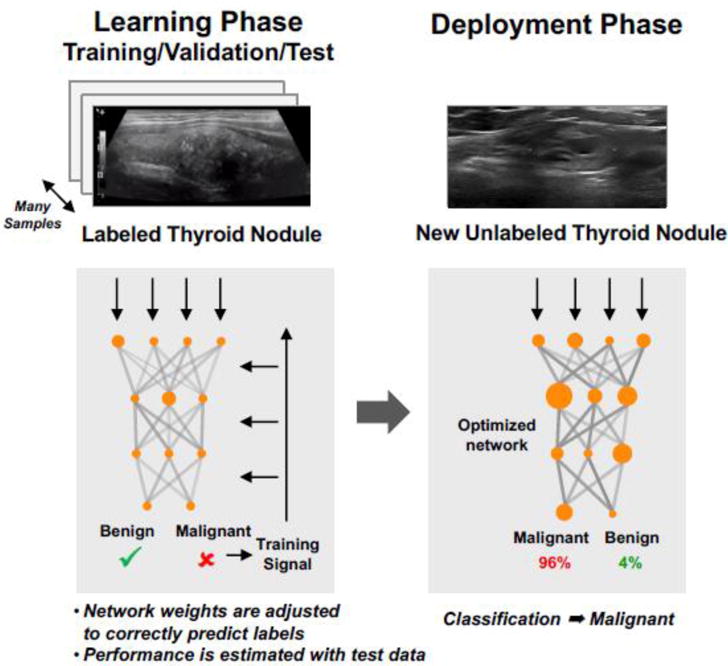
Supervised learning with deep neural networks.
The multiple processing layers have been demonstrated to learn features of the data with multiple levels of hierarchy and abstraction [49]. For example, in imagery of humans, a low level of abstraction is edges; higher levels are body parts. A variety of deep learning structures have been explored. Among them, convolutional neural networks (CNNs) are one of the most popular choices for classifying images, due to unprecedented classification accuracy [50], [51] in applications such as object detection [52]–[54], face detection [55]–[57], and segmentation [58], [59]. In a typical CNN, convolutional filters are applied in each CNN layer to automatically extract features from the input image at multiple scales (e.g., edges, colors, and shapes), and a pooling process (termed ‘max pooling’) is often used between CNN layers in order to progressively reduce the feature map size. The last two layers are typically fully connected layers, from which classification labels are predicted (Fig. 5).
Fig. 5.
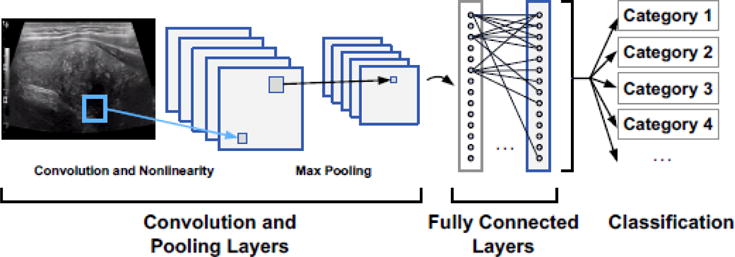
Example convolutional neural network (CNN).
3.4 Testing and Performance Metrics
As mentioned in Section 3.3, classifier development and testing typically involves splitting the randomized labeled data into training/test sets or training/validation/test sets. A validation set is used to determine the best network structure and other classifier variations based on several training runs, and an independent test set held aside to evaluate performance until the classifier has been completed.
When a database is sufficiently large, it can be partitioned a priori into these distinct sets above. For smaller data sets such as those commonly seen in US research, k-fold cross validation is often used. Cross-fold testing can be performed up to a maximum of N times for a database of N samples. In this case, termed leave-one-out testing, all of the samples in the database are used for training except for one sample data (e.g., one image), which is used for testing. Details of these techniques and other cross validation techniques can be found in [60], [61].
Classifier performance is reported in a variety of ways. The most common for two classes is area under the receiver operating characteristic curve (AUROC), often simplified as “area under the curve” (AUC). An operating characteristic is formed by measuring true positive and false positive rates as the decision threshold applied to the classifier output is varied [62]. The AUC is then computed from the operating characteristic.
IV. ML for US
Principal applications of ML to US include classification or computer-aided diagnosis, regression, and tissue segmentation. Other applications include image registration and content retrieval. Each of these applications is surveyed in the following subsections, with an aim to provide insights into progress and best approaches. In particular, advances in approaches using deep learning are highlighted, compared to approaches that use handcrafted features. Table 1 provides a summary of the applications in the papers surveyed.
Table 1.
List of applications for papers surveyed
| Organ or Body Location | Modality | Application | # of papers | References |
|---|---|---|---|---|
| Breast | US | Classification (Lesions) | 10 | [69], [71], [74], [93], [114]–[119] [120] |
| Liver | US | Classification (Lesions, cirrhosis, other focal and diffuse conditions) | 4 | [64], [121]–[123] |
| Lung | US | Classification (Several diseases, B-lines) | 2 | [124], [125] |
| Muscle | US | Classification (Atrophy, myositis) | 2 | [126], [127] |
| Intravascular | US | Classification (Plaque) | 2 | [72], [128] |
| Spine | US | Classification (Needle placement) | 1 | [129] |
| Ovary/Uterus | US | Classification (masses) | 2 | [130], [131] |
| Kidney | US | Classification (Renal disease, cysts) | 1 | [132] |
| Spleen | US | Classification (Lesion) | 1 | [133] |
| Eye | US | Classification (Cataracts) | 1 | [134] |
| Abdominal cavity | US | Classification (Free fluid from trauma) | 1 | [135] |
| Thyroid | US | Classification (Nodules) | 1 | [70] |
| Other | US | Classification (Abdominal organs) | 1 | [73] |
| Heart | US | Classification (viewing planes, congenital heart disease) Content retrieval |
8 | [95–102] |
| Heart | US | Segmentation | 1 | [88] |
| Placenta | US | Segmentation | 1 | [91] |
| Lymph Node | US | Segmentation | 1 | [90] |
| Brain | US | Segmentation | 1 | [92] |
| Prostate | US | Segmentation | 2 | [82], [83] |
| Carotid Artery | US | Segmentation | 1 | [89] |
| Breast | US | Segmentation | 1 | [81] |
| Gastrocnemius Muscle | US | Regression (Orientation) | 1 | [76] |
| Fetal brain | US | Regression (Gestational age) | 1 | [77] |
| Spine | US | Registration | 1 | [93] |
| Prostate | US (TRUS) | Registration | 1 | [94] |
| Liver | Elastography | Classification (fibrosis, chronic liver disease) | 2 | [109][106] |
| Thyroid | Elastography | Classification (Nodule) | 1 | [108] |
| Breast | Elastography | Classification (Tumor) | 3 | [107], [110], [111] |
| Liver | CEUS | Classification (Lesions) | 1 | [112] |
| Total | 56 |
4.1 Classification
Computer-aided disease diagnosis and classification in radiology have received extensive attention and have benefited greatly from the recent advances in ML. A variety of applications have been addressed in computer-aided diagnosis, but primarily for detecting or classifying lesions, mainly in the breast and liver. Most of the recent papers surveyed follow the classic approach of computing handcrafted features, applying a feature selection algorithm, and training a classifier on the reduced feature set. This basic approach has been investigated for over 20 years, e.g., [63] [64]. Specific feature and algorithm choices for each step vary. Preprocessing includes despeckling.
