Abstract
Hypermethylated-in-Cancer 1 (Hic1) is a tumor suppressor gene frequently inactivated by epigenetic silencing and loss-of-heterozygosity in a broad range of cancers. Loss of HIC1, a sequence-specific zinc finger transcriptional repressor, results in deregulation of genes that promote a malignant phenotype in a lineage-specific manner. In particular, upregulation of the HIC1 target gene SIRT1, a histone deacetylase, can promote tumor growth by inactivating TP53. An alternate line of evidence suggests that HIC1 can promote the repair of DNA double strand breaks through an interaction with MTA1, a component of the nucleosome remodeling and deacetylase (NuRD) complex. Using a conditional knockout mouse model of tumor initiation, we now show that inactivation of Hic1 results in cell cycle arrest, premature senescence, chromosomal instability and spontaneous transformation in vitro. This phenocopies the effects of deleting Brca1, a component of the homologous recombination DNA repair pathway, in mouse embryonic fibroblasts. These effects did not appear to be mediated by deregulation of Hic1 target gene expression or loss of Tp53 function, and rather support a role for Hic1 in maintaining genome integrity during sustained replicative stress. Loss of Hic1 function also cooperated with activation of oncogenic KRas in the adult airway epithelium of mice, resulting in the formation of highly pleomorphic adenocarcinomas with a micropapillary phenotype in vivo. These results suggest that loss of Hic1 expression in the early stages of tumor formation may contribute to malignant transformation through the acquisition of chromosomal instability.
Introduction
Hypermethylated-in-Cancer-1 (HIC1) was discovered during a screen for highly methylated tumor suppressor genes on chromosome 17p [1]. HIC1 resides immediately telomeric to TP53 at 17p13.3, and encodes a BTB/POZ domain zinc finger transcriptional repressor closely related to the PLZF family of proteins [1–5]. The locus also contains a Tp53 response element, and two major CpG rich promoters that generate different alternatively spliced transcripts [1, 6–8]. Over the last decade, several groups have shown that epigenetic gene silencing and/or loss of heterozygosity of HIC1 is one of the most common events in human cancer [9–12], and this is associated with poor outcomes in a wide variety of tumor types [12–17], including lung cancer [18].
Homozygous deletion of Hic1 with a conventional mouse knockout approach results in mid-gestation embryonic lethality [19], whereas heterozygous mutants develop a range of spontaneous tumors in an age-dependent manner [7]. Furthermore, Hic1 mutant mice demonstrate an accelerated tumor phenotype when crossed into established genetic models of colorectal cancer [20], medulloblastoma [21], and osteosarcoma [22].
The conventional model of Hic1 function is based on the identification of transcriptional targets through a combination of Hic1 re-expression in cancer cells and gene expression profiling [5, 21, 23–25]. Based on this premise, loss of Hic1 expression through promoter hypermethylation results in aberrant overexpression of lineage-specific genes that promote or maintain the malignant phenotype [5]. In addition, Hic1 also has been shown to attenuate the effect of oncogenic transcription factors complexes that mediate WNT [26] or STAT3 [24, 27] signaling.
One important transcriptional target of Hic1 is the Sirtuin ortholog Sirt1 [28]. Sirtuins belong to the NAD-dependent, trichostatin-insensitive class III histone deacetylases that mediate life span extension in response to caloric restriction in yeast [29]. Importantly, Sirt1 actively deacetylates and inactivates p53 [30], thus establishing a direct link between loss of Hic1 and attenuation of Tp53 signaling [28]. By contrast, mice carrying heterozygous mutations in both Tp53 and Hic1 in cis rapidly develop osteosarcomas in which the wild type copies of both genes are deleted [22], suggesting that loss of both genes predisposes to tumor development that cannot be fully explained through a Hic1-Sirt1-p53 dependent mechanism.
To better define the function of Hic1 in suppressing tumor initiation, we developed a conditional deletion mouse mutant to overcome the embryonic lethality of the Hic1 knockout mouse, and bypass the reliance on stochastic promoter methylation of the wild type allele in Hic1 heterozygous mutants [7].
Results and discussion
Growth arrest and premature senescence in Hic1-deficient mouse embryonic fibroblasts
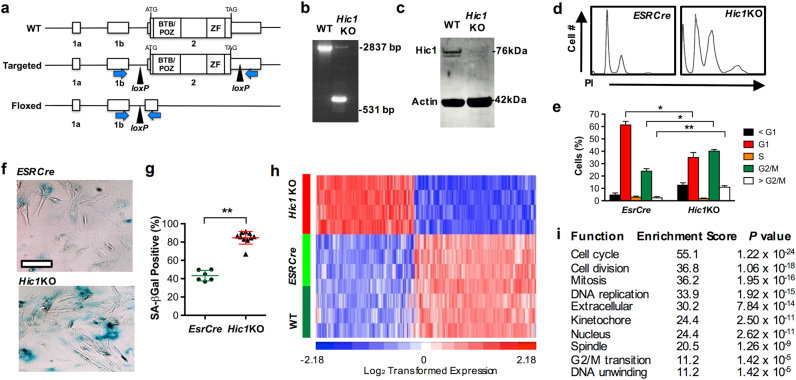
Using targeted homologous recombination in mouse embryonic stem cells, we introduced loxP sites flanking Exon 2 of the Hic1 locus (Fig. 1a). Mice homozygous for the conditional allele (hereafter Hic1lox/lox) demonstrated no developmental phenotype, were not cancer prone, and were fertile (data not shown). To generate a conditional mutant model in mouse embryonic fibroblasts (MEFs), we crossed the Hic1lox/lox mouse with a line in which a Cre recombinase transgene fused to the estradiol receptor has been introduced into the constitutively expressed ROSA locus (hereafter EsrCre) [31]. Ex-vivo treatment with tamoxifen induces activation and translocation of Cre recombinase resulting in loxP recombination with high efficiency.
