Abstract
Background: Non-alcoholic fatty liver disease (NAFLD) is a global health issue. Dietary methyl donor restriction is used to induce a NAFLD/non-alcoholic steatohepatitis (NASH) phenotype in rodents, however the extent to which this model reflects human NAFLD remains incompletely understood. To address this, we undertook hepatic transcriptional profiling of methyl donor restricted rodents and compared these to published human NAFLD datasets.
Methods: Adult C57BL/6J mice were maintained on control, choline deficient (CDD) or methionine/choline deficient (MCDD) diets for four weeks; the effects on methyl donor and lipid biology were investigated by bioinformatic analysis of hepatic gene expression profiles followed by a cross-species comparison with human expression data of all stages of NAFLD.
Results: Compared to controls, expression of the very low density lipoprotein (VLDL) packaging carboxylesterases ( Ces1d, Ces1f, Ces3b) and the NAFLD risk allele Pnpla3 were suppressed in MCDD; with Pnpla3 and the liver predominant Ces isoform, Ces3b, also suppressed in CDD. With respect to 1-carbon metabolism, down-regulation of Chka, Chkb, Pcty1a, Gnmt and Ahcy with concurrent upregulation of Mat2a suggests a drive to maintain S-adenosylmethionine levels. There was minimal similarity between global gene expression patterns in either dietary intervention and any stage of human NAFLD, however some common transcriptomic changes in inflammatory, fibrotic and proliferative mediators were identified in MCDD, NASH and HCC.
Conclusions: This study suggests suppression of VLDL assembly machinery may contribute to hepatic lipid accumulation in these models, but that CDD and MCDD rodent diets are minimally representative of human NAFLD at the transcriptional level.
Keywords: non-alcoholic fatty liver disease, triglycerides, methyl donor, methionine, choline, lipid
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the predominant cause of chronic liver disease in the developed world, with an estimated prevalence of between 20–68% 1. The accumulation of hepatic fat in the form of triglycerides and other lipid species in NAFLD has two major clinical consequences. Firstly, a subgroup of patients with hepatic steatosis will progress to an inflammatory hepatitis, hepatic cirrhosis and in some cases hepatocellular carcinoma (HCC) 2. Secondly, almost all patients with NAFLD also exhibit hepatic insulin resistance, which can be associated with impaired glucose uptake, increased gluconeogenesis and type 2 diabetes, possibly as a direct consequence of the increased hepatic lipid load 1, 3, 4. Together, these conditions are responsible for significant morbidity and mortality and represent a substantial burden for health resources 5.
The molecular mechanisms underpinning NAFLD pathology are incompletely understood, and as such there is a need for accurately representative rodent models in which to investigate this common disease and to trial novel therapeutics. Given the association of NAFLD with human obesity, the use of high fat diet feeding in rodents remains a popular model in which to investigate mechanisms. However whilst high fat feeding generates a NAFLD-like picture, the disadvantages with this model include the protracted time required to induce even mild non-alcoholic steatohepatitis (NASH), the lack of malignant transformation to HCC even with prolonged exposure, and the variation in the histological and transcriptional changes due to the behavioural characteristics of mice in social groups 6– 8. Thus, a number of alternative models have been employed. In rodents, dietary restriction of the methyl donors methionine and/or choline rapidly and reliably induces a spectrum of liver injury histologically similar to human NAFLD, within weeks of instigation 9, 10. Although the precise biological mechanisms responsible for the predictable phenotypic changes are poorly understood, the histological similarity to human steatosis (choline deficient diets; CDD) and NASH (methionine and choline deficient diets; MCDD) means that these models have been used in mechanistic and therapeutic studies for a number of years 11– 13. Since impaired metabolism of the key methyl donor S-adenosylmethionine (SAMe) is a well documented feature of chronic liver disease regardless of aetiology 14– 16, there may be common molecular mechanisms which may present an opportunity for therapeutic intervention. Although a number of transcriptional changes have been reported during the progression of human NAFLD, no detailed transcriptional comparisons have been performed to identify similarities or differences between human disease and the CDD and MCDD models of NAFLD, despite their widespread use. In this study, we set out firstly to dissect potential mechanisms underpinning the development of liver pathology in CDD and MCDD models by mapping pathways of lipid and one-carbon metabolism, and secondly to evaluate their potential usefulness as models of human disease. To address these aims, we have examined in detail the transcriptional profiles in liver from mice maintained on CDD and MCDD, and compared these with published human NAFLD transcriptome data series.
Materials and methods
Animals
All experiments were carried out under a UK Home Office Licence (PPL 70/7874), and with local ethical committee approval and adhering to the ‘Animal Research: Reporting In Vivo Experiments’ (ARRIVE) guidelines. Adult C57BL/6J mice (purchased from Charles River, Tranent, UK) were maintained under controlled conditions in social groups of 5 animals per cage; n=10/group. A 12-hour light cycle (07.00h to 19.00h) and twelve hour dark cycle was implemented throughout. The temperature was maintained at 22°C +/- 2°C. Mice were maintained on control, CDD or MCDD diets (Dyets, Bethlem, PA) for 4 weeks. All efforts were made to ameliorate any suffering of animals, in particular, since MCDD diets can result in weight loss, mice were weighed and their health status checked daily by experienced technicians for the 4 weeks they remained on the diets. All mice remained well, with no concerns about health other than weight loss in the MCDD group. After 4 weeks mice were killed by Schedule 1 (CO 2 inhalation), and tissues were collected and used for histology or snap-frozen and stored at -80C. The diet composition can be found in Supplementary Table 1.
Histology staining, triglyceride and SAMe quantification
Livers were removed and sections were fixed in methacarn solution (methanol:chloroform:glacial acetic acid; ratio 6:3:1) and mounted in paraffin blocks prior to staining with haematoxylin and eosin or picosirius red. Hepatic triglyceride concentration was determined by spectrophotometric analysis (BioVision, Milpitas, USA) as previously described 17. Image analysis for fibrosis content was performed in ImageJ ( http://imagej.nih.gov/ij/). SAMe was quantified by matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI) using the 12T SolariX MALDI-FTICR-MS (Bruker Daltonics, MA, US), as previously described 18.
Reverse transcription and qPCR
RNA was extracted from snap frozen liver tissue using the RNeasy kit (Qiagen, Manchester, UK). 800ng of RNA was DNAse treated using Promega RQ1 DNAase (Promega, Southampton, UK) and reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Paisley, UK). Quantitive real time PCR was performed using Roche Universal Probe Library assays or TaqMan qPCR assays (Life Technologies, Paisley, UK) (please see Supplementary Table 2 for the primer sequences), using the Roche Lightcycler 480 and associated Lightcycler 480 software: release 1.5.1.62 (Roche, West Susssex, UK). Gene expression is displayed relative to mean of three housekeeping genes (Gapdh, Ppia, Ldha).
Transcript analysis
RNA labelling was performed on 500ng RNA using the Illumina Total Prep RNA amplification kit (Life Technologies. Paisley, UK) and subsequently hybridised to Illumina Mouse-ref6 expression bead arrays as per the manufacturer’s instructions, at the Edinburgh Clinical Research Facility, Western General Hospital, Edinburgh, UK. Intensity data were generated using a HiScan array scanner (Illumina, San Diego, USA) and analysed using iScan Illumina software. Data analysis and generation of plots were perfomed in RStudio ( http://www.rstudio.com), with R version 3.1.2. Data import, quality control, normalisation and between array adjustment was performed using the Lumi package, and differential expression was determined using the Limma package (Bioconductor.org). Unsupervised clustering was performed using Euclidean distance. Where multiple probes mapped to the same gene, the median result was used. Data have been uploaded to EBI-Array Express, accession number E-MTAB-3943.