Features considered are primarily texture-based or morphological. The largest number of publications has been on classifying breast lesions. A review of breast image analysis [65] places US in the context of several imaging modalities. For classifying breast lesions, computerized methods have been developed to automatically extract features from the BI-RADS (Breast Imaging Reporting and Data System) lexicon, relating to shape, margin, orientation, echo pattern and acoustic shadowing [66]. These features are standardized and readily understandable by radiologists. Typically, a large number of features is reduced in dimension by either selecting the most informative features or by linearly combining features with principal components analysis [41]. Commonly used classifiers include multilayer networks (neural networks) [67], support vector machines [43], and random forests [68], the details of which extend beyond the scope of this review.
Although these papers indicate the promise of US computer-aided diagnosis, the reported studies have several limitations. These classification studies typically rely on manual region-of-interest (ROI) selection of the portion of the image that includes candidate pathology; that subimage is then classified. Manual ROI selection assumes significant involvement by a radiologist in practice, or at least neglects the problem of ROI selection. The number of patients and images available for training and testing is typically small; in nearly all cases, the number of images was less than 300. In addition, the US images were often collected at a single location by a single type of US device. Each paper reports results obtained on a different validation database, making results difficult or impossible to compare.
Two recent papers have compared the performance of commercial diagnosis systems vs. radiologists. In [69], performance of a system from ClearView Diagnostics (Piscataway, New Jersey, USA) for diagnosing breast lesions was compared to that of three certified radiologists. At the time of publication, the system was being reviewed for FDA clearance. The study was co-authored by ClearView Diagnostics employees and thus was not an independent evaluation. Ground truth for 1300 images was determined based on biopsy or one-year follow-up. Likelihood of malignancy and the preliminary BI-RADS assessment were assessed. The comparison focused on images; the reading radiologists did not have access to other information, such as patient history and previous imaging studies. Based on likelihood of malignancy, the computer system was determined to have outperformed the radiologists. Fusing the radiologist and computer assessments was also found to improve sensitivity and specificity over radiologist assessments alone.
In [70], performance of a system from Samsung (Seoul, South Korea) for assessing malignancy of thyroid nodules was compared to that from an experienced radiologist. One hundred two nodules with a definitive diagnosis from 89 patients were included in the study. The system’s performance was lower than that of the radiologist’s. It was speculated that improved segmentation would improve the performance.
The number of papers that have applied deep learning techniques to US disease classification has dramatically increased in the last two to three years [71], [72]. For deep learning, it has been unclear until very recently whether CNNs that have been trained on nonmedical color images can be used as a starting point and partially retrained to classify US images that do not resemble optical images. However, recent work, such as [73], have shown that this method, referred to as “transfer learning,” can be effective. This technique avoids overfitting on small data sets, which is often the case for US imagery. Fusing handcrafted features with those computed with deep learning has been shown to further improve performance [74]. Weakly supervised learning has also been successfully applied to US [75].
4.2 Regression
Regression involves estimating continuous values as opposed to discrete classes of data. Deep learning has been applied to regression, for example by [76] to estimate muscle fiber orientation from US imagery. Deep learning was found to improve over previous approaches using handcrafted features, specifically a well-established wavelet-based method. However, another regression application provides an example of how handcrafted features may still be the preferred approach. In this application, gestational age is estimated from 3D US images of the fetal brain [77]. A semi-automated approach based on deformable surfaces is used to compute standard biometric features, e.g., head circumference, as well as information on local structural changes in the brain.
4.3 Segmentation
Segmentation is the delineation of structural boundaries. Automated US segmentation is challenging in that US data is often affected by speckle, shadow, and missing boundaries, as well as by tradeoffs between US frequency, depth, and resolution during image acquisition.
Many US segmentation approaches have been developed, including methods based on intensity thresholding, level sets, active contours [78] and other model-based methods. These techniques are reviewed in [79], [80]. Intensity-based approaches are sensitive to the noise and image quality. Active contour and level sets require initialization, which can affect the results. Most conventional approaches are not fully automated.
Segmentation methods based on ML typically involve two steps: first, a pixel-wise classification of the desired structure, followed by a clean-up or smoothing step since the pixel-wise classification is noisy. In recent papers, several classification approaches have been investigated involving handcrafted features [81] [82], [83] [84]–[87], and various types of neural networks, including deep-learning [88]–[92]. Three papers [82], [91], [92] made use of 3D US.
4.4 Additional Applications of Machine Learning to US
In addition to US segmentation, ML has also been applied to US registration, for example for imagery of vertebrae [93] and transrectal US [94].
One key advantage of US over some other modalities is that it is well suited for real-time guidance (e.g., needle guidance, intra-cardiac procedures, and robotic surgeries), but the real-time performance has not been fully realized due to the limitations in US image processing, including lack of robust content retrieval from US video clips. A number of very recent papers focus on using deep learning techniques for frame labeling or content interpretation [95]–[98]. One approach [99] was evaluated on a database of about 30,000 images, which is very large for US. Techniques that integrate spatiotemporal information have started to emerge, particularly in dealing with echocardiograms acquired from different views, to capture key information of the motion of heart [100]–[102]. We predict ML will play a major role in the near future in enabling US guided interventions.
4.5 US Elastography and CEUS
Elastography, particularly SWE, is being increasingly used in conjunction with US as a quantitative measurement to characterize tissue lesions [103]. Key limitations of SWE, as summarized in [13], include variability in stiffness cutoff thresholds, lack of image quality control, and variability in ROI selection. It has been shown that SWE measurements depend greatly on the quality of the acquired data [104] [105]. Using liver fibrosis staging as an example, Fig. 6(a) illustrates the existing clinical workflow and challenges. As such, the current clinical protocol requires multiple image acquisitions as a way to mitigate measurement variability. Fig. 6(b) presents a potential solution to improving the clinical workflow. It includes algorithms to automatically check image quality and ML methods to quantify SWE and classify disease stages. In addition, algorithms can also assist with assessing additional useful biomarkers (e.g., subcutaneous fat content, steatosis, inflammation), which are currently not used because of the time-intensive manual interpretation required.
Fig. 6a.