Fig. 1. Conditional deletion of Hic1 in mouse embryonic fibroblasts (MEFs).
Experiments in this Figure were analyzed by an unblinded observer unless otherwise stated. Female mice used in this study were housed under SPF conditions with a standard day/night cycle and fed ad libitum on a pure C57Bl6 background following approval by the Monash Animal Ethics Committee (MMCA/2012/23; MMCA/2012/24; MMCA/2013/26) in accordance with current National Health and Medical Research Council guidelines. All mice were obtained from Jackson Laboratories except for the conditional Hic1lox/lox transgenic line, which was generated by Ozgene (Perth, WA, Australia). Genotyping was performed with Jackson Laboratory protocols except for the Hic1lox/lox line which was genotyped using primers as follows: Fwd 5′-cgcagaccacgcacttcct-3′, Rev 5′-cccaggctaaggcactaaacag-3′, 486 wt; 312 mutant. MEFs were generated and cultured as described [65]. a Targeting strategy, showing exons 1a, 1b and 2 and loxP sites in the wild type (WT), targeted and floxed locus following Cre-mediated excision. PCR primers to detect excision of exon 2 are shown as blue arrows. Primers: Fwd 5′- caacctgtacgtgtgcatcc-3′ and Rev 5′- cagctaaagttgggctcagg-3′. b Genomic PCR using the primers indicated in Fig. 1a. from wild type (WT) or EsrCre-Hic1lox/lox (Hic1KO) MEFs treated with tamoxifen to induce Cre-mediated recombination. c Western blot analysis of Hic1 and Actin expression in MEF cell lysates from the same experiment shown in Fig. 1b. To generate the Hic1 antibody, full length human HIC1 was cloned into the pET-15b vector and soluble recombinant full length HIC1 protein. Antiserum against full length HIC1 protein was raised in rabbits by the Antibody Facility at Flinders University of South Australia. Hic1 antibodies were purified from serum using a NAb Protein A Plus Spin Kit (Thermo Scientific, Waltham, MA, USA, #89978). Validation experiments relating to this antibody are shown in Supplementary Fig. S1. The Actin antibody was obtained from Abcam, Cambridge, UK (#abactn05). d Representative DNA histograms from EsrCre and Hic1KO MEFs 48 h following treatment with tamoxifen. e Quantitiative analysis of the data shown in Fig. 1f. n = 4, mean + SEM, **P < 0.01, *P < 0.05, one-way ANOVA with Bonferonni correction. Cell cycle analysis and sample sizewas performed as described [66, 67]. f Phase contrast photomicrographs of senescence-associated β-Galactosidase (β-Gal) staining in EsrCre or EsrCre-Hic1lox/lox (Hic1KO) MEFs 5 days following tamoxifen treatment. Scale bar = 50 μm. Cells were stained and scored by an observed blinded to the MEF genotype as described [68]. g Quantitative assessment of β-Gal staining in the same experiment shown in Fig. 1h. n = 5–10, **P < 0.01 unpaired t-test. h A heat map depicting differentially expressed genes from preimmortal wild type (WT), EsrCre and Hic1KO MEFs 48 h following treatment with tamoxifen, performed by the Australia Genome Research Facility (Melbourne, VIC, Australia) using the MouseWG-6 v2.0 Expression BeadChip (Illumina, San Diego, CA, USA) as previously described [69]. Detailed bioinformatic methods are described in Supplementary Information. Array data are available through GEO, GSE104394. i Gene ontology analysis of differentially expressed genes in Hic1KO MEFs compared with WT and EsrCre MEFs 2 days following tamoxifen treatment
Treatment of MEFs generated from Hic1lox/lox EsrCre embryos (hereafter Hic1KO) with tamoxifen, followed by a 48 h incubation period confirmed deletion of Exon 2, which contains the entire coding region of the Hic1 gene (Fig. 1b). Loss of Hic1 protein expression in the same MEF model was also confirmed by Western blot analysis (Fig. 1c).
In contrast to EsrCre MEFs, tamoxifen treated Hic1KO MEFs displayed a marked G2/M arrest (Figs. 1d, e), and premature activation of senescence-associated beta-galactosidase (SA-βGal) (Figs. 1f, g). This was unexpected, since Hic1 is a known tumor suppressor gene, and because its capacity to prevent Tp53 deacetylation through transcriptional repression of Sirt1 would have predicted that Hic1 deletion would phenocopy p53 deletion in this model. Equally surprising was the pattern of gene expression in Hic1KO MEFs 48 h after tamoxifen treatment (Fig. 1h). Although an increase in the expression of the known Hic1 target genes Efna1 [24] and Tlr2 [32] was observed, no changes in Sirt1 were seen (Supplementary Table S1). Unexpectedly, gene expression microarray and gene ontology analysis revealed highly significant enrichment for pathways involved in the regulation of cell cycle, mitosis, and DNA replication consistent with the induction of cell cycle arrest (Figs. 1h, i; Supplementary Table S2). These data are resemble a phenomenon in MEFs known as “tumor suppressor inactivation-induced senescence”, best exemplified by inactivation of the breast cancer tumor suppressor gene Brca1, which results in overwhelming DNA damage as the result of replication-dependent double-strand break (DSB) formation, chromosomal instability and Tp53-dependent senescence [33, 34].
