Abstract
To determine the nature and cellular localization of amino acid transport in pea seeds, two cDNA clones belonging to the AAP family of H+/amino acid co-transporters (PsAAP1 and PsAAP2) were isolated from a cotyledon cDNA library of pea (Pisum sativum L.). Functional expression in the yeast amino acid uptake mutants 22Δ6AAL and 22Δ8AA showed that PsAAP1 mediates transport of neutral, acidic, and basic amino acids. RNA-blot analyses showed that PsAAP1 is expressed in seeds and vegetative organs, including amino acid sinks and sources, whereas PsAAP2 could not be detected. For developing seeds, transcripts of PsAAP1 were detected in coats and cotyledons, with seed coats giving a weak signal. In cotyledons, expression was highest in epidermal-transfer-cell-enriched tissue. RNA in situ hybridization analysis showed that PsAAP1 was predominantly present in epidermal transfer cells forming the outer surface of cotyledons, which abuts the seed coats. Overall, our observations suggest that this transporter, which is localized in transfer cells of cotyledons, might play a role in the uptake of the full spectrum of amino acids released from seed coats.
Seeds of grain legumes are nutritionally important due to their relatively high content of essential amino acids, which are accumulated in the cotyledons as storage proteins (Müntz, 1982). Because of its large seeds, we used pea (Pisum sativum L.) as a model system for studying amino acid import into cotyledons. The symplasmic discontinuity between maternal and filial tissues in seeds necessitates membrane efflux from the maternal tissues and subsequent uptake by filial tissues such as the endosperm or embryo. Amino acids are delivered to developing grain legume seeds almost exclusively in the phloem, leading to the proposal that transfer occurs along the path from xylem to phloem (Pate et al., 1975, 1977; Pate, 1980; Van Bel, 1984).
Phloem unloading in the seed coats is considered to be symplasmic (Offler and Patrick, 1984, 1993; Grusak and Minchin, 1988; Offler et al., 1989). De Jong and Wolswinkel (1995) found that release of amino acids from seed coats occurs by a facilitated membrane transport mechanism, probably through non-selective pores (De Jong et al., 1997). The released nutrients are taken up from the seed apoplasm by the developing embryos. In pea cotyledons, a saturable transport system supplemented by passive transport was demonstrated by uptake studies with l-Val (Lanfermeijer et al., 1990). The saturable uptake component for Suc and amino acids seems to be proton motive force coupled (Lanfermeijer et al., 1990; Tegeder et al., 1999). The molecular mechanism of efflux from the maternal tissue and uptake of amino acids by the filial tissue has not to our knowledge been studied until now.
In developing fava bean (Vicia faba) and pea seeds, transfer cells form the inner surface of the seed coats and external epidermis of the enclosed cotyledons (Hardham, 1976; Offler et al., 1989; Tegeder et al., 1999). These transfer cells are probably the principal sites for the exchange of Suc across the extracellular space between seed coats and cotyledons (Harrington et al., 1997a, 1997b; Weber et al., 1997; Tegeder et al., 1999). It is unknown if the observed amino acid transport processes in developing legume seeds are also associated with these specialized cells.
Physiological and genetic evidence demonstrates the existence of multiple, carrier-mediated transport systems responsible for the uptake and transfer of amino acids within plants (Fischer et al., 1998). Studies using whole tissues, individual cells, or protoplasts have shown that amino acid transport in plants is mediated by specific membrane transport proteins that couple the electrochemical potential gradient of H+ to secondary active accumulation of amino acids (Despeghel and Delrot, 1983; Reinhold and Kaplan, 1984; Borstlap and Schuurmans, 1988; Bush and Langston-Unkefer, 1988; Li and Bush, 1990, 1992; Williams et al., 1990; Weston et al., 1995). A number of amino acid transporter genes have been identified in Arabidopsis (for review, see Fischer et al., 1998). Based on sequence homology, these plant transporters are classified into two major families: the ATF family and APC family (Fischer et al., 1998).
The ATF family is comprised of the broad-specificity AAP transporters, the basic amino acid LHT transporters, the compatible solute ProT transporters, and the putative auxin AUX transporters. Each of the seven AAP genes identified to date show a distinct expression pattern, suggesting that the carriers fulfill specific functions within plants (Fischer et al., 1995, 1998). Promoter-GUS studies showed that AtAAP1 expression was exclusively found in seeds, suggesting a role in supplying the developing seeds with amino acids (Hirner et al., 1998). Aside from Arabidopsis, AAPs have been identified in only a few other species: Solanum tuberosum, Nicotiana silvestris, Ricinus communis (Fischer et al., 1998; accession no. AJ132228), Nepenthes alata (Schulze et al., 1999), and fava bean (accession no. Y09591). Apart from Arabidopsis, only a R. communis amino acid symporter clone has been functionally described (Marvier et al., 1998).
