Abstract
In cancer immunology, somatic missense mutations have been mostly studied regarding their role in the generation of neoantigens. However, growing evidence suggests that mutations in certain genes, such as CASP8 or TP53, influence the immune response against a tumor by other mechanisms. Identifying these genes and mechanisms is important because, just as the identification of cancer driver genes led to the development of personalized cancer therapies, a comprehensive catalog of such cancer immunity drivers will aid in the development of therapies aimed at restoring antitumor immunity. Here we present an algorithm, domainXplorer, that can be used to identify potential cancer immunity drivers. To demonstrate its potential, we used it to analyze a dataset of 5,164 tumor samples from TCGA and to identify protein domains whose mutation status correlates with the presence of immune cells in cancer tissue (immune infiltrate). We identified 122 such protein regions including several that belong to proteins with known roles in immune response, such as C2, CD163L1, or FCγR2A. In several cases we show that mutations within the same protein can be associated with more or less immune cell infiltration, depending on the specific domain mutated. These results expand the catalog of potential cancer immunity drivers and highlight the importance of taking into account the structural context of somatic mutations when analyzing their potential association with immune phenotypes.
Keywords: Mutations, domains, cancer immunology, algorithm, cancer immunity driver, CTNNB1, POLR3B, CDH11, complement pathway
Introduction
The immune system detects abnormal cells and destroys potential tumors in a process called immunosurveillance. Although this process protects the host from many nascent tumors, it also leads to the selection of cells with genetic alterations that provide them with mechanisms to escape or modulate the immune response, leading to development of immune resistant tumors (1). Human cells use multiple mechanisms to interact with their microenvironment and most of these mechanisms can also be exploited by cancer cells to evade host immune responses. Many somatic molecular alterations in cancer cells can alter the host immune response, such as overexpression as a result of increased somatic copy number (2), somatic missense mutations that create neoantigens that make the tumor more immunogenic and elicit immune responses (3, 4) and can be exploited to treat some cancer patients (5, 6), or mutations that help cancer cells avoid the cytotoxic immune responses (7). Most extant analyses of mutations in cancer genomes look for correlations between mutations in a gene and some immune-related metric. This approach, although successful in some cases (7), is limited by the assumption that all the mutations in a given gene will lead to the same phenotype. However, it is known that mutations can lead to drastically different phenotypes depending on the specific protein region mutated (8-12). The reason is that genes are not monolithic entities; they consist of different regions, coding for specific protein domains that are usually responsible for different functions. Accounting for this fact in the analysis of cancer-driving mutations and drug sensitivity biomarkers led to discovery of many “domain drivers” or “domain biomarkers” (13, 14).
Here, we explore this approach in the immune response to cancer, searching for “cancer immunity drivers” on the domain level. To this end we analyzed the somatic cancer genomes of 5,164 cancer patients with domainXplorer, an expanded version of our earlier e-Driver algorithm (14) that identifies correlations between any phenotype (including immune responses) and mutations in individual protein regions. We compared the mutation patterns in individual tumors to the predicted presence of the immune infiltrate, as measured by ESTIMATE (15), and found 122 protein regions, mutations in which correlated with the presence of immune cells in cancer tissues. Some of these regions were located in proteins involved in immune response–related pathways, such as the antibody receptor FCγR2A, the complement protein C2 or the scavenger receptor CD163L1, confirming that our method is able to identify known cancer immune-evading mechanisms. Others represent potentially novel mechanisms of cancer cells influencing host immune response and would have to be studied in more detail.
Materials and Methods
Code and data availability
All the raw data and the algorithms used in this manuscript, as well as the raw results used for figures, can be downloaded from www.github.com/eduardporta/domainXplorer. All the statistical calculations were done using R 3.1.0(16). All figures have been generated using the R package ggplot2 (17).
Mutation data
Level 3 mutation data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov) for 5,164 tumor samples that belong to 21 different cancer types. Variant Effect Predictor tool was used to map mutations from their genomic to their protein coordinates (18) using gene and protein annotations from ENSEMBL version 72(19). We identified a total of 636,399 missense mutations in 19,106 proteins. Note that we only analyzed the longest isoform of each gene in order to minimize problems related to multiple testing. Also, we excluded from the analysis proteins that are suspected to be prone to false positives in cancer genomics analysis, such as olfactory receptors or TTN.
Protein regions
We defined protein functional regions as protein domains defined by Pfam(20) or intrinsically disordered regions as identified by Foldindex (21) with a score below −0.1. We have also included a set of 1,300 potential domains identified by AIDA (22), an algorithm based on iterative recognition of domains by homology recognition algorithms with various sensitivities.