Immortalized Hic1KO MEFs are phenotypically distinct from p53KO MEFs
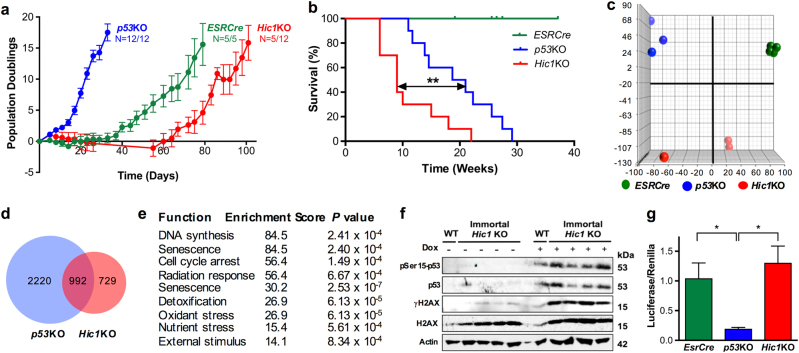
To determine whether the effects of Hic1 deletion could be functionally separated from downregulation of Tp53 function through upregulation of Sirt1, we employed a conditional Tp53 knockout allele (hereafter p53lox/lox) to generate MEFs derived from p53lox/lox EsrCre embryos (hereafter p53KO). Embryos heterozygous for the EsrCre allele were used as controls (hereafter EsrCre). Serial passaging of EsrCre, p53KO, and Hic1KO MEFs using a modified 3T3 protocol demonstrated that deletion of Hic1 can replicate the effect of loss of Brca1 function in the MEF model (Fig. 2a). As expected, p53KO MEFs spontaneously immortalized with high efficiency, while control EsrCre cells escaped senescence with similar efficiency but in a much longer timeframe. By contrast, Hic1KO MEFs immortalized with low efficiency after a period of dormancy lasting over 60 days (Fig. 2a) in similar fashion to the Brca1 knockout model [33].
Fig. 2. Immortalization of MEFs lacking Hic1 results in a phenotype distinct to MEFs lacking p53.
a Growth of MEFs (shown as cumulative population doublings) following tamoxifen treatment using the 3T3 protocol [65]. Data shown as mean+SEM. Sample size was determined by the number of available immortalized MEF lines. Cell lines were checked for Mycoplasma and genotype every 6 months. b Kaplan–Meier survival analysis of athymic nude mice injected with 1 × 106 immortalized MEFs with the genotypes indicated. **P < 0.005, log-rank analysis. 1 × 106 MEFs were resuspended in 50 µl media+50 µl Matrigel and injected subcutaneously in the right flank and observed for 26 weeks, until the tumor reached 800 mm3 measured by an observed blinded to the MEF genotype. c Principal component (PC) analysis of gene expression in immortalized MEFs generated from embryos with the genotypes indicated compared to control MEFs. Detailed bioinformatic methods are described in Supplementary Information. Array data are available through GEO, GSE104394. d A Venn diagram depicting differentially expressed genes in the immortalized MEF lines shown in Fig. 1c when compared with control MEFs. e Gene ontology analysis of differentially expressed genes when comparing immortalized p53KO vs. Hic1KO MEFs. The analysis was performed by an observer with no a priori knowledge of the cellular phenotype. f Western blot analysis of lysates from wild type (WT) or Hic1 KO MEFs showing the expression of p53 phosphorylated at serine 15 (pSer15-p53, Cell Signaling Technology, Danvers, MA, USA, #9284S), p53 (Santa Cruz Biotechnology, Dallas, TX, USA, sc-6243), phosphorylated-γH2AX (γH2AX, Novus Biologicals, Littleton, CO, USA #NB100-74435), total H2AX (Cell Signaling Technology, 2595 S) and Actin [66, 67]. Cells were treated with vehicle or doxorubicin (Dox; 1 μM, 6 h). g Activity of a p53-responsive luciferase reporter (Qiagen, Hilden, Germany, #CCS-004L) in MEFs 24 h after treatment with doxorubin, 1 μM, for 6 h. n = 4 indepdent cell lines performed, each performed in triplicate, mean + SEM, *P < 0.05
One of the most rigorous tests of malignant transformation in the MEF model is the capacity of immortalized cells to grow as allografts in athymic nude mice [35]. To asses this, early passage immortalized MEF lines from each genotype were injected subcutaneously into the flanks of nude mice and observed. Fast growing Hic1KO tumors rapidly appeared within 3 weeks, whereas tumors derived from p53KO MEFs developed with a longer latency period (Fig. 2b). No tumors developed from EsrCre MEFs
Gene expression profiling of immortalized EsrCre, p53KO, and Hic1KO MEFs revealed a highly divergent transcriptional signature (Fig. 2c, d; Supplementary Tables S3,4,5). When compared with p53KO cells, Hic1KO MEFs upregulated gene sets strongly associated with DNA synthesis, senescence, oxidative and nutrient stress (Fig. 2e). As expected, expression of Cdkn1a (which encodes p21) was markedly downregulated in p53KO cells while immortalization in response to deletion in Hic1 was associated with dramatic loss of Cdkn1c gene expression (which encodes p57) (Supplementary Table S5). Genomic and RT-PCR sequencing also showed that immortalized Hic1KO MEFs retained intact Tp53 and Cdkn2a genes (data not shown), both of which are commonly inactivated in MEFs that spontaneously escape senescence [36]. To further define the status of Tp53 in this model, we treated WT or immortalized Hic1KO MEFs with doxorubicin to induce DNA damage. As shown in Fig. 2f, this resulted in robust upregulation of Tp53 and phosphorylated Tp53 expression. Using a Tp53-resposive reporter system, we further showed that both EsrCre and Hic1KO MEFs were able to induce a p53-dependent transcriptional response when compared to p53KO MEFs (Fig. 2g).