The aim of this work was to isolate amino acid transporter genes involved in transferring amino acids between coats and cotyledons of developing pea seeds. Yeast complementation was used to test whether the isolated genes function as an amino acid transport system and to determine the substrate specificity of the amino acid transporters. The expression patterns of these genes were studied by northern-blot analysis, and cellular localization was examined by in situ hybridization. It was concluded that epidermal transfer cells are the main sites of amino acid transport into pea cotyledons.
MATERIALS AND METHODS
Plant Material
Pea (Pisum sativum L. cv Greenfeast) plants were raised in 1.5-L pots under greenhouse conditions (partial temperature control of 20°C–26°C by day, 15°C–17°C by night; supplementary lighting with metal halide lamps to ensure a minimum photosynthetically active radiation [PAR] on the uppermost leaves of 200 μmol m−2 s−1, and a 14-h photoperiod) in a potting mix of coarse sand, peat, and perlite (3:1:1), with the addition of lime (4 g L−1) and slow-release fertilizer (6 g L−1, Nutricote, Chuso Asaki Fertiliser, Tokyo). Mineral nutrition of plants was supplemented with full-strength Hoagland no. 1 solution (Hoagland and Snyder, 1933). Developing seeds were harvested for observation during their linear phase of cotyledon dry weight gain. At this developmental stage, the relative water content of cotyledons was between 68% and 75%.
Isolation of Amino Acid Symporter by cDNA Library Screening
A pea cotyledon cDNA library in UniXR-ZAP (Stratagene, La Jolla, CA) was kindly provided by T. Wang (John Innes Centre, Norwich, UK). For library construction, cotyledons of the early developmental stages were used. The cDNA library was screened using Arabidopsis amino acid/H+ symporters (AtAAP1, AtAAP2, AtAAP3, AtAAP5, and AtAAP6) as probes (for review, see Fischer et al., 1998). Hybridization was performed for 16 h at 42°C in 5× SSC, 5× Denhardt's solution, 0.5% (w/v) SDS, 0.1 mg mL−1 tRNA, and 25% (v/v) formamide. Subsequently, filters were washed three times for 20 min in 2× SSC (0.15 m NaCl and 0.015 m sodium citrate), and 0.1% (w/v) SDS. After two rounds of screening, plaque-purified phages were converted to pBluescript SK− derivatives by in vivo excision. DNA sequence analysis was performed with an automated DNA sequencer (Perkin-Elmer, Foster City, CA).
Yeast Growth and Transformation
Saccharomyces cerevisiae strains 22Δ8AA (MATα, ura3-1, gap-1, put4-1, uga4-1, can1::HisG, lyp/alp::HisG, hip1::HisG, and dip5::HisG) and 22Δ6AAL (MATα, ura3-1, gap-1, put4-1, uga1; can1::HisG, lyp/alp::HisG, and lys2::HisG) (W.-N. Fischer, unpublished data) were used to investigate substrate specificity of PsAAP1. S. cerevisiae strain 22574d (MATα, ura3-1, gap-1, put4-1, and ad uga1; Jauniaux et al., 1987) served as genetic background to create both strains. 22Δ6AAL can be used to select for Lys transport, since it is deficient in Lys uptake and the lys2 gene encoding for Lys biosynthesis is interrupted. Growth of 22Δ6AAL is dependent on either a high concentration of Lys (1.5 g/L) or on dipeptides containing Lys (such as LysAsp; 0.3g/L).
PsAAP1 cDNA was cloned in pFL61 expression vector (Minet et al., 1992), and yeast strains 22Δ8AA and 22Δ6AAL were transformed according to the method of Dohmen et al. (1991). AtAAP2 and AtAAP3 in pFL61 and the empty vector pFL61 were used as controls (Fischer et al., 1995). 22Δ8AA transformants were selected on nitrogen-free minimal medium containing 0.5 mg/L l-Pro, l-citrulline, l-Glu, or γ-amino butyric acid (GABA) as the sole nitrogen source. For non-selective conditions, 0.5 g/L ammonium sulfate was added to the medium. Medium for strain 22Δ6AAL was supplemented with 0.5 g/L urea and 0.3 g/L Lys-Asp. To select for a basic amino acid transporter, transformants were plated on medium containing 0.5 g/L ammonium sulfate and 1.5 μg/mL Lys.
