Abstract
Sleep is crucial for survival and well-being. This behavioral and physiological state has been studied in all major genetically accessible model animals, including rodents, fish, flies, and worms. Genetic and optogenetic studies have identified several neurons that control sleep, making it now possible to compare circuit mechanisms across species. The “motor” of sleep across animal species is formed by neurons that depolarize at the onset of sleep to actively induce this state by directly inhibiting wakefulness. These sleep-inducing neurons are themselves controlled by inhibitory or activating upstream pathways, which act as the “drivers” of the sleep motor: arousal inhibits “sleep-active” neurons whereas various sleep-promoting “tiredness” pathways converge onto sleep-active neurons to depolarize them. This review provides the first overview of sleep-active neurons across the major model animals. The occurrence of sleep-active neurons and their regulation by upstream pathways in both vertebrate and invertebrate species suggests that these neurons are general and ancient components that evolved early in the history of nervous systems.
Keywords: Caenorhabditis elegans, Drosophila melanogaster, behavioral genetics, Danio rerio, model organisms, Mus musculus, optogenetics, sleep
Sleep Is a Conserved Behavioral and Physiological State
LACK of sleep or low sleep quality causes tiredness, lowers productivity, and decreases mood. Sleeping problems are widespread in human societies across the globe with severe health and economic consequences (Colten and Altevogt 2006). Because of its vital role, this behavior can be found in most animals that have a nervous system and that have been studied carefully (Campbell and Tobler 1984; Cirelli and Tononi 2008). To fulfill its essential functions, sleep is exquisitely controlled by the brain and other organs to ensure that enough of this behavioral and physiological state takes place (Saper et al. 2010). It is necessary to understand how sleep is controlled if we are to develop strategies to intervene with sleeping disorders and to harness the power of sleep to improve human health.
Sleep is defined and can be identified by behavioral criteria such as reversible behavioral quiescence with a reduced responsiveness to sensory stimulation and homeostatic control (Campbell and Tobler 1984). Using these behavioral criteria, sleep has been detected in various species including mammals and birds, and also in all other major animal model systems, such as the zebrafish Danio rerio, the fruit fly Drosophila melanogaster, and the nematode Caenorhabditis elegans (Hendricks et al. 2000; Shaw et al. 2000; Zhdanova et al. 2001; Raizen et al. 2008; Kayser and Biron 2016; Trojanowski and Raizen 2016; Artiushin and Sehgal 2017; Oikonomou and Prober 2017). Sleep has even been detected in basal metazoans that have a nervous system such as cnidarians, suggesting that sleep evolved together with a nervous system before its centralization (Figure 1) (Nath et al. 2017).
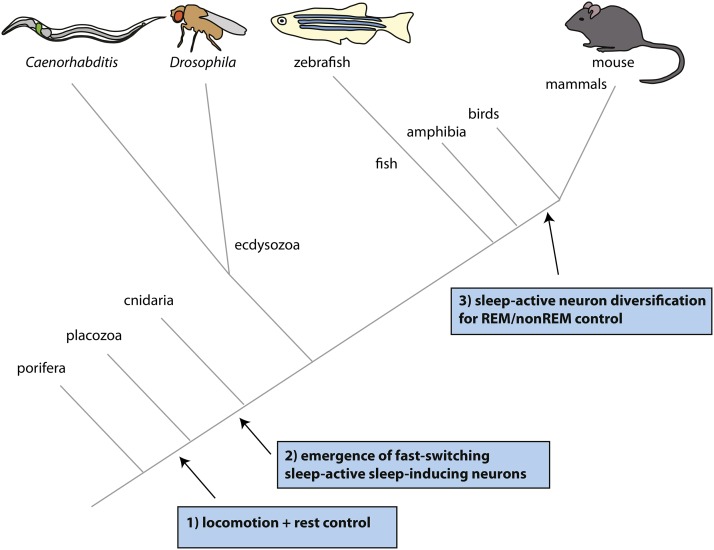
Figure 1.
Three key steps in the evolution of sleep-active neurons. (1) As soon as locomotion behavior evolved, a need for rest led to the (likely endocrine) control of active and inactive states. (2) With the invention of a nervous system, the need for a fast-switching sleep system arose, leading to the evolution of inhibitory sleep-active sleep-inducing neurons and circuits controlling these. Cnidarians sleep and presumably possess sleep-active neurons, even though these neurons have not been identified yet. (3) To cope with demands for higher brain function complex brains diversified sleep-active neurons to generate both non-REM (rapid eye movement) and REM sleep.
Electrophysiological parameters that match behavioral characteristics are often used to detect and characterize sleep. For example, in the mammalian and avian brain, electrical activity measurements called electroencephalograms (EEGs) are typically used to classify and quantify different sleep stages. The EEG measures local field potentials of the cortex (Jackson and Bolger 2014). Electrophysiological measurements in humans led to the discovery of two types of sleep: rapid eye movement (REM) sleep, which is also called active sleep, and non-REM sleep, which is also called quiet sleep. REM sleep is characterized by asynchronous brain activity that is similar to cerebral firing patterns during wakefulness. Body muscles are paralyzed, with the notable exception of certain muscles required for respiration as well as muscles required for eye movement, allowing the continuation of breathing and causing complex eye movements (Aserinsky and Kleitman 1953; Orem et al. 2000; Peever and Fuller 2017). REM sleep is present in mammals and birds, and evidence for REM sleep also exists in nonavian reptiles, suggesting an evolutionary emergence of REM sleep at the level of the reptiles (Libourel and Herrel 2016; Shein-Idelson et al. 2016). Non-REM sleep in mammals and birds is characterized by strongly reduced muscle tone and brain activity, as well as slow oscillatory EEG patterns called slow waves. The extent of slow waves, called slow-wave activity, was found to correlate with the difficulty of arousing a subject during sleep across many conditions and is thus used as a proxy for sleep depth (Dijk 2009; Vorster and Born 2015). While EEG slow-wave activity is an established marker for non-REM sleep in mammals and birds, such electrical activity is a product of specific brain architecture and thus cannot necessarily be detected in other species. Similar patterns can still be observed in amphibians and reptiles (Libourel and Herrel 2016; Shein-Idelson et al. 2016). However, in zebrafish, Drosophila, and C. elegans, slow-wave activities have not been described. Nevertheless, even invertebrates show brain activity characteristic for quiet sleep, such as an overall reduction of neural activity (Nitz et al. 2002; Schwarz et al. 2011; Bushey et al. 2015; Nichols et al. 2017). Thus, the type of sleep that is found most widely across species is non-REM or quiet sleep, which likely occurs in most or all animals that have a nervous system. By contrast, there is currently no compelling evidence for the existence of REM or active sleep outside of the amniotes. The restricted distribution of REM sleep and the wide occurrence of quiet sleep have shaped ideas as to the functions of sleep. REM sleep is believed to be required for higher brain functions, perhaps learning or forgetting. Its importance is suggested by a rebound after deprivation, but there is no consensus as to how important REM sleep is for survival (Kushida et al. 1989; Siegel 2001; Peever and Fuller 2017). While non-REM/quiet sleep functions are only just beginning to be understood, there is a broad consensus that sleep is required to restore fundamental cellular processes, manage energy metabolism and behavior, and is vital for animal life (Figure 1) (Cirelli and Tononi 2008; Siegel 2009; Joiner 2016).
