Abstract
Objectives
To investigate the frequency of Human papillomavirus (HPV)-driven head and neck squamous cell carcinoma (HNSCC) among patients living in North-East Italy, by assessing HPV-DNA positivity in all tumors and additional markers whenever possible.
Material and methods
HPV types, viral load, viral RNA, HPV16/18 E6 protein and p16INK4a and pRb expression were determined in primary tumor tissues from 247 HNSCC patients. Tumor-specific HPV seropositivity was analyzed in 102 patients.
Results
Tumor HPV-DNA prevalence was 8.5% overall (21/247) and 27% in oropharynx (17/63). HPV16 accounted for 95% of all HPV types found. Among HPV-DNA+ tumors, type-concordant HPV E6*I RNA prevalence was 79%. HPV DNA+ RNA+ tumors showed high viral load, up-regulated p16INK4a, down-regulated pRb and presence of HPV16 E6 protein. Eight cases showed tumor-specific HPV seropositivity, all type-concordant with the tumor. Tumors were defined as HPV-driven when positive for HPV-DNA plus 2 additional HPV transformation-related markers.
Conclusion
Relative prevalence of HPV-driven tumors (14 HPV16, 1 HPV58) was 6% overall and 20% among oropharyngeal cancers. In the oropharynx the HPV-driven group showed a trend for better survival versus the HPV-negative group.
The relative prevalence of HPV-driven oropharyngeal cancer is low in North-East Italy as compared to Western and Northern Europe.
Abbreviations: HNSCC, head and neck squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma; HPV, Human Papilloma Virus; CxCa, cervical carcinoma; FF, fresh-frozen; FFPE, formalin-fixed paraffin-embedded; PCR, polymerase chain reaction; MS, multiplex serology; GST, glutathione S-transferase; OS, overall survival; PFS, progression free survival
Keywords: Head and neck cancer, Oropharyngeal cancer, HPV-driven, HPV-related markers, HPV antibodies, Prevalence
Highlights
-
•
HPV DNA alone is not sufficient to demonstrate causality in HNSCC.
-
•
Additional transformation markers are needed to correctly identify HPV-driven tumors.
-
•
HPV prevalence in HNSCC shows large geographical variation, even within Europe.
-
•
Few small studies investigated the relative prevalence of HPV in HNSCC in Italy.
-
•
In North-East Italy 6% of HNSCC and 20% of OPSCC are HPV-driven.
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) accounts for approximately 5% of all cancers worldwide [1]. Globally, human papillomavirus (HPV) DNA was found in 30% of HNSCC, with a higher prevalence in oropharyngeal SCC (OPSCC) (46%) than oral SCC (24%) or laryngeal and hypopharyngeal SCC (22%) [2].
Among the 12 HPV types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59) defined as carcinogenic in cervical carcinoma (CxCa) by the International Agency for Research on Cancer (IARC), only type 16 is currently recognized as carcinogenic in OPSCC [3].
A wide variation of HPV DNA prevalence in OPSCC has been reported in the literature between regions and countries and within regions and countries, with overall figures higher in North America (60.4%) than in Europe (41.4%), and ranging from 56.5% for Northern to 24.2% for Southern countries within Europe [2].
While in the last decades HNSCC showed declining or unchanged incidence, an increase of HPV-associated HNSCC, particularly for the oropharynx, was observed [4]. In countries like USA, Denmark, Norway and Spain a 5% yearly increase (not statistically significant in Spain) of new OPSCC or tonsillar carcinoma cases was reported [5], [6], [7], [8]. In other surveys the increase was not observed [9], while for several areas data are still scarce.
However, only a subset of HPV DNA positive HNSCC display HPV carcinogenic activity in the tumor tissue, i.e. are HPV-driven. Thus, the presence of HPV DNA alone is not a sufficient proof for causal involvement of HPV in HNSCC tissue [10]. The HPV carcinogenic model derived from CxCa [11] was also recently confirmed for OPSCC [12].
Truly HPV-driven tumor cells contain at least one viral genome copy per cell and express the viral oncogenes E6 and E7 [11]. Interaction of the viral E7 and E6 proteins with key regulators of cell cycle and apoptosis leads to up-regulation of cellular protein p16INK4a [13] and down-regulation of tumor suppressor proteins pRb and p53. Patients with invasive HPV-driven cervical [14], penile [15] and oropharyngeal [16] SCC frequently show strong antibody responses against HPV E6 and E7 proteins while such antibodies are rare in tumor-free populations. Clinically, patients with HPV DNA positive HNSCC in comparison to those with HPV DNA negative HNSCC present a smaller tumor size, early nodal involvement, and are more frequently nonsmokers and non drinkers [17]. Controversial data were reported regarding younger age of HPV-positive HNSCC patients [18].
In Italy, HNSCC accounts for approximately 5% of all cancers [19]. The prevalence of HPV-DNA positive HNSCC cases reported so far has ranged from 10% [20] to 46% [21]. Most of the studies relied on qualitative HPV DNA markers alone, only a few included detection of viral transcripts [20], [22], [23] or the expression of the p16INK4a protein [24], [25].
In the present study, we provide new data on prevalence of HPV-driven HNSCC in North-East Italy, evaluate their clinical outcome and suggest a combination of markers to define the HPV-driven cases.
2. Patients and methods
2.1. Patients and samples
A total of 247 patients diagnosed with primary HNSCC located in oral cavity (International Classification of Diseases for Oncology (ICD-O): C02–04, C05.0, C06), oropharynx (ICD-O: C01, C05.1, C05.2, C09, C10), larynx (ICD-O: C32, C10.1) and hypopharynx (ICD-O C12–13) from 2003 to 2012 in the ENT (Ear Nose and Throat) Units of Treviso Regional Hospital, Hospital of Mirano and Hospital of Cattinara (Trieste), North-East Italy, were enrolled in the study. The first 77 enrolled patients have been already reported [20]. The whole study was approved by the ethic committee for clinical experimentation (CEP) of Treviso (Ethic votes: 345/AULSS9 and 421/AULSS9). All patients signed an informed consent.
Of the enrolled patients, 75% were males, 36% never tobacco smokers and 30% never alcohol drinkers. Of the tumors, 39% were in the larynx, 26% each in the oropharynx and the oral cavity and 10% in the hypopharynx. Small-sized T category (T1-T2) was present in 50% of the patients, with regional lymph nodes being involved in 52%. Only 1% of patients had distant metastases at diagnosis date.