Immune scores
We used the ESTIMATE algorithm (15) to evaluate the presence of immune cell infiltrates in each tumor sample. The ESTIMATE score is based on the expression of a gene signature of 140 genes related to the immune system and correlates well with data obtained by cell sorting. We downloaded the pan-cancer normalized gene expression data from the UCSC Cancer Genomics Browser (23) on February 25, 2015. We then analyzed this data with ESTIMATE to evaluate the presence of immune cells in each cancer sample.
domainXplorer algorithm
This algorithm compares the immune and mutation profiles of cancer samples to identify protein regions that, when mutated, correlate with presence of immune cells within the tumor infiltrate. In brief, domainXplorer first seeks a correlation between mutations in a specific domain and immune scores using an analysis of variance (ANOVA, type I) test on a multivariate linear modeling (Test 1, Equation 1). The linear model also takes into account potential biases caused by differences in the immune responses between the tissues of origin of the tumors as covariates. The model is:
| (Equation 1) |
where “E” is the ESTIMATE score of each sample, “T” is the tissue of origin of each sample, and “D” is a binary variable showing whether the domain is mutated. This first step assesses whether any correlation between the “D” term of the linear model and the ESTIMATE score is statistically significant and gives the P value that we report.
In the next step, domainXplorer compares the E score in tumors with mutations in the domain being analyzed against those with mutations in other regions of the same protein (Test 2) or no mutations in the protein at all (Test 3) using a Wilcoxon test. Finally, domainXplorer identifies all regions in which Tests 1 and 2 have a P value below 0.01 and Test 3 has a P value below 0.05. We are less stringent with Test 3 because it intrinsically has a smaller sample size (only those samples with mutations in the analyzed proteins). We do not use multiple-testing corrections (such as Bonferroni or Benjamini-Hochberg algorithms (24)) because, since we use protein regions instead of whole genes, the number of samples with mutations in each region is relatively small. This effectively reduces our statistical power for most regions below that needed to detect significant effect with multiple-hypothesis testing corrections. However, in other applications of a similar protocol we found that the framework with three different tests is stringent enough to reduce the number of false positives while identifying true positives (10). Another reason not to correct for multiple-hypothesis testing is that this setting leads to a manageable list of candidates for further analysis, which is the goal of domainXplorer. Downstream analysis of such candidates is then up to the users, who should rely upon their biological insights and expert opinion.
Neoantigen analysis
We downloaded data on the predicted number of neoantigens for 4,592 TCGA samples from a previous study(7). The HLA alleles of these samples were identified using Polysolver(25) and the neoantigens predicted using NetMHCpan(26). The subset of these samples that was common to our cohort (n = 3,421) was used to repeat the first step of the domainXplorer analysis, but now adding the number of neoantigens per sample in our linear model (Equation 2):
| (Equation 2) |
where “E”, “T” and “D” represent the same variables as in Equation 1 and “N” is the number of predicted neoantigens in each sample. We used the P value of the “domain” variable in this type I ANOVA test to evaluate whether a domain was still significant beyond what can be explained by tissue of origin or the presence of neoantigens.
Gene expression analysis
As explained above, we downloaded normalized gene expression data from the UCSC Cancer Genomics Browser. Then, in the case of the CTNNB1 analysis, we used an ANOVA test on a linear regression model using ESTIMATE as the response and the tissue of origin and CTNNB1 expression as covariates (Equation 3):
| (Equation 3) |
where, again, “E” and “T” have the same values as in Equation 1, and C is a variable containing the expression of CTNNB1 in either mRNA or reverse phase protein array (RPPA) experiments.
For the analysis of the STING pathway, we also used an ANOVA test of a linear model. In this test, expression of each gene in the pathway was treated as a response variable. As covariates we used a categorical variable indicating whether or not the sample had a mutation in the POLR3B domains and either (I) the ESTIMATE score of the sample (Equation 4), (II) its tissue of origin (Equation 5) or (III) both (Equation 6).
| (Equation 4) |
| (Equation 5) |
| (Equation 6) |
where “E”, “D” and “T” have the same values as in Equation 1 and “G” represents the expression levels of the different genes in the pathway.
Proteomics analysis
We used the Pancan16 normalized RPPA file from The Cancer Proteome Atlas (TCPA) website(27). This file contains RPPA data from 4768 samples from 16 different cancer types and is already normalized to correct for batch effects. Again, we used an ANOVA on a linear regression model where either the gene expression or ESTIMATE scores were the response variables and the tissue of origin and beta-catenin protein levels were the covariates (Equation 7):
| (Equation 7) |
where “T” and “C” have the same value as in Equation 3 and “V” represents the expression levels of the different genes tested (CDH11, CDH1, CD8A and CD3E) or the ESTIMATE immune score.
