Abstract
Histone deacetylases (HDACs) catalyze the removal of acetyl groups from acetylated histone tails that consequently interact more closely with DNA, leading to chromatin state refractory to transcription. Zea mays HDA108 belongs to the Rpd3/HDA1 HDAC family and is ubiquitously expressed during development. The newly isolated hda108/hda108 insertional mutant exhibited many developmental defects: significant reduction in plant height, alterations of shoot and leaf development, and alterations of inflorescence patterning and fertility. Western blot analyses and immunolocalization experiments revealed an evident increase in histone acetylation, accompanied by a marked reduction in H3K9 dimethylation, in mutant nuclei. The DNA methylation status, in the CHG sequence context, and the transcript level of ribosomal sequences were also affected in hda108 mutants, while enrichment in H3 and H4 acetylation characterizes both repetitive and nonrepetitive transcriptional up-regulated loci. RNA-Seq of both young leaf and anthers indicated that transcription factor expression is highly affected and that the pollen developmental program is disrupted in hda108 mutants. Crosses between hda108/hda108 and epiregulator mutants did not produce any double mutant progeny indicating possible genetic interactions of HDA108 with distinct epigenetic pathways. Our findings indicate that HDA108 is directly involved in regulation of maize development, fertility, and epigenetic regulation of genome activity.
Keywords: histone acetylation/deacetylation, histone deacetylases, HDA108, transcriptomic analysis, Zea mays
HISTONE acetyltransferases (HATs) and histone deacetylases (HDAs) are the enzymes required to perform histone acetylation and deacetylation, respectively. Acetylation, together with other covalent modifications of the histone N-terminal tails in nucleosomes, plays important roles in chromatin assembly. Acetylation of conserved lysine residues in the N-terminal tails of histones neutralizes their positive charge, decreasing histone affinity for negatively charged DNA and resulting in chromatin conformation and gene promoter accessibility changes. As a general rule, hyper-acetylated histones are associated with gene activation, while hypoacetylated histones are related to gene repression. HDAs interact with various corepressors in different large multi-protein chromatin modifying complexes (Mehdi et al. 2016; Perrella et al. 2016). However, some reports have shown that HDAC complexes are involved in both repression and activation of transcription in yeast as well as in human cells (Wang et al. 2002; Z. Wang et al. 2009; Greer et al. 2015; Jian et al. 2017). Histone modification patterns are also thought to generate a “histone code” that provides signals for the recruitment of specific protein complexes, which alter chromatin states and affect transcription (Jenuwein and Allis 2001).
HDAs are grouped into three families based on their similarity to known yeast histone deacetylases (Pandey et al. 2002); the first family of HDAs includes proteins that carry homology to the yeast RPD3 (Reduced Potassium Deficiency 3) and HDA1 (Histone Deacetylase 1) proteins, which are present in all eukaryotes. The second family comprises the plant-specific HD-tuins, the first member of which was identified in maize (Lusser et al. 1997), whereas the third contains proteins that are homologous to the Sir2 (Silent Information Regulator Protein 2) NAD-dependent proteins.
In plants, the RPD3/HDA1 family is further divided into three distinct groups: class I, class II, and class IV (Alinsug et al. 2009; Aiese Cigliano et al. 2013). In Arabidopsis there are six genes that belong to class I of the RPD3/HDAI family, including HDA19 and HDA6, which share similar expression profiles and biological processes (Hollender and Liu 2008). HDA19 acts as a global transcriptional regulator in response to changes in developmental programs, physiological processes, and pathogen response (Tian et al. 2005; Zhou et al. 2005). In transgenic Arabidopsis plants, the up- and down-regulation of HDA19 was associated with histone H4 hypoacetylation and hyper-acetylation, respectively (Tian and Chen 2001; Tian et al. 2005; Zhou et al. 2005).
The Arabidopsis HDA6 is responsible for the silencing of transgenes, transposable elements, and ribosomal RNA, as demonstrated through the characterization of several hda6 mutant alleles (Murfett et al. 2001; Aufsatz et al. 2002; Lippman et al. 2003; Probst et al. 2004). HDA6 is also required for inactivation of Nuclear Organizing Regions in Arabidopsis (NORs; Earley et al. 2006) and is a crucial player in Arabidopsis growth and development: it can interact with ASYMETRIC LEAVES1 MYB domain proteins in vivo and in vitro, being part of the AS1 repression complex that regulates the expression of KNOX genes in leaf primordia (Luo et al. 2012). Identification and characterization of the epigenetic control1 (epic1) Arabidopsis HDA6 mutant allele (renamed hda6-8) led to the conclusion that this histone deacetylase has independent euchromatic and heterochromatic functions and may inhibit de novo DNA methylation in the CG sequence context (Hristova et al. 2015). The characterization of two further mutant alleles, namely hda6-9 and hda6-10, confirmed that mutations in different domains of the HDA6 protein may have different impact on DNA methylation and histone modifications (Zhang et al. 2015). Very recently, it has been reported that HDA6 can deacetylate BIN2 to repress kinase activity and enhance brassinosteroids signaling in Arabidopsis (Hao et al. 2016). This observation opens a new scenario on a possible role of HDA6 in deacetylation of nonhistone substrates.
Intriguingly, HDA6 knockout mutations do not confer a drastic phenotype on Arabidopsis plants: the hda6/rts1 mutant plants display a very mild phenotype (Aufsatz et al. 2002) and a delay in flowering was reported for other hda6 mutant alleles (Probst et al. 2004).
AtHDA7, another HDA member of the RPD3 superfamily, was shown to be crucial for female gametophyte development and embryogenesis in Arabidopsis: its silencing causes both a degeneration of micropilar nuclei at the stage of four-nucleate embryo sac and a delay in embryo development (Cigliano et al. 2013).
Together with Arabidopsis, rice and maize are the best-characterized systems for studying plant HDACs. Three genes for class I type histone deacetylases (OsHDAC1-3, renamed OsjHDA702, OsjHDA710, and OsjHDA703) were first characterized in rice (Jang et al. 2003). Interestingly, down-regulation of the three rice orthologs of AtHDA19 affect different developmental processes, suggesting that rice HDA genes may have a divergent developmental function compared to the related Arabidopsis genes (Hu et al. 2009).
During plant development and differentiation, maize Rpd3-like HDA genes, HDA101, HDA102, and HDA108, are ubiquitously expressed in all plant organs (Varotto et al. 2003). These three Rpd3-like HDAs were shown to interact with the maize retinoblastoma-related protein RBR1 and with RbAp, the histone-binding protein involved in nucleosome assembly, indicating a possible involvement in the cell cycle G1/S transition (Rossi et al. 2003).
In maize, down-regulation and overexpression of the HDA101 histone deacetylase gene induced morphological and developmental defects as well as variations in nuclear distributions and total levels of acetylated histones. Characterization of transgenic mutants indicated that HDA101 affects gene transcription and provided evidence of its involvement in setting the histone code and, as a consequence, tuning developmental programs (Rossi et al. 2007). In addition, investigations on the role of HDA101 during early seed development demonstrated that HDA101 is mainly targeted to genes with high and intermediate levels of expression and it represses the expression of a small subset of its direct target genes: these repressed genes must be kept at low expression levels to allow proper seed development (Yang et al. 2016).
This paper reports the characterization of the HDA108 gene, a member of the maize Rpd3/HDA1 family of histone deacetylases through the analysis of a Mutator insertional mutant line. The phenotype of maize hda108/hda108 mutant plants indicates that the HDA108 gene knockout is correlated with many developmental defects, spanning from a significant reduction in plant height, alterations of shoot and leaf development to alterations of both male and female inflorescence patterning and fertility. An evident increase in acetylated histone H3 and H4 in homozygous mutant nuclei compared with wild type was observed; H3K9me2-modified histones instead showed a marked reduction in mutants. Transcriptomic analysis of young leaf and anther tissues at two developmental stages identified specific gene families and biological processes misregulated in the mutant tissues compared to wild type. Chromatin immunoprecipitation assays revealed an enrichment in acetylated histones together with a decrease in H3K9me2 modification at the level of several genes with increased expression in the mutant. DNA methylation, histone acetylation, and transcript level of ribosomal repeats were also affected in mutant plants. Moreover, three double mutant lines, hda101/hda108, zmet2/hda108, and rmr6/hda108, were generated: the phenotypic characterization of these lines indicated that the coexistence of hda108 with other epiregulator mutations affects proper seed development and germination. Taken together these results showed that HDA108 is involved in plant development and epigenetic regulation of genome activity.
Materials and Methods
Isolation and characterization of the hda108 Mu mutant
hda108 Mu insertion event (G1755) was isolated at Biogemma within their Mutator transposon insertional maize mutant collection (Martin et al. 2006). The screening for Mu insertion events was performed on F1 progeny through a PCR-based approach, using a Mu-specific primer called OmuA: 5′-CTTCGTCCATAATGGCAATTATCTC-3′. OmuA, which is complementary to the edges of the element, the so-called terminal inverted repeat, was used in combination with the PCR primer AEN11_F02, 5′-TGCCGATTGCCTAAACCC-3′. After receiving the seed from Biogemma, five rounds of crosses were performed with the maize B73 line. At each generation, there was a genetic segregation for the mutant allele and the presence of the Mu insertion in the hda108 gene was verified by PCR, for each plant of each generation, using the two gene-specific primers AEN11_F03 and AEN11_R01 (Supplemental Material, File S9), each in combination with the OmuA primer. After introgression in the B73 genetic background, 40 BC5 heterozygous mutant plants were self-pollinated to produce the segregating progeny Back Cross 5 Selfed 1 (BC5S1). BC5S1 seeds were sown in the field and genomic DNA was extracted from leaf tissue for genotyping. The primer combination AEN11_F01 and AEN11_R01 (File S9) amplified an 800-bp-long fragment from both wild-type and heterozygous plants, while the primer combination AEN11_F03 and OmuA amplified a 300-bp fragment in heterozygous and homozygous plants. On the basis of amplification results, we selected BC5S1 hda108/hda108 homozygous plants that were selfed to produce the Back Cross 5 Selfed 2 (BC5S2) generation. Due to the extremely low fertility of hda108 homozygous plants, only 20 BC5S2 plants were obtained from 10 hda108 BC5S1 parent plants. In addition, a sibling cross between BC5S1 homozygous and heterozygous mutant plants was performed to obtain a Back Cross 5 full-Sib progeny 2 (BC5σ2). About 100 BC5σ2 seeds were obtained from 15 BC5S1 homozygous plants pollinated with pollen collected from heterozygous plants. Segregation of the mutation in BC5S2 and BC5σ2 progenies was followed as above.
Phenotypic characterization of hda108 mutant plants
BC5S1, BC5S2, and BC5σ2 progenies were grown in the field for the phenotypic characterization. Plant height (at the base of the tassel), tassel length, and number of branches were measured after genotyping in 36 plants of BC5S1, 12 plants of BC5S2, and 22 plants of BC5σ2 during field trials in 2013. Further trials were conducted on 49 plants of the BC5S1 population in 2015, measuring a larger number of traits: plant height, number of leaves, length of internodes below and above the node bearing the uppermost ear, tassel length and number of tassel ramifications, ear and axis length, sheath length, blade length, and blade width of ear-bearing leaf. Collected data were analyzed using ANOVA with a threshold P value of 0.05. Means were compared by Tukey’s test using the Daniel’s XL Toolbox (http://xltoolbox.sourceforge.net). Four further BC5S1 and BC5σ2 progenies were grown in a greenhouse or in the field for plant material collection and molecular characterization.
Pollen analysis
Microgametogenesis was analyzed in anthers at different developmental stages and mature grain pollen collected from BC5S1 and BC5σ2 plants, squashed on glass slides, and stained with carminic acid or DAPI for cytological observations under light and fluorescence microscopes, respectively.
Expression analyses by RT-PCR and Real-Time RT-PCR
The RT-PCR and quantitative Real-Time PCR expression analyses were carried out in the meristematic enriched area of sixth leaf stage plants (MA; Mascheretti et al. 2013), the basal third of the expanded 11th leaf, tassel, and ear at the stage of 1 cm of BC5S1 and BC5σ2 plants. In addition, the transcription level of ribosomal DNA loci (5S, 26S, and 18–26 intergenic spacer) was analyzed by RT-PCR in 2-week-old seedlings of BC5S1 wild-type and homozygous mutant plants and BC5σ2 homozygous mutant plants. rDNA loci expression was investigated also in Zmet2 and Rmr6 wild-type and mutant 2-week-old seedlings. Plant tissues were collected from at least three plants per genotype and two independent biological replicates were analyzed.
Total RNA was extracted from maize tissues (two biological replicates) using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) and subjected to on-column DNase treatment (QIAGEN). Complementary DNA (cDNA) synthesis was performed with the SuperScript III reverse transcriptase kit (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. Quantitative Real-Time PCR expression analysis was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) and the POWER SYBR GREEN PCR Master Mix (Applied Biosystems) following the manufacturer’s guidelines. Three replicates were carried out for each primer combination in each biological sample and a relative quantification of gene expression (normalized to GAPC2 transcript quantities) was performed with the RealTime StatMiner 5.0 software (https://www.integromics.com/) using previously determined amplification efficiencies for each target gene. Further detailed methods for expression analyses are fully provided in Supplemental Materials and Methods in File S16, while primer sequences are reported in File S9.
