Abstract
Currently available vaccines prevent HPV infection and development of HPV-associated malignancies, but do not cure existing HPV infections and dysplastic lesions. Persistence of infection(s) in immunocompetent patients may reflect induction of local immunosuppressive mechanisms by HPV, providing a target for therapeutic intervention. We have proposed that a mouse, expressing HPV16 E7 oncoprotein under a Keratin 14 promoter (K14E7 mice), and which develops epithelial hyperplasia, may assist with understanding local immune suppression mechanisms that support persistence of HPV oncogene-induced epithelial hyperplasia. K14E7 skin grafts recruit immune cells from immunocompetent hosts, but consistently fail to be rejected. Here, we review the literature on HPV-associated local immunoregulation, and compare the findings with published observations on the K14E7 transgenic murine model, including comparison of the transcriptome of human HPV-infected pre-malignancies with that of murine K14E7 transgenic skin. We argue from the similarity of i) the literature findings and ii) the transcriptome profiles that murine K14E7 transgenic skin recapitulates the cellular and secreted protein profiles of high-grade HPV-associated lesions in human subjects. We propose that the K14E7 mouse may be an appropriate model to further study the immunoregulatory effects of HPV E7 expression, and can facilitate development and testing of therapeutic vaccines.
1. Introduction
Cervical cancer is responsible for 5% of the global cancer burden and it is the second most diagnosed cancer in women [1]. Cervical and other anogenital cancers are typically associated with infection by a subset of human papillomaviruses (HPVs). The ‘high-risk’ oncogenic HPV genotypes 16 and 18 are responsible for about 70% of all cervical cancer cases worldwide and approximately 50% of the cervical cancers are caused by HPV16 [2], [3]. Prevention of infection by high-risk oncogenic HPVs (HPV16 and 18) has been effective (~95%) with the development of virus-like particle-based vaccines [4]. Currently, there are three approved multi-valent (Gardasil® and Gardasil® 9, Merck) and bivalent (Cervarix®, GlaxoSmithKline) commercially available efficacious vaccines. However, the adoption of these vaccines in resource-limited countries has been hindered by high costs of the vaccines, jeopardising the prevention of HPV infection despite proven clinical efficacy [1], [5]. Furthermore, current vaccines do not protect against existing infections, offer an undetermined capacity in maintaining protection over time and will not have a major impact on cancer incidence for at least another 30 years. Thus, it remains a research priority to discover improved therapies that can resolve HPV-associated disease in patients who do not benefit from currently available vaccines.
HPVs are epitheliotropic double-stranded DNA viruses that infect the basal keratinocytes on surface epithelia of skin and mucous membranes. Replication of virus is dependent on the expression of non-structural viral genes E1, E2, E6, E7 and the structural L1 and L2 capsid proteins. The E1 and E2 proteins regulate viral DNA replication, while E6 and E7 deregulate cell cycle control and promote epithelial proliferation and delay epithelial differentiation. The late structural viral proteins L1 and L2 are responsible for virion assembly and lytic release and form the basis of currently available prophylactic vaccine formulations. Most HPV infections are cleared rapidly by the immune system, but extended virus persistence increases the risk of progressive dysplastic transformation of normal epithelium, eventually into cancer [6]. The clinical stages of cervical pre-malignancy are classified into worsening grades of dysplasia (cervical intraepithelial neoplasia (CIN) grades 1, 2, and 3; CIN1, CIN2, CIN3). CIN1/2 lesions frequently spontaneously regress in immune-competent individuals, but progression to CIN3 gives a high risk of eventual cancer development [4]. The spontaneous clearance of lower grade CINs is associated with an effective cellular immune response. Lower CIN grades are marked by increased CD4+ T cell infiltration [7] whereas a higher CD8+/CD4+ ratio is observed in CIN3 and cancer stages [8]; dysregulation of immune cell dynamics is requisite for lesion persistence and progression.
To determine why the immune system fails to clear persistent HPV infections and associated dysplastic epithelial lesions, we need better understanding of the cellular and molecular mechanisms regulating immunity in HPV-infected skin, which is challenging as HPV infects only human epithelia. Canine, bovine and rabbit papillomavirus lesions are generally self-limiting and rarely promote malignancy, similar to ‘low risk’ HPV types in humans. A mouse papillomavirus (MmuPV1) can replicate in some lines of immunocompromised laboratory mice [9], [10], [11], [12]. Susceptible animals are otherwise outbred, and reagents are limited. Nevertheless, animal models for canine oral papillomavirus (COPV) [13], rabbit oral papillomavirus (ROPV) [14] and MmuPV1 infections [10], [11] confirm that regression of lesions requires functioning cellular immunity. Further, rejection of transplanted tumours that express HPV16 E6 and E7 proteins can be achieved in mice if an effective CD8+ cytotoxic T cell lysis response is mounted [15], [16], [17], [18]. However, immunotherapies that are effective against murine tumours expressing HPV antigens generally have not been effective in human HPV-associated diseases [4].
To investigate the immune microenvironment resulting from persistent HPV oncoprotein expression, we have utilised a mouse model in which keratinocytes within murine skin expresses the HPV16 E7 oncoprotein as a transgene driven by keratin 14 promoter (henceforth referred to as K14E7 mice), with E7 protein levels comparable to those in infected human cervix [16]. The HPV16 E7 transgene is also expressed in the thymus, producing transgenic mice that tolerate the HPV E7 protein [19]. While keratinocytes express only the E7 oncoprotein, rather than E6 and E7 as in HPV-infected tissues in humans, they develop epithelial hyperplasia and dysplasia despite not fully replicating the viral oncogene expression kinetics during cervical pre-malignancy. Furthermore, K14E7 skin presents with an elevated immune cell infiltrate similar to those observed in human CIN lesions [20], [21]. Therefore, we believe that the K14E7 mouse provides a valid comparison to persistent HPV oncogene expression in human HPV-related epithelial disease. We have utilised a skin grafting/transplantation model based around the K14E7 mouse model to evaluate factors that may influence immunotherapy efficacy in the context of therapeutic vaccination against HPV-related cervical cancer and CINs. Further, K14E7 skin grafts consistently fail to be rejected when transplanted onto immunocompetent syngeneic recipients, with or without immunisation; E7-immunized mice are able to reject E7-transduced transplantable tumours [22], [23], [24], [25] but not E7-expressing skin grafts, a finding contrasting with the rejection of skin grafts that express other non-self proteins as transgenes, including chicken ovalbumin and human growth hormone [26], [27], [28], [29]. K14E7 skin grafts can be induced to be rejected by administration of live or killed Listeria monocytogenes [23], or by adoptive transfer of large numbers (>105) of immunisation-activated E7 peptide-specific CD8 T cells [25], excluding the possibility that E7 antigen presentation by target keratinocytes is insufficient to enable graft rejection. The primary aim of this paper is to provide a review of the literature on immune responses in HPV-associated human lesions, and to correlate the observations with those in the K14E7 transgenic mouse.
2. Materials and methods
2.1. Literature search
Search terms used for identifying articles on the PubMed®/MEDLINE® database were: HPV AND (cervical cancer OR cervical intraepithelial neoplasia) AND (immune OR cytokines OR chemokines OR lymphocytes OR dendritic cells OR T cells OR myeloid cells OR NK cells OR NKT cells OR MAIT cells OR IEL OR ILC OR macrophages OR monocytes OR neutrophils) NOT (cell line OR HIV). The literature search was performed on 18/07/2017.
2.2. Human cervical cancer progression microarray and K14E7 mouse RNA-sequencing data
The publically available microarray data from the den Boon et al. manuscript [30] was downloaded from the GEO database (GSE63514) and used to test for enrichment of the K14E7 signatures in cervical cancer and CIN cohorts. Procedures involved in murine skin grafting, sample collection, RNA extraction, cDNA library preparation, sequencing and data pre-processing for the mouse skin RNA-sequencing (RNA-seq) data have been previously described [20], [31]. Primary data are available via NCBI sequence read archive (SRA ID: SRP113560, BioProject accession ID: PRJNA395772). The ‘UP’ and ‘DOWN’ gene signatures (gene sets) of K14E7 mice were curated using the top-ranked ~200–300 genes up/down-regulated in K14E7 skin versus age-matched C57BL/6 skin from the RNA-seq analysis. The ranking of genes was performed using standardised differential gene expression testing based on Empirical Bayes moderated t-statistics embedded within the Linear Models for Microarray Data (LIMMA) R package [32]. The gene expression heat maps were generated using HeatMapImage module within the GenePattern public server [33].
2.3. Gene set testing procedures
Gene symbols from the murine gene sets (GRCm38.p5/mm10) were converted to human orthologous symbols (GRCh38.p5) for the gene set tests with Gene Set Enrichment Analysis (GSEA) [34]. GSEA is a popular analysis tool that can be used to assess whether samples in a microarray or an RNA-seq experiment would display correlation with biological phenotypes defined by a priori classified gene sets/signatures, such as gene sets contained in the Molecular Signatures Database (MSigDB) [34]. It is a competitive gene set test that uses sample permutation/re-sampling to detect the enrichment of genes at the top or bottom of gene sets, correlating the enrichment between either of two phenotypes. This is typically performed a large database of gene sets where a false discovery rate (FDR) is applied to the nominal p-value calculations to adjust for multiple testings. In this paper, we used GSEA on a small list of gene sets (n=2 for K14E7 ‘UP’ and ‘DOWN’ gene sets, and n=4 ‘UP’ and ‘DOWN’ gene sets from K14E7 grafting data) to broadly evaluate if the mouse K14E7 gene sets would be enriched in the human microarray data. To supplement this approach, we used two additional gene set tests: Correlation Adjusted MEan RAnk (CAMERA) [35] and Rotation (ROAST) gene set tests [36]. These two methods are included in the LIMMA R package and can be applied to data fitted to a linear model, such as RNA-seq count data analysed using the LIMMA-Voom pipeline [32], [37], displaying improved statistical power even for small data sets [35]. CAMERA is a competitive gene set test that evaluates whether ranking of genes in a gene set is highly ranked relative to all other genes not in the gene set, while accounting for inter-gene correlation in a linear model fit [35]. ROAST is a self-contained gene set test that tests whether genes in the gene set are differentially expressed, returning the proportion of genes that are up- or down-regulated in the gene set [36]. A pre-ranked GSEA analysis was also performed where we tested for enrichment of CIN3 or Cancer ‘UP’ and ‘DOWN’ gene sets in a pre-ranked list of genes from K14E7 skin versus C57BL/6 skin, ranked according to t-statistics values assigned by LIMMA. The CIN3 and cancer gene signatures were curated using the top ~200–300 significant differentially expressed genes identified via LIMMA.
CAMERA and ROAST were also used for gene set testing for enrichment of immunologic gene sets curated at the MSigDB. The gene sets were downloaded from the Walter and Eliza Hall Institute of Medical Research Bioinformatics Resources website: http://bioinf.wehi.edu.au/software/MSigDB/ where complete human and converted murine versions of the MSigDB C7 gene sets (annotated with Entrez Gene IDs) are available.
2.4. CIBERSORT
The CIBERSORT gene expression deconvolution package was used to estimate the immune cell composition in the grafting RNA-seq data [38]. Read counts (per million), equalised for library size using edgeR [39], were used as input for the analysis. The LM22 signature was used as the immune cell gene signature and the settings for the run were: 1000 permutation with quantile normalisation disabled. Two-way ANOVA with Tukey's multiple corrections was used to infer the statistical significance of the predicted immune cell populations where P<0.05 was considered significant.
3. Results
3.1. Comparing the immune cell infiltrate and cytokine profile presented in the literature with the K14E7 mouse model
A literature search on the PubMed®/MEDLINE® database was used to identify recent (within the last 10 years) relevant literature investigating the immune landscape of cervical cancers and CINs in human studies. Excluding review articles and articles that were not written in English, there were 490 primary research articles identified. Of these, 42 articles reported on the local changes within cervical lesions or draining lymph nodes (Supplementary Table 1) and 38 articles reported on genetic variants in genes encoding for cytokines, chemokines and other immune-related molecules associated with CIN and cancer progression. The first group of articles were chosen for further review as they were deemed to be more relevant to reports of changes to immune cell composition or cytokine/chemokine profiles in human cervical cancer and CIN lesions. It should be noted that, despite stringent search terms and unbiased review of the results returned by the search, a literature review is by nature limited and therefore our list does not claim to be exhaustive. We focus the subsequent section on the most frequently named cell types and markers in the list of articles reviewed that were reported to be associated with disease outcome in ≥5 papers.
