A variety of neoplasias of the skin and mucous membranes in humans and animals can be ascribed to papillomavirus infections. Some of these papillomavirus induced neoplasias can undergo malignant transformation, while most remain benign. Papillomavirus infections are often even asymptomatic. Therefore the understanding of these viruses is important for appropriate prevention, diagnostic and therapeutic approaches. The entire genomic sequence of a new canine papillomavirus was determined from DNA detected in pigmented plaques in a pug. The novel canine papillomavirus type 18 (CPV18) falls into the genus Chipapillomavirus whose members have all been isolated form dogs with pigmented plaques, some of them also with squamous cell carcinomas. A small partial sequence of a 2009 isolate from a dog in California is identical with CPV18.
Papillomaviruses (PVs) are non-enveloped, double-stranded DNA viruses with a circular genome of about 8.000 base pairs (bp). They are generally host species-specific with some exceptions. Numerous of the known PVs are associated with benign and malignant neoplasias of the skin and mucous membranes in humans and animals, but there is evidence, that asymptomatic infections are more common [1], [9], [14]. More than 200 human and 140 animal PVs have been characterized, illustrating broad genetic diversity (http://pave.niaid.nih.gov/) [2], [6], [9]. The identification and study of new animal PVs are important because new PVs are potential pathogens in veterinary medicine, some animal PVs may serve as models for human disease and tools for basic biology, and the analysis of animal PVs helps provide insights into PV evolution. Canine PVs (CPVs) have been found associated with classical exophytic papillomas such as the common canine oral papillomatosis, with endophytic papillomas, with pigmented plaques and in rare cases with squamous cell carcinomas (Table 1) [13]. Pigmented plaques are most often seen in pugs where CPV4 seems to be the most prevalent type involved [17], [18], [22], [23], [26]. Overall seventeen complete genomes and several partial sequences of different CPVs have been communicated. The different CPVs belong to either of three different papillomavirus genera Lambda, Tau or Chi [2], [5], [11], [15], [16], [17], [18], [19], [20], [21], [22], [23], [25], [26], [28], [29], [30], [31]. These three genera are not monophyletic and hence only distantly related. The aim of this study was the analysis and description of a new potentially pathogenic CPV.
Table 1.
List of carnivore papillomaviruses and associated lesions.
| Virus | Genus | Host | Associated lesions reported | Genebank ID |
|---|---|---|---|---|
| CPV1 | Lambda | Dog | Oral papillomatosis, endophytic papillomas, exophytic papillomas, squamous cell carcinoma | D55633 |
| CPV6 | Lambda | Dog | Endophytic papillomas | FJ492744 |
