Polygenic adaptation occurs when natural selection changes the average value of a complex trait in a population, via small shifts in allele frequencies at many loci. Here, Racimo, Berg, and Pickrell present a method...
Keywords: polygenic adaptation, selection, admixture, complex traits, GWAS
Abstract
An open question in human evolution is the importance of polygenic adaptation: adaptive changes in the mean of a multifactorial trait due to shifts in allele frequencies across many loci. In recent years, several methods have been developed to detect polygenic adaptation using loci identified in genome-wide association studies (GWAS). Though powerful, these methods suffer from limited interpretability: they can detect which sets of populations have evidence for polygenic adaptation, but are unable to reveal where in the history of multiple populations these processes occurred. To address this, we created a method to detect polygenic adaptation in an admixture graph, which is a representation of the historical divergences and admixture events relating different populations through time. We developed a Markov chain Monte Carlo (MCMC) algorithm to infer branch-specific parameters reflecting the strength of selection in each branch of a graph. Additionally, we developed a set of summary statistics that are fast to compute and can indicate which branches are most likely to have experienced polygenic adaptation. We show via simulations that this method—which we call PolyGraph—has good power to detect polygenic adaptation, and applied it to human population genomic data from around the world. We also provide evidence that variants associated with several traits, including height, educational attainment, and self-reported unibrow, have been influenced by polygenic adaptation in different populations during human evolution.
THERE is much interest in identifying the individual genetic variants that have experienced natural selection during recent human evolution. Many popular methods tackle this problem by identifying alleles that have changed frequency faster than can be explained by genetic drift alone, and that can instead be explained by selective processes. These methods exploit patterns like haplotype homozygosity (Sabeti et al. 2002, 2007; Voight et al. 2006) and extreme population differentiation (Chen et al. 2010; Yi et al. 2010; Racimo 2016), and have yielded several important candidates for human adaptation: for example, LCT (Bersaglieri et al. 2004), EDAR (Sabeti et al. 2007), EPAS1 (Yi et al. 2010), and the FADS region (Fumagalli et al. 2015). In order for these signals to be detectable at the level of an individual locus, the historical changes in allele frequency must have been large and rapid. Therefore, they can only be produced by alleles that confer a strong selective advantage.
With the advent of large-scale genome-wide association studies (GWAS) for a variety of measurable traits, however, it has now become possible to detect a more subtle mechanism of adaptation. If a trait is polygenic, positive selection may instead occur by concerted shifts at many loci that all contribute to the variation in a trait. Over short time scales, these shifts are expected to be small [but see Barton and de Vladar (2009) for polygenic dynamics under longer time scales]. They are also expected to occur in consistent directions, such that alleles that increase the trait will systematically rise in frequency (if selection favors the increase of the trait) or fall in frequency (if selection operates in the opposite direction). None of the allele frequency changes need to be large on their own for the phenotypic change to be large. This process is called polygenic adaptation, and may underlie major evolutionary processes in recent human history (Pritchard et al. 2010; Pritchard and Di Rienzo 2010; Mathieson et al. 2015; Field et al. 2016).
A number of methods have been developed to detect polygenic adaptation using loci identified from GWAS. Turchin et al. (2012) was the first such study. They developed a test for polygenic adaptation between two populations, and showed that there were systematic frequency differences at height-associated loci between northern and southern Europeans, which could not be explained by genetic drift alone. Berg and Coop (2014), Berg et al. (2017) developed a more general method to detect polygenic adaptation by testing for over-dispersion of mean genetic values among several populations, using a genome-wide population covariance matrix to predict how alleles should behave under neutrality. Robinson et al. (2015) used theory by Ovaskainen et al. (2011) to develop a similar population differentiation method to detect polygenic adaptation with GWAS. They also made use of the genome-wide covariance matrix, but, in contrast to Berg et al. (2017), their method is implemented in a Bayesian linear mixed model.
None of these methods require a detailed model of human history to detect polygenic adaptation. Their use of the genome-wide covariance matrix allows them to capture patterns of genetic drift among populations without having to infer their history. While this makes them quite powerful, it also means that they are not very useful at determining where and when polygenic adaptation took place in the past.
Here, we develop a method to detect polygenic adaptation that uses a more parameter-rich model of historical population structure: an admixture graph, which is a simplified representation of the history of divergences and admixture events among populations (Patterson et al. 2012; Pickrell and Pritchard 2012). An explicit model allows us to infer where particular bouts of polygenic adaptation took place in human history, so as to better understand how selection on trait-associated variants has occurred over past generations.
Materials and Methods
Model
Assume we have measured genotypes at a single-nucleotide polymorphism (SNP) that influences a trait in a set of M populations. Let be the count of the derived allele in population m, and let be the vector across the M populations of each observation. Let be the total number of chromosomes observed in population m (together ). Assume we have an admixture graph G relating these populations, and that this graph consists of an accurate topology, as well as accurate branch lengths and admixture rates. Branch lengths are in units of drift, which are approximately equal to where, for each branch, t is the number of generations and is the effective population size, assuming (Crow and Kimura 1970). In practice, we can estimate such a graph from neutral genome-wide data, and we use the program MixMapper (Lipson et al. 2013) when applying our method to human genomic data below.
We wish to model the changes in frequency of a trait-associated allele over the graph G. At each node in the graph, we introduce a parameter that corresponds to the allele frequency of the variant at that node. Let be the derived allele frequency at the root of the graph, be the vector of allele frequencies at all the other internal nodes of the graph, and be the vector of allele frequencies at the tips of the graph.
The probability of the parameter values and the data can then be decomposed as follows:
| (1) |
We now take each of the terms above in turn. First, the probability of the observed counts is simply a product of binomial probabilities:
| (2) |
where is the element of that corresponds to population m.
To get the probabilities of the changes in allele frequency across different nodes, consider a single branch of G. Assuming the branch is relatively short (such that the allele does not approach fixation or extinction in this time period) and there is no natural selection, we can use the Normal approximation to the Wright-Fisher diffusion (Cavalli-Sforza et al. 1964; Felsenstein 1973; Nicholson et al. 2002) to model the allele frequency at the descendant node of the branch () as a function of the allele frequency at the ancestral node ():
| (3) |
where c corresponds to the amount of drift that has occurred in the branch. In practice, we use a truncated Normal distribution with point masses at 0 and 1, to account for the possibility of fixation or extinction of the allele (Coop et al. 2010).