3.1.1. Regulatory T lymphocytes and cytokines
There is general agreement that cervical cancer and severe CIN lesions are frequently infiltrated with large numbers of T cells, characterised as CD3+ [40], [41], [42] and/or CD4+ [42], [43], [44], [45] and CD8+ T cells [42], [43], [45], [46], [47], and that T cell infiltrates increase with disease severity. Expression of the Forkhead box P3 (FOXP3) transcription factor, an accepted marker for T regulatory (Treg) cells, was positively associated with disease severity in 10 of 11 reported studies [42], [43], [44], [45], [48], [49], [50], [51], [52], [53]. Interestingly, reduced Treg cells (FOXP3+ cells) and interleukin 17 (IL-17)+ cells were associated in one study with poorer disease free survival in cervical adenocarcinoma [54]. Recruitment of Treg cells to tumours is held to induce a locally immunosuppressive environment, confirmed by association with increased expression of IL-10, an immunoregulatory cytokine that is frequently associated to be produced by Treg cells, and with increasing disease severity in cervical lesions (11/13 papers reviewed) [43], [46], [50], [52], [55], [56], [57], [58], [59], [60], [61]. Increased IL-10 production in HPV-associated cancer was detected as mRNA or protein in local and circulating T cells, dendritic cells (DCs) and macrophages. No association of IL-10 with disease progression was seen using IL-10 mRNA from cytobrush biopsies of cervical mucosa cells [51], or protein levels from low-grade CIN lesions [62]. Scott et al. monitored the rate of HPV clearance upon infection in a longitudinal study and they detected increased levels of several cytokines, including IL-10, in cervicovaginal lavage samples from HPV positive patients. This observation was associated with reduced likelihood of HPV clearance for both high-risk HPVs and low-risk HPVs [63]. In keeping with the expectation that Treg cells and IL-10 should suppress effector T cell and antigen presenting cell functions within tumours, and thus prevent appropriate anti-tumour cytolytic responses, cervical cancer patients without lymph node metastases tend to display increased CD8+T cells/Treg ratios and a more favourable outcome [45]. Van der Burg et al. observed HPV-specific FOXP3- CD4+ T cells displaying suppressor functions [64]. Kojima et al. observed an increased percentage of CD4+ that express the inhibitory checkpoint programmed cell death 1 (PD-1) receptor in persisting HPV-associated cervical lesions, when compared to those that regressed [44]. These T cells may arrive in the tissue with regulatory functions such as are expressed by conventional Treg cells from the thymus and peripheral lymphoid organs, or might be induced locally to become regulatory. However, regression of low or high grade CIN lesions have also been associated with increased CD4+ and CD8+ T cells and CD4/CD25 or CD8/CD25 ratios [65], [66], [67], highlighting the lack of clarity about immune factors/markers that would favour regression of CIN lesions.
The role of interferon gamma (IFN-γ) produced by T cells, natural killer (NK) cells and NKT cells in regression of HPV-associated lesions is uncertain. IFN-γ can promote anti-tumour responses by further activating effector immune cells. However, chronic IFN-γ secretion in cancer has been associated with immunosuppression [50] and can promote resistance to immune checkpoint blockade therapy [68]. Of 8 reviewed articles addressing IFN-γ expression and HPV-associated disease progression, three reported that elevated IFN-γ expression was associated with disease progression of high-grade CIN lesions and cancer [41], [50], [69]. Infiltrating CD4+, CD8+ and invariant NKT (iNKT) cells were the main sources of IFN-γ in these three studies [50]. Hu et al. [41] further demonstrated that increased iNKT cells were increased in high-grade lesions (CIN2 and CIN3) compared to normal cervix controls. McKenzie et al. similarly reported that CIN2/3 tissue attracted cells expressing T cell and NK markers (CD3+, CD8+, CD56+, CD16+) [70]. Yang et al. and Scott et al. reported reduced IFN-γ levels in cervical exudates [60] or cytobrush biopsies of cells from the mucosal layers of the cervix [51], in contrast to mRNA or protein measurements of larger tissue biopsies that would have a more complex tissue architecture with infiltrated immune cells, or to isolated immune cells [41], [50], [69]. Song et al. reported that elevated IFN-γ expression was associated with regression of low-grade lesions (CIN1) and did not investigate higher-grade lesions [71]. Sharma et al. reported that IFN-γ secretion by PBMCs was decreased in cervical cancer patients compared to normal controls. However, the levels of the cytokines within the lesions were not examined [58]; while van der Burg et al. showed that HPV-specific CD4+ T cells isolated from draining lymph node biopsies of cervical cancer patients (n=3) displayed regulatory/suppressor T cell functions, and that co-culturing these T cells with CD4+CD25- responder T cells challenged with cognate HPV antigens induced suppression of the responder T cells, resulting in reduced IFN-γ production [64].
3.1.2. Antigen presenting cells
Tumour-associated macrophages and professional antigen presenting cells (APCs) have been implicated in the regulation of immunosuppression during cervical cancer progression. Four of four reviewed studies on macrophages in CIN reported a positive correlation between numbers of macrophages and increasing severity of CIN lesions and cancer [40], [46], [50], [72]. Dendritic cells represent another population of APCs that may play a role in this setting, but their contributions to the disease is less clear. Two studies reported decrease in CD1a+ DCs classified as immature DCs [73] or CD1a+ Langerhans cells [74] in the cervical stroma and/or epithelium of CIN3 and cancer patients. Other studies reported increase in CD1a+ DCs [50], or were inconclusive due to variability in their quantification [45]. Conversely, Hayati et al. [73] and Piersma et al. [45] reported that proportions of mature DCs (CD83+ or CD208+) were higher in cancer and CIN3 versus healthy controls. The presence of CD83+ DCs was also increased in cancer biopsies as a consequence of inhibited CCR7-dependent migration of mature DCs towards draining lymph nodes, which would hinder adaptive immune responses against the tumour [75]. Secreted products of the innate immune system, including colony stimulating factor 1 (CSF-1), IL-10 and vascular endothelial growth factor (VEGF) are thought to play a role in recruiting and conditioning macrophages and DCs, as well as being produced by these cells themselves. CD14+CD33-CD163- matured proinflammatory ‘M1′ macrophages are significantly associated with improved prognostic outcomes for disease survival [76]. APCs were also required to appropriately prime allogenic T cells for anti-tumour effects for cervical cancer therapy [77]. Conversely, DCs in cervical cancers can display ‘tolerogenic’ behaviour, and have been shown to be potent producers of the immunosuppressive indoleamine 2,3-dioxygenase 1 (IDO1) molecule [50]. They can also be a source of inhibitory ligands for immune checkpoint molecules on Treg or dysfunctional T cells [60], [78].
Generally, the reviewed articles support the hypothesis that heightened inflammatory responses are associated with chronicity and persistency of disease. How the transformed epithelium, or HPV proteins, induce this inflammatory response remains to be elucidated; expression of HPV oncoproteins during early events of HPV infection can however potentiate chronic inflammatory responses. Cervical cancer derived IL-6 was shown to be an important signal for induction of expression of CCL20, a chemokine associated with recruitment of CD4+/IL-17+ T cells, by fibroblasts, maintaining chronic inflammation in severe CIN lesions [79].
3.2. Consensus in cellular and cytokine changes between human cervical cancers, high-grade CIN lesions and K14E7 mouse model
Disease models that can emulate the immune environment in CIN may be useful for evaluating immunotherapy options for HPV-associated disease. We have proposed that a transgenic C57BL/6 mouse, where the HPV16 E7-protein is expressed from a keratin promoter in epithelial cells (K14E7), may be a preclinical tool to assess the biology and immunology of chronic/persistent HPV-related disease. K14E7 mouse skin displays epithelial hyperplasia and is markedly infiltrated with immune cells [20]. The mechanisms of lymphocyte recruitment to CIN lesions is similar to that of the K14E7 mice – for example, CCL20 is significantly enriched in K14E7 skin and results in elevated recruitment of CCR6+ CD4 T cells [20], similar to the findings in human CIN [79]. There are also markers of local immune suppression within the K14E7 skin, including significantly up-regulated expression of Treg and/or immunosuppression-related genes, identified in the list of differentially expressed genes deemed to be statistically significant from RNA-seq analysis of the K14E7 mouse ear skin [31] (Fig. 1), and this findin is consistent with human studies. Up-regulated genes include Foxp3, Il10, Ctla4, Pdcd1 (PD-1), Pdcd1lg2 (PD-1 ligand 2; PD-L2) and molecules such as Icos, Gzmb, Il2rb2 and Il2ra, which have been reported to be expressed on subpopulations of cutaneous Treg cells [80]. However, it is important to note that previous attempts to neutralise regulatory lymphocytes in K14E7 mice in our hands did not yield significant alleviation of immunosuppression; while CD8 effector T cell suppression could be rescued when CD4+CD25+ T cells are neutralised in K14E7 mice [81], specific depletion of conventional Foxp3+ Treg cells in either K14E7 donors or recipients did not enable graft rejection [82]. Taken together, this suggests that additional suppressive factors contribute to the immunosuppressive environment in K14E7 skin.
Fig. 1.
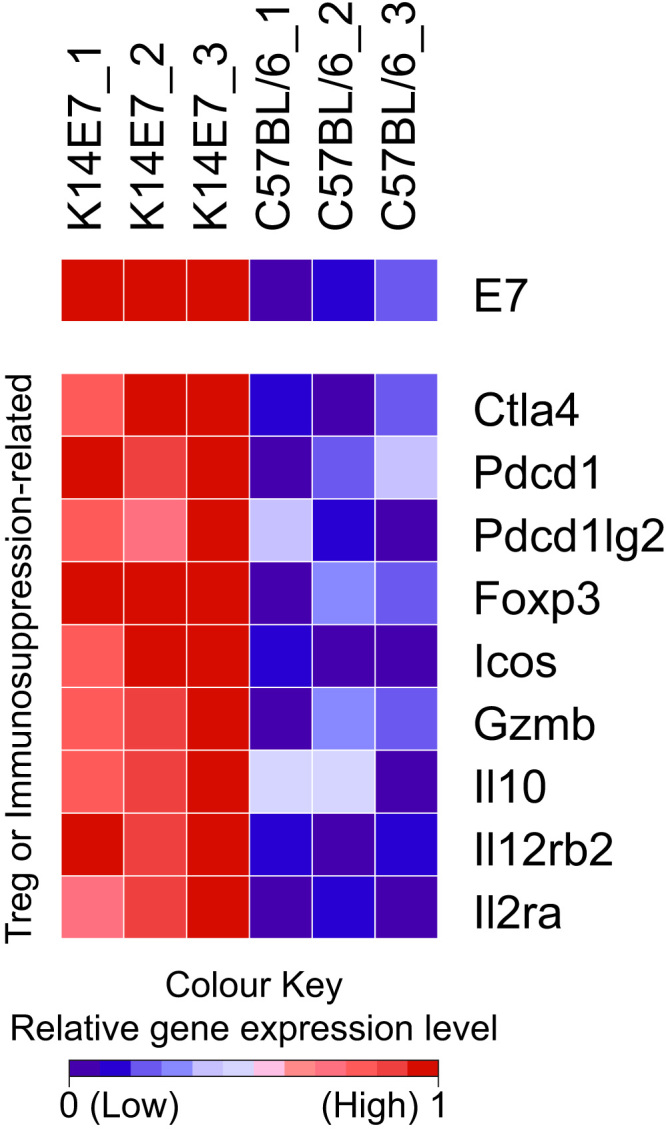
Relative gene expression heat map of selected genes associated with immune cell dysfunction and Treg markers. The colour scheme of the heat map corresponds to the relative expression level normalised to a range from 0 to 1 using the minimum and maximum values within each row/gene and represented as a colour gradient from blue to red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We have identified that rejection of K14E7 transgenic skin from immunocompetent mice can be achieved through genetic or pharmacological ablation/depletion of select infiltrating and/or resident immune cell types (e.g. T cells and iNKT cells) and soluble factors (e.g. IFN-γ, IL-17, IL-1β and IDO1) in the E7 transgenic skin (summarised in Table 1). Elevated IFN-γ production from iNKT cells was identified to be a major determinant of immunosuppression, and a critical component that contributes to graft tolerance; K14E7 skin grafts that are NKT-deficient or IFN-γ-deficient were robustly rejected from immunogenic recipients [83]. The immunosuppression mediated by infiltrating NKT cells in K14E7 skin grafts was independent of IFN-γ, IL-10 and IL-17, as NKT cell reconstitution to skin grafts with or without the expression of either of the three cytokines completely inhibited graft rejection [82]. CD8 effector T cell functions were suppressed by the presence of NKT cells, as well as chronic IFN-γ expression, in the K14E7 skin environment [82]. MHCII+CD11c+ DCs in the skin expressed the highest level of IFN-γ-receptor (IFN-γR) and the increased expression of the immunosuppressive IDO1 in K14E7 mice was dependent on IFN-γR expression. Further, inhibition of IDO1 activity via oral administration of 1-Methyl-DL-tryptophan (1-D/L-MT) to K14E7 graft donor and C57BL/6 recipient mice was able to induce partial graft rejection (~50%) [84]. We reason that the K14E7 immunosuppressive environment alters the ability of APCs to react against the persistent E7 antigen. Consistent with this hypothesis, Langerhans cells from K14E7 skin expressed higher levels of immunosuppressive molecules such as IDO1, arginase 1 and IL-10, but paradoxically expressed lesser major histocompatibility complex II (MHCII) and PD-1 ligand 1 (PD-L1) [85]. Additionally, APCs displayed impaired antigen presenting capabilities [86], [87]; and although K14E7 skin displayed higher absolute numbers of DCs compared to wild-type controls, there was an increase in relative proportions of CD103+ and CD11b+ DCs but reduced Langerhans cells [87]. This was supported in another mouse model where the authors demonstrated that epidermal Langerhans cells were depleted from the epidermis upon expression of HPV E7, using an inducible lentiviral system [88].