Results
Using ESTIMATE to evaluate tumor immune infiltrate
Algorithms that can deduce the cell composition of heterogeneous samples by analysis of gene expression data offer an alternative to direct cell quantification methods and have been benchmarked against methods such as immunostaining or histological analysis (15, 28, 29). Here we used ESTIMATE (15) to infer the presence of immune cells in 5,164 cancer samples from 21 different projects of The Cancer Genome Atlas (TCGA) (30).
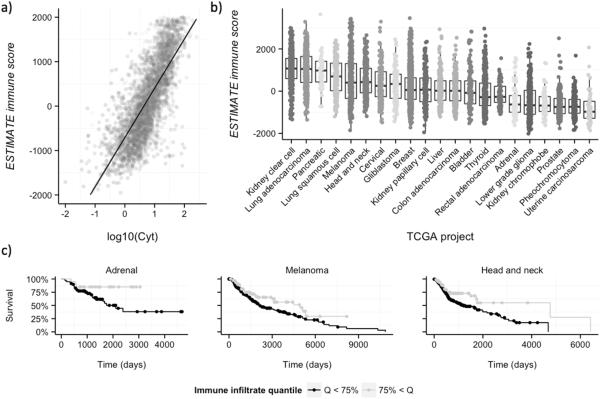
Although using a single measure of immune response is an obvious oversimplification, ESTIMATE scores correlate well with several important features of host immune response, such as the cytolytic activity [measured as the combined expression of GZMA and PRF1 (7)] of the immune infiltrate (P < 1e-16, Fig. 1A), which can be attributed mostly to active CD8 lymphocytes and natural killer cells (7). Therefore, cancer cells from samples with high ESTIMATE immune scores are immunogenic, and yet must have some molecular mechanisms that allow them to survive in the presence of lymphocytes. In contrast, lower immune scores could be due to having few neoantigens and/or some genomic alteration that suppresses the immune response, such as expression of CTNNB1 (31).
Figure 1. ESTIMATE immune scores of TCGA samples.
A, The ESTIMATE immune score correlates with cytolytic activity (see the main text for details). B, Distribution of immune scores across the different TCGA samples. Each dot corresponds to an individual cancer sample. The immune score obtained with ESTIMATE is shown in the y-axis, and samples are grouped according to the TCGA project (tissue origin). Projects are sorted according to their average immune score, from higher (left) to lower (right). C, Immune scores correlate with survival in some cancer types. There is a correlation between higher ESTIMATE immune scores and better outcomes in the Pancancer dataset (Cox P < 0.01, adjusted by tissue of origin). This correlation can also be identified in some individual cancer types, such as adrenocortical carcinoma, melanoma or head and neck cancer.
The overall results were in line with previous studies (7), with some cancer types showing more immune infiltration than others (Fig. 1B), especially those with a potential viral origin (head and neck or cervical cancers) or a strong inflammatory component (lung cancers) or those considered to be immunogenic (kidney clear cell carcinoma or melanoma). However, the immune scores among different samples of the same cancer type were highly variable(Fig. 1B), implicating unique properties of each specific tumor.
Immune scores derived from ESTIMATE correlate with the survival of cancer patients (Fig. 1C). Patients with higher immune scores had better outcomes in the Pan-cancer dataset, which includes all 5,164 samples (Cox P < 0.01, adjusted by tissue of origin), as well as in some individual cancer types, such as adrenocortical carcinoma, melanoma, or head and neck cancer (Cox P < 0.05 in all three cases).
domainXplorer reveals potential immune drivers
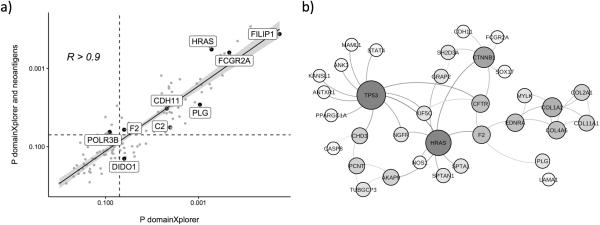
Our goal was to identify potential mechanisms used by cancer cells to influence the immune response, following the hypothesis that mutations in cancer cells influence the immune response in various ways beyond creation of neoantigens. Such effects were demonstrated for some proteins, for instance for the cadherin-associated protein CTNNB1(31) or CASP8(7). To expand this list, we used domainXplorer to analyze ESTIMATE immune scores. Our analysis yielded a total of 122 protein regions that are potential cancer immunity drivers. Several of these regions are discussed in detail below, and the full list is presented in Supplementary Materials Table S1. Given the importance of neoantigens in modulating the immune response against tumors(32, 33), we determined whether cancer immunity driver regions and neoantigens represent independent signals or are mutually redundant. We repeated the linear regression step of domainXplorer on a subset of patients whose neoantigens had been previously predicted (n = 3421). Because of a smaller sample size of this second group, and due to the inclusion of the neoantigen variable, we lowered the significance threshold to P < 0.05 for this specific analysis. In this second analysis 64 of the original 122 domains were identified with a standard domainXplorer linear regression model (Equation 1), reflecting the effects of the smaller sample size. Of these, 52 were also identified when adding the number of neoantigens in each sample to the linear regression model (Equation 2, Fig. 2A). Therefore, at least for these 52 protein regions, the correlation between mutations and the immune scores seems to be independent of the presence of neoantigens. Several of the cancer immunity driver regions identified in our analysis belonged to proteins with known roles in the immune system (such as CD163L1, FCγR2A or C2). Several others were located in the extracellular matrix (COL11A1, LAMA1 or VWA2 for example) where they could interact with immune cells, immediately suggesting a potential mechanism that could mediate the correlation. The remaining group was remarkably diverse with no apparent dominant function, cellular localization, or pathway.