| CcrPV1 | Lambda | Spotted Hyena | Oral papillomatosis | HQ585856 |
| FcaPV1 | Lambda | Cat | Exophytic papillomas | AY057109 |
| ElPV1 | Lambda | Sea Otter | Oral papillomatosis | KJ410351 |
| LrPV1 | Lambda | Bobcat | Oral papillomatosis | AY904722 |
| PcPV1 | Lambda | Puma | Oral papillomatosis | AY904723 |
| PlPV1 | Lambda | Raccoon | Exophytic papillomas | AY763115 |
| PlpPV1 | Lambda | Asiatic Lion | Oral papillomatosis | AY904724 |
| UuPV1 | Lambda | Snow Leopard | Oral papillomatosis | DQ180494 |
| CPV2 | Tau | Dog | Endophytic papillomas, exophytic papillomas, squamous cell carcinoma | AY722648 |
| CPV7 | Tau | Dog | Exophytic papillomas, squamous cell carcinoma | FJ492742 |
| CPV13 | Tau | Dog | Oral papillomatosis | JX141478 |
| CPV17 | Tau | Dog | Oral squamous cell carcinoma | KT272399 |
| FcaPV3 | Tau | Cat | Bowenoid in situ carcinoma | JX972168 |
| FcaPV4 | Tau | Cat | Gingivitis | KF147892 |
| MpPV1 | Tau | European Polecat | No lesion | KF006988 |
| CPV3 | Chi | Dog | Pigmented plaques, squamous cell carcinoma | DQ295066 |
| CPV4 | Chi | Dog | Pigmented plaques | EF584537 |
| CPV5 | Chi | Dog | Pigmented plaques | FJ492743 |
| CPV8 | Chi | Dog | Pigmented plaques | HQ262436 |
| CPV9 | Chi | Dog | Pigmented plaques | JF800656 |
| CPV10 | Chi | Dog | Pigmented plaques | JF800657 |
| CPV11 | Chi | Dog | Pigmented plaques | JF800658 |
| CPV12 | Chi | Dog | Pigmented plaques | JQ754321 |
| CPV14 | Chi | Dog | Pigmented plaques | JQ701802 |
| CPV15 | Chi | Dog | No details reported | JX899359 |
| CPV16 | Chi | Dog | Pigmented plaques, squamous cell carcinoma | KP099966 |
| CPV18 | Chi | Dog | Pigmented plaques | KT326919 |
| UmPV1 | Omega | Polar Bear | Oral papillomatosis | EF536349 |
| FcaPV2 | Dyotheta | Cat | Bowenoid in situ carcinoma | EU796884 |
| VvPV1 | Treiseta | Fox | No lesion | KF857586 |
| ZcPV1 | Dyonu | Sea lion | Exophytic papillomas | HQ293213 |
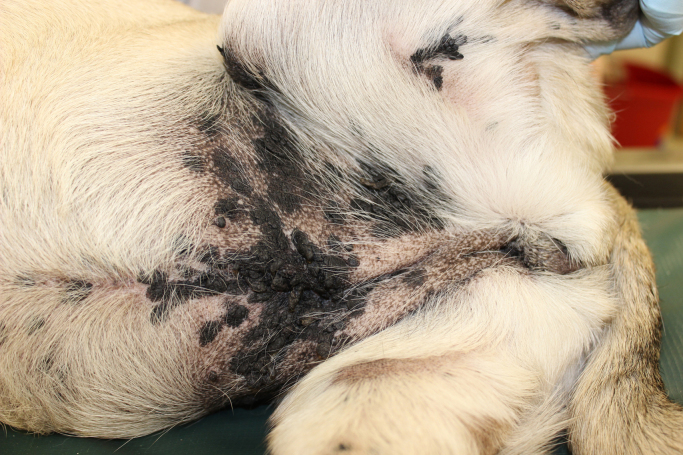
A 3 year old, female spayed pug was presented to Tufts Cummings Veterinary Medical Center due to the development of multiple pigmented plaques on the medioventral abdomen and inner thigh (Fig. 1). The dog was the only pet in the household but had frequent contact with other dogs. After thorough physical examination two 8 mm punch biopsies for histopathological analysis and a cytobrush sample for microbiological assessment were taken from the lesions. The skin biopsies were fixed in 4% buffered formalin. After embedding in paraffin, 4 µm sections were stained with haematoxylin and eosin (HE) for histopatholgical examination. The cytobrush was moistened with sterile 0.9% NaCl solution, rubbed for thirty seconds on the affected area and stored in an 1.5 ml Eppendorf tube at −20 °C until DNA extraction.
Fig. 1.
Three year old female pug with numerous coalescing pigmented plaques and exophytic nodules (from 0.5 cm to 3 cm) on the medioventral abdomen and inner thighs.