Table 1.
Summary of factors that influence K14E7 graft fate.
| Inducing molecules | Infiltrating and residing cells | Secreted molecules | Donor skin graft | Skin grafts rejected? | References |
|---|---|---|---|---|---|
| IL-23 and IL-1β | CD4 T cells and γδ T cells | IL-17 | K14E7xIL-17-/- | Yes | [28], [120] |
| K14E7 and K14E7xIL-1Ra-/- on IL-1Ra-/- recipients | Yes | ||||
| K14E7xIL−12/23p40-/- | No | ||||
| IL-18 | NKT cells | IFN-γ | K14E7xJα18-/- | Yes | [82], [83], [121] |
| K14E7xCD1d-/- | |||||
| K14E7xIFN-γ-/- | |||||
| IFN-γ | IFN-γ-R+ migratory dendritic cells | IDO1 | K14E7 skin graft administered with inhibitor of IDO1 | Yes | [84] |
| (1-D/L-MT) | |||||
| CCL2 and CCL5 | Mast cells | K14E7xKitw-sh/w-sh | Yes | [122] | |
| CCR6 | CD4 T cells | K14E7xRAG-/- | Yes | [20] | |
| CD8 T cells | |||||
| CD4+CD25+ | K14E7xFoxp3-/- (K14E7xDEREG) | No | [81], [82] | ||
| Treg cells |
We hypothesise that these factors, when present in the K14E7 skin environment, co-ordinately induce ineffective CD8 effector T cell responses against K14E7 skin grafts. More recently, the overarching hypothesis has been refined in that E7-induced hyperplasia rather than expression of E7, is responsible for the recruitment, programming and/or retention of these immunosuppressive cells and molecules in the K14E7 hyperplastic epithelium milieu, as mice expressing E7 protein from the Keratin 14 promoter, and with a mutated retinoblastoma protein that is functionally effective but cannot bind E7, have no hyperplasia and no inflammatory infiltrates [20], [31], [89]. Hyperplastic murine K14E7 transgenic skin thus models some important aspects of the cellular infiltrate and immunosuppressive cytokine secretion profile in cervical cancer and high-grade CIN lesions in human patients.
3.3. K14E7 skin shares transcriptional features with human high-grade squamous intraepithelial lesions
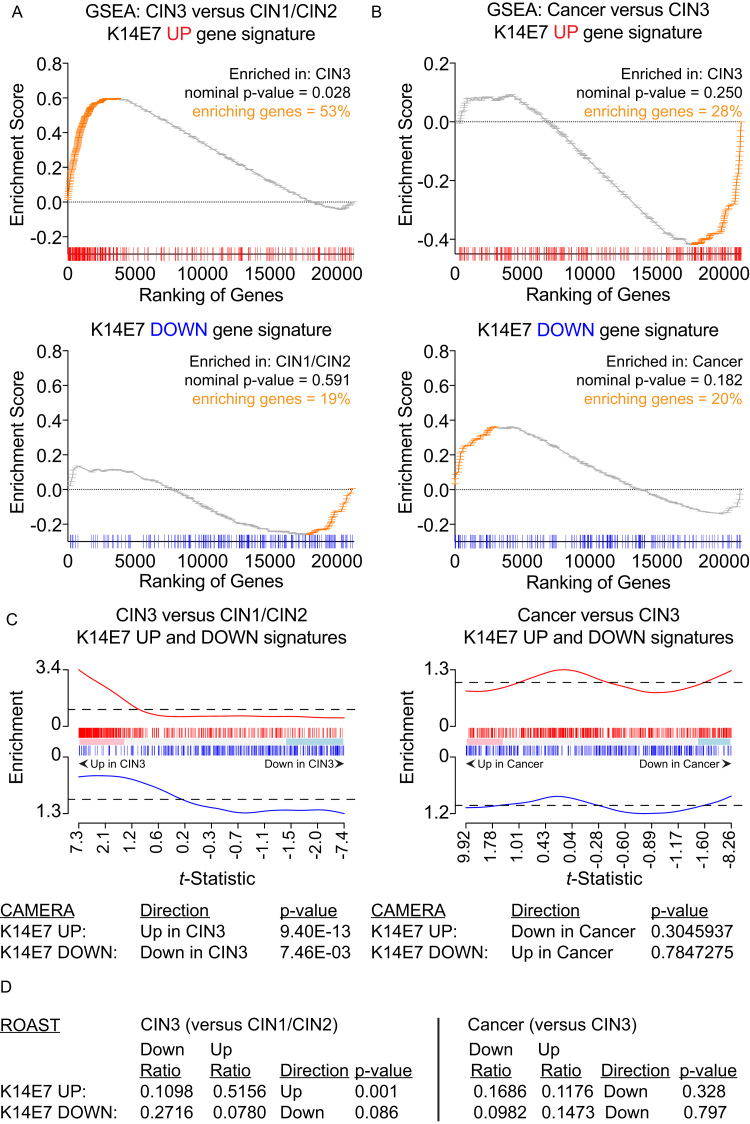
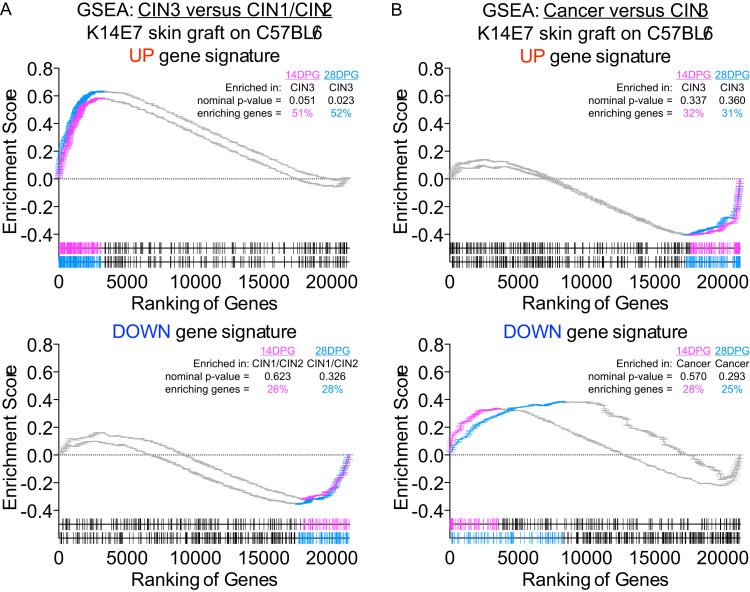
Defining the molecular changes associated with the establishment of cervical cancer pre-malignancy can reveal underlying mechanisms and highlight potential means to improve cancer and pre-cancer risk assessment, diagnosis, prognosis, and treatment. To elaborate on our initial RNA-seq analysis generated from K14E7 and C57BL/6 skin [31], we performed gene set tests to broadly evaluate whether gene changes found in K14E7 mice would be similarly observed in microarray data of the clinically recognised stages of cervical cancer progression [30]. K14E7 ‘UP’ or ‘DOWN’ gene signatures (gene sets) were curated from the top-ranked differentially expressed genes in K14E7 skin (versus C57BL/6 skin) and murine gene symbols from the K14E7 gene sets were mapped to orthologous symbols in human. 270 of the 293 genes contained in the ‘UP’ gene set (~92%) and 181 of the 203 genes contained in the ‘DOWN’ gene set (~92%) were successfully matched with human gene symbols/probes found on the cervical cancer progression microarray data set. The classic method for gene set enrichment analysis (GSEA) [34] was used to test the enrichment of the K14E7 gene signatures in the following phenotype comparisons: i) CIN3 versus CIN1/CIN2 and ii) cancer versus CIN3. In this analysis, human CIN3 displayed significant positive enrichment of the K14E7 ‘UP’ gene set where ~53% of genes in the K14E7 ‘UP’ gene set were highly ranked and correlated with CIN3 phenotype and not CIN1/CIN2 (Fig. 2A). While the enrichment did not achieve statistical significance, the ~28% of genes in the K14E7 ‘UP’ gene set were positively enriched in CIN3 and not in invasive cancer (Fig. 2B). The K14E7 ‘DOWN’ gene sets did not attain statistical significance in the GSEA but displayed a general trend of being positively enriched in CIN1/CIN2 or cancer versus CIN3 (Fig. 2A and B); up-regulation of ‘K14E7′ down genes were observed in CIN1/CIN2 and cancer but these genes were down-regulated in CIN3, similar to the regulation pattern in K14E7 skin versus C57BL/6 skin.
Fig. 2.
Gene set testing of K14E7 ‘UP’ and ‘DOWN’ gene signatures in human CIN3 versus CIN1/CIN2 and cervical cancer versus CIN3. (A-B) GSEA analysis of (A) CIN3 versus CIN1/CIN2 or (B) cancer versus CIN3 of K14E7 ‘UP’ or ‘DOWN’ signatures. Ranking of genes in the ‘UP’ gene signatures (red) or ‘DOWN’ gene signatures (blue) are shown at the bottom of each GSEA plot. Position and frequency of genes contributing the most to each enrichment are highlighted in orange. (C) Barcode enrichment plot of t-statistic ranked genes from LIMMA for CAMERA gene set testing. The enrichment value corresponds to the relative ‘weight’ value in LIMMA. (D) ROAST gene set testing of CIN3 versus CIN1/CIN2 or cancer versus CIN3. Proportion of genes corresponding to up or down regulated and assigned direction and p-value are shown. Genes are assigned to the up or down bins according to the probability formula specified in ROAST. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
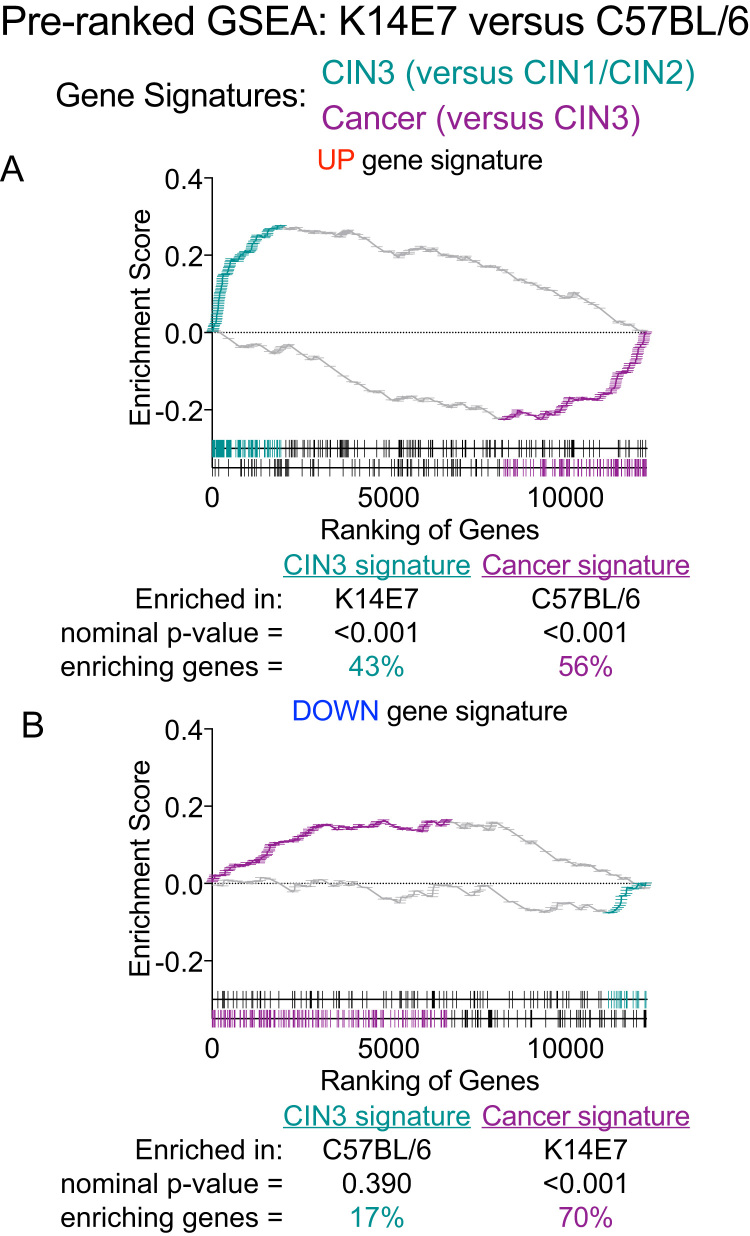
Gene set testing using Correlation Adjusted MEan RAnk gene set tests (CAMERA) [35] supports the results from GSEA; human CIN3 displayed positive enrichment of both K14E7 ‘UP’ and ‘DOWN’ signatures (Fig. 2C). In addition, Rotation gene set test (ROAST) identified that a significant proportion of genes contained in the K14E7 ‘UP’ gene set (~50%) was up-regulated in CIN3 versus CIN1/CIN2 (Fig. 2D). Approximately 27% of genes in the K14E7 ‘DOWN’ gene set were also observed to be down-regulated in CIN3 versus CIN1/CIN2 but this did not attain statistical significant (Fig. 2D). The enrichment of the K14E7 gene sets by cancer (versus CIN3) was less clear; gene sets were inversely associated with cancer in CAMERA analysis (Fig. 2C) and although ROAST identified that genes contained in both gene sets were identified to be down-regulated in cancer, there is a large proportion of genes displaying ‘mixed’ regulation pattern (~80%) (Fig. 2D). The GSEA was also tested in a reciprocal fashion with a pre-ranked gene list ordered by t-statistics values (assigned after LIMMA linear modelling) of genes in the K14E7 skin versus C57BL/6 skin comparison/phenotypes. The pre-ranked gene list was tested with gene signatures curated from the top ~200–300 up- or down-regulated genes in i) CIN3 versus CIN1/CIN2 and ii) cancer versus CIN3 accordingly. It is important to note that although mice and human share many orthologous genes, the pre-ranked gene list of K14E7 skin versus C57BL/6 skin was truncated to ~70% after conversion to do this comparison. Nevertheless, pre-ranked GSEA using this truncated ranked list still displayed similar association as before; K14E7 skin displayed significant positive enrichment of the CIN3 ‘UP’ gene set where ~43% of genes in the CIN3 ‘UP’ gene set were highly ranked and correlated with K14E7 phenotype and not C57BL/6 mice (Fig. 3A). Conversely, ~56% of genes in the cancer ‘UP’ gene set positively enriched the C57BL/6 phenotype in a significant fashion and not K14E7 skin (Fig. 3A). The CIN3 ‘DOWN’ gene set did not attain statistical significance in the GSEA but displayed a general trend of being positively enriched in C57BL/6 skin whereas the Cancer ‘DOWN’ gene set was significantly enriched in the K14E7 phenotype (Fig. 3B).