In our model there may have also been selection on the allele on the branch, such that it was pushed to either higher or lower frequency because of its influence on a trait. We can model the selected allele frequency by modifying the infinitesimal mean of the Wright-Fisher diffusion and approximating the diffusion with a Normal distribution that now includes some additional terms (Coop et al. 2010; Turchin et al. 2012; Günther and Coop 2013) (C. Bhérer, personal communication):
| (4) |
where is the effect size estimate at that site (defined with respect to the derived allele), is some function that relates the effect size estimate to the selective pressure, and α is our positive selection parameter, which is approximately equal to the product of the selection coefficient for the advantageous allele and the duration of the selective process (Turchin et al. 2012; Mathieson and McVean 2013). In practice, we will set to be equal to the sign ( or ) of so as to avoid giving too much weight to variants of strong effect. We will model selection only on SNPs that are associated with a trait in a particular GWAS.
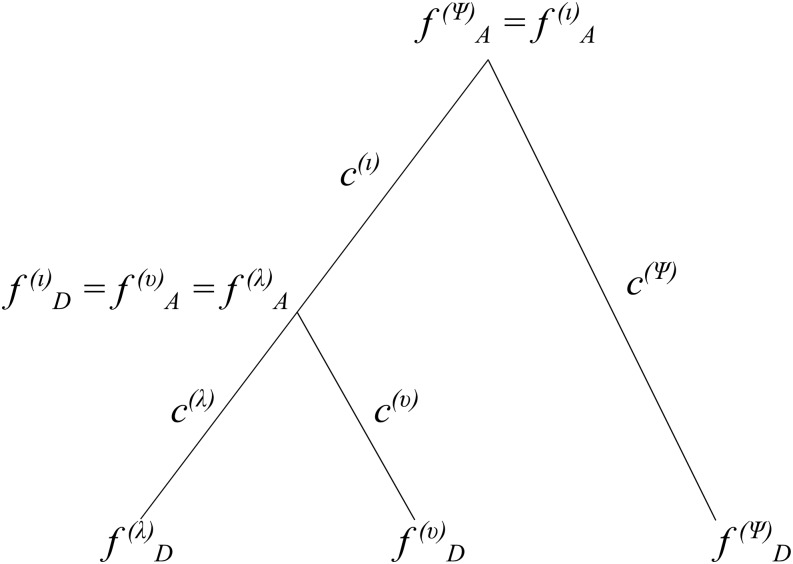
We can calculate the probability of these parameters at a particular site as the product of the Normal probability densities that correspond to the evolution of allele frequencies down each branch times a binomial probability density to account for sampling error. Let us denote the Normal density that corresponds to a particular branch λ as where we suppress notation of for clarity. Then, for example, the probability of a given pattern of allele frequencies and sample counts over a rooted three-leaf tree with four branches λ, ι, υ, and ψ (Figure 1) can be computed as follows:
| (5) |
when the α parameters and the allele frequency at the root of the tree are known. Note that some of the symbols here correspond to the same allele frequencies. For example, if the υ branch is one of the immediate descendant branches of the ι branch, then Assuming SNPs are unlinked, we can compute the probability of the allele frequency configurations at all N trait-associated SNPs as a product over the probabilities at each of the SNPs:
| (6) |
where is the probability of the parameters of interest at SNP i under our tree model.
Figure 1.
Schematic of drift values (c) and allele frequencies (f) for a three-leaf population tree with no admixture.
More generally, we can also compute the probability of our parameters in an admixture graph, containing nodes with more than one parent. In that case, the probability of an allele frequency of an admixed node is a weighted sum of the probability paths corresponding to its two parents, where the weights are the admixture rates for each of the two contributions.
MCMC algorithm
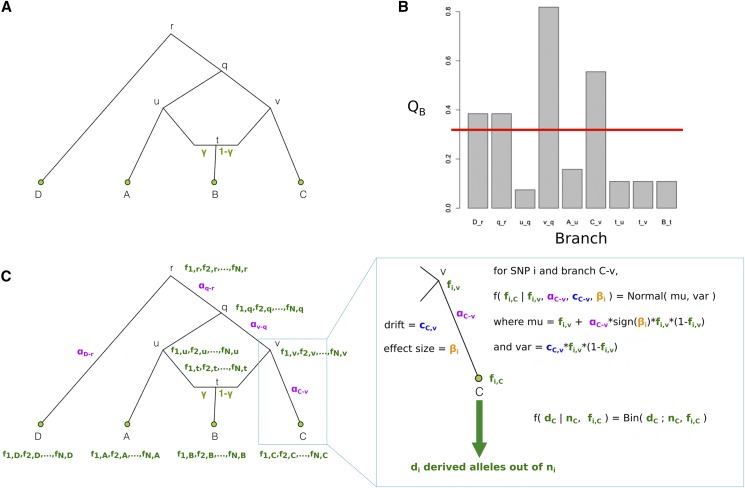
In practice, for a given SNP, we know neither the allele frequencies at the inner nodes , at the tip nodes and at the root node (), nor the α parameters in each branch (). We want to obtain a posterior distribution of these parameters, given the data and the known graph: We aim to do this for all trait-associated SNPs. We therefore developed an MCMC sampler to transition between the states of these variables and estimate their posterior distribution (Figure 2).
Figure 2.
Schematic of PolyGraph estimation procedure for a four-population graph with one admixture event. (A) The first step is the estimation of the admixture graph topology using neutral SNP data, via an admixture-graph fitting program like MixMapper. (B) Then, we use the statistic to determine which branches to explore in the MCMC. Selection parameters whose corresponding branches have a statistic that is smaller than a specific cutoff (red line) are set to a fixed value of 0 in the MCMC. (C) Model for MCMC sampling. The SNP frequencies in the nodes of the graph are shown in green, while the selection parameters for each candidate branch are shown in purple. For each SNP, the likelihood of each branch of the graph is a Normal distribution. To model the sampling of derived alleles in the leaves of the graph, we use a binomial distribution.
We set the prior for the frequencies at the root to be a uniform distribution. For a given SNP i,
| (7) |
As there are many combinations of α parameters that generate almost equivalent likelihoods in a complex admixture graph (Supplemental Material, Figure S1 in File S1), we use a “spike-and-slab” prior for the α parameters, so as to promote sparsity. For a given branch j,
| (8) |
This is a mixture of two Normal distributions centered at 0: one of the distributions has a wide standard deviation (SD) (τ), while the other has a much narrower SD, which is a fraction () of τ, and approximates a point mass at 0 (Mitchell and Beauchamp 1988; George and McCulloch 1993). Here, κ is the mixture probability of drawing from the narrower Normal distribution, and we model it with a uniform hyperprior. The idea behind this is that our assumed prior belief is that only a few of the branches in the admixture graph have experienced bouts of polygenic adaptation, so we reward α parameters that tend to stay in the neighborhood of 0 during the MCMC run.