Immunostaining of nuclei
Interphase nuclei prepared from squashed root tips, as described in Forestan et al. (2013), were immunostained with specific antibodies against histone modifications and Alexa Fluor 488-conjugated secondary antibodies (Life Technologies). Immunolocalization experiments were carried out in triplicate and for each genotype × antibody combination at least eight squashed root tips were analyzed. Nuclei were counterstained with propidium iodide in Vectashield Mounting Medium (Vector) and observed with a Leica TCS SP2 laser confocal microscope (Leica Microsystems, Heidelberg, Germany). Microscope images were then analyzed and quantified using CellProfiler automated image analysis software (Carpenter et al. 2006). Nuclei intensity for both antibody and propidium iodide signal was calculated from at least 200 nuclei (on different slides of the three replicates) for each antibody × genotype combination. Further detailed methods for nuclei isolation, antibody dilutions, and Western blot analysis are fully provided in Supplemental Materials and Methods in File S16.
RNA sequencing (RNA-Seq) and differential expression analysis
Transcriptome analyses were performed on total RNA isolated from developing 11th leaves collected from V8 stage plants (three biological replicates, each composed by pooling five plants per genotype) and postmeiotic and mitotic anthers (two biological replicates for stage, each obtained by pooling anthers from three tassels per genotype). Paired-end sequencing libraries were prepared with the TruSeq Stranded Total RNA Library Preparation Kit with Ribo-Zero Plant (Illumina), and sequenced on a HiSeq2500 (Illumina).
The raw reads were first processed for adapter clipping, quality score trimming, and contaminant rRNA filtering and then mapped to the maize B73 reference genome (RefGen ZmB73 Assembly AGPv3 and Zea_mays.AGPv3.31.gtf transcript annotation) with Tophat 2.0.13 (Kim et al. 2013). Multi-mapped reads assigned to >10 different genomic positions were filtered using Samtools (Li et al. 2009).
Pairwise differential expression analyses were performed with Cuffdiff v2.2.1 (Trapnell et al. 2013): genes showing an absolute value of log2 (fold change; hda108/wild type) ≥|1| and FDR (false discovery rate)-adjusted P value ≤ 0.05 were considered as differentially expressed genes (DEG). Further detailed description of RNA-Seq data production and analysis are fully provided in Supplemental Materials and Methods in File S16, while complete results of differential expression tests are reported in File S11, File S12, File S13, File S14, and File S15. Differential expression of a subset of target genes was verified by quantitative Real-Time PCR expression analysis as previously described; primer sequences are reported in File S9.
Gene Ontology (GO) enrichment and functional analysis
GO term enrichment was determined by comparing the number of GO terms in DEGs to the number of GO terms in the expressed genes via agriGO2 Singular Enrichment Analysis tool (Tian et al. 2017) with default parameters and a critical cut-off value of FDR ≤ 0.05 (http://systemsbiology.cau.edu.cn/agriGOv2/, Genome version Zea mays AGPv3.30, last accessed on 7/10/2017). Expressed genes were defined as all genes with FPKM > 0.8 in at least one analyzed sample.
Functional analysis of differential expression (DE) genes was done using MapMan (Thimm et al. 2004; Usadel et al. 2009): overrepresentation of categories was determined using Fisher’s exact test and resulting P-values were adjusted according to Benjamini and Hochberg. A critical cut-off value of 0.05 (corresponding to a Z-score ≥ 1.96) was applied to select enriched categories.
Transcription factors (TFs) annotation was retrieved from GRASSIUS database (http://grassius.org/), while gene name and description were obtained from MAIZEGDB (http://maizegdb.org/) and EnsemblPlant (http://archive.plants.ensembl.org/index.html) archives.
Pathways and gene expression were visualized using heat maps generated by the Morpheus software (https://software.broadinstitute.org/morpheus/).
Chromatin immuno-precipitation (ChIP) assay
Chromatin was extracted and purified from developing 11th leaves (the same samples used for RNA-Seq analysis; the three biological replicates were pooled together) and from the basal part of fully expanded 11th leaves (collected from five plants per genotype) of BC5S1 wild-type and homozygous mutant plants. For the ChIP assay 10 µg of chromatin was immunoprecipitated overnight at 4° with the appropriate antibody and used for Real-Time PCR analyses as reported in Rossi et al. (2007). Additional detailed methods for chromatin purification, immunoprecipitation, antibody dilutions, and Real-Time PCR analysis are fully provided in Supplemental Materials and Methods in File S16.
Further detailed methods for phylogenetic, Southern blot, and X-ray microtomography analyses and double mutant production are fully provided in Supplemental Materials and Methods in File S16.
Data availability
Raw Illumina RNA-Seq reads and the summaries of FPKM values in the different samples have been deposited in the NCBI Gene Expression Omnibus repository under accessions GSE101943 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101943) and GSE102036 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102036), while File S11, File S12, File S13, File S14, and File S15 contain the complete results of differential expression tests.
Results
Mu G1755 insertion knocked out the HDA108 gene in maize
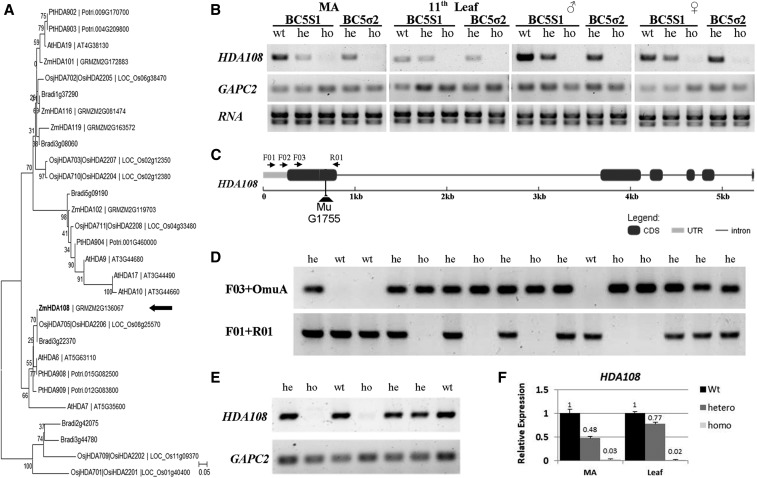
Maize histone deacetylase 108 (HDA108) belongs to the class I RPD3/HDA1 family and represents the ortholog of Arabidopsis HDA6 and HDA7 proteins (Figure 1A). Based on the expression atlas of maize inbred B73 and the qTeller tool (http://qteller.com/), the HDA108 gene (GRMZM2G136067) is ubiquitously expressed in maize plants, with the highest expression levels detectable in leaves and young meristem-enriched tissues (embryos, shoot apex, and inflorescence primordia; Figure S1 in File S16). RT-PCR experiments in the B73 line confirmed that HDA108 is actively expressed in different tissues and organs, such as shoot meristem enriched area (MA), leaves, and inflorescences (Figure 1B).
Figure 1.
HDA108 gene structure, diagram of MuG175 insertion, plant genotyping, and expression analysis. (A) Neighbor-Joining phylogenetic tree of RPD3/HDA1 histone deacetylases of Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa (spp. japonica and indica-cultivar group), Populus trichocarpa, and Zea mays (accessions and sequences of the HDA proteins are reported in File S10). The tree is based on a Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) multiple alignment and was reconstructed using MEGA7; bootstrap was inferred from 1000 replicates. (B) RT-PCR expression analysis of HDA108 in the two segregating progenies BC5S1 and BC5σ2 with different genotypes: wild-type (wt) heterozygous (he) homozygous (ho) in meristematic enriched tissues (MA), basal part of 11th leaf, male (♂) and female (♀) inflorescence. HDA108 is expressed in the wild-type tissues but is not expressed in homozygous genotypes. A lower level of expression characterized the heterozygous genotypes. (C) HDA108 gene structure showing exon/intron structure, Mu G1755 insertion, and primer position and orientation. (D) Genotyping of a BC5S1 progeny showing the different PCR amplification patterns of the three possible genotypes (wt: wild-type; he: heterozygous; and ho: homozygous) identified with two primer combinations: F03-OmuA (OmuA is designed on the Mu inverted repeats) and F01-R01. (E) RT-PCR expression analysis on the sixth leaf showing that HDA108 transcript is not detected in homozygous mutant plants. (F) Quantitative Real-Time PCR expression analysis in meristematic enriched tissues (MA) and leaves confirms the lack of HDA108 transcript in homozygous tissues (34- and 55-fold decrease in transcript level, respectively, as compared to wild type) and the intermediate expression level in the heterozygous samples.
A knockout insertional mutant for the HDA108 gene was isolated in the Mutator-induced Biogemma mutant collection (insertional event Mu G1755; Martin et al. 2006) during a forward genetic screening aimed at the identification of mutations of maize epiregulators. The Mu element is inserted in the first exon of the HDA108 gene, 351 bp downstream of the transcription starting site (Figure 1C and Figure S2 in File S16).
The hda108::Mu insertion was introgressed in the B73 background through five backcrosses of heterozygous plants, as determined by a genotyping PCR-based approach (see Materials and Methods, Figure 1D). Forty BC5 plants carrying the heterozygous hda108::Mu allele were then selfed to produce the B5S1 segregant population. Out of 414 BC5S1 plants used for the phenotypic and molecular characterization, and for immunolocalization studies, 105 homozygous mutants, 213 heterozygous, and 96 wild-type plants were detected: a value that fits with a 1:2:1 segregation (chi-square test probability: P = 0.69). The hda108 mutation behaved as a monogenic recessive trait when BC5S1 plants were grown in the field or in controlled conditions. All the hda108/hda108 homozygous mutant plants lack the HDA108 transcript (Figure 1E) and showed evident developmental alterations. BC5S2 plants were generated by self-pollination of BC5S1 homozygous mutants. As expected, all the 20 BC5S2 plants obtained were homozygous for the mutation. In the BC5Sσ2 progeny, generated by crossing BC5S1 homozygous with BC5S1 heterozygous plants, we obtained a 1:1 segregation ratio (18 heterozygous and 17 homozygous mutants), as expected in the case of a simple recessive mutation. The phenotypic and molecular characterization of plants was done after the genotyping and the expression analysis of HDA108 in BC5S1, BC5σ2, and BC5S2 populations (see Materials and Methods and Figure 1D). RT-PCR experiments indicated that the mutation determined the absence of the HDA108 transcript in MA, leaves, and inflorescences of homozygous mutant plants of each progeny analyzed. Moreover, the lower HDA108 expression level observed in heterozygous plants compared with wild-type ones indicated a dosage-effect (Figure 1B). Further quantitative Real-Time PCR expression analysis confirmed the lack of HDA108 transcript in homozygous MA and leaf tissues (34- and 55-fold decrease in transcript level, respectively, as compared to wild-type tissues) and the intermediate expression level in the heterozygous samples (Figure 1F).
hda108 knockout mutants present many phenotypic alterations
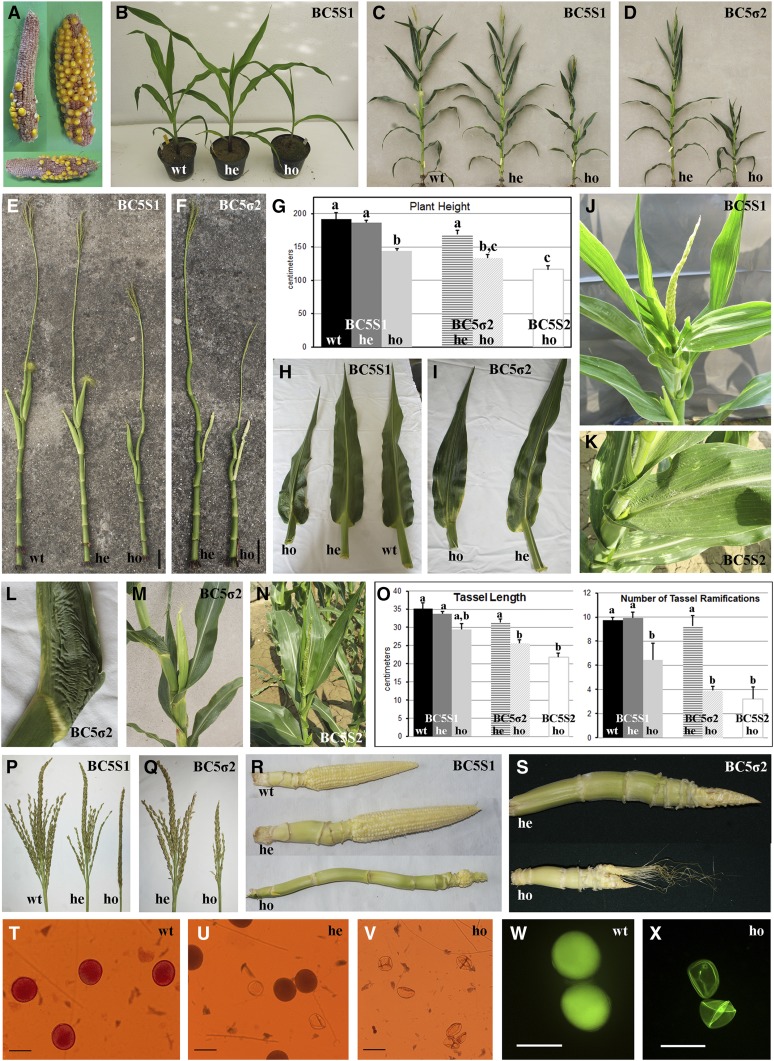
Phenotypic analysis of the BC5S1, BC5S2, and BC5σ2 progenies revealed that the lack of HDA108 expression is associated with developmental defects affecting many organs and tissues of homozygous plants (Figure 2 and Table 1). Due to their reduced fertility (Figure 2A), only a few BC5S2 plants were generated by self-pollination of BC5S1 homozygous mutants. Compared to wild-type and heterozygous plants, homozygous mutant plants of the three progenies presented a significant reduction in height (Figure 2, B, D, and G and Table 1) that was determined by a combination of internodes shortening and reduction in leaf number (Figure 2, E and F and Table 1). In addition, homozygous mutant plants showed phenotypic alterations of leaf development, tassel length and its number of ramifications, anther maturation, and ear development. In detail, the main defects observed in BC5S1, BC5S2, and BC5σ2 mutant plants were shoot curvatures, leaf blade length reduction, leaf twisting, leaf knots, and disorganized differentiation of the blade–sheath boundary (Figure 2, H–N and Table 1). All of these morphological alterations were more evident in homozygous mutant plants of the BC5S2 and BC5σ2 progenies than BC5S1 plants (Figure 2, C–G). The heterozygous genotypes, which have a lower hda108 expression level compared with wild-type plants (Figure 1, B and F), did not have a wild-type phenotype but presented some intermediate developmental defects (Table 1).