RNA-Blot Analysis and in Situ Hybridization
Total RNA of different plant organs was extracted from a pool of 10 plants according to the method of Frommer et al. (1994). RNA (20 μg) was electrophoresed and then transferred to Hybond N nylon membranes (Amersham-Pharmacia Biotech, Uppsala) by standard procedures. Filters were hybridized with PsAAP1 and PsAAP2 (full-length clone or 200 bp of 3′ untranslated region) as 32P-labeled cDNA probes for 16 h at 42°C in a hybridization solution containing 5× SSC, 2× Denhardt's, 0.5% (w/v) SDS, 0.1 mg mL−1 tRNA, and 50% (v/v) formamide buffered at pH 6.4 with 200 mm 1,4-piperazinediethanesulfonic acid (PIPES). Post-hybridization filters were washed in 2×, 0.5×, and 0.1× SSC containing 0.1% (w/v) SDS for 20 min at 65°C.
Digoxigenin (DIG)-labeled PsAAP1 sense and antisense riboprobes were synthesized by in vitro transcription using the pGEM-T vector (Promega, Madison, WI) that contained the 3′-untranslated region of cDNA template of pea H+/amino acid symporter (see above). The plasmid was linearized and DIG-11-UTP was incorporated with either SP6 or T7 RNA polymerase according to the manufacturer's instructions (Boehringer Mannheim/Roche, Basel).
Cotyledons were fixed for 6 h at 4°C in 4% (v/v) paraformaldehyde buffered at pH 7.2 with 50 mm Na-phosphate. Following dehydration in ethanol and substitution with tertiary-butyl alcohol, tissue was infiltrated with Paraplast-saturated tertiary-butyl alcohol prior to embedding in 100% (w/v) Paraplast (Sherwood Medical, St. Louis) (for details, see Harrington et al., 1997a). Sections (8 μm) were mounted on 3-aminopropylethoxysilane-coated slides (Digene Diagnostics, Beltsville, MD) and probed with a sense or antisense riboprobe as detailed in Harrington et al. (1997a).
RESULTS
Molecular Cloning of Amino Acid/H+ Symporters
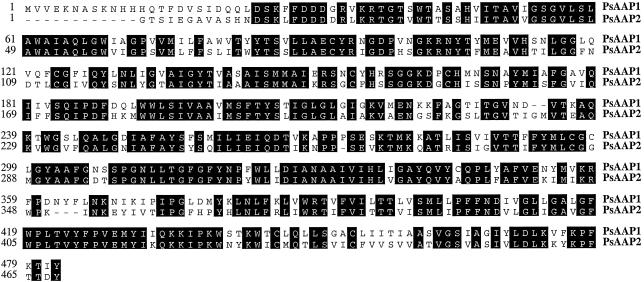
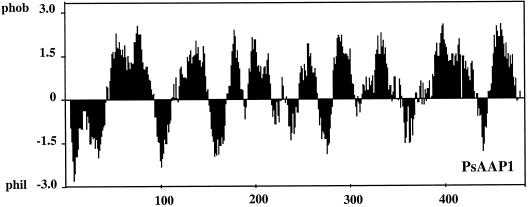
Screening of a pea cDNA library resulted in the isolation of an amino acid transporter gene, PsAAP1, and a partial cDNA clone, PsAAP2 (Fig. 1). PsAAP1 and PsAAP2 share approximately 66% identical amino acids. A hydropathy analysis algorithm demonstrated their highly hydrophobic nature, with nine to 11 putative transmembrane spanning regions (Fig. 2). PsAAP1 (2,034 bp) encodes an open reading frame of 1,575 bp, which corresponds to a protein of 53 kD. The sequence shares between 71% and 74% identical amino acids with the amino acid transporters RcAAP1 (R. communis; Bick et al., 1998) and AtAAP3 (Arabidopsis, Fischer et al., 1995). PsAAP2 represents a partial clone lacking the translation start and, in addition, contains two intron sequences (in Fig. 1 shown without introns). Compared with other AAPs, the intron positions correspond to the first and second intron in genomic AAPs (e.g. AtAAP2, accession no. X95623).
Figure 1.
Amino acid sequence alignment of amino acid transporters PsAAP1 and PsAAP2 from pea.
Figure 2.
Prediction of putative membrane-spanning regions in PsAAP1 using LASERGENE software (DNASTAR, Madison, WI).
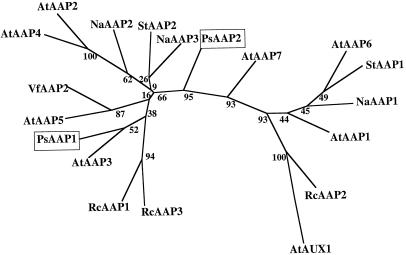
Amino acid sequence comparison between AAP amino acid transporters (Fig. 3) shows that PsAAP1 falls in the same group with AtAAP3, indicating a closer relationship with each other than with other AAPs. PsAAP2 seems to be more closely related to AtAAP7 than to any other transporter protein.