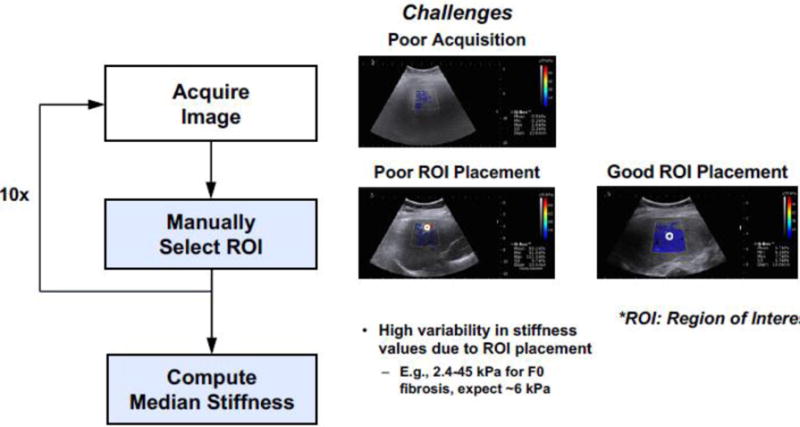
Example pipeline of using SWE for liver fibrosis staging.
Fig. 6b.
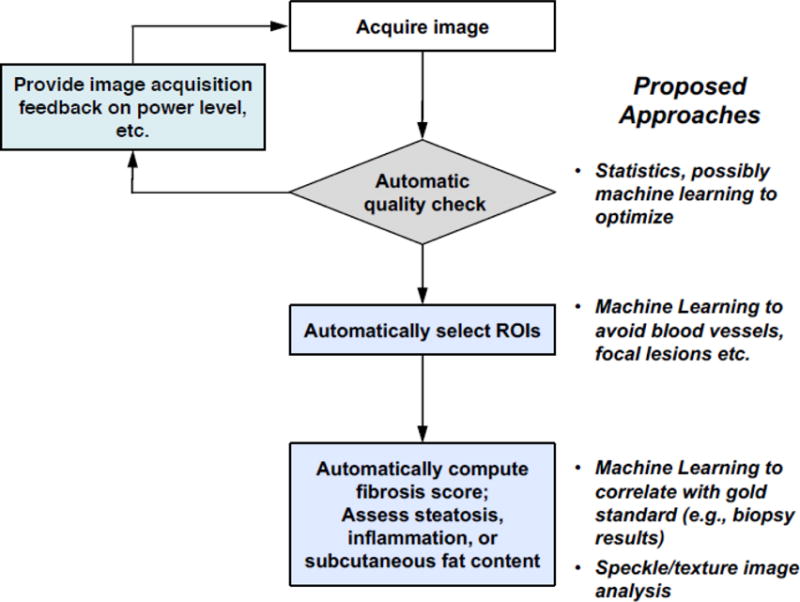
Proposed semi-automated SWE acquisition and analysis workflow.
Among the surveyed papers from the past 5 years, the most common ML approach is to extract statistical features from the SWE images and then apply a classifier [106]–[110].
SWE images often contain irrelevant patterns (e.g., artifacts, noise, areas absence of SWE information), which can be difficult for both handcrafted feature extraction approaches and for typical DL methods such as CNN. Very recently, [111] reported using a two-layer DL network for automated feature extraction from SWE breast data. The work focuses on differentiating task-relevant (i.e. patterns of interest) vs. task-irrelevant patterns (i.e. distracting patterns).
CEUS is a noninvasive diagnostic tool for focal liver lesion evaluation. Typically, time intensity curves (TICs) are extracted from manually selected ROI in CEUS. Results are often subjected to operator variability, motion sensitivity, and speckle noise. Recently, DL has been applied to CEUS to improve the classification of benign and malignant focal liver lesions from automatically extracted TICs with respiratory compensation [112]. DL shows higher accuracy than conventional ML methods.
V. Discussion and Outlook
While the use of medical US is becoming ubiquitous, advanced US image analysis techniques lag behind other modalities such as CT and MRI. As with CT and MRI, ML is a promising approach to improve US image analysis, disease classification, and computer-aided diagnosis.
Overall, application of ML to US is at an early stage, but is rapidly progressing, as evidenced by the large number of 2016 and 2017 surveyed papers. Most of the recent papers surveyed use databases of a few hundred images. Only a few papers use databases of at least one thousand images, which is three orders of magnitude smaller than large challenge databases of optical images. On the other hand, it is unrealistic to expect US databases will reach that size in the foreseeable future, pointing to the need for ML techniques that can train on smaller databases. In many cases, databases are generated from a single device type and a single collection site, limiting the generalizability of ML classification models derived from these databases. Large, publicly accessible challenge databases such as ImageNet that have significantly advanced conventional image classification performance are currently unavailable for US. Most of the present US ML research has concentrated on single functions within an overall system, such segmentation or classification.
Within the past few years, deep learning approaches have been shown to significantly improve performance when compared with classifiers operating on handcrafted features. Transfer learning, which involves retraining a portion of a network originally trained on other images, has been shown to be effective for classifying the relatively small databases that are currently available. These results address early skepticism that transfer learning would not be useful for US because US images appear to be quite different than the optical color imagery on which the networks were originally trained. Deep learning approaches have also obviated the need for sophisticated preprocessing, such as despeckling. On the other hand, certain applications have based on sophisticated handcrafted features are unlikely to be surpassed by deep learning with currently available databases. Moreover, surveyed papers combining classifiers with both deep learning and handcrafted features have shown improved results over either approach, indicating Deep Learning techniques alone are unlikely to achieve the potential of ML in US.
There are several challenges in applying ML to US and other medical imaging modalities: (1) because US is often used as a first-line imaging modality, there is often an imbalance with an excess of normal “no-disease” images, and (2) obtaining consistently annotated data is a challenge as there is significant inter-operator and inter-observer variability among expert US physicians. The variability that this subjectivity adds to the annotations requires a larger database so the classifier can be trained to smooth over the variations. Transfer learning has been widely adopted to address the challenge of operating with relatively small databases. Weakly supervised learning was also successfully used in one surveyed paper; its use is likely to increase, although challenges have been found in unpublished work. In addition to these techniques, other approaches commonly used by the deep learning community to address small, annotated databases are unsupervised learning, database augmentation and active learning. Interestingly, these techniques have been rarely used in US, and are likely to be promising approaches. Active learning requires an interactive annotation tool that is somewhat more complex than a static tool, but once developed, has the value of focusing the expert’s time on images most important to annotate. Another approach to annotating images would be to apply natural language processing tools to extract annotations from the patient reports. This is still an area of research that has its own challenges to address.
Another algorithmic challenge is the need for the results to be interpretable by radiologists, as opposed to a “black box” result that might suffice in domains other than clinical medicine. Although interpretability is not an intrinsic characteristic of deep learning, it is an active area of research. Within the past few years, new techniques for interpreting CNNs have emerged, and other classification techniques are being developed that are intrinsically interpretable [113].