Taken together, these data show that immortalization following the loss of Hic1 occurs independent of Tp53. Although we cannot exclude the possibility that attenuation of Tp53 function plays a role in maintenance of the immortalization phenotype in Hic1KO MEFs, the weight of evidence strongly suggests that a significant component of the tumor suppressive activity of Hic1 in the context of tumor initiation outside the previously described Hic1-Tp53-Sirt1 regulatory loop [28].
Loss of Hic1 in MEFs leads to chromosomal instability
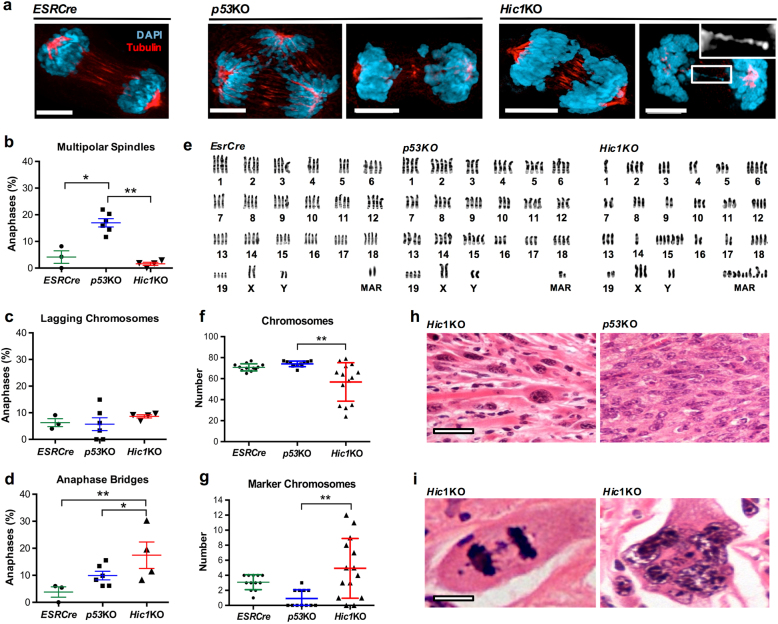
Chromosomal instability is a cardinal feature of tumors with defective HR, typified by cancers with mutations in BRCA1 and BRCA2 [37]. Considering the similarities between Hic1KO and Brca1 mutant MEFs, we asked whether loss of Hic1 function in immortalized MEFs would result in chromosomal instability. Quantitative anaphase analysis of immortalized MEFs revealed largely normal chromosomal morphology in EsrCre MEFs, whereas multipolar spindle formation was the predominant defect in p53KO MEFs (Fig. 3a, b), in keeping with previous reports [38]. The prevalence of lagging chromosomes was similar across all genotypes. By contrast, mitoses in Hic1KO MEFs were characterized by frequent anaphase bridges (Fig. 3a, b), consistent with a defect in HR mediated repair of DSB [39].
Fig. 3. Deletion of Hic1 results in chromosomal instability.
a Representative confocal photomicrographs showing anaphases in MEFs with the genotypes indicated. Cells were stained for tubulin (Red) and DNA (DAPI) (Blue) as described [66]. Scale bar = 5 μm. Anaphases in cell culture were scored as described previously by an observer blinded to the MEF genotype [66]. b–d Quantitative analysis of aberrant anaphase events in MEFs with the genotypes from the same experiment shown in Fig. 1a, n = 3 (ESRCre), 6 (p53KO) and 4 (Hic1KO) cell lines, average of 55 anaphases per cell line. Mean+SEM. *P < 0.05; **P < 0.01, one-way ANOVA with Bonferonni correction. Sample size was chosen by the number of available MEF lines. e Examples of karyotypes from immortalized MEFs with the genotypes shown. Karyotyping was performed as previously described [70]. f, g Quantification of chromosome number and the number of marker chromosomes, n = 4 ESRCre, n = 6 p53KO and n = 5 Hic1KO cell lines, average of 15 metaphase cells per genotype. **P < 0.01, one-way ANOVA with Bonferonni correction. h Representative photomicrographs of hematoxylin and eosin (H&E) stained sections from formalin-fixed, paraffin-embedded nude mouse allograft tumors with the genotypes indicated. Scale bar = 20 μm. i Representative high-powered photomicrographs of H&E stained sections of Hic1KO nude mouse allograft tumors. Scale bar = 5 μm. MAR marker chromosomes
We next analyzed the consequences of this ongoing chromosomal instability with karyotype analysis in each of the immortalized MEF lines. Both EsrCre and p53KO cell lines were near tetraploid, resembling the stable karyotype of NIH-3T3 cells, a well-characterized MEF line generated by spontaneous immortalization (Fig. 3e, f) [40]. In keeping with the degree of anaphase bridge formation, numerical and segmental aneuploidy with large numbers of marker chromosomes was seen in Hic1KO MEFs (Fig. 3e, f). Histologic analysis of the MEF allograft tumors described in Fig. 2b showed that Hic1KO MEFs formed aggressive, pleomorphic sarcomas with some features of skeletal muscle differentiation resembling adult pleomorphic rhabdomyosarcoma (Fig. 3h). By contrast, p53KO MEFs form sarcomas more consistent with pediatric rhabdomyosarcoma, with a more regular nuclear morphology. Consistent with our in vitro findings, Hic1KO MEF nude mouse allograft tumors exhibited marked nuclear pleomorphism, as well as numerous anaphase bridges and multinucleated giant cells (Fig. 3i), These histological findings are consistent with marked chromosomal instability [41–43].