Figure 3.
Computer-aided analysis of homologies between PsAAP1 and PsAAP2 and related proteins. The analysis was performed using PHYLIP (J. Felsenstein, 1995, Phylogenetic Interference Package, version 3.5, distributed by the author: Department of Genetics, University of Washington, Seattle, WA) with aligned sequences of PsAAP1 and PsAAP2 and amino acid transporters from Arabidopsis (AtAAPs), S. tuberosum (StAAPs), R. communis (RcAAPs; RcAAP3-accession no. AJ132228) (Fischer et al., 1998), N. alata (NaAAPs; Schulze et al., 1999), and V. faba (VfAAP2, accession no. Y09591). The numbers indicate the occurrence of a given branch in 100 bootstrap replicates of the given data set. AtAUX1 was used as the outgroup.
Functional Analysis of PsAAP1
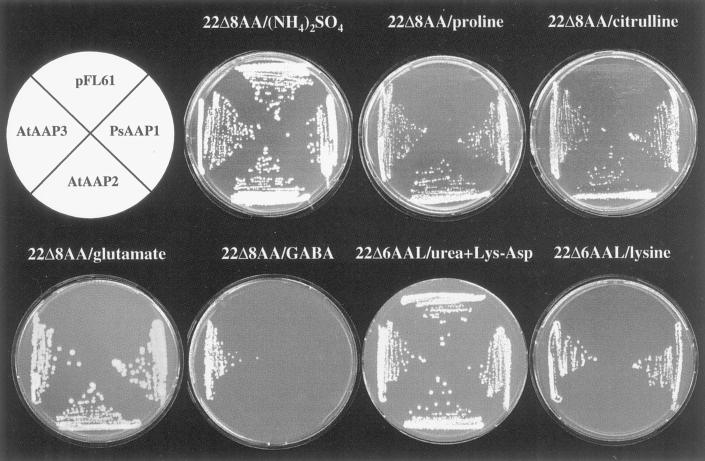
To analyze the substrate specificity of PsAAP1, the cDNA was expressed in two yeast strains lacking uptake systems for different amino acids. The yeast strain 22Δ8AA is deficient in the uptake of citrulline, Pro, Glu, and GABA, whereas 22Δ6AAL is unable to grow on medium containing Lys as a sole nitrogen source (W.-N. Fischer, unpublished data). PsAAP1 was able to mediate the efficient uptake of Pro, citrulline, and Glu, but not of GABA (Fig. 4). AtAAP2 and AtAAP3 and the empty vector pFL61 were used as controls. Like AtAAP3, PsAAP1 mediates growth on selective concentrations of Lys; AtAAP2 did not recognize Lys.
Figure 4.
Complementation of growth of yeast strains 22Δ8AA and 22Δ6AAL, which are deficient in the uptake of amino acids, by PsAAP1, AtAAP2, and AtAAP3. Growth of 22Δ8AA and 22Δ6AAL expressing PsAAP1, AtAAP2, and AtAAP3 and the strains transformed with the vector pFL61 is shown. For selection, the medium was supplemented with Pro, citrulline, Gaba, Glu (22Δ8AA), or Lys (22Δ6AAL) as the sole nitrogen source. For non-selective conditions, the medium contained either ammonium sulfate (22Δ8AA) or urea and the dipeptide Lys-Asp as the sole nitrogen source (22Δ6AAL).
Expression Analysis of PsAAP1 in Developing Pea Seeds
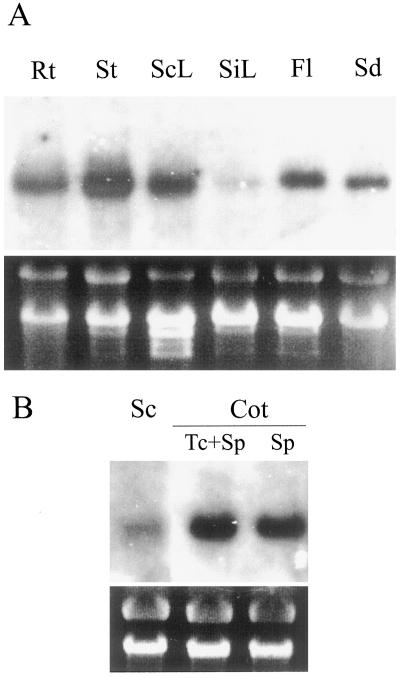
Northern-blot analysis showed that PsAAP1 is expressed in all organs, with the strongest expression in stems and the weakest in sink leaves (Fig. 5A). Cross-hybridization with other AAPs did not occur, which was confirmed by using the 3′-untranslated region of PsAAP1. Expression of PsAAP2 was not detected in any of the organs tested (data not shown).