One key strength of US is its ability to produce real-time video. ML applied to echocardiography and obstetrics has increasingly exploited the advantages of spatiotemporal data to improve results. Even in the case of detecting tumors and other pathologies, video clips provide more information than a single image frame. None of the surveyed papers about classifying pathologies exploited video data. This is an aspect that will likely advance in future work.
Returning to the system view in Fig. 1, advances across the workflow are needed. ML enables part of the system solution, but not all of it. For example, a unique challenge of US is the expertise required for image acquisition, which currently contributes to variable interpretations. Operating on freehand US is preferred. In the future, it will be important for ML systems to provide real-time feedback to the sonographer during image acquisition, and not only to interpret freehand US post hoc. Also, manual ROI selection and caliper placement for measurements are still common, which also result in significant variability. Image quality control, automatic ROI selection, and attention to computer-human integration are needed to replace manual ROI selection and caliper placement for measurements.
Based on recent rapid progress summarized in this review, we expect ML for US will continue to progress, and will be one of the most important trends in diagnostic US in the coming years. Broadly speaking, US will likely become one of the many inputs of a ML-based intelligent diagnostic assistant system, where multimodal and multiscale observations are learned over time and are turned into clinical viable quantitative models (Fig. 7), The aggregated machine intelligence will have the ability to observe data, orient the end user, assess new information, and assist with decision making. Such a system has the potential to greatly improve not only the clinical workflow but also the overall outcome of care.
Figure 7.
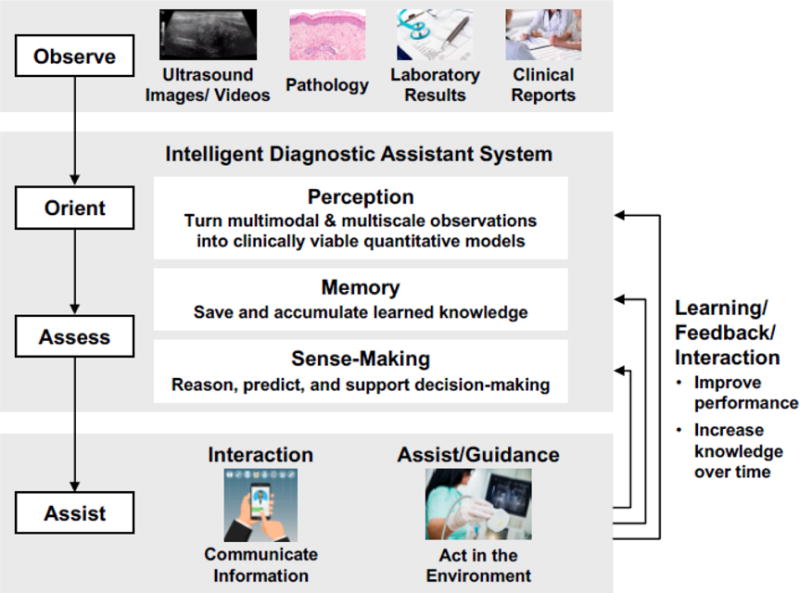
Proposed framework for machine learning-based intelligent diagnostic assistant
Acknowledgments
Funding: This material is based upon work supported by the Assistant Secretary of Defense for Research and Engineering under Air Force Contract No. FA8721-05-C-0002 and/or FA8702-15-D-0001. Any opinions, findings, conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Assistant Secretary of Defense for Research and Engineering. This work is also supported by the NIBIB of the National Institutes of Health under award numbers HHSN268201300071 C and K23 EB020710. The authors are solely responsible for the content and the work does not represent the official views of the National Institutes of Health.
Footnotes
Laura J. Brattain declares that she has no conflict of interest.
Brian A. Telfer declares that he has no conflict of interest.
Manish Dhyani declares that he has no conflict of interest.
Joseph R. Grajo declares that he has no conflict of interest.
Anthony E. Samir declares that he has no conflict of interest.
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest: All the authors declare that they have no conflict of interest.
Contributor Information
Laura J. Brattain, Technical Staff, MIT Lincoln Laboratory, Lexington, MA 02420.
Brian A. Telfer, Senior Staff, MIT Lincoln Laboratory, Lexington, MA 02420.
Manish Dhyani, Director for Clinical Translation, Center for Ultrasound Research & Translation, Department of Radiology, Massachusetts General Hospital, Boston, MA 02114.
Joseph R. Grajo, Assistant Professor of Radiology, Abdominal Imaging Division, Vice Chair for Research, University of Florida College of Medicine, Gainesville, FL.
Anthony E. Samir, Co-Director, Center for Ultrasound Research & Translation, Division of Ultrasound, Department of Radiology, Massachusetts General Hospital, Boston, MA 02114.