Figure 2. domainXplorer reveals novel players in cancer immunology.
A, Re-analysis with domainXplorer of a subset of TCGA data with data regarding the number of neo-antigens. The P values obtained with (y-axis), and without (x-axis), neoantigens in the model were highly correlated. In this smaller subset, 64 of the original 122 domains still show a statistically significant correlation using the standard domainXplorer (P < 0.05, vertical black dashed line). A total of 52 of these domains, are also statistically significant when adding the number of neoantigens in the model (P < 0.05, horizontal black dashed line). B, Many proteins containing the regions identified by domainXplorer interact with each other or with proteins known to influence the immune response, such as TP53 or CTNNB1.
We tested the hypothesis that proteins we identified may form a network. We analyzed several different protein interaction databases to identify physical or functional interactions either between these proteins themselves or between them and proteins known to modulate the tumor immune infiltrate (13). A tight interaction cluster was formed by 33 proteins (Fig. 2B) and several of them interact with either β-catenin (cadherin-associated protein, CTNNB1) or TP53, two proteins known to alter the anticancer immune response(7, 31). Thus, we hypothesized that some of the regions we identified correlated with immune infiltrates because they alter their interaction with these two proteins.
CDH11 is a potential CTNNB1 inhibitor
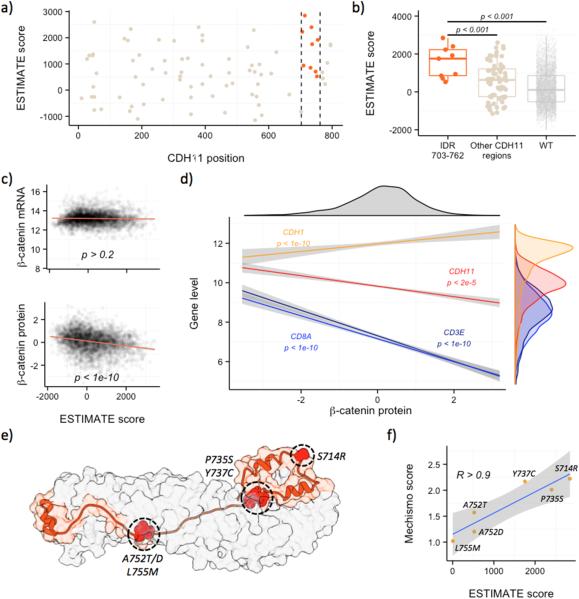
CDH11 is one of the proteins in the 33-protein interaction cluster, and its study highlights many of the advantages of domainXplorer over gene-centric analysis. Correlations between mutations in this protein and the ESTIMATE score were not picked up by gene-centric analysis (P > 0.2, Wilcoxon test), but were recognized by a domain-based analysis (Fig. 3A and B). This protein interacts with CTNNB1, through the region identified by domainXplorer (prediction by homology), immediately suggesting a molecular mechanism for the correlation: mutations in this region correlate with the immune infiltrate because they are altering the interaction between these two proteins.
Figure 3. Exploring the influence of CTNNB1 and CDH11 in cancer immune infiltrate.
A, domainXplorer identified the C-terminal disordered region of CDH11 (aminoacids 703-762, between vertical dashed lines) as correlating with higher ESTIMATE immune scores. B, Immune infiltration by mutated CDH11 region C, Although ESTIMATE immune scores and the expression of CTNNB1 measured with RNAseq did not correlate (top), CTNNB1 protein measured by RPPA (reverse phase protein array) had a negative correlation (bottom). D, CTNNB1 protein (x-axis) also negatively correlated with CDH11, CD8A, and CD3E. E, Structural model (based on PDB 1I7W) of the interaction between the CDH11 disordered region (in orange) and CTNNB1 (in grey). The residues highlighted in red are those with mutations scored by MECHISMO. F, MECHISMO interaction scores (y-axis) predicted for each mutation and the ESTIMATE immune score of the samples (x-axis) were highly correlated (R > 0.9). Higher MECHISMO scores indicate stronger interactions. .