Figure 2.
Knockout of HDA108 expression induces developmental defects in maize vegetative and reproductive organs. (A) Reduced fertility of cobs produced by self-pollination of BC5 plants. (B and C) Wild-type (wt), heterozygous (he), and homozygous (ho) plants, respectively, for the hda108 mutated allele at the V5 stage grown in the greenhouse (B) and at flowering in the field (C) belonging to BC5S1 progeny. (D) BC5σ2 heterozygous and homozygous plants grown in the field. Homozygous BC5σ2 plants are smaller than heterozygous plants of the same segregating progeny. (E and F) Comparison of stalks of different genotypes of BC5S1 (E) and B5σ2 (F) after complete removal of leaf sheaths revealed a reduction of node number and internode length in hda108 mutants, accompanied by stalk curvatures. These defects are more evident in B5σ2 progenies, in both heterozygous and homozygous plants. Bar, 10 cm. (G) Histograms of plant height at flowering in different genotypes and progenies; bars represent SE. Data were collected from at least eight plants per genotype during the field trial in 2013. Different letters indicate means with significant difference (P < 0.05; Tukey’s test). (H–N) In BC5S1 homozygous and BC5σ2 heterozygous and homozygous plants, the hda108 mutation resulted in leaf blade reduction (H and I), leaf twisting (J and M), leaf knots and wrinkles (L), and disorganized differentiation of the blade–sheath boundary (H and I). The plant and leaf phenotypes are even more altered in BC5S2 mutant homozygous plants (K and N). (O) Histograms of tassel length and number of tassel ramifications in different genotypes and progenies. Data were collected from at least eight tassels per genotype during the field trial in 2013. Bars represent SE and different letters indicate significantly different means (P < 0.05; Tukey’s test). (P) Tassels of wild-type, heterozygous, and homozygous plants of BC5S1 progeny in which the reduction in number of ramifications is evident comparing the different genotypes. (Q) Tassels of heterozygous and homozygous BC5σ2. (R) Ears of wild-type, heterozygous, and homozygous plants of BC5S1 progeny: in homozygous mutant plants the ear does not differentiate along the axis. (S) Ear of heterozygous and homozygous BC5σ2 with evident developmental defects and displaced tissues. (T–X) Microscope images of mature pollen collected from wild-type (T and W), heterozygous (U), and homozygous mutant (V and X) BC5S1 plants. In homozygous plants the pollen grains are shrunken, collapsed, do not stain with carminic acid, and present reduced autofluorescence (V and X), while the heterozygous genotypes produce ∼50% of shrunken pollen (U). Bar, 100 µm.
Table 1. Phenotypic variation of BC5S1 segregating population.
| Plant height (cm) | Number of leaves | Length of internode below ear (cm) | Length of internode above ear (cm) | Tassel length (cm) | Number of tassel ramifications | Ear length (cm) | Ear axis length (cm) | Leaf sheath length (cm) | Leaf blade length (cm) | Leaf blade width (cm) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 176 ± 2.9a | 20.6 ± 0.2a | 15.3 ± 0.2a | 13.1 ± 0.7a | 42.2 ± 2.5a | 9.4 ± 0.5a | 11.1 ± 0.9a | 3.2 ± 0.4a | 13.8 ± 0.3 | 80.8 ± 2.1a | 10 ± 0.4 |
| Heterozygous | 156.7 ± 2.6b | 19.9 ± 0.1b | 14.3 ± 0.6a,b | 12.8 ± 0.2a,b | 36.4 ± 1.1b | 8.1 ± 0.5a,b | 7.3 ± 0.5b | 5.4 ± 0.2b | 14 ± 0.1 | 77.5 ± 1.7a | 9.9 ± 0.1 |
| Homozygous | 131.3 ± 5.4c | 18 ± 0.1c | 13.2 ± 0.2b | 11 ± 0.2b | 28.7 ± 1.8c | 5.7 ± 0.9b | 3.9 ± 0.7c | 6.2 ± 0.5c | 12.9 ± 0.2 | 65.2 ± 1.9b | 9.1 ± 0.6 |
Phenotypic traits were measured in BC5S1 wild-type, heterozygous, and hda108 homozygous mutant plants at flowering. Traits analyzed are: plant height, number of leaves, length of internodes below and above the node bearing the uppermost ear, tassel length and number of tassel ramifications, ear and axis length, sheath length, blade length and blade width of ear-bearing leaf. Data were collected from at least 10 plants per genotype during the field trial in 2015. Mean and SE for each trait are reported; different letters indicate significantly different means (P < 0.05; Tukey’s test).
During reproductive development, abnormalities affected the whole tassel development (Figure 2, O–Q and Table 1): the reduction of tassel length and branch numbers in mutant plants was accompanied by a failure of anther dehiscence and defective pollen differentiation, with the accumulation of shrunken pollen grains inside the anthers of mutant plants (Figure 2, T–X). To assess the reason why pollen appeared shrunken at maturity in homozygous mutant plants (Figure 2, V and X), we observed squashed anthers stained with DAPI or carminic acid at different developmental stages during microsporogenesis and microgametogenesis: no alterations in microsporogenesis were detected until meiosis, as demonstrated by the presence of regular tetrads inside the anthers (data not shown). However, after release from the tetrads the young microspores collapsed and did not stain with carminic acid (compare Figure 2, V and X with T and W). This, together with the observation that heterozygous plants segregated for pollen phenotype, producing ∼50% of shrunken pollen grains (Figure 2U), indicated that the haploid microspore carrying the hda108 mutation failed to develop properly.
An unusual ear axis elongation was observed along with an abnormal ear differentiation pattern that led to a reduction of ear fertility in homozygous mutant plants (Figure 2, E, F, R, and S and Table 1). In general, heterozygous plants showed intermediate phenotypes compared to the wild type (Figure 2R and Table 1), while all the observed developmental defects appeared emphasized in the BC5σ2 population compared to BC5S1 (Figure 2, R and S).
Levels and nuclear distribution of histone H3 and H4 post-translational modifications are affected in hda108 mutant
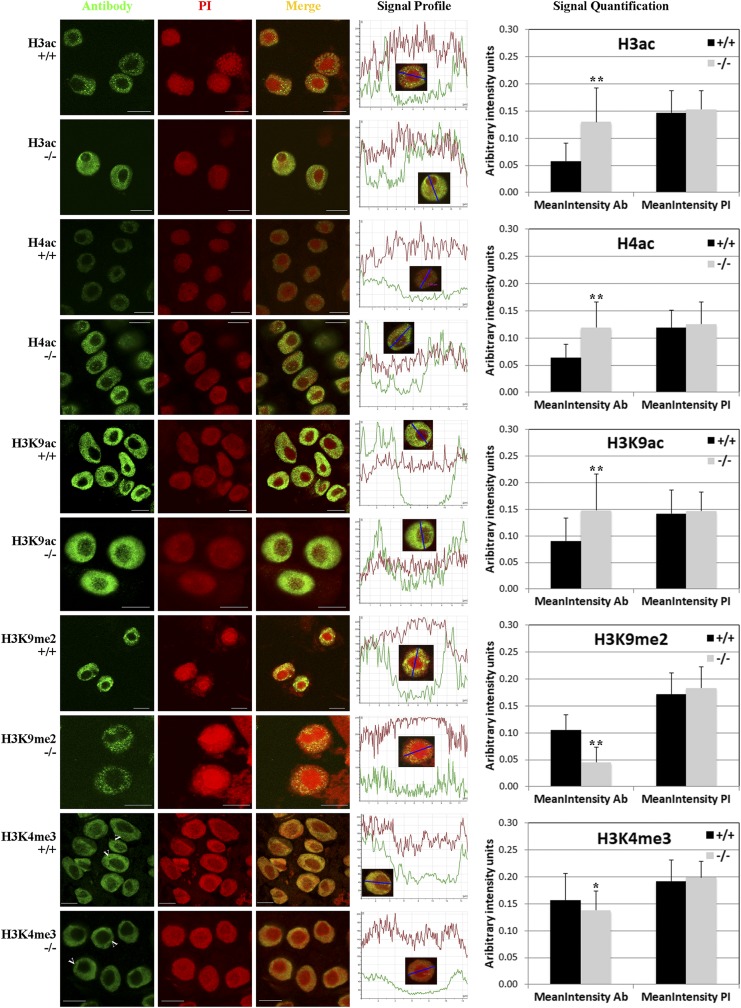
To evaluate the effect of hda108 gene knockout on the levels and distribution of modified histones in cell nuclei, we coupled immunolocalization experiments on interphase nuclei from wild-type and homozygous hda108 root tip of BC5S1 seedlings with Western blot analyses on leaf protein extracts. The following histone modifications were analyzed: acetylation of histone H3 (H3ac, pan-acetylated) and histone H4 (H4ac, pan-acetylated); acetylation of lysine 9 in histone H3 (H3K9ac); di-methylation of lysine 9 in histone H3 (H3K9me2); trimethylation of lysine 4 in histone H3 (H3K4me3) and trimethylation of lysine 27 in histone H3 (H3K27me3). Furthermore, an antibody against the C-terminal portion of histone H3 (H3 C-term) and one against unmodified histone H4 (H4 pAB) were used as internal controls. Confocal microscope observations of immuno-labeled nuclei showed an increase in acetylated H3 and H4 histones in homozygous nuclei (Figure 3): this increase might be the consequence of hda108 knockout, which causes a reduction in deacetylation activity. In mutant leaves, the alteration of H4ac levels was confirmed also by the immunoblots experiments (Figure S3A in File S16), while no differences in unmodified H3 and H4 histones were detected following both approaches (Figure S3, A and B in File S16 and File S1).
Figure 3.
Effect of had108 knockout on total levels and nuclear distribution of histone H3 and H4 modifications. Nuclei from wild-type (+/+) and homozygous (−/−) hda108 mutant plants were immunostained with anti-H3ac, H4ac, H3K9ac, H3K9me2, and H3K4me3 antibodies to analyze the nuclear distribution and levels of modified histones (green signal) and counterstained with propidium iodide (PI; red signal). A minimum of 50 nuclei from wild-type and homozygous mutant root apexes in three replicated immunolocalizations were observed. For each antibody × genotype combination, the signal profiles of fluorescence emission throughout the nucleus, obtained by Leica Confocal Software, are reported for both antibody and propidium iodide. Profile y-axis shows the fluorescence signal intensity (arbitrary units) measured along the line segment (reported on x-axis; µm) shown in the inset nucleus. On the right, signal quantification graphs report the mean nucleus intensities (arbitrary units) calculated for both antibody and propidium iodide signal in each genotype, using the CellProfiler automated image analysis software. For each antibody × genotype combination, at least 200 nuclei (on different slides of the three replicates) were selected and the mean nucleus intensities calculated for both antibody and propidium iodide signal. Summary data from image analyses are reported in File S1. Bars represent SD. Asterisks indicate statistically significant changes: * P ≤ 0.05, ** P ≤ 0.01; Student’s t-test. Arrowheads point to the chromocenters. Bar, 10 µm.
Considering the modifications of single histone H3 residues, in homozygous mutant nuclei compared with wild-type nuclei, an increase of the H3K9 acetylation signal was observed that intrudes upon the nucleolus (Figure 3). This observation was confirmed by blot analysis (Figure S3A in File S16), while a marked reduction of H3K9me2 was detected in mutants’ root nuclei and leaves (Figure 3 and Figure S3A in File S16). The observed increase in histone H3 and H4 acetylation and decrease in H3K9me2 in the nuclei of mutant roots was confirmed by quantifying the fluorescence levels in the wild-type and homozygous nuclei (Figure 3 and File S1).
An increase in histone acetylation is often correlated with an alteration of H3K4me3 distribution. Confocal images and fluorescence measurements of nuclei immunostained with an anti-H3K4me3 antibody revealed a weak reduction of this modification in homozygous nuclei compared to wild-type nuclei (Figure 3 and File S1). Anyway, chromocenters visible as unstained aggregates in wild-type nuclei are less evident in the homozygous nuclei (arrowheads in Figure 3). On the contrary, nonmutant-specific differences were observed in the abundance of H3K27me3 (Figure S3C in File S16). Variation of fluorescence levels for this facultative heterochromatin mark strongly depends on the nuclei observed, irrespective of genotype (Figure S3C in File S16 and File S1).