References
- 1.Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012 Jul;16(5):933–951. doi: 10.1016/j.media.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;(0) doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litjens G, et al. A survey on deep learning in medical image analysis. ArXiv Prepr ArXiv170205747. 2017 doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Ravi D, et al. Deep Learning for Health Informatics. IEEE J Biomed Health Inform. 2017 Jan;21(1):4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 5.Cassinotto C, et al. Non-invasive assessment of liver fibrosis with impulse elastography: comparison of Supersonic Shear Imaging with ARFI and FibroScan®. J Hepatol. 2014;61(3):550–557. doi: 10.1016/j.jhep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 6.Ferraioli G, Parekh P, Levitov AB, Filice C. Shear wave elastography for evaluation of liver fibrosis. J Ultrasound Med. 2014;33(2):197–203. doi: 10.7863/ultra.33.2.197. [DOI] [PubMed] [Google Scholar]
- 7.Poynard T, et al. Liver fibrosis evaluation using real-time shear wave elastography: applicability and diagnostic performance using methods without a gold standard. J Hepatol. 2013;58(5):928–935. doi: 10.1016/j.jhep.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Samir AE, et al. Shear-wave elastography for the estimation of liver fibrosis in chronic liver disease: determining accuracy and ideal site for measurement. Radiology. 2014;274(3):888–896. doi: 10.1148/radiol.14140839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, et al. Breast lesions: quantitative diagnosis using ultrasound shear wave elastography—a systematic review and meta-analysis. Ultrasound Med Biol. 2016;42(4):835–847. doi: 10.1016/j.ultrasmedbio.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, et al. Differential Diagnosis of Breast Category 3 and 4 Nodules Through BI-RADS Classification in Conjunction with Shear Wave Elastography. Ultrasound Med Biol. 2017;43(3):601–606. doi: 10.1016/j.ultrasmedbio.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZL, Li Y, Wan WB, Li N, Tang J. Shear-Wave Elastography: Could it be Helpful for the Diagnosis of Non-Mass-Like Breast Lesions? Ultrasound Med Biol. 2017;43(1):83–90. doi: 10.1016/j.ultrasmedbio.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Anvari A, Dhyani M, Stephen AE, Samir AE. Reliability of Shear-Wave elastography estimates of the young modulus of tissue in follicular thyroid neoplasms. Am J Roentgenol. 2016;206(3):609–616. doi: 10.2214/AJR.15.14676. [DOI] [PubMed] [Google Scholar]
- 13.Dhyani M, Li C, Samir AE, Stephen AE. Elastography: Applications and Limitations of a New Technology. Advanced Thyroid and Parathyroid Ultrasound, Springer. 2017:67–73. [Google Scholar]
- 14.Ding J, Cheng HD, Huang J, Zhang Y, Liu J. An improved quantitative measurement for thyroid cancer detection based on elastography. Eur J Radiol. 2012 Apr;81(4):800–805. doi: 10.1016/j.ejrad.2011.01.110. [DOI] [PubMed] [Google Scholar]
- 15.Park AY, Son EJ, Han K, Youk JH, Kim JA, Park CS. Shear wave elastography of thyroid nodules for the prediction of malignancy in a large scale study. Eur J Radiol. 2015;84(3):407–412. doi: 10.1016/j.ejrad.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Eby SF, et al. Shear wave elastography of passive skeletal muscle stiffness: Influences of sex and age throughout adulthood. Clin Biomech. 2015;30(1):22–27. doi: 10.1016/j.clinbiomech.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pass B, Jafari M, Rowbotham E, Hensor E, Gupta H, Robinson P. Do quantitative and qualitative shear wave elastography have a role in evaluating musculoskeletal soft tissue masses? Eur Radiol. 2017;27(2):723–731. doi: 10.1007/s00330-016-4427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taljanovic MS, et al. Shear-Wave Elastography: Basic Physics and Musculoskeletal Applications. RadioGraphics. 2017;37(3):855–870. doi: 10.1148/rg.2017160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubry S, Nueffer JP, Tanter M, Becce F, Vidal C, Michel F. Viscoelasticity in Achilles tendonopathy: quantitative assessment by using real-time shear-wave elastography. Radiology. 2014;274(3):821–829. doi: 10.1148/radiol.14140434. [DOI] [PubMed] [Google Scholar]
- 20.Zhang ZJ, Ng GY, Lee WC, Fu SN. Changes in morphological and elastic properties of patellar tendon in athletes with unilateral patellar tendinopathy and their relationships with pain and functional disability. PloS One. 2014;9(10):e108337. doi: 10.1371/journal.pone.0108337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouvière O, et al. Stiffness of benign and malignant prostate tissue measured by shear-wave elastography: a preliminary study. Eur Radiol. 2017;27(5):1858–1866. doi: 10.1007/s00330-016-4534-9. [DOI] [PubMed] [Google Scholar]
- 22.Sang L, Wang X, Xu D, Cai Y. Accuracy of shear wave elastography for the diagnosis of prostate cancer: A meta-analysis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-02187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo S, Suh CH, Kim SY, Cho JY, Kim SH. Shear-Wave Elastography for Detection of Prostate Cancer: A Systematic Review and Diagnostic Meta-Analysis. Am J Roentgenol. 2017:1–9. doi: 10.2214/AJR.17.18056. [DOI] [PubMed] [Google Scholar]
- 24.D’Onofrio M, Crosara S, De Robertis R, Canestrini S, Mucelli RP. Contrast-enhanced ultrasound of focal liver lesions. Am J Roentgenol. 2015;205(1):W56–W66. doi: 10.2214/AJR.14.14203. [DOI] [PubMed] [Google Scholar]
- 25.Kim TK, Jang HJ. Contrast-enhanced ultrasound in the diagnosis of nodules in liver cirrhosis. World J Gastroenterol. 2014 Apr;20(13):3590–3596. doi: 10.3748/wjg.v20.i13.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strobel D, et al. Contrast-enhanced ultrasound for the characterization of focal liver lesions–diagnostic accuracy in clinical practice (DEGUM multicenter trial) Ultraschall Med Stuttg Ger 1980. 2008 Oct;29(5):499–505. doi: 10.1055/s-2008-1027806. [DOI] [PubMed] [Google Scholar]
- 27.Westwood M, et al. Contrast-enhanced ultrasound using SonoVue® (sulphur hexafluoride microbubbles) compared with contrast-enhanced computed tomography and contrast-enhanced magnetic resonance imaging for the characterisation of focal liver lesions and detection of liver metastases: a systematic review and cost-effectiveness analysis. Health Technol Assess Winch Engl. 2013 Apr;17(16):1–243. doi: 10.3310/hta17160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh TH, Lee YH, Seo IY. Diagnostic efficacy of contrast-enhanced ultrasound for small renal masses. Korean J Urol. 2014 Sep;55(9):587–592. doi: 10.4111/kju.2014.55.9.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Z, Quan J, Yunxiao Z, Jian C, Zhu H. Contrast-enhanced ultrasound in the diagnosis of solitary thyroid nodules. J Cancer Res Ther. 2015 Mar;11(1):41–45. doi: 10.4103/0973-1482.147382. [DOI] [PubMed] [Google Scholar]