Fig. 3.
Pre-ranked GSEA of CIN3 or Cancer ‘UP’ and ‘DOWN’ gene signatures in K14E7 versus C57Bl/6. Ranking of genes in the ‘UP’ gene signatures or ‘DOWN’ gene signatures in the K14E7 ranked list are shown at the bottom of each pre-ranked GSEA plot. Position and frequency of genes contributing the most to each enrichment are highlighted in teal for genes in CIN3 signatures or purple for genes in Cancer signatures. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
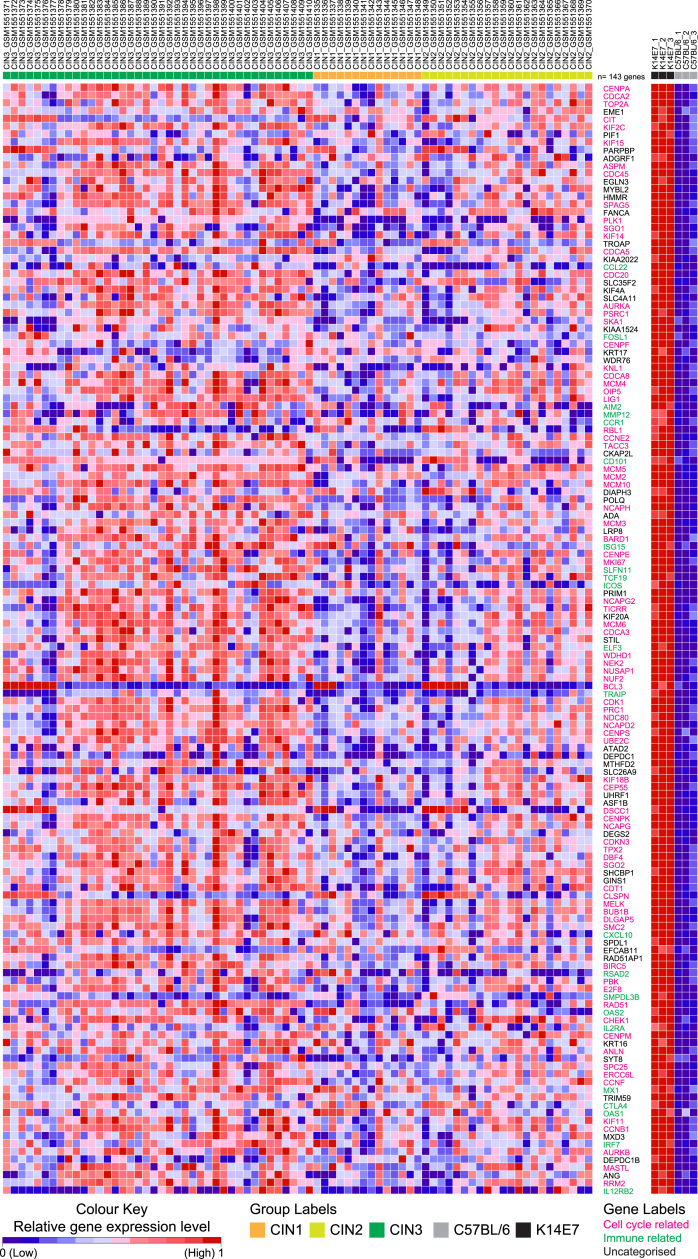
The 143 genes that significantly contributed to enrichment of the K14E7 ‘UP’ gene set (~53%) in CIN3 in the GSEA analysis in Fig. 2A showed a general trend of increased expression in CIN3 and decreasing expression intensity in CIN2 followed by CIN1 (Fig. 4, left panel). The relative expression level of these genes in the K14E7 and C57BL/6 mice are shown on the right panel of Fig. 4 (all genes were significantly up-regulated in K14E7 mice versus wild-type controls). In this list, ~50% are genes related to cell division and proliferation, kinetochore assembly, regulation of mitosis and DNA repair machinery (Fig. 4, highlighted in pink). Several of these cell cycle related genes have been previously associated with HPV-positive cervical cancers and some are used as biomarkers for diagnostic purposes in this setting. For example, overexpression of the nuclear markers – topoisomerase (DNA) II alpha (TOP2A), marker of proliferation Ki-67 (MKI67), baculoviral inhibitor of apoptosis repeat-containing protein 5 (BIRC5), centromere protein F (CENPF) and minichromosome maintenance complex component 2 and 5 (MCM2 and MCM5) – can indicate increased cell division in the context of tumour development. They have each been previously reported to be overexpressed in HPV-positive CIN or cancer and some have been evaluated for use as biomarkers [90], [91], [92]. Other genes, such as the genes encoding for aurora kinase A and B (AURKA and AURKB), have been implicated as survival signals for HPV-infected cells and inhibition of AURKA/B using Alisertib induced cell death in HPV-transformed cervical cancer cell lines [93], [94]. It important to note that the utility of this drug was also evaluated in the K14E7 skin grafting model where Alisertib-treated E7-expressing skin grafts displayed increased apoptotic bodies compared to untreated controls [93]. Immune response-related genes constitute ~30% of remaining list of genes (~15% of total/143 genes) (Fig. 4, highlighted in green). While several of these immune response-related genes are typically associated with pro-inflammatory function, the increased expression of the cytotoxic T-lymphocyte associated protein 4 (CTLA4), interleukin 2 receptor subunit alpha (IL2RA), inducible T-cell costimulator precursor (ICOS), C-C motif chemokine 22 (CCL22), C-C motif chemokine receptor 1 (CCR1) and C-X-C motif chemokine ligand 10 (CXCL10) have been associated to be expressed on conventional Treg, effector Treg [53], [95], [96] and adipose tissue-derived Treg cells [97], supporting increased frequency/infiltration of regulatory cells in HPV-associated high-grade squamous intraepithelial lesions and K14E7 skin.
Fig. 4.
Heat map of relative gene expression of genes that contribute the most to the enrichment of K14E7 ‘UP’ gene set in CIN3 versus CIN1/CIN2. Relative gene expression in K14E7 mice and C57BL/6 mice are provided in the right panel. The colour scheme for both heat maps correspond to the relative expression level normalised to a range from 0 to 1 using the minimum and maximum values within each row/gene and represented as a colour gradient from blue to red. Sample groups for each are additionally provided with coloured annotations. Gene symbols that correspond to cell cycle-related genes are annotated in pink and gene symbols that correspond to immune-related genes are annotated in green. Uncategorised gene symbols are in black. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Top 50 immunologic gene signatures are similarly enriched in human CIN3 and murine K14E7 skin but not human cervical cancer
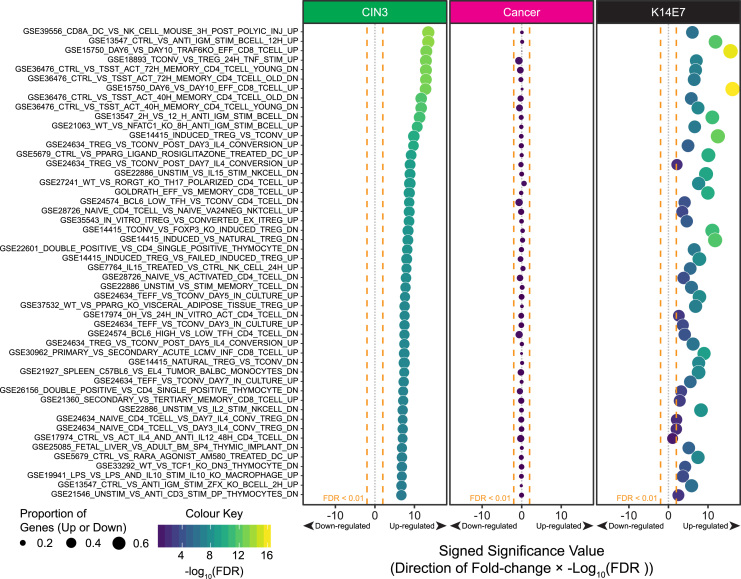
To further investigate the changes to immune response related genes highlighted from the K14E7 signatures gene set testing, we expanded the analysis to test for significant enrichment of the immunological related gene signatures derived from the ~4800 immunological gene sets curated in the Molecular Signature Database (MSigDB) [34]. The following phenotype comparisons were tested: i) CIN3 versus CIN1/CIN2, ii) Cancer versus CIN3 and iii) K14E7 versus C57BL/6. Because the classic GSEA approach would require large sample numbers to be efficient for their sample permutation statistics (>10 per phenotype), and hence are only applicable for the CIN and cervical cancer microarray data, we used CAMERA and ROAST to perform the gene set testing for the three comparisons as they are reported to retain good statistical power and control for type 1 error rates correctly even for small sample sizes, provided that the data used for the contrasts are fitted to a linear model (e.g. linear models embedded in LIMMA applied to microarray and RNA-seq data) [35], [36]. The full table for the CAMERA analysis is provided in Supplementary Table 2. The top 50 enriched immunologic gene sets ranked in CIN3, with the corresponding false discovery rate adjusted significance, are shown in Fig. 5; the top 50 enriched immunologic gene sets were also significantly enriched in K14E7 mice skin versus C57BL/6 mice. In CIN3 versus CIN1/CIN2, 673 immunologic gene sets were significantly enriched in CIN3 at FDR<0.01, with only 1 gene set predicted to be down-regulated. In K14E7 mice, 330 gene sets were significantly enriched and 287/330 were predicted to be up-regulated. Importantly, a total of 235 immunologic gene sets were commonly significantly enriched (FDR<0.01) in CIN3 and K14E7, and all 235 were predicted to be up-regulated. In cancer versus CIN3, none of the top 50 immunologic gene sets were significant enriched (Fig. 5) and surprisingly, only 1 gene set (GSE45837_WT_VS_GFI1_KO_PDC_DN; genes down-regulated in wild-type plasmacytoid DCs) reached statistical significance of FDR<0.01. This gene set was predicted to be down-regulated in cancer versus CIN3 but up-regulated in CIN3 versus CIN1/CIN2 (Supplementary Table 2). In summary, the immunologic gene signatures that correspond to the immunological architecture within CIN3 versus CIN1/CIN2 and K14E7 versus C57BL/6 are maintained/shared, but not in cancer versus CIN3. This might reflect that immunological changes that occur in high-grade squamous intraepithelial lesions are largely conserved during cancer, and indirectly implies that immunotherapies that may be developed may be effective for both CIN3 and cancer.
Fig. 5.