Drivers and Motors of Sleep
Sleep-promoting neurons are important regulators of sleep. These neurons can act in two different ways to promote sleep, either directly or indirectly. The first type typically depolarizes strongly at the onset of sleep, which leads to the release of inhibitory neurotransmitters such as neuropeptides and γ-aminobutyric acid (GABA) directly onto wake-promoting circuits. This then causes an inhibition of arousal and alertness, and thus an induction of sleep. This type of sleep-promoting neuron is called sleep-active, because the depolarization of these neurons precisely coincides with sleep induction (Saper et al. 2005). Sleep-active neurons that directly inhibit arousal circuits could thus be viewed as the motor that causes sleep.
A second group of sleep-promoting neurons acts indirectly by increasing the activity of sleep-active neurons (Schwartz and Roth 2008; Artiushin and Sehgal 2017). The neurons that activate the sleep motor could be viewed as the drivers that control sleep-active neurons. While it is plausible to assume that a sleep-promoting “driver” neuron should also depolarize in relation to sleep, physiological data are scarce, in part because calcium imaging and electrophysiology is technically difficult in some systems, and because the circuitry is only partially known. Thus, our understanding of the physiology of sleep-promoting driver neurons still is highly incomplete. Hypothetically, sleep-promoting driver neurons should depolarize prior to sleep-active neurons, before sleep induction, when the system is still in wakefulness.
The key defining behavioral characteristics of sleep—namely reduced responsiveness, reversibility, and homeostasis—can be viewed and understood from the point of neural circuits consisting of positive and negative drivers controlling a sleep motor. According to this interpretation, tiredness is generated by sleep-promoting driver pathways that can lead to depolarization of the sleep-active neurons of the sleep motor. Reduced responsiveness is generated by the inhibition of arousal and sensory circuits through the activation of sleep-active neurons. Wake-promoting pathways inhibit sleep, either by directly inhibiting the sleep motor or its activators. Sleep reversibility can be triggered by a stimulus to the brain that involves the activation of arousal circuits plus the inhibition of sleep-inducing neurons. Homeostasis could involve pathways leading to increased sleep drive, i.e., would present an overactivation of upstream driver pathways leading to increased depolarization of sleep-active neurons, leading to increased sleep.
Similar to sleep neurons, stop neurons function to interrupt locomotion sequences. Locomotion stop is, for instance, required for episodic locomotion. To regulate locomotion patterns, central pattern generators in the spinal cord are controlled to start, maintain, or halt locomotion. In vertebrates, brain stem stop neurons activate at the end of a movement period to inhibit central pattern generators required for locomotion, resulting in the cessation of a locomotion sequence (Bouvier et al. 2015; Juvin et al. 2016). Sleep neurons and stop neurons can be distinguished by the scope of their actions. Whereas sleep neurons induce a systemic inhibition of wakefulness, stop neurons halt locomotion without impairing arousal and alertness.
Upstream driver pathways include timers such as the circadian clock that control at what time of the day the organism engages in this behavior. In addition, sleep is controlled by homeostatic and allostatic mechanisms that reflect the sleep need of the system. Whereas homeostatic mechanisms detect sleep deprivation or extended wakefulness, allostatic mechanisms respond to physiological insult and increase sleep propensity to regain physiological equilibrium (Saper et al. 2005). In summary, understanding sleep control requires understanding the activation and fast switching of sleep-active neurons of the sleep motor through driver pathways.
The use of model organisms has greatly contributed to a molecular mechanistic understanding of sleep. In particular, the study of non-REM or quiet sleep is possible in all major animal models, and the focus of this review will be on the control of sleep though sleep-active neurons from vertebrates to invertebrates. Existing studies, though still giving a highly incomplete picture of sleep regulation, show a high level of conservation of molecular pathways and reoccurrence of circuit mechanisms for sleep control. The high level of similarity and conservation suggests that not only sleep, but also the mechanisms that control it, sleep-active neurons forming a sleep motor and diverse drivers regulating their activity, evolved early in the history of nervous systems.
Sleep-Active Neurons Across Model Animals: Mammals
The first evidence for an active role of a specialized brain region in controlling sleep came from the brains of patients, which, as a consequence of infectious encephalitis, experienced insomnia. There were brain lesions in the anterior hypothalamus in all of the insomnia patients, providing the first evidence of a dedicated brain area that controls sleep, which was called the “center for regulation of sleep” (von Economo 1930). While several mammals, including cats and rats, have been used for anatomical sleep studies, mice have become the most used model mammal for molecular dissections of sleep. Using various models, a sleep-inducing region was narrowed down to a subset of GABAergic/peptidergic neurons in the Preoptic Area (POA) (Figure 2A). It turned out that POA neurons depolarize at the onset of sleep, and were hence called sleep-active. Lesions in this region led to reduced sleep, implying that these neurons are sleep promoting (Nauta 1946; McGinty and Sterman 1968; Lu et al. 2000). Activating all GABAergic neurons of the POA leads to increased wakefulness. However, a subset of these neurons, specifically those that are projecting to the tuberomammillary nucleus (TMN), a wake-promoting region, can induce sleep acutely when activated optogenetically, suggesting that this population of neurons is required for the loss of sleep after POA lesion (Chung et al. 2017).
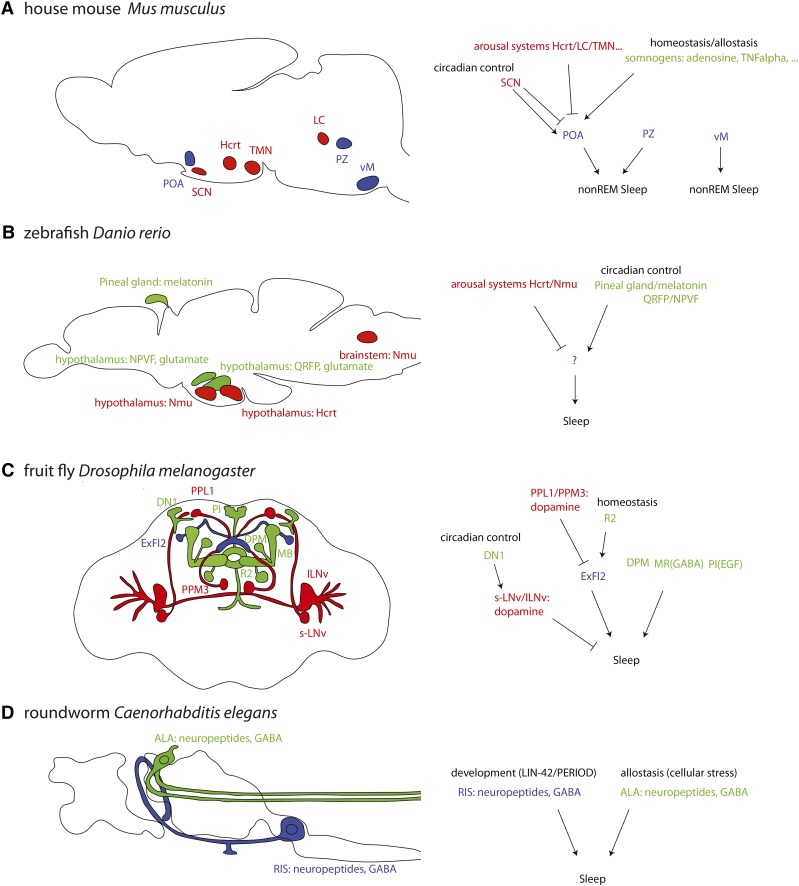
Figure 2.