- 30.Li W, et al. Real-time contrast enhanced ultrasound imaging of focal splenic lesions. Eur J Radiol. 2014 Apr;83(4):646–653. doi: 10.1016/j.ejrad.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Baur ADJ, et al. A direct comparison of contrast-enhanced ultrasound and dynamic contrast-enhanced magnetic resonance imaging for prostate cancer detection and prediction of aggressiveness. Eur Radiol. 2017 Dec; doi: 10.1007/s00330-017-5192-2. [DOI] [PubMed] [Google Scholar]
- 32.Bishop CM. Pattern recognition and machine learning springer. 2006 [Google Scholar]
- 33.Mitchell TM. Machine learning WCB McGraw-Hill Boston, MA. 1997 [Google Scholar]
- 34.De Mantaras RL, Armengol E. Machine learning from examples: Inductive and Lazy methods. Data Knowl Eng. 1998;25(1–2):99–123. [Google Scholar]
- 35.Dutton DM, Conroy GV. A review of machine learning. Knowl Eng Rev. 1997;12(4):341–367. [Google Scholar]
- 36.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artif Intell Rev. 2006;26(3):159–190. [Google Scholar]
- 37.Torresani L. Weakly supervised learning. Computer Vision, Springer. 2014:883–885. [Google Scholar]
- 38.Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning MIT press. 2012 [Google Scholar]
- 39.Soh LK, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans Geosci Remote Sens. 1999;37(2):780–795. [Google Scholar]
- 40.Materka A, Strzelecki M, et al. Texture analysis methods–a review. Tech Univ Lodz Inst Electron COST B11 Rep Bruss. 1998:9–11. [Google Scholar]
- 41.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst. 1987;2(1–3):37–52. [Google Scholar]
- 42.Liaw A, Wiener M, et al. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 43.Cortes C, Vapnik V. Support vector machine. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 44.Suykens JA, Vandewalle J. Least squares support vector machine classifiers. Neural Process Lett. 1999;9(3):293–300. [Google Scholar]
- 45.White H. Connectionist nonparametric regression: Multilayer feedforward networks can learn arbitrary mappings. Neural Netw. 1990;3(5):535–549. [Google Scholar]
- 46.Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989;2(5):359–366. [Google Scholar]
- 47.Leshno M, Lin VY, Pinkus A, Schocken S. Multilayer feedforward networks with a nonpolynomial activation function can approximate any function. Neural Netw. 1993;6(6):861–867. [Google Scholar]
- 48.Geisser S. Predictive inference: an introduction New York: Chapman & Hall. 1993 [Google Scholar]
- 49.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 50.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–1105. [Google Scholar]
- 51.Szegedy C, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015:1–9. [Google Scholar]
- 52.Sermanet P, Kavukcuoglu K, Chintala S, LeCun Y. Pedestrian detection with unsupervised multi-stage feature learning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2013:3626–3633. [Google Scholar]
- 53.Szegedy C, Toshev A, Erhan D. Deep neural networks for object detection. Advances in neural information processing systems. 2013:2553–2561. [Google Scholar]
- 54.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv Prepr ArXiv14091556. 2014 [Google Scholar]
- 55.Lawrence S, Giles CL, Tsoi AC, Back AD. Face recognition: A convolutional neural-network approach. IEEE Trans Neural Netw. 1997;8(1):98–113. doi: 10.1109/72.554195. [DOI] [PubMed] [Google Scholar]
- 56.Matsugu M, Mori K, Mitari Y, Kaneda Y. Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Netw. 2003;16(5):555–559. doi: 10.1016/S0893-6080(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 57.Farfade SS, Saberian MJ, Li LJ. Multi-view face detection using deep convolutional neural networks. Proceedings of the 5th ACM on International Conference on Multimedia Retrieval. 2015:643–650. [Google Scholar]
- 58.Turaga SC, et al. Convolutional networks can learn to generate affinity graphs for image segmentation. Neural Comput. 2010;22(2):511–538. doi: 10.1162/neco.2009.10-08-881. [DOI] [PubMed] [Google Scholar]
- 59.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015:3431–3440. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 60.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. 1995 [Google Scholar]
- 61.Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010;4:40–79. [Google Scholar]
- 62.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 63.Garra BS, Krasner BH, Horii SC, Ascher S, Mun SK, Zeman RK. Improving the distinction between benign and malignant breast lesions: the value of sonographic texture analysis. Ultrason Imaging. 1993;15(4):267–285. doi: 10.1177/016173469301500401. [DOI] [PubMed] [Google Scholar]
- 64.Maclin PS, Dempsey J. Using an artificial neural network to diagnose hepatic masses. J Med Syst. 1992;16(5):215–225. doi: 10.1007/BF01000274. [DOI] [PubMed] [Google Scholar]
- 65.Giger ML, Karssemeijer N, Schnabel JA. Breast Image Analysis for Risk Assessment, Detection, Diagnosis, and Treatment of Cancer. Annu Rev Biomed Eng. 2013 Jul;15(1):327–357. doi: 10.1146/annurev-bioeng-071812-152416. [DOI] [PubMed] [Google Scholar]
- 66.Shan J, Alam SK, Garra B, Zhang Y, Ahmed T. Computer-Aided Diagnosis for Breast Ultrasound Using Computerized BI-RADS Features and Machine Learning Methods. Ultrasound Med Biol. 2016;42(4):980–988. doi: 10.1016/j.ultrasmedbio.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 67.Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Carbonell JG, Michalski RS, Mitchell TM. An overview of machine learning. Machine learning, Springer. 1983:3–23. [Google Scholar]
- 69.Barinov L, Jairaj A, Paster L, Hulbert W, Mammone R, Podilchuk C. Decision quality support in diagnostic breast ultrasound through artificial Intelligence [Google Scholar]
- 70.Choi YJ, et al. A Computer-Aided Diagnosis System Using Artificial Intelligence for the Diagnosis and Characterization of Thyroid Nodules on Ultrasound: Initial Clinical Assessment. Thyroid. 2017;27(4):546–552. doi: 10.1089/thy.2016.0372. [DOI] [PubMed] [Google Scholar]
- 71.Hiramatsu Y, Muramatsu C, Kobayashi H, Hara T, Fujita H. Automated detection of masses on whole breast volume ultrasound scanner: False positive reduction using deep convolutional neural network. 2017;10134 [Google Scholar]
- 72.Lekadir K, et al. A Convolutional Neural Network for Automatic Characterization of Plaque Composition in Carotid Ultrasound. IEEE J Biomed Health Inform. 2017;21(1):48–55. doi: 10.1109/JBHI.2016.2631401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng PM, Malhi HS. Transfer Learning with Convolutional Neural Networks for Classification of Abdominal Ultrasound Images. J Digit Imaging. 2017;30(2):234–243. doi: 10.1007/s10278-016-9929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antropova N, Huynh BQ, Giger ML. A Deep Feature Fusion Methodology for Breast Cancer Diagnosis Demonstrated on Three Imaging Modality Datasets. Med Phys. 2017 Jul; doi: 10.1002/mp.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi H, Collins S, Noble A. Weakly Supervised Learning of Placental Ultrasound Images with Residual Networks. Annual Conference on Medical Image Understanding and Analysis. 2017:98–108. doi: 10.1007/978-3-319-60964-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cunningham R, Harding P, Loram I. Deep Residual Networks for Quantification of Muscle Fiber Orientation and Curvature from Ultrasound Images. In: ValdésHernández M, González-Castro V, editors. Medical Image Understanding and Analysis. Vol. 723. Cham: Springer International Publishing; 2017. pp. 63–73. [Google Scholar]