CAMERA gene set testing of immunologic gene sets in MSigDB C7. Differential gene expression testing for CIN3 versus CIN1/CIN2 (green column), cancer versus CIN3 (pink column) and K14E7 versus C57BL/6 (black column) were performed using LIMMA. The Empirical Bayesian t-statistics from the analysis was used for the ranking of genes in each comparison for CAMERA gene set testing. The significance value of the CAMERA gene set test, Log10 transformed false discovery rate (FDR) adjusted p-values, for each gene set are plotted along the viridis colour scale (purple, blue, green to yellow) in each bubble as well as on the x-axis where the direction regulation of the of the gene set (up or down; +1 or −1) is multiplied to the significance value. The size of the bubbles correspond to the proportion of genes that are up or down-regulated in each gene set calculated by ROAST where up-regulated pathways were plotted with proportion of up-regulated genes and vice versa. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Transcriptomic changes associated with persistence of murine K14E7 skin grafting procedure is also found in human in high-grade squamous intraepithelial lesions
The transcriptomic changes associated in our skin grafting model was also evaluated. This was achieved by performing RNA-seq on RNA extracted from the skin grafts of K14E7 or C57BL/6 donor ear skins that were transplanted to the flank of recipient C57BL/6 mice 14 or 28 days post-grafting (DPG). These time points were empirically chosen as time-points where we expect skin grafts to display a potential ‘successful’ or ‘failed’ rejection effect as with grafting E7-expressing skin grafts depleted of CD1d-restricted NKT cells [83]. GSEA comparisons were performed using the top ~300 up/down-regulated genes from 14DPG or 28DPG K14E7 skin graft RNA-seq data (compared to 14DPG or 28DPG C57BL/6 skin grafts) as the gene signatures with the cervical cancer progression microarray dataset, as per Fig. 2. This comparison showed the same trend as before, with CIN3 enriching for both 14DPG and 28DPG skin grafts ‘UP’ gene signatures (Fig. 6A and B). Up-regulation of ‘DOWN’ signatures were enriched in the opposing phenotypes (CIN1/2 and Cancer) (Fig. 6A and B).
Fig. 6.
Gene set testing of K14E7 on C57BL/6 14DPG and 28DPG ‘UP’ and ‘DOWN’ gene signatures in human CIN3 versus CIN1/CIN2 and cervical cancer versus CIN3. (A-B) GSEA analysis of (A) CIN3 versus CIN1/CIN2 or (B) cancer versus CIN3 of K14E7 on C57BL/6 14DPG and 28DPG ‘UP’ or ‘DOWN’ signatures. Ranking of genes in the ‘UP’ gene signatures or ‘DOWN’ gene signatures are shown at the bottom of each GSEA plot. Position and frequency of genes contributing the most to each enrichment are highlighted (14DPG, pink; 28DPG, blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
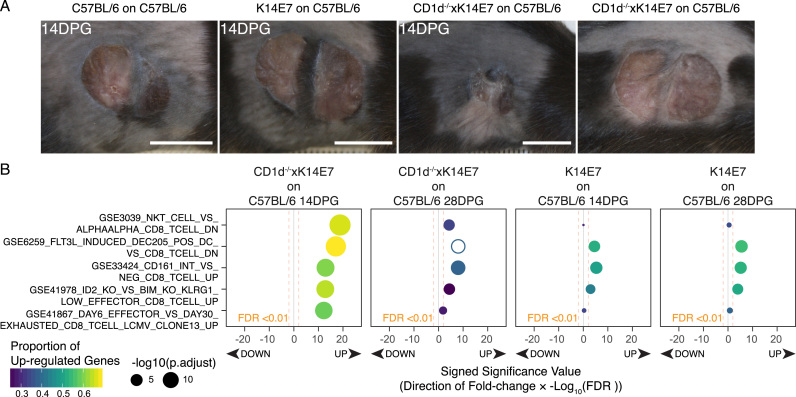
The skin grafting model, based around the K14E7 mouse, has been used to discover and understand which factors influences graft persistence, and which of these may be targeted to promote effective immunotherapy. As mentioned previously, we have identified one such instance where NKT cells derived from K14E7 donor skin grafts inhibit priming of CD8 T cells by APCs in the draining lymph nodes [82], [83], presumably preventing effective adaptive immunity priming against the E7-expressing skin grafts. However, whether this effect is directly translated to functional defects in CD8 T cell activity in the skin grafts is unknown, and this hypothesis has been difficult to assess using flow cytometry based approaches. Thus, we decided to determine via RNA-seq whether there is a distinct molecular pattern associated with graft rejection within E7-expressing skin grafts undergoing rejection. We have previously showed that depletion of NKT or iNKT cells from E7-expressing donor skin was sufficient to induce graft rejection in immunocompetent C57BL/6 recipients [27], [83]. Hence, we performed skin grafting of CD1d-/-xK14E7 donor ear skin onto the flank of C57BL/6 recipient mice and performed RNA-seq on RNA collected from the skin grafts at the same time points, 14DPG and 28DPG. In this experiment, we collected CD1d-/-xK14E7 skin grafts that have begun to display signs of rejection at 14DPG, as well as CD1d-/-xK14E7 skin grafts that have yet to display graft shrinkage at 28DPG (Fig. 7A). As per Fig. 5, we used CAMERA to broadly associate the immunological signatures (contained within MSigDB) that may be altered in the CD1d-/-xK14E7 skin grafts undergoing rejection (versus K14E7 skin grafts at the same time points). The full table for this analysis is also included in Supplementary Table 2. The top 50 gene sets enriched in CD1d-/-xK14E7 14DPG and 28DPG skin grafts were largely shared between the two time points although the significance values in the 28DPG list were generally lower than 14DPG (Supplementary Table 2). In contrast, the top 50 immunological gene sets enriched by 14DPG and 28DPG K14E7 skin grafts (versus 14DPG and 28DPG C57BL/6 skin grafts respectively) were similar to the top 50 gene sets enriched by CIN3 versus CIN1/CIN2 and K14E7 skin versus C57BL/6 skin as per Fig. 5 (Supplementary Table 2). The top 5 CD8 T cell related gene sets within the top 50 enriched by 14DPG CD1d-/-xK14E7 skin grafts are shown Fig. 7B. Interestingly, CD1d-/-xK14E7 14DPG skin grafts displayed significant enrichment of gene sets that suggests activation of effector T cells at 14DPG, such as GSE33424_CD161_INT_VS_NEG_CD8_TCELL_UP (genes upregulated in highly functional memory CD8+ T cells) and GSR41867_DAY6_EFFECTOR_VS_DAY30_EXHAUSTED_CD8_TCELL_LCMV_CLONE13_UP (genes up regulated in functional effector CD8 T cells).
Fig. 7.
CAMERA gene set testing of CD8 T cell immunologic gene sets from MSigDB C7 in grafting comparisons. (A) Photographs of representative grafts from the various donors and recipients prior to sample collection at 14DPG or 28DPG. (B) Top 5 CD8 T cell related immunological gene sets from CAMERA gene set testing. The significance value of the CAMERA gene set test, Log10 transformed false discovery rate (FDR) adjusted p-values, for each gene set are plotted along the viridis colour scale (purple, blue, green to yellow) in each bubble as well as on the x-axis where the direction regulation of the of the gene set (up or down; +1 or −1) is multiplied to the significance value. The size of the bubbles correspond to the proportion of genes that are up-regulated in each gene set calculated by ROAST. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
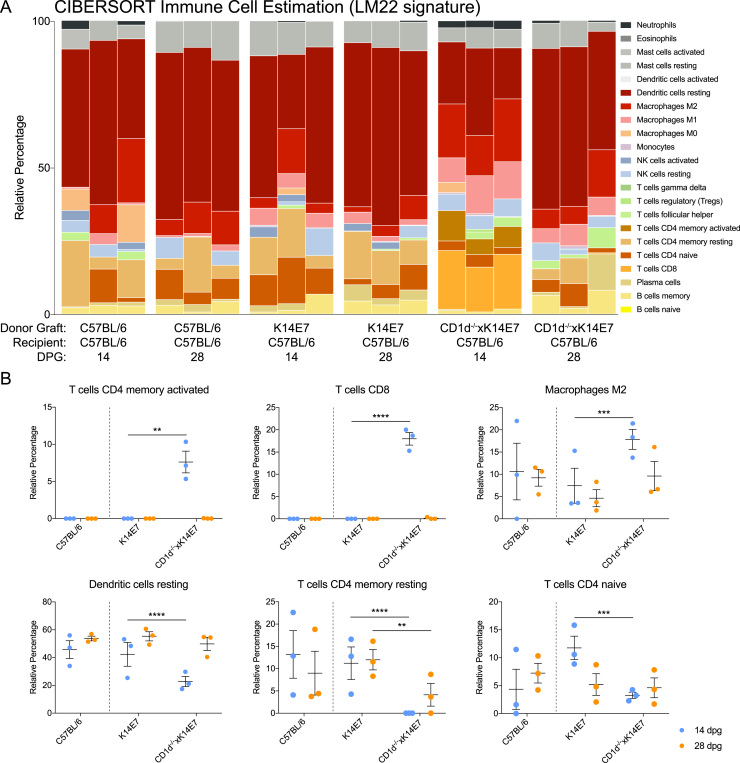
Further, we used CIBERSORT [38], a gene expression deconvolution package that estimates the composition of immune cell types in a bulk sample using defined gene signatures, to probe whether immune cell compositions are altered in the different grafting groups. The LM22 gene signature provided with the package was used in this analysis and it contains gene signatures of 22 human immune cell subtypes rigorously curated and validated by the developers [38]. We note that the analysis is limited, because the LM22 gene signature was curated for microarray data and some genes in the LM22 gene signature were not found in our mouse RNA-seq data. However, the developers have noted that they have “observed significant correlation for specific LM22 populations on paired microarray/RNA-seq TCGA datasets, indicating reasonably robust cross-platform performance”, and 505 of the 547 genes were successfully matched to orthologous symbols for the analysis. Fig. 8A shows the relative percentage of the 22 immune cell subtypes estimated to be present within the various skin graft samples in a stacked bar plot. Of note, there is a dramatically increased proportion of CD8 T cells (light orange) and CD4 memory activated T cells (brown) in CD1d-/-K14E7 14DPG samples, and not the other groups analysed (Fig. 8A). Upon closer examination, we can identify significant decrease in resting dendritic cells, naïve CD4 T cells and resting memory T cells in CD1D-/-xK14E7 skin grafts compared to K14E7 skin grafts (Fig. 8B), as well as significant increase in M2 macrophages, activated memory CD4 T cells and CD8 T cells in CD1D-/-xK14E7 skin grafts compared to K14E7 skin grafts (Fig. 8B).
Fig. 8.
CIBERSORT analysis of immune cell composition in grafting groups. (A) Relative percentage of the estimated immune cells in the various grafting groups. Each of the 22 immune cell types are assigned a different colour on the plot. (B) Relative fractions from (A) are plotted as a dot plot and two-way ANOVA analysis with Bonferonni's post-test was performed, specifically comparing CD1d-/-xK14E7 on C57BL/6 skin grafts versus K14E7 on C57BL/6 skin grafts at 14DPG or 28DPG separately. Significance is denoted by *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
From the published literature and the transcription data analyses described in this paper, the K14E7 transgenic mouse provides a model to study the immunological consequences of persistent/chronic high-risk HPV-related high-grade squamous intraepithelial lesions. Both cellular and molecular observations from these mice correlate well with human clinical literature. However, we acknowledge that there are differences between mouse and human immune systems and between cervical epithelia and squamous skin. Consequently, there may be subtle differences in the local immune response to E7 protein and induced hyperplasia. Our analysis is limited to the comparison of the transcriptomic profiles of the human and mouse data sets – while all tested mRNA differences between normal and E7 transgenic mouse skin have been reflected in protein levels, some untested differences might not correlate. We also note that gene changes might be due to technical differences in ranking metrics employed by the various gene set tests and differential gene expression analysis tools. Lastly, our K14E7 mouse model does not mimic a natural course of the human carcinogenic process, which involves the ongoing acquisition of damaging genetic mutations in tumours in addition to the chronicity of the disease and altered immune response. Nevertheless, the analysis has provided us with an insight into the transcriptional regulation that is maintained/shared between the mouse model and human high-grade squamous intraepithelial lesions. The analysis has additionally revealed local molecular changes associated with graft rejection of E7-expressing skin.
An alternative HPV transgenic mouse model exists in the form of the K14-HPV16 model, which expresses all HPV16 early genes (E1, E2, E4, E5, E6 and E7) [98], [99], in contrast to the single or dual E6/E7-expressing transgenic mice from which we derived our K14E7 mice. The rationale for this cloning strategy was to factor in the roles that the other early genes might play in early events of cell transformation. These transgenic mice also display severe dysplastic squamous epithelial lesions with increasing age, similar to observations in human cervical disease [99]. However, the same study also reported that transgenic mice that do not express E1 or E2, but still expressing the other HPV early genes, were phenotypically identical to the transgenic mice that expressed all HPV16 early genes [98]. This mutant model did not display a more severe phenotype, suggesting that early events due to expression of E1 and E2 play very minor roles for disease progression in a chronic setting. In contrast, single oncogene expression of E5, E6 or E7, or dual expression of E5/E6, E5/E7 or E6/E7 can result in epithelial hyperplasia; in some instances, this led to induction of spontaneous squamous cell carcinoma-like pathology in the mouse skin [100]. However, triple transgenic expression of E5, E6 and E7 induced less pronounced tumours than the dual HPV oncogene expressing counterparts, suggesting an inhibitory role of E5 on E6/E7-induced carcinogenesis [101]. This discrepancy on the role of E5 in the carcinogenic process may be due to differences in the natural kinetics of E5, E6 and E7 expression in an infected cell. Hence, their cooperation for carcinogenesis may be time and stage dependent in different individuals. While the co-expression of E5/E6, E5/E7 and E6/E7 do have synergistic effects, E7 is the most damaging of the three and consequently the most critical for cervical cancer development [reviewed in [102]]. Parenthetically, expression of HPV16 E7 is likely a major contributor of the immunological consequences accompanying high-grade squamous intraepithelial lesions.