Figure 5.
Profiles of organ expression of PsAAP1. Gel blots of total RNA (20 μg per lane) were hybridized with 32P-labeled 3′-untranslated region cDNA probe of PsAAP1. Ethidium bromide staining of rRNA is shown at bottom. A, Northern-blot analyses of RNA extracted from different pea organs. Rt, Roots; St, stems; ScL, source leaves; SiL, sink leaves; Fl, flowers; and Sd, developing seeds. B, Tissue-specific expression of PsAAP1 in developing pea seeds. Northern-blot analyses of RNA extracted from seed coat (Sc) and from cotyledon (Cot) epidermal-transfer-cell-enriched (Tc+Sp) and cotyledon storage parenchyma (Sp) tissues.
A more detailed study of PsAAP1 expression in seeds was performed. Transcripts of PsAAP1 were detected in all seed tissues, with the weakest signal in seed coats (Fig. 5B). Expression was highest in fractions enriched in cotyledonary transfer cells, and weaker signal strengths were detected in extracts from cotyledon storage parenchyma cells. PsAAP2 could not be detected throughout the seed.
Localization of PsAAP1
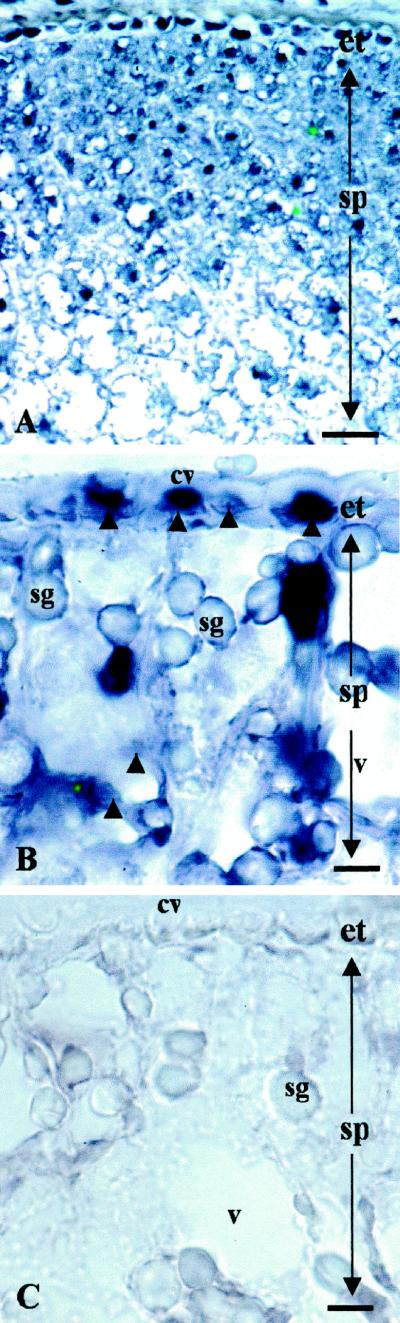
Cellular localization of PsAAP1 gene expression in cotyledons was examined by in situ hybridization using identical cDNA fragments as described for the northern-blot analyses. Antisense riboprobe strongly labeled epidermal transfer cells with a noticeably decreasing radial gradient of expression through consecutive rows of storage parenchyma cells (Fig. 6, A and B). Sections hybridized with a sense riboprobe showed no color reaction (Fig. 6C).
Figure 6.
Light micrographs illustrating in situ hybridization of DIG-labeled PsAAP1 antisense (A and B) and sense (C) riboprobes to transverse sections of developing pea cotyledons. Probe binding in the cytoplasm of the epidermal transfer cells and the storage parenchyma cells is evident as a strong blue color indicated by darts in B (compare with the sense control, C). The dense color reactant in the epidermal transfer cells and five to six rows of storage parenchyma cells inward of the epidermis declines through subsequent consecutive rows of storage parenchyma cells (A). In A, the bar = 100 μm (×120); in B and C, the bar = 30 μm (×300). cw, Cell wall; et, epidermal transfer cell; sg, starch granule; sp, storage parenchyma cell; and v, vacuole.
DISCUSSION
In Arabidopsis, a number of amino acid transporter genes have been identified, but little is known about the amino acid transporter in other plant species (Fischer et al., 1998; Marvier et al., 1998). During development of legume seeds, large amounts of protein are accumulated in cotyledons. The amino acids necessary for these storage compounds are imported through the phloem strands in the funiculus and seed coat and finally taken up into cotyledons from the seed apoplasm. For pea, physiological and biochemical data of these transport processes are available, but molecular analysis is still lacking (De Ruiter et al., 1984; Lanfermeijer et al., 1990, 1992; De Jong et al., 1997).