From all the enrolled patients we obtained fresh frozen (FF) specimens of the neoplastic lesions. Formalin-fixed paraffin-embedded (FFPE) tumor tissues of 54 (22%) cases were retrieved from one of the three participating centres (Treviso), and used to complete molecular analyses of HPV-DNA-positive cases with small FF samples, and to perform immunohistochemistry. Sera were collected starting in 2010 from a total of 102 (41%) patients. Anatomical and clinical data were collected and available from the clinical database for each patient. No differences were observed between the above-mentioned subgroups and the whole case series. Histological diagnoses were determined by the local pathologists. Management decisions for the patients were not guided by knowledge of HPV status or other tested markers.
2.2. Tissues sectioning and DNA analyses
HPV analyses were carried out partly at the Veneto Institute of Oncology and partly at the German Cancer Research Center; homogenization in liquid nitrogen of the cryosections [26] was performed only in the German Center. The mean tumor content on adjacent hematoxylin-and-eosin stained sections was 55% (range 25–80%).
DNA was isolated from all 247 FF tumor tissues by Phenol-Chlorophorm (PC9) method as described in [27], [28]. One-hundred-fourteen samples were re-sectioned and DNA was re-extracted by MagNA Pure 96 DNA and viral NA Large Volume Kit (Roche, Penzberg, Germany) according to the manufacturer´s recommendations [26], [29].
FFPE sectioning and genomic DNA extraction were performed as described in [29].
PC9-extracted DNA was tested by MY09/MY11 PCR and restriction fragment length polymorphism (RFLP) analysis of the amplification products [30].
MagNA Pure-extracted DNA was analyzed by BSGP5+/6+-PCR/MPG51 assay, a broad-spectrum PCR for 51 mucosal HPV types followed by hybridization to type-specific probes on Luminex beads [31], [32].
DNA quality of samples tested by MY09/MY11 was verified by amplification of the β-globin gene; the analytical sensitivity ranges from 102 to 104 HPV copies for any of the 49 HPV mucosal types [33]. All samples were also analyzed by HPV16 type-specific PCR [20].
The analytical sensitivity for any of the 51 HPV types detected by the BSGP5+/6+-PCR/MPG51 assay is equal or less than 100 viral genome copies [29].
HPV16 viral loads were determined by quantitative (q)PCR targeting E6 gene sequences (Schmitt et al. submitted) and expressed as HPV16 genome copy-number per cell. A cut-off of 0.1 copies per cell defined high (HPV16high≥0.1) or low (HPV16low<0.1) viral load. Real-time qPCR for β-globin was used to verify the DNA quality and to measure the amount of input DNA. The analytical sensitivity for both HPV16 and β-globin detection is below 100 plasmid copies.
HPV58 viral load was estimated from the quantitative BSGP5+/6+- PCR/MPG51 data as described in [32].
2.3. RNA isolation and E6*I mRNA reverse transcription (RT)-PCR
RNA was isolated from FF sections using the RNeasy Mini kit (Qiagen, Hilden, Germany) and from FFPE sections using the Pure Link FFPE Total RNA Isolation Kit (Invitrogen, Carlsbad, CA). DNase I digestion (RNase-free DNase Set, Qiagen) was included to ensure an exclusive amplification of RNA, and was carried out on the RNA purifying columns during sample processing.
All samples HPV DNA positive by either method and a group of HPV DNA negatives were analyzed for ubiquitin C and HPV16 E6*I RNA and for E6*I RNA of additional HPV types positive by HPV genotyping [29].
2.4. Immunohistochemistry for cellular proteins p16INK4a and pRb
Immunostaining was performed on FFPE sections to evaluate the expression of the HPV-targeted cellular proteins p16INK4a and pRb, as previously described in detail [12]. p16INK4a protein was detected using the primary antibody CINtec for V-kit (MTM laboratories, Heidelberg, Germany) while pRb protein was detected using the NCL-RB (Novocastra, Newcastle upon Tyne, United Kingdom) antibody. Stained sections were evaluated by two investigators (L.B. and D.H.), applying two protein expression categories, low and high. The cut-off for protein up- or down-regulation was determined by evaluation of the staining intensity and stain distribution in sections of healthy uvula (nuclear and cytoplasmic signal absent for p16INK4a and abundant for pRb) analyzed in parallel with cervical carcinoma tissues (nuclear and cytoplasmic signal present for p16INK4a and <25% for pRb). For initially discordant cases a consensus was reached after joined review and discussion.
2.5. Multiplex serology
The presence of HPV antibodies against the E6 and E7 proteins of high-risk HPV types 16, 18, 31, 33, 45, 52 and 58 and low-risk types 6 and 11 was investigated by multiplex serology (MS), a glutathione S-transferase (GST) capture immunosorbent-based assay combined with fluorescent bead technology as previously described [34].
2.6. HPV16 E6 protein detection
E6 protein of HPV16 and HPV18 was analyzed in lysates obtained from FF tumor sections by the OncoE6™ test (Arbor Vita Corporation, Fremont) following the manufacturer's instructions [35]. The assay is an immunographic assay which consists in the capture of the E6 protein with monoclonal antibodies and its detection through colorimetric visualization. The analytical sensitivity of the assay, as determined in 10-fold dilution series of the HPV16 positive cervical carcinoma cell line MRI-H 186, was less than 5×103 cells (Holzinger et al., manuscript in preparation).
2.7. Statistical analyses
Patient and tumor characteristics were evaluated in relation to the HPV DNA status. Follow-up was determined as the time-difference between the date of primary tumor diagnosis and the last clinical examination.
Overall survival (OS) was determined as time interval from date of diagnosis until date of death (by any cause). Progression-free survival (PFS) time was determined as time interval from the date of primary tumor diagnosis to the date of the first negative event (local or regional recurrence, metastasis, second primary HNSCC) or death from any cause.
The Kaplan-Meier method was used to estimate survival distributions. Patients with HPV-driven and HPV non-driven tumors were compared using the log-rank test. In all statistical tests a P value of 0.05 or below was considered as statistically significant. The statistical analyses were carried out using SAS as well as the Sigmaplot softwares.