Other K14-restricted HPV oncoprotein transgenic mouse models have been described and these models have been used to investigate the oncogenic potential of HPV proteins expressed by other HPV types, such as HPV8 [103] and HPV18 [104]. Specifically, although long-term overexpression of HPV8 E2 gene in K14E2 transgenic mice induced transformation of skin keratinocytes, the rate was very low without addition of further carcinogens [103]. In the HPV18-K14E7 mouse model, there was no demonstrable effect on cervical cancer development although all transgenic mice developed severe cataracts [104], similar to initial observation of mice expressing HPV16 E6 and E7 in the ocular lenses (under the αA-crystallin promoter; αA-HPV16-E6/E7) [105]. While our analyses performed in this study have been restricted to the comparison of K14E7 murine skin expressing E7, the mouse model has been used for studying cervical disease. K14E7 mice have been shown to be able to develop cervical carcinoma when other carcinogens are present, such as administration of oestrogen as a cocarcinogen in the mouse cervix [106]. The oestrogen-induced carcinogenesis model has been used to evaluate the utility of therapy options such as hormone replacement therapy in the form of a progestin drug (medroxyprogesterone acetate) [107]. Xenografts of HPV16 E7 plasmid transfected human foreskin on immunodeficient recipient mice have also been used as a humanised mouse model of HPV-associated disease [108]. Other tumour xenograft models in humanised mice are being explored as models for head and neck HPV-related malignancies and for testing therapeutic vaccine modalities in these settings [109], [110]. We emphasise that there are other non-murine animal models that have been used in studying HPV-associated carcinogenesis and therapeutic strategies, such as experimentally infected cattle, dogs, and rabbits with bovine- canine- and rabbit papillomaviruses respectively. However, the cost and ethical issue of these models limit their clinical utility [111], [112]. More recently, immunocompromised laboratory mice infected by MmuPV1 have shown promising results for investigating early stages of papillomavirus infection but infection in immunocompetent mice did not develop papillomavirus-associated malignancy, limiting the applicability of this model at this stage [9], [10], [11], [12].
Currently, there is considerable attention towards the development of therapeutic vaccination as a mode of anti-HPV immunotherapy for targeting pre-existing infections. In a recent study, Brown et al., used live intra-vital imaging to elegantly demonstrate that healthy skin cells promote the spontaneous regression of cells harbouring damaging mutations at a surprisingly high rate [113], supporting the hypothesis that a healthy tissue microenvironment can efficiently recognise and eliminate transformed cells. Certainly, a baseline level of immune competence by the epithelium is required to help reject cancer. In an ideal situation, therapeutic vaccines against HPV should augment this effect and amplify the interactions between the immune system and the epithelia for efficient removal of infected cells. However, there have been considerable challenges in this space; clinical trials evaluating protein/peptide-, viral vector-, and E7-DNA-based therapeutic vaccines have been inconclusive. For example, protein/peptide vaccines linking the E7 antigen to different pathogen proteins, such as Haemophilus influenza protein D [114], and HSP65 from Mycobacterium bovis [115], [116] were able to invoke a greater immune response from subjects as a result of increased immunogenicity, showing increased vaccine-induced IFNγ production in CD4 and CD8 T cells and ~50% lesion shrinkage [116]. However, the rate of spontaneous regression was similar between the vaccinated and unvaccinated cohort. Promising results from a clinical trial that administered HPV16 E6/E7 peptide vaccines to recruited stage 3 vulvar intraepithelial neoplasia patients demonstrated good induction of CD4 T and CD8 T cell responses and long lasting clinical protection [117]. However, a similar approach utilised for CIN2 and CIN3 therapeutic vaccine development was accompanied with a concomitant induction of CD4+FOXP3+CD25+ regulatory T cells, dampening the response [118]. This highlights a lack of understanding of the crosstalk between the immune system and the HPV-infected skin microenvironment.
To our knowledge, there have been two reports on the use of combination immunotherapy involving PD-L1 blockade with HPV16 therapeutic vaccination in mouse models, eliciting improved anti-tumour responses in both cases [85], [119]. However, vaccine studies showing efficacy against the transplantable TC-1 tumour cell line expressing HPV16 E6 and E7, despite promising results in mouse studies, have failed to predict effective responses in human clinical trials [4]. While these are useful models to assess the efficacy of tumour rejection or HPV infection clearance in an acute setting, the mechanisms involved may be different for pre-malignant lesions on immunocompetent individuals where chronic changes induced by HPV16 are dominant. This is evident from the fact that transplanted TC-1 tumours are very aggressive but can be efficiently prevented and treated in mice with an immunisation protocol. In contrast, we argue that mouse models that chronically express HPV16 oncoproteins on basal keratinocytes can better emulate the persistence of HPV-associated dysplasia and allow for insights into factors that promote lesion or HPV clearance. We propose that the K14E7 transgenic mouse and K14E7 skin grafting model may be of particular use for preclinical testing of HPV immunotherapies.
Acknowledgements
We thank Janin Chandra, Rohit Sinha, Samal Zhussupbekova and the staff of the Biological Research Facility (Brisbane, QLD, Australia) for excellent technical assistance and animal care. This work was supported by the Australian Cancer Research Foundation (ACRF) for the Comprehensive Cancer Genomics Facility at The University of Queensland Diamantina Institute. This work is funded from public competitive grant awarding bodies with no involvement in the conduct of the research, or production of the manuscript. Specifically, Z. K. T is a recipient of the Advance Queensland Early Career Research Fellowship and this work was supported by research project grants from the National Health and Medical Research Council (NHMRC) and the Merchant Charitable Foundation. No payment was received from any pharmaceutical company or other agency to write this manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pvr.2017.10.001.
Appendix A. Supplementary material
Supplementary material
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Song D., Li H., Li H., Dai J. Effect of human papillomavirus infection on the immune system and its role in the course of cervical cancer. Oncol. Lett. 2015;10:600–606. doi: 10.3892/ol.2015.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walhart T. Human papillomavirus biology, pathogenesis, and potential for drug discovery: a literature review for HIV nurse clinical scientists. J. Assoc. Nurses AIDS Care: Janac. 2015;26:693–702. doi: 10.1016/j.jana.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazer I.H., Leggatt G.R., Mattarollo S.R. Prevention and treatment of papillomavirus-related cancers through immunization. Annu. Rev. Immunol. 2011;29:111–138. doi: 10.1146/annurev-immunol-031210-101308. [DOI] [PubMed] [Google Scholar]
- 5.Frazer I. Correlating immunity with protection for HPV infection. Int. J. Infect. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2007;11(Suppl 2):S10–S16. doi: 10.1016/S1201-9712(07)60016-2. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez A.C., Schiffman M., Herrero R., Wacholder S., Hildesheim A., Castle P.E. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J. Natl. Cancer Inst. 2008;100:513–517. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.E. Deligeoroglou, A. Giannouli, N. Athanasopoulos, V. Karountzos, A. Vatopoulou, K. Dimopoulos, et al. HPVinfection: immunological aspects and their utility in future therapy Infectious diseases in obstetrics and gynecology;2013:540850, 2013. [DOI] [PMC free article] [PubMed]
- 8.Monnier-Benoit S., Mauny F., Riethmuller D., Guerrini J.S., Capilna M., Felix S. Immunohistochemical analysis of CD4+ and CD8+ T-cell subsets in high risk human papillomavirus-associated pre-malignant and malignant lesions of the uterine cervix. Gynecol. Oncol. 2006;102:22–31. doi: 10.1016/j.ygyno.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Ingle A., Ghim S., Joh J., Chepkoech I., Bennett Jenson A., Sundberg J.P. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 2011;48:500–505. doi: 10.1177/0300985810377186. [DOI] [PubMed] [Google Scholar]
- 10.Handisurya A., Day P.M., Thompson C.D., Bonelli M., Lowy D.R., Schiller J.T. Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1. PLoS Pathog. 2014;10:e1004314. doi: 10.1371/journal.ppat.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundberg J.P., Stearns T.M., Joh J., Proctor M., Ingle A., Silva K.A. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PloS One. 2014;9:e113582. doi: 10.1371/journal.pone.0113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joh J., Jenson A.B., Proctor M., Ingle A., Silva K.A., Potter C.S. Molecular diagnosis of a laboratory mouse papillomavirus (MusPV) Exp. Mol. Pathol. 2012;93:416–421. doi: 10.1016/j.yexmp.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Jain S., Moore R.A., Anderson D.M., Gough G.W., Stanley M.A. Cell-mediated immune responses to COPV early proteins. Virology. 2006;356:23–34. doi: 10.1016/j.virol.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 14.Wilgenburg B.J., Budgeon L.R., Lang C.M., Griffith J.W., Christensen N.D. Characterization of immune responses during regression of rabbit oral papillomavirus infections. Comp. Med. 2005;55:431–439. [PubMed] [Google Scholar]
- 15.Chen L.P., Thomas E.K., Hu S.L., Hellstrom I., Hellstrom K.E. Human papillomavirus type 16 nucleoprotein E7 is a tumor rejection antigen. Proc. Natl. Acad. Sci. USA. 1991;88:110–114. doi: 10.1073/pnas.88.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazer I.H. Prevention of cervical cancer through papillomavirus vaccination. Nat. Rev. Immunol. 2004;4:46–54. doi: 10.1038/nri1260. [DOI] [PubMed] [Google Scholar]
- 17.Peng S., Song L., Knoff J., Wang J.W., Chang Y.N., Hannaman D. Control of HPV-associated tumors by innovative therapeutic HPV DNA vaccine in the absence of CD4+ T cells. Cell Biosci. 2014;4:11. doi: 10.1186/2045-3701-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng S., Ji H., Trimble C., He L., Tsai Y.C., Yeatermeyer J. Development of a DNA vaccine targeting human papillomavirus type 16 oncoprotein E6. J. Virol. 2004;78:8468–8476. doi: 10.1128/JVI.78.16.8468-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazer I.H., Fernando G.J., Fowler N., Leggatt G.R., Lambert P.F., Liem A. Split tolerance to a viral antigen expressed in thymic epithelium and keratinocytes. Eur. J. Immunol. 1998;28:2791–2800. doi: 10.1002/(SICI)1521-4141(199809)28:09<2791::AID-IMMU2791>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Choyce A., Yong M., Narayan S., Mattarollo S.R., Liem A., Lambert P.F. Expression of a single, viral oncoprotein in skin epithelium is sufficient to recruit lymphocytes. PloS One. 2013;8:e57798. doi: 10.1371/journal.pone.0057798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herber R., Liem A., Pitot H., Lambert P.F. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn L.A., Evander M., Tindle R.W., Bulloch A.L., de Kluyver R.L., Fernando G.J. Presentation of the HPV16E7 protein by skin grafts is insufficient to allow graft rejection in an E7-primed animal. Virology. 1997;235:94–103. doi: 10.1006/viro.1997.8650. [DOI] [PubMed] [Google Scholar]
- 23.Fernando G.J., Stewart T.J., Tindle R.W., Frazer I.H. Th2-type CD4+ cells neither enhance nor suppress antitumor CTL activity in a mouse tumor model. J. Immunol. 1950;1998(161):2421–2427. [PubMed] [Google Scholar]
- 24.Frazer I.H., De Kluyver R., Leggatt G.R., Guo H.Y., Dunn L., White O. Tolerance or immunity to a tumor antigen expressed in somatic cells can be determined by systemic proinflammatory signals at the time of first antigen exposure. J. Immunol. 1950;2001(167):6180–6187. doi: 10.4049/jimmunol.167.11.6180. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto K., Leggatt G.R., Zhong J., Liu X., de Kluyver R.L., Peters T. Impaired antigen presentation and effectiveness of combined active/passive immunotherapy for epithelial tumors. J. Natl. Cancer Inst. 2004;96:1611–1619. doi: 10.1093/jnci/djh301. [DOI] [PubMed] [Google Scholar]