Sleep-controlling neural circuits already identified in genetic model animals. Sleep-active sleep-promoting inhibitory neurons are shown in blue, wake-promoting neurons are shown in red, and indirectly sleep-promoting neurons are shown in green. For several neurons the classification is tentative. (A) Sleep-active sleep-promoting neurons that inhibit wake-promoting neurons were defined in mammals. These key regulators are themselves controlled by inhibitory and activating pathways. (B) The zebrafish contains sleep-promoting and wake-promoting circuits that are highly similar to mammalian sleep circuits. Sleep-active neurons have not yet been identified in this system. (C) Drosophila possesses several sleep-promoting and wake-promoting circuits. Similar to mammals, it appears to have fast-switching sleep-promoting neurons that are controlled by positive and negative upstream circuits. (D) C. elegans has two key sleep neurons: a single sleep-active sleep-promoting neuron that confers the fast switching kinetics typical for sleep, plus a sleep-promoting neuron that detects allostatic sleep need.
Counteracting the sleep-promoting system is the so-called ascending arousal system, which is formed by several brain areas expressing activating neurotransmitters, an example being the noradrenergic locus ceruleus (LC) in the brain stem. The ascending arousal system causes wakefulness by activation of the thalamus and cerebral cortex (Fuller et al. 2011). Sleep-active neurons of the POA innervate and inhibit wake-promoting areas of the ascending arousal system and thus directly induce sleep by inhibiting wakefulness. By analogy, the sleep-inducing system has been called the descending system (Saper et al. 2010).
While these sleep-active neurons appear to control sleep by inhibiting wake-promoting centers such as the TMN, they are themselves inhibited by the wake-promoting ascending arousal system. This mutual inhibition acts like a “flip–flop” switch, which ensures that sleep and wake exist as discrete states, and allows fast switching between the states, which is a defining characteristic of sleep (Saper et al. 2001, 2010). The importance of stable states is illustrated in narcolepsy patients, who suddenly fall asleep. Narcolepsy can be caused by the loss of a switch-stabilizing neuropeptide called orexin/hypocretin, which is expressed in an area of the hypothalamus and presents a part of the ascending arousal system (Chemelli et al. 1999; Lin et al. 1999; Sakurai 2007).
The fact that lesions in the POA produced only partial sleep loss suggested that additional sleep-active areas exist. Sleep-active neurons were subsequently found in several other brain areas, including the basal forebrain, lateral hypothalamus, cortex, and, importantly, in the medulla of the brain stem (Gerashchenko et al. 2008; Anaclet et al. 2014; Luppi et al. 2017). Inhibitory neurons of the parafacial zone (PZ) of the brain stem were shown to be both required and sufficient for sleep induction. The PZ targets and inhibits neurons of the parabrachial nucleus, a major arousal-controlling center (Weber and Dan 2016; Anaclet and Fuller 2017). REM sleep is controlled by GABAergic neurons of the ventral Medulla. These neurons are active during REM sleep and also during wakefulness, and their optogenetic activation can potently induce the switch from non-REM to REM sleep and maintenance of REM sleep (Weber et al. 2015). Thus, both REM sleep and non-REM sleep induction is mediated by inhibitory sleep-active neurons. Similar to the flip–flop switch controlling sleep and wakefulness, non-REM and REM sleep appear to be mutually exclusive and inhibit each other (Lu et al. 2006). Several centers for sleep induction have been identified in the mammalian brain, with each consisting of inhibitory sleep-active sleep-promoting neurons. Thus, a likely scenario is that, under physiological conditions, sleep in mammals is induced by the concerted action of several regions containing sleep-active neurons.
Circadian control ensures the timing of sleep: for instance, rodents sleep mostly during the day, whereas humans typically show consolidated sleep during the night. Circadian rhythms are generated by conserved factors generating an oscillating transcriptional translational feedback loop. A master oscillator in the suprachiasmatic nucleus (SCN) controls peripheral clocks in cells of other brain parts or organs (Moore and Eichler 1972; Ralph et al. 1990). The SCN provides an arousal cue to consolidate sleep. Ablation of the SCN increases sleep and alters its timing, but most of the daily sleep and homeostasis is still present (Easton et al. 2004). The SCN controls the activity of sleep-active neurons indirectly via a three-step circuit. This indirect connectivity likely allows adaptation of the sleep-wake rhythm to different circadian times: Pacemaker neurons of the SCN are active during the light phase in a wide range of mammals, but circadian sleeping times differ among species and environmental conditions (Chou et al. 2003; Saper et al. 2010). While the SCN provides a wake-promoting signal, it can also be sleep promoting: for example, the SCN rhythmically expresses TGF α, which was reported to promote sleep through EGF signaling (Kramer et al. 2001).
Homeostatic processes play a role in controlling sleep amount and depth. As a part of sleep homeostasis, an increased activity of sleep-active neurons in the POA has been found after sleep deprivation when the animal returned to sleep, indicating that sleep-inducing neurons convey a homeostatic process (Saper et al. 2005; Alam et al. 2014). But what are the mechanisms upstream of sleep-inducing neurons and how do they measure sleep need? It was proposed that sleep-promoting molecules called somnogens are produced as a function of wakefulness. The somnogens then lead to increased sleep drive. One such somnogenic modulator of sleep need is extracellular adenosine, which accumulates in the forebrain during wakefulness (Porkka-Heiskanen et al. 1997). It was suggested that adenosine nucleotides accumulate as a function of neural activity. Thus, increased wakefulness would lead to an increase of extracellular adenosine (Porkka-Heiskanen et al. 2002), which could then lead to the inhibition of wake neurons and the activation of sleep-inducing neurons, perhaps by a disinhibitory mechanism (Morairty et al. 2004; Weber and Dan 2016). Astrocytes control neuronal activity though the secretion of neurotransmitters, a phenomenon that is called gliotransmission. Inhibition of gliotransmission impairs adenosine-mediated aspects of sleep homeostasis. A potential mechanism underlying extracellular adenosine accumulation could be that neuronal activity leads to astrocyte activation, and then astrocytes in turn secrete ATP to the extracellular space, where it is hydrolyzed to adenosine (Halassa et al. 2009; Frank 2013).
Sleep is important for learning and memory as sleep deprivation impairs memory formation (Diekelmann and Born 2010; Vorster and Born 2015). Conversely, learning or stimulating environments appear to increase sleep, consistent with the view that wakefulness promotes sleep though neural use. This effect is region-specific, as those brain areas that are heavily used show a subsequent local increase in sleep. (Datta 2000; Huber et al. 2004; Donlea et al. 2009; Vyazovskiy et al. 2011; Siclari and Tononi 2017). If sleep is under a local use-dependent control and assuming that this local sleep is induced by sleep-active neurons, this would predict that sleep-inducing neurons exist that target local areas specifically. It would be interesting to find out whether global sleep is generated by the concerted action of many parallel local sleep inducers or by systemic sleep generators that exist in addition to the local systems.