For the MCMC transition probabilities of the α parameters, we use a Normal distribution with constant variance. For the transition probabilities of the ancestral allele frequencies, we use a truncated Normal distribution with point masses at 0 and 1, with variance equal to a constant (input by the user) times where is the frequency of an allele in its current state. This allows for the proposed transitions to be larger for SNPs at medium frequencies and smaller for SNPs at high or low frequencies. We apply a Hastings correction in the acceptance ratio to account for this asymmetric proposal distribution.
For all applications below, we ran our MCMC sampler for 1 million steps with a burn-in period of 10,000, and obtained posterior samples every 1000 steps. The variance of the transition probabilities of the ancestral nodes and the α parameters were chosen so that the acceptance rate was close to 23% for each set of parameters. For the spike-and-slab prior, we set τ to be equal to 0.1, and ζ to be equal to 25. The lower and upper boundaries of the uniform hyperprior for κ were set to be 60 and 80%, respectively, unless otherwise stated. We note that all of these parameters can be adjusted by the user as needed.
A statistic for prioritizing branches
We observed via simulations that different combinations of α parameters can produce very similar likelihood values. This causes the MCMC sampler to explore different combinations of α values in the same posterior run, when only one such combination is actually correct (Figure S1 in File S1). The aforementioned spike-and-slab prior serves to partially ameliorate this problem, but we aimed to find a way to further encourage sparsity by reducing the possible number of candidate branches that are explored in the MCMC.
We therefore devised a set of summary statistics that can be computed before starting an MCMC run and are meant to detect branch-specific deviations from neutrality. Let F be the empirical population covariance matrix, which—under ideal neutral conditions—should be determined by the admixture graph connecting all the populations. Let be the mean-centered vector of estimated mean genetic values for each of the M populations, computed from the N SNPs that are known to be associated with a trait. For a specific population m:
| (9) |
Here, is the sample frequency of SNP i in a panel of population m, and is its effect size estimate. The vector therefore contains the values for all M populations.
Furthermore, let be the additive genetic variance of the ancestral population for a given character. We compute by taking the ancestral frequency for each SNP i to be the mean sample frequency over all populations ():
| (10) |
Following Wright (1949), Berg and Coop (2014), if we are willing to assume that
| (11) |
then, by the definition of the multivariate normal distribution, for any choice of and
| (12) |
| (13) |
where we have set for notational convenience. It follows that
| (14) |
and this holds for all choices of
Now, let:
| (15) |
where is a scaled version of so that is has unit length. is the square of and has a distribution under the null. Importantly, one is free to choose such that it represents a branch (j) in an admixture graph. Let us then define the vector for a particular branch j to be equal to the contribution w of that branch to each of the leaves of the graph. For example, branch v-q in Figure 2 is a full ancestral branch to leaf C, a partial ancestral branch to leaf B (due to the admixture event), but not an ancestral branch to either A or D. Therefore, the vector is equal to By the same reasoning, and
If we scale such that it has unit length:
| (16) |
then
| (17) |
has an interpretation as the amount of among population additive genetic variance that we would expect to come about because of drift which occurs down branch j. In turn, is the actual amount of variance in genetic values observed along the axis consistent with that branch. In other words, the denominator of gives the expectation of the numerator under the neutral model.
The statistic reflects how much of the deviations from neutrality among the population mean genetic values for a trait is due to branch j. Excessively large values of (j) represent evidence suggesting non-neutral evolution down branch j. Note that, by design, branches with the exact same child nodes have equal statistics, as do branches at the root of the graph.
We used this summary statistic (computed for each branch) to prioritize which branches to explore in our MCMC. We applied a prior point mass of zero to all branches whose corresponding statistic were smaller than a particular cutoff. The choice of cutoff was based on simulations (see below). The MCMC only produces posterior samples for branches that pass this cutoff, and are therefore highly deviated from their expectation under neutrality. We also update the α parameters of these latter branches in our MCMC with a frequency proportional to their values, in a similar fashion to Zhou and Stephens (2012).
Choosing a cutoff for
We aimed to find a cutoff for that would serve to minimize the number of candidate branches to be explored in the MCMC while at the same time trying to ensure that the true selected branches would be included among these candidates. One choice would be to select the cutoff of a distribution that would correspond to a P-value of where k is the number of branches tested. We find, however, that a constant value for this cutoff is not the most desirable choice as a way to prioritize branches for exploring the strength of selection in each of the branches in the MCMC, as graphs of different sizes (i.e., amounts of drift) result in quite different sensitivity values, as well as number of candidate branches included, when (Figure S2 and Figure S3 in File S1 for two cases where each branch has drift length = 0.02 and Figures S4 and S5 in File S1 for two cases where each branch has drift length = 0.05). We observe the same issues when simulating under (Figures S6–S9 in File S1).
A more stable strategy across graphs of different sizes that also works better at minimizing the number of candidates is to choose the cutoff to be a fraction of the largest statistic among all branches in the graph (Figure S2 and Figure S5 in File S1). For all analyses below, we chose this to be of the maximum statistic. We note that this is a less conservative strategy than the fixed cutoff. However, it is important to remember that we do not aim to formally test for selection here, but merely obtain a set of likely candidate branches for the MCMC to explore downstream, some of which may end up producing a posterior mean estimate of α that is consistent with neutrality (i.e., ).
Simulations
To assess the performance of our method, we simulated different demographic scenarios. For each simulated SNP, we sampled the root allele frequency () from a Beta(2,2) distribution, to emulate the fact that, in the real data, variants in the leaf populations tend to be further away from the boundaries of fixation and extinction than under a uniform distribution. We evolved the SNPs throughout an admixture graph forward in time using Wright-Fisher binomial sampling, and used binomial distributions to sample panel allele frequencies from the leaf populations.