Pathway analysis
Gene Ontology and pathway enrichment was performed using the GOstats package (Bioconductor.org). Investigation of fat handling was performed by interrogating relevant pathways of lipid metabolism and insulin signalling from the Kyoto encyclopedia for genes and genomes (KEGG) module database ( http://www.genome.jp/kegg/module.html). KEGG module sets ‘M00003 gluconeogenesis’, ‘M00086 beta-Oxidation, acyl-CoA synthesis’, ‘M00083 Fatty acid biosynthesis, elongation’ and ‘mmu_M00089 Triacylglycerol biosynthesis’, ‘mmu00071 Fatty acid degradation’, and KEGG pathways ‘Fat Digestion and Absorption’ and ‘Insulin Signalling Pathway’ and ‘Glycerolipid Metabolism’ were used. In addition, the family of carboxylesterase (Ces) genes were analysed due to their recently discovered role in triglyceride hydrolysis 19– 21. Finally, to dissect the link between lipid metabolism and one carbon metabolism, relevant mediators were analysed and mapped to known biochemical pathways.
Cross species comparison
A comparison of transcriptional data from both CDD and MCDD was made with published human expression sets of normal liver, simple steatosis and NASH (GSE48452, E-MEXP-3291), and HCC (GSE63898) 22– 24. Datasets were retrieved from the ArrayExpress archive ( http://www.ebi.ac.uk/arrayexpress/). Predicted gene orthologues were determined using homologene via the Hugo Gene Nomenclature Committee server ( http://www.genenames.org). For all human NASH, HCC and all mouse datasets, a transcriptional threshold of 1.5 with an adjusted p value of <0.05 was applied. For human steatosis gene sets alone, the fold change transcriptional threshold was reduced to 1.2 to allow comparison with the relatively mild transcriptional derangement observed. All genes found to be dysregulated at each stage of human NAFLD were then examined in the CDD and MCDD datasets to determine if expression was altered, and if so, what was the direction of change. Genes dysregulated in human and mouse models were then depicted in scatter plot analyses with linear regression used to compare datasets.
Statistical analysis
Animal model and qPCR statistical analysis was performed using Prism GraphPad software (GraphPad Software Inc.). Data were routinely analysed for outliers, normalisation and sphericity where required. Non-parametric data were either log transformed or a non-parametric test used as indicated.
Results
Phenotype
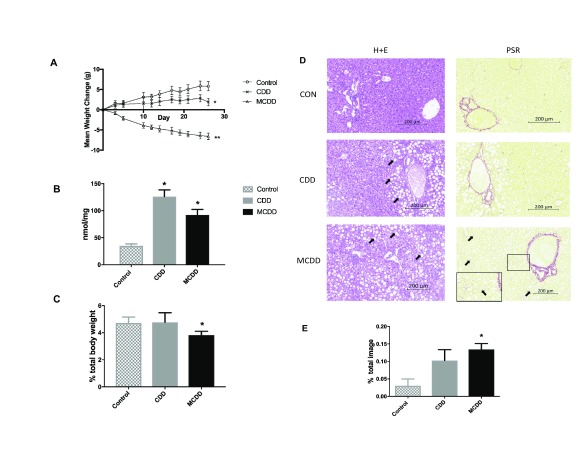
Mice on CDD gained significantly less weight than animals on a control diet, whereas MCDD fed mice lost weight from the outset, consistent with previous observations 17, 25 ( Figure 1A). At the end of the experiment, hepatic triglyceride content was significantly higher in both CDD and MCDD groups compared to controls, but there was no significant difference between the two interventions ( Figure 1B). MCDD liver weights were lower in MCDD fed mice but not CDD ( Figure 1C). Histological analysis revealed severe hepatic steatosis in both groups ( Figure 1D), with a significant increase in hepatic fibrosis in the MCDD group ( Figure 1E).
Figure 1. Body weight and hepatic lipid content.
A) In comparison with control animals, mice on CDD gain less weight whereas those on MCDD lose weight. B) Hepatic liver triglyceride content was increased on both CDD and MCDD diets. C) Liver weight was reduced in MCDD fed mice but not CDD. D) Liver histology from control, CDD and MCDD mice. H+E = Hematoxylin and Eosin staining showing marked macrovesicular steatosis in both CDD and MCDD mice (white arrows). PSR = Picosirius Red stain staining for new collagen formation showing increased periportal and interstitial fibrosis in MCDD animals (black arrows). In MCDD liver stained with PSR, insert at higher magnification shows increased fibrosis more clearly. E) Quantified fibrosis content (PSR positive staining as a percentage of entire image). n = 10 per group for all figures. * P < 0.05 ** P < 0.01 (one way ANOVA with Tukey post hoc test versus control animals). Error bars = +/- SEM.
Gene expression changes
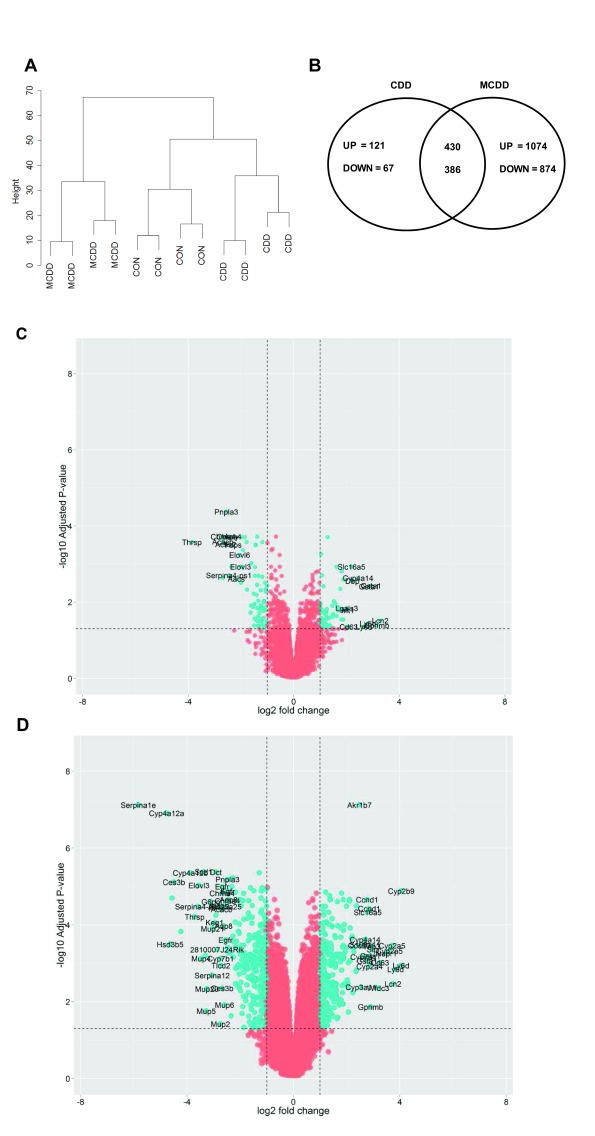
Next we carried out analysis of the transcriptome in control, CCD or MCDD mouse livers, each n=4. This approach allowed us to interrogate ~18,000 transcripts per mouse liver and analysis of total datasets revealed a number of transcriptional differences between animals. Unsupervised clustering of the 500 most variable transcripts between all animals was sufficient to cluster into the different dietary interventions ( Figure 2A). Both interventions induced a >1.5-fold differential expression in multiple transcripts when corrected for multiple testing (adjusted P value <0.05, Benjamini-Hochberg test) ( Figure 2B–D). The top 100 up- and down-regulated genes in each group are shown in Supplementary Table 3 and Supplementary Table 4. Of the 234 genes differentially expressed in CDD, 194 (82.4%) were also differentially expressed in MCDD. The additional restriction of methioine over and above choline induced the differential expression of a further 1032 transcripts ( Figure 2B).
