Abstract
The ability of high-risk HPV E6 oncoproteins to target cellular proteins which harbor PDZ domains is believed to play an important role in the virus life cycle and to influence the ability of these viruses to bring about malignant transformation. Whilst many of these PDZ proteins are potential tumour suppressors, involved in the control of cell polarity and cell-contact, recent studies suggest that mislocalisation or overexpression might result in the emergence of oncogenic functions. This has been shown most clearly for two E6 targets, hDlg and hScrib. In this study we show that hScrib plays such a role in HeLa cells, where its expression is required for maintaining high levels of HPV-18 E6 protein. Loss of hScrib has no effect on E6 stability but results in lower levels of E6 transcription and a reduced rate of E6 translation. We further show that, in the context of cervical tumour-derived cell lines, both hScrib and E6 cooperate in the activation of the S6 kinase signaling pathway, and thereby contribute towards maintaining high rates of protein translation. These results indicate that the residual hScrib that is present within HPV transformed cells is pro-oncogenic, and highlights the dual functions of E6 cell polarity targets.
Keywords: HPV E6, hScrib, S6 kinase, Protein translation
1. Introduction
Human papillomaviruses (HPVs) infect cutaneous and mucosal epithelial sites, where the vast majority of HPV types are associated with the formation of benign and self-remissive warts. However, infection with a small group of high-risk HPV types can result in tumour formation. HPV-16 and HPV-18 account for the majority of these HPV-positive cancer cases, with cervical cancer being the most common HPV-associated malignancy [1], [2].
Although almost 100% of cervical cancers are associated with high-risk HPV infection, only a small proportion of HPV infections progress to cancer, with the major risk factor for cervical cancer progression being the long-term persistence of the infection. The development of malignancy over long periods of time is often accompanied by elevated expression of the two major viral oncoproteins E6 and E7. Indeed, loss of expression of either protein results in the cessation of transformed cell growth, highlighting the crucial role played by these two viral gene products in the malignant process [3], [4]. An essential feature of the high-risk E6 and E7 proteins is the ability to interact with and modulate the function of key cellular components that regulate cell cycle progression and apoptosis, including the tumour-suppressors p53 and pRB [5], [6].
A distinctive feature of the high-risk HPV E6 proteins is the presence of a highly conserved class I PDZ (PSD-95, Disc-Large, Zonula-Occludens-1)-binding motif (PBM) at their C-termini. This motif enables E6 to interact with a number of cellular PDZ domain-containing proteins, and this has been shown to play a role in the ability of E6 to bring about malignant changes in a variety of different assays [7], [8], [9], [10], [11]. So far, 14 PDZ domain-containing proteins have been verified as interacting partners for the high-risk HPV E6 oncoproteins [12], although a recent high throughput screen would suggest the possibility of many more potential interacting partners [13]. Some of these interacting partners include hDlg, the MAGI family of proteins and hScrib, all of which are potential tumour suppressor proteins and whose levels of expression are potentially perturbed by the presence of E6 [14], [15], [16]. hDlg and hScrib together with Hugl, form the scribble polarity complex, which assembles at adherens junctions of polarised cells and plays a crucial role in the maintenance of cell polarity and tissue homeostasis [17]. Consistent with this, loss of hScrib and hDlg is a common event in the later stages of cancer progression, although at earlier stages of disease the two proteins are frequently expressed at extremely high levels and often mislocalised [18], [19]. Furthermore, recent studies have suggested that the tumour-suppressive potential of hDlg in human keratinocytes is highly context-dependent [20], and the residual hDlg in HeLa cells has been shown to play a direct role in maintaining the invasive potential of these cells through its ability to activate RhoG [21]. Indeed, whilst hScrib is normally believed to down-regulate growth promoting pathways [22], [23], recent studies have shown that mislocalised or overexpressed hScrib can directly activate the PI3kinase signaling pathway in breast cancer [24]. Furthermore, in the case of Tip-1, another E6 PDZ-domain containing target, E6 also appears to promote gain of function activity [25], [26]. Taken together, these studies suggest that the consequences of E6-PDZ interactions might not only result in a perturbation of tumour suppressor functions, but might also promote the pro-oncogenic functions of the same targets.
In organotypic raft cultures of human foreskin keratinocytes (HFKs) the PDZ-binding activity of HPV-31 E6 is critical for the induction of hyperplasia in suprabasal layers [27] and loss of the PBM in the context of the whole HPV-31 genome increases the likelihood for viral integration into the host genome [27]. Similar results have also been observed with HPV-16 and HPV-18 [28], [29], although in the case of HPV-16 E6ΔPBM mutant genomes, the increased propensity to lose viral episomes appeared to correlate with reduced stability of the E6 oncoprotein [28]. Indeed overexpression studies suggested that HPV-16 E6 was stabilised when co-expressed with certain PDZ-containing binding partners, including MAGI-1, hDlg and hScrib, whilst siRNA ablation of hScrib expression in HPV-18 positive HeLa cells also resulted in a decrease in the levels of HPV-18 E6 expression [28]. These studies raise the intriguing possibility that certain PDZ domain containing substrates of HPV E6 might contribute towards maintaining high levels of E6 expression. Obviously this has important consequences for their potential effects upon malignant progression. In this study we have performed a detailed analysis of the impact of different cellular PDZ proteins upon the levels of expression of endogenously expressed HPV-16 and HPV-18 E6 proteins. We demonstrate that only hScrib appears to contribute directly towards the regulation of E6 expression levels. Furthermore, this is not due to an effect on E6 turnover, but rather is a reflection of the ability of hScrib to modulate E6 transcription and translation, and this correlates in part with the ability of hScrib to control the function of the mTORC/S6K pathway in HPV-18 containing cells.