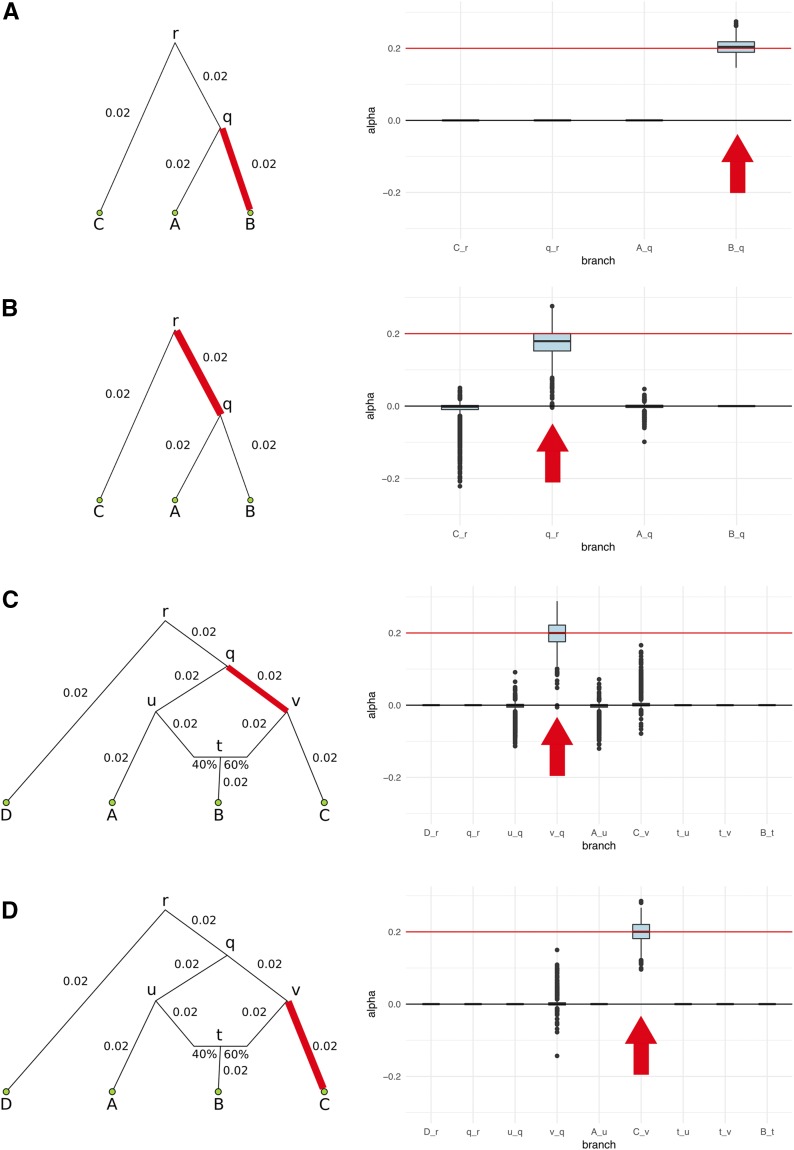
We first simulated a simple three-leaf tree with four branches, in which the sampled panels in the leaves were each composed of 100 diploid individuals (Figure 3, A and B). We tested scenarios of different branch lengths: each of the branches was simulated to be either of length 0.02 or of length 0.05. We also tested different types of branch under selection (either a terminal branch or an internal branch). We additionally tested a four-leaf admixture graph with one admixture event (Figure 3, C and D) in these same scenarios. For comparison, the amount of genetic drift between Spanish and French human populations is 0.016, and the amount of drift between French and Han Chinese human populations is 0.22 (Haak et al. 2015). The latter is approximately equal to the drift separating populations A and C in our four-population graph, when each branch has length equal to 0.05.
Figure 3.
We simulated 400 SNPs affecting a trait under polygenic adaptation, and then used our MCMC to obtain posterior distributions of the α parameters for each branch. The red arrows denote the selected branch. The red line in the box-plots denotes the simulated value of α (in this case, 0.2). The lower, middle, and upper hinges denote the 25th, 50th, and 75th percentiles, respectively. The upper whisker extends to the highest value that is within 1.5 * IQR of the upper hinge, where IQR is the inter-quartile range. The lower whisker extends to the lowest value within 1.5 * IQR of the lower hinge. Data beyond the whiskers are plotted as points. The numbers in each graph denote the drift lengths and admixture proportions. (A) Three-leaf tree with selection in a terminal branch (B-q). (B) Three-leaf tree with selection in an internal branch (q-r). (C) Four-population admixture graph with selection in an internal branch (v-q). (D) Four-population admixture graph with selection in a terminal branch (C-v).
In all cases, we set a constant effective population size of 10,000 and adjusted the number of generations in each branch, depending on how much drift we specified in each scenario. We used Equation 2 from the Supplemental Note of Turchin et al. (2012) to simulate selection. We set a constant time during which selection operates () to be equal to 100 generations, and adjusted the selection coefficient (s) to obtain the selection parameter specified in each scenario (). If a branch had a larger number of generations than the selective phase was simulated to occur during the ancestral-most generations, followed by a neutrality period until reaching the end of the branch. The effect sizes of the SNPs were drawn from a Normal distribution with mean 0, and we simulated polygenic adaptation in a particular branch using only the sign of the effect size of each SNP. We also simulated an additional 10,000 SNPs that evolved neutrally under the same demography, so as to estimate the neutral population covariance matrix.
For all simulation scenarios, we tested five 400-SNP replicates. We ran the MCMC four times on each simulation to check that it was behaving consistently. We observed that the runs for each simulation were very similar to each other, so we only show one run for each simulation.
Building admixture graphs
We fitted admixture graphs using genome-wide data from Phase 3 of the 1000 Genomes Project (Auton et al. 2015), and a more broadly sampled SNP chip dataset of present-day humans from 203 populations genotyped with the Human Origins array (Patterson et al. 2012; Lazaridis et al. 2014). The latter dataset was imputed using SHAPEIT (Delaneau et al. 2013) on the Michigan Imputation Server (Das et al. 2016) with the 1000 Genomes data as the reference panel (C. Bhérer, personal communication).
We excluded SNPs that were deemed to be associated with the trait of interest when fitting population graphs. We used MixMapper (v1.02) (Lipson et al. 2013) to build best-fitting scaffold trees, and placed putatively admixed populations as mixtures originating from different branches in these trees. We pruned both datasets by sampling one out of every 100 SNPs before feeding them as input into MixMapper. To account for any residual linkage disequilibrium (LD), we also performed 100 bootstrap replicates of the computed statistics, computed over 500 blocks along the genome. Because MixMapper cannot distinguish between the two drift values corresponding to two admixing branches with a common child node and the drift value specific to the immediate descendant branch of the child node (Figure 2 in Lipson et al. 2013), we forced the drift in the two admixing branches to be equal to 0.001 and assigned the drift estimated by MixMapper to the branch immediately descending from the child node. We also verified that the topologies we found in MixMapper were the same topologies as the ones inferred by TreeMix (Pickrell and Pritchard 2012) (Figure S10 in File S1).