Cooperation between inactivation of Hic1 and oncogenic KRas in vivo
Our data suggest that loss of Hic1 in the setting of the replication stress and impending senescence induced by culture of MEFs [44] can lead to chromosomal instability and spontaneous transformation. To test the functional importance of this observation in vivo, we made use of a conditional mouse model of lung adenocarcinoma in which an oncogenic mutant KRasG12D allele is knocked into the endogenous KRas locus downstream of a loxP-STOP-loxP cassette (hereafter KRas) [45]. When treated with an inhaled recombinant adenovirus expressing Cre recombinase (Ad5Cre), these mice develop multiple lung adenocarcinomas within 6–8 weeks [46]. We chose this model based on (i) the ability of mutant KRas to trigger replication stress and senescence in the absence of a cooperating mutations in Tp53 or p16 [47, 48]; (ii) the cooperating effect in this model on tumor progression due to combined deletion of Tp53 [49]; (iii) the prevalence of HIC1 hypermethylation in human lung adenocarcinoma [50]; and (iv) the clinical significance of HIC1 methylation in non-small cell lung cancer [18].
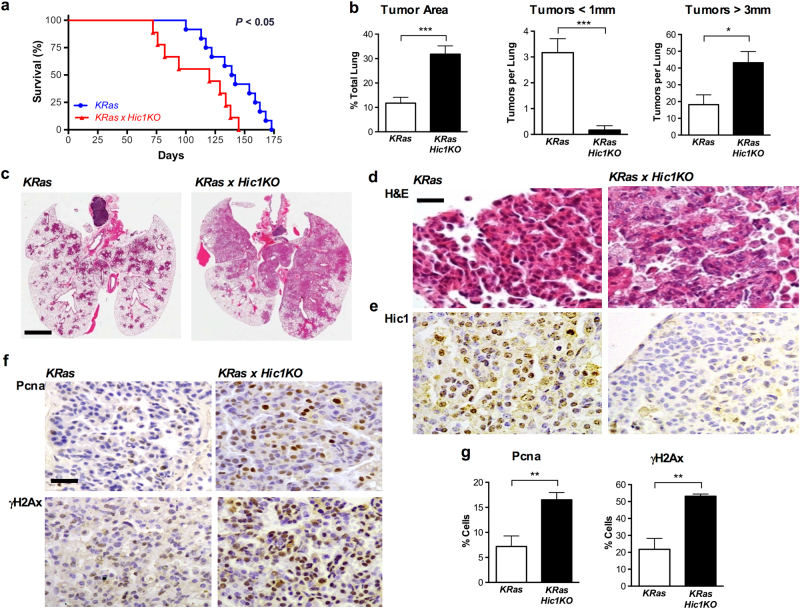
We crossed KRas and Hic1lox/lox mice to generate wild type, Hic1KO, KRas, and KRas× Hic1KO experimental cohorts, and administered inhaled Ad5Cre at 6–8 weeks of age. Wild type and Hic1KO mice treated with Ad5Cre showed no abnormalities and did not develop lung lesions by 12 months of age (data not shown). As shown in Fig. 4a, KRas × Hic1KO mice treated with inhaled Ad5Cre had a shorter lifespan compared to KRas littermates, consistent with a reduced tumor latency. Consistent with this observation, quantitative histologic analysis showed a marked increase in tumor size (Fig. 4b, c). Immunohistochemical staining for Pancytokeratin, Surfactant Protein C and Thyroid Transcription Factor-1 confirmed the identity of the tumors as primary lung adenocarcinomas (Supplementary Fig. S1). Early KRas tumors retained Hic1 expression, whereas this was lost in the KRas × Hic1KO tumors (Fig. 4e). KRas × Hic1KO tumors retained intact Cdkn2a and Tp53 alleles (data not shown).
Fig. 4. Effects of Hic1 deletion (Hic1KO) on the development of lung tumors induced by the conditional activation of an activating KRasG12D (KRas) mutant in the airway epithelium of adult mice.