2. Materials and methods
2.1. Cell culture and transfection
HeLa, CaSki and SiHa cells were maintained in Dulbecco's modified Eagles Medium (DMEM) supplemented with 10% fetal bovine serum, penicillin-streptomycin (100 U/ml) and glutamine (300 µg/ml). For all siRNA (Dharmacon) delivery, the cells were seeded on 6 cm dishes at a confluence of 1.2×105 and transfected using Lipofectamine 2000 (Invitrogen) with siRNA against luciferase, HPV-18 E6/E7 (5’CAUUUACCAGCCCGACGAG), HPV-18 E6 (5’CUCUGUGUAUGGAGACACA), or siRNA against the different PDZ-proteins (relevant Dharmacon Smart Pools).
2.2. Antibodies
The following antibodies were used: mouse monoclonal anti-human pRB (BD Pharmingen), mouse monoclonal anti-p84 (5E10; Abcam), mouse monoclonal anti-α-tubulin (T6199; Sigma). The following antibodies were purchased from Santa Cruz Biotechnology: mouse monoclonal anti-p53 (DO-1), mouse monoclonal anti-α-actinin (H-2), mouse monoclonal anti-Dlg (2D11), goat polyclonal anti-Scribble (C-20), goat polyclonal anti-TIP2 (N-19), rabbit polyclonal anti-E-cadherin (H-108) and mouse monoclonal anti-vimentin (V-9). The following antibodies were from Cell Signaling Technology: rabbit polyclonal anti-PDK1 (3062), rabbit monoclonal anti-phosphorylated PDK1 (S241) (C49H2), rabbit monoclonal anti-p70 S6 kinase (49D7), mouse monoclonal anti-phosphorylated p70 S6 kinase (T389) (1A5). The mouse monoclonal antibody anti HPV-18 E6 (N-terminus #399) was generated and generously provided by the Arbor Vita Corporation. For detection of HPV-16 E6 expression we used the Arbor Vita E6 detection kit according to the manufacturer's instructions (supplied by AB Analitica srl).
2.3. Western blotting
For western blot sample preparation, cells were lysed in 2x SDS sample buffer (100 mM Tris HCl pH 6.8; 200 mM DTT, 4% SDS, 20% glycerol, 0.2% bromophenol blue) and the whole cell extracts were separated by SDS-PAGE and blotted on 0.22 nitrocellulose membranes (Schleicher and Schuell). The membranes were blocked at 37 °C for 1 h in 10% milk/PBS, except for membranes probed with the antibodies from Cell Signaling Technology, which were incubated in 5% milk/TBS 0.1% TWEEN 20. The membranes probed with the anti-HPV-18 E6 antibody were incubated in 2% BSA/5% milk in 1xTBS 0,1% TWEEN 20. The membranes were incubated with the appropriate primary antibodies diluted in 10% milk/PBS 0.5% TWEEN 20; all the antibodies from Cell Signaling Technology were diluted in 5% BSA/1xTBS 0.1% TWEEN except for the anti-phosphorylated p70 S6 kinase, which was incubated in 5% milk/1xTBS 0.1% TWEEN 20. The HPV-18 E6 antibody was incubated in 1% BSA/2.5% milk in 1xTBS 0.1% TWEEN 20. The incubation times were 2 h at room temperature for all antibodies, except for anti E-cadherin, the Cell Signaling Technology antibodies and the anti-HPV-18 E6 antibody, which were incubated overnight at 4 °C. After several washes the membranes were incubated with the appropriate HRP-conjugated secondary antibody (DAKO) for 1 h at room temperature. After extensive washing the blots were developed with ECL or ECL plus reagent (GE Healthcare) according to the manufacturer's instructions. Protein band intensities were quantitated where possible using the OptiQuant quantification program.
2.4. Half-life experiments
72 h post transfection, cells were treated for different time points as indicated with cycloheximide (50 μg/ml in DMSO) to block protein synthesis. DMSO treated cells were used as the control. Total cellular extracts were then analyzed by Western blot and the intensity of the bands was measured using the Optiquant program. The standard deviation was calculated from three independent assays.
2.5. Analysis of E6 mRNA levels
72 h post transfection of the siRNAs, Hela cells were harvested and RNA extracted using Quick RNA Miniprep Plus (Zymo Research) and subject to reverse transcription using the Quantitect Reverse Transcription Kit (Qiagen). qPCR was performed in duplicate 12ul reactions on undiluted cDNA using 10uM primers and Maxima SYBR Green/Rox PCR master mix in the StepOneplus real time PCR system (Applied Biosystems). The primers for the GAPDH control were 5’ AAGGTCGGAGTCAACGGATTT 3’ (Forward) and 5’ ACCAGAGTTAAAAGCAGCCCTG 3’ (Reverse) and the primers for HPV-18 E6 were 5’ CCAGAAACCGTTGAATCCAG 3’ (Forward) and 5’ GTTGGAGTCGTTCCTGTCGT 3’ (Reverse).
2.6. Determination of HPV-18 E6 and p53 translation efficiency
HeLa cells were seeded on 6 cm dishes at a confluence of 1.2×105and transfected using Lipofectamine 2000 (Invitrogen) with siRNA against Luciferase or hScrib (Dharmacon). 72 h after transfection cells were treated with cycloheximide (50 μg/ml in DMSO) for an additional 6 h. Cells treated with DMSO alone were used as control for the expression of HPV-18 E6 and p53. In order to monitor the recovery of E6 and p53 protein translation cycloheximide was removed and, after several washes in PBS, cells were left to grow for different time points in fresh DMEM supplemented with 10% fetal bovine serum, penicillin-streptomycin (100 U/ml) and glutamine (300 µg/ml). Total cellular extracts were prepared by harvesting cell in 2X SDS sample buffer, and then analyzed by Western blotting.