Figure 2. Differential gene expression.
A) Transcript profiles of each dietary intervention were sufficiently consistent to cluster by Euclidean distance. B) Venn diagram demonstrating high degree of overlap in dysregulated transcripts (≥2 fold change) in each group and the direction of transcriptional change. C and D) Volcano plots demonstrating transcriptional changes in CDD ( C) and MCDD ( D) mice. Dotted lines and blue colour represent adjusted P values < 0.05 and two fold differential expression change.
Transcripts showing at least a 2-fold change were segregated into up-regulated and down-regulated gene lists and examined for over-representation within all detected transcripts on the array platform. GO-terms for lipid, sterol, fatty acid and organic acid biosynthesis were markedly over-represented in the list of suppressed genes. Over-expressed pathways in CDD mice included ‘immune system process’ and ‘inflammatory response’. Up-regulated pathways in mice on the MCDD diet included ‘positive regulation of mitotic cell cycle’ and ‘negative regulation of cell cycle arrest’ ( Supplementary Figure 1).
The most up-regulated genes in CDD mice included immune mediators ( Gpnmb, Ly6d); fibrosis mediators ( Mmp12, Mmp13); and the detoxification enzymes Gsta1 and Gsta2 and the microsomal enzyme Cyp4a14. The most suppressed genes in CDD included lipid synthesis genes ( Sqle, Elovl3, Elovl6, Aacs, Acly, Acss2, Acacb), the endopeptidase inhibitor Serpina4-ps1 and the multifunctional triglyceride metabolism enzyme Pnpla3. Whilst these genes were similarly differentially expressed in MCDD mice, volcano plots revealed considerably more severe transcriptional derangement in MCDD compared with CDD, both in the number and fold change of differentially expressed genes ( Figures 2C and 2D). Additional genes up-regulated in MCDD included the mitotic proteins Cdc20, Nupr1, the metalloproteinase Adam32, the inflammatory mediator Slpi and the aldo-ketoreductase Akrb7.
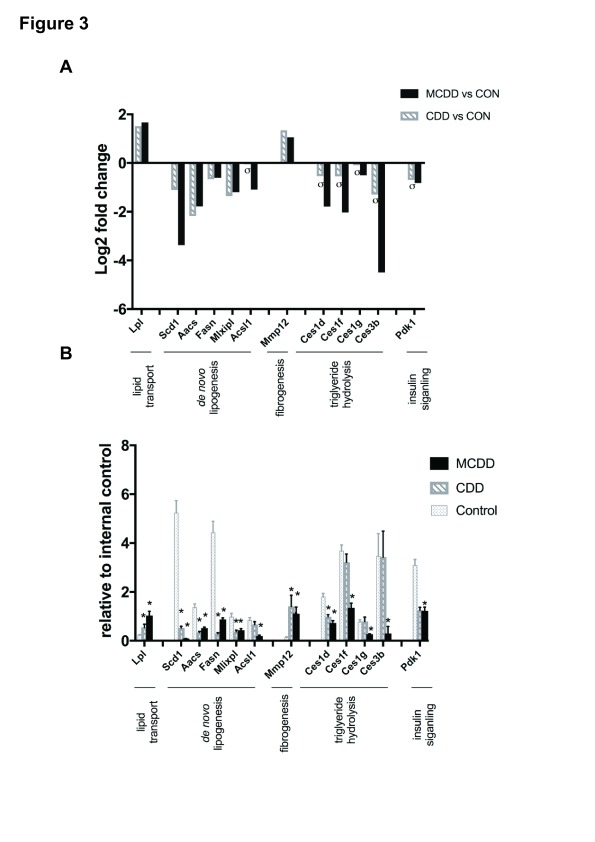
qPCR validation of array findings was performed for known mediators of lipid uptake ( Lpl), putative contributary genes to hepatic fat accumulation in NAFLD ( Scd1, Aacs, Fasn, Mlxipl, Acsl1) and hepatic fibrogenesis ( Mmp12), and Pdk1 which is an important link between insulin signaling and HCC. In addition, we analysed gene expression of four members of the Ces family due to their functional role in triglyceride hydrolysis. Gene expression by qPCR was consistent with array findings for all genes ( Figures 3A and B).
Figure 3. CDD and MCDD induce changes in the hepatic expression of genes important in lipid metabolism and storage.
( A) Microarray analysis and ( B) qPCR validation of selected genes important in lipid transport, de novo lipogenesis, fibrogenesis, triglyceride hydrolysis and insulin signalling. ( A) Adjusted P value < 0.05 (FDR) for all samples except those marked σ. ( B) * = P < 0.05 versus control, one-way ANOVA with Bonferroni post hoc analysis. Error bars = +/- SEM.
Lipid pathway analysis
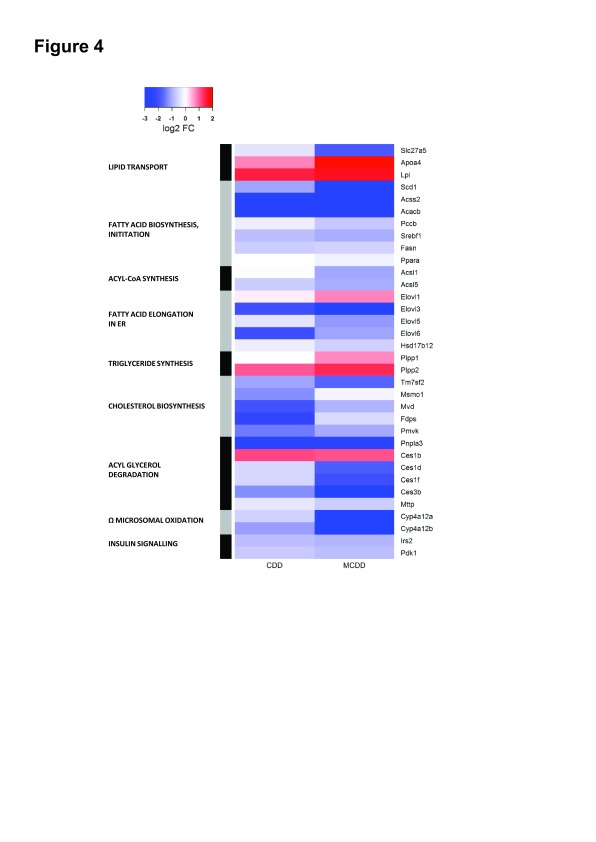
We then proceeded to examine pathways of lipid uptake, synthesis and disposal. Differentially expressed genes in relevant KEGG pathways in either group are depicted in Figure 4. Expression changes were generally greater in MCDD than CDD although they occurred in the same direction. Previous studies have suggested an increase in lipid uptake with MCDD, with upregulation of some of the FATP/solute carrier family 27 genes in association with increased sequestration of isotope labelled fatty acids 17, 26, however we noted only the down-regulation of the fatty acid translocase Scla27a5 (FATP5) with no change in the other FATP isoforms. We did identify marked upregulation of lipoprotein lipase, a key mediator in triglyceride hydrolysis from lipoproteins. In keeping with the gene ontology analysis, the global picture suggests suppressed hepatic lipid synthesis. Perturbed genes in pathways of fatty acid biosynthesis initiation, Acyl-CoA synthesis, fatty acid elongation and cholesterol synthesis were almost universally down-regulated.