Mice from each geneotype were anesthetized using Avertin at a dose of 0.5 mg/gram of body weight, and administered 5 × 108 PFU Ad5CMVCre virus (Viral Vector Core Facility, University of Iowa, Iowa City, IA) by intranasal inhalation at 8 weeks of age and observed for 9 months or until ethical endpoint. a Kaplan–Meier survival analysis of KRas and KRas × Hic1 KO mice following inhalation of adenoviral Cre recombinase. n = 12 (KRas) and 9 (KRas× Hic1KO). Sample size was based on previous published studies [71]. b Quantitative analysis of lung tumor area and size in mice from the same experiment depicted in Fig. 4a. n = 6 per genotype, ***P < 0.001, *P < 0.05, unpaired t-test. Mouse lungs were inflated with 10% buffered formalin and fixed overnight before paraffin embedding. For quantitation of tumor burden and number, sections were scanned using the Aperio Scanscope XT (Leica Biosystems, Buffalo Grove, IL, USA) and analysed using Aperio Imagescope software by a blinded observer as described [71]. Sample size was chosen based on previous studies [71]. c Representative photomicrographs of hematoxylin and eosin (H&E) stained sections of lungs from the experiment depicted in Fig. 4a. Scale bar = 5 mm. d Representative high-powered photomicrographs of hematoxylin and eosin (H&E) stained sections of lungs from the same experiment. Scale bar = 20 μm. e Representative photomicrographs of sections from the same tumors stained with immunoperoxidase (brown) for Hic1, and countersained with hematoxylin (blue). Immunohistochemistry was performed as described [71]. f Immunstaining for Proliferating cell nuclear antigen (Pcna, Dako, Troy, MI, #M087901-2) or phospho-γH2AX (γH2AX, Abcam, Cambridge, UK) in the same tumors shown in Fig. 4d. Immunohistochemistry and quantitifcation of staining was performed as described [71]. Scale bar = 100 μm. g Quantitative analysis of Pcna and γH2AX staining from the same experiment depicted in Fig. 4f. n = 5 per genotype, 5 fields of view at 40× counted per animal. **P < 0.01, unpaired t-test
Tumors in the KRas animals displayed a well differentiated adenocarcinoma morphology consistent with previous reports [45]. In contrast, KRas × Hic1KO mice developed adenocarcinomas with prominent micropapillary and pleomorphic features (Fig. 4d), both of which are known to be associated with a poor prognosis in human lung cancer [51, 52]. To our knowledge, this is the first description of mouse model in which both these histologic features have been identified, and are distinct from tumors arising in KRas/p53 mutant animals [49]. In keeping with this aggressive phenotype, tumors from KRas× Hic1KO animals were more proliferative, evidenced by quantitative analysis of Pcna immunohistochemistry (Fig. 4f, g). Despite this proliferative advantage, KRas × Hic1KO tumors displayed prominent expression of nuclear γH2AX (Fig. 4f, g), indicative of spontaneous DSB formation. Although we cannot exclude the possibility that deregulation of Hic1 target genes may contribute to the micropapillary lung adenocarcinoma phenotype seen in KRas × Hic1KO mice, these data support our findings in the MEF model, and demonstrate that loss of Hic1 in the setting of oncogenic KRas activation in the adult airway epithelium leads to chromosomal instability and a distinct mouse lung adenocarcinoma phenotype.
A novel role for Hic1 as a tumor suppressor
Using a conditional mutant mouse model, we have identified a new and unexpected function for Hic1 as tumor suppressor by maintaining chromosomal stability in the setting of sustained DNA replication stress. Although our model may not be broadly applicable to the models in which the functions of Hic1 have been described previously, our results do suggest that loss of Hic1 function in the early phases of tumor initiation may have a major impact on the subsequent tumor phenotype. These data support the notion that the acquisition of chromosomal instability during the early phases of tumor evolution can have a major effect on tumor phenotype and genotype through deletion of tumor suppressors, amplification of oncogenes, and transcriptional deregulation [53–55]. Our results are also consistent with the idea that chromosomal instability can drive tumor evolution, as well as genomic, epigenetic and phenotypic heterogeneity [53–55].
Several lines of evidence support a potential role for Hic1 in maintaining chromosomal stability independent of Tp53. In a human cell line model, Dehennaut et al. showed that HIC1 promotes the response to DNA double strand breaks through an ATM-SIRT1-HDAC4 dependent mechanism [56]. Importantly, this was dependent on the interection between HIC1 and MTA1, a component of the nucleosome remodeling and deacetylase (NuRD) complex. Interestingly, this complex is required for the effective DNA repair during S phase [57–59]. By contrast, Paget et al. recently showed that in response to repairable DNA damage, HIC1 was more important in mounting a transcriptional response to DSB than directly promoting DNA repair [60]. Although our data do not resolve this apparent contradiction, our findings are consistent with both papers in they support a role for Hic1 in mediating DNA repair and/or in triggering cell death in response to DNA damage.
Interestingly, HIC1 is also known to interact with the tumor suppressor gene ARID1A [61], a component of the SWI/SNF chromatin remodeling complex that also has important roles in maintaining genome stability during DNA replication [62, 63]. This concept is strengthened by the observation that mouse embryonic stem cells lacking SWI/SNF protein Brg1 undergo cell cycle arrest associated with chromatin bridge formation and a defective decatenation checkpoint [64]. Taken together, these observations suggest that genome-wide interactions between Hic1 and both the NuRD and SWI/SNF complexes may play a role in maintaining chromosomal instability during DNA replication.
Electronic supplementary material
Acknowledgements
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia (Project Grant GNT10838550, The Victorian Cancer Agency (TS10-01), The National Institutes of Health (NCI-RO1 NS054085), The Petre Foundation, and the Victorian Government’s Operational Infrastructure Support Program. Dr Gough is supported by an NHMRC Career Development Fellowship (GNT1063914). Dr Burgess is supported by Cancer Institute NSW Fellowship (10/FRL/3-02) and a Patricia Helen Guest Fellowship. The contents of this manuscript are solely the responsibility of the participating institutions and individual authors, and do not reflect the views of these funding agencies. We thank Dr Dominique LePrince for the gift of the Hic1-FLAG expression vector. National Health and Medical Research Council of Australia (GNT1083855): J.E.C., D.N.W.; (GNT1063914): D.J.G. National Institutes of Health (NINDS RO1 NS054085): D.N.W., S.B.B. The Petre Foundation: D.N.W. Cancer Institute of NSW Fellowship (10/FRL/ 3-02) and the Patricia Helen Guest Fellowship: A.B.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Contributor Information
Jason E. Cain, Email: jason.cain@hudson.org.au
D. Neil Watkins, Email: n.watkins@garvan.org.au.
Electronic supplementary material
The online version of this article (10.1038/s41388-017-0022-1) contains supplementary material, which is available to authorized users.