3. Results
3.1. hScrib is required for high level expression of the HPV-18 and HPV-16 E6 proteins
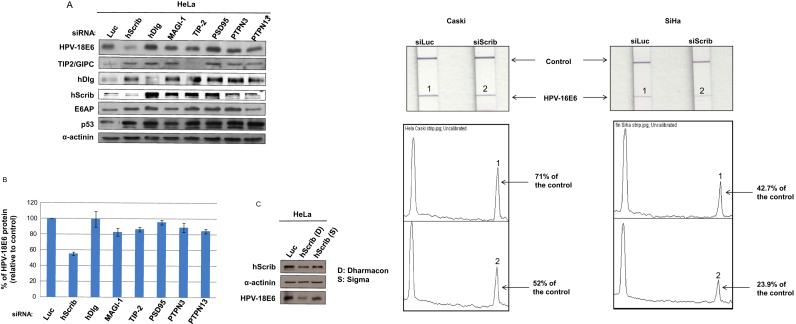
Previous studies have shown that loss of hScrib in HeLa cells resulted in lower levels of HPV-18 E6 expression [28]. We were therefore first interested in determining whether this effect was specific for hScrib, or whether loss of other E6 PDZ domain-containing substrates would exert a similar effect. To do this, HeLa cells were transfected with either control siRNA (Luciferase) or siRNAs directed against the following HPV E6 PDZ domain-containing target proteins: hScrib, hDlg, TIP2, PSD95, MAGI-1, PTPN3 and PTPN13. 72 h after transfection, the cells were extracted and the expression levels of HPV-18 E6, p53 and the set of silenced PDZ proteins were monitored by western blot analysis. In addition, since recent studies had shown that the stability of HPV-18 E6 in HeLa cells is dependent upon E6AP expression [30], we also assessed the levels of E6AP to determine whether PDZ proteins might affect the expression of E6 indirectly through the modulation of E6AP. The results are shown in Fig. 1A, and the quantifications of HPV-18 E6 levels from multiple experiments are shown in Fig. 1B. As can be seen, control siLuciferase HeLa cells express readily detectable levels of endogenous HPV-18 E6, p53 and E6AP. Among the E6 PDZ targets assayed, only the ablation of hScrib consistently reduced the levels of HPV-18 E6 expression by between 50% and 60% (Fig. 1A and B). Consistent with this reduction in E6 levels, the ablation of hScrib also resulted in increased levels of known E6 targets, hDlg, E6AP and p53.
Fig. 1.
hScrib regulates the expression of HPV-18 E6 in HeLa cells. Panel A. HPV-positive HeLa cells were transfected with siRNA Luciferase or siRNA against the indicated E6 PDZ substrates. Cells were grown for 72 h prior to harvesting and the expression patterns of HPV-18 E6, hDlg, hScrib, TIP2, p53, E6AP and α-actinin (to monitor the protein loading) were assessed by western blot analysis. Note that Lanes 1 and 2 have been spliced but all samples were run on the same gel. Panel B. Band intensities were determined using the OptiQuant quantification program. E6 levels were normalized to 100% relative to siLuciferase-transfected HeLa cells. Standard deviations are also shown. Panel C. The silencing of hScrib was performed as in A but using two different siRNAs specific for hScrib. The expression levels of HPV-18 E6, hScrib and α-actinin to monitor the protein loading, were assessed by western blot analysis. Panel D. Strips showing levels of HPV16 E6 detection in CaSki and SiHa cells following transfection with siRNA luciferase (si Luc) or si Scrib. Arrows indicate the position of the internal positive control and the HPV-16 E6 specific band. The bottom panels show the quantification of the band intensities.
To determine whether loss of hScrib in HPV-16 positive cells could also affect the levels of E6 expression, we transfected CaSki cells with siRNAs against luciferase and hScrib. After 72 h the cells were extracted and the levels of E6 expression were ascertained. The results obtained in Fig. 1D show that the loss of hScrib in CaSki cells also results in a marked decrease in the levels of HPV-16 E6 expression.
We then proceeded to confirm that the effect of hScrib loss on E6 protein levels was indeed due to the specific ablation of hScrib and not to any off-target effect of the hScrib-specific siRNA. To do this we repeated the analysis with an alternative hScrib-specific siRNA obtained from a different supplier. As can be seen in Fig. 1C, the two hScrib siRNAs reduced the expression of hScrib, although with slightly different efficiencies. Consistent with the results in Fig. 1A, the two hScrib siRNAs produced a similar reduction in HPV-18 E6 levels, and this reduction correlated directly with the efficiency with which hScrib levels were reduced. Taken together these data demonstrate that loss of hScrib expression in HeLa and CaSki specifically leads to a reduction in the levels of E6 protein in these cells.
3.2. Ablation of hScrib reduces the levels of expression of E6 in all cellular compartments
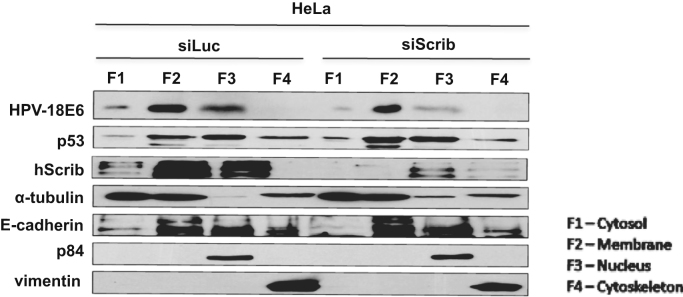
Previous studies have shown that E6 has quite a diverse pattern of expression, with nuclear, cytoplasmic and membrane-bound pools being reported [31], [32]. We were therefore interested in determining whether the ablation of hScrib expression might impact specifically upon a specific subcellular pool of E6. To do this HeLa cells were transfected with Luciferase or hScrib-specific siRNAs and after 72 h the cells were harvested and divided into cytosolic, membrane, nuclear and cytoskeletal fractions. The levels of E6 expression in these different fractions was then analysed by western blotting. The results in Fig. 2 demonstrate that endogenously expressed HPV-18 E6 is found predominantly within the membrane fraction of HeLa cells, with smaller amounts present within the cytosolic and nuclear compartments. Interestingly loss of hScrib appears to reduce the levels of E6 expression to the same degree in all three locations, suggesting that hScrib loss has a general effect on the total levels of E6 expression.