Identification of genes regulated by HDA108 during leaves and anthers development
To better address the pleiotropic effects of the hda108 mutant and to identify genes and biological processes directly or indirectly regulated by HDA108, a genome-wide transcriptomic analysis was undertaken. Our aim was to identify the genes differentially expressed between the wild-type and the hda108 mutant during development of the vegetative and reproductive tissues that appear phenotypically altered in mutant plants.
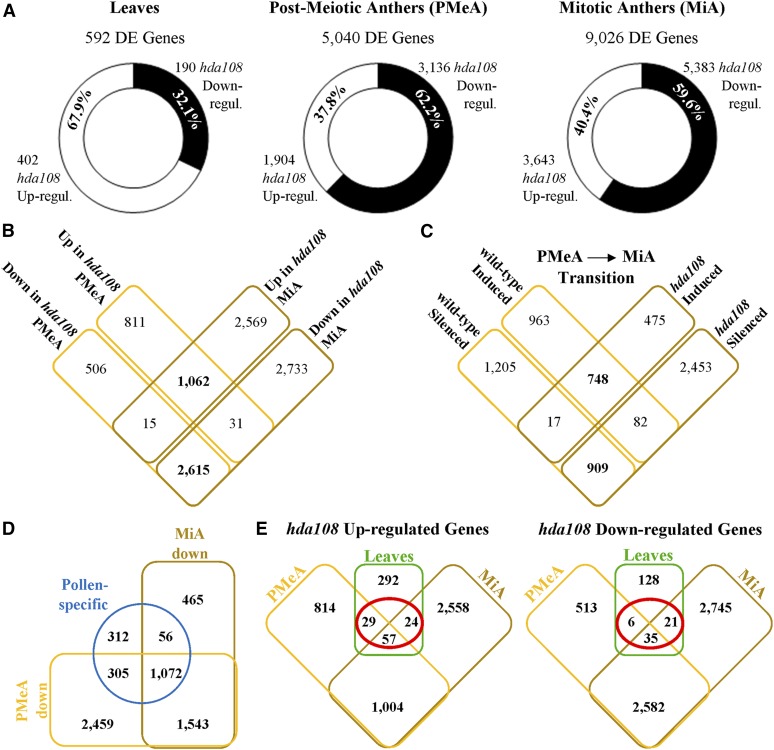
The 11th wrapped leaf collected from V8 plants (three biological replicates, each composed by pooling five plants per genotype) and anthers at postmeiotic and mitotic stages (two biological replicates for stage, each obtained by pooling anthers from three tassels per genotype) were collected from wild-type and hda108 homozygous BC5S1 plants and used for RNA-Seq analysis. A total of 576 million 100-bp reads were generated, with a minimum of 80 million per sample (File S2). These paired-end reads were mapped to the maize reference genome (B73 Refgen-V3) and used for DE analysis, comparing hda108 mutant to the wild type. DE analysis resulted in 592, 5040, and 9026 DE genes (log2FC > |1|; FDR < 0.05) in leaves, postmeiotic (PMeA), and mitotic anthers (MiA), respectively (Figure 4A), suggesting a greater overall impact of HDA108 on gene expression in anthers than in leaves. Among DE genes, opposite proportions of up- and down-regulated genes were observed in the two analyzed tissues: ∼68% of the leaf-DE genes were up-regulated in mutant plants, while down-regulated genes were clearly predominant in both anther stages (Figure 4A).
Figure 4.
Summary of hda108 differentially expressed genes. (A) Number and proportion of differentially expressed genes in hda108 mutant leaf and anther at postmeiotic (PMeA) and mitotic (MiA) stages. (B) Venn diagram representing the shared and unique differentially expressed genes in hda108 mutant PMeA and MiA. (C) Venn diagram representing the shared and unique genes induced (log2FC > 1; FDR < 0.05) or silenced (log2FC < −1; FDR < 0.05) during transition from PMeA and MiA in wild-type and hda108 mutant. (D) Venn diagram representing the proportion of pollen-specific genes identified by Chettoor et al. (2014) and down-regulated in hda108 mutant PMeA and MiA compared to wild type. (E) Venn diagrams representing the shared and unique differentially expressed genes in hda108 mutant leaves, and postmeiotic (PMeA) and mitotic (MiA) anthers.
Identification of pathway and processes affected by hda108 mutation in leaf and anthers
The GO functional annotation of DEG revealed that leaf up-regulated genes were highly enriched in terms related to regulation of gene expression, transcription factor activity, calcium binding, and regulation of cellular and biosynthetic processes, while those down-regulated were enriched in GO terms related to chloroplast assembly and functioning (File S3 and Table S1 in File S16).
Conversely, GO terms related to sexual reproduction, pollen development, and pollination terms were overrepresented in genes down-regulated in mutant anthers (in both PMeA and MiA), together with genes involved in cell wall modification (prevalently genes with pectinesterase, pectate lyase, and polygalacturonase activity). Hydrolase activity and cell wall macromolecule metabolic processes were also enriched in genes up-regulated in mutant PMeA, but in this case the enrichment is mainly due to genes encoding for chitinases, involved in the biotic stress response against fungi. Genes encoding for β 1,3 glucan hydrolases resulted as up- and down-regulated in PMeA, indicating opposite regulation of members of this gene family (File S4). Genes up-regulated in MiA were enriched in RNA processing, nuclease activity, ncRNA processing, chromatin structure, and methyltransferase-activity-related GO terms (File S3 and Table S1 in File S16).
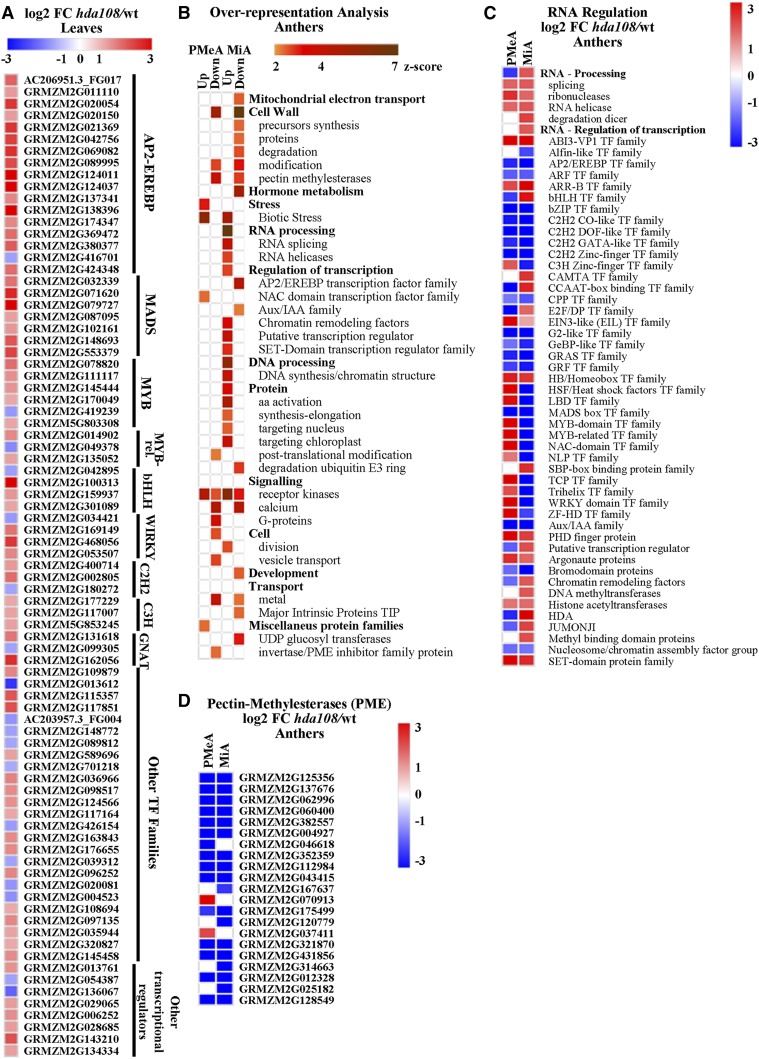
To further investigate the cellular pathways or processes affected by hda108, we tested the enrichment of MapMan categories (Thimm et al. 2004; Usadel et al. 2009) on DE genes. We found a unique functional category overrepresented among leaf misregulated genes: the expression of APETALA2/Ethylene-responsive element binding protein TFs (AP2/EREBP) was significantly increased in mutant leaf. Sixteen TFs belonging to this family (MapMan annotation was confirmed with the annotation available in the GRASSIUS database; http://grassius.org/) were significantly up-regulated while only one was down-regulated (Figure 5A).
Figure 5.
Pathway and gene family analysis of hda108 differentially expressed genes. (A) Log2 fold change heat map of transcription factor coding genes differentially expressed in mutant leaf. TFs were identified according to MapMan (Thimm et al. 2004; Usadel et al. 2009) and Grassius database (http://grassius.org/) annotations. (B) MapMan functional categories enriched in hda108 up- and down-regulated genes in postmeiotic (PMeA) and mitotic (MiA) anthers. Z-scores automatically calculated from P-values (e.g., 1.96 corresponds to a P-value of 0.05) are plotted in an orange to brown color scale. (C) Log2 fold change heat map of gene families included in the “RNA processing” and “RNA regulation of transcription” categories that are differentially expressed in hda108 PMeA or MiA. The Morpheus (https://software.broadinstitute.org/morpheus/) “Collapse” tool was used to aggregate DE genes belonging to the same gene family and to produce the heat map. (D) Log2 fold change heat map of gene annotated as pectin-methylesterases (PME) and differentially expressed in hda108 PMeA or MiA.
In addition to the AP2/ERBP genes that represent key regulators of numerous plant growth processes, from cell identity determination to response to various types of biotic and environmental stresses (Riechmann and Meyerowitz 1998), several other TFs were strongly up-regulated in hda108 mutant leaves: the 16% of the 402 mutant up-regulated genes are indeed annotated as TFs or transcriptional regulators. In this context, MADS box and MYB TF families resulted as overrepresented among hda108 up-regulated genes (Figure 5A and Table S2 in File S16).
Several pathways were found enriched in genes differentially expressed in the two analyzed anther developmental stages (Figure 5B). The MapMan category “Cell Wall” was highly enriched among hda108 down-regulated transcripts, in particular in MiA. In this category, genes involved in precursor synthesis, cell wall modification, and degradation were included, with many pectin methylesterase coding genes strongly down-regulated in mutant anthers (Figure 5D).
Also, genes involved in plant hormone metabolism and responses were strongly enriched in MiA down-regulated genes (File S5 and Figure S4 in File S16). Investigating this category more deeply, we found that many genes related to abscisic acid (ABA) biosynthesis, signal transduction, and perhaps relatedly, many ABA responsive proteins, were down-regulated (Figure S4B in File S16). Similarly, many IAA-responsive and ethylene induced genes were switched off by the hda108 mutation (Figure S4, A and C in File S16). Conversely, the down-regulation of genes involved in cytokinin (CK) degradation may be related to the up-regulation of a few CK-signal transduction genes (Figure S4G in File S16). In addition, many genes related to jasmonic acid and brassinosteroid biosynthesis were misregulated (File S5 and Figure S4, D and E in File S16).
Together with this strong perturbation of hormone metabolism, hda108 down-regulated genes in MiA showed enrichment for the “Development” category (Figure 5B); not only genes specifically expressed during inflorescence and pollen development (Zea floricaula leafy2 – GRMZM2G180190 and aberrant pollen transmission1 – GRMZM2G448687, for example), but also sugar transporters and genes encoding storage and late embryogenesis abundant (LEA) proteins are included in this annotation. Conversely, the MapMan categories “RNA processing” and “Regulation of transcription” were highly enriched among MiA hda108 up-regulated transcripts (Figure 5B). Investigations on the genes included in the RNA processing category revealed that RNAses, RNA helicases, and RNA splicing factors (such as the Rough Endosperm3 α and ζ isoforms, GRMZM2G128228 and GRMZM5G849788) were up-regulated in MiA and to a lesser extent in PMeA, while Dicer-like ribonuclease III family proteins (that are required for endogenous RDR2-dependent siRNA, but not microRNA formation) were overexpressed exclusively in hda108 mutant MiA (Figure 5C).
Despite the “Regulation of transcription” category being globally enriched in MiA hda108 up-regulated transcripts, different gene subcategories or TF families behaved differently. AP2/EREBP TFs and Aux/IAA transcriptional regulators were strongly repressed in PMeA (Figure 5, B and C), while ABI3/VP1, ARR-B, bHLH, CAMTA, CCAAT-box, E2F/DP, and HB homeobox families resulted as globally up-regulated in mutant mitotic anthers. Of these, ABI3/VP1, ARR-B, and HB families were also up-regulated at an earlier developmental stage (Figure 5C). Transcription of several TF families, such as bZIP, C2H2, G2, GeBP-like, GRAS, GRF, and MADS box instead resulted as globally down-regulated in mutant anthers at both developmental stages, while LBD, MYB and MYB-related, NAC, TCP, Trihelix, WRKY, and ZF-HD family members presented the opposite behavior in PMeA and MiA (Figure 5C).