Figure 4. Heatmap depiction of expression changes in KEGG pathways of lipid transport, lipid synthesis and degradation and insulin signalling.
Up- and down-regulated genes in each dietary intervention versus control animals are demonstrated by colour key. Pathways of lipid and cholesterol synthesis are globally suppressed.
We then examined dominant pathways of hepatic fatty acid fate (triglyceride synthesis, β-oxidation, oxidation and peroxisomal oxidation). These were found to be relatively unaffected, apart from an upregulation of the phospatidic acid phosphatases Pap2a and Pap2c (which convert phospatidic acids to diacylglycerol) and the peroxisomal fatty acid elongation enzyme Elovl1 in MCDD. The Cyp4a family of enzymes broadly catalyse the microsomal (ω) oxidation of saturated and unsaturated fatty acids and have reported to be upregulated by dietary and drug-induced hepatic inflammation 27– 29. Interestingly, isoforms of Cyp4a enzymes demonstrated marked bi-directional differential expression with Cyp4a12a and Cyp4a12b strongly suppressed in CDD and MCDD whilst Cyp4a14 was upregulated by 4-fold with both diets. Other microsomal oxidation Cyp450 isoforms (Cyp4a10, Cyp4a32, Cyp4a29, Cyp4a30b) were unchanged. Cyp2e, which has previously been reported to be upregulated in MCDD was also unchanged 29. In keeping with the theory of impaired very-low-density lipoprotein (VLDL) secretion in NAFLD 17, 30, endoplasmic reticulum (ER)-associated mediators of triglyceride hydrolysis and VLDL assembly ( Pnpla3, Mttp and the carboxylesterase enzymes: Ces1d, Ces1f, Ces3b) were markedly suppressed in MCDD, with Pnpla3 also suppressed in CDD. Interestingly, expression of the Ces1b isoform was clearly up-regulated in both groups, in contrast to the other members of this class. Apoa4, a lipid binding protein involved in the expansion and secretion of VLDL particles was significantly induced. Finally, two key intermediaries in hepatic insulin signal transduction ( Irs2 and Pdk1) were also down-regulated.
One-carbon metabolism
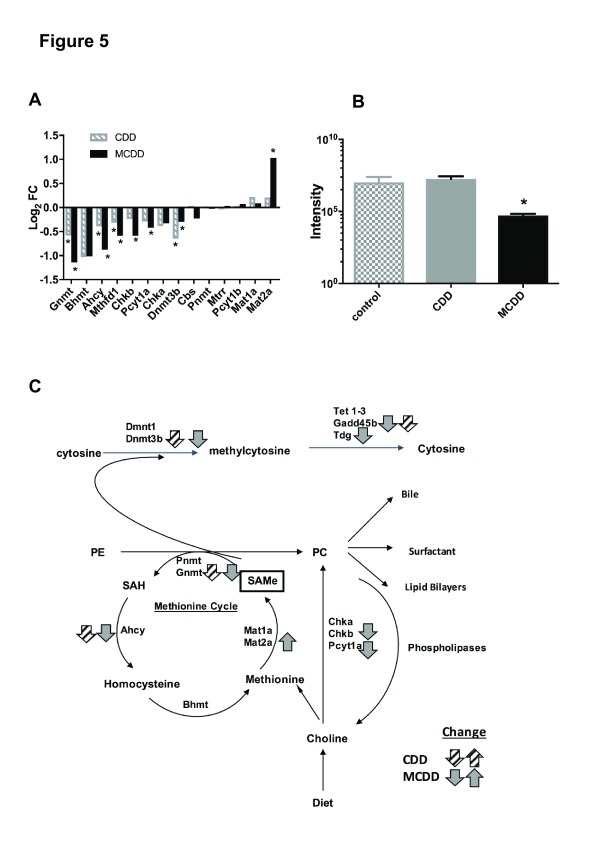
Given the importance of choline and methionine as methyl donors, we then proceeded to examine the expression of genes important in one-carbon metabolism. There were striking changes in the expression of genes associated with choline, methionine and phosphatidylcholine (PC) metabolism in both interventions, broadly in the same direction ( Figure 5A). Two pathways demonstrated significant down-regulation of key enzymes in MCDD: including genes important in the synthesis of PC from choline ( Chkb, Pcyt1a) and in the conversion of SAMe to homocysteine ( Gnmt, Ahcy). Furthermore, the expression of enzymes that contribute to the clearance of methionine, SAMe and S-adenosylhomocysteine ( Mthfd1, Gnmt, Achy, Dnmt3b) were suppressed in both groups. In MCDD mice, there was also marked upregulation of expression of Mat2a, an enzyme necessary for the synthesis of SAMe from methionine. In the light of these results, we measured SAMe concentrations in each group using MALDI-MSI. This demonstrated a significant reduction in SAMe in MCDD mice with no change in CDD ( Figure 5B). A summary of pathways showing changes in enzyme expression is shown in Figure 5C.
Figure 5. CDD and MCDD induce changes in the hepatic expression of genes important in one-carbon metabolism.
( A) Transcriptional changes in one-carbon metabolism enzymes induced by CDD and MCDD diets (* = adjusted P value < 0.05, Benjamini – Hochberg analysis). ( B) SAMe levels as measured by MALDI analysis. (* = P< 0.01 one-way ANOVA with Bonferroni post hoc analysis). C) Metabolic interactions between methionine cycle, phosphatidylcholine synthesis and DNA methylation. Grey (CDD) and black (MCDD) arrows depict expression changes in each pathway. PE=phosphatidylethanolamine, PC=phosphatidylcholine, SAH=S-adenosylhomocysteine, SAMe = S-adenosylmethionine. Adapted from Li and Vance 2008.
Comparison with human NAFLD datasets
We then proceeded to compare our transcriptional findings with large published expression sets of three stages of NAFLD: simple steatosis, NASH and HCC 22– 24 ( Figure 6). While a number of microarray studies have been performed in NAFLD 31– 33 we selected datasets from Ahrens et al and Lake et al (accession numbers: GSE48452 and E-MEXP-3291), due to the detailed patient and histological descriptors (including Kleiner NAFLD activity score) confirming NAFLD stage 22, 23. In addition, these datasets include all three NAFLD stages and control samples in the same data series, reducing assay variation, and are directly available from the ArrayExpress repository. This allowed direct comparison of obese subjects with simple steatosis (n=22, NAS score <3), and patients with NASH (n = 24, NAS score 3-5) with well characterised controls (n = 37). Details of subject numbers and arrays used are in Supplementary Table 5. There are no current datasets available from exclusively NAFLD-induced HCC. We therefore used a large dataset of mixed Hepatitis C and alcohol-induced HCC samples (n=228), which are directly compared with cirrhotic liver samples (n=168) (accession number: GSE63898). In this way, we aimed to identify the transcriptional changes associated with HCC malignant transformation and compared these with our findings in murine methyl donor deficiency.