Fig. 2.
Loss of hScrib reduces total HPV-18 E6 protein levels. HeLa cells were transfected with siRNA Luciferase or siRNA hScrib and after 72 h the cells were harvested and fractionated into cytosolic (F1), membrane (F2), nuclear (F3) and cytoskeletal (F4) fractions. These were then analysed by western blotting for HPV-18 E6, p53 and hScrib. Also shown are the loading controls for each fraction.
3.3. Ablation of hScrib does not affect HPV-18 E6 turnover
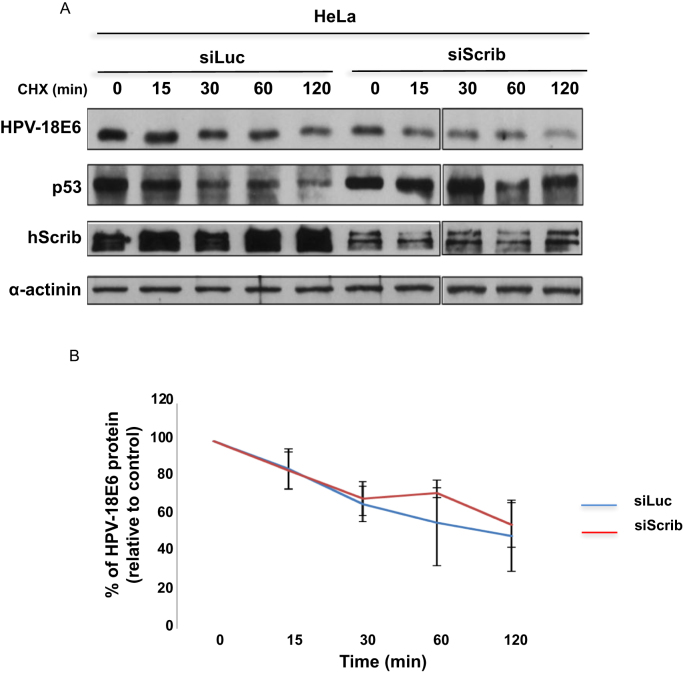
Having shown that loss of hScrib results in decreased levels of HPV-18 E6, we were interested in investigating whether this was due to an increase in the rate of E6 turnover, since previous studies had shown that the lack of PDZ binding capacity could affect E6 stability [28]. To do this, we monitored the E6 half-life in HeLa cells transfected with siRNA against Luciferase or hScrib. 72 h after transfection, the cells were treated with cycloheximide for different times. The cells were then harvested and the levels of E6 protein were determined by western blotting. The results are shown in Fig. 3A, with the quantifications from multiple experiments shown in Fig. 3B. In agreement with previous studies, the half-life of HPV-18 E6 in control siRNA-transfected HeLa cells was between 60 and 120 min [30], [31]. The silencing of hScrib however did not produce any significant change in the E6 half-life, suggesting that loss of hScrib expression does not affect E6 turnover. It is interesting to note, however, that although the turnover of E6 was not affected in the hScrib silenced HeLa cells, p53 was nonetheless stabilized, which is consistent with the overall decrease in E6 protein levels.
Fig. 3.
Loss of hScrib does not affect HPV-18 E6 protein stability. Panel A. HeLa cells were transfected with siRNA against Luciferase or siRNA against hScrib, 72 h after transfection, cells were treated with cycloheximide for 5 different time points: 0, 15, 30, 60 and 120 minutes prior to harvesting. The expression levels of HPV-18 E6, p53, hScrib, and α-actinin to monitor the protein loading, were assessed by western blot. The collated results from 3 independent experiments are shown in panel B. Band intensities were determined using the OptiQuant quantification program. The E6 levels in siLuciferase and siScrib transfected cells were normalized to 100% at time 0. Standard deviations are also shown.
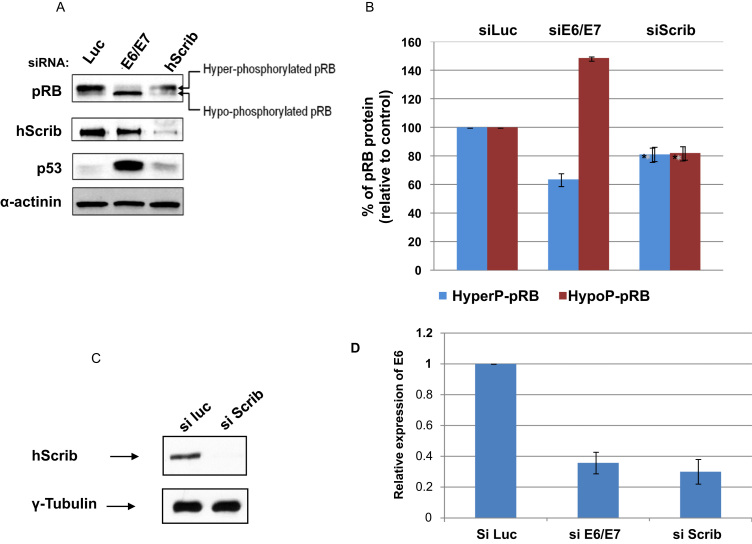
We were then interested in investigating whether hScrib might alter the levels of E6 gene expression. Since E6 and E7 are expressed from a bicistronic mRNA [33], [34], we reasoned that any reduction in E6 transcription in HeLa cells would also be reflected by lower levels of E7 expression. The high-risk HPV E7 oncoprotein promotes the proteasome-mediated degradation of hypophosphorylated, E2F binding-competent forms of pRB [35]. Therefore, analysis of the expression pattern of differentially phosphorylated forms of pRB in HPV-positive cells can be used as a surrogate marker to monitor the levels of E7 expression [36]. HeLa cells were transfected with control siLuciferase, siRNA against hScrib or siRNA against HPV-18 E6/E7 as a reference for the silencing of E7. After 72 h, cells were harvested and the expression pattern of pRB was monitored by western blot analysis. The results are presented in Fig. 4A, and the quantifications of pRB levels from multiple experiments are shown in Fig. 4B. In agreement with previous studies, our results demonstrate that pRb is expressed as differentially phosphorylated forms, and that HeLa cells predominantly express hyper-phosphorylated pRB [36]. Following ablation of E6 and E7, the expression of p53 and the ratio between the hypo- and hyper-phosphorylated pRB levels are significantly increased, demonstrating the efficient silencing of both oncoproteins. In contrast, whilst loss of hScrib expression led to an increase in p53 levels, the ratio between the hypo- and hyper-phosphorylated forms of pRB was intermediate between that obtained in the control and siE6/E7 cells. Furthermore, loss of hScrib resulted in an overall reduction of both pRB forms.