We tried to validate the association between CTNNB1 and lower immune responses(31) using the ESTIMATE immune score, as this association had been reported for melanoma, but we did not know if the correlation was true for other cancer types. Although the correlation between CTNNB1 and ESTIMATE scores at the mRNA level was not statistically significant (P > 0.2, Fig. 3C, top), using protein expression data revealed a significant correlation (P < 1e-10, Fig. 3C, bottom). The correlation was negative, suggesting that CTNNB1 inhibits the immune response, in agreement with previous results. β-catenin protein amounts also were negatively correlated with CD3ε and CD8α (Fig. 3D). We measured CD3ε and CD8α transcripts instead of protein, as neither gene has proteomics data in TCGA; one should be cautious when interpreting these correlations. However, the negative correlation of CTNNB1 protein with CD8α, CD3ε and ESTIMATE immune scores, suggests that the effect of β-catenin in suppressing the homing of lymphocytes (and particularly CD8 T cells) at tumor sites seems to extend beyond melanoma and may be a mechanism common to many cancer types.
The expression of CDH11 also negatively correlates to the concentration of CTNNB1 (P < 1e-5, Fig. 3C, red regression line), leading to the hypothesis that CDH11 could be a β-catenin inhibitor. If that were the case, since all the cancer samples with mutations in the disordered region of CDH11 have higher levels of immune infiltrate, the mutations should be strengthening the CDH11/CTNNB1 interaction. To test this hypothesis, we used MECHISMO(34) to predict the consequences of six different CDH11 mutations. MECHISMO is a method that, among other things, uses an empirical score of how likely it is for two amino acids to be together in an interface, to estimate the effect of a specific mutation. All the mutations in the IDR region of CDH11 were located on the predicted interface between CDH11and CTNNB1 (Fig. 3E) and the MECHISMO interaction score for specific mutations and the ESTIMATE immune scores are strongly correlated (R > 0.9, Fig. 3F), meaning that samples with CDH11 mutations that cause stronger interactions between these two proteins also had more immune infiltration in agreement with our original hypothesis. Overall, CTNNB1 levels correlated with anticancer immune responses beyond melanoma, CDH11 is likely a CTNNB1 inhibitor, and mutations, specifically in the disordered c-terminal region of CDH11, probably alter the immune infiltrate by influencing the activity of β-catenin.
Mutations in regions of complement cascade proteins
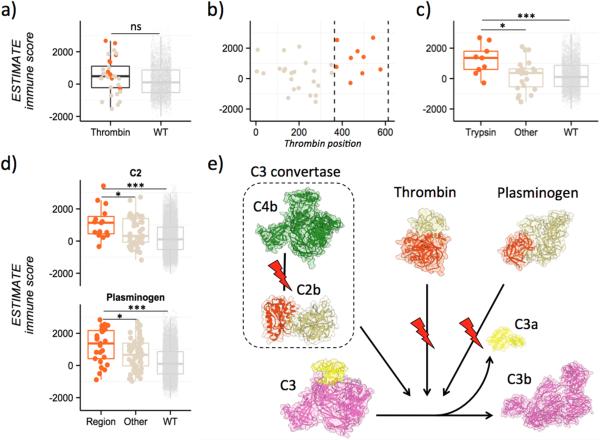
The gee-centric analysis of thrombin, shown in detail in Fig. 4, as in the case of CDH11, compares samples with and without mutations in the entire gene and leads to the wrong conclusion: that mutations in this protein do not correlate with the amount of immune cell infiltrates (Fig. 4A). However, by analyzing each domain individually, domainXplorer identified an association between mutations in the trypsin domain of thrombin responsible for its proteolytic activity and greater immune cell infiltrate (Fig. 4B and C). A potential connection between thrombin and the immune response against cancer may come from the role of thrombin in the complement cascade, whose role in anticancer immunity is beginning to be recognized (35-37). Thrombin is known to cleave C3 and C5 into their corresponding subunits, activating the complement pathway (38, 39).
Figure 4. Several domains identified by domainXplorer can be linked to the complement pathway.