3. Results
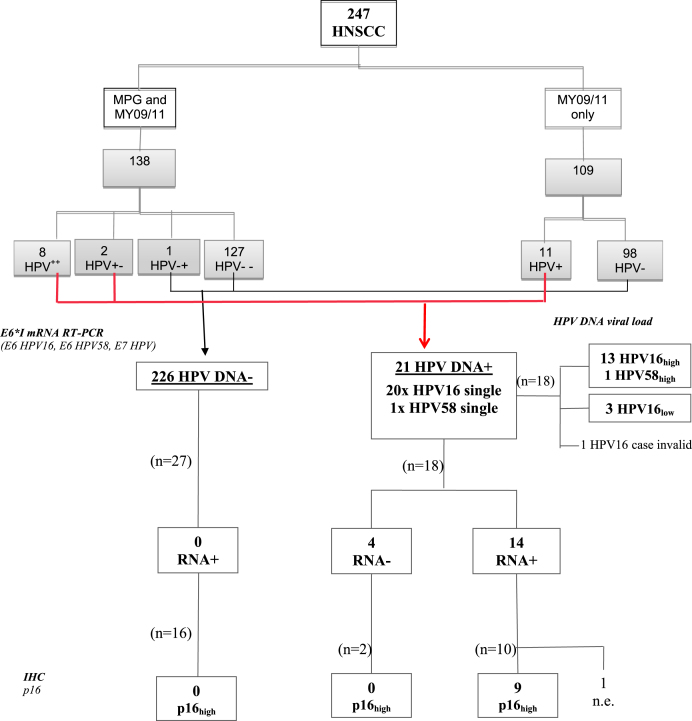
The anatomo-clinical characteristics of the 247 HNSCC enrolled patients (age range 27–95 years) are described in Table 1. The main results for HPV-DNA, HPV-RNA and p16 IHC are illustrated in Fig. 1.
Table 1.
Clinicopathologic characteristics of the 247 study participants stratified by HPV DNA and by HPV-driven status.
| Total | HPVDNA_ | HPVDNA+ | HPV-driven | ||
|---|---|---|---|---|---|
| Characteristicsa | N=247 | N=226 | N=21 | N=15/19 tested | |
| Age (Median, y) | 65 | 65 | 68 | 68 | |
| N (%) | N (%) | N (%) | N (%) | ||
| Gender | Male | 188 (76) | 173 (76) | 15 (71) | 9 (60) |
| Female | 59 (24) | 53 (24) | 6 (29) | 6 (40) | |
| Tobaccob | Never | 79 (32) | 69 (31) | 10 (48) | 8 (53) |
| Former | 34 (15) | 30 (13) | 4 (19) | 3 (20) | |
| Current | 124 (50) | 118 (52) | 6 (28) | 3 (20) | |
| Missing | 10 (4) | 9 (4) | 1 (5) | 1 (7) | |
| Alcoholb | Never | 75 (30) | 69 (31) | 6 (28) | 5 (33) |
| Former | 29 (12) | 26 (11) | 3 (14) | 1 (7) | |
| Current | 131 (53) | 120 (53) | 11 (53) | 8 (53) | |
| Missing | 12 (5) | 11 (5) | 1 (5) | 1 (7) | |
| Localization | Oral cavity | 63 (26) | 62 (28) | 1 (5) | 1 (7) |
| Oropharynx | 63 (26) | 46 (20) | 17 (81) | 13 (86) | |
| Larynx | 96 (39) | 95 (42) | 1 (5) | 1 (7) | |
| Hypopharynx | 25 (9) | 23 (10) | 2 (9) | 0 | |
| Tumor size | T1-T2 | 124 (50) | 113 (50) | 11 (52) | 8 (53) |
| T3-T4 | 123 (50) | 113 (50) | 10 (48) | 7 (47) | |
| Lymph nodes | N0 | 128 (52) | 123 (54) | 5 (24) | 4 (27) |
| N+ | 119 (48) | 103 (46) | 16 (76) | 11 (73) | |
| Metastasis | M0 | 244 (99) | 223 (99) | 21 (100) | 15 (100) |
| M+ | 3 (1) | 3 (1) | 0 | 0 | |
P-value (by chi-square analyses) referred to the HPV-DNA+ and HPV-DNA- groups was statistically significant (0.0001) only for oropharyngeal localization.
For statistical analyses, tobacco and alcohol categories were simplified in never and current (which includes current and former) tobacco/alcohol consumers.
Fig. 1.
Flow chart of the most significant analyses (i.e. E6*I mRNA RT-PCR,HPV viral load by HPV16 ultrashort-qPCR or MPG MFI, and p16 IHC) performed on the tumors of the 247 HNSCC enrolled patients, by HPV-DNA status. 138/247 (56%) samples were double tested for HPV-DNA by BSGP5+/6+-PCR/MPG51 (MPG) and MY09/MY11-RFLP (MY), and 109/247 (44%) by MY only. HPV++ and HPV−−: sample positive or negative by both methods; HPV+ and HPV−: sample positive or negative by one method only. HPV+− and HPV−+: sample positive/negative or negative/positive by MPG and MY, respectively. Overall, 3 cases were discordant (2 HPV+− and 1 HPV−+); considering that the MPG sensitivity is higher compared to the MY assay, the 2 HPV+− and 1 HPV−+ were considered as positive and negative, respectively. HPV16high: >0.1 HPV16 copies per cell; HPV16low: <0.1 HPV16 copies per cell. p16 high: nuclear and cytoplasmic signal present in most tumor cells.
3.1. Human papillomavirus DNA and RNA prevalence
All DNA samples extracted from FF and FFPE tissues were β-globin positive in the genotyping assays and thus valid for HPV DNA analyses. All the 247 DNA samples were tested by MY09/MY11 PCR-RFLP, and 114 were additionally tested by BSGP5+/6+ PCR/MPG51. Agreement between the two methods calculated for the latter group resulted in a k-value of 0.8 (good agreement).