Fig. 4.
Loss of hScrib decreases the levels of HPV-18 E6 mRNA. Panel A. HeLa cells were transfected with siRNA against Luciferase or siRNA HPV-18 E6/E7 or siRNA hScrib. 72 h after transfection, cells were harvested and the expression patterns of: pRB as a surrogate marker for E7 expression, p53 to monitor E6/E7 loss, hScrib and α-actinin, to monitor the protein loading, were assessed by western blot. Panel B. Band intensities of hypo-phosphorylated and hyper-phosphorylated pRB were determined using the OptiQuant quantification program. Levels of pRB expression are expressed as the % of hyper- and hypo-phosphorylated pRB relative to siLuciferase-transfected control cells. Standard deviations are also shown. Panel D. The graph shows the result of quantitative real time pcr for the levels of E6 mRNA, using GAPDH as the control. The bars show the relative levels of E6 expression where siLuc is used as the reference. Note the greater than 60% reduction in E6 mRNA following transfection with either siE6/E7 or siScrib. The numbers represent the means from 3 independent experiments and standard deviations are shown. Panel C shows the accompanying western blot verifying ablation of hScrib expression in a representative assay.
To directly assess the effects of loss of hScrib upon the levels of E6 mRNA expression we performed quantitative real time PCR following transfection of HeLa cells with siLuciferase control, siE6/E7 and siScrib. The results in Fig. 4D show that knockdown of hScrib results in a dramatic decrease in the levels of E6 mRNA levels, which is comparable to that seen following siRNA ablation of E6/E7 expression. Taken together, these results indicate that hScrib can directly affect the levels of E6 mRNA expression.
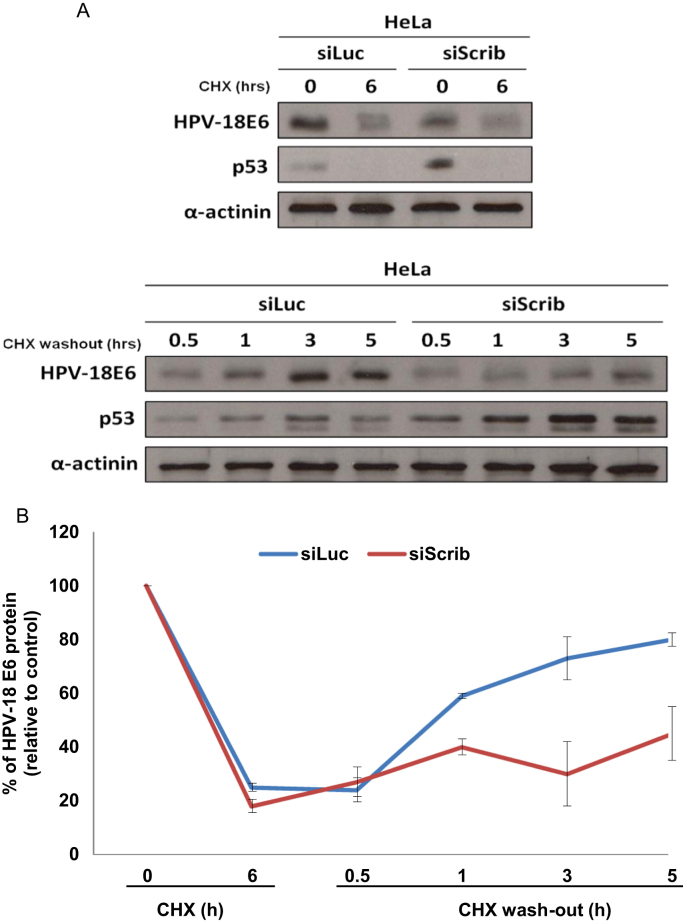
3.4. Loss of hScrib also decreases HPV-18 E6 translation
The above results indicate that loss of hScrib reduces E6 mRNA but also reduces pRb levels. We therefore decided to investigate whether hScrib could also have an effect on E6 translation. To do this, HeLa cells were transfected with siLuciferase or siScrib, and after 72 h the cells were treated for an additional 6 h with cycloheximide to block protein translation. Cells were then washed several times with PBS to remove the cycloheximide, and the recovery in the levels of HPV-18 E6 and p53 protein expression was monitored over time by western blotting. The results are shown in Fig. 5A, and the quantifications of multiple experiments are shown in Fig. 5B. As can be seen, prolonged treatment with cycloheximide decreased the levels of E6 by over 80% in the control and siScrib transfected HeLa cells. Even more apparent was the drop in p53 levels, which upon cycloheximide treatment became undetectable in both the siLuciferase and siScrib transfected cells. Upon removal of cycloheximide, the levels of HPV-18 E6 protein progressively recovered after the 30 min time-point. It is interesting to note that in control HeLa cells the bulk of E6 oncoprotein was translated between 1 and 3 h after the cycloheximide wash-out, suggesting that the accumulation of E6-encoding mRNAs during translation inhibition led to the rapid synthesis of newly translated E6 upon removal of cycloheximide. In contrast, loss of hScrib greatly reduced the rate of recovery in E6 protein levels upon translation re-initiation. Interestingly, the pattern of p53 recovery was opposite to that of E6, being more rapidly upregulated in siScrib cells after cycloheximide wash-out, which is consistent with the lower levels of E6 expression in these cells. Taken together these data suggest that the expression of hScrib in HeLa cells contributes towards maintaining high levels of HPV-18 E6 expression also through the modulation of its rate of translation.