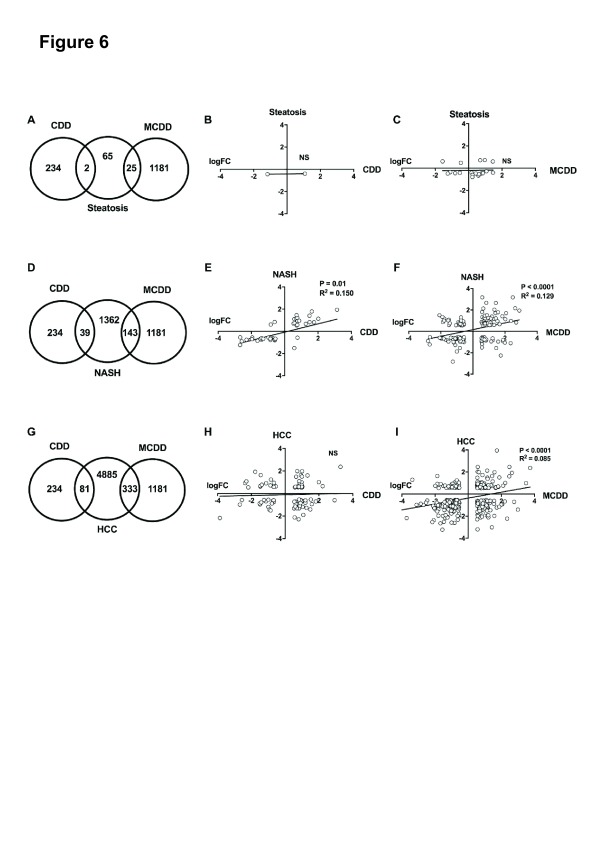
Figure 6. Cross-species comparison of CDD and MCDD transcriptional changes with human stages of NAFLD.
Venn diagrams showing common and distinct dysregulated gene orthologues between CDD and MCDD livers and human hepatic steatosis ( A), human NASH ( D) and human HCC ( G). Scatter plots demonstrate Log2 fold change in common dysregulated orthologues between CDD and MCDD mouse livers and corresponding orthologues in human steatosis ( B+ C), human NASH ( E+ F) and human HCC ( H+ I). Trend line, P value and R 2 value calculated by linear regression analysis.
The CDD and MCDD transcriptomes demonstrated very limited similarity to all stages of human NAFLD. Only 2 (3%) of genes identified as dysregulated in human steatosis were also dysregulated in CDD livers ( Figure 6A and B). 26 (40%) of genes identified as dysregulated in human steatosis were also altered in MCDD mice, however changes in expression of the most commonly dysregulated genes were not in the same direction and there was no significant correlation on linear regression analysis (P = 0.9) ( Figure 6C). There was a greater but still comparatively small overlap in dysregulated gene sets from human NASH studies and CDD and MCDD mice (39 (2.8%) and 143 (10.4%) respectively ( Figure 6D). Those transcripts that were dysregulated in NASH and CDD or MCDD mice did demonstrate a weak but significant correlation in terms of directional change (P 0.01, R 2 0.150 for CDD and P < 0.001, R 2 0.129 for MCDD) ( Figure 6E and F). Upregulated transcripts common to both CDD mice and NASH were almost exclusively involved in inflammatory (Lgals3, Cd52, Clec7a) and malignant processes (Tm4sf4, S100A11, Gpnmb). These genes were also upregulated in MCDD and NASH, in which there were additional changes in fibrosis regulators ( Lum, Osbpl3, Col6a3, Tgfbi, Tmsb10, Tpm1) and oncogenes ( Golm1 and Emp1). Down-regulated genes common to CDD, MCDD and NASH were overrepresented in the GO terms “GO:0006629 Lipid metabolic process” and included the master lipid regulator Mlxipl and the lipid synthesis enzymes Acat2, Agpat2, Lss, Acacb and Mvd.
When comparing the transcriptome of each dietary intervention to HCC, there was again only minimal overlap in perturbed transcripts (81 (1.6%) in CDD and 333 (7.6%) in MCDD) ( Figure 6G). There was no correlation in terms of directional change between CDD and HCC ( Figure 6H). Overlapping transcripts between MCDD mice and HCC did show a weak and highly significant agreement in direction of transcriptional change (R 2 0.085, P < 0.0001, Figure 6I). Genes which were dysregulated in both MCDD and CDD datasets and in HCC were overrepresented in GO terms ‘GO:0000278 mitotic cell cycle’, ‘GO:0007599 Haemostasis’ and ‘GO:0070373 negative regulation of ERK1 and ERK2 cascade’ and include putative HCC oncogenes Cdc20, Osgin1 and Cdk1.
Discussion
It is widely assumed that the steatosis induced by CDD and MCDD results from impaired export of VLDLs, which are required for triglyceride clearance from hepatocytes, perhaps because deficiency of choline and methionine results in an inability to synthesise the major lipid bilayer component phosphatidylcholine (PC) required for VLDL synthesis 30, 34. Our study supports the concept that both decreased PC synthesis and impaired VLDL secretion may play a role in the hepatic pathology in these models and suggest a potential role for the carboxylesterase (Ces) enzymes in mediating the reduction in VLDL secretion.
The importance of reduced hepatic lipid clearance in MCDD is supported by studies demonstrating i) reduced clearance of radiolabelled hepatic fatty acids, ii) a decrease in serum VLDL concentrations and iii) reduced serum triglyceride accumulation in the context of the peripheral lipase inhibitor typoxalol 17, 26. Additionally, increased hepatic sequestration of radiolabelled fatty acids and increased incorporation of 14C into hepatic triglycerides suggest that increased lipid uptake and/or increased de novo lipogenesis may also occur with MCDD 17, 26, 34. These effects have not been reported in rodents exposed to CDD alone 17, 35; indeed ex vivo studies using primary hepatocytes isolated from rats maintained on CDD has shown that the presence of methionine is sufficient to maintain normal levels of PC synthesis and VLDL export into culture media 34 and similar experiments in mouse primary hepatocytes demonstrated only a minor reduction in triglyceride export and no change in apolipoprotein secretion in choline deficient media 9. These findings may be due to the presence of an accessory pathway for PC synthesis which is only present in liver, where in the absence of choline, PC can be directly synthesised from phosphatidylethanolamine (PE) by the enzyme phosphatidylethanolamine N-methyltransferase (PEMT) using methionine as a methyl donor. Indeed ~30% of PC is synthesised in this way in rodent liver 36.