Implementation
We implemented both the MCMC and the statistic computation in a program that we call PolyGraph. We use the R packages admixturegraph (Leppälä et al. 2017) and igraph (Csardi and Nepusz 2006) to visualize and manipulate various aspects of a graph. The R scripts to run PolyGraph can be downloaded from https://github.com/FerRacimo/PolyGraph.
Visualizing poly-graphs
To facilitate the visualization of posterior distributions for α parameters, we developed a new way to plot polygenic adaptation in a graph: a “poly-graph,” which simultaneously depicts the structure of the studied graph and the marginal posterior mean of each α parameter, in the form of different colorings for each branch.
In a poly-graph, the vertical component of a nonadmixing branch is proportional to the amount of genetic drift that it experienced (calculated via MixMapper). The position of admixed nodes is determined based on the drift value of one randomly chosen parent branch. The colors indicate the marginal posterior mean estimate of the selection parameter for variants associated with the corresponding trait (with red indicating an increase in trait-increasing variant frequency, and blue indicating a decrease in trait-increasing variant frequency). When plotting poly-graphs in this study, we impose a minimum branch height (= 0.075) for clarity, as otherwise the selection parameters of some branches with very short drift lengths are impossible to visualize.
Results
Performance on simulations
When the α parameter is large (), the MCMC performed very well (Figure 3). For a tree (Figures S11 and S12 in File S1) or a graph (Figures S13 and S14 in File S1) with small branch lengths (0.02), the branch simulated to be under selection was included as a potential candidate branch in the MCMC in all simulations, indicating that the cutoff was not overly stringent. PolyGraph then consistently converged on the appropriate joint distribution of selection parameters. When the branches were simulated to be longer (0.05), the MCMC performed well, but, in a few simulations, it produced positive estimates for α parameters in neutral branches, or failed to find evidence for selection in any branch (Figures S15–S18 in File S1). This occurred more often when the α parameter was simulated to be smaller (), but, again, this was less of a problem with short-branch graphs (Figures S19–S22 in File S1) than with long-branch graphs (Figures S23–S26 in File S1). In general, we conclude that the method performs best when selective pressures are strong and/or exerted over long time periods (i.e., large α), and when drift parameters are small. We also observe that using a nonsparse prior (i.e., setting κ to 0) leads to strong misestimation of parameters when selection is concentrated on a single branch (Figure S27 in File S1). To verify that the MCMC chain was mixing well, we also built autocorrelation plots of the α parameters (Figure S28 in File S1 for the chain corresponding to simulation 1 of Figure S14 in File S1).
We were concerned about false positive estimates of selection when the graph is misspecified. To assess this, we simulated a graph like the one shown in Figure 3C but with no selection. We first run PolyGraph while correctly specifying the topology and the branch lengths (of length equal to 0.02) as input (Figure S29 in File S1), and observed that all posterior α estimates are tightly centered at 0, as expected. Then, we simulated a graph with the same topology but with each branch having length equal to 0.03, while incorrectly underestimating the length of each branch to still be equal to 0.02 (Figure S30 in File S1). Finally, we simulated the same graph but with each branch having length equal to 0.04 while incorrectly underestimating the branch lengths to all be equal to 0.02 (Figure S31 in File S1). With increasingly stronger misspecification of the branch lengths, we observe that the behavior of some of the posterior estimates becomes more erratic. Visual inspection of the MCMC trace indicates that underestimation of the branch lengths makes the chain to become more “sticky,” causing some parameters to get stuck at incorrect areas of parameter space for long periods of time.
We also simulated a neutral graph as in Figure 3C but pretended that population A had not been sampled, and that the graph was (incorrectly) estimated to be a three-population tree like the one in Figure 3A. This topological misspecification slightly affected the inference of neutrality in one of the five simulations (Figure S32 in File S1), and we do not discard the possibility of other incorrect types of topologies that could also generate wrong inferences. We therefore stress that the admixture graph—especially the branch lengths—relating the populations under study should be correctly estimated before running PolyGraph. We also advise to run the MCMC only when there is significant evidence for selection based on the statistic (Berg and Coop 2014), which does not need an admixture graph as input, as it uses the full genome-wide covariance matrix to model the expected amount of drift separating each of the populations.
Application to 1000 Genomes data
We tested our method on sets of associated variants from 43 GWAS on 42 different traits (Table S1 in File S1; two of the GWAS are for age at menarche) that were previously assembled as part of a meta-analysis studying the genetic correlations between such traits (Pickrell et al. 2016). The meta-analysis split the genome into approximately independent linkage disequilibrium blocks (Berisa and Pickrell 2016). The blocks were computed using the European populations in Phase 1 of the 1000 Genomes Project, but we observe virtually no differences in genetic scores when using the East Asian blocks instead. For each block with a posterior probability of containing an association [obtained from fgwas (Pickrell 2014)], the SNP with the maximum posterior probability of being the causative variant was extracted. These SNPs can be found on github (https://github.com/PickrellLab/gwas-pw-paper/tree/master/all_single), and we downloaded them to build our candidate trait-associated SNPs. To fit admixture graphs and test for polygenic adaptation, we used the allele frequency data found in the VCFs from Phase 3 of the 1000 Genomes Project (Auton et al. 2015). We excluded SNPs for which the ancestral allele in the 1000 Genomes data were unknown or unsure (lower case in VCF file).
We started by finding the most additive fitted tree that included diverse populations sampled across the world (Figure S33A in File S1): Nigerian Esan (ESN), Sierra Leone Mende (MSL), individuals of Northern European ancestry from Utah (CEU), Southern Europeans from Tuscany (TSI), Dai Chinese (CDX), Japanese (JPT), and Peruvians (PEL). For our first analysis, we did not attempt to model any admixture events.
We took trait-associated variants to be under polygenic adaptation if the P-value for the corresponding (Berg and Coop 2014) statistic (testing for overall selection among the populations) was where n is the number of assessed GWASs. Traits with associated variants that passed this criterion are shown in Table 1. These were: height (Wood et al. 2014), educational attainment (Okbay et al. 2016), self-reported unibrow, self-reported male-pattern baldness (Pickrell et al. 2016), and schizophrenia (SWGPGC 2014). To account for possible artifacts arising from the ascertainment scheme for each GWAS, we also generated 1000 samples in which we randomly switched the sign of the estimated effect size for all trait-associated SNPs. This serves to preserve the genetic architecture of each trait, while removing the effect of selection. We computed a second P-value of the observed () by comparing it to these samples (Table 1).