We isolated PsAAP1 and PsAAP2 amino acid transporters from a pea cotyledon cDNA library using known AtAAPs as probes. Transcripts of PsAAP1 (Fig. 5B) and of a H+/ATPase (Tegeder et al., 1999) were detected in coats of developing pea seeds, implying that transporters for active uptake of amino acids are present in these organs, even if they are not highly expressed. This is in contrast to the absence of such transporter activities using uptake experiments (De Jong et al., 1997). In seed coats the uptake of Lys was not inhibited by the protonophore carbonylcyanide m-chloro-phenylhydrazone, indicating that no H+ symporter is involved. For amino acids, uptake is envisaged to occur by passive diffusion, possibly through non-selective pores (De Jong and Wolswinkel, 1995; De Jong et al., 1997). It is likely that this pathway is also used for efflux of endogenous amino acids to make the nitrogen available to the embryo (De Jong and Wolswinkel, 1995; De Jong et al., 1997). PsAAP1 might operate to retrieve basic amino acids leaked to the seed apoplasm during symplasmic passage through the seed coats.
Northern-blot analysis showed that PsAAP1 is expressed in pea cotyledons, with the strongest expression confined to the epidermal-transfer-cell-enriched tissue. These observations indicate that carrier-mediated amino acid transport takes place throughout developing pea seeds, with cotyledons exhibiting the greatest potential for transport. The amino acid transport capacity of cotyledons is consistent with the appearance of a saturable, protonophore-inhibitable component for l-Val transport during cotyledon development (Lanfermeijer et al., 1990).
Differences in substrate specificity of amino acid transporters can reflect different physiological functions of the transporters within the plant, and allow different cell types within species or even different plant species to adapt to specific amino acid profiles. Sequence analysis indicated a close relationship of PsAAP1 to AtAAP3, which is expressed in roots. Functional expression of AtAAP3 in yeast and Xenopus oocytes have shown that AtAAP3 mediates transport of basic amino acids as well as recognizing acidic and neutral amino acids (Fischer et al., 1995; W.-N. Fischer, unpublished data). The close relationship of AtAAP3 to PsAAP1 (Fig. 3) could indicate a similar physiological function. Indeed, expression studies in yeast demonstrated that PsAAP1 mediates transport of the basic amino acid Lys. Additionally, PsAAP1 recognizes acidic and neutral amino acids and citrulline. The low substrate selectivity allows PsAAP1 to respond to the wide spectrum of amino acids present in pea plants (Urquhart and Joy, 1981). In this context, it is relevant to note that pea seed coats release a mixture of amides and amino acids to the seed apoplasm, including Glu, Gln, Ala, Thr, Lys, and Arg, for uptake by developing cotyledons (De Ruiter et al., 1984; Lanfermeijer et al., 1992).
Using DIG-labeled antisense and sense probes, PsAAP1 was localized to the epidermal transfer cells, with decreasing levels of expression through consecutive radial layers of storage parenchyma cells. The observed co-localization with H+/ATPase (Tegeder et al., 1999) to epidermal transfer cells provides strong evidence that these regions are the principal sites of energy-coupled uptake of amino acids from the seed apoplasm. Similar conclusions have been reached for Suc import in cotyledons of fava bean (Harrington et al., 1997a, 1997b; Weber et al., 1997) and pea (Tegeder et al., 1999). Subsequent transport of accumulated amino acids to the storage parenchyma cells is likely to occur through a symplasmic route, as was found for Suc (McDonald et al., 1995; Tegeder et al., 1999). The amino acid transporter expressed in the storage parenchyma cells could function as a retrieval mechanism for amino acids leaked to the cotyledon apoplasm. AtAAP1 from Arabidopsis could also be detected in cotyledons, indicating a function in import of amino acids into the embryo (Hirner et al., 1998).
PsAAP1 and PsAAP2 could have different transport functions, as indicated by an amino acid sequence homology of 66% and a markedly different expression pattern. It is interesting that PsAAP2 transcripts could not be detected even in seeds, the source material for construction of the cDNA library. This outcome suggests that the expression of PsAAP2 might be differently regulated than that of PsAAP1. Therefore, the expression of PsAAP2 under different environmental and nutritional conditions (e.g. various nitrogen sources), as well as at various stages during seed development, merits further examination.
In conclusion, a full-length clone of PsAAP1 isolated from a pea cotyledon cDNA library is grouped together with AtAAP3. Expression studies in yeast proved that PsAAP1 functions as an amino acid transport system mediating transport of a broad spectrum of amino acids. The gene is expressed throughout the plant body, suggesting a general role in amino acid transport, including the uptake of amino acids by developing cotyledons.