In addition to homeostatic drivers, allostatic mechanisms exist that also lead to the production of somnogens. Sickness is known to alter sleep patterns and increase sleepiness. Examples of allostatic somnogens are cytokines produced by infection and inflammation. For example, interleukin 1 and Tumor Necrosis Factor α were shown to increase sleep amount, which in turn was found to improve the functions of the nervous system (Toth et al. 1993; Bryant et al. 2004). While somnogens certainly play a role in sleep regulation, they cannot explain all aspects of sleep homeostasis. Activation of sleep-inducing POA neurons also contributes to the effectiveness of isoflurane anesthesia (Moore et al. 2012). Thus, the amount and quality of sleep are under the control of multiple pathways that include circadian, homeostatic, and allostatic mechanisms. In a likely scenario, these pathways converge onto sleep-inducing neurons and cause their depolarization. Identifying and understanding pathways leading to sleep-inducing neuron depolarization will probably present the most promising strategy for developing treatments for sleeping disorders.
Sleep in Nonmammalian Models: Zebrafish
The appeal of using the zebrafish as a model resides in its bridging position between complex and expensive rodent models, and simpler and cheaper invertebrate models. Zebrafish have smaller brains than mammals yet have a higher conservation of neural architecture compared with invertebrates. Thus, this species allows the dissection of genetic and neural circuits likely relevant to human non-REM sleep. Unlike rodents and like humans, zebrafish are a diurnal species. Sleep in zebrafish can be studied in both the larva and the adult. While adult zebrafish sleep only relatively little, larvae show substantial sleeping behavior and the use of larvae also speeds up the analysis, as large numbers of individuals can be produced (Zhdanova et al. 2001; Yokogawa et al. 2007). The system allows genetic and pharmacological screening and shows a high level of conservation of neuropeptide systems. Sleep in fish is typically detected by behavioral criteria rather than electrophysiological criteria. The main readout is locomotion and its quiescence is used to score sleep. This appears to be a valid approach as locomotion quiescence has been shown to be sleep-like by additional behavioral tests, such as reduced arousal thresholds and reversibility. Homeostatic control of sleep appears to be less pronounced in zebrafish compared with other species and quiescence behavior can be potently suppressed by light (Yokogawa et al. 2007; Rihel and Schier 2013; Barlow and Rihel 2017; Oikonomou and Prober 2017).
Zebrafish brains contain sleep- and wake-promoting neurons. Among the wake-promoting systems are the orexin/hypocretin system that controls the flip–flop switch in mammals, as well as neuromedin U (Chiu et al. 2016; Elbaz et al. 2017) (Figure 2B). Among the best-characterized sleep-promoting systems are melatonin, which is produced by the pineal gland and presents the major sleep-promoting output of the circadian rhythm at night (Zhdanova et al. 2001; Gandhi et al. 2015), and neuropeptides of the RFamide (Arg-Phe-NH2 motif at their C-terminus) family called QRFP. These peptides are expressed in a subset of glutamatergic neurons in the hypothalamus. Gain- and loss-of-function experiments of components of this system suggest that QRFP-expressing neurons are sleep-promoting specifically during the day, and that loss of the peptide receptors leads to hyperactivity. The glutamatergic neurotransmitter expression suggests that these neurons are activating (Chen et al. 2016). It is not known when these neurons are active, but current evidence suggest that they do not present inhibitory sleep-active sleep-promoting neurons, but rather constitute an arousal-controlling circuit that may be linked to the activation of sleep-active neurons, but this classification is tentative. Irrespective of their classification, QRFP neurons cannot explain the majority of sleep, which occurs during the night.
Another sedating peptide in the zebrafish that promotes sleep and inhibits activity is the vertebrate hypothalamic RFamide neuropeptide VF (NPVF). As NPVF-expressing neurons are also glutamatergic, this could mean that they act indirectly by controlling other sleep neurons (Lee et al. 2017). The discovery of sleep promotion by a RFamide peptide in a vertebrate presents an important finding, as RFamide neuropeptides were previously implicated in invertebrate sleep (see below). While it is often difficult to identify homologies for the short and often unstructured neuropeptides between invertebrates and vertebrates, this suggests that RFamide neuropeptides are ancient and conserved regulators of sleep.
In summary, wake- and sleep-promoting systems clearly exist in zebrafish, but sleep-active neurons are as yet unidentified. Because of the high level of conservation and the presence of conserved sleep-regulatory components in this system, it can be expected that sleep-active neurons also exist. Functional imaging of sleep in a strain expressing calcium indicators in inhibitory neurons (Ahrens et al. 2013), focusing on regions such as the hypothalamus and brain stem, may allow the identification of sleep-active neurons also in this species.
Sleep-Promoting Neurons in Drosophila
Drosophila, along with C. elegans, is a major invertebrate model for the molecular dissection of behavior. This diurnal species has a short generation time and has the advantage of allowing facile genetic dissections. It is an instructive model for solving the molecular mechanisms and neural circuits underlying sleep, and results from the fly have been shown to be relevant also for understanding vertebrate sleep (Artiushin and Sehgal 2017).
There are four main regions of inhibitory (GABAergic/peptidergic) sleep-promoting neurons described in the fly (Figure 2C) (Artiushin and Sehgal 2017). One of the first brain structures to have been implicated in sleep control in the fly was the mushroom body. This brain part is complex and contains several populations of cells, including both wake-promoting as well as sleep-promoting neurons (Joiner et al. 2006; Sitaraman et al. 2015; Artiushin and Sehgal 2017). A second sleep-promoting structure is the single pair of dorsal paired medial neurons that innervate the mushroom body (MB). These neurons are GABAergic and serotonergic, and inhibit wake-promoting neurons. Intriguingly, these neurons are also involved in memory consolidation (Haynes et al. 2015). Third, peptidergic neurons in the PI, which may be the counterpart of the mammalian hypothalamus, were shown to be sleep promoting. This process depends on EGF signaling, which also plays a role in mammalian and worm sleep, underscoring the high level of conservation of sleep-regulating pathways (Foltenyi et al. 2007). Fourth are ExFl2 neurons of the dorsal fan-shaped body, which clearly fulfil the criteria for sleep-promoting neurons, with artificial activation leading to sleep induction and ablation leading to sleep reduction (Donlea et al. 2011; Liu et al. 2012). These neurons were not shown to be GABAergic, but are peptidergic. Similar to mammalian sleep-active neurons, they modulate the anesthetic effects of isoflurane (Kottler et al. 2013). Reminiscent of a flip–flop switch, these neurons can be inhibited by wake-promoting dopaminergic PPL1 and PPM3 neurons, switching them from an electrically excitable ON state to a silenced OFF state (Pimentel et al. 2016). As part of a homeostatic compensation of sleep loss and extended wakefulness, ExFl2 cells show increased firing after sleep deprivation. Key to the overactivation of ExFl2 cells during rebound sleep are R2 neurons of the ellipsoid body that are activators of ExFl2 neurons, though the circuitry of this interaction is not known. It was proposed that wakefulness leads to the potentiation of presynaptic zones of R2 neurons, likely dependent on N-methyl-d-aspartate receptors that confer an increased excitability of these neurons (Liu et al. 2016). The circadian rhythm controls the timing of sleep so that most sleep occurs during the night. In addition, flies show daytime sleep that is called a siesta and is more pronounced in males. Thus, there are two activity phases, one in the morning and one in the evening. The sLNvs (small ventrolateral neurons) and fifth sLNv pacemaker arousal neurons are under circadian control to inhibit sleep and trigger activity in the morning and evening, respectively. These pacemakers are themselves inhibited by sleep-promoting glutamatergic dorsal clock (DN1) neurons, resulting in the control of a sleep–wake rhythm across the day (Guo et al. 2016).