A, Thrombin analysis. Standard analysis ESTIMATE data of thrombin, comparing samples with mutations over the whole thrombin gene to samples without mutations. B, Analysis of ESTIMATE data by location of mutation in the thrombin amino acid sequence. Highlighted in orange between dashed vertical lines is the region coding for the trypsin domain (between positions 364 and 613).. C, Immune infiltration segmented by location of thrombin mutations in the sample. Trypsin domain, orange (left); mutations in other regions, brown (center), and no mutations in this protein, light brown (right). D, Immune infiltration and mutation data assessed by domainXplorer identified the Von Willebrand domain of C2 (top) and the catalytic domain of plasminogen (bottom). E, A plausible hypothesis that emerges from these results is that mutations altering the complement cascade at C3 influence antitumor immunity by blocking cleavage of C3 to C3b. For thrombin and plasminogen, mutations in their catalytic domains (orange) could be lowering the rate of conversion from C3 to C3a (yellow) and C3b (pink). Similarly, mutations identified in the C2 region (shown in orange), mediating the interaction with C4b, also influence the same step (C3 to C3b conversion).
Given the complexity of the complement pathway and its controversial role in cancer, showing both pro and anticancer effects (40, 41) it is difficult to decipher the specific mechanism underlying this association. One possibility is that mutations inactivate the catalytic function of thrombin, limiting the local activation of complement. Lower activation could lead to lower cytolytic activity of the immune infiltrate, allowing cancer cells to survive the numerous leukocytes found in these samples. Lower complement-mediated cell death (42, 43), or changes in anaphylatoxins C3a and C5a concentrations could alter the types of immune cells recruited to the tumor. If mutations instead caused instead a gain-of-function in thrombin's catalytic activity, more complement would be activated and the tolerance of these cancer cells to immune cells could be explained by the recruitment of myeloid-derived suppressor cells (MDSCs) by C3a and C5a (44). However, the lack of a mutation cluster in the catalytic domain suggests that these are inactivating mutations, although direct experimental evidence is needed to support either scenario.
domainXplorer identified other elements of the complement cascade, such as C2 protein and plasminogen (Fig. 4D). The results for plasminogen were similar to those for thrombin, as the region identified as significantly mutated in cancer cells is also the peptidase domain, and plasminogen activates the complement cascade by cleaving C3 and C5 (38, 39) (Fig. 4E). For the C2 protein, the Von Willebrand domain was the most relevant: TCGA samples with mutations in this region have unusually high numbers of immune cells. This domain is likely responsible for the interaction with C4b; therefore, these mutations could disrupt the formation of C3 convertase by blocking the interaction between C2b and C4b, ultimately leading, like thrombin, to the downregulation or blockade of the complement cascade. Regardless of the specific mechanism underlying each of these three associations, they all suggest a key role of the complement cascade in helping tumor cells evade the immune response (i.e., making them capable of surviving in the presence of high numbers of immune cells), in full agreement with studies that highlight the relevance of the complement cascade in tumor immunology (37) (Fig. 4D).
Mutations in the same protein can be associated with opposite immune phenotypes
Another example of the importance of domain-level data analysis is POLR3B, the second-largest subunit of the RNA polymerase III complex. This complex is responsible for the transcription of noncoding RNAs and affects innate immunity by inducing the expression of type I IFN after detecting cytosolic DNA (45, 46). It also plays a role in the response of patients to cancer drugs and in antitumor immunity (47).
In a gene-centric analysis, patients with POLR3B mutations have higher immune scores than those with no mutations in this protein (P < 0.05). However, results from domainXplorer show that this effect is domain-dependent. Patients with mutations in the hybrid-binding domain have more immune cell infiltration than POLR3B wild-type patients (P < 0.01), as well as patients with mutations in other domains of POLR3B (P < 0.01) (Fig. 5A and B).
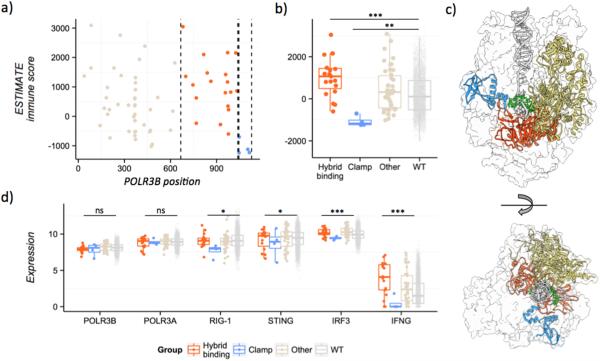
Figure 5. Mutations in POLR3B can have opposite effects on the host immune response depending on which domain they alter.
A, Scatterplot showing the ESTIMATE immune scores (y-axis) in different samples depending on the position of the POLR3B mutation (x-axis). Mutations in the hybrid-binding domain (red), samples with mutations in the clamp region (blue). B, Boxplot comparing the immune scores in different TCGA samples depending on their POLR3B mutation status. Hybrid-binding domain (orange), clamp region (blue), other POLR3B mutations (light brown), or no POLR3B mutations (gray). C, Structure model of the RNA polymerase III, highlighting the different POLR3B regions. A ribbon diagram is shown for the clamp region (blue), the hybrid-binding domain (orange) and the rest of POLR3B (light brown), transcribed DNA (gray), and the nascent RNA molecule (green). Only the surface is shown for the rest of the RNA polymerase III complex. Model based on PDB coordinates file 4Y52. D, Expression of different genes involved in the STING pathway. Samples with mutations in the clamp region show consistently lower expression of many genes downstream in the pathway (RIG-I, STING, IRF3) and the type II interferon gene IFNG.