- 77.Namburete AI, Stebbing RV, Kemp B, Yaqub M, Papageorghiou AT, Noble JA. Learning-based prediction of gestational age from ultrasound images of the fetal brain. Med Image Anal. 2015;21(1):72–86. doi: 10.1016/j.media.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cary TW, Reamer CB, Sultan LR, Mohler ER, Sehgal CM. Brachial artery vasomotion and transducer pressure effect on measurements by active contour segmentation on ultrasound: Brachial artery vasomotion and transducer pressure effect. Med Phys. 2014 Jan;41(2):022901. doi: 10.1118/1.4862508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imaging. 2006 Aug;25(8):987–1010. doi: 10.1109/tmi.2006.877092. [DOI] [PubMed] [Google Scholar]
- 80.Noble JA. Ultrasound image segmentation and tissue characterization. Proc Inst Mech Eng [H] 2010 Feb;224(2):307–316. doi: 10.1243/09544119JEIM604. [DOI] [PubMed] [Google Scholar]
- 81.Torbati N, Ayatollahi A, Kermani A. An efficient neural network based method for medical image segmentation. Comput Biol Med. 2014;44:76–87. doi: 10.1016/j.compbiomed.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 82.Yang X, Rossi PJ, Jani AB, Mao H, Curran WJ, Liu T. 3D transrectal ultrasound (TRUS) prostate segmentation based on optimal feature learning framework. 2016:97842F. doi: 10.1117/12.2216396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghose S, et al. A supervised learning framework of statistical shape and probability priors for automatic prostate segmentation in ultrasound images. Med Image Anal. 2013;17(6):587–600. doi: 10.1016/j.media.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 84.Sultan LR, Xiong H, Zafar HM, Schultz SM, Langer JE, Sehgal CM. Vascularity Assessment of Thyroid Nodules by Quantitative Color Doppler Ultrasound. Ultrasound Med Biol. 2015 May;41(5):1287–1293. doi: 10.1016/j.ultrasmedbio.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 85.Chauhan A, Sultan LR, Furth EE, Jones LP, Khungar V, Sehgal CM. Diagnostic accuracy of hepatorenal index in the detection and grading of hepatic steatosis: Factors Affecting the Accuracy Of HRI. J Clin Ultrasound. 2016 Nov;44(9):580–586. doi: 10.1002/jcu.22382. [DOI] [PubMed] [Google Scholar]
- 86.Noe MH, et al. High frequency ultrasound: a novel instrument to quantify granuloma burden in cutaneous sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2017 Aug;34(2):136–141. doi: 10.36141/svdld.v34i2.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.“The diagnostic performance of leak-plugging automated segmentation versus manual tracing of breast lesions on ultrasound imagesUltrasound – Hui Xiong, Laith R Sultan, Theodore W Cary, Susan M Schultz, Ghizlane Bouzghar, Chandra M Sehgal, 2017.” [Online]. Available: http://journals.sagepub.com/doi/pdf/10.1177/1742271X17690425#articleCitationDownloadContainer. [Accessed: 17-Jan-2018]
- 88.Carneiro G, Nascimento JC, Freitas A. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans Image Process. 2012;21(3):968–82. doi: 10.1109/TIP.2011.2169273. [DOI] [PubMed] [Google Scholar]
- 89.Menchón-Lara RM, Sancho-Gómez JL. Fully automatic segmentation of ultrasound common carotid artery images based on machine learning. Neurocomputing. 2015;151(P1):161–167. [Google Scholar]
- 90.Zhang Y, Ying MT, Yang L, Ahuja AT, Chen DZ. Coarse-to-Fine Stacked Fully Convolutional Nets for lymph node segmentation in ultrasound images. Bioinformatics and Biomedicine (BIBM), 2016 IEEE International Conference on. 2016:443–448. [Google Scholar]
- 91.Looney P, et al. Automatic 3D ultrasound segmentation of the first trimester placenta using deep learning. Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on. 2017:279–282. [Google Scholar]
- 92.Milletari F, et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput Vis Image Underst. 2017 Apr; [Google Scholar]
- 93.Chen F, Wu D, Liao H. Registration of CT and Ultrasound Images of the Spine with Neural Network and Orientation Code Mutual Information. In: Zheng G, Liao H, Jannin P, Cattin P, Lee S-L, editors. Medical Imaging and Augmented Reality. Vol. 9805. Cham: Springer International Publishing; 2016. pp. 292–301. [Google Scholar]
- 94.Yang X, Fei B. 3D prostate segmentation of ultrasound images combining longitudinal image registration and machine learning. Proceedings of SPIE. 2012;8316:83162O. doi: 10.1117/12.912188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao Y, Maraci MA, Noble JA. Describing ultrasound video content using deep convolutional neural networks. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 2016:787–790. [Google Scholar]
- 96.Baumgartner CF, Kamnitsas K, Matthew J, Smith S, Kainz B, Rueckert D. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Vol. 9901. LNCS; 2016. Real-time standard scan plane detection and localisation in fetal ultrasound using fully convolutional neural networks; pp. 203–211. [Google Scholar]
- 97.Kumar A, et al. Plane identification in fetal ultrasound images using saliency maps and convolutional neural networks. 2017 Jun;2016:791–794. [Google Scholar]
- 98.Chen H, et al. Automatic fetal ultrasound standard plane detection using knowledge transferred recurrent neural networks. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2015;9349:507–514. [Google Scholar]
- 99.Yaqub M, Kelly B, Papageorghiou AT, Noble JA. Guided random forests for identification of key fetal anatomy and image categorization in ultrasound scans. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015:687–694. [Google Scholar]
- 100.Gao X, Li W, Loomes M, Wang L. A fused deep learning architecture for viewpoint classification of echocardiography. Inf Fusion. 2017 Jul;36:103–113. [Google Scholar]
- 101.Sundaresan V, Bridge CP, Ioannou C, Noble JA. Automated characterization of the fetal heart in ultrasound images using fully convolutional neural networks. Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on. 2017:671–674. [Google Scholar]