Chromatin organization and epigenetic related gene families were also affected, showing a general trend of increased expression in mutant anthers (Figure 5B). Argonaute protein coding genes and histone acetyl transferases together with SET-domain protein families were up-regulated in both PMeA and MiA, while histone deacetylases (including HDA119, another member of the Rpd3/HDA family), JUMONJII histone demethylases, DNA methyltransferases, and several chromatin remodeling factors were instead up-regulated exclusively in MiA. On the contrary, nucleosome assembly factors coding genes were significantly down-regulated in both analyzed mutant stages (Figure 5C, Figure S5A in File S16, and File S5).
These alterations of transcriptional regulation pathways result in an evident enrichment in protein synthesis and targeting-related genes in MiA and in a parallel down-regulation of the ubiquitin-protein degradation category (Figure 5B). Pathway analysis also suggests a strong effect of hda108 mutation on signaling pathways, with receptor kinases coding genes being significantly up- and down-regulated in both PMeA and MiA and a significant enrichment of calcium-signaling and G-proteins in down-regulated genes (Figure 5B).
Finally, cell division-related genes were enriched among hda108 MiA up-regulated genes while the “vesicle transport” category was down-regulated in PMeA (Figure 5B, Figure S5B in File S16, and File S5). Among transporter proteins, aquaporins of the tonoplast intrinsic protein subfamily (TIP) were significantly down-regulated in MiA, whereas metal transporter expression was reduced in hda108 anthers at both developmental stages (Figure 5B).
hda108 mutation causes disruption of the male gametophytic transcriptional program
Given the huge number of genes differentially expressed in mutant anthers compared to wild type, and the complexity of the cellular pathways and processes in which they are included, we further characterized the genes commonly misregulated in PMeA and MiA to better describe the molecular mechanisms affected by hda108 mutation. A total of 3677 genes were DE between the mutant and wild type in both developmental stages: 2615 down-regulated and 1062 up-regulated (Figure 4B). GO functional annotation on these core sets of DE genes confirmed that the up-regulated genes were enriched in defense-related GO terms, in particular in chitin and cell wall catabolic/degradation processes, while those down-regulated were enriched for regulation of cell development, differentiation and morphogenesis, reproduction, and pollen differentiation, including cell wall modification (pectinesterase activity resulted as the strongest enriched GO term) and the dehydration process (File S3).
To better evaluate the impact of hda108 mutation on the developmental program of anthers and pollen we reanalyzed our sequencing data comparing PMeA vs. MiA in wild-type and hda108 mutant samples independently. This approach identified 3924 and 4684 genes differentially expressed (log2FC > |1|; FDR < 0.05) during the transition from the postmeiotic to mitotic stage in the wild-type and hda108 mutant, respectively. Despite the larger number of DE genes modulated in the hda108 mutant, only 42% of those modulated during the PMeA to MiA progression in wild type were similarly regulated in hda108 mutant anthers (Figure 4C).
These results seem to indicate that the loss of a functional HDA108 resulted in the misregulation of the core set of genes necessary for proper anther and pollen development. Evaluation of the expression levels of a set of 1745 pollen-specific genes identified by Chettoor et al. (2014) revealed that >80% of them were significantly down-regulated in the hda108 mutant (Figure 4D), perhaps causing the observed male sterile phenotype.
HDA108 loss of function results in the misregulation of genes normally expressed in young and meristematic tissues
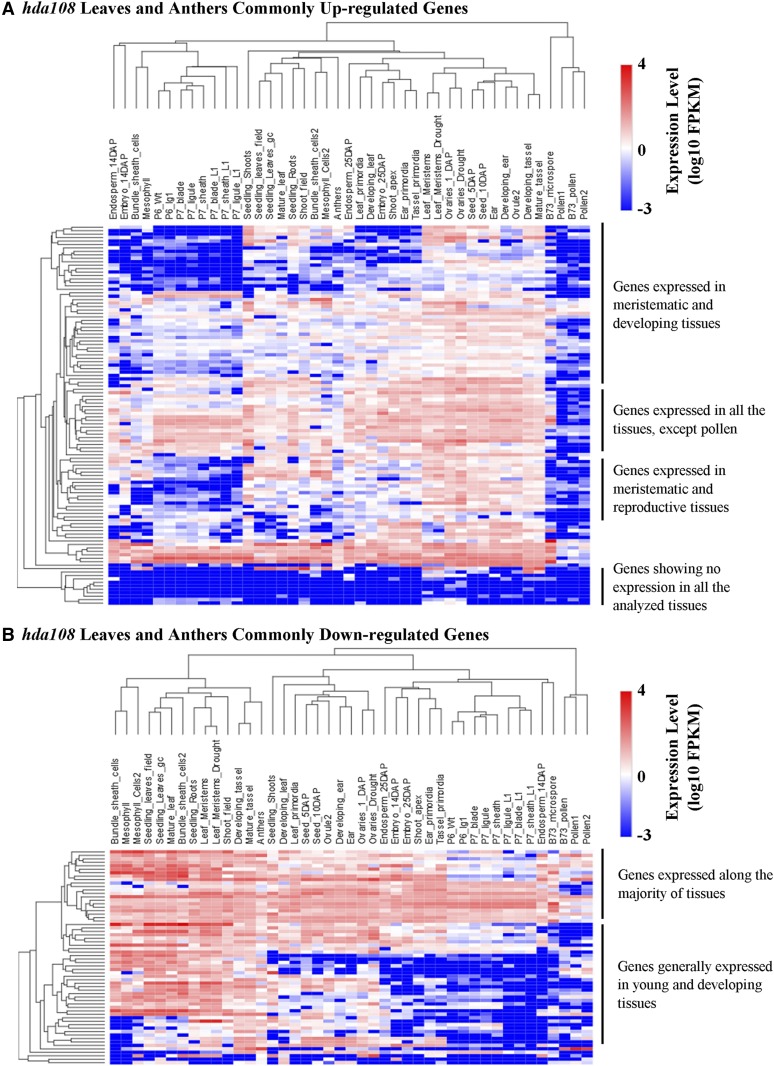
To conclude the exploitation of HDA108 putative targets, we selected the up- or down-regulated genes in leaf and at least one anther sample. In this way we identified 110 and 62 commonly up- and down-regulated genes, respectively (Figure 4E).
We evaluated the expression profile of these two groups of genes in 40 different maize tissues using publicly available data from the qTeller database (http://qteller.com/). The heat map of the 110 commonly up-regulated genes indicated that the majority of them are normally expressed at high levels in young, meristematic tissues while they show low expression in leaves and are completely silenced in anthers and pollen. More than 10% of these genes were not expressed in any of the tissues included in the database (Figure 6A). Among the 62 commonly down-regulated genes (the HDA108 gene was down-regulated in the three samples), two distinctive expression groups could be identified in the heat map (Figure 6B): the first group includes genes expressed along the majority of tissues and the second genes generally expressed in young and developing tissues.
Figure 6.
Expression profiles of genes commonly misregulated in hda108 mutant anther and leaf. (A and B) Heat maps showing gene expression profiles in 40 different maize tissues (http://qteller.com/qteller3/) for the two subsets of genes commonly up- (A) or down- (B) regulated in both mutant leaf and anther, selected as shown in Figure 4E.
Histone acetylation levels are altered in hda108 up-regulated genes
To determine whether the transcriptional changes identified by RNA-Seq are linked to altered histone acetylation levels, Chromatin ImmunoPrecipitation (ChIP) assays were performed using antibodies against H3ac, H3K9ac, and H4ac on the same leaf tissues used for transcriptomic analysis. In addition to acetylated histones, the level of H3K9me2 and H3K4me3 histone modifications were evaluated on a subset of DE genes. Immunoprecipitation was followed by quantification of immunoprecipitates by Real-Time qPCR analysis, which was used also to confirm the transcriptional changes highlighted by RNA-Seq (Figure 7 and Figures S6 and S7 in File S16). For each analyzed gene, primers for ChIP-qPCR assays were designed to specifically amplify a 150- to 300-bp fragment corresponding to the 5′-end genomic region. Primers for expression analysis were preferentially designed on the 3′-end of the resulting transcript (primer sequences are reported in File S9).
Figure 7.
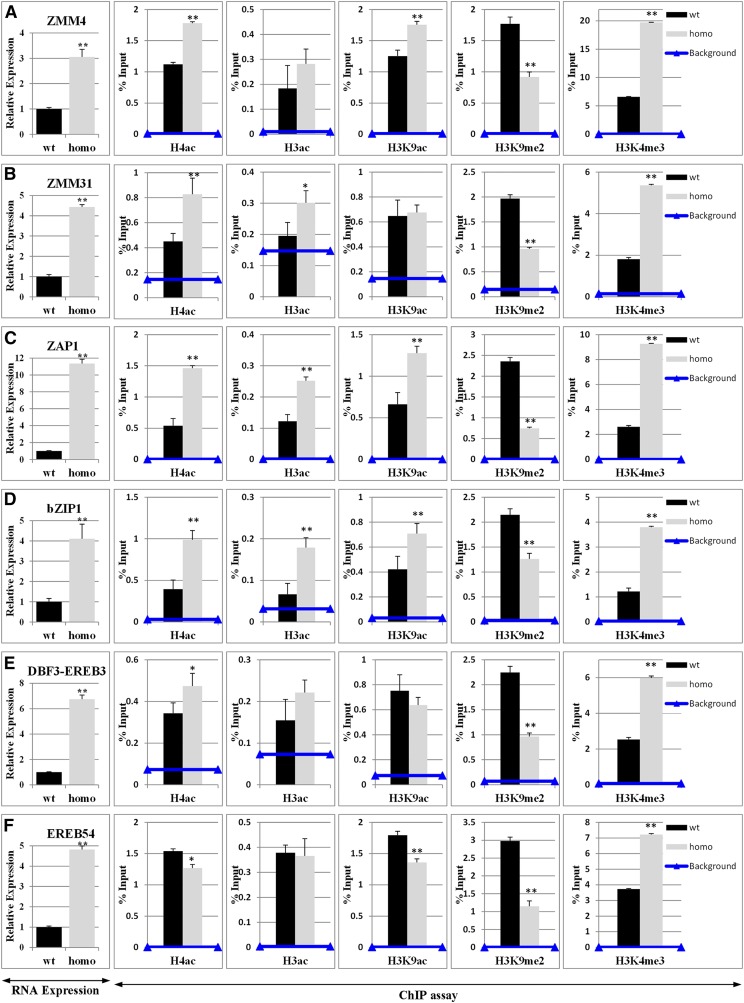
Expression validation and chromatin analysis of hda108 up-regulated genes. Real-Time PCR expression analysis and ChIP assay at (A) ZMM4, (B) ZMM31, (C) ZAP31, (D) bZIP1, (E) DBF3/EREB3, and (F) EREB54 loci in developing 11th leaf of BC5S1 wild-type and homozygous mutant plants. Expression analysis confirmed the transcriptional up-regulation revealed by the RNA-Seq assay. Graphs represent the values of transcript amount standardized to the transcript amount of GAPC2 gene (for details see Materials and Methods). The analyses were performed using two independent cDNA preparations and three technical replicates. The relative fold changes are calculated comparing them to the arbitrary unit, which is the transcript level in wild-type plants. Bars represent SD. Expression of all of the analyzed genes resulted in significant up-regulation in hda108 mutant (P ≤ 0.01; Student’s t-test) as indicated by RNA-Seq analysis. At the selected loci, histone modification levels were analyzed by Real-Time PCR quantification of ChIP immunoprecipitated with α-H4ac, α-H3c, α-H3K9ac, α-H3K9me2, and α-H3K4me3 antibodies on chromatin extracted from the developing 11th leaf of BC5S1 wild-type and homozygous mutant plants. Data are reported as percentage of chromatin input and are average values from two independent ChIP assays and three PCR repetitions for each ChIP assay. SD are reported. Horizontal line indicates the background signal, measured by omitting antibody during ChIP procedure. Asterisks indicate statistically significant changes: * P ≤ 0.05, ** P ≤ 0.01; Student’s t-test.
Comparison of chromatin immunoprecipitates and background signal revealed that mutant up-regulation of MADS box TFs ZMM4, ZMM31, ZAP1, and ZMM24 was associated with a significant increase in H3 and/or H4 acetylation levels (Figure 7, A–C and Figure S6 in File S16); H4 hyper-acetylation was detectable at all four MADS gene loci, while gene-specific H3ac and H3K9ac enrichment patterns were observed. Similarly, hyper-acetylation of both H3 and H4 histones was detected in mutant leaves also at GBP15 and bZIP1 loci (both transcriptionally induced in the hda108 mutant), together with an enrichment in H3K9ac in the latter one (Figure 7D and Figure S6 in File S16). Conversely, histone acetylation within three up-regulated AP2/EREBP genes (DBF3/EREB3, EREB54, and EREB209) resulted as not substantially increased by the hda108 mutation: a very weak increase in H4ac and H3ac characterized DBF3/EREB3 and EREB209, respectively, together with a decrease in H3K9ac level within EREB54 and EREB209 (Figure 7, E and F and Figure S6 in File S16). Similarly, histone acetylation was not affected at NAC134 and MYBR69 mutant up-regulated TF loci, while a decrease in both H3K9ac and H4ac was detectable at the SAUR11 auxin responsive gene locus (Figure S6 in File S16). Histone hypoacetylation, mainly at the level of H4 and H3K9, was instead observed at the level of mutant down-regulated loci (Figure S7 in File S16).