Thus, the circuit understanding of sleep control in Drosophila has advanced in the past few years, with current evidence pointing to several centers thought to be sleep promoting. Several of these are inhibitory and present counterparts of mammalian sleep-active neurons. Nevertheless, the Drosophila literature is largely devoid of the term “sleep-active” as a descriptor of fly sleep-inducing neurons. For instance, the sleep-promoting dFB neurons were described as the “output branch” of sleep, which means that these neurons could actually present a counterpart to sleep-active neurons in the mammalian POA. While important sleep centers have been identified in the fly, the future challenge will be to figure out how the different sleep circuits work together to induce sleep systemically.
Sleep-Active Neurons in C. elegans
Like Drosophila, C. elegans is an invertebrate species that belongs to the ecdysozoa, or molting animals, and allows straightforward genetic dissections. It has the smallest nervous system among genetically accessible sleep models with only 302 neurons in the hermaphrodite adult. Together with the mapped and invariant connectivity, and the transparency of this organism, dissection of sleep circuits by functional neuronal imaging and optogenetics should be straightforward.
Best characterized is sleep under two conditions. The first is during development and preceding the shedding of the old cuticle at the end of each larval molting cycle. The second condition is cellular stress, such as is experienced through heat shock, and is typically studied in the adult worm. A series of studies showed that these types of sleep fulfill the behavioral criteria that define sleep (Raizen et al. 2008; Kayser and Biron 2016; Trojanowski and Raizen 2016). For each of the two types of sleep, a crucial single neuron has been identified (Figure 2D). The sensory neuron ALA is required for sleep following stressful conditions, involving its activation via a conserved EGF signaling pathway (Van Buskirk and Sternberg 2007; Hill et al. 2014). ALA is GABAergic and produces a set of peptides to induce sleep, likely through an endocrine mechanism (Nelson et al. 2014; Nath et al. 2016). ALA has neither been clearly shown to depolarize specifically at the onset of a sleep bout nor has it been shown to be inhibited by a waking stimulus. Thus, it is unclear whether it exhibits the activity pattern and fast switching kinetics typical for mammalian sleep-active neurons.
Developmental sleep crucially requires the RIS neuron. Deletion of the aptf-1 transcription factor, which determines neuropeptide expression in RIS, or ablation of the RIS neuron using a laser completely abolishes developmental sleep behavior. RIS is GABAergic and peptidergic, and induces sleep through peptides including the inhibitory RFamide neuropeptide FLP-11. Calcium imaging has shown that RIS depolarizes at the onset of sleep. Optogenetic activation in waking animals induces acute quiescence and inhibits command interneurons. A waking stimulus can inhibit RIS, suggesting that a regulatory mechanism exists that is similar to the flip–flop switch in mammals (Turek et al. 2013, 2016). Thus, RIS is a bona fide sleep-active neuron that appears to work like the mammalian sleep motor, including fast switching kinetics. It provides a highly simplified model system of a sleep-active sleep-inducing neuron, as it is only one cell in a small nervous system. Even though the sleep pattern does not follow a 24 hr pattern, developmental sleep is timed by the homolog of the circadian protein PERIOD called LIN-42 (Jeon et al. 1999; Monsalve et al. 2011). Other than this, little is known about the circuits and pathways driving RIS activation, and the relationship of ALA and RIS.
The discovery of sleep-active neurons in both mammals and simple invertebrates raises the question of how sleep-inducing neurons evolved. Are they the product of convergent evolution or do they present homologous structures? While the mechanisms of action of RIS and POA sleep-promoting neurons are strikingly similar, molecular information on these neurons is required to definitively solve this question. APTF-1 is a member of the highly conserved group of AP2 transcription factors. In Drosophila, AP2 has also been shown to be required for sleep and, in humans, mutations in TFAP2β cause Char syndrome, which is linked to insomnia (Mani et al. 2005; Kucherenko et al. 2016). Several additional genes have been found to be conserved in sleep control, including neurotransmitter systems, but also specific regulators such as salt-inducible kinase, which plays a role in sleep in worms and mice, suggesting that sleep in C. elegans and mammals has a common evolutionary history, making the worm a valuable model for the study of sleep control (Singh et al. 2014; Funato et al. 2016).
What Are the Origins of Sleep-Active Neurons?
Sleep has been detected by behavioral criteria in most animals that possess a nervous system and that have been studied carefully. Among basal metazoans, behavioral quiescence was found in some species of cnidarians that possess a nerve net containing GABAergic and peptidergic neurons. Box jellyfish medusae were found to display cessation of locomotion and rest on the sea floor (Seymour et al. 2004). Even more impressive, quiescence in medusae of Cassiopea spp. has recently been characterized as sleep (Nath et al. 2017). Additionally, planula larvae of Hydractinia echinata larvae were shown to have locomotion that is controlled by neuropeptides to create periods of activity and rest. Intriguingly, locomotion cessation in H. echinata larvae can be induced by RF peptides (Katsukura et al. 2004). As RFamide peptides are also required for sleep induction in other systems, it is imaginable that cnidarians may possess sleep-active neurons (Lenz et al. 2015; Nath et al. 2016; Turek et al. 2016; Lee et al. 2017). Thus, sleep-active sleep-inducing neurons could well have evolved prior to a centralized nervous system. Obviously, additional studies would be necessary to test this. Studying even more basal metazoans could shed light on how sleep-active neurons evolved. Trichoplax lacks neurons and muscles, but still shows locomotion behaviors that are mediated by cilia and also has rest and activity phases. Interestingly, Trichoplax contains gland cells. While these secretory cells are not neurons themselves, as they do not seem to be connected by synapses, they might control locomotion behavior including active and quiescence phases by endocrine signaling (Smith et al. 2014). Thus, precursors of sleep-active neurons may even be found in animals that lack nervous systems. Hypothetically, as a part of evolving locomotion, mechanisms controlling activity and rest were needed. Thus, sleep-active neurons may have emerged from simpler activity–rest-controlling systems together with the emergence of a nervous system (Figure 1). While this is speculative, it may stimulate the search for sleep-active neurons or their precursors in basal metazoans.
While sleep is believed to be essential for the survival of most species, some animals can survive with little sleep. For example, large herbivores, migrating or mating birds, and postpartum whales have been found to get away with little sleep (Campbell and Tobler 1984; Lyamin et al. 2005; Cirelli and Tononi 2008; Lesku et al. 2012; Rattenborg et al. 2016). It would be interesting to conclusively identify animals that have a nervous system but do not sleep, but this has not been achieved yet. Such animals could provide insight into how to live without sleep. In a sleep-neuron centric view, it would be interesting to see whether these animals possess sleep-active neurons or whether these exist but are not activated. Because sleep-active neurons generate the fast-switching dynamic behaviors that define sleep, there appears to be no sleep without such neurons. While convergent evolution of sleep-active neurons is possible, the currently most likely scenario is that sleep evolved once through the emergence of sleep-active neurons early in the evolution of metazoans, perhaps at the base of bilaterians, but maybe even earlier, for instance shortly after or with the development of a nervous system (Figure 1).