Total DNA was isolated from the cytobrush sample using a Qiagen DNeasy Blood and Tissue kit. The extract was tested for the presence of host and papillomavirus DNA by PCR using the primer combinations dogGAPDHf/dogGAPDHr, canPVf/FAP64 and CP4/CP5 [10], [12], [14]. Circular DNA was amplified from the total DNA extract by rolling circle amplification (RCA) using a TempliPhi Amplification kit. The RCA product was digested and entirely cloned into the SpeI site of a pBluescript II KS+ vector [17]. The sequences of the pure and uncloned RCA product and the genomic clones were determined independently, and the primary sequences assembled in DNA Star (DNASTAR, Inc.). Comparison of RCA sequence and clone sequence ensured correctness and completeness especially around the SpeI cloning site. Pairwise sequence alignments were performed with the open reading frames (ORFs) of the obtained sequence using the Needleman-Wunsch algorithm. A phylogenetic tree for 80 PVs including the sequences of the CPVs as well as representatives of all thus far characterized PV genera was calculated (Supplement 1). For that purpose the predicted amino acid sequences of E1, E2, L2 and L1 were translated and aligned by using MAFFT before being back-translated to DNA sequences [17]. GBLOCK (version 0.91b) was used to remove regions of poor similarity and the resulting shortened sequences were combined into a single multiple sequence alignment by concatenating the sequences from each virus [3]. The optimal model of DNA evolution was evaluated for best fit of the dataset using MODELTEST (version 1.4.4); using the default settings [4], [8]. Bayesian phylogeny of the E1-E2-L2-L1 concatenate was inferred using MRBAYES (version 3.2); Markov Chain Monte Carlo with GTR substitution matrix, variable gamma rates, invariant sites, two runs, four chains of 10 000 000 generations [26]. FcPV1 was defined as outgroup, to root the tree. Trees were sampled after every 1000 steps during the process to monitor phylogenetic convergence. The average standard deviation of split frequencies was below 0.0053 (MrBayes recommended final average <0.01). This was performed on the Orchestra High Performance Compute Cluster at Harvard Medical School. The first 10% of the trees were discarded and the remaining ones combined using TreeAnnotator (version 1.8.2; http://beast.bio.ed.ac.uk) and displayed with FIGTREE (1.4.2; http://tree.bio.ed.ac.uk/) [24], [25]. The code to run MrBayes as well as the files produced during this analysis are stored on GitHub: https://github.com/alosdiallo/Phylogeny_model_papillomavirus.
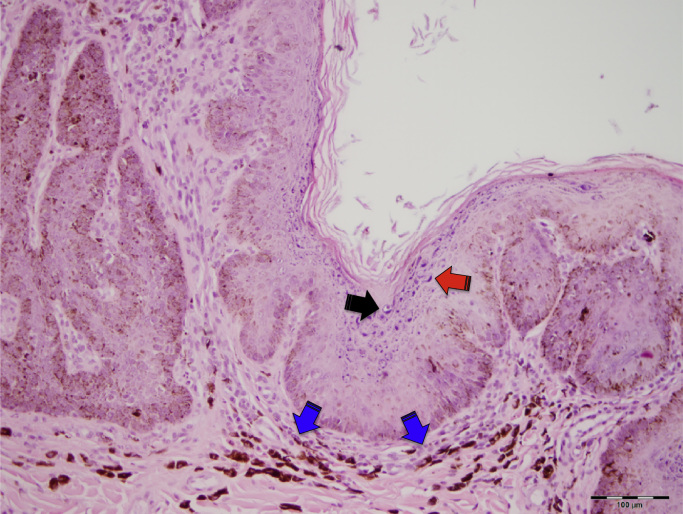
The histological examination revealed severe papillated epidermal hyperplasia, with melanosis, large clumped keratohyalin granules as well as a few single scattered clear cells reminiscent of koilocytes. The superficial dermis presented a mild inflammatory infiltrate and pigmentary incontinence (Fig. 2). The lesions were classified as canine viral pigmented plaques without signs of malignant transformation.
Fig. 2.
Histologic section of one of the pigmented plaques. Epidermal hyperplasia, with melanosis, large keratohyalin granules (red arrow), as well as a few single scattered clear cells reminiscent of koilocytes (black arrow). The superficial dermis presented a mild inflammatory infiltrate and pigmentary incontinence (blue arrows).