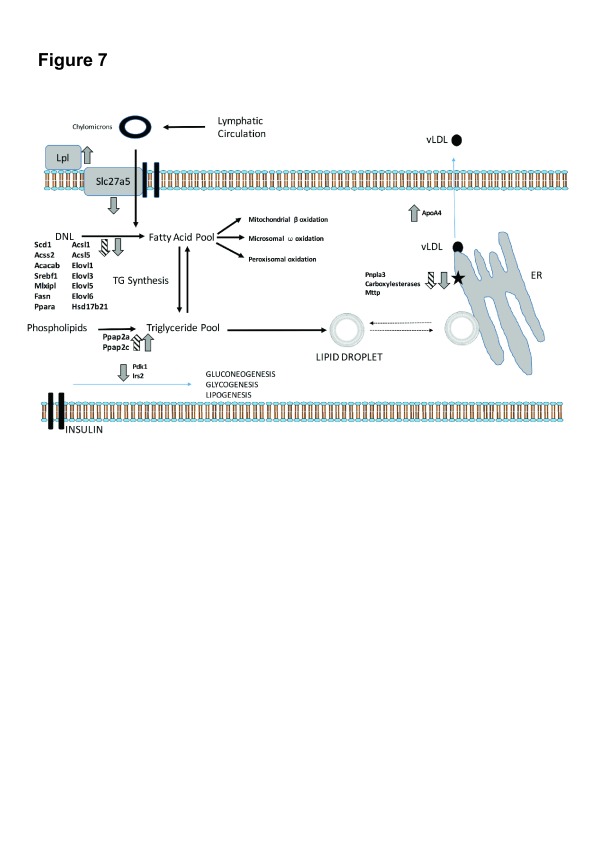
In our study, detailed analysis of the expression of genes in de novo lipogenesis pathways in CDD and MCDD strongly suggest an appropriate compensatory response to the high hepatic triglyceride content, with a clear suppression of key mediators of fatty acid synthesis and elongation and cholesterol synthesis ( Figure 7). This supports the concept that impaired lipid clearance rather than impaired de novo lipogenesis is responsible for the hepatic fat accumulation that occurs with both diets. Consistent with this, the expression of the Ces enzymes (Ces1d, Ces1f, Ces3b) was markedly suppressed in MCDD and the expression of the liver predominant Ces isoform, Ces3b, was also suppressed in CDD. These enzymes are important regulators of VLDL lipid packaging and assembly in the hepatic endoplasmic reticulum (ER), and as such a reduction in expression would be expected to result in reduced hepatic lipid clearance 20. Mice lacking liver specific Ces3 (also known as triacylglycerol hydrolase) have a reduction in circulating VLDL triglycerides and cholesterol levels on a standard chow diet with altered hepatic lipid droplet morphology 20, 37. Furthermore, Ces1 overexpression in mice reduces hepatic triglyceride content and plasma glucose levels whereas liver specific knock-down results in increased hepatic triglyceride 19. Whilst it is unclear why the expression of these genes is suppressed in the presence of an increased hepatic lipid load (notably in MCDD), we suggest that these models may present an opportunity for investigating the mechanism of action of these important hepatic lipid clearance enzymes and the screening of therapeutics that exploit these molecular targets. The expression of Patatin-like phospholipase domain containing 3 (Pnpla3) was also suppressed in both CDD and MCDD models. The human PNPLA3 I148M variant is strongly associated with human NAFLD 38; humans homozygous for the PNPLA3 I148M allele are reported to have ~73% more hepatic triglyceride when compared with matched heterozygote controls, and both in vivo and in vitro studies suggest that this is due to impaired triglyceride hydrolysis and VLDL export 39– 42. Thus, Pnpla3 suppression may also contribute to triglyceride accumulation in methyl donor deficiency.
Figure 7. Transcriptional dysregulation mapped to pathways of lipid transport and metabolism in hepatocytes.
Arrows demonstrate direction of transcriptional change in CDD (hatched) and MCDD (grey) diets. Pnpla3 and Ces suppression suggest impaired packaging of lipid into VLDL particles on the surface of the ER (black star), which may represent a novel site of lipid accumulation.
Dissection of the interacting pathways involved in choline, methionine and PC metabolism in these models provides further insights into the mechanisms by which the murine liver responds to dietary choline and methionine deficiency. Whilst choline has a major role as a substrate for PC synthesis 36, the essential amino acid methionine is also necessary for the methylation of a large variety of substrates including DNA, proteins and lipids and for the synthesis of polyamines, and it is also crucial for normal hepatocyte function 43. Both substrates are important for the maintenance of hepatic SAMe levels, which are normally tightly regulated to maintain normal hepatic function 15, and the direction of transcriptional changes with choline and methoinine deficiency strongly suggest a drive to maintain hepatic SAMe concentrations. The down-regulation of Chkb and Pcyta which are involved in the synthesis of PC from choline, coupled with the upregulation of methionine adenosyl-transferase (Mat2a), which synthesises SAMe from methionine, suggest a forward drive to maintain SAMe levels. The concurrent down-regulation of Gnmt and Ahcy (which metabolise SAMe and SAH respectively) may act as a further cellular buffer to maintain SAMe concentrations 44. Nevertheless, despite these changes, we found reduced levels of hepatic SAMe in MCDD mice in agreement with other studies 45, suggesting an inability to maintain SAMe levels with severe deficiency of both substrates. Thus, the cumulative effect of the observed transcriptional changes in MCDD mice is directed at maintaining SAMe concentrations at the expense of PC synthesis, with the potential to result in decreased VLDL synthesis. Further evidence in support of the importance of SAMe deficiency in the pathogenesis of liver disease in MCDD mice is supported by the fact that the deleterious effects of MCDD diets can be rescued by the administration of SAMe 46.
Whilst there are some clear biological similarities between the hepatic pathology induced by methyl donor deficiency in rodents and human NAFLD/NASH 35, 47– 49, 50, there are also a number of major differences. In humans, NAFLD is closely associated with obesity and insulin resistance, whereas in rodents, CDD results in profound hepatic steatosis without insulin resistance 17, 51 and MCDD causes an inflammatory steatohepatitis with fibrogenesis and significant weight loss with an increase in peripheral insulin sensitivity 30, 52. Our transcriptomic analysis also suggests that CDD and MCDD produce a hepatic phenotype which is markedly dissimilar to human NAFLD in terms of lipid handling. Whereas human NAFLD is associated with an upregulation of genes important in de novo lipogenesis (FASN, MLPXL, ACACA, SREB-1c) 53, 54, this is either not seen, or indeed the reverse is observed in mice maintained on CDD/MCDD diets. Furthermore, although some findings in human NASH support the concept that NAFLD may result at least in part in from an inability to synthesise PC 55, 56, none of the genes dysregulated in the one carbon metabolism pathways of interest in CDD/MCDD were also altered in the human simple steatosis or NASH datasets. Although SAMe depletion is a feature of human NAFLD and correlates with severity of disease in NAFLD biopsies, and oral SAMe preparations are currently under review as a treatment for chronic liver disease 14, 16, mediators of SAMe metabolism were not altered the human statosis or NASH datasets. Some enzymes important in SAMe metabolism were dysregulated in the human HCC data (upregulated AHCY (LogFC 0.58); down-regulated BHMT (logFC-1.35), GNMT (LogFC -1.05) and MAT1a (logFC -0.97)), however these did not reflect the changes seen in the mouse model apart from a similar change in the expression of GNMT.
In conclusion, our data suggest a novel alternative mechanism for methyl donor deficient liver injury involving impaired VLDL particle assembly due to suppression of key triglyceride hydrolysis proteins. Although these CDD and MCDD models are widely used for the study of NAFLD, their translational impact in studies of NAFLD/NASH is likely to be limited by fundamental differences in the global transcriptional profiles between these models and human disease states. Our data do suggest that MCDD may be a useful model for studying the development of HCC secondary to the premalignant inflammatory steatohepatitis NASH. We suggest that there remains an urgent need for novel, more representative models of the full spectrum of NAFLD pathology.