Fig. 5.
hScrib regulates the translation of HPV-18 E6 in HeLa cells. Panel A. HeLa cells were transfected with siRNA against Luciferase or siRNA against hScrib. 72 h after transfection, cells were treated with cycloheximide for 6 h. Prior to harvesting the cells were washed three times with PBS to remove the cycloheximide and protein translation was left to recover in complete medium for 4 different time points: 0.5, 1, 3 and 5 h. The collated results from 3 independent experiments are shown in panel B. Band intensities were determined using the OptiQuant quantification program. The E6 levels in siLuciferase and siScrib transfected cells were normalized to 100% at time 0. Standard deviations are also shown.
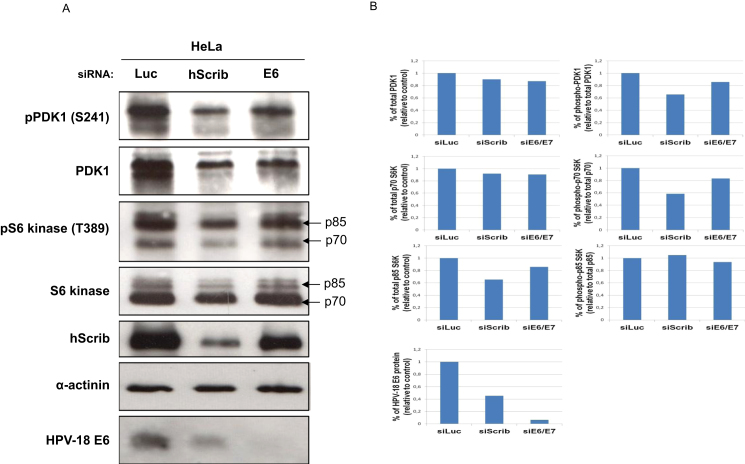
3.5. hScrib activates the mTORC1 pathway in HeLa cells
Previous studies suggested that the E6 mRNA is translated using canonical cap-dependent ribosome scanning [36], [37], in which the rate-limiting step is translation initiation, regulated through the mammalian target of rapamycin complex-1 (mTORC1) pathway [38]. It has also been shown that mislocalised hScrib can activate mTORC1 signaling [24], and HPV-16 E6 can activate PDK1 and S6 kinase, an activity that requires E6AP [39], [40]. Thus E6-induced alteration of hScrib function might be responsible for the ability of hScrib to activate the mTORC1 pathway in HeLa cells, and thereby act in a feedback loop to maintain high levels of protein expression, including that of E6. To investigate these possibilities we monitored the effects of loss of hScrib or E6 upon the levels of two essential components of the mTORC1 signaling pathway, PDK1 and S6 Kinase. Hela cells were transfected with luciferase, hScrib or HPV-18 E6 specific siRNAs and after 72 h the cells were harvested and the extracts analysed by western blotting. As can be seen from Fig. 6, loss of hScrib results in a marked down-regulation of both total and active phosphorylated PDK1. A similar decrease in S6 kinase levels is also obtained. In the case of E6 ablation there is also a marked decrease in these two components of the mTORC1 pathway. These results demonstrate that both E6 and hScrib can cooperate in HeLa cells to maintain the activity of the mTORC pathway and hence maintain high levels of protein translation.
Fig. 6.
The expression of hScrib in HeLa cells maintains high levels of total PDK1 and S6 kinase. HeLa cells were transfected with siRNA Luciferase, siRNA hScrib or siRNA E6. 72 h after transfection cells were harvested and the patterns of expression of total and phosphorylated levels of PDK1 and S6 kinase as well as those of hScrib, HPV-18 E6 and α-actinin as a loading control, were assessed by western blot. Panel B. Protein band intensities relative to the experiment shown in panel A were determined using the OptiQuant quantification program. Levels of the indicated proteins were normalized to 100% relative to siLuciferase-transfected HeLa cells.
4. Discussion
Whilst much attention has focused on the potential tumour suppressive properties of the high-risk HPV E6 PDZ domain-containing targets, there is mounting evidence that certain of these proteins might harbor pro-oncogenic activities. This was first suggested for hDlg, where there is evidence for very high levels of mislocalised hDlg protein expression in the early stages of tumour development [18], [19]. This was then shown directly for certain hDlg isoforms in their ability to cooperate with Adenovirus E4ORF1 for cell transformation [41], [42], and in HPV positive tumour cell lines, where it was found to be required for full invasive potential [21]. More recently, studies with hScrib have also highlighted the importance of the correct level of protein expression and localization, since perturbation of either can result in pro-oncogenic signaling through the mTORC1 pathway [24]. In our current study we also find that hScrib can directly affect the levels of E6 mRNA expression and by maintaining active mTORC1 signaling can also contribute towards enhancing the rates of E6 protein translation.
Previous studies had shown that the maintenance of HPV episomes in keratinocytes required an intact E6 PBM [27], [28], [29]. In addition, over-expression analyses implicated certain PDZ domain containing substrates of E6 as being required for maintaining stable levels of E6 expression [28], whilst siRNA ablation studies highlighted a potential role for hScrib in this activity in HPV-18 containing HeLa cells [28]. Therefore we first embarked upon a screen of other known E6 PDZ domain-containing targets in order to ascertain whether their loss could also impact negatively upon the levels of E6 expression. In this analysis we monitored the effects of ablating hScrib, hDlg, MAGI-1, TIP2, PSD95, PTPN3 and PTPN13. Of these only ablation of hScrib has a significant negative impact upon the levels of HPV-18 E6 protein. This indicates that, of these PDZ substrates of E6, only hScrib appears to have a role in helping maintain high levels of E6 protein expression.