Table 1. Trait-associated variants with Bonferroni-corrected significant evidence of being under polygenic adaptation in the 1000 Genomes data, using the statistic: where n is the number of GWAS tested, assuming a distribution.
| Graph | Trait | P | ||
|---|---|---|---|---|
| Seven-leaf tree | Height** | |||
| Educational attainment** | ||||
| Unibrow* | 0.012 | |||
| Male-pattern baldness | 0.000679 | 0.177 | 0.007 | |
| Five-leaf graph | Height** | |||
| Educational attainment** | ||||
| Schizophrenia* | 0.000608 | 0.01 | 0.007 | |
| Unibrow | 0.056 | |||
| Male-pattern baldness | 0.000562 | 0.213 | 0.003 |
We also computed P-values from 1000 samples in which we randomly switched the sign of effect size estimates, to simulate neutrality while preserving the genetic architecture of the traits (). Additionally, we computed P-values from an empirical null distribution produced using 1000 samples, each containing SNPs that were frequency-matched to the trait-associated SNPs, using their allele frequency in CEU, to account for each GWAS’s ascertainment scheme ().
Trait-associated variants for which
Trait-associated variants for which .
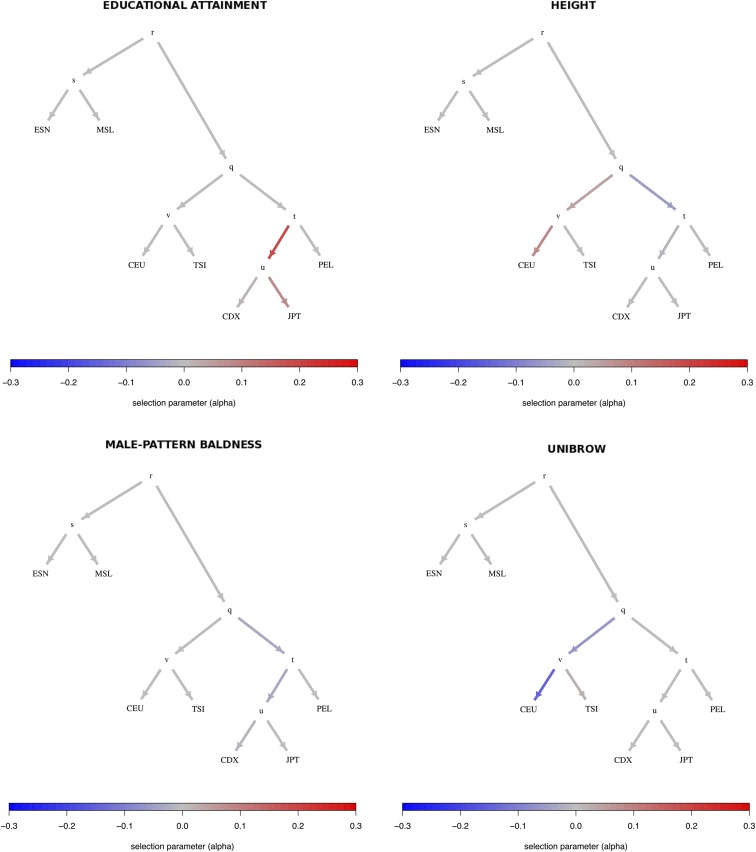
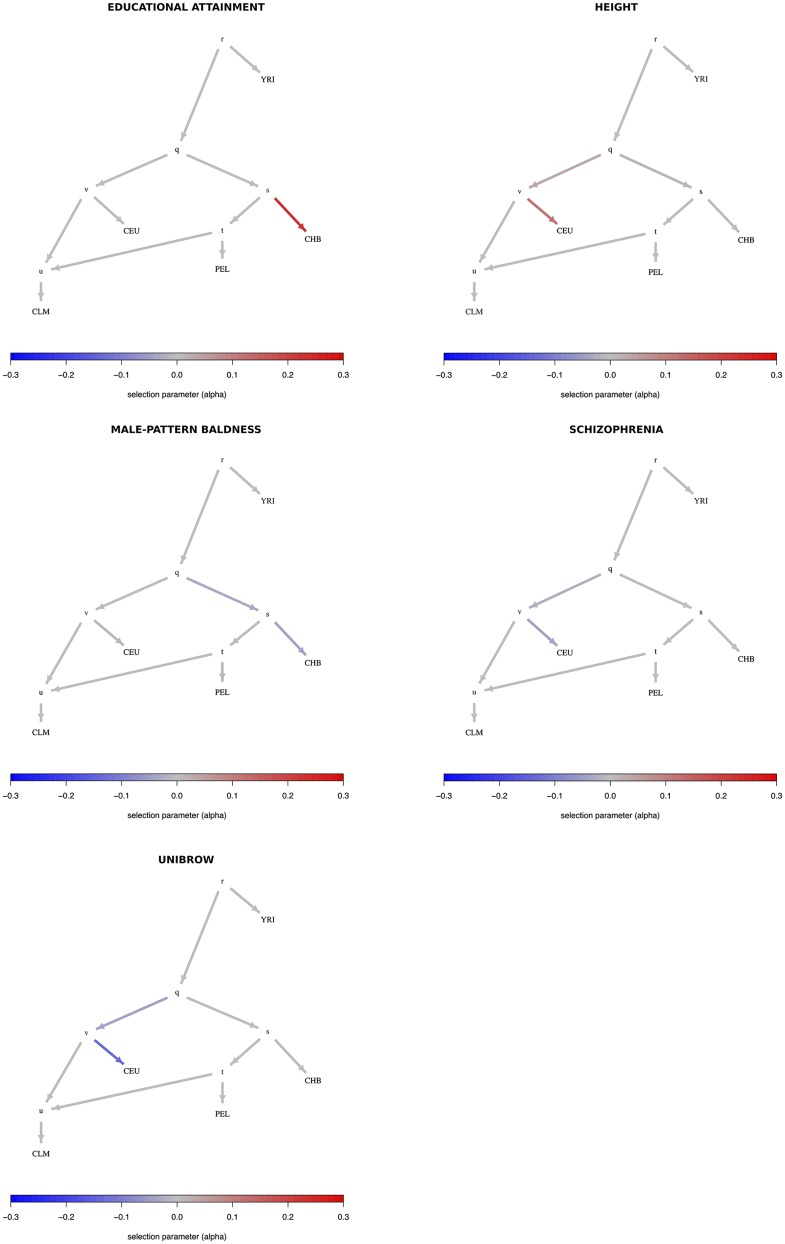
We then ran our MCMC on these trait-associated variants, using the genome-wide-fitted graph, and obtained posterior distributions for the α parameters with the strongest evidence for selection, prioritizing branches as explained above (Figure S34 in File S1). The P-values of the statistic (obtained from a distribution) for each branch are shown in Table S2 in File S1. We find strong evidence for selection on variants associated with height, educational attainment, and self-reported unibrow, but little or no evidence for variants associated with male-pattern baldness or schizophrenia: even though these trait-associated variants are significant under the and frameworks; all their α parameters are approximately centered at 0. For height, we observe both selection for variants increasing height in the ancestral European branch, and for variants decreasing height in the ancestral East Asian/Native American branch. However, this is only a consequence of the MCMC showing alternate strong support for selection in either one or the other branch at different points in the run, but only weak support for selection in both branches simultaneously (Figure S35 in File S1), suggesting we are unable to discern which among these is the correct configuration. We plotted poly-graphs for all traits that passed the significance criterion in the seven-leaf tree (Figure 4).