References
- 1.Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–7. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 2.Deltour S, Guerardel C, Stehelin D, Leprince D. The carboxy-terminal end of the candidate tumor suppressor gene HIC-1 is phylogenetically conserved. Biochim Biophys Acta. 1998;1443:230–2. doi: 10.1016/S0167-4781(98)00219-X. [DOI] [PubMed] [Google Scholar]
- 3.Deltour S, Guerardel C, Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc Natl Acad Sci Usa. 1999;96:14831–6. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinte S, Stankovic-Valentin N, Deltour S, Rood BR, Guérardel C, Leprince D. The tumor suppressor gene HIC1 (hypermethylated in cancer 1) is a sequence-specific transcriptional repressor: definition of its consensus binding sequence and analysis of its DNA binding and repressive properties. J Biol Chem. 2004;279:38313–24. doi: 10.1074/jbc.M401610200. [DOI] [PubMed] [Google Scholar]
- 5.Briggs KJ, Eberhart CG, Watkins DN. Just say no to ATOH: how HIC1 methylation might predispose medulloblastoma to lineage addiction. Cancer Res. 2008;68:8654–6. doi: 10.1158/0008-5472.CAN-08-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerardel C, Deltour S, Pinte S, Monte D, Bégue A, Godwin AK, et al. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J Biol Chem. 2001;276:3078–89. doi: 10.1074/jbc.M008690200. [DOI] [PubMed] [Google Scholar]
- 7.Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 8.Pinte S, Guérardel C, Deltour-Balerdi S, Godwin AK, Leprince D. Identification of a second G-C-rich promoter conserved in the human, murine and rat tumor suppressor genes HIC1. Oncogene. 2004;23:4023–31. doi: 10.1038/sj.onc.1207504. [DOI] [PubMed] [Google Scholar]
- 9.Britschgi C, Jenal M, Rizzi M, Mueller BU, Torbett BE, Andres AC, et al. HIC1 tumour suppressor gene is suppressed in acute myeloid leukaemia and induced during granulocytic differentiation. Br J Haematol. 2008;141:179–87. doi: 10.1111/j.1365-2141.2008.06992.x. [DOI] [PubMed] [Google Scholar]
- 10.Fujii H, Biel MA, Zhou W, Weitzman SA, Baylin SB, Gabrielson E. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–64. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 11.Melki JR, Vincent PC, Clark SJ, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–40. [PubMed] [Google Scholar]
- 12.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–7. [PubMed] [Google Scholar]
- 13.Chen WY, Baylin SB. Inactivation of tumor suppressor genes: choice between genetic and epigenetic routes. Cell Cycle. 2005;4:10–12. doi: 10.4161/cc.4.1.1361. [DOI] [PubMed] [Google Scholar]
- 14.Aggerholm A, Holm MS, Guldberg P, Olesen LH, Hokland P. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur J Haematol. 2006;76:23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 15.Stephen JK, Chen KM, Shah V, Havard S, Kapke A, Lu M, et al. DNA hypermethylation markers of poor outcome in laryngeal cancer. Clin Epigenet. 2010;1:61–9. doi: 10.1007/s13148-010-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Liu X, Dong ZM, Ling ZQ. Epigenetic silencing of HIC1 promotes epithelial-mesenchymal transition and drives progression in esophageal squamous cell carcinoma. Oncotarget. 2015;6:38151–65. doi: 10.18632/oncotarget.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng G, Sun X, Wang J, Xiao G, Wang X, Fan X, et al. HIC1 silencing in triple-negative breast cancer drives progression through misregulation of LCN2. Cancer Res. 2014;74:862–72. doi: 10.1158/0008-5472.CAN-13-2420. [DOI] [PubMed] [Google Scholar]
- 18.Tseng RC, Lee CC, Hsu HS, Tzao C, Wang YC. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11:763–70. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter MG, Johns MA, Zeng X, Zhou L, Zink MC, Mankowski JL, et al. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum Mol Genet. 2000;9:413–9. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- 20.Mohammad HP, Zhang W, Prevas HS, Leadem BR, Zhang M, Herman JG, et al. Loss of a single Hic1 allele accelerates polyp formation in Apc(Δ716) mice. Oncogene. 2011;30:2659–69. doi: 10.1038/onc.2010.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs KJ, Corcoran-Schwartz IM, Zhang W, Harcke T, Devereux WL, Baylin SB, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–85. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, Cooper TK, Zahnow CA, Overholtzer M, Zhao Z, Ladanyi M, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6:387–98. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guérardel C, et al. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284:20927–35. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Zeng X, Briggs KJ, Beaty R, Simons B, Chiu Yen RW, et al. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29:2467–76. doi: 10.1038/onc.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foveau B, Boulay G, Pinte S, Van Rechem C, Rood BR, Leprince D. The receptor tyrosine kinase EphA2 is a direct target gene of hypermethylated in cancer 1 (HIC1) J Biol Chem. 2012;287:5366–78. doi: 10.1074/jbc.M111.329466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25:2326–37. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YM, Wang CM, Jeng JC, Leprince D, Shih HM. HIC1 interacts with and modulates the activity of STAT3. Cell Cycle. 2013;12:2266–76. doi: 10.4161/cc.25365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- 30.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 31.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–22. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeckova L, Pospichalova V, Fafilek B, Vojtechova M, Tureckova J, Dobes J, et al. HIC1 tumor suppressor loss potentiates TLR2/NF-κB signaling and promotes tissue damage-associated tumorigenesis. Mol Cancer Res. 2015;13:1139–48. doi: 10.1158/1541-7786.MCR-15-0033. [DOI] [PubMed] [Google Scholar]
- 33.Cao L. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–13. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–97. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 35.Sun H, Taneja R. Analysis of transformation and tumorigenicity using mouse embryonic fibroblast cells. Method Mol Biol. 2007;383:303–10. doi: 10.1007/978-1-59745-335-6_19. [DOI] [PubMed] [Google Scholar]