The hda108 mutation provoked also changes in histone modification other than acetylation: the mutant showed an enrichment in H3K4me3 and depletion in H3K9me2 at the level of up-regulated gene loci, independently of the specific combination of acetylation changes observed at these loci (Figure 7).
Transcription and histone acetylation levels of additional patterning genes are tissue-specifically altered in hda108 mutant progenies
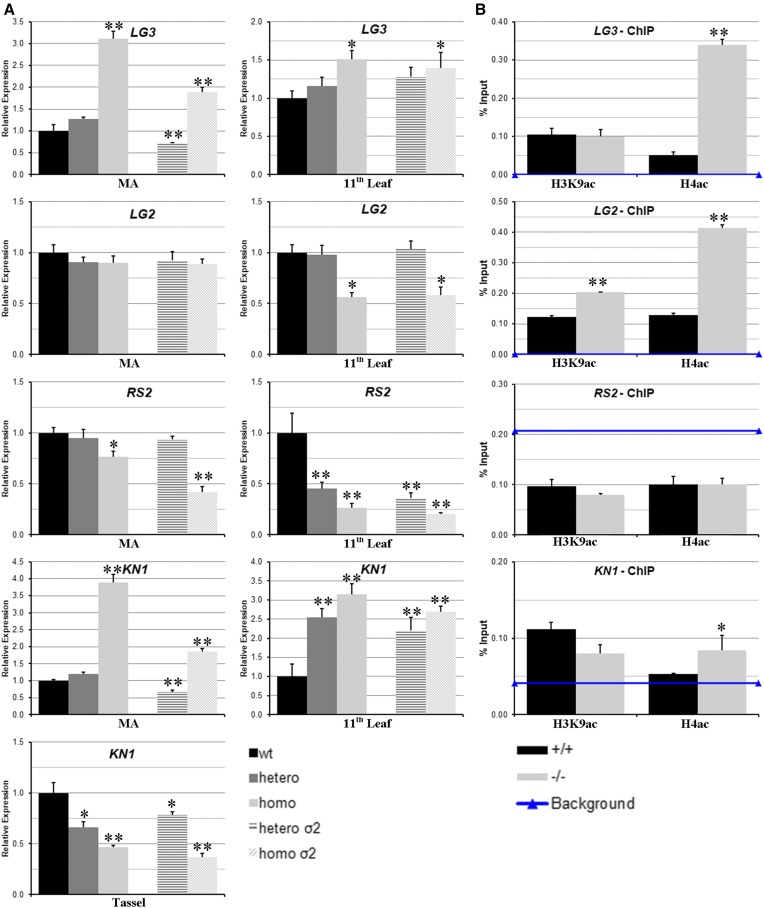
Considering the phenotypic alterations observed in hda108 mutant leaves and the revealed role for HDA108 in regulating the expression of TFs and other genes normally expressed in young, meristematic tissues, we analyzed the expression level of genes encoding important regulators of leaf development in MA and the basal part of fully expanded 11th leaf of BC5S1 and BC5σ2 progenies. Real-Time PCR analyses were carried out to verify whether hda108 knockout could affect the expression of homeobox genes Knotted1 (KN1; Kerstetter et al. 1997) and Liguleless3 (LG3; Muehlbauer et al. 1999) that promote the maintenance of meristematic fate; Liguleless2 (LG2; Walsh et al. 1998) that acts in the pathway that specifies ligule and auricle tissues; and Rough Sheath2 (RS2; Schneeberger et al. 1998) that negatively regulates expression of KNOX genes (Figure 8A). In addition, histone acetylation levels at these loci were analyzed by ChIP, on chromatin extracted from the basal part of expanded 11th leaf of BC5S1 wild-type and mutant plants (Figure 8B). LG3 expression was highly induced in MA of the homozygous mutant plants of BC5S1 and BC5σ2, while a slight up-regulation was observed in the basal part of the 11th leaf of both mutant homozygous genotypes, accompanied by hyper-acetylation of histone H4. No differential expression of LG2 was observed in MA of BC5S1 and BC5σ2 mutant plants, whereas there was a transcript down-regulation in leaves of both progenies. Despite the lower expression level, an increase of both acetylated H3K9 and H4 at 5′ of the LG2 gene was detected in the mutant. RS2 expression resulted as down-regulated in homozygous MA and strongly down-regulated in both heterozygous and homozygous 11th leaf compared to the wild type. On this locus, both chromatin marks were absent, with an amplification signal of the immunoprecipitate lower than the background. Finally, KN1 was up-regulated in MA and in the 11th leaf of homozygous plants of both progenies, together with a hyper-acetylation of histone H4. Conversely, when analyzed in the tassel, KN1 was strongly down-regulated in both mutant progenies.
Figure 8.
Expression and chromatin analysis of hda108 target genes controlling patterning in maize leaves. (A) Real-Time PCR analysis of LG2, LG3, RS2, and KN1 expression levels in MA and basal part of the 11th leaf. Wild-type, heterozygous, and homozygous mutant plants of BC5S1 and heterozygous and homozygous mutant plants of BC5σ2 segregating progeny were analyzed. Expression of KN1 was tested also in developing tassels of both progenies. Diagrams represent the values of transcript amount standardized to the transcript amount of GAPC2 gene (for details see Materials and Methods). The analyses were performed using two independent cDNA preparations and three technical replicates. The relative fold changes are calculated comparing them to the arbitrary unit, which is the transcript level in wild-type plants. Bars represent SD. Asterisks indicate statistically significant changes: * P ≤ 0.05, ** P ≤ 0.01; Student’s t-test. (B) Histone modification analysis at selected loci by Real-Time PCR quantification of ChIP DNA performed with α-H4ac and α-H3K9ac antibodies on chromatin extracted from basal part of the 11th leaf of wild-type and homozygous mutant plants. Data are reported as percentage of chromatin input and are average values from two independent ChIP assays and three PCR repetitions for each ChIP assay. SD are reported. Horizontal line indicates the background signal, measured by omitting antibody during ChIP procedure. Asterisks indicate statistically significant changes: * P ≤ 0.05, ** P ≤ 0.01; Student’s t-test.
hda108 knockout affects expression level, histone acetylation, and DNA methylation of rDNA loci
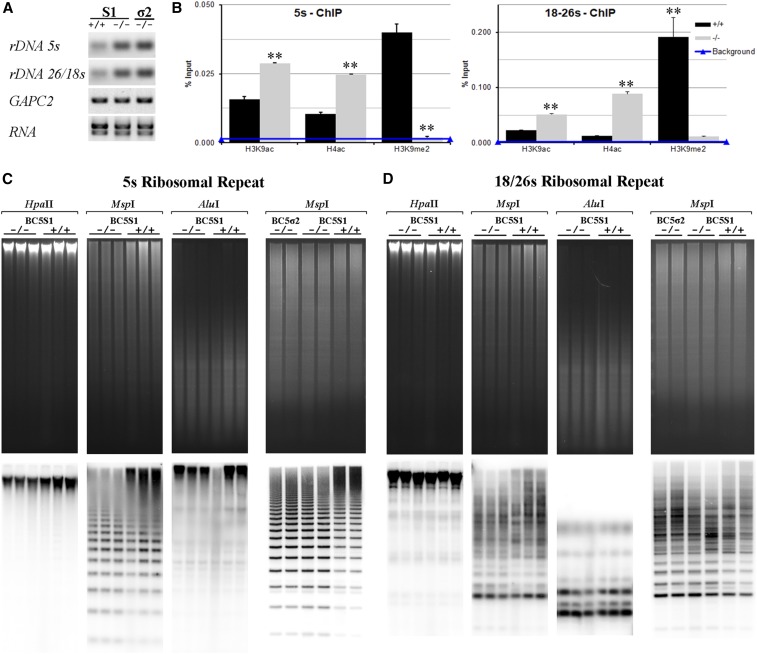
To determine whether hda108 knockout also affects the expression of specific repetitive sequences, an expression analysis by RT-PCR was performed to analyze the transcript level of rDNA loci (5s, intergenic spacer 26/18s, and 26s) in BC5S1 wild-type and homozygous mutant plants. An increased transcript level in the 5s and 26/18s regions was observed in the homozygous mutants if compared with wild-type plants (Figure 9A). Both rDNA repeats were overexpressed also in BC5σ2 homozygous seedlings (Figure 9A). On the contrary, no difference in transcript level between wild-type and homozygous genotypes was detected for the 26s gene because its high expression level resulted in a saturated signal in PCR reactions (data not shown).
Figure 9.
Expression, chromatin acetylation, and DNA methylation patterns at rDNA loci are altered in hda108 mutant. (A) RT-PCR experiments show the up-regulation of 5s locus and 18–26s intergenic spacer expression in homozygous (−/−) seedlings of BC5S1 and BC5σ2 progenies compared to BC5S1 wild-type (+/+). (B) Real-Time PCR quantification of ChIP DNA performed with α-H4ac, α-H3K9ac, and α-H3K9me2 antibodies on chromatin extracted from basal part of the 11th leaf revealed enrichment of both H3K9ac and H4ac together with depletion of H3K9me2 at 5s and 18–26s ribosomal repeats in homozygous hda108 plants. Data are reported as percentage of the chromatin input and are average values from two independent ChIP assays and three PCR repetitions for each ChIP assay. SE are reported. Horizontal line indicates the background signal, measured by omitting antibody during ChIP procedure. Asterisks indicate statistically significant changes (P ≤ 0.01; Student’s t-test). (C and D) DNA methylation was analyzed on genomic DNA from homozygous wild-type (+/+) and mutant plants (−/−) from segregating BC5S1and BC5σ2 progenies. The DNA was digested with methylation-sensitive restriction enzymes HpaII, MspI, and AluI, blotted and probed with 5S (C) and 18–26S (D) rDNA probes.
ChIP assays revealed an evident enrichment of both H3K9 and H4 acetylation level and a strong reduction of H3K9me2 level at the chromatin of the 5S ribosomal sequence and the 26–18S intergenic spacer. The enrichment in chromatin acetylation is positively correlated with their increased expression level (Figure 9B).
Furthermore, to understand whether the increased level of H3 and H4 acetylation in the nucleolus of hda108 plants is accompanied by changes in DNA methylation, we used Southern blot analysis to profile DNA methylation at the rDNA genes of wild-type and homozygous mutant plants (BC5S1 and BC5σ2). Genomic DNA was extracted from seedling tissues and digested with the methylation-sensitive enzymes HpaII, MspI, and AluI, which are sensitive to CG/CHG, CHG, and CHG/CHH methylation, respectively. The analysis revealed a clear, although moderate, reduction of DNA methylation in the 5s and 18/26s regions in homozygous mutant plants in both types of progeny (Figure 9C). Loss of DNA methylation at these regions was only observed in the MspI digests, whereas no hybridization differences were detectable among samples in the HpaII and AluI digests. Because, HpaII and MspI recognize the same target sequence (CCGG) but have different methylation sensitivities, the hybridization differences observed between wild-type and mutant samples suggest that loss of function of the HDA108 gene may impair the deposition of CHG methylation.
Inability to recover hda108/hda101, hda108/cmt3, or hda108/rmr6 double mutants
To explore the functional redundancy between HDA108 and HDA101 genes, BC5S1 hda108 homozygous plants were crossed with plants of the AS33 antisense mutant line of the hda101 gene (Rossi et al. 2007). The AS33 antisense line is characterized by a down-regulation of hda101 transcript and protein, which determines plant developmental defects and alteration of both total level of histone acetylation and transcription of genes responsible for vegetative to reproductive transition. hda108/+;AShda101 plants were selfed to obtain hda108/hda108;AShda101 kernels. Strong defective kernel phenotype segregation was observed in the cobs. Approximately 25% of the selfing progeny presented a small collapsed nonviable kernel, with completely or partially empty pericarp (Figure S8A in File S16). To analyze the alteration that affected both embryo and endosperm development in more detail, X-ray microtomographs were taken of mature normal and defective seeds collected from different segregant ears. Reconstructed axial, sagittal, and coronal microtomography images show different degrees of aberration in defective kernels (Figure S8, B–K in File S16, File S6, File S7, and File S8). Compared to normal kernels, starchy endosperm failed to accumulate starch or developed only partially in defective kernels, while the embryo also showed abnormalities that varied from the presence of an undifferentiated aborted embryo to a defective embryo blocked at the coleoptilar stage.
Maize rpd1-1/rmr6 mutant is a PolIV subunit mutant involved in siRNA biogenesis and in the RNA-directed DNA methylation RdDM pathway (Parkinson et al. 2007; Erhard et al. 2009, 2015). To investigate a possible interaction between HDA108 and the RdDM pathway, we attempted to produce hda108/hda108;rmr6/rmr6 double mutants: rmr6/rmr6 mutant plants were used as male parents in the crosses with hda108 homozygous or heterozygous BC5S1 plants and the resulting double heterozygous plants (hda108/+; rmr6/+) were selfed to produce double mutant segregating populations. The same approach was applied in the attempt to produce double mutants between HDA108 and Zmet2, the maize CMT3 ortholog that catalyzes methylation in genome CHG and CHH sequence context (Papa et al. 2001; Q. Li et al. 2014). In both cases no specific kernel phenotypes were observed in the cobs, but, after genotyping of 93 hda108/rmr6 and 75 hda108/zmet2 progenies, no double mutants were recovered (Table S3 in File S16). χ2 tests revealed a significant deviation from the expected segregation in both progenies (P < 0.001 and P < 0.05 for hda108/rmr6 and hda108/zmet2 progenies, respectively). In addition to the failure to recover double mutants in both crosses, we observed, compared to the expected segregation ratios, a lower number of all genotypes carrying the hda108/hda108 homozygous mutation (Table S3 in File S16). This could be due either to reduced transmission or viability of mutant gametes or reduced viability for the double mutant zygote. Even if we observed a slight reduction of hda108 mutant allele transmission in both crosses (Table S3 in File S16), the segregation ratios do not significantly deviate from 1:1 (P = 0.21 and P = 0.16, respectively, χ2). Furthermore, we did not observe a substantial decrease in germination rates for the offspring of these crosses, suggesting that double mutant individuals failed to complete seed development and that HDA108 might display genetic epistasis on both RdDM and CHG methylation pathways.