These results suggest that POLR3B mutations should lead to cancer cells that resist the immune response. However, domainXplorer identified mutations in the clamp region (C-terminal domain) that have the opposite effect. Patients with clamp region mutations have less immune cell infiltration (Fig. 5A and B). Thus, the specific position of the POLR3B mutations seems to determine the amount of immune cell infiltration, an effect that would have been missed in gene-centric analyses. The predicted three-dimensional model of the RNA polymerase III, based on PDB coordinates file of S. cerevisiae RNA polymerase II elongation complex (PDB ID 4Y52 (48)), highlights the positions of the clamp region versus the hybrid-binding domain and the rest of POLR3B (Fig. 5C).
To better understand the role of mutations in the clamp region, we analyzed expression data for the different genes involved in the RNA polymerase III/Interferon pathway (POLR3A, POLR3B, RIG-I, STING, and IRF3) as well as several interferon genes. Only patients with mutations in this region of POLR3B had the equivalent amounts of both POLR3A and POLR3B (Fig. 5D), with significantly less expression of the rest of the proteins in the pathway and several type I interferon genes (IFNA1, IFNE, IFNRA1, and IFNRA2), although we observed no differences in IFNA2 or IFNB1. We observed these differences when either the tissue of origin or the ESTIMATE score of each sample was included as a covariate, but not when including both, probably due to lack of statistical power. Thus, RNA polymerase III with mutations in the POLR3B clamp domain could not activate the full type I interferon response, leading to reduced IFNG expression and a less immunogenic environment (Fig. 5D).
Discussion
In this work we have explored the landscape of cancer immunity drivers focusing on protein domains. The role of somatic mutations in cancer immunology has usually been interpreted in the context of neoantigens. Although they play a key role in determining the immunogenicity of a tumor and can be used in clinical settings to try to personalize immune-based therapy (6, 32, 33), few analyses have focused on whether cancer somatic mutations can influence host immune response through other means. Our results suggest that many proteins, when mutated in a specific domain, could affect the host immune response to tumors. Interpreting these associations is not trivial and is additionally complicated because ESTIMATE scores use a single value to measure presence of immune cells but do not provide any details about the composition of the immune infiltrate. Nevertheless, as shown by the examples discussed, domain analysis not only finds correlations, but can also be used to develop specific hypotheses on the mechanisms of how these mutations affect the immune cells attacking the tumor.
The full list of 122 immune domain drivers identified are in Supplementary Table S1. Given the limited statistics available in even largest current databases, this table should be treated as a preliminary list of candidates for mutation immunity drivers that needs further validation using other knowledge of biological data and direct experimentation. Although we have used ESTIMATE scores, this approach is amenable to other cancer immunology data, such as profiles of different types of immune infiltrating cells. Given the importance of the composition of the immune infiltrate in influencing, among other things, the overall survival of patients (49, 50), we foresee that domainXplorer analysis of more detailed immune profiles that include information about specific cell types (29) will reveal further details of tumor immunology. The only requirement is that the samples have data regarding both a certain immune phenotype and protein-coding variations (somatic or germline). Also, the functionality of domainXplorer and similar analysis can be extended by including functional regions that are defined in three dimensions, such as protein interaction interfaces. Just as in the case of cancer-driver genes (13), analyses focusing on three-dimensional regions will likely reveal more features that influence the immune infiltrate in cancer. Finally, these 122 associations between protein domains and the amount of immune infiltration, as well as the algorithm used to discover them, show that domainXplorer could become a valuable resource to improve our understanding of the complex relationships between cancer and immune cells.