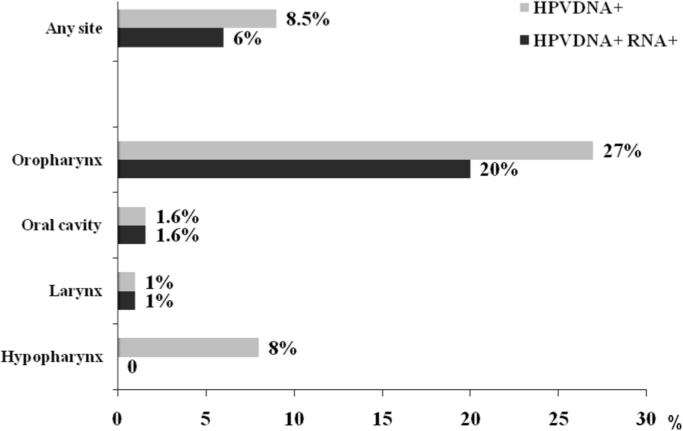
HPV DNA prevalence was 8.5% (21/247) (Fig. 2). HPV type 16 accounted for 95% (20/21) of HPV DNA positive cases, and HPV58 was identified once (5%). No multiple infections were identified.
Fig. 2.
Overall and site specific HPV DNA and RNA prevalence among all head and neck tumors and stratified by anatomical site. Overall, 247 tumors were analyzed for HPV DNA, 63 from the oropharynx, 63 from the oral cavity, 96 from the larynx, and 25 from the hypopharynx. Of the 21 HPVDNA positive tumors, 18 were analyzed for HPV RNA, while 3 (2 from oropharynx and 1 from larynx) could not be analyzed.
HPV DNA prevalence was highest in the oropharynx (27%, 17/63) (P<0.0001) and low in hypopharynx (8%, 1/25), oral cavity (2%, 1/63) and larynx (1%, 1/96).
A total of 45 RNA samples from 18 HPV DNA positive (17 for HPV16 and 1 for HPV58) and 27 HPV DNA negative cases were prepared for HPV16 E6*I and ubiquitin C RNA analyses. Of 3 HPV DNA positive cases (2 oropharynx, 1 larynx) no sufficient tumor sample was available for RNA extraction. All RNA samples were valid (ubiquitin C RNA positive). All 27 HPV DNA negative tumors analyzed showed no presence of HPV16 transcripts.
Among the 17 HPV16 DNA positive samples, 4 were HPV16 RNA negative and 13 showed expression of type-concordant HPV16 E6*I RNA; the single case identified as HPV58 DNA positive showed presence of HPV58 E6*I transcripts and absence for HPV16 E6*I transcripts (Fig. 2). Therefore, 14/18 (77.8%) HPV-DNA-positive samples were also RNA-positive.
Compared to the other head and neck anatomical sites, the oropharynx with a figure of 20% (12/61) showed the highest prevalence of HPV DNA and RNA positive tumors, followed by 1.6% (1/63) in the oral cavity, 1% (1/94) in the larynx, and 0% (0/25) in the hypopharynx.
3.2. Additional markers of HPV transformation (Table 2)
Table 2.
Definition of HPV-driven HNSCC. From left: anatomic site, HPV types identified, HPV type of E6*I RNA, viral load by HPV16 qPCR (copies/cell), p16INK4a up-regulation, pRb down-regulation, antibodies to HPV16 and HPV58 E6 and E7 proteins, HPV16 E6 protein, final HPV status. Positivity for individual HPV transformation markers highlighted in bold.
| Nr. | Anatomic site |
HPV type |
Viral load | p16INK4aup-regulation | pRb down-regulation |
HPV16/58 |
HPV16 E6 protein | HPV-driven | ||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA+ | RNA+ | Ab E6+ | Ab E7+ | |||||||
| 1 | Oropharynx | 16 | 16 | 1.3 | yes | yes | ||||
| 2 | Oropharynx | 16 | 16 | 1000 | no | yes | ||||
| 3 | Oropharynx | 16 | 16 | 26 | yes | yes | yes | |||
| 4 | Oropharynx | 16 | 16 | 24 | yes | no | yes | yes | ||
| 5 | Oropharynx | 16 | 16 | 40 | yes | yes | ||||
| 6 | Oropharynx | 16 | 16 | 3 | yes | no | yes | |||
| 7 | Oropharynx | 16 | 16 | 36 | yes | yes | yes | yes | yes | yes |
| 8 | Larynx | 16 | 16 | 25 | yes | yes | yes | yes | yes | yes |
| 9 | Oropharynx | 16 | 16 | 285 | yes | yes | yes | yes | yes | |
| 10 | Oropharynx | 16 | 16 | 145 | yes | no | yes | yes | yes | |
| 11 | Oropharynx | 16 | 16 | 0.6 | yes | yes | yes | yes | ||
| 12 | Oropharynx | 16 | 16 | 37 | yes | yes | yes | yes | ||
| 13 | Oropharynx | 58 | 58 | higha | yes | yes | yes | yes | yes | yes |
| 14 | Oral cavity | 16 | 16 | invalid | yes | yes | yes | |||
| 15 | Oropharynx | 16 | yes | yes | yes | |||||
| 16 | Hypopharynx | 16 | no | 208 | no | no | no | |||
| 17 | Oropharynx | 16 | no | <0.001 | no | no | ||||
| 18 | Hypopharynx | 16 | no | <0.001 | no | no | ||||
| 19 | Oropharynx | 16 | no | <0.001 | no | no | ||||
| 20b | Oropharynx | 16 | n.d | |||||||
| 21b | Oropharynx | 16 | n.d | |||||||
High=tested by quantitative BSGP5+/6+-PCR MPG51 assay [32]; empty=sample not tested; Ab=antibody; n.d=not defined.
Tumor cases 20 and 21 are not defined since no additional HPV transformation-associated data could be obtained.
3.2.1. HPV16 viral load
One out of 18 HPV DNA positive samples was β-globin DNA negative (invalid). A high viral load (>0.1 HPV16 genome copies/tumor cell and/or high MPG MFI) was present in 82% (14/17) of HPV DNA positive valid samples (median =33.7 HPV16 genome copies per cell, range 0.6–1000). The other 3 carcinomas contained <0.002 HPV genome copies/tumor cell. All the HPV RNA positive tumors had a high viral load. One HPV16 high viral load tumor (Nr. 16) was HPV RNA negative and showed no p16INK4a up-regulation nor pRb down-regulation.
3.2.2. p16INK4a and pRb expression
Of the 54 tumors stained for p16INK4a, 2 were excluded (1 case not evaluable for tumor section loss during staining and no additional material available, and the other for lack of tumor cells in the stained section). Out of 11 HPV DNA positive tumors with valid results analyzed, p16INK4a up-regulation was recorded in 9, all HPV RNA positives, while the 2 HPV RNA negatives were p16-negative. Among the 41 HPV DNA negative tumors, one case showed p16INK4a up-regulation.
pRb expression was analyzed in 48 cases; 1 case was excluded due to lack of tumor cells in the stained section. Of the 10 HPV DNA and RNA positive tumors, only 6 showed pRb down-regulation. No pRb down-regulation was observed in the HPV DNA positive RNA negative tumor and in 35 of 36 HPV DNA negative tumors.