- 36.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600–29. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarapore P, Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene. 2002;21:6234–40. doi: 10.1038/sj.onc.1205707. [DOI] [PubMed] [Google Scholar]
- 39.Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–81. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leibiger C, Kosyakova N, Mkrtchyan H, Glei M, Trifonov V, Liehr T. First molecular cytogenetic high resolution characterization of the NIH 3T3 cell line by murine multicolor banding. J Histochem Cytochem. 2013;61:306–12. doi: 10.1369/0022155413476868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens JB, Horne SD, Abdallah BY, Ye CJ, Heng HH. Chromosomal instability and transcriptome dynamics in cancer. Cancer Metastas- Rev. 2013;32:391–402. doi: 10.1007/s10555-013-9428-6. [DOI] [PubMed] [Google Scholar]
- 42.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–9. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Wallace MD, Southard TL, Schimenti KJ, Schimenti JC. Role of DNA damage response pathways in preventing carcinogenesis caused by intrinsic replication stress. Oncogene. 2014;33:3688–95. doi: 10.1038/onc.2013.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weyemi U, Lagente-Chevallier O, Boufraqech M, Prenois F, Courtin F, Caillou B, et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2012;31:1117–29. doi: 10.1038/onc.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mannava S, Moparthy KC, Wheeler LJ, Natarajan V, Zucker SN, Fink EE, et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am J Pathol. 2013;182:142–51. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The Differential Effects of Mutant p53 Alleles on Advanced Murine Lung Cancer. Cancer Res. 2005;65:10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 50.Cancer Genome Atlas Research Network. Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell PA, Barnett SA, Walkiewicz M, Wainer Z, Wainer Z, Conron M, et al. Correlation of mutation status and survival with predominant histologic subtype according to the new IASLC/ATS/ERS lung adenocarcinoma classification in Stage III (N2) Patients. J Thorac Oncol. 2013;8:461–8. doi: 10.1097/JTO.0b013e3182828fb8. [DOI] [PubMed] [Google Scholar]
- 52.Ung M, Rouquette I, Filleron T, Taillandy K, Brouchet L, Bennouna J, et al. Characteristics and clinical outcomes of sarcomatoid carcinoma of the lung. Clin Lung Cancer. 2016;17:391–7. doi: 10.1016/j.cllc.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Heng HH, Bremer SW, Stevens JB, Horne SD, Liu G, Abdallah BY, et al. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastas- Rev. 2013;32:325–40. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 54.Thomas A, Pommier Y. Small cell lung cancer: Time to revisit DNA-damaging chemotherapy. Sci Transl Med. 2016;8:fs12. doi: 10.1126/scitranslmed.aaf6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dehennaut V, Loison I, Dubuissez M, Nassour J, Abbadie C, Leprince D. DNA double-strand breaks lead to activation of hypermethylated in cancer 1 (HIC1) by SUMOylation to regulate DNA repair. J Biol Chem. 2013;288:10254–64. doi: 10.1074/jbc.M112.421610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–96. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li DQ, Kumar R. Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle. 2010;9:2071–9. doi: 10.4161/cc.9.11.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell. 2011;22:3094–102. doi: 10.1091/mbc.e11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paget S, Dubuissez M, Dehennaut V, Nassour J, Harmon BT, Spruyt N, et al. HIC1 (hypermethylated in cancer 1) SUMOylation is dispensable for DNA repair but is essential for the apoptotic DNA damage response (DDR) to irreparable DNA double-strand breaks (DSBs) Oncotarget. 2017;8:2916–35. doi: 10.18632/oncotarget.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Rechem C, Boulay G, Leprince D. HIC1 interacts with a specific subunit of SWI/SNF complexes, ARID1A/BAF250A. Biochem Biophys Res Commun. 2009;385:586–90. doi: 10.1016/j.bbrc.2009.05.115. [DOI] [PubMed] [Google Scholar]
- 62.Cohen SM, Chastain PD, Rosson GB, Groh BS, Weissman BE, Kaufman DG, et al. BRG1 co-localizes with DNA replication factors and is required for efficient replication fork progression. Nucl Acid Res. 2010;38:6906–19. doi: 10.1093/nar/gkq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mermoud JE, Rowbotham SP, Varga-Weisz PD. Keeping chromatin quiet: how nucleosome remodeling restores heterochromatin after replication. Cell Cycle. 2011;10:4017–25. doi: 10.4161/cc.10.23.18558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–7. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005; Chapter 28: Unit 28.1. [DOI] [PubMed]
- 66.Caldon CE, Sergio CM, Burgess A, Deans AJ, Sutherland RL, Musgrove EA. Cyclin E2 induces genomic instability by mechanisms distinct from cyclin E1. Cell Cycle. 2013;12:606–17. doi: 10.4161/cc.23512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–12. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 69.Cain JE, McCaw A, Jayasekara WSN, Rossello FJ, Marini KD, Irving AT, et al. Sustained low-dose treatment with the histone deacetylase inhibitor LBH589 induces terminal differentiation of osteosarcoma cells. Sarcoma. 2013;2013:608964. doi: 10.1155/2013/608964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagy A., Gertsenstein M., Vintersten K., Behringer R. Karyotyping Mouse Cells. Cold Spring Harbor Protocols. 2008;2008(6):pdb.prot4706-pdb.prot4706. doi: 10.1101/pdb.prot4706. [DOI] [PubMed] [Google Scholar]
- 71.Szczepny A, Rogers S, Jayasekara WSN, Park K, McCloy RA, Cochrane CR, et al. The role of canonical and non-canonical Hedgehog signaling in tumor progression in a mouse model of small cell lung cancer. Oncogene. 2017;23:2314. doi: 10.1038/onc.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.