Considering the detected variation in DNA methylation at rDNA loci in the hda108 mutant and the involvement of both Zmet2 and Rmr6 in context-specific DNA methylation, we evaluated the rDNA expression and methylation patterns in these mutants. If compared to the appropriate wild-type control, RT-PCR analysis revealed an increased transcript level of the 5s locus only in zmet2 seedlings, while no consistent expression changes were detected for the 18/26s region in both mutants (Figure S9A in File S16). DNA methylation profiles at the rDNA genes of wild-type and homozygous mutant plants confirmed the previously described reduction of CHG DNA methylation (MspI digest) at 5s and in 18/26s regions in the zmet2 homozygous mutant (Figure S9B in File S16; Papa et al. 2001), while no significant alterations were observed in the rmr6 mutant (Figure S9C in File S16). No general differences in CHG cytosine methylation at centromeric or 45S rDNA repeats were previously observed in the rmr6 mutant (Parkinson et al. 2007), which is instead impaired in CHH methylation at the edges between genes and transposable elements (CHH island; Gent et al. 2013; Li et al. 2015).
Discussion
The maize hda108 null mutant line shows an increased level of acetylated histone H3 and H4 and a variation of additional epigenetic marks, such as H3K9me2, H3K4me3, and CHG methylation at ribosomal repeats. hda108/hda108 plants display a bunch of altered phenotypes that negatively affect both the vegetative growth and differentiation of male and female inflorescences, causing male sterility. Although only one knockout mutant allele was identified and described, multiple generations, including BC5S1, BC5S2, and BC5Sσ2, were generated and characterized at both phenotypic and molecular level. BC5S1 progeny were generated by selfing of hda108::Mu heterozygous plants after the introgression of the insertion in the B73 background through five backcrosses, while BC5S2 and BC5Sσ2 are the progeny of BC5S1. In all of the analyzed tissues of different progenies, no expression of the HDA108 gene was observed in hda108/hda108 mutants, which always showed altered phenotypes, while heterozygous plants showed intermediate HDA108 transcript levels and defects.
When combined with other mutations affecting either histone acetylation or other epigenetic pathways, more severe defects than those of the single mutant were detected. The kernels bearing the double mutation hda108/hda108;AShda101 were impaired in both embryogenesis and endosperm differentiation. Considering that both HDA101 and HDA108 are expressed during seed development (Varotto et al. 2003; Sekhon et al. 2013; Stelpflug et al. 2016), but kernels of the two single mutants present a weak phenotype, the defective kernel phenotypes of the double mutant hda108/hda108;AShda101 indicate that the two histone deacetylases may act synergistically on common targets. This finding, together with the inability to recover hda108/hda108;rmr6/rmr6 and hda108/hda108;zmet2/zmet2 double mutant seeds, indicates that HDA108 has an essential role in modulating plant development and reproduction, by controlling transcription through histone deacetylation and altering both the histone code setting and whole genome stability.
HDA108 is an important regulator of maize plant development and reproduction
HDA108 ubiquitous expression indicates a general function of this gene in the regulation of transcription during development: the gene knockout has a pleiotropic effect on plant development and organ differentiation. The effect of the mutation is particularly strong in homozygous plants, whereas in heterozygous genotypes the phenotypic alterations are more evident in BC5σ2 plants that have experienced a generation of homozygosity for the mutation, suggesting that the alteration in epigenetic marks induced by the mutation and affecting development might persist through generations and cannot be rescued by heterozygosity. The drastic male sterile phenotype and persistence of the mutation effect through generations of the maize hda108 knockout line differ from what is observed in Arabidopsis hda6/rts1 mutant plants, which display a very mild phenotype, even in plants that have been homozygous for four generations (Aufsatz et al. 2002).
At leaf development level, the main defects observed were blade length reduction, twisting, knots, and disorganized differentiation of the blade–sheath boundary. By comparing the expression profiles of wild-type and mutant leaves we observed that the majority of the DE genes were up-regulated and annotated as TFs. The AP2/EREBP family of TFs, which is involved in several plant developmental processes and in responses to various types of biotic and abiotic stresses (Riechmann and Meyerowitz 1998; Sakuma et al. 2002; Kaufmann et al. 2010), has 16 members overexpressed in hda108 plants, and together with other families of TFs, such as MYB and MADS, represent the most affected gene family in our mutant leaf transcriptome analysis. The misregulation of these patterning genes seems to only slightly and probably indirectly affect leaf physiology and metabolism through the down-regulation of genes related to chloroplast assembly and functioning.
The hda108 mutation affects plant reproduction by altering ear and tassel development and also microgametogenesis in the anthers. Because shrunken pollen is visible after meiosis in ∼50 and 100% of heterozygous and homozygous anthers, respectively, we supposed that pollen degeneration is associated to its haploid status. However, the results of our plant crosses demonstrated that pollen lacking the HDA108 transcript, in spite of its shrunken phenotype, can successfully fertilize wild-type, heterozygous, and homozygous mutant plants (and even zmet2 and rmr6 epiregulator mutant plants) without distortion of Mendelian expected segregation. Indeed, homozygous plant male sterility seems to be mainly due to anther indehiscence rather than pollen infertility. Anther dehiscence is a complex and multistage process that leads to the breakdown of the septum in the anther wall and is known to involve cell-wall-degrading enzymes, such as polygalacturonases (PGs), b-1,4-glucanases, pectin methylesterases, and expansins (Futamura et al. 2000; Micheli 2001; Wilson et al. 2011). Interestingly, transcripts of these genes related to cell wall degradation are among the strongly down-regulated genes in both the analyzed stages of mutant anthers, while other degrading enzymes, such as chitinases, are up-regulated, indicating the lack of a correct synchronization between anther maturation and pollen release. In mutant plants, the disruption of regular anther development is confirmed by the down-regulation of genes related to hormone metabolism and storage of different compounds at the meiotic stage. After meiosis, young microspores initiate the gametophytic development: intriguingly in meiotic mutant anthers we observed the up-regulation of genes related to the activation and regulation of gene transcription, including genes involved in chromatin regulation, RNA processing, protein synthesis, and cell division. We can speculate that following the activation of these pathways, which are normally silenced, the male gametophyte might be induced by the mutation to potentially change its canonical developmental pathway toward an unknown differentiation pathway that alters pollen phenotype but preserves its fertilization capacity. Indeed, hyper-acetylation of histone H3 and H4 and related alteration of H3K4me3 and H3K9me2 marks could provoke genome instability, compromising correct pollen development in a gametophytic selection context. Other plant HDAs are known to affect either female or male gametogenesis (Cigliano et al. 2013; Liu et al. 2014), and it has been reported that blocking HDAC activity with trichostatin A (TSA) in cultured male gametophytes of Brassica napus and Arabidopsis thaliana leads to an increase in the proportion of cells that switch from pollen to embryogenic growth (H. Li et al. 2014).
As a general regulator of transcription, HDA108 might control the expression of its target genes directly in different plant organs. In view of the fact that histone deacetylation is commonly associated to repression of transcription in eukaryotic organisms (Liu et al. 2014), HDA108 is supposed to act in transcription repressive complexes and regulate specific patterning genes spatially and temporarily. It is therefore predictable that in hda108 mutant plants, the direct targets of this deacetylase activity could be up-regulated as a consequence of an increase in acetylated histones in the chromatin at their loci. Histone H3 and/or H4 hyper-acetylation at ZMM4, ZMM24, ZMM31, and ZAP1 loci, four MADS box TFs up-regulated in mutant leaves (ZAP1 was up-regulated also in mutant PMeA and MiA), suggested that they could represent HDA108 direct targets, together with GBP15, bZIP1 (both up-regulated in mutant leaves and anthers), and LG3 loci. Interestingly, histone H4 was hyper-acetylated at all these loci, while gene-specific H3ac and H3K9ac enrichment patterns were highlighted. In expanded leaves, an enrichment in H4ac level positively correlates also with the up-regulation of the KN1 gene, even if its strong up-regulation could also be due to the equally strong down-regulation of its negative regulator RS2 (Timmermans et al. 1999).
Although histone acetylation levels were generally not increased or even decreased in mutant tissues at NAC134, MYBR69, and EREB loci analyzed, indicating that the mutant up-regulation of these TFs could represent secondary effects, only ChIP experiments with an anti-HDA108 antibody would supply the direct evidence for primary target identification. Considering the observed phenotypical defects of hda108 mutant plants, it is indeed possible that many of the genes that are differentially expressed will be indirectly misregulated as a consequence of the altered development. Altogether, these results indicated that ZmHDA108, similarly to AtHDA6 (Krogan et al. 2012), controls the expression of important leaf patterning genes that could represent its main likely direct targets and whose misregulation disrupts the genetic leaf regulatory networks, drastically altering leaf phenotype from the early differentiation stage to complete development.
Furthermore, the role of HDACs is not restricted to gene expression repression by removal of the acetyl group from histone tails: genome-wide mapping of HDAC binding sites in yeast, mammals, and maize (Kurdistani and Grunstein 2003; Z. Wang et al. 2009; Yang et al. 2016) showed that these enzymes are indeed preferentially targeted to transcriptionally active genes. In addition, HDACs participate in protein complexes as transcriptional corepressors and coactivators or are associated with chromatin remodelers as modulators of the accessibility of DNA to different machinery. Particularly in maize, it has been reported that during early kernel development stages, the lack of HDA101 does not affect the transcript level of the majority of genes directly bound by HDA101 (Yang et al. 2016). This observation indicates that an unambiguous association between HDACs and gene expression cannot be ruled out and that further studies are necessary in plants to identify the direct targets of HDACs in different tissues and during their development, although, as in our case, at some specific loci a clear correlation between histone acetylation, transcript level, and phenotypic effect can be observed.
HDA108 participates in setting the histone code and interacts with DNA methylation pathways
In maize hda108 plants, we observed an increase in acetylated histone H3 and H4 in the nuclei of somatic cells as well as a decrease in H3K9me2 and of H3K4me3. Histone H3K4 trimethylation that marks promoter and the 5′ end of highly expressed genes was indeed increased in hda108 mutant up-regulated genes, confirming the observations made in Arabidopsis hda6 mutants, where a replacement of H3K9 dimethylation with H3K4 trimethylation, H3K9 acetylation, H3K14 acetylation, and histone H4 tetra-acetylation was observed (Earley et al. 2006). On the contrary, the slight decrease in H3K4me3 observed in root mutant interphase nuclei could suggest that HDA108 plays a role in genome-wide tissue-specific epigenetic alteration, taking also into account the demonstrated lower abundance of H3K4me3 in maize roots compared to shoots (X. Wang et al. 2009).
In Arabidopsis, the hda6 mutation determines the loss of repressive chromatin modifications, such as histone deacetylation and CG and CHG methylation, which are required for developmental regulation of rRNA genes (Earley et al. 2010). Importantly, the characterization of many HDA6 allelic variant mutations has clarified that alteration of both CG and CHG methylation depends on a tight interaction between HDA6 and MET1 (Hristova et al. 2015; Zhang et al. 2015).
In maize, the HDA101 overexpression and antisense transgenic lines, which affect histone acetylation, and also methylation (e.g., H3K4me2, H3K9me2) did not show any relevant changes in the DNA methylation at DNA repeats (Rossi et al. 2007). Conversely, in our mutant, an enrichment in H3K9ac, H4 acetylated histone, and a concomitant decrease in H3K9me2 and CHG DNA methylation was observed at ribosomal 5s rDNA and 26/18s intergenic spacer loci. This increase in histone acetylation and loss in H3K9me2 and DNA methylation was associated with an increase in 5s and 26/18s ribosomal transcript accumulation. These observations suggest that in maize also there may exist either a direct or indirect relationship between histone deacetylation mediated by HDA108 and the CHG methylation pathway. Moreover, the lack of a clear alteration of CG methylation at the 5S rDNA repeats suggests that DNA methylases other than MET1 maize ortholog enzymes, such as Zmet2 (Papa et al. 2001) or Zmet5 (Q. Li et al. 2014), might be involved in this cross talk. Because we observed a decrease in H3K9me2 histone marks in hda108 mutants, it may be possible that this histone modification plays a major role in guiding DNA methylation at rDNA repeats. Indeed, the results on the effect of zmet2 mutation on maize methylation at genomic level suggested that Zmet2 might be recruited to heterochromatic regions by H3K9me2 to perform CHG methylation and concomitantly catalyze some CHH methylation at a lower rate (Q. Li et al. 2014).