3.2.3. HPV16 E6 protein expression
HPV16 E6 protein was detected in lysates of 7/7 analyzed tumors positive for high viral load HPV16 DNA and for E6*I RNA. Two low viral load HPV16 DNA positive but HPV16 RNA negative tumors showed no HPV16 E6 protein.
3.2.4. HPV antibodies
All the 7 HPV DNA and HPV RNA positive tumors analyzed by serology showed antibodies to both HPV E6 and E7 proteins of the tumor-infecting HPV type. Anti-E6 and -E7 double seropositivity was also observed in one additional case with HPV16 DNA positive tumor for which the other tissue markers could not be analyzed and among 92 HPV DNA negative tumors.
3.3. Definition of HPV-driven HNSCC
The full spectrum of HPV transformation-associated markers analyzed includes high viral load, HPV RNA expression, p16INK4a up-regulation, pRb down-regulation, HPV E6 protein and seropositivity to HPV E6 and E7 of the tumor infecting HPV type (Table 2). Cases were defined as HPV-driven when positive for HPV DNA and at least 2 additional HPV transformation-associated markers.
Due to limited tissue availability, 2 out of 21 HPV DNA positive cases could not be further analyzed and therefore could not be defined.
Of 19 HPV DNA positive tumors analyzed for additional HPV transformation markers, 15 (79%) were HPV-driven and 4 non-HPV-driven.
3.4. Patients’ survival after HNSCC diagnosis
The cut-off date for follow-up was June 30, 2013 and follow-up time was 1–121 months (median 20 months).
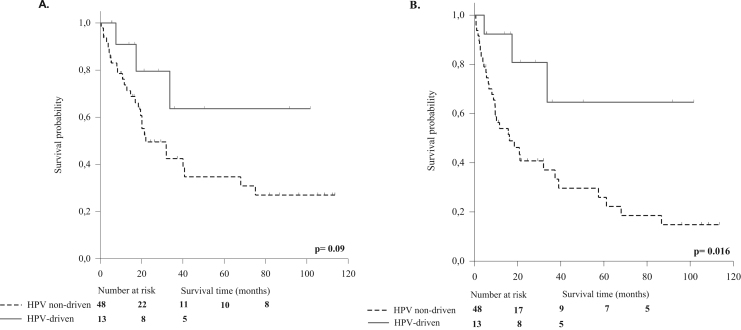
Of the 247 patients, 152 (61.5%) were alive at time of analysis; death had occurred in overall 95 (38.5%) patients, in 53 due to the primary tumor, in 20 due to a second malignancy, while in 22 was not tumor-related. A negative event (local, regional or distant recurrent HNSCC or second primary tumor) was recorded for 93 (37.7%) patients. Overall, a difference in clinical outcome was recorded between patients with HPV-driven and HPV-non driven tumors (statistically not significant, data not shown). Restricting the analyses to the oropharynx site (i.e., 61 cases on which HPV causality was assessed), a trend of better survival (OS and PFS, statistically significant for PFS) was observed in patients with HPV-driven tumors (13 cases) compared to the group (48 cases) with HPV non-driven tumors (Fig. 3, A and B).
Fig. 3.
A. Overall survival (OS) and B. Progression free survival (PFS) of OPSCC patients by HPV-driven and HPV non-driven status (assessed in 61/63 cases). The continuous line refers to patients with HPV-driven OPSCC, the dotted line refers to patients with HPV-non driven OPSCC. The patients with HPV-driven tumors showed a trend (P=0.09) for better OS (A) and a significant (P=0.016) difference for PFS (B).
4. Discussion
We enrolled 247 patients with primary HNSCC and analyzed the neoplastic tissue of all cases for HPV DNA. All available HPV DNA positive tumors and a randomly selected group of HPV DNA negative tumors were further tested for presence of HPV-transformation-associated markers, i.e. high viral load, HPV RNA expression, p16INK4a up-regulation, pRb down-regulation, HPV E6 protein, and seropositivity to HPVE6 and E7 of the tumor infecting HPV type.
The overall HPV DNA prevalence was low for all HNSCC and also among the oropharyngeal cancers (8.5% and 27%, respectively) compared to the reported global HPV DNA prevalences (30% and 46%, respectively) [2], but similar to the rates for HNSCC reported in Spain (6%) [8] and France (13%) [36].
HPV prevalence among patients with HNSCC in Italy was analyzed in a low number of studies, that reported a highly variable range (from 10% [20] to 46% (by nested PCR) [21]).
The low HPV DNA prevalence observed in our HNSCC series is probably due to the actual main role of tobacco smoking and alcohol consumption in this area [37], [38], where programmes for tobacco use and alcohol abuse prevention have been less efficient than in Northern America and Northern Europe. In fact, in our series most (>60%) of the patients were current tobacco smokers and/or alcohol drinkers. In relation to these risk factors, we could see a significantly higher prevalence of HPV in never tobacco smokers (12.4% (14/113) vs 4.8% (6/124); P<0.05) but not in never alcohol drinkers (P>0.05) (data not shown). A study conducted in Germany [39] showed a similar proportion of smokers and a 40% HPV prevalence among OPSCC, with a statistically significant higher HPV prevalence among never/ex-smokers compared to current smokers.
With 93%, HPV16 was identified as the most prevalent type in HPV-driven HNSCC in contrast to CxCa in which HPV16 contributes to about 60% of the HPV-driven tumors, with other HPV types such as 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59 driving the remaining CxCa [2], [29]. In our case series, we identified one case with HPV58 as single type infection and could demonstrate its causal involvement by a broad variety of viral and cellular markers, as described in detail previously [40].
The presence of HPV DNA alone is not sufficient to determine causality in HNSCC. For cervical carcinoma [29], and recently for oropharyngeal [16] and laryngeal cancer [41], several markers, used alone or combined, were established as stringent criteria to discriminate HPV-driven from HPV non-driven cases. For HNSCC in Italy, the analysis of HPV prevalence by using at least one additional HPV transformation-specific marker is reported only in a few studies [20], [22].