- 102.Khamis H, Zurakhov G, Azar V, Raz A, Friedman Z, Adam D. Automatic apical view classification of echocardiograms using a discriminative learning dictionary. Med Image Anal. 2017 Feb;36:15–21. doi: 10.1016/j.media.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 103.Sigrist RM, Liau J, El Kaffas A, Chammas MC, Willmann JK. Ultrasound Elastography: Review of Techniques and Clinical Applications. Theranostics. 2017;7(5):1303. doi: 10.7150/thno.18650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rouze NC, Wang MH, Palmeri ML, Nightingale KR. Parameters affecting the resolution and accuracy of 2-D quantitative shear wave images. IEEE Trans Ultrason Ferroelectr Freq Control. 2012;59:8. doi: 10.1109/TUFFC.2012.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pellot-Barakat C, Lefort M, Chami L, Labit M, Frouin F, Lucidarme O. Automatic Assessment of Shear Wave Elastography Quality and Measurement Reliability in the Liver. Ultrasound Med Biol. 2015 Apr;41(4):936–943. doi: 10.1016/j.ultrasmedbio.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 106.Wang J, Guo L, Shi X, Pan W, Bai Y, Ai H. Real-time elastography with a novel quantitative technology for assessment of liver fibrosis in chronic hepatitis B. Eur J Radiol. 2012;81(1):e31–e36. doi: 10.1016/j.ejrad.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 107.Xiao Y, et al. Computer-aided diagnosis based on quantitative elastographic features with supersonic shear wave imaging. Ultrasound Med Biol. 2014;40(2):275–286. doi: 10.1016/j.ultrasmedbio.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 108.Bhatia KSS, Lam ACL, Pang SWA, Wang D, Ahuja AT. Feasibility study of texture analysis using ultrasound shear wave elastography to predict malignancy in thyroid nodules. Ultrasound Med Biol. 2016;42(7):1671–1680. doi: 10.1016/j.ultrasmedbio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 109.Gatos I, et al. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. doi: 10.1016/j.ultrasmedbio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 110.Zhang Q, Xiao Y, Chen S, Wang C, Zheng H. Quantification of elastic heterogeneity using contourlet-based texture analysis in shear-wave elastography for breast tumor classification. Ultrasound Med Biol. 2015;41(2):588–600. doi: 10.1016/j.ultrasmedbio.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Q, et al. Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics. 2016 Dec;72:150–157. doi: 10.1016/j.ultras.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Wu K, Chen X, Ding M. Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Opt-Int J Light Electron Opt. 2014;125(15):4057–4063. [Google Scholar]
- 113.Zeng J, Ustun B, Rudin C. Interpretable classification models for recidivism prediction. J R Stat Soc Ser A Stat Soc. 2017;180(3):689–722. [Google Scholar]
- 114.Shi J, Zhou S, Liu X, Zhang Q, Lu M, Wang T. Stacked deep polynomial network based representation learning for tumor classification with small ultrasound image dataset. Neurocomputing. 2016 Jun;194:87–94. [Google Scholar]
- 115.Singh BK, Verma K, Thoke AS. Fuzzy cluster based neural network classifier for classifying breast tumors in ultrasound images. Expert Syst Appl. 2016;66:114–123. [Google Scholar]
- 116.Wu WJ, Lin SW, Moon WK. An Artificial Immune System-Based Support Vector Machine Approach for Classifying Ultrasound Breast Tumor Images. J Digit Imaging. 2015;28(5):576–85. doi: 10.1007/s10278-014-9757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shan J, Cheng HD, Wang Y. Completely automated segmentation approach for breast ultrasound images using multiple-domain features. Ultrasound Med Biol. 2012;38(2):262–75. doi: 10.1016/j.ultrasmedbio.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 118.Nascimento CDL, Silva SDS, da Silva TA, Pereira WCA, Costa MGF, Costa Filho CFF. Breast tumor classification in ultrasound images using support vector machines and neural networks. Rev Bras Eng Biomed. 2016;32(3):283–292. [Google Scholar]
- 119.Marcomini KD, Carneiro AAO, Schiabel H. Application of artificial neural network models in segmentation and classification of nodules in breast ultrasound digital images. Int J Biomed Imaging. 2016;2016 doi: 10.1155/2016/7987212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jamieson AR, Giger ML, Drukker K, Li H, Yuan Y, Bhooshan N. Exploring nonlinear feature space dimension reduction and data representation in breast CADx with Laplacian eigenmaps and t-SNE: Nonlinear dimension reduction and representation in breast CADx. Med Phys. 2009 Dec;37(1):339–351. doi: 10.1118/1.3267037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng. 2015;26:S1599–S1611. doi: 10.3233/BME-151459. [DOI] [PubMed] [Google Scholar]
- 122.Suganya R, Kirubakaran R, Rajaram S. Classification and retrieval of focal and diffuse liver from ultrasound images using machine learning techniques. 2014;264:253–261. [Google Scholar]
- 123.Kalyan K, Jakhia B, Lele RD, Joshi M, Chowdhary A. Artificial Neural Network Application in the Diagnosis of Disease Conditions with Liver Ultrasound Images. Adv Bioinforma. 2014;2014 doi: 10.1155/2014/708279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brattain LJ, Telfer BA, Liteplo AS, Noble VE. Automated B-Line Scoring on Thoracic Sonography. J Ultrasound Med. 2013;32(12):2185–2190. doi: 10.7863/ultra.32.12.2185. [DOI] [PubMed] [Google Scholar]
- 125.Veeramani SK, Muthusamy E. Detection of abnormalities in ultrasound lung image using multi-level RVM classification. J Matern Fetal Neonatal Med. 2016;29(11):1844–52. doi: 10.3109/14767058.2015.1064888. [DOI] [PubMed] [Google Scholar]
- 126.Konig T, Steffen J, Rak M, Neumann G, von Rohden L, Tonnies KD. Ultrasound texture-based CAD system for detecting neuromuscular diseases. Int J Comput Assist Radiol Surg. 2015;10(9):1493–503. doi: 10.1007/s11548-014-1133-6. [DOI] [PubMed] [Google Scholar]
- 127.Srivastava T, Darras BT, Wu JS, Rutkove SB. Machine learning algorithms to classify spinal muscular atrophy subtypes. Neurology. 2012;79(4):358–64. doi: 10.1212/WNL.0b013e3182604395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sheet D, et al. Joint learning of ultrasonic backscattering statistical physics and signal confidence primal for characterizing atherosclerotic plaques using intravascular ultrasound. Med Image Anal. 2014;18(1):103–17. doi: 10.1016/j.media.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 129.Yu S, Tan KK, Sng BL, Li S, Sia AT. Lumbar Ultrasound Image Feature Extraction and Classification with Support Vector Machine. Ultrasound Med Biol. 2015;41(10):2677–89. doi: 10.1016/j.ultrasmedbio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 130.Pathak H, Kulkarni V. Identification of ovarian mass through ultrasound images using machine learning techniques. 2015:137–140. [Google Scholar]
- 131.Aramendía-Vidaurreta V, Cabeza R, Villanueva A, Navallas J, Alcázar JL. Ultrasound Image Discrimination between Benign and Malignant Adnexal Masses Based on a Neural Network Approach. Ultrasound Med Biol. 2016;42(3):742–752. doi: 10.1016/j.ultrasmedbio.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 132.Subramanya MB, Kumar V, Mukherjee S, Saini M. SVM-Based CAC System for B-Mode Kidney Ultrasound Images. J Digit Imaging. 2015;28(4):448–58. doi: 10.1007/s10278-014-9754-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takagi K, Kondo S, Nakamura K, Takiguchi M. Lesion type classification by applying machine-learning technique to contrast-enhanced ultrasound images. IEICE Trans Inf Syst. 2014;E97D(11):2947–2954. [Google Scholar]
- 134.Caxinha M, et al. Automatic cataract classification based on ultrasound technique using machine learning: A comparative study. 2015;70:1221–1224. [Google Scholar]
- 135.Sjogren AR, Leo MM, Feldman J, Gwin JT. Image Segmentation and Machine Learning for Detection of Abdominal Free Fluid in Focused Assessment With Sonography for Trauma Examinations: A Pilot Study. J Ultrasound Med. 2016;35(11):2501–2509. doi: 10.7863/ultra.15.11017. [DOI] [PMC free article] [PubMed] [Google Scholar]