ACKNOWLEDGMENTS
We gratefully acknowledge the supply of a pea cotyledon cDNA library from Dr. Trevor Wang (John Innes Centre, Norwich, UK). We also thank Dr. Wolf-Nicolas Fischer (Zentrum für Molekularbiologie der Pflanzen, Tübingen, Germany) for providing the yeast strains. The expert technical expertise offered by Britta Kanzok, Dr. Xin-Ding Wang, and Kevin Stokes is much appreciated.
Footnotes
This work was financially supported by an Australian Research Council Large Grant (no. A19530955 to J.W.P. and C.E.O.) and by the Deutsche Forschungsgemeinschaft (contract no. FR989/2–3 to W.B.F.).
LITERATURE CITED
- Bick JA, Neelam A, Hall JL, Williams LE. Amino acid carriers of Ricinus communis expressed during seedling development: molecular cloning and expression analysis of two putative amino acid transporters, RcAAP1 and RcAAP2. Plant Mol Biol. 1998;36:377–385. doi: 10.1023/a:1005903229458. [DOI] [PubMed] [Google Scholar]
- Borstlap AC, Schuurmans J. Kinetics of l-valine uptake in tobacco leaf discs: comparison of wild type, the digenic mutant Valr-2 and its monogenic derivatives. Planta. 1988;176:42–50. doi: 10.1007/BF00392478. [DOI] [PubMed] [Google Scholar]
- Bush DR, Langston-Unkefer PJ. Amino acid transport into membrane vesicles isolated from zucchini. Plant Physiol. 1988;88:487–490. doi: 10.1104/pp.88.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong A, Koerselman-Kooij JW, Schuurmans JAMJ, Borstlap AC. The mechanisms of amino acid efflux from seed coats of developing pea seeds as revealed by uptake experiments. Plant Physiol. 1997;114:731–736. doi: 10.1104/pp.114.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong A, Wolswinkel P. Differences in release of endogenous sugars and amino acids from attached and detached seed coats of developing pea seeds. Physiol Plant. 1995;94:78–86. [Google Scholar]
- De Ruiter H, Schuurmans JAMJ, Kollöffel C. Amino acid leakage from cotyledons of developing and germinating pea seeds. J Plant Physiol. 1984;116:47–57. doi: 10.1016/S0176-1617(84)80083-8. [DOI] [PubMed] [Google Scholar]
- Despeghel JP, Delrot S. Energetics of amino acid uptake by Vicia faba leaf tissue. Plant Physiol. 1983;71:1–6. doi: 10.1104/pp.71.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AWM, Höner CB, Hollenberg C. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- Fischer WN, André B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB. Amino acid transport in plants. Trends Plant Sci. 1998;3:188–195. [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem. 1995;270:16315–16320. doi: 10.1074/jbc.270.27.16315. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D. Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett. 1994;347:185–189. doi: 10.1016/0014-5793(94)00533-8. [DOI] [PubMed] [Google Scholar]
- Grusak MA, Minchin PEH. Seed coat unloading in Pisum sativum: osmotic effects in attached versus excised empty ovules. J Exp Bot. 1988;39:543–559. [Google Scholar]
- Hardham AR. Structural aspects of the pathway of nutrient flow to the developing embryo and cotyledons of Pisum sativum L. Aust J Bot. 1976;24:711–721. [Google Scholar]
- Harrington GN, Franceschi VR, Offler CE, Patrick JW, Tegeder M, Frommer WB, Harper JF, Hitz WD. Cell specific expression of three genes involved in plasma membrane sucrose transport in developing Vicia faba seed. Protoplasma. 1997a;197:160–173. [Google Scholar]
- Harrington GN, Nussbaumer Y, Wang X-D, Tegeder M, Franceschi VR, Frommer WB, Patrick JW, Offler CE. Spatial and temporal expression of sucrose transport-related genes in developing cotyledons of Vicia faba L. Protoplasma. 1997b;200:35–50. [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB. Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 1998;14:535–544. doi: 10.1046/j.1365-313x.1998.00151.x. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Snyder WC. Nutrition of strawberry plants under controlled conditions. Proc Am Soc Hortic. 1933;30:288–296. [Google Scholar]
- Jauniaux JC, Vandenbol M, Vissers S, Broman K, Grenson M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae: cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur J Biochem. 1987;164:601–606. doi: 10.1111/j.1432-1033.1987.tb11169.x. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Koerselman-Kooij JW, Borstlap AC. Changing kinetics of l-valine uptake by immature pea cotyledons during development: an unsaturable pathway is supplemented by a saturable system. Planta. 1990;181:576–582. doi: 10.1007/BF00193013. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, van Oene MA, Borstlap AC. Compartmental analysis of amino-acid release from attached and detached pea seed coats. Planta. 1992;187:75–82. doi: 10.1007/BF00201626. [DOI] [PubMed] [Google Scholar]