The most accurate marker to demonstrate viral activity and the transforming capability of HPV in the tissues is considered to be the detection of viral mRNA encoding the two viral oncoproteins E6 and E7 [26].
RNA of sufficient quality is often difficult to obtain especially when no fresh frozen material is available. We could use an ultra-short E6*I RNA assay adequate to successfully analyze also FFPE specimens. To increase transformation-specificity, in addition to RNA analysis, we analyzed several HPV-transformation specific (direct and/or indirect) markers that allowed us to robustly discriminate HPV-driven from HPV non-driven tumors, and allowed for rare technical failures of one method in individual cases. Depending on the availability of the sample, we could not further analyze two HPV DNA positive samples.
We defined as HPV-driven only tumors positive for HPV DNA and at least 2 additional HPV-transformation associated markers. With such cut-off, 79% of DNA positive tumors were confirmed as HPV-driven [2], [26].
In line with the literature [26], we observed a good concordance between presence of viral transcripts and high viral load (at the cut-off of 0.1 HPV genome copies per tumor cell) and with detection of the newly introduced marker E6 protein (93% and 100%, respectively).
E6 protein could represent a good marker to identify HPV-driven carcinomas. Although we could test the presence of the E6 protein only in a limited number (9) of cases, we observed a 100% concordance with HPV RNA positivity. In a larger set of HPV-driven and HPV non-driven HPV16 DNA positive OPSCC (Holzinger et al., manuscript in preparation), sensitivity and specificity of HPV16 E6 detection to classify HPV16-driven OPSCC was 97% and 96%, respectively, showing an excellent agreement with the HPV RNA status (κ=0.94).
Expression of HPV E6 oncoprotein reduces the steady state level of p53 that induces upregulation of p16INK4a protein expression via a negative feedback mechanism. Conversely, expression of the HPV E7 protein leads to pRb degradation. Therefore, in HPV-driven OPSCC p16INK4a is almost always up-regulated while pRb is frequently albeit not always down-regulated, making these 2 proteins of potential use as indirect markers [12].
In our series, p16INK4a protein showed a higher sensitivity (100%) but a lower specificity (75%) compared to pRb protein (60% and 97.3%, respectively) [42]. Comparing up-regulation of p16INK4a and down-regulation of pRb (as single markers) with HPV RNA positivity, we had a concordance of 100% and 60%, respectively. p16INK4a protein is considered to be a reliable surrogate marker in CxCa [43], and other HPV-associated anogenital cancers [44]. In HNSCC, p16INK4a was a more reliable marker in tumors arising from the oropharynx [10], [45] and larynx [41] than in hypopharyngeal [46] and oral cavity carcinoma [47]. Nevertheless, a fraction of oropharyngeal tumors have up-regulated p16INK4a but lack HPV DNA positivity, and are similar to the non-HPV associated oropharyngeal tumors [48]. In our study we could not test all the HPV-negative samples, so we cannot affirm that all the HPV-negative tumors were also p16 negative.
HPV type concordant E6 and E7 double antibody positivity has been demonstrated to be highly associated and specific for invasive HPV-associated cancers with an extremely low prevalence of about 0.1% in tumor-free individuals [16] and to be strongly associated (odds ratios of 44-180) with invasive cancer of the cervix [14], the penis [15] and the upper aerodigestive tract [16]. Interestingly, among the 102 patients for whom anti-HPV antibodies could be assessed, the sera from all the 7 patients with HPV RNA positive tumors showed strong positive antibody responses against both type-concordant HPVE6 and E7 proteins. In contrast, none of the sera from 92 patients with HPV DNA negative tumors reacted positive with both E6 and E7 for any of the 7 high-risk and 2 low-risk HPV types tested. In this study, double HPVE6 and E7 seropositivity was the best marker to identify the type concordant HPV RNA positive cases in the tumor, with a 100% sensitivity and 100% specificity.
Based on the correlations found for each of the analyzed markers, we defined as HPV-driven all the samples that were positive for HPV DNA and 2 additional HPV-transformation related markers.
Among the 19 cases with HPV DNA positive tumors evaluated by this algorithm, 15 (79%) were defined as HPV-driven, confirming that HPV is not a causal pathogenic factor in all HPV DNA positive HNSCC [10], [12].
Considering the whole HNSCC cohort, we could not see a better survival in the HPV-driven group, probably due to the low number of HPV-driven cases that might influence the statistical power. Nonetheless, survival analyses restricted to patients with OPSCC showed a trend of better survival for patients with HPV-driven tumors.
Due to the low number of tumors from the oropharyngeal site, the analyses of a multivariate Cox regression and HPV prevalence over time in the Italian northern region could not be performed.
The results of our study confirm the low prevalence of HPV-driven HNSCC in North-East-Italy, in line with data from neighboring countries (like Spain), with a significant role of HPV16 only in the oropharynx. Moreover, we highlight the importance of good classification of the truly HPV-driven tumors. Although the additional markers could not be analyzed in all HNSCC patients (representing a limitation of the study), the correlations are strong and point to the use of an algorithm to correctly identify the truly HPV-driven HNSCC.
Author's disclosures of potential conflicts of interest
The authors L. Baboci, D. Holzinger, P. Boscolo-Rizzo, G. Tirelli, R. Spinato, R. Fuson, V. Lupato, A. Michel, G. Halec, M.C Da Mosto, A. Del Mistro, disclosed no potential conflicts of interest. M. Pawlita and M. Schmitt have received royalties for patents owned by DKFZ and research support through cooperation contracts of DKFZ with Roche and Qiagen in the field of HPV diagnostics.
Acknowledgments
This work was supported by Grant no. FP7-HEALTH-2011-282562 from the European Commission (HPV-AHEAD), by Veneto Institute of Oncology (IOV) - IRCCS fellowship and by Lega Italiana per la Lotta contro i Tumori (LILT). The funders had no role in study design, data collection, data analysis, data interpretation or writing of the report, nor in the decision to submit for publication.
We appreciate the excellent technical assistance of R. Trevisan, A. Leischwitz, U. Koch and M. Oppenländer.
Contributor Information
Lorena Baboci, Email: lorena.baboci@gmail.com.
Dana Holzinger, Email: d.holzinger@dkfz.de.
Paolo Boscolo-Rizzo, Email: paolo.boscolorizzo@unipd.it.
Giancarlo Tirelli, Email: tirelli@units.it.