- 26.Broom J.K., Lew A.M., Azukizawa H., Kenna T.J., Leggatt G.R., Frazer I.H. Antigen-specific CD4 cells assist CD8 T-effector cells in eliminating keratinocytes. J. Invest. Dermatol. 2010;130:1581–1589. doi: 10.1038/jid.2010.17. [DOI] [PubMed] [Google Scholar]
- 27.Mattarollo S.R., Yong M., Tan L., Frazer I.H., Leggatt G.R. Secretion of IFN-gamma but not IL-17 by CD1d-restricted NKT cells enhances rejection of skin grafts expressing epithelial cell-derived antigen. J. Immunol. 1950;2010(184):5663–5669. doi: 10.4049/jimmunol.0903730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadis U., Leggatt G.R., Thomas R., Frazer I.H., Kovacs E.M. IL-1 signalling determines the fate of skin grafts expressing non-self protein in keratinocytes. Exp. Dermatol. 2010;19:723–729. doi: 10.1111/j.1600-0625.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong J., Hadis U., De Kluyver R., Leggatt G.R., Fernando G.J., Frazer I.H. TLR7 stimulation augments T effector-mediated rejection of skin expressing neo-self antigen in keratinocytes. Eur. J. Immunol. 2008;38:73–81. doi: 10.1002/eji.200737599. [DOI] [PubMed] [Google Scholar]
- 30.den Boon J.A., Pyeon D., Wang S.S., Horswill M., Schiffman M., Sherman M. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc. Natl. Acad. Sci. USA. 2015;112:E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhussupbekova S., Sinha R., Kuo P., Lambert P.F., Frazer I.H., Tuong Z.K. A mouse model of hyperproliferative human epithelium validated by keratin profiling shows an aberrant cytoskeletal response to injury. EBioMedicine. 2016;9:314–323. doi: 10.1016/j.ebiom.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law C.W., Alhamdoosh M., Su S., Smyth G.K., Ritchie M.E. RNA-seq analysis is easy as 1-2-3 with limma. Glimma edgeR. F1000Research. 2016;5:1408. doi: 10.12688/f1000research.9005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., Smyth G.K. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrero Y., Callejas D., Alana F., Silva C., Mindiola R., Mosquera J. Increased vascular endothelial growth factor expression, CD3-positive cell infiltration, and oxidative stress in premalignant lesions of the cervix. Cancer. 2009;115:3680–3688. doi: 10.1002/cncr.24411. [DOI] [PubMed] [Google Scholar]
- 41.Hu T., Yang P., Zhu H., Chen X., Xie X., Yang M. Accumulation of invariant NKT cells with increased IFN-gamma production in persistent high-risk HPV-infected high-grade cervical intraepithelial neoplasia. Diagn. Pathol. 2015;10:20. doi: 10.1186/s13000-015-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaafar F., Righi E., Lindstrom V., Linton C., Nohadani M., Van Noorden S. Correlation of CXCL12 expression and FoxP3+ cell infiltration with human papillomavirus infection and clinicopathological progression of cervical cancer. Am. J. Pathol. 2009;175:1525–1535. doi: 10.2353/ajpath.2009.090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adurthi S., Krishna S., Mukherjee G., Bafna U.D., Devi U., Jayshree R.S. Regulatory T cells in a spectrum of HPV-induced cervical lesions: cervicitis, cervical intraepithelial neoplasia and squamous cell carcinoma. Am. J. Reprod. Immunol. 2008;60:55–65. doi: 10.1111/j.1600-0897.2008.00590.x. [DOI] [PubMed] [Google Scholar]
- 44.Kojima S., Kawana K., Tomio K., Yamashita A., Taguchi A., Miura S. The prevalence of cervical regulatory T cells in HPV-related cervical intraepithelial neoplasia (CIN) correlates inversely with spontaneous regression of CIN. Am. J. Reprod. Immunol. 2013;69:134–141. doi: 10.1111/aji.12030. [DOI] [PubMed] [Google Scholar]
- 45.Piersma S.J., Jordanova E.S., van Poelgeest M.I., Kwappenberg K.M., van der Hulst J.M., Drijfhout J.W. High number of intraepithelial CD8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage cervical cancer. Cancer Res. 2007;67:354–361. doi: 10.1158/0008-5472.CAN-06-3388. [DOI] [PubMed] [Google Scholar]
- 46.Li L., Ma Y., Liu S., Zhang J., Xu X.Y. Interleukin 10 promotes immune response by increasing the survival of activated CD8+ T cells in human papillomavirus 16-infected cervical cancer. Tumour Biol. 2016 doi: 10.1007/s13277-016-5466-3. [DOI] [PubMed] [Google Scholar]
- 47.Trimble C.L., Clark R.A., Thoburn C., Hanson N.C., Tassello J., Frosina D. Human papillomavirus 16-associated cervical intraepithelial neoplasia in humans excludes CD8 T cells from dysplastic epithelium. J. Immunol. 1950;2010(185):7107–7114. doi: 10.4049/jimmunol.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Q., Wei H., Morihara J., Stern J., Yu M., Kiviat N. Th2 type inflammation promotes the gradual progression of HPV-infected cervical cells to cervical carcinoma. Gynecol. Oncol. 2012;127:412–419. doi: 10.1016/j.ygyno.2012.07.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peghini B.C., Abdalla D.R., Barcelos A.C., Teodoro L., Murta E.F., Michelin M.A. Local cytokine profiles of patients with cervical intraepithelial and invasive neoplasia. Human. Immunol. 2012;73:920–926. doi: 10.1016/j.humimm.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi A., Weinberg V., Darragh T., Smith-McCune K. Evolving immunosuppressive microenvironment during human cervical carcinogenesis. Mucosal Immunol. 2008;1:412–420. doi: 10.1038/mi.2008.33. [DOI] [PubMed] [Google Scholar]
- 51.Scott M.E., Ma Y., Kuzmich L., Moscicki A.B. Diminished IFN-gamma and IL-10 and elevated Foxp3 mRNA expression in the cervix are associated with CIN 2 or 3. Int. J. Cancer. 2009;124:1379–1383. doi: 10.1002/ijc.24117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang X., Zhang Y., Wei H., Zhang J., Luo Q., Huang C. Expression and effects of high-mobility group box 1 in cervical cancer. Int. J. Mol. Sci. 2014;15:8699–8712. doi: 10.3390/ijms15058699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao M., Li Y., Wei X., Zhang Q., Jia H., Quan S. Negative immune factors might predominate local tumor immune status and promote carcinogenesis in cervical carcinoma. Virol. J. 2017;14:5. doi: 10.1186/s12985-016-0670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punt S., van Vliet M.E., Spaans V.M., de Kroon C.D., Fleuren G.J., Gorter A. FoxP3+ and IL-17+ cells are correlated with improved prognosis in cervical adenocarcinoma. Cancer Immunol. Immunother. 2015;64:745–753. doi: 10.1007/s00262-015-1678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali K.S., Ali H.Y., Jubrael J.M. Concentration levels of IL-10 and TNFalpha cytokines in patients with human papilloma virus (HPV) DNA(+) and DNA(-) cervical lesions. J. Immunotoxicol. 2012;9:168–172. doi: 10.3109/1547691X.2011.642419. [DOI] [PubMed] [Google Scholar]
- 56.Bermudez-Morales V.H., Gutierrez L.X., Alcocer-Gonzalez J.M., Burguete A., Madrid-Marina V. Correlation between IL-10 gene expression and HPV infection in cervical cancer: a mechanism for immune response escape. Cancer Invest. 2008;26:1037–1043. doi: 10.1080/07357900802112693. [DOI] [PubMed] [Google Scholar]
- 57.Prata T.T., Bonin C.M., Ferreira A.M., Padovani C.T., Fernandes C.E., Machado A.P. Local immunosuppression induced by high viral load of human papillomavirus: characterization of cellular phenotypes producing interleukin-10 in cervical neoplastic lesions. Immunology. 2015;146:113–121. doi: 10.1111/imm.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma A., Rajappa M., Saxena A., Sharma M. Cytokine profile in Indian women with cervical intraepithelial neoplasia and cancer cervix. Int. J. Gynecol. Cancer.: Off. J. Int. Gynecol. Cancer. Soc. 2007;17:879–885. doi: 10.1111/j.1525-1438.2007.00883.x. [DOI] [PubMed] [Google Scholar]
- 59.Syrjanen S., Longhato-Filho A., Sarian L.O., Naud P., Derchain S., Rottelli-Martins C. Competing-risks regression models in analysis of biomarkers as predictors of high-risk human papillomavirus (HPV) infection outcomes and incident CIN in the LAMS cohort. Int. J. Gynecol. Pathol.: Off. J. Int. Soc. Gynecol. Pathol. 2013;32:406–415. doi: 10.1097/PGP.0b013e31826739b1. [DOI] [PubMed] [Google Scholar]
- 60.Yang W., Song Y., Lu Y.L., Sun J.Z., Wang H.W. Increased expression of programmed death (PD)-1 and its ligand PD-L1 correlates with impaired cell-mediated immunity in high-risk human papillomavirus-related cervical intraepithelial neoplasia. Immunology. 2013;139:513–522. doi: 10.1111/imm.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniilidis A., Koutsos J., Oikonomou Z., Nasioutziki M., Hatziparadisi K., Tantanasis T. Cytokines of cervical mucosa and human papilloma virus infection of the cervix: a descriptive study. Acta Cytol. 2016;60:58–64. doi: 10.1159/000445161. [DOI] [PubMed] [Google Scholar]
- 62.Ortiz-Sanchez E., Chavez-Olmos P., Pina-Sanchez P., Salcedo M., Garrido E. Expression of the costimulatory molecule CD86, but not CD80, in keratinocytes of normal cervical epithelium and human papillomavirus-16 positive low squamous intraepithelial lesions. Int. J. Gynecol. Pathol.: Off. J. Int. Soc. Gynecol. Pathol. 2007;17:571–580. doi: 10.1111/j.1525-1438.2007.00904.x. [DOI] [PubMed] [Google Scholar]
- 63.Scott M.E., Shvetsov Y.B., Thompson P.J., Hernandez B.Y., Zhu X., Wilkens L.R. Cervical cytokines and clearance of incident human papillomavirus infection: Hawaii HPV cohort study. Int. J. Cancer. 2013;133:1187–1196. doi: 10.1002/ijc.28119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Burg S.H., Piersma S.J., de Jong A., van der Hulst J.M., Kwappenberg K.M., van den Hende M. Association of cervical cancer with the presence of CD4+ regulatory T cells specific for human papillomavirus antigens. Proc. Natl. Acad. Sci. USA. 2007;104:12087–12092. doi: 10.1073/pnas.0704672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Origoni M., Parma M., Dell'Antonio G., Gelardi C., Stefani C., Salvatore S. Prognostic significance of immunohistochemical phenotypes in patients treated for high-grade cervical intraepithelial neoplasia. BioMed. Res. Int. 2013:831907. doi: 10.1155/2013/831907. (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ovestad I.T., Gudlaugsson E., Skaland I., Malpica A., Kruse A.J., Janssen E.A. Local immune response in the microenvironment of CIN2-3 with and without spontaneous regression. Mod. Pathol.: Off. J. U. S. Can. Acad. Pathol., Inc. 2010;23:1231–1240. doi: 10.1038/modpathol.2010.109. [DOI] [PubMed] [Google Scholar]
- 67.Ovestad I.T., Gudlaugsson E., Skaland I., Malpica A., Munk A.C., Janssen E.A. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. J. Clin. Pathol. 2011;64:303–307. doi: 10.1136/jcp.2010.083626. [DOI] [PubMed] [Google Scholar]
- 68.Benci J.L., Xu B., Qiu Y., Wu T.J., Dada H., Twyman-Saint Victor C. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(1540–54):e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L., Li H., Liang F., Hong Y., Jiang S., Xiao L. Examining IL-33 expression in the cervix of HPV-infected patients: a preliminary study comparing IL-33 levels in different stages of disease and analyzing its potential association with IFN-gamma. Med. Oncol. 2014;31:143. doi: 10.1007/s12032-014-0143-0. [DOI] [PubMed] [Google Scholar]
- 70.McKenzie J., King A., Hare J., Fulford T., Wilson B., Stanley M. Immunocytochemical characterization of large granular lymphocytes in normal cervix and HPV associated disease. J. Pathol. 1991;165:75–80. doi: 10.1002/path.1711650112. [DOI] [PubMed] [Google Scholar]
- 71.Song S.H., Lee J.K., Lee N.W., Saw H.S., Kang J.S., Lee K.W. Interferon-gamma (IFN-gamma): a possible prognostic marker for clearance of high-risk human papillomavirus (HPV) Gynecol. Oncol. 2008;108:543–548. doi: 10.1016/j.ygyno.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 72.Hammes L.S., Tekmal R.R., Naud P., Edelweiss M.I., Kirma N., Valente P.T. Macrophages, inflammation and risk of cervical intraepithelial neoplasia (CIN) progression--clinicopathological correlation. Gynecol. Oncol. 2007;105:157–165. doi: 10.1016/j.ygyno.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 73.Hayati A.R., Zulkarnaen M. An immunohistochemical study of CD1a and CD83-positive infiltrating dendritic cell density in cervical neoplasia. Int. J. Gynecol. Pathol.: Off. J. Int. Soc. Gynecol. Pathol. 2007;26:83–88. doi: 10.1097/01.pgp.0000225850.90115.bc. [DOI] [PubMed] [Google Scholar]
- 74.Jiang B., Xue M. Correlation of E6 and E7 levels in high-risk HPV16 type cervical lesions with CCL20 and Langerhans cells. Genet. Mol. Res.: GMR. 2015;14:10473–10481. doi: 10.4238/2015.September.8.8. [DOI] [PubMed] [Google Scholar]