The inability to recover double mutants and to produce seeds from plants that were homozygous for hda108 mutation and segregating for Zmet2 prevents the study of the combined effect of alteration in histone acetylation/deacetylation and CHG DNA methylation in maize. One could speculate that summing up the CHG losses caused by each single mutation may result in the misregulation of many genes and genome destabilization, impairing seed development as proposed for the zmet2/zmet5 double mutant (Q. Li et al. 2014).
Interestingly, specific transposon sequences were transcriptionally reactivated in the Arabidopsis hda6 mutant (Liu et al. 2012), while our transcriptome analysis did not highlight a significant activation of TE transcription either in developing leaves or anthers. The unchanged levels of CG methylation observed at rRNA loci level could explain this lack of TE transcriptional activation, most of the maize TEs being silenced through both histone modifications and CG DNA methylation (Q. Li et al. 2014; West et al. 2014).
Recent work in Arabidopsis also proposed a role for HDA6 in the specification of loci subjected to silencing: acting upstream of the RdDM pathway, HDA6 is necessary for the establishment, maintenance, and inheritance of chromatin states responsible for silent locus identity (Blevins et al. 2014). Comparing HDA6 and RdDM targets, the authors identified RdDM targets at which HDA6 and Pol IV display genetic epistasis and showed that at these loci, hda6 mutants are defective for siRNA biogenesis, cytosine methylation, and histone deacetylation. Importantly, they observed that at these epialleles, transgene-mediated restoration of HDA6 activity does not restore siRNA biogenesis, cytosine methylation, or histone deacetylation, indicating that the epigenetic memory required for silent locus identity is lost in the absence of HDA6 and is not easily regained (Blevins et al. 2014). We could speculate that this important function exerted by HDA6 in epigenetic inheritance of chromatin silenced states is in accordance with our findings: an interesting aspect of hda108 mutant phenotypes concerns their severity in progenies bearing the mutant allele. Mutant homozygous plants are almost sterile because they do not release viable pollen grains and do not differentiate normal ears. Homozygous mutant plants produced by selfing (BC5S2) or crossing (BC5σ2) BC5S1 homozygous mutants have even more severe phenotypes than the S1 parents. The onset of similar stable epialleles in the maize hda108 mutant could explain also the stronger developmental defect observed in BC5σ2 heterozygous plants compared to BC5S1 ones: once experienced, the homozygosity for the mutation, epigenetic marks, gene expression, and related phenotypes cannot be rescued by heterozygosity.
A progressive transgenerational degradation in plant quality is typical of plant epigenetic mutations affecting genome stability (Mathieu et al. 2007; Parkinson et al. 2007; Erhard et al. 2013) and is thought to be the principal cause of the identification of a reduced number of naturally occurring mutations in maize epiregulators.
Also, in this case, the failure to recover viable double mutants of hda108 and rmr6 indicates that mutations in epiregulators have a stronger phenotypic effect in maize than in Arabidopsis and, as also observed in rice (Hu et al. 2014; Yamauchi et al. 2014), that major perturbations of DNA methylation disrupt development causing embryonic lethality. Alternative approaches and more investigations are therefore needed to identify HDA108 interacting proteins and shared targets between epiregulator mutants and our newly characterized hda108 maize mutant, which can evidently represent a valid tool for future genome-wide studies.
Further experiments to fully address the role of HDA108 in transcriptional regulation during maize plant development and genome stability are necessary. All the presented results are indeed based on a single null mutant; therefore, the isolation and characterization of additional mutant alleles, together with their introgression in different genetic backgrounds, are essential for fully understanding the function of HDA108 in epigenetic phenomena. Alternatively, genome editing techniques could be used to edit the HDA108 protein in different domains, to assess the different impact of these mutations on DNA methylation and histone modifications. Finally, the production of an antiHDA108 antibody could be useful for genome-wide direct target mapping and also to identify the interacting proteins in chromatin-modifying complexes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300625/-/DC1.
Acknowledgments
The authors thank C. Conicella for critical reading of the manuscript, V. Rossi for sharing the ChIP protocol, A. Masi and A.R. Trentin for helping with Western blot experiments, and A. Garside for revision of the English. This work was supported by special grants from the European Commission (FP7 Project KBBE 2009 226477 – “AENEAS”: Acquired Environmental Epigenetics Advances: from Arabidopsis to maize) and Italian Consiglio Nazionale delle Ricerche (CNR) Flagship project “Progetto Bandiera Epigenomica (EPIGEN)”. The authors declare no conflict of interest.
Author contributions: CF, SF, and SV designed and managed the experiments. JR and HL isolated the mutant. CF and SF introgressed and characterized the mutant, collected the samples, and performed the expression analyses. CF carried out microscope analyses and analyzed RNA-Seq data. SF performed ChIP analysis. ML performed DNA methylation analysis; NDF performed X-ray microtomography. CF, SF, and SV conceived and wrote the manuscript with the contribution of all authors.
Footnotes
Communicating editor: N. Springer
Literature Cited
- Aiese Cigliano R., Sanseverino W., Cremona G., Ercolano M. R., Conicella C., et al. , 2013. Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alinsug M. V., Yu C. W., Wu K., 2009. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W., Mette M. F., van der Winden J., Matzke M., Matzke A. J., 2002. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21: 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T., Pontvianne F., Cocklin R., Podicheti R., Chandrasekhara C., et al. , 2014. A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., et al. , 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettoor A. M., Givan S. A., Cole R. A., Coker C. T., Unger-Wallace E., et al. , 2014. Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 15: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigliano R. A., Cremona G., Paparo R., Termolino P., Perrella G., et al. , 2013. Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol. 163: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Lawrence R. J., Pontes O., Reuther R., Enciso A. J., et al. , 2006. Erasure of histone acetylation by arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K. W., Pontvianne F., Wierzbicki A. T., Blevins T., Tucker S., et al. , 2010. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic pol II transcription and differential effects on maintenance vs. siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard K. F., Stonaker J. L., Parkinson S. E., Lim J. P., Hale C. J., et al. , 2009. RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205. [DOI] [PubMed] [Google Scholar]
- Erhard K. F., Jr., Parkinson S. E., Gross S. M., Barbour J. E., Lim J. P., et al. , 2013. Maize RNA polymerase IV defines trans-generational epigenetic variation. Plant Cell 25: 808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard K. F., Jr., Talbot J. E., Deans N. C., McClish A. E., Hollick J. B., 2015. Nascent transcription affected by RNA polymerase IV in Zea mays. Genetics 199: 1107–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C., Carraro N., Varotto S., 2013. Protein immunolocalization in maize tissues. Methods Mol. Biol. 959: 207–222. [DOI] [PubMed] [Google Scholar]
- Futamura N., Mori H., Kouchi H., Shinohara K., 2000. Male flower-specific expression of genes for polygalacturonase, pectin methylesterase and beta-1,3-glucanase in a dioecious willow (Salix gilgiana Seemen). Plant Cell Physiol. 41: 16–26. [DOI] [PubMed] [Google Scholar]
- Gent J. I., Ellis N. A., Guo L., Harkess A. E., Yao Y., et al. , 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. B., Tanaka Y., Kim Y. J., Xie P., Zhang M. Q., et al. , 2015. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 13: 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Wang H., Qiao S., Leng L., Wang X., 2016. Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. USA 113: 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C., Liu Z., 2008. Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Hristova E., Fal K., Klemme L., Windels D., Bucher E., 2015. HISTONE DEACETYLASE6 controls gene expression patterning and DNA methylation-independent euchromatic silencing. Plant Physiol. 168: 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Li N., Xu C., Zhong S., Lin X., et al. , 2014. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 111: 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., et al. , 2009. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388: 266–271. [DOI] [PubMed] [Google Scholar]
- Jang I. C., Pahk Y. M., Song S. I., Kwon H. J., Nahm B. H., et al. , 2003. Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1–3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. Plant J. 33: 531–541. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D., 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jian W., Yan B., Huang S., Qiu Y., 2017. Histone deacetylase 1 activates PU.1 gene transcription through regulating TAF9 deacetylation and transcription factor IID assembly. FASEB J. 31: 4104–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muino J. M., Ferrier T., Wuest S. E., et al. , 2010. Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kerstetter R. A., Laudencia-Chingcuanco D., Smith L. G., Hake S., 1997. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. T., Hogan K., Long J. A., 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M., 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4: 276–284. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Soriano M., Cordewener J., Muino J. M., Riksen T., et al. , 2014. The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 26: 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Eichten S. R., Hermanson P. J., Zaunbrecher V. M., Song J., et al. , 2014. Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Gent J. I., Zynda G., Song J., Makarevitch I., et al. , 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R., 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C. W., Duan J., Luo M., Wang K., et al. , 2012. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang S., Zhao M., Luo M., Yu C. W., et al. , 2014. Transcriptional repression by histone deacetylases in plants. Mol. Plant 7: 764–772. [DOI] [PubMed] [Google Scholar]
- Luo M., Yu C. W., Chen F. F., Zhao L., Tian G., et al. , 2012. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in aArabidopsis. PLoS Genet. 8: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A., Brosch G., Loidl A., Haas H., Loidl P., 1997. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277: 88–91. [DOI] [PubMed] [Google Scholar]
- Martin A., Lee J., Kichey T., Gerentes D., Zivy M., et al. , 2006. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18: 3252–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti I., Battaglia R., Mainieri D., Altana A., Lauria M., et al. , 2013. The WD40-repeat proteins NFC101 and NFC102 regulate different aspects of maize development through chromatin modification. Plant Cell 25: 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J., 2007. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862. [DOI] [PubMed] [Google Scholar]
- Mehdi S., Derkacheva M., Ramstrom M., Kralemann L., Bergquist J., et al. , 2016. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F., 2001. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6: 414–419. [DOI] [PubMed] [Google Scholar]
- Muehlbauer G. J., Fowler J. E., Girard L., Tyers R., Harper L., et al. , 1999. Ectopic expression of the maize homeobox gene liguleless3 alters cell fates in the leaf. Plant Physiol. 119: 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J., Wang X. J., Hagen G., Guilfoyle T. J., 2001. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C. A., Selinger D. A., Pikaard C. S., et al. , 2002. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa C. M., Springer N. M., Muszynski M. G., Meeley R., Kaeppler S. M., 2001. Maize chromomethylase zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson S. E., Gross S. M., Hollick J. B., 2007. Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308: 462–473. [DOI] [PubMed] [Google Scholar]
- Perrella G., Carr C., Asensi-Fabado M. A., Donald N. A., Paldi K., et al. , 2016. The histone deacetylase complex 1 protein of Arabidopsis has the capacity to interact with multiple proteins including histone 3-binding proteins and histone 1 variants. Plant Physiol. 171: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A. V., Fagard M., Proux F., Mourrain P., Boutet S., et al. , 2004. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Meyerowitz E. M., 1998. The AP2/EREBP family of plant transcription factors. Biol. Chem. 379: 633–646. [DOI] [PubMed] [Google Scholar]
- Rossi V., Locatelli S., Lanzanova C., Boniotti M. B., Varotto S., et al. , 2003. A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51: 401–413. [DOI] [PubMed] [Google Scholar]
- Rossi V., Locatelli S., Varotto S., Donn G., Pirona R., et al. , 2007. Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 19: 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., et al. , 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290: 998–1009. [DOI] [PubMed] [Google Scholar]
- Schneeberger R., Tsiantis M., Freeling M., Langdale J. A., 1998. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125: 2857–2865. [DOI] [PubMed] [Google Scholar]
- Sekhon R. S., Briskine R., Hirsch C. N., Myers C. L., Springer N. M., et al. , 2013. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug S. C., Sekhon R. S., Vaillancourt B., Hirsch C. N., Buell C. R., et al. , 2016. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9. [DOI] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., et al. , 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Tian L., Chen Z. J., 2001. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M. P., Wang J. J., Wei N. E., Jiang H., et al. , 2005. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., et al. , 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45: W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M. C., Hudson A., Becraft P. W., Nelson T., 1999. ROUGH SHEATH2: a myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Poree F., Nagel A., Lohse M., Czedik-Eysenberg A., et al. , 2009. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ. 32: 1211–1229. [DOI] [PubMed] [Google Scholar]
- Varotto S., Locatelli S., Canova S., Pipal A., Motto M., et al. , 2003. Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 133: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J., Waters C. A., Freeling M., 1998. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 12: 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Kurdistani S. K., Grunstein M., 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang X., Elling A. A., Li X., Li N., Peng Z., et al. , 2009. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D. E., Barski A., et al. , 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West P. T., Li Q., Ji L., Eichten S. R., Song J., et al. , 2014. Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Z. A., Song J., Taylor B., Yang C., 2011. The final split: the regulation of anther dehiscence. J. Exp. Bot. 62: 1633–1649. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Johzuka-Hisatomi Y., Terada R., Nakamura I., Iida S., 2014. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol. Biol. 85: 219–232. [DOI] [PubMed] [Google Scholar]
- Yang H., Liu X., Xin M., Du J., Hu Z., et al. , 2016. Genome-wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhan X., Xu X., Cui P., Zhu J. K., et al. , 2015. Two domain-disrupted hda6 alleles have opposite epigenetic effects on transgenes and some endogenous targets. Sci. Rep. 5: 17832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K., 2005. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina RNA-Seq reads and the summaries of FPKM values in the different samples have been deposited in the NCBI Gene Expression Omnibus repository under accessions GSE101943 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101943) and GSE102036 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102036), while File S11, File S12, File S13, File S14, and File S15 contain the complete results of differential expression tests.