Roberto Spinato, Email: robertospinato@hotmail.com.
Valentina Lupato, Email: valentinalupato@gmail.com.
Roberto Fuson, Email: roberto.fuson@gmail.com.
Markus Schmitt, Email: markus-schmitt@dkfz-heidelberg.de.
Angelika Michel, Email: a.michel@dkfz-heidelberg.de.
Gordana Halec, Email: g.halec@dkfz.de.
Maria Cristina Da Mosto, Email: mariacristina.damosto@unipd.it.
Michael Pawlita, Email: m.pawlita@dkfz-heidelberg.de.
Annarosa Del Mistro, Email: annarosa.delmistro@ioveneto.it.
References
- 1.J. Ferlay, E. Steliarova-Foucher and J. Lortet-Tieulent et al., Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012, Eur. J. Cancer 49, 2013, 1374–1403 [DOI] [PubMed]
- 2.Ndiaye C., Mena M., Alemany L. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: A systematic review and meta-analysis. Lancet Oncol. 2014;15(12):1319–1331. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 3.IARC. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum 2011; 100B: 1–475. A review of human carcinogens—Part B: biological agents, vol. 100B, 2011. IARC Monogr Eval Carcinog Risks Hum 100B (2011) 1–475.
- 4.Chaturvedi A.K., Anderson W.F., Lortet-Tieulent J. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturgis E.M., Ang K.K. The epidemic of HPV-associated oropharyngeal cancer is here: Is it time to change our treatment paradigms? J. Natl. Compr. Cancer Netw. 2011;9(6):665–673. doi: 10.6004/jnccn.2011.0055. [DOI] [PubMed] [Google Scholar]
- 6.Blomberg M., Nielsen A., Munk C. Trends in head and neck cancer incidence in Denmark, 1978–2007: Focus on human papillomavirus associated sites. Int. J. Cancer. 2011;129(3):733–741. doi: 10.1002/ijc.25699. [DOI] [PubMed] [Google Scholar]
- 7.Mork J., Moller B., Dahl T. Time trends in pharyngeal cancer incidence in Norway 1981–2005: a subsite analysis based on a reabstraction and recoding of registered cases. Cancer Causes Control. 2010;21(9):1397–1405. doi: 10.1007/s10552-010-9567-9. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigo J.P., Heideman D.A., Garcia-Pedrero J.M. Time trends in the prevalence of HPV in oropharyngeal squamous cell carcinomas in northern Spain (1990–2009) Int. J. Cancer. 2014;134(2):487–492. doi: 10.1002/ijc.28355. [DOI] [PubMed] [Google Scholar]
- 9.van Monsjou H.S., Schaapveld M., van den Brekel M.W.M., Balm A.J.M. The epidemiology of head and neck squamous cell carcinoma in The Netherlands during the era of HPV-related oropharyngeal squamous cell carcinoma. Is there really evidence for a change? Oral. Oncol. 2015;51:901–907. doi: 10.1016/j.oraloncology.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Castellsagué X., Alemany L., Quer M. HPV involvement in head and neck cancers: Comprehensive assessment of biomarkers in 3680 patients. J. Natl. Cancer Inst. 2016;108(6) doi: 10.1093/jnci/djv403. djv403. [DOI] [PubMed] [Google Scholar]
- 11.zur Hausen H. Perspectives of contemporary papillomavirus research. Vaccine. 2006;24(Suppl. 3) doi: 10.1016/j.vaccine.2006.06.054. S3/iii-iv. [DOI] [PubMed] [Google Scholar]
- 12.Holzinger D., Flechtenmacher C., Henfling N. Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int. J. Cancer. 2013;133(6):1389–1399. doi: 10.1002/ijc.28142. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M., Tribius S., Quabius E.S. HPV DNA, E6*I-mRNA expression and p16INK4A immunohistochemistry in head and neck cancer - how valid is p16INK4A as surrogate marker? Cancer Lett. 2012;323(1):88–96. doi: 10.1016/j.canlet.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Meschede W., Zumbach K., Braspenning J. Antibodies against early proteins of human papillomaviruses as diagnostic markers for invasive cervical cancer. J. Clin. Microbiol. 1998;36(2):475–480. doi: 10.1128/jcm.36.2.475-480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heideman D.A., Waterboer T., Pawlita M. Human papillomavirus-16 is the predominant type etiologically involved in penile squamous cell carcinoma. J. Clin. Oncol. 2007;25(29):4550–4556. doi: 10.1200/JCO.2007.12.3182. [DOI] [PubMed] [Google Scholar]
- 16.Anantharaman D., Gheit T., Waterboer T. Human papillomavirus infections and upper aero-digestive tract cancers: the ARCAGE study. J. Natl. Cancer Inst. 2013;105(8):536–545. doi: 10.1093/jnci/djt053. [DOI] [PubMed] [Google Scholar]
- 17.Lassen P. The role of Human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother. Oncol. 2010;95(3):371–380. doi: 10.1016/j.radonc.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Sethi S., Ali-Fehmi R., Franceschi S. Characteristics and survival of head and neck cancer by HPV status: a cancer registry-based study. Int. J. Cancer. 2012;131(5):1179–1186. doi: 10.1002/ijc.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AIOM. Linee guida tumori della testa e del collo, 2013 (in Italian).