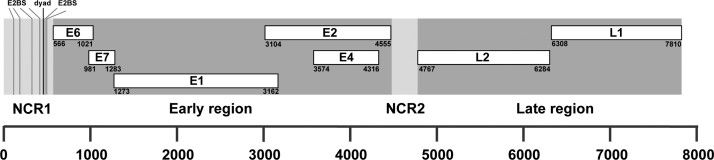
No differences between RCA product and clone were found upon comparison of the sequencing results. The circular sequence of the novel isolate consists of 7810 base pairs, which falls into the typical size range of Chi-PVs and has a GC content of 51% (Supplement 2) [11]. It contains the PV typical early (E1, E2, E4, E6, E7) and late (L1, L2) ORFs as well as non-coding region one between L1 and E6 (NCR1, 565 bp) and a second one between E2 and L2 (NCR2, 211 bp) (Fig. 3). Various characteristic PV motifs can be identified: Dyad symmetry repeats (TTGTTGTTAACAACAA) flanked by four E2 binding sites (ACC-N6-GGT) upstream and two downstream are located in an AT-rich area in the NCR1 putatively marking the origin of replication (Fig. 3) [9], [27]. Polyadenylation signals (AATAAA) were found within L2 near the 5′ end and overlapping with the 3′ end of the L1 ORF. The E1 amino acid (aa) sequence contains a modified ATP-dependent helicase motif (GPPDTGKS). A pRB-binding motif (LXCXE) is present in E7. E7 also contains one zinc-binding motif (CX2CX29CX2C), whereas E6 contains two.
Fig. 3.
Schematic representation of the linearized genome of CPV18 indicating the genome size and the organization of the open reading frames. Position 1 defined by the first base after the L1 stop codon.
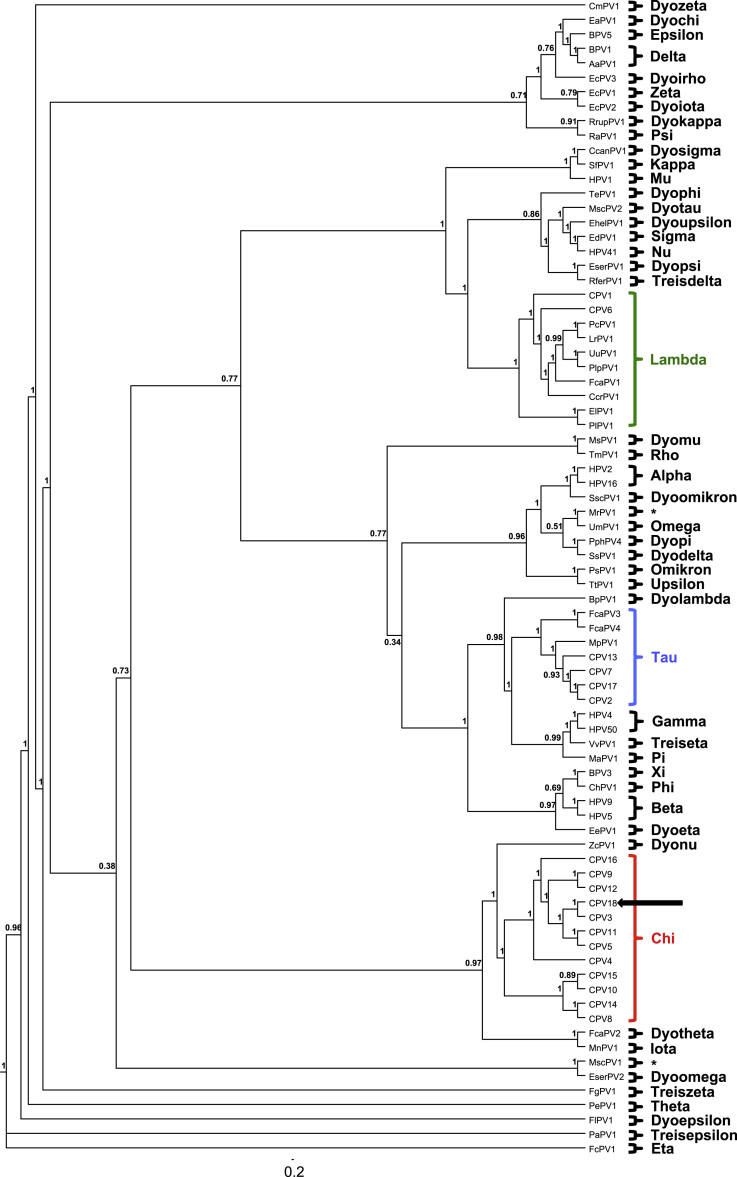
The L1 nucleotide sequence, which is used for classification, shares 85% identity with CPV3 and 74% with CPV5. The sequence of L1 of this novel isolate is 100% identical with a 355 bp partial L1 sequence (JQ040499). The sequence derived from a sample of a pug with pigmented plaques collected in California in 2009 [18]. The highest degree of identity in the E1 aa sequence was found with CPV3 (92%), CPV5 and CPV11 (82%). At the aa level for E6 and E7, the highest similarity is observed with CPV3 (83%, 98%), CPV9 (77%, 85%) and CPV12 (77%, 83%). Upon the phylogenetic analysis based on joined the E1-E1-L2-L1 ORFs the novel papillomavirus sequence clustered with these Chi-CPVs (Fig. 4). Based on the overall analysis, the isolate was designated as a new PV type and recognized by the Papillomavirus Study Group of the ICTV as CPV18.