In summary, sleep-active neurons are key inhibitors of wakefulness and thus crucial triggers of sleep. They appear to be widespread in animals and may share a common evolutionary origin. Important steps toward understanding sleep control will be the dissection of neural circuits and molecular players for sleep induction across species, to identify all drivers and motors of sleep. Given the high level of conservation of sleep across species, the use of model organisms to solve sleep is valid, and findings from various models can be merged to obtain a holistic view of sleep control.
Acknowledgments
Part of the manuscript and figures were modified, with permission, from Bringmann (2017), copyright Wiley-VCH Verlag GmbH & Co., Weinheim, Germany. The Max Planck Society, the Deutsche Forschungs Gemeinschaft, and a European Research Council Starting grant (SLEEPCONTROL) supported work in the laboratory of H.B.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Ahrens M. B., Orger M. B., Robson D. N., Li J. M., Keller P. J., 2013. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10: 413–420. 10.1038/nmeth.2434 [DOI] [PubMed] [Google Scholar]
- Alam M. A., Kumar S., McGinty D., Alam M. N., Szymusiak R., 2014. Neuronal activity in the preoptic hypothalamus during sleep deprivation and recovery sleep. J. Neurophysiol. 111: 287–299. 10.1152/jn.00504.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaclet C., Fuller P. M., 2017. Brainstem regulation of slow-wave-sleep. Curr. Opin. Neurobiol. 44: 139–143. 10.1016/j.conb.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaclet C., Ferrari L., Arrigoni E., Bass C. E., Saper C. B., et al. , 2014. The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat. Neurosci. 17: 1217–1224 (erratum: Nat Neurosci. 17: 1841). 10.1038/nn.3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G., Sehgal A., 2017. The Drosophila circuitry of sleep-wake regulation. Curr. Opin. Neurobiol. 44: 243–250. 10.1016/j.conb.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aserinsky E., Kleitman N., 1953. Regularly occurring periods of eye motility, and concomitant phenomena, during sleep. Science 118: 273–274. 10.1126/science.118.3062.273 [DOI] [PubMed] [Google Scholar]
- Barlow I. L., Rihel J., 2017. Zebrafish sleep: from geneZZZ to neuronZZZ. Curr. Opin. Neurobiol. 44: 65–71. 10.1016/j.conb.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Bouvier J., Caggiano V., Leiras R., Caldeira V., Bellardita C., et al. , 2015. Descending command neurons in the brainstem that halt locomotion. Cell 163: 1191–1203. 10.1016/j.cell.2015.10.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., 2017. Motoren des Schlafs. Biol. Unserer Zeit 47: 370–377. 10.1002/biuz.201710637 [DOI] [Google Scholar]
- Bryant P. A., Trinder J., Curtis N., 2004. Sick and tired: does sleep have a vital role in the immune system? Nat. Rev. Immunol. 4: 457–467. 10.1038/nri1369 [DOI] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C., 2015. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc. Natl. Acad. Sci. USA 112: 4785–4790. 10.1073/pnas.1419603112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. S., Tobler I., 1984. Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 8: 269–300. 10.1016/0149-7634(84)90054-X [DOI] [PubMed] [Google Scholar]
- Chemelli R. M., Willie J. T., Sinton C. M., Elmquist J. K., Scammell T., et al. , 1999. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98: 437–451. 10.1016/S0092-8674(00)81973-X [DOI] [PubMed] [Google Scholar]
- Chen A., Chiu C. N., Mosser E. A., Kahn S., Spence R., et al. , 2016. QRFP and its receptors regulate locomotor activity and sleep in zebrafish. J. Neurosci. 36: 1823–1840. 10.1523/JNEUROSCI.2579-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. N., Rihel J., Lee D. A., Singh C., Mosser E. A., et al. , 2016. A zebrafish genetic screen identifies neuromedin U as a regulator of sleep/wake states. Neuron 89: 842–856. 10.1016/j.neuron.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. C., Scammell T. E., Gooley J. J., Gaus S. E., Saper C. B., et al. , 2003. Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J. Neurosci. 23: 10691–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Weber F., Zhong P., Tan C. L., Nguyen T. N., et al. , 2017. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545: 477–481. 10.1038/nature22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Tononi G., 2008. Is sleep essential? PLoS Biol. 6: e216 10.1371/journal.pbio.0060216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colten H. R., Altevogt B. M. (Editors), 2006. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem, 4, Functional and Economic Impact of Sleep Loss and Sleep-Related Disorders. Institute of Medicine (US) Committee on Sleep Medicine and Research, Washington (DC). National Academies Press, Washington, D.C. [PubMed] [Google Scholar]
- Datta S., 2000. Avoidance task training potentiates phasic pontine-wave density in the rat: a mechanism for sleep-dependent plasticity. J. Neurosci. 20: 8607–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekelmann S., Born J., 2010. The memory function of sleep. Nat. Rev. Neurosci. 11: 114–126. [DOI] [PubMed] [Google Scholar]
- Dijk D. J., 2009. Regulation and functional correlates of slow wave sleep. J. Clin. Sleep Med. 5: S6–S15. [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Ramanan N., Shaw P. J., 2009. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324: 105–108. 10.1126/science.1166657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L., Shaw P. J., 2011. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332: 1571–1576. 10.1126/science.1202249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton A., Meerlo P., Bergmann B., Turek F. W., 2004. The suprachiasmatic nucleus regulates sleep timing and amount in mice. Sleep. 27: 1307–1318. [DOI] [PubMed] [Google Scholar]
- Elbaz I., Levitas-Djerbi T., Appelbaum L., 2017. The hypocretin/orexin neuronal networks in zebrafish. Curr. Top. Behav. Neurosci. 33: 75–92. 10.1007/7854_2016_59 [DOI] [PubMed] [Google Scholar]
- Foltenyi K., Greenspan R. J., Newport J. W., 2007. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10: 1160–1167. 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- Frank M. G., 2013. Astroglial regulation of sleep homeostasis. Curr. Opin. Neurobiol. 23: 812–818. 10.1016/j.conb.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Fuller P. M., Sherman D., Pedersen N. P., Saper C. B., Lu J., 2011. Reassessment of the structural basis of the ascending arousal system. J. Comp. Neurol. 519: 933–956. 10.1002/cne.22559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H., Miyoshi C., Fujiyama T., Kanda T., Sato M., et al. , 2016. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539: 378–383. 10.1038/nature20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi A. V., Mosser E. A., Oikonomou G., Prober D. A., 2015. Melatonin is required for the circadian regulation of sleep. Neuron 85: 1193–1199. 10.1016/j.neuron.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko D., Wisor J. P., Burns D., Reh R. K., Shiromani P. J., et al. , 2008. Identification of a population of sleep-active cerebral cortex neurons. Proc. Natl. Acad. Sci. USA 105: 10227–10232. 10.1073/pnas.0803125105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yu J., Jung H. J., Abruzzi K. C., Luo W., et al. , 2016. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536: 292–297. 10.1038/nature19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa M. M., Florian C., Fellin T., Munoz J. R., Lee S. Y., et al. , 2009. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 61: 213–219. 10.1016/j.neuron.2008.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes P. R., Christmann B. L., Griffith L. C., 2015. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4: e03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks J. C., Finn S. M., Panckeri K. A., Chavkin J., Williams J. A., et al. , 2000. Rest in Drosophila is a sleep-like state. Neuron 25: 129–138. 10.1016/S0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. 10.1016/j.cub.2014.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R., Ghilardi M. F., Massimini M., Tononi G., 2004. Local sleep and learning. Nature 430: 78–81. 10.1038/nature02663 [DOI] [PubMed] [Google Scholar]