Data availability
Data are available on Open Science Framework at DOI 10.17605/OSF.IO/CY5PH
Acknowledgements
Our thanks go the the Edinburgh Clinical Research Facility Genetics Core, Western General Hospital, Edinburgh, UK for technical expertise with microarray processing.
Funding Statement
This work was supported by the Wellcome Trust [102839 and 092494]; the UK Medical Research Council [MC_PC_U127574433 and G0801924], CEFIC and the Innovative Medicine Initiative Joint Undertaking (IMI JU) [115001] and a Scottish Senior Clinical Fellowship [SCD/09].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
Supplementary material
Supplementary Figure 1: Gene Ontology analysis of CDD and MCDD mouse liver. Overrepresented pathways of up (black) and down (hatched) dysregulated genes in CDD (A+B) and MCDD (C+D) mouse liver microarray analysis.
Supplementary Table 1: Dietary constituents for CDD and MCDD diets. Mice were maintained on control, Choline deficient (CDD) or methionine and choline deficient (MCDD) diets (Dyets, Bethlem, PA) for 4 weeks.
Supplementary Table 2: Primer sequences for qPCR validation. Candidate genes identified as differentially expressed on the microarray were validated using qPCR.
References
- 1. Loomba R, Sanyal AJ: The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10(11):686–690. 10.1038/nrgastro.2013.171 [DOI] [PubMed] [Google Scholar]
- 2. Leung C, Yeoh SW, Patrick D, et al. : Characteristics of hepatocellular carcinoma in cirrhotic and non-cirrhotic non-alcoholic fatty liver disease. World J Gastroenterol. 2015;21(4):1189–96. 10.3748/wjg.v21.i4.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smits MM, Ioannou GN, Boyko EJ, et al. : Non-alcoholic fatty liver disease as an independent manifestation of the metabolic syndrome: results of a US national survey in three ethnic groups. J Gastroenterol Hepatol. 2013;28(4):664–670. 10.1111/jgh.12106 [DOI] [PubMed] [Google Scholar]
- 4. Perry RJ, Samuel VT, Petersen KF, et al. : The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510(7503):84–91. 10.1038/nature13478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blachier M, Leleu H, Peck-Radosavljevic M, et al. : The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol. 2013;58(3):593–608. 10.1016/j.jhep.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 6. Ito M, Suzuki J, Tsujioka S, et al. : Longitudinal analysis of murine steatohepatitis model induced by chronic exposure to high-fat diet. Hepatol Res. 2007;37(1):50–57. 10.1111/j.1872-034X.2007.00008.x [DOI] [PubMed] [Google Scholar]
- 7. Gallou-Kabani C, Vigé A, Gross MS, et al. : C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring). 2007;15(8):1996–2005. 10.1038/oby.2007.238 [DOI] [PubMed] [Google Scholar]
- 8. Duval C, Thissen U, Keshtkar S, et al. : Adipose tissue dysfunction signals progression of hepatic steatosis towards nonalcoholic steatohepatitis in C57Bl/6 mice. Diabetes. 2010;59(12):3181–3191. 10.2337/db10-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kulinski A, Vance DE, Vance JE: A choline-deficient diet in mice inhibits neither the CDP-choline pathway for phosphatidylcholine synthesis in hepatocytes nor apolipoprotein B secretion. J Biol Chem. 2004;279(23):23916–24. 10.1074/jbc.M312676200 [DOI] [PubMed] [Google Scholar]
- 10. Leclercq IA, Lebrun VA, Starkel P, et al. : Intrahepatic insulin resistance in a murine model of steatohepatitis: effect of PPARgamma agonist pioglitazone. Lab Invest. 2007;87(1):56–65. 10.1038/labinvest.3700489 [DOI] [PubMed] [Google Scholar]
- 11. Lyman RL, Giotas C, Medwadowski B, et al. : Effect of low methionine, choline deficient diets upon major unsaturated phosphatidyl choline fractions of rat liver and plasma. Lipids. 1975;10(3):157–167. 10.1007/BF02534154 [DOI] [PubMed] [Google Scholar]
- 12. Collin de l'Hortet A, Zerrad-Saadi A, Prip-Buus C, et al. : GH administration rescues fatty liver regeneration impairment by restoring GH/EGFR pathway deficiency. Endocrinology. 2014;155(7):2545–2554. 10.1210/en.2014-1010 [DOI] [PubMed] [Google Scholar]
- 13. Okubo H, Kushiyama A, Sakoda H, et al. : Involvement of resistin-like molecule β in the development of methionine-choline deficient diet-induced non-alcoholic steatohepatitis in mice. Sci Rep. 2016;6: 20157. 10.1038/srep20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martinez-Chantar ML, García-Trevijano ER, Latasa MU, et al. : Importance of a deficiency in S-adenosyl-L-methionine synthesis in the pathogenesis of liver injury. Am J Clin Nutr. 2002;76(5):1177s–82s. [DOI] [PubMed] [Google Scholar]
- 15. Mato JM, Lu SC: Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–1312. 10.1002/hep.21650 [DOI] [PubMed] [Google Scholar]
- 16. Anstee QM, Day CP: S-adenosylmethionine (SAMe) therapy in liver disease: a review of current evidence and clinical utility. J Hepatol. 2012;57(5):1097–1109. 10.1016/j.jhep.2012.04.041 [DOI] [PubMed] [Google Scholar]
- 17. Macfarlane DP, Zou X, Andrew R, et al. : Metabolic pathways promoting intrahepatic fatty acid accumulation in methionine and choline deficiency: implications for the pathogenesis of steatohepatitis. Am J Physiol Endocrinol Metab. 2011;300(2):E402–409. 10.1152/ajpendo.00331.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swales JG, Tucker JW, Strittmatter N, et al. : Mass spectrometry imaging of cassette-dosed drugs for higher throughput pharmacokinetic and biodistribution analysis. Anal Chem. 2014;86(16):8473–80. 10.1021/ac502217r [DOI] [PubMed] [Google Scholar]
- 19. Xu J, Li Y, Chen WD, et al. : Hepatic carboxylesterase 1 is essential for both normal and farnesoid X receptor-controlled lipid homeostasis. Hepatology. 2014;59(5):1761–1771. 10.1002/hep.26714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lian J, Wei E, Wang SP, et al. : Liver specific inactivation of carboxylesterase 3/triacylglycerol hydrolase decreases blood lipids without causing severe steatosis in mice. Hepatology. 2012;56(6):2154–2162. 10.1002/hep.25881 [DOI] [PubMed] [Google Scholar]
- 21. Quiroga AD, Li L, Trötzmüller M, et al. : Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia. Hepatology. 2012;56(6):2188–2198. 10.1002/hep.25961 [DOI] [PubMed] [Google Scholar]
- 22. Ahrens M, Ammerpohl O, von Schönfels W, et al. : DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. 10.1016/j.cmet.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 23. Lake AD, Novak P, Fisher CD, et al. : Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39(10):1954–60. 10.1124/dmd.111.040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Villanueva A, Portela A, Sayols S, et al. : DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–1956. 10.1002/hep.27732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raubenheimer PJ, Nyirenda MJ, Walker BR: A choline-deficient diet exacerbates fatty liver but attenuates insulin resistance and glucose intolerance in mice fed a high-fat diet. Diabetes. 2006;55(7):2015–2020. 10.2337/db06-0097 [DOI] [PubMed] [Google Scholar]