Supplementary Material
Acknowledgments
We thank the reviewers for their thoughtful comments and suggestions, The Cancer Genome Atlas team for making their results publicly available for further analysis, as well as our colleagues from the SBP Inflammation and Infectious Diseases Center (IIDC) and the Cancer Center (CC), especially Drs. Carl Ware and Marcus Kaul, for providing advice and guidance on cancer immunity and comments and edits to the manuscript. This research was partly supported by the SBP CC grant (P30 CA030199)
Footnotes
Conflict of interest: The authors declare no conflict of interest
References
- 1.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nature immunology. 2002;3(11):991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 2.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Upregulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Science translational medicine. 2013;5(200):200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. The Journal of experimental medicine. 2014;211(11):2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer immunology research. 2014;2(6):522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer immunology research. 2013;1(1):11–5. doi: 10.1158/2326-6066.CIR-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453–62. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(40):E5486–95. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosca R, Tenorio-Laranga J, Olivella R, Alcalde V, Ceol A, Soler-Lopez M, et al. dSysMap: exploring the edgetic role of disease mutations. Nature methods. 2015;12(3):167–8. doi: 10.1038/nmeth.3289. [DOI] [PubMed] [Google Scholar]
- 10.Porta Pardo E, Godzik A. Analysis of individual protein regions provides novel insights on cancer pharmacogenomics. PLoS computational biology. 2015;11(1):e1004024. doi: 10.1371/journal.pcbi.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nature biotechnology. 2012;30(2):159–64. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, et al. Edgetic perturbation models of human inherited disorders. Molecular systems biology. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porta-Pardo E, Garcia-Alonso L, Hrabe T, Dopazo J, Godzik A. A Pan-Cancer Catalogue of Cancer Driver Protein Interaction Interfaces. PLoS computational biology. 2015;11(10):e1004518. doi: 10.1371/journal.pcbi.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porta-Pardo E, Godzik A. e-Driver: a novel method to identify protein regions driving cancer. Bioinformatics. 2014;30(21):3109–14. doi: 10.1093/bioinformatics/btu499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Team RC. R: A language and environment for statistical computing. 2015 [Google Scholar]
- 17.Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J R Stat Soc a Stat. 2011;174:245. [Google Scholar]
- 18.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic acids research. 2016;44(D1):D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic acids research. 2012;40(Database issue):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, et al. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21(16):3435–8. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 22.Xu D, Jaroszewski L, Li Z, Godzik A. AIDA: ab initio domain assembly for automated multi-domain protein structure prediction and domain-domain interaction prediction. Bioinformatics. 2015;31(13):2098–105. doi: 10.1093/bioinformatics/btv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, et al. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Scientific reports. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 25.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nature biotechnology. 2015;33(11):1152–8. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nature methods. 2013;10(11):1046–7. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29(17):2211–2. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 29.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature methods. 2015;12(5):453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–5. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 32.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 33.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betts MJ, Lu Q, Jiang Y, Drusko A, Wichmann O, Utz M, et al. Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic acids research. 2015;43(2):e10. doi: 10.1093/nar/gku1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Molecular cancer research : MCR. 2010;8(11):1453–65. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 36.Janelle V, Langlois MP, Tarrab E, Lapierre P, Poliquin L, Lamarre A. Transient complement inhibition promotes a tumor-specific immune response through the implication of natural killer cells. Cancer immunology research. 2014;2(3):200–6. doi: 10.1158/2326-6066.CIR-13-0173. [DOI] [PubMed] [Google Scholar]
- 37.Pio R, Corrales L, Lambris JD. The role of complement in tumor growth. Advances in experimental medicine and biology. 2014;772:229–62. doi: 10.1007/978-1-4614-5915-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barthel D, Schindler S, Zipfel PF. Plasminogen is a complement inhibitor. The Journal of biological chemistry. 2012;287(22):18831–42. doi: 10.1074/jbc.M111.323287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borkowska S, Suszynska M, Mierzejewska K, Ismail A, Budkowska M, Salata D, et al. Novel evidence that crosstalk between the complement, coagulation and fibrinolysis proteolytic cascades is involved in mobilization of hematopoietic stem/progenitor cells (HSPCs). Leukemia. 2014;28(11):2148–54. doi: 10.1038/leu.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, et al. Modulation of the antitumor immune response by complement. Nature immunology. 2008;9(11):1225–35. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campa MJ, Gottlin EB, Bushey RT, Patz EF., Jr. Complement Factor H Antibodies from Lung Cancer Patients Induce Complement-Dependent Lysis of Tumor Cells, Suggesting a Novel Immunotherapeutic Strategy. Cancer immunology research. 2015;3(12):1325–32. doi: 10.1158/2326-6066.CIR-15-0122. [DOI] [PubMed] [Google Scholar]
- 42.Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, et al. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. Journal of immunology. 2012;189(9):4674–83. doi: 10.4049/jimmunol.1201654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunn L, Ding C, Liu M, Ma Y, Qi C, Cai Y, et al. Opposing roles for complement component C5a in tumor progression and the tumor microenvironment. Journal of immunology. 2012;189(6):2985–94. doi: 10.4049/jimmunol.1200846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annual review of immunology. 2015;33:445–74. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 45.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–91. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cellular and molecular life sciences : CMLS. 2011;68(7):1157–65. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STINGDependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–52. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, et al. Molecular basis for 5- carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523(7562):621–5. doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature medicine. 2015;21(8):938–45. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.