- 20.Boscolo-Rizzo P., Da Mosto M.C., Fuson R. HPV-16 E6 L83V variant in squamous cell carcinomas of the upper aerodigestive tract. J. Cancer Res. Clin. Oncol. 2009;135(4):559–566. doi: 10.1007/s00432-008-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rittà M., De Andrea M., Mondini M. Cell cycle and viral and immunologic profiles of head and neck squamous cell carcinoma as predictable variables of tumor progression. Head Neck. 2009;31(3):318–327. doi: 10.1002/hed.20977. [DOI] [PubMed] [Google Scholar]
- 22.Venuti A., Badaracco G., Rizzo C. Presence of HPV in head and neck tumours: high prevalence in tonsillar localization. J. Exp. Clin. Cancer Res. 2004;23(4):561–566. [PubMed] [Google Scholar]
- 23.Perrone F., Suardi S., Pastore E. Molecular and cytogenetic subgroups of oropharyngeal squamous cell carcinoma. Clin. Cancer Res. 2006;12(22):6643–6651. doi: 10.1158/1078-0432.CCR-06-1759. [DOI] [PubMed] [Google Scholar]
- 24.Pannone G., Rodolico V., Santoro A. Evaluation of a combined triple method to detect causative HPV in oral and oropharyngeal squamous cell carcinomas: P16 Immunohistochemistry, Consensus PCR HPV-DNA, and In Situ Hybridization. Infect. Agent. Cancer. 2012;7:4. doi: 10.1186/1750-9378-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morbini P., Dal Bello B., Alberizzi P. Oral HPV infection and persistence in patients with head and neck cancer. Oral Surg, Oral Med, Oral Pathol. Oral Radiol. 2013;116(4):474–484. doi: 10.1016/j.oooo.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Holzinger D., Schmitt M., Dyckhoff G. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res. 2012;72(19):4993–5003. doi: 10.1158/0008-5472.CAN-11-3934. [DOI] [PubMed] [Google Scholar]
- 27.Kirby K.S. A new method for the isolation of ribonucleic acids from mammalian tissues. Biochem. J. 1956;64(3):405–408. doi: 10.1042/bj0640405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Mistro A., Bonaldi L., Bertorelle R. Genital human papillomavirus types in immunocompetent and immunodepressed women in northeast Italy: Prevalence and cytomorphological correlations. J. Low Genit. Tract. Dis. 2001;5(1):12–20. [PubMed] [Google Scholar]
- 29.Halec G., Schmitt M., Dondog B. Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. Int. J. Cancer. 2013;132(1):63–71. doi: 10.1002/ijc.27605. [DOI] [PubMed] [Google Scholar]
- 30.Nobre R.J., de Almeida L.P., Martins T.C. Complete genotyping of mucosal human papillomavirus using a restriction fragment length polymorphism analysis and an original typing algorithm. J. Clin. Virol. 2008;42(1):13–21. doi: 10.1016/j.jcv.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt M., Bravo I.G., Snijders P.J. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 2006;44(2):504–512. doi: 10.1128/JCM.44.2.504-512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt M., Dondog B., Waterboer T. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers. J. Clin. Microbiol. 2008;46(3):1050–1059. doi: 10.1128/JCM.02227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Depuydt C.E., Boulet G.A., Horvath C.A., Benoy I.H., Vereecken A.J., Bogers J.J. Comparison of MY09/11 consensus PCR and type-specific PCRs in the detection of oncogenic HPV types. J. Cell. Mol. Med. 2007;11:881–891. doi: 10.1111/j.1582-4934.2007.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreimer A.R., Johansson M., Waterboer T. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. J. Clin. Oncol. 2013;31(21):2708–2715. doi: 10.1200/JCO.2012.47.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao F.H., Jeronimo J., Qiao Y.L. An evaluation of novel, lower-cost molecular screening tests for human papillomavirus in rural China. Cancer Prev. Res. 2013;6(9):938–948. doi: 10.1158/1940-6207.CAPR-13-0091. [DOI] [PubMed] [Google Scholar]
- 36.Jung A.C., Briolat J., Millon R. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer. 2010;126(8):1882–1894. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 37.Gallus S., Muttarak R., Martinez-Sanchez J.M. Smoking prevalence and smoking attributable mortality in Italy, 2010. Prev. Med. 2011;52(6):434–438. doi: 10.1016/j.ypmed.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 38.E. Scafato, C. Gandin, L. Galluzzo, et al., Epidemiologia e monitoraggio alcol-correlato in Italia e nelle Regioni - Istituto Superiore di Sanita. In: Epidemiologia e monitoraggio alcol-correlato in Italia e nelle Regioni. Valutazione dell’Osservatorio Nazionale Alcol – CNESPS sull’impatto del consumo di alcol ai fini dell’implementazione delle attività del Piano Nazionale Alcol e Salute. Rapporti ISTISAN 13/3, 2013 (in Italian).
- 39.Tinhofer I., Johrens K., Keilholz U. Contribution of human papilloma virus to the incidence of squamous cell carcinoma of the head and neck in a European population with high smoking prevalence. Eur. J. Cancer. 2015;51(4):514–521. doi: 10.1016/j.ejca.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Baboci L., Boscolo-Rizzo P., Holzinger D. Evidence of the causal role of human papillomavirus type 58 in an oropharyngeal carcinoma. Virol. J. 2013;10(1):334. doi: 10.1186/1743-422X-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halec G., Holzinger D., Schmitt M. Biological evidence for a causal role of HPV16 in a small fraction of laryngeal squamous cell carcinoma. Br. J. Cancer. 2013;109(1):172–183. doi: 10.1038/bjc.2013.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hafkamp H.C., Speel E.J., Haesevoets A. A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5-8. Int. J. Cancer. 2003;107(3):394–400. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 43.Vinokurova S., Wentzensen N., von Knebel Doeberitz M. Analysis of p16INK4a and integrated HPV genomes as progression markers. Methods Mol. Med. 2005;119:73–83. doi: 10.1385/1-59259-982-6:073. [DOI] [PubMed] [Google Scholar]
- 44.Pirog E.C., Quint K.D., Yantiss R.K. P16/CDKN2A and Ki-67 enhance the detection of anal intraepithelial neoplasia and condyloma and correlate with human papillomavirus detection by polymerase chain reaction. Am. J. Surg. Pathol. 2010;34(10):1449–1455. doi: 10.1097/PAS.0b013e3181f0f52a. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann M., Ihloff A.S., Gorogh T. p16(INK4a) overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int. J. Cancer. 2010;127(7):1595–1602. doi: 10.1002/ijc.25174. [DOI] [PubMed] [Google Scholar]
- 46.Wendt M., Romanitan M., Nasman A. Presence of human papillomaviruses and p16 expression in hypopharyngeal cancer. Head and Neck. 2014;36(1):107–112. doi: 10.1002/hed.23394. [DOI] [PubMed] [Google Scholar]
- 47.Reuschenbach M., Kansy K., Garbe K. Lack of evidence of human papillomavirus-induced squamous cell carcinomas of the oral cavity in southern Germany. Oral. Oncol. 2013;49(9):937–942. doi: 10.1016/j.oraloncology.2013.03.451. [DOI] [PubMed] [Google Scholar]
- 48.Rietbergen M.M., Snijders P.J., Beekzada D. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int. J. Cancer. 2014;134(10):2366–2372. doi: 10.1002/ijc.28580. [DOI] [PubMed] [Google Scholar]