- 26. Rinella ME, Elias MS, Smolak RR, et al. : Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J Lipid Res. 2008;49(5):1068–1076. 10.1194/jlr.M800042-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ip E, Farrell GC, Robertson G, et al. : Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology. 2003;38(1):123–132. 10.1053/jhep.2003.50307 [DOI] [PubMed] [Google Scholar]
- 28. Johnson EF, Hsu MH, Savas U, et al. : Regulation of P450 4A expression by peroxisome proliferator activated receptors. Toxicology. 2002;181–182:203–206. 10.1016/S0300-483X(02)00282-2 [DOI] [PubMed] [Google Scholar]
- 29. Leclercq IA, Farrell GC, Field J, et al. : CYP2E1 and CYP4A as microsomal catalysts of lipid peroxides in murine nonalcoholic steatohepatitis. J Clin Invest. 2000;105(8):1067–1075. 10.1172/JCI8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rinella ME, Green RM: The methionine-choline deficient dietary model of steatohepatitis does not exhibit insulin resistance. J Hepatol. 2004;40(1):47–51. 10.1016/j.jhep.2003.09.020 [DOI] [PubMed] [Google Scholar]
- 31. Rubio A, Guruceaga E, Vázquez-Chantada M, et al. : Identification of a gene-pathway associated with non-alcoholic steatohepatitis. J Hepatol. 2007;46(4):708–718. 10.1016/j.jhep.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 32. Younossi ZM, Baranova A, Ziegler K, et al. : A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42(3):665–674. 10.1002/hep.20838 [DOI] [PubMed] [Google Scholar]
- 33. Starmann J, Fälth M, Spindelböck W, et al. : Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One. 2012;7(10):e46584. 10.1371/journal.pone.0046584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yao ZM, Vance DE: The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J Biol Chem. 1988;263(6):2998–3004. [PubMed] [Google Scholar]
- 35. Veteläinen R, van Vliet A, van Gulik TM: Essential pathogenic and metabolic differences in steatosis induced by choline or methione-choline deficient diets in a rat model. J Gastroenterol Hepatol. 2007;22(9):1526–1533. 10.1111/j.1440-1746.2006.04701.x [DOI] [PubMed] [Google Scholar]
- 36. Li Z, Vance DE: Phosphatidylcholine and choline homeostasis. J Lipid Res. 2008;49(6):1187–1194. 10.1194/jlr.R700019-JLR200 [DOI] [PubMed] [Google Scholar]
- 37. Wang H, Wei E, Quiroga AD, et al. : Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol Biol Cell. 2010;21(12):1991–2000. 10.1091/mbc.E09-05-0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He S, McPhaul C, Li JZ, et al. : A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285(9):6706–15. 10.1074/jbc.M109.064501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pirazzi C, Adiels M, Burza MA, et al. : Patatin-like phospholipase domain-containing 3 ( PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol. 2012;57(6):1276–82. 10.1016/j.jhep.2012.07.030 [DOI] [PubMed] [Google Scholar]
- 40. Huang Y, Cohen JC, Hobbs HH: Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286(43):37085–93. 10.1074/jbc.M111.290114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chamoun Z, Vacca F, Parton RG, et al. : PNPLA3/adiponutrin functions in lipid droplet formation. Biol Cell. 2013;105(5):219–33. 10.1111/boc.201200036 [DOI] [PubMed] [Google Scholar]
- 42. Sookoian S, Rosselli MS, Gemma C, et al. : Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52(6):1992–2000. 10.1002/hep.23927 [DOI] [PubMed] [Google Scholar]
- 43. Mato JM, Martínez-Chantar ML, Lu SC: S-adenosylmethionine metabolism and liver disease. Ann Hepatol. 2013;12(2):183–189. [PMC free article] [PubMed] [Google Scholar]
- 44. Luka Z, Mudd SH, Wagner C: Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2009;284(34):22507–22511. 10.1074/jbc.R109.019273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caballero F, Fernández A, Matías N, et al. : Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: Impact on mitochondrial S-adenosyl-L-methionine and glutathione. J Biol Chem. 2010;285(24):18528–18536. 10.1074/jbc.M109.099333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dahlhoff C, Worsch S, Sailer M, et al. : Methyl-donor supplementation in obese mice prevents the progression of NAFLD, activates AMPK and decreases acyl-carnitine levels. Mol Metab. 2014;3(5):565–80. 10.1016/j.molmet.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schattenberg JM, Wang Y, Singh R, et al. : Hepatocyte CYP2E1 overexpression and steatohepatitis lead to impaired hepatic insulin signaling. J Biol Chem. 2005;280(11):9887–9894. 10.1074/jbc.M410310200 [DOI] [PubMed] [Google Scholar]
- 48. Emery MG, Fisher JM, Chien JY, et al. : CYP2E1 activity before and after weight loss in morbidly obese subjects with nonalcoholic fatty liver disease. Hepatology. 2003;38(2):428–435. 10.1053/jhep.2003.50342 [DOI] [PubMed] [Google Scholar]
- 49. Weltman MD, Farrell GC, Hall P, et al. : Hepatic cytochrome P450 2E1 is increased in patients with nonalcoholic steatohepatitis. Hepatology. 1998;27(1):128–133. 10.1002/hep.510270121 [DOI] [PubMed] [Google Scholar]
- 50. Fisher CD, Lickteig AJ, Augustine LM, et al. : Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos. 2009;37(10):2087–2094. 10.1124/dmd.109.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raubenheimer PJ, Nyirenda MJ, Walker BR: A choline-deficient diet exacerbates fatty liver but attenuates insulin resistance and glucose intolerance in mice fed a high-fat diet. Diabetes. 2006;55(7):2015–2020. 10.2337/db06-0097 [DOI] [PubMed] [Google Scholar]
- 52. Rizki G, Arnaboldi L, Gabrielli B, et al. : Mice fed a lipogenic methionine-choline-deficient diet develop hypermetabolism coincident with hepatic suppression of SCD-1. J Lipid Res. 2006;47(10):2280–2290. 10.1194/jlr.M600198-JLR200 [DOI] [PubMed] [Google Scholar]
- 53. Auguet T, Terra X, Quintero Y, et al. : Liver lipocalin 2 expression in severely obese women with non alcoholic fatty liver disease. Exp Clin Endocrinol Diabetes. 2013;121(2):119–124. 10.1055/s-0032-1331696 [DOI] [PubMed] [Google Scholar]
- 54. Higuchi N, Kato N, Shundo Y, et al. : Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol Res. 2008;38(11):1122–9. 10.1111/j.1872-034X.2008.00382.x [DOI] [PubMed] [Google Scholar]
- 55. Li Z, Agellon LB, Allen TM, et al. : The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3(5):321–331. 10.1016/j.cmet.2006.03.007 [DOI] [PubMed] [Google Scholar]
- 56. Arendt BM, Ma DW, Simons B, et al. : Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl Physiol Nutr Metab. 2013;38(3):334–340. 10.1139/apnm-2012-0261 [DOI] [PubMed] [Google Scholar]