Fig. 4.
Phylogenetic tree of 80 papillomaviruses including all currently assigned genera and carnivore infecting virus types. Tree based on the sequence information of E1, E1, L2 and L1. Genera containing canine papillomaviruses are highlighted in red (Chi), blue (Tau) and green (Lambda). Unclassified viruses are marked with an asterisk.
The clinical and histopathological findings strongly indicate a PV induced disorder. The detection of CPV18 and no other PV DNA suggests that this virus might be causative. The type of lesion found in the here-described case has repeatedly been associated with CPV infections. In all cases of canine pigmented plaques that were tested Chi-PV DNA was found and Chi-PVs were only found in respective pigmented lesions (Table 1). Viral gene expression and replication has been demonstrated for some Chi-PVs and a causal relationship between Chi-PV infection and the development of pigmented plaques seems likely based on the overall data [15], [17]. Considering, that the DNA of CPV18 and CPV4 have previously or repeatedly been found in pugs with pigmented plaques, a genetic predisposition in a lineage of this breed might be plausible. So far however, there is no data regarding this matter.
CPV18 also aligns well with the other Chi-PVs in terms of genome size and organization (Fig. 3, Supplement 2). Chi-PVs belong to the smallest known PVs overall in terms of their genome size and do neither have an E5 ORF nor a large second NCR. Within the Chi-PVs CPV18 falls into the Chi−1 species group with CPV3 being its closest relative. This holds true upon phylogenetic analysis of all six ORFs individually (Supplement 3). While the species allocation within the Chi-PVs is very robust in the analysis, the exact position of the individual types within them varies somewhat from gene to gene; a phenomenon that has previously been observed in PVs in general [7]. Especially the trees based on E6 and E7 vary from the others and from each other (Supplement 3, Supplement 4). No signs of recombination events were found within the known Chi-PVs.
Taken together the findings identify CPV18 as a new member of the papillomavirus genus Chi (Chi−1) that is putatively involved in the development of pigmented plaques in dogs.
Nucleotide sequence accession number. The nucleotide sequence of CPV18 is deposited in GenBank under accession no. KT326919.
Acknowledgments
The authors thank Peter M. Howley of the Department of Microbiology and Immunobiology, Harvard Medical School for his support and Kurt Tobler of the University of Zurich for advice. This work was partially financed by a grant to CEL from the Swiss Foundation for Grants in Biology and Medicine donated by Novartis.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.pvr.2016.08.001.
Appendix A. Supplementary material
Supplement 1 List of papillomaviruses included in the phylogenetic analysis.
.
Supplement 2 Genome size range with mean and standard deviation for canine papillomaviruses belonging to different genera.
.
Supplement 3 Phylogenetic trees of the individual ORFs E6, E7, E1, E2, L2 and L1 of the Chipapillomaviruses including ZcPV1, FcaPV2 and MnPV1 as outgroups. Obtained as described in material and methods.
.