- 75.Pahne-Zeppenfeld J., Schroer N., Walch-Ruckheim B., Oldak M., Gorter A., Hegde S. Cervical cancer cell-derived interleukin-6 impairs CCR7-dependent migration of MMP-9-expressing dendritic cells. Int J. Cancer. 2014;134:2061–2073. doi: 10.1002/ijc.28549. [DOI] [PubMed] [Google Scholar]
- 76.de Vos van Steenwijk P.J., Ramwadhdoebe T.H., Goedemans R., Doorduijn E.M., van Ham J.J., Gorter A. Tumor-infiltrating CD14-positive myeloid cells and CD8-positive T-cells prolong survival in patients with cervical carcinoma. Int J. Cancer. 2013;133:2884–2894. doi: 10.1002/ijc.28309. [DOI] [PubMed] [Google Scholar]
- 77.van Meir H., Nout R.A., Welters M.J., Loof N.M., de Kam M.L., van Ham J.J. Impact of (chemo)radiotherapy on immune cell composition and function in cervical cancer patients. Oncoimmunology. 2017;6:e1267095. doi: 10.1080/2162402X.2016.1267095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mezache L., Paniccia B., Nyinawabera A., Nuovo G.J. Enhanced expression of PD L1 in cervical intraepithelial neoplasia and cervical cancers. Mod. Pathol.: Off. J. U. S. Can. Acad. Pathol., Inc. 2015;28:1594–1602. doi: 10.1038/modpathol.2015.108. [DOI] [PubMed] [Google Scholar]
- 79.Walch-Ruckheim B., Mavrova R., Henning M., Vicinus B., Kim Y.J., Bohle R.M. Stromal fibroblasts induce CCL20 through IL6/C/EBPbeta to support the recruitment of Th17 cells during cervical cancer progression. Cancer Res. 2015;75:5248–5259. doi: 10.1158/0008-5472.CAN-15-0732. [DOI] [PubMed] [Google Scholar]
- 80.Ikebuchi R., Teraguchi S., Vandenbon A., Honda T., Shand F.H., Nakanishi Y. A rare subset of skin-tropic regulatory T cells expressing Il10/Gzmb inhibits the cutaneous immune response. Sci. Rep. 2016;6:35002. doi: 10.1038/srep35002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Narayan S., Choyce A., Linedale R., Saunders N.A., Dahler A., Chan E. Epithelial expression of human papillomavirus type 16 E7 protein results in peripheral CD8 T-cell suppression mediated by CD4+CD25+ T cells. Eur. J. Immunol. 2009;39:481–490. doi: 10.1002/eji.200838527. [DOI] [PubMed] [Google Scholar]
- 82.Mattarollo S.R., Yong M., Gosmann C., Choyce A., Chan D., Leggatt G.R. NKT cells inhibit antigen-specific effector CD8 T cell induction to skin viral proteins. J. Immunol. 1950;2011(187):1601–1608. doi: 10.4049/jimmunol.1100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mattarollo S.R., Rahimpour A., Choyce A., Godfrey D.I., Leggatt G.R., Frazer I.H. Invariant NKT cells in hyperplastic skin induce a local immune suppressive environment by IFN-gamma production. J. Immunol. 1950;2010(184):1242–1250. doi: 10.4049/jimmunol.0902191. [DOI] [PubMed] [Google Scholar]
- 84.Mittal D., Kassianos A.J., Tran L.S., Bergot A.S., Gosmann C., Hofmann J. Indoleamine 2,3-dioxygenase activity contributes to local immune suppression in the skin expressing human papillomavirus oncoprotein e7. J. Invest. Dermatol. 2013;133(2686–94) doi: 10.1038/jid.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chandra J., Dutton J.L., Li B., Woo W.P., Xu Y., Tolley L.K. DNA vaccine encoding HPV16 oncogenes E6 and E7 induces potent cell-mediated and Humoral immunity which protects in tumor challenge and drives E7-expressing skin graft rejection. J. Immunother. 1997;2017(40):62–70. doi: 10.1097/CJI.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chandra J., Miao Y., Romoff N., Frazer I.H. Epithelium expressing the E7 oncoprotein of HPV16 attracts immune-modulatory dendritic cells to the skin and suppresses their antigen-processing capacity. PLoS One. 2016;11:e0152886. doi: 10.1371/journal.pone.0152886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abd Warif N.M., Stoitzner P., Leggatt G.R., Mattarollo S.R., Frazer I.H., Hibma M.H. Langerhans cell homeostasis and activation is altered in hyperplastic human papillomavirus type 16 E7 expressing epidermis. PLoS One. 2015;10:e0127155. doi: 10.1371/journal.pone.0127155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jemon K., Leong C.M., Ly K., Young S.L., McLellan A.D., Hibma M.H. Suppression of the CD8 T cell response by human papillomavirus type 16 E7 occurs in Langerhans cell-depleted mice. Sci. Rep. 2016;6:34789. doi: 10.1038/srep34789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jazayeri S.D., Kuo P.T., Leggatt G.R., Frazer I.H. HPV16-E7-specific activated CD8 T cells in E7 transgenic skin and skin grafts. Front. Immunol. 2017;8:524. doi: 10.3389/fimmu.2017.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Del Pino M., Svanholm-Barrie C., Torne A., Marimon L., Gaber J., Sagasta A. mRNA biomarker detection in liquid-based cytology: a new approach in the prevention of cervical cancer. Mod. Pathol. 2015;28:312–320. doi: 10.1038/modpathol.2014.106. [DOI] [PubMed] [Google Scholar]
- 91.Peres A.L., Paz E.S.K.M., de Araujo R.F., de Lima Filho J.L., de Melo Junior M.R., Martins D.B. Immunocytochemical study of TOP2A and Ki-67 in cervical smears from women under routine gynecological care. J. Biomed. Sci. 2016;23:42. doi: 10.1186/s12929-016-0258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buitrago-Perez A., Garaulet G., Vazquez-Carballo A., Paramio J.M., Garcia-Escudero R. Molecular signature of HPV-induced carcinogenesis: pRb, p53 and gene expression profiling. Curr. Genom. 2009;10:26–34. doi: 10.2174/138920209787581235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gabrielli B., Bokhari F., Ranall M.V., Oo Z.Y., Stevenson A.J., Wang W. Aurora A is critical for survival in HPV-transformed cervical cancer. Mol. Cancer Ther. 2015;14:2753–2761. doi: 10.1158/1535-7163.MCT-15-0506. [DOI] [PubMed] [Google Scholar]
- 94.Martin D., Fallaha S., Proctor M., Stevenson A., Perrin L., McMillan N. Inhibition of Aurora A and Aurora B is required for the sensitivity of HPV-driven cervical cancers to Aurora kinase inhibitors. Mol. Cancer Ther. 2017 doi: 10.1158/1535-7163.MCT-17-0159. [DOI] [PubMed] [Google Scholar]
- 95.Liang D., Xiao-Feng H., Guan-Jun D., Er-Ling H., Sheng C., Ting-Ting W. Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim. Biophys. Acta. 2015;1852:2494–2503. doi: 10.1016/j.bbadis.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 96.Cretney E., Xin A., Shi W., Minnich M., Masson F., Miasari M. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 97.Cipolletta D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arbeit J.M., Munger K., Howley P.M., Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 1994;68:4358–4368. doi: 10.1128/jvi.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith-McCune K., Zhu Y.H., Hanahan D., Arbeit J. Cross-species comparison of angiogenesis during the premalignant stages of squamous carcinogenesis in the human cervix and K14-HPV16 transgenic mice. Cancer Res. 1997;57:1294–1300. [PubMed] [Google Scholar]
- 100.Maufort J.P., Williams S.M., Pitot H.C., Lambert P.F. Human papillomavirus 16 E5 oncogene contributes to two stages of skin carcinogenesis. Cancer Res. 2007;67:6106–6112. doi: 10.1158/0008-5472.CAN-07-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maufort J.P., Shai A., Pitot H.C., Lambert P.F. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res. 2010;70:2924–2931. doi: 10.1158/0008-5472.CAN-09-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lambert P.F. Transgenic mouse models of tumor virus action. Annu. Rev. Virol. 2016;3:473–489. doi: 10.1146/annurev-virology-100114-054908. [DOI] [PubMed] [Google Scholar]
- 103.Pfefferle R., Marcuzzi G.P., Akgul B., Kasper H.U., Schulze F., Haase I. The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J. Invest. Dermatol. 2008;128:2310–2315. doi: 10.1038/jid.2008.73. [DOI] [PubMed] [Google Scholar]
- 104.Ghim S., Jenson A.B., Bubier J.A., Silva K.A., Smith R.S., Sundberg J.P. Cataracts in transgenic mice caused by a human papillomavirus type 18 E7 oncogene driven by KRT1-14. Exp. Mol. Pathol. 2008;85:77–82. doi: 10.1016/j.yexmp.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Griep A.E., Herber R., Jeon S., Lohse J.K., Dubielzig R.R., Lambert P.F. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol. 1993;67:1373–1384. doi: 10.1128/jvi.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Riley R.R., Duensing S., Brake T., Munger K., Lambert P.F., Arbeit J.M. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 107.Mehta F.F., Baik S., Chung S.H. Recurrence of cervical cancer and its resistance to progestin therapy in a mouse model. Oncotarget. 2017;8:2372–2380. doi: 10.18632/oncotarget.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buitrago-Perez A., Hachimi M., Duenas M., Lloveras B., Santos A., Holguin A. A humanized mouse model of HPV-associated pathology driven by E7 expression. PLoS One. 2012;7:e41743. doi: 10.1371/journal.pone.0041743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ahn J., Bishop J.A., Akpeng B., Pai S.I., Best S.R. Xenograft model for therapeutic drug testing in recurrent respiratory papillomatosis. Ann. Otol., Rhinol. Laryngol. 2015;124:110–115. doi: 10.1177/0003489414546400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kimple R.J., Harari P.M., Torres A.D., Yang R.Z., Soriano B.J., Yu M. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clin. Cancer Res. 2013;19:855–864. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Campo M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- 112.Peh W.L., Middleton K., Christensen N., Nicholls P., Egawa K., Sotlar K. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 2002;76:10401–10416. doi: 10.1128/JVI.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brown S., Pineda C.M., Xin T., Boucher J., Suozzi K.C., Park S. Correction of aberrant growth preserves tissue homeostasis. Nature. 2017;548:334–337. doi: 10.1038/nature23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Corona Gutierrez C.M., Tinoco A., Navarro T., Contreras M.L., Cortes R.R., Calzado P. Therapeutic vaccination with MVA E2 can eliminate precancerous lesions (CIN 1, CIN 2, and CIN 3) associated with infection by oncogenic human papillomavirus. Hum. Gene Ther. 2004;15:421–431. doi: 10.1089/10430340460745757. [DOI] [PubMed] [Google Scholar]
- 115.Hallez S., Simon P., Maudoux F., Doyen J., Noel J.C., Beliard A. Phase I/II trial of immunogenicity of a human papillomavirus (HPV) type 16 E7 protein-based vaccine in women with oncogenic HPV-positive cervical intraepithelial neoplasia. Cancer Immunol. Immunother. 2004;53:642–650. doi: 10.1007/s00262-004-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Einstein M.H., Kadish A.S., Burk R.D., Kim M.Y., Wadler S., Streicher H. Heat shock fusion protein-based immunotherapy for treatment of cervical intraepithelial neoplasia III. Gynecol. Oncol. 2007;106:453–460. doi: 10.1016/j.ygyno.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kenter G.G., Welters M.J., Valentijn A.R., Lowik M.J., Berends-van der Meer D.M., Vloon A.P. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 2009;361:1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- 118.Welters M.J., Kenter G.G., Piersma S.J., Vloon A.P., Lowik M.J., Berends-van der Meer D.M. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin. Cancer Res. 2008;14:178–187. doi: 10.1158/1078-0432.CCR-07-1880. [DOI] [PubMed] [Google Scholar]
- 119.Liu Z., Zhou H., Wang W., Fu Y.X., Zhu M. A novel dendritic cell targeting HPV16 E7 synthetic vaccine in combination with PD-L1 blockade elicits therapeutic antitumor immunity in mice. Oncoimmunology. 2016;5:e1147641. doi: 10.1080/2162402X.2016.1147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gosmann C., Mattarollo S.R., Bridge J.A., Frazer I.H., Blumenthal A. IL-17 suppresses immune effector functions in human papillomavirus-associated epithelial hyperplasia. J. Immunol. 1950;2014(193):2248–2257. doi: 10.4049/jimmunol.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gosmann C., Frazer I.H., Mattarollo S.R., Blumenthal A.I.L.-18. but not IL-12, induces production of IFN-gamma in the immunosuppressive environment of HPV16 E7 transgenic hyperplastic skin. J. Invest. Dermatol. 2014;134:2562–2569. doi: 10.1038/jid.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bergot A.S., Ford N., Leggatt G.R., Wells J.W., Frazer I.H., Grimbaldeston M.A. HPV16-E7 expression in squamous epithelium creates a local immune suppressive environment via CCL2- and CCL5- mediated recruitment of mast cells. PLoS Pathog. 2014;10:e1004466. doi: 10.1371/journal.ppat.1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material