Figure 4.
Poly-graphs for trait-associated variants that show significant evidence for polygenic adaptation in the seven-leaf tree built using 1000 Genomes allele frequency data. ESN, Nigerian Esan; MSL, Sierra Leone Mende; CEU, Northern Europeans from Utah; TSI, Southern Europeans from Tuscany; CDX, Dai Chinese; JPT, Japanese; PEL, Peruvians.
Given that PEL has European admixture (which can be observed when adding one migration event to the seven-leaf tree in TreeMix, Figure S10 in File S1), we also replaced PEL with an East Asian population (CHB) to verify that selection signals observed in the Eurasian branches were not dependent on poor modeling of PEL as a simple sister group to East Asians (Figure S36 in File S1). Additionally, we tested an alternative set of panels, in which we kept PEL in the tree, but replaced the two European populations—CEU and TSI—by Finnish (FIN) and Iberians (IBS), and the two East Asian populations—JPT and CDX—by Han Chinese (CHB) and Southern Han (CHS) (Figure S37 in File S1). The results from both alternative sets of panels are very similar to our original tree. We also find very similar results when using a Beta(2,2) prior for the root allele frequency in our MCMC (Figure S38 in File S1), instead of the default Uniform[0,1] prior.
We then proceeded to explore a graph with an admixture event (Figure S33B in File S1). This graph contained Yoruba (YRI), Colombians (CLM), CEU, CHB, and PEL. To build this five-leaf graph, we first fit the most additive tree containing CEU, CHB, YRI, and PEL, and then attempted to fit CLM as a putative mixture of branches in the tree (MixMapper residual norm = 1.91e−07). The best-fitting combination was a mixture of the terminal branch leading to CEU () and the terminal branch leading to PEL (), the latter of which is the panel with the highest amount of Native American ancestry in the 1000 Genomes Project (Auton et al. 2015). Here, we recapitulated many of our previous findings from the seven-leaf tree (Figure S39 in File S1 and Table 1), like selection on variants associated with height and educational attainment. We list the P-values of the statistic for each branch in Table S3 in File S1. Poly-graphs of the five-leaf admixture graph are shown in Figure 5.
Figure 5.
Poly-graphs for trait-associated variants that show significant evidence for polygenic adaptation in the five-leaf admixture graph built using 1000 Genomes allele frequency data. CEU, Northern Europeans from Utah; TSI, Southern Europeans from Tuscany; PEL, Peruvians; CLM, Colombians; YRI, Yoruba; CHB, Han Chinese.
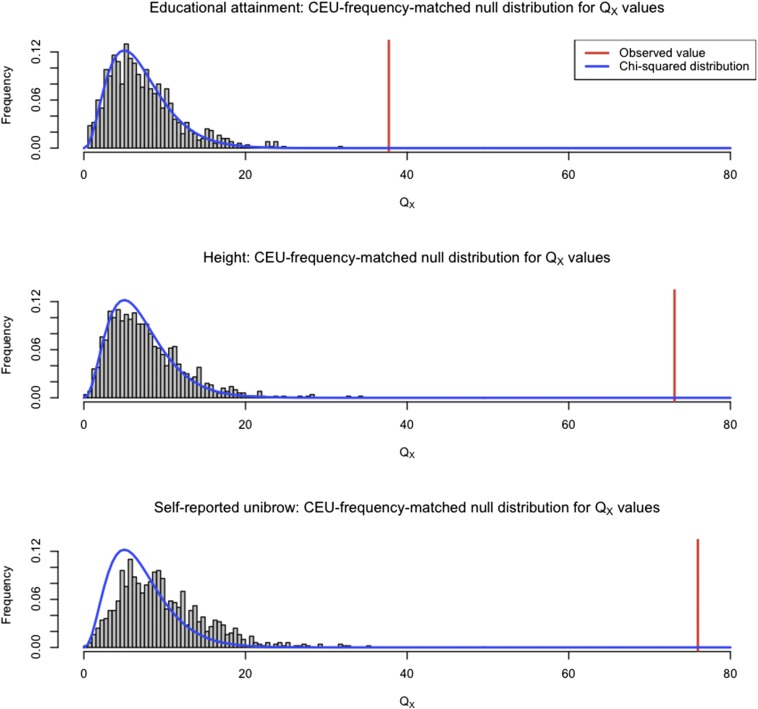
To make sure there were no artifacts due to GWAS ascertainment (Berg and Coop 2014), we also generated an empirical null distribution produced using 1000 samples, each containing SNPs that were frequency-matched to the trait-associated SNPs, using their allele frequency in CEU. We computed the statistic for each of these samples, to obtain an empirical P-value ( in Table 1). We do not observe a value of as high as the one observed in the real data, for either height, educational attainment, or self-reported unibrow (Figure 6).
Figure 6.
We generated an empirical null distribution by sampling SNPs from the genome that matched the CEU allele frequency of the SNPs associated with educational attainment, self-reported unibrow, and height. We generated 1000 samples this way, and computed the statistic for each sample, using the population panels from Figure 4. The value observed in the real data are depicted with a red line. We also plot the density of the corresponding distribution (blue line) for comparison.
To test how robust our results were to our modeling assumptions, we also performed a simpler two-tailed binomial sign test between every pair of 1000 Genomes panels. The assumption here is that—for every panel X and Y—we should observe roughly equal number of trait-increasing alleles at higher frequency in X than in Y as trait-decreasing alleles at higher frequency in X than in Y, under a model of neutrality with respect to the effect size sign (Orr 1998). This test only uses information about the sign of the effect estimates of each SNP, not their magnitudes, and does not use information about genome-wide drift parameters between each population. Thus, it is bound to have less power than the or MCMC tests. The P-values for these pairwise binomial tests are shown in Tables S4–S8 in File S2 for all traits that were found to have significant evidence of selection using the statistic. The top 10 most significant pairwise comparisons are shown in Tables S9–S13 in File S1. For ease of visualization, we also plotted, for each panel, the number of pairwise tests involving that panel that resulted in a P-value (Figures S40–S44 in File S1).