Analysis of the mechanisms by which hScrib can modulate the levels of E6 expression appears to rule out an effect on E6 protein turnover, since the half-life of E6 was unchanged upon ablation of hScrib. However loss of hScrib clearly results in a marked decrease in the levels of E6 mRNA, suggesting that reduced levels of E6 protein expression occur in part through a decrease in viral gene expression. At the same time we also noted a marked change in the pattern of pRb expression following hScrib ablation, with a clear reduction in total protein level and a marked decrease in the level of hyper-phosphorylated pRb. Previous studies have shown that inhibition of mTOR signaling also results in decreased levels of pRb phosphorylation [43], suggesting that loss of hScrib might have multiple consequences. In agreement with this, by using a cycloheximide block and release analysis, we found compelling evidence that loss of hScrib also decreases the rate of E6 translation. Intriguingly, we also found that upon loss of hScrib the half-life of p53 was significantly increased in HeLa cells, consistent with reduced levels of E6 expression, whereas its translation efficiency was largely unaffected. This is consistent with the fact that p53 is translated through both cap-dependent and cap-independent mechanisms due to the presence of an internal ribosome entry site (IRES) in its 5’-UTR [44], whilst in contrast E6 translation is largely cap-dependent. These studies therefore demonstrate that the loss of hScrib in HPV-positive tumour-derived cell lines helps maintain E6 expression at both the transcriptional and post-translational levels. Currently we have no information on the mechanism by which hScrib might modulate viral gene expression, but this will be an aim of future studies.
In an attempt to define the molecular mechanism by which loss of hScrib could affect protein translation, we analysed the effects on the mTORC1 pathway, as recent studies had indicated that under certain conditions hScrib could directly contribute toward enhanced mTORC signaling [24]. We assessed two major components of the mTORC pathway, PDK1 and S6 Kinase. Both proteins were expressed at high levels in HeLa cells with significant levels of phosphorylation indicative of pathway activation. In contrast, loss of hScrib resulted in dramatically reduced levels of expression of PDK1, and a corresponding decrease in its levels of phosphorylation. Likewise there was also a similar reduction in the levels of active S6 kinase. This suggests that loss of hScrib in HeLa cells directly downregulates mTORC1 signaling, thereby resulting in decreased rates of protein translation. Interestingly, loss of E6 also reduced the levels of mTORC pathway activation, which is in agreement with previous studies [39], [40], albeit not to the same degree as loss of hScrib. Taken together these results demonstrate that E6 and hScrib can act cooperatively in HPV-18 positive HeLa cells to maintain active mTORC signaling and hence maintain high rates of protein translation. This suggests that some PDZ targets of E6 might be subject to proteasome mediated degradation during certain stages of the virus life cycle or during malignant progression, but it also raises the intriguing prospect that mislocalisation of certain PDZ targets, such hScrib, might directly contribute towards increased cell proliferation and oncogenic progression in HPV-induced malignancies.
Acknowledgements
We are most grateful to Miranda Thomas for valuable comments on the manuscript. This work was supported in part by a research grant provided by the Associazione Italiana per la Ricerca sul Cancro.
References
- 1.zur Hausen H. Papillomaviruses in the causation of human cancer – a brief historical account. Virology. 2009;384:26–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Hartwig S., Baldauf J.-J., Dominiak-Felden G., Simondon F., Alemany L., de Sanjose S., Castellsague X. Estimation of the epidemiological burden of HPV-related anogenital cancers, precancerous lesions and gential warts in women and men in Europe: potential additional benefits of nine-valent second generation HPV vaccine compared to first generation HPV vaccines. Papillomavirus Res. 2015;1:90–100. [Google Scholar]
- 3.Yoshinouchi M., Yamada T., Kizaki M., Fen J., Koseki T., Ikeda Y., Nishihara T., Yamato K. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by E6 siRNA. Mol Ther. 2003;8:762–768. doi: 10.1016/j.ymthe.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Butz K., Ristriani T., Hengstermann A., Denk C., Scheffner M., Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22:5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- 5.Dyson N., Howley P., Munger K., Harlow E. The human papillomavirus- 16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 6.Werness B., Levine A., Howley P. Association of human papillomavirus type 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 7.Kiyono T., Hiraiwa A., Fujita M., Hayashi Y., Akiyama T., Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S.S., Weiss R., Javier R. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson R., Thomas M., Banks L., Roberts S. Activity of the human papillomavirus E6 PDZ-binding motif correlates with an enhanced morphological transformation of immortalized human keratinocytes. J. Cell Sci. 2003;115:4925–4934. doi: 10.1242/jcs.00809. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen M.L., Nguyen M.M., Lee D., Griep A., Lambert P. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6's induction of epithelial hyperplasia in vivo. J. Virol. 2003;77:6957–6964. doi: 10.1128/JVI.77.12.6957-6964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shai A., Brake T., Somoza C., Lambert P. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res. 2007;67:1626–1635. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pim D., Bergant M., Boon S.S., Ganti K., Kranjec C., Massimi P., Subbaiah V.K., Thomas M., Tomaic V., Banks L. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J. 2012;279:3530–3537. doi: 10.1111/j.1742-4658.2012.08709.x. [DOI] [PubMed] [Google Scholar]
- 13.Vincentelli R., Luck K., Poirson J., Polanowska J., Abdat J., Blemont M., Turchetto J., Iv F., Ricquier K., Straub M., Forster A., Cassonnet P., Borg J.P., Jacob Y., Masson M., Nomine Y., Reboul J., Wolff N., Charbonnier S., Trave G. Quantifying domain-ligand affinities and specificities by high-throughput holdup assay. Nat. Methods. 2015;12:787–793. doi: 10.1038/nmeth.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiol D., Kuhne C., Glaunsinger B., Lee S.S., Javier R., Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa S., Huibregtse J. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomaviruses E6 proteins and the E6AP ubiquitn-protein ligase. Mol. Cell Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas M., Laura R., Hepner K., Guccione E., Sawyers C., Lasky L., Banks L. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene. 2002;21:5088–5096. doi: 10.1038/sj.onc.1205668. [DOI] [PubMed] [Google Scholar]