- Jackson A. F., Bolger D. J., 2014. The neurophysiological bases of EEG and EEG measurement: a review for the rest of us. Psychophysiology 51: 1061–1071. 10.1111/psyp.12283 [DOI] [PubMed] [Google Scholar]
- Jeon M., Gardner H. F., Miller E. A., Deshler J., Rougvie A. E., 1999. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286: 1141–1146. 10.1126/science.286.5442.1141 [DOI] [PubMed] [Google Scholar]
- Joiner W. J., 2016. Unraveling the evolutionary determinants of sleep. Curr. Biol. 26: R1073–R1087. 10.1016/j.cub.2016.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J., Crocker A., White B. H., Sehgal A., 2006. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757–760. 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- Juvin L., Gratsch S., Trillaud-Doppia E., Gariepy J. F., Buschges A., et al. , 2016. A specific population of reticulospinal neurons controls the termination of locomotion. Cell Rep. 15: 2377–2386. 10.1016/j.celrep.2016.05.029 [DOI] [PubMed] [Google Scholar]
- Katsukura Y., Ando H., David C. N., Grimmelikhuijzen C. J., Sugiyama T., 2004. Control of planula migration by LWamide and RFamide neuropeptides in Hydractinia echinata. J. Exp. Biol. 207: 1803–1810. 10.1242/jeb.00974 [DOI] [PubMed] [Google Scholar]
- Kayser M. S., Biron D., 2016. Sleep and development in genetically tractable model organisms. Genetics 203: 21–33. 10.1534/genetics.116.189589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler B., Bao H., Zalucki O., Imlach W., Troup M., et al. , 2013. A sleep/wake circuit controls isoflurane sensitivity in Drosophila. Curr. Biol. 23: 594–598. 10.1016/j.cub.2013.02.021 [DOI] [PubMed] [Google Scholar]
- Kramer A., Yang F. C., Snodgrass P., Li X., Scammell T. E., et al. , 2001. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294: 2511–2515. 10.1126/science.1067716 [DOI] [PubMed] [Google Scholar]
- Kucherenko M. M., Ilangovan V., Herzig B., Shcherbata H. R., Bringmann H., 2016. TfAP-2 is required for night sleep in Drosophila. BMC Neurosci. 17: 72 10.1186/s12868-016-0306-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida C. A., Bergmann B. M., Rechtschaffen A., 1989. Sleep deprivation in the rat: IV. Paradoxical sleep deprivation. Sleep. 12: 22–30. [DOI] [PubMed] [Google Scholar]
- Lee D. A., Andreev A., Truong T. V., Chen A., Hill A. J., et al. , 2017. Genetic and neuronal regulation of sleep by neuropeptide VF. Elife 6: e25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O., Xiong J., Nelson M. D., Raizen D. M., Williams J. A., 2015. FMRFamide signaling promotes stress-induced sleep in Drosophila. Brain Behav. Immun. 47: 141–148. 10.1016/j.bbi.2014.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesku J. A., Rattenborg N. C., Valcu M., Vyssotski A. L., Kuhn S., et al. , 2012. Adaptive sleep loss in polygynous pectoral sandpipers. Science 337: 1654–1658. 10.1126/science.1220939 [DOI] [PubMed] [Google Scholar]
- Libourel P. A., Herrel A., 2016. Sleep in amphibians and reptiles: a review and a preliminary analysis of evolutionary patterns. Biol. Rev. Camb. Philos. Soc. 91: 833–866. 10.1111/brv.12197 [DOI] [PubMed] [Google Scholar]
- Lin L., Faraco J., Li R., Kadotani H., Rogers W., et al. , 1999. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98: 365–376. 10.1016/S0092-8674(00)81965-0 [DOI] [PubMed] [Google Scholar]
- Liu S., Liu Q., Tabuchi M., Wu M. N., 2016. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165: 1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liu S., Kodama L., Driscoll M. R., Wu M. N., 2012. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol. 22: 2114–2123. 10.1016/j.cub.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Greco M. A., Shiromani P., Saper C. B., 2000. Effect of lesions of the ventrolateral preoptic nucleus on NREM and REM sleep. J. Neurosci. 20: 3830–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Sherman D., Devor M., Saper C. B., 2006. A putative flip-flop switch for control of REM sleep. Nature 441: 589–594. 10.1038/nature04767 [DOI] [PubMed] [Google Scholar]
- Luppi P. H., Peyron C., Fort P., 2017. Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med. Rev. 32: 85–94. 10.1016/j.smrv.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Lyamin O., Pryaslova J., Lance V., Siegel J., 2005. Animal behaviour: continuous activity in cetaceans after birth. Nature 435: 1177 10.1038/4351177a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A., Radhakrishnan J., Farhi A., Carew K. S., Warnes C. A., et al. , 2005. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc. Natl. Acad. Sci. USA 102: 2975–2979. 10.1073/pnas.0409852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty D. J., Sterman M. B., 1968. Sleep suppression after basal forebrain lesions in the cat. Science 160: 1253–1255. 10.1126/science.160.3833.1253 [DOI] [PubMed] [Google Scholar]
- Monsalve G. C., Van Buskirk C., Frand A. R., 2011. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21: 2033–2045. 10.1016/j.cub.2011.10.054 [DOI] [PubMed] [Google Scholar]
- Moore J. T., Chen J., Han B., Meng Q. C., Veasey S. C., et al. , 2012. Direct activation of sleep-promoting VLPO neurons by volatile anesthetics contributes to anesthetic hypnosis. Curr. Biol. 22: 2008–2016. 10.1016/j.cub.2012.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. Y., Eichler V. B., 1972. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42: 201–206. 10.1016/0006-8993(72)90054-6 [DOI] [PubMed] [Google Scholar]
- Morairty S., Rainnie D., McCarley R., Greene R., 2004. Disinhibition of ventrolateral preoptic area sleep-active neurons by adenosine: a new mechanism for sleep promotion. Neuroscience 123: 451–457. 10.1016/j.neuroscience.2003.08.066 [DOI] [PubMed] [Google Scholar]
- Nath R. D., Chow E. S., Wang H., Schwarz E. M., Sternberg P. W., 2016. C. elegans stress-induced sleep emerges from the collective action of multiple neuropeptides. Curr. Biol. 26: 2446–2455. 10.1016/j.cub.2016.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath R. D., Bedbrook C. N., Abrams M. J., Basinger T., Bois J. S., et al. , 2017. The jellyfish Cassiopea exhibits a sleep-like state. Curr. Biol. 27: 2984–2990.e3. 10.1016/j.cub.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauta W. J., 1946. Hypothalamic regulation of sleep in rats; an experimental study. J. Neurophysiol. 9: 285–316. 10.1152/jn.1946.9.4.285 [DOI] [PubMed] [Google Scholar]