Supplement 4 Comparison of phylogenetic trees based on the individual ORFs E6, E7, E1, E2, L2 and L1 of 15 papillomaviruses. Trees were obtained as described in material and methods and converted into Newick format (using FigTree software). For the comparison pairs of trees were imported into SpitsTree4 (version 4.14.4, Huson DH, Bryant D. 2006. Application of Phylogenetic Networks in Evolutionary Studies. Molecular Biology and Evolution. 23, 254–267) software and a consensus network analysis performed with default settings..
.
References
- 1.Antonsson A., Forslund O., Ekberg H., Sterner G., Hansson B. The ubiquity and impressive genomic diversity of human papillomaviruses suggests a commensalic nature of these viruses. J. Virol. 2000;74:11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard H.U., Burk R., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 4.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delius H., Van Ranst M., Jenson A., zur Hausen H., Sundberg J. Canine oral papillomavirus genomic sequence: a unique 1.5-kb intervening sequence between the E2 and L2 open reading frames. Virology. 1994;204:447–452. doi: 10.1006/viro.1994.1552. [DOI] [PubMed] [Google Scholar]
- 6.de Villiers E.M. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Vallve S., Alonso A., Bravo I.G. Papillomaviruses: different genes have different histories. Trends Microbiol. 2006;13:514–521. doi: 10.1016/j.tim.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Guindon S., Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood”. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 9.Howley P.M., Schiller J.S., Lowy D.R. Papillomaviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wikins; Philadelphia: 2013. pp. 1662–1703. [Google Scholar]
- 10.Iftner A., Klug S.J., Garbe C., Blum A., Stancu A., Wilczynski S.P., Iftner T. The prevalence of human papillomavirus genotypes in nonmelanoma skin cancers of nonimmunosupressed individuals identifies high-risk genital types as possible risk factors. Cancer Res. 2003;63:7515–7519. [PubMed] [Google Scholar]
- 11.Lange C.E., Tobler K., Ackermann M., Panakova L., Thoday K.L., Favrot C. Three novel canine papillomaviruses support taxonomic clade formation. J. Gen. Virol. 2009;90:2615–2621. doi: 10.1099/vir.0.014498-0. [DOI] [PubMed] [Google Scholar]
- 12.Lange C.E., Tobler K., Brandes K., Breithardt K., Ordeix L., von Bomhard W., Favrot C. Canine inverted papillomas associated with DNA of four different papillomaviruses. Vet. Dermatol. 2010;21:287–291. doi: 10.1111/j.1365-3164.2009.00817.x. [DOI] [PubMed] [Google Scholar]
- 13.Lange C.E., Favrot C. Canine Papillomaviruses. Vet. Clin. North AM: Small Anim. Pract. 2011;41:1183–1195. doi: 10.1016/j.cvsm.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Lange C.E., Zollinger S., Tobler K., Ackermann M., Favrot C. The clinically healthy skin of dogs is a potential reservoir for canine papillomaviruses. J. Clin. Microbiol. 2011;49:707–709. doi: 10.1128/JCM.02047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange C.E., Tobler K., Lehner A., Vetsch E., Favrot C. A case of a canine pigmented plaque associated with the presence of a Chi-papillomavirus. Vet. Dermatol. 2012;23 doi: 10.1111/j.1365-3164.2011.01007.x. 76-e19. [DOI] [PubMed] [Google Scholar]
- 16.Lange C.E., Ackermann M., Favrot C., Tobler K. Entire genomic sequence of novel canine papillomavirus type 13. J. Virol. 2012;86:10226–10227. doi: 10.1128/JVI.01553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange C.E., Tobler K., Schraner E.M., Vetsch E., Fischer N.M., Favrot C. Complete canine papillomavirus life cycle in pigmented lesions. Vet. Microbiol. 2013;162:388–395. doi: 10.1016/j.vetmic.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Luff J.A., Affolter V.K., Yeargan B., Moore P.F. Detection of six novel papillomavirus sequences within canine pigmented plaques. J. Vet. Diagn. Invest. 