- 17.Banks L., Pim D., Thomas M. Human tumour viruses and the deregulation of cell polarity in cancer. Nat. Rev. Cancer. 2012;12:877–886. doi: 10.1038/nrc3400. [DOI] [PubMed] [Google Scholar]
- 18.Watson R., Rollason T., Reynolds G., Murray P., Roberts S. Changes in expression of the human homologue of the Drosophila discs large tumour suppressor protein in high-grade premalignant cervical neoplasias. Carcinogenesis. 2002;23:1791–1796. doi: 10.1093/carcin/23.11.1791. [DOI] [PubMed] [Google Scholar]
- 19.Cavatorta A., Fumero G., Chouhy D., Aguirre R., Nocito A., Giri A., Banks L., Gardiol D. Differential expression of the human homologue of drosophila discs large oncosuppressor in histologic samples from human papillomavirus-associated lesions as a marker for progression to malignancy. Int. J. Cancer. 2004;111:373–380. doi: 10.1002/ijc.20275. [DOI] [PubMed] [Google Scholar]
- 20.Massimi P., Zori P., Roberts S., Banks L. Differential regulation of cell-cell contact, invasion and anoikis by hScrib and hDlg in keratinocytes. PLoS One. 2012;7:e40279. doi: 10.1371/journal.pone.0040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subbaiah V., Massimi P., Boon S.S., Myers M., Sharek L., Garcia-Mata R., Banks L. The invasive capacity of HPV transformed cells requires the hDlg-dependent enhancement of SGEF/RhoG activity. PLoS Pathog. 2012;8:e1002543. doi: 10.1371/journal.ppat.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan L., Rosenberg A., Bergami K., Yu M., Xuan Z., Jaffe A., Allred C., Muthuswamy S. Deregulation of Scribble promotes mammary tumnourigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsum I., Martin C., Humbert P. Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J. Cell Sci. 2013;126:3990–3999. doi: 10.1242/jcs.129387. [DOI] [PubMed] [Google Scholar]
- 24.Feigin M., Akshinthala D., Araki K., Rosenberg A., Muthuswamy L., Martin B., Lehmann B., Berman H., Pietenpol J., Cardiff R., Muthuswamy S. Mislocalization of the cell polarity protein Scribble promotes mammary tumourigensis and is associated with basal breast cancer. Cancer Res. 2014;74:3180–3194. doi: 10.1158/0008-5472.CAN-13-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampson L., Li C., Oliver A., Kitchener H., Hampson I. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. Int. J. Oncol. 2004;25:1249–1256. [PubMed] [Google Scholar]
- 26.Oliver A., He X., Borthwick K., Donne A., Hampson L., Hampson. I. The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. British J. Cancer. 2011;104:324–331. doi: 10.1038/sj.bjc.6606026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C., Laimins L. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J. Virol. 2004;78:12366–12377. doi: 10.1128/JVI.78.22.12366-12377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolaides L., Davy C., Raj K., Kranjec C., Banks L., Doorbar J. Stabilization of HPV16 E6 protein by PDZ proteins, and potential implications for genome maintenance. Virology. 2011;414:137–145. doi: 10.1016/j.virol.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Delury C., Marsh E., James C., Boon S.S., Banks L., Knight G., Roberts S. The role of protein kinase A regulation of the E6 PDZ-binding domain during the differentiation-dependent life cycle of human papillomavirus type 18. J. Virol. 2013;87:9463–9472. doi: 10.1128/JVI.01234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaic V., Pim D., Banks. L. The stability of the human papillomavirus E6 oncoprotein is E6AP dependent. Virology. 2009;393:7–10. doi: 10.1016/j.virol.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Grossman S., Mora R., Laimins L. Intracellular localization and DNA-binding properties of human papillomavirus type 18 E6 protein expressed with a baculovirus vector. J. Virol. 1989;63:366–374. doi: 10.1128/jvi.63.1.366-374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guccione E., Massimi P., Bernat A., Banks L. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology. 2002;293:20–25. doi: 10.1006/viro.2001.1290. [DOI] [PubMed] [Google Scholar]
- 33.Schneider-Gadicke A., Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus 18 early genes. EMBO J. 1986;5:2285–2292. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smotkin D., Wettstein F. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer S., Wazer D., Band V. E7 protein of human papillomavirus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 36.S. Tang, M. Tao, J. McCoy, ZM Zheng. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16-or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80 (2006) 4249–4263. [DOI] [PMC free article] [PubMed]
- 37.Stacey S., Jordan D., Williamson A., Brown M., Coote J., Arrand J. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 2000;74:7284–7297. doi: 10.1128/jvi.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X., Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell. Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 39.Spangle J., Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol. 2010;84:9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spangle J., Ghosh-Choudhury N., Munger K. Activation of cap-dependent translation by mucosal human papillomavirus E6 proteins is dependent on the integrity of the LXXLL binding motif. J. Virol. 2012;86:7466–7472. doi: 10.1128/JVI.00487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar M., Kong K., Javier R. Hijacking Dlg1 for oncogenic phosphatidylinositol 3-kinase activation in human epithelial cells is a conserved mechanism of human adenovirus E4-ORF1 proteins. J. Virol. 2014;88:14268–14277. doi: 10.1128/JVI.02324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong K., Kumar M., Taruishi M., Javier R. The human adenovirus E4-ORF1 protein subverts discs large 1 to mediate membrane recruitment and dysregulation of phosphatidylinositol 3-kinase. PLoS Pathog. 2014;10:e1004102. doi: 10.1371/journal.ppat.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popowski M., Ferguson H., Sion A., Koller E., Knudsen E., Van Den Berg C. Stress and IGF-1 differently control cell fate through mammalian target of rapamycin (mTOR) and retinoblastoma protein (pRB) J. Biol. Chem. 2008;283:28265–28273. doi: 10.1074/jbc.M805724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray P., Grover R., Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]