- Nelson M. D., Lee K. H., Churgin M. A., Hill A. J., Van Buskirk C., et al. , 2014. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol. 24: 2406–2410. 10.1016/j.cub.2014.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A. L. A., Eichler T., Latham R., Zimmer M., 2017. A global brain state underlies C. elegans sleep behavior. Science 356: eaam6851. [DOI] [PubMed] [Google Scholar]
- Nitz D. A., van Swinderen B., Tononi G., Greenspan R. J., 2002. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr. Biol. 12: 1934–1940. 10.1016/S0960-9822(02)01300-3 [DOI] [PubMed] [Google Scholar]
- Oikonomou G., Prober D. A., 2017. Attacking sleep from a new angle: contributions from zebrafish. Curr. Opin. Neurobiol. 44: 80–88. 10.1016/j.conb.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orem J., Lovering A. T., Dunin-Barkowski W., Vidruk E. H., 2000. Endogenous excitatory drive to the respiratory system in rapid eye movement sleep in cats. J. Physiol. 527: 365–376. 10.1111/j.1469-7793.2000.00365.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever J., Fuller P. M., 2017. The biology of REM sleep. Curr. Biol. 27: R1237–R1248. 10.1016/j.cub.2017.10.026 [DOI] [PubMed] [Google Scholar]
- Pimentel D., Donlea J. M., Talbot C. B., Song S. M., Thurston A. J., et al. , 2016. Operation of a homeostatic sleep switch. Nature 536: 333–337. 10.1038/nature19055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka-Heiskanen T., Strecker R. E., Thakkar M., Bjorkum A. A., Greene R. W., et al. , 1997. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science 276: 1265–1268. 10.1126/science.276.5316.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka-Heiskanen T., Alanko L., Kalinchuk A., Stenberg D., 2002. Adenosine and sleep. Sleep Med. Rev. 6: 321–332. 10.1053/smrv.2001.0201 [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y. J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572 (erratum: Nature 453: 952). 10.1038/nature06535 [DOI] [PubMed] [Google Scholar]
- Ralph M. R., Foster R. G., Davis F. C., Menaker M., 1990. Transplanted suprachiasmatic nucleus determines circadian period. Science 247: 975–978. 10.1126/science.2305266 [DOI] [PubMed] [Google Scholar]
- Rattenborg N. C., Voirin B., Cruz S. M., Tisdale R., Dell’Omo G., et al. , 2016. Evidence that birds sleep in mid-flight. Nat. Commun. 7: 12468 10.1038/ncomms12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J., Schier A. F., 2013. Sites of action of sleep and wake drugs: insights from model organisms. Curr. Opin. Neurobiol. 23: 831–840. 10.1016/j.conb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T., 2007. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat. Rev. Neurosci. 8: 171–181. 10.1038/nrn2092 [DOI] [PubMed] [Google Scholar]
- Saper C. B., Chou T. C., Scammell T. E., 2001. The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci. 24: 726–731. 10.1016/S0166-2236(00)02002-6 [DOI] [PubMed] [Google Scholar]
- Saper C. B., Scammell T. E., Lu J., 2005. Hypothalamic regulation of sleep and circadian rhythms. Nature 437: 1257–1263. 10.1038/nature04284 [DOI] [PubMed] [Google Scholar]
- Saper C. B., Fuller P. M., Pedersen N. P., Lu J., Scammell T. E., 2010. Sleep state switching. Neuron 68: 1023–1042. 10.1016/j.neuron.2010.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Lewandrowski I., Bringmann H., 2011. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr. Biol. 21: R983–R984. 10.1016/j.cub.2011.10.046 [DOI] [PubMed] [Google Scholar]
- Schwartz J. R., Roth T., 2008. Neurophysiology of sleep and wakefulness: basic science and clinical implications. Curr. Neuropharmacol. 6: 367–378. 10.2174/157015908787386050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour J. E., Carrette T. J., Sutherland P. A., 2004. Do box jellyfish sleep at night? Med. J. Aust. 181: 707. [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Shein-Idelson M., Ondracek J. M., Liaw H. P., Reiter S., Laurent G., 2016. Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 352: 590–595. 10.1126/science.aaf3621 [DOI] [PubMed] [Google Scholar]
- Siclari F., Tononi G., 2017. Local aspects of sleep and wakefulness. Curr. Opin. Neurobiol. 44: 222–227. 10.1016/j.conb.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. M., 2001. The REM sleep-memory consolidation hypothesis. Science 294: 1058–1063. 10.1126/science.1063049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. M., 2009. Sleep viewed as a state of adaptive inactivity. Nat. Rev. Neurosci. 10: 747–753. 10.1038/nrn2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Ju J. Y., Walsh M. B., Dilorio M. A., Hart A. C., 2014. Deep conservation of genes required for both Drosophila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep 37: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Jin X., Chen N., Felix M., et al. , 2015. Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the drosophila mushroom body. Curr. Biol. 25: 2915–2927. 10.1016/j.cub.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Varoqueaux F., Kittelmann M., Azzam R. N., Cooper B., et al. , 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24: 1565–1572. 10.1016/j.cub.2014.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth L. A., Tolley E. A., Krueger J. M., 1993. Sleep as a prognostic indicator during infectious disease in rabbits. Proc. Soc. Exp. Biol. Med. 203: 179–192. 10.3181/00379727-203-43590 [DOI] [PubMed] [Google Scholar]
- Trojanowski N. F., Raizen D. M., 2016. Call it worm sleep. Trends Neurosci. 39: 54–62. 10.1016/j.tins.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek M., Lewandrowski I., Bringmann H., 2013. An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans. Curr. Biol. 23: 2215–2223. 10.1016/j.cub.2013.09.028 [DOI] [PubMed] [Google Scholar]
- Turek M., Besseling J., Spies J. P., Konig S., Bringmann H., 2016. Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife 5: e12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2007. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 10: 1300–1307. 10.1038/nn1981 [DOI] [PubMed] [Google Scholar]
- von Economo C., 1930. Sleep as a problem of localization. J. Nerv. Ment. Dis. 71: 249–259. 10.1097/00005053-193003000-00001 [DOI] [Google Scholar]
- Vorster A. P., Born J., 2015. Sleep and memory in mammals, birds and invertebrates. Neurosci. Biobehav. Rev. 50: 103–119. 10.1016/j.neubiorev.2014.09.020 [DOI] [PubMed] [Google Scholar]
- Vyazovskiy V. V., Olcese U., Hanlon E. C., Nir Y., Cirelli C., et al. , 2011. Local sleep in awake rats. Nature 472: 443–447. 10.1038/nature10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Dan Y., 2016. Circuit-based interrogation of sleep control. Nature 538: 51–59. 10.1038/nature19773 [DOI] [PubMed] [Google Scholar]
- Weber F., Chung S., Beier K. T., Xu M., Luo L., et al. , 2015. Control of REM sleep by ventral medulla GABAergic neurons. Nature 526: 435–438. 10.1038/nature14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T., Marin W., Faraco J., Pezeron G., Appelbaum L., et al. , 2007. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 5: e277 10.1371/journal.pbio.0050277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova I. V., Wang S. Y., Leclair O. U., Danilova N. P., 2001. Melatonin promotes sleep-like state in zebrafish. Brain Res. 903: 263–268. 10.1016/S0006-8993(01)02444-1 [DOI] [PubMed] [Google Scholar]