2012;24:576–580. doi: 10.1177/1040638712443360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luff J., Moore P., Wang J., Usuda Y., Affolter V., Schlegel R., Yuan H. Complete Genome Sequence of Canine Papillomavirus Type 10. J. Virol. 2012;86:11407. doi: 10.1128/JVI.01982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luff J., Mader M., Britton M., Fass J., Rowland P., Orr C., Schlegel R., Yuan H. Complete genome sequence of canine papillomavirus type 16. Genome Announc. 2015;3:e00404–e00415. doi: 10.1128/genomeA.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munday J.S., Dunowska M., Laurie R.E., Hills S. Genomic characterization of canine papillomavirus type 17, a possible rare cause of canine oral squamous cell carcinoma. Vet. Microbiol. 2016;182:135–140. doi: 10.1016/j.vetmic.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Nagata M., Nanko H., Moriyama A., Washizu T., Ishida T. Pigmented plaques associated with papillomavirus infection in dogs. Is this epidermodysplasia verruciformis? Vet. Dermatol. 1995;6:179–181. doi: 10.1111/j.1365-3164.1995.tb00063.x. [DOI] [PubMed] [Google Scholar]
- 23.Narama I., Kobayashi Y., Yamagami T., Ozaki K., Ueda Y. Pigmented cutaneous papillomatosis (pigmented epidermal nevus) in three pug dogs; histopathology, electron microscopy and analysis of viral DNA by the polymerase chain reaction. J. Comp. Pathol. 2005;132:132–138. doi: 10.1016/j.jcpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchadr M.A., Huelsenbeck J.P. MeBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobler K., Favrot C., Nespeca G., Ackermann M. Detection of the prototype of a potential novel genus in the family Papillomaviridae in association with canine epidermodysplasia verruciformis. J. Gen. Virol. 2006;87:3551–3557. doi: 10.1099/vir.0.82305-0. [DOI] [PubMed] [Google Scholar]
- 26.Tobler K., Lange C., Carlotti D.N., Ackermann M., Favrot C. Detection of a novel papillomavirus in pigmented plaques of four pugs. Vet. Dermatol. 2008;19:21–25. doi: 10.1111/j.1365-3164.2007.00640.x. [DOI] [PubMed] [Google Scholar]
- 27.Ustav M., Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan H., Ghim S., Newsome J., Apolinario T., Olcese V., Martin M., Delius H., Felsburg P., Jenson B., Schlegel R. An epidermotropic canine papillomavirus with malignant potential contains an E5 gene and establishes a unique genus. Virology. 2007;359:28–36. doi: 10.1016/j.virol.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 29.Yuan H., Luff J., Zhou D., Wang J., Affolter V., Moore P., Schlegel R. Complete Genome Sequence of Canine Papillomavirus Type 9. J. Virol. 2012;86:5966. doi: 10.1128/JVI.00543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou D., Luff J., Usada Y., Affolter V., Moore P., Schlegel R., Yuan H. Complete Genome Sequence of Canine Papillomavirus Type 11. Genome Announc. 2014;2 doi: 10.1128/genomeA.00529-14. e00529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou D., Luff J., Paul S., Alkhilaiwi, Usada Y., Wang N., Affolter V., Moore P., Schlegel R., Yuan H. Complete Genome Sequence of Canine Papillomavirus Type 12. Genome Announc. 2015;3 doi: 10.1128/genomeA.00294-15. e00294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1 List of papillomaviruses included in the phylogenetic analysis.
Supplement 2 Genome size range with mean and standard deviation for canine papillomaviruses belonging to different genera.
Supplement 3 Phylogenetic trees of the individual ORFs E6, E7, E1, E2, L2 and L1 of the Chipapillomaviruses including ZcPV1, FcaPV2 and MnPV1 as outgroups. Obtained as described in material and methods.
Supplement 4 Comparison of phylogenetic trees based on the individual ORFs E6, E7, E1, E2, L2 and L1 of 15 papillomaviruses. Trees were obtained as described in material and methods and converted into Newick format (using FigTree software). For the comparison pairs of trees were imported into SpitsTree4 (version 4.14.4, Huson DH, Bryant D. 2006. Application of Phylogenetic Networks in Evolutionary Studies. Molecular Biology and Evolution. 23, 254–267) software and a consensus network analysis performed with default settings..