- Li ZC, Bush DR. ΔpH-dependent amino acid transport into plasma membranes vesicles isolated from sugar beet leaves. I. Evidence for carrier-mediated, electrogenic flux through multiple transport systems. Plant Physiol. 1990;94:268–277. doi: 10.1104/pp.94.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZC, Bush DR. Structural determinants in substrate recognition by proton-amino acid symporters in plasma membrane vesicles isolated from sugar beet leaves. Arch Biochem Biophys. 1992;294:519–526. doi: 10.1016/0003-9861(92)90719-d. [DOI] [PubMed] [Google Scholar]
- Marvier AC, Neelam A, Bick JA, Hall JL, Williams LE. Cloning of a cDNA coding for an amino acid carrier from Ricinus communis (RcAAP1) by functional complementation in yeast: kinetic analysis, inhibitor sensitivity and substrate specificity. Biochim Biophys Acta. 1998;1373:321–331. doi: 10.1016/s0005-2736(98)00117-5. [DOI] [PubMed] [Google Scholar]
- McDonald R, Wang HL, Patrick JW, Offler CE. The cellular pathway of sucrose transport in developing cotyledons of Vicia faba L. and Phaseolus vulgaris L.: a physiological assessment. Planta. 1995;196:659–667. [Google Scholar]
- Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Müntz K. Seed development: nucleic acids and proteins in plants. In: Boulter D, Parthier B, editors. Encyclopedia of Plant Physiology, New Series. 14a. New York: Springer; 1982. pp. 505–558. [Google Scholar]
- Offler CE, Nerlich SM, Patrick JW. Pathway of photosynthate transfer in the developing seed of Vicia faba L.: transfer in relation to seed anatomy. J Exp Bot. 1989;40:769–780. [Google Scholar]
- Offler CE, Patrick JW. Cellular structures, plasma membrane surface areas and plasmodesmatal frequencies of seed coats of Phaseolus vulgaris L. in relation to photosynthate transfer. Aust J Plant Physiol. 1984;11:79–99. [Google Scholar]
- Offler CE, Patrick JW. Pathway of photosynthate transfer in the developing seed of Vicia faba L.: a structural assessment of the role of transfer cells in unloading from the seed coat. J Exp Bot. 1993;44:711–724. [Google Scholar]
- Pate JS. Transport and partitioning of nitrogenous solutes. Annu Rev Plant Physiol. 1980;31:313–340. [Google Scholar]
- Pate JS, Sharkey PJ, Atkin CA. Nutrition of a developing legume fruit. Plant Physiol. 1977;59:506–510. doi: 10.1104/pp.59.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Lewis OAM. Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta. 1975;122:11–26. doi: 10.1007/BF00385400. [DOI] [PubMed] [Google Scholar]
- Reinhold L, Kaplan A. Membrane transport of sugars and amino acids. Annu Rev Plant Physiol Plant Mol Biol. 1984;35:45–83. [Google Scholar]
- Schulze W, Frommer WB, Ward JM. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. Plant J. 1999;17:637–646. doi: 10.1046/j.1365-313x.1999.00414.x. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Wang X-D, Frommer WB, Offler CE, Patrick JW. Sucrose transport into developing seeds of Pisum sativum L. Plant J. 1999;18:151–161. doi: 10.1046/j.1365-313x.1999.00439.x. [DOI] [PubMed] [Google Scholar]
- Urquhart AA, Joy KW. Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiol. 1981;68:750–754. doi: 10.1104/pp.68.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel AJE. Quantification of the xylem-to-phloem transfer of amino acids by use of inulin (14C) carboxylic acid as xylem transport marker. Plant Sci Lett. 1984;15:285–291. [Google Scholar]
- Weber H, Borisjuk L, Sauer N, Wobus U. A role for sugar transporters during seed development: molecular characterisation of a hexose and a sucrose carrier in fava bean seeds. Plant Cell. 1997;9:895–908. doi: 10.1105/tpc.9.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K, Hall JL, Williams LE. Characterization of amino-acid transport in Ricinus communis roots using isolated membrane vesicles. Planta. 1995;196:166–173. [Google Scholar]
- Williams LE, Nelson SJ, Hall JL. Characterization of a solute transport in plasma membrane vesicles isolated from cotyledons of Ricinus communis. II. Evidence for a proton-coupled mechanism for sucrose and amino acid uptake. Planta. 1990;182:540–545. doi: 10.1007/BF02341029. [DOI] [PubMed] [Google Scholar]