We were interested in verifying how sensitive different proportions of missing data (i.e., removal of SNPs) or erroneous effect size estimates would be to our three strongest signals of polygenic adaptation, on variants associated with height, educational attainment and self-reported unibrow. For this purpose, we focused on the comparison between CEU and CHB. First, we simulated different proportions of missing trait-associated SNPs, ranging from to with step sizes of For each of 10,000 simulations under each missing data scenario, we assessed how often the polygenic score for unibrow and educational attainment in CHB was higher than the polygenic score for CEU, like we observe in the 1000 Genomes data. Height follows the opposite pattern (with CEU having a higher polygenic score than CHB), so in that case we assessed how often its polygenic score in CEU was higher than in CHB, across the 10,000 simulations for each scenario. Note that we built these scores using only the SNPs used in our selection tests. The results are in Figure S45 in File S1. For example, we see that—even with missing data—the polygenic scores for either of the three traits preserve the observed relationship of inequality between CEU and CHB almost of the time. Finally, we simulated a situation in which some proportion of the signs of the effect size estimates were misassigned. We then assessed how often we could replicate the signal we see between CEU and CHB, but this time under different proportions of sign misassignment (Figure S46 in File S1).
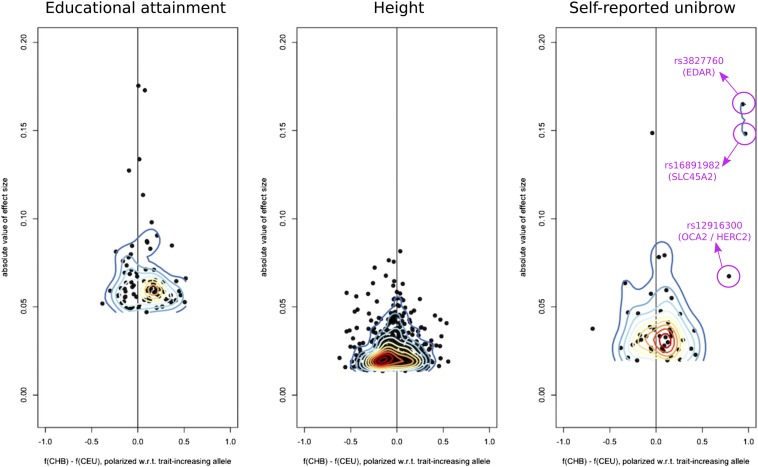
To understand how the signal of selection was distributed among our SNPs, we plotted the absolute value of the effect sizes of trait-associated SNPs for height, educational attainment, and self-reported unibrow, as a function of the difference in frequency observed between CHB and CEU, polarized with respect to the trait-increasing allele in each SNP (Figure 7). We find that, in the case of self-reported unibrow, there are three variants of large effect with large frequency differences contributing to a higher polygenic score in CHB: rs3827760, rs16891982, and rs12916300. These SNPs are located in the genes EDAR, SLC45A2, and OCA2/HERC2. These are genes involved in pigmentation and skin development, and all three have documented signatures of selective sweeps causing strong allele frequency differences between Europeans and East Asians (Bersaglieri et al. 2004; Voight et al. 2006; Sabeti et al. 2007; Williamson et al. 2007; Mathieson et al. 2015). After removing SNPs with large absolute effect size values (), the P-value of the statistic for these variants remains significant (). When looking at the other two sets (variants associated with height and educational attainment), the signal of selection is more uniformly distributed among the SNPs, with no strong outliers of large effect with large frequency differences (Figure 7).
Figure 7.
We plotted the absolute value of the effect sizes of trait-associated SNPs for height, educational attainment, and self-reported unibrow, as a function of the difference in frequency observed between CHB and CEU. The panel frequencies are polarized with respect to the trait-increasing allele. We then overlaid a contour plot over each scatter-plot.
Application to Human Origins SNP chip data
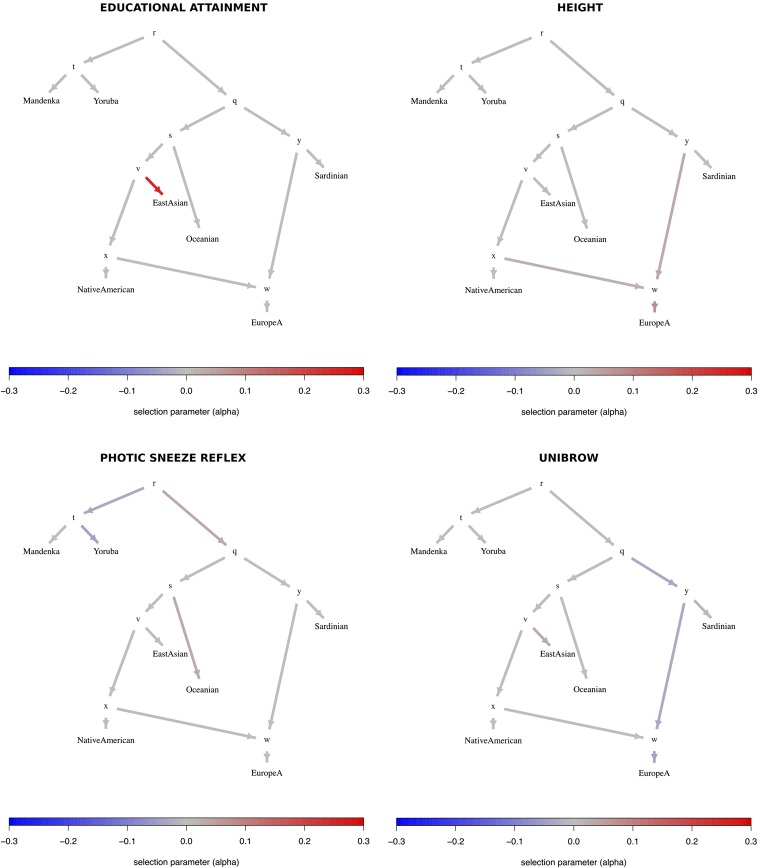
We also applied our method to the imputed Human Origins SNP chip dataset (Patterson et al. 2012; Lazaridis et al. 2014). We tested for polygenic adaptation in a seven-leaf admixture graph. This graph contains the panels Yoruba, Mandenka, and Sardinian, along with the following four combinations of panels, which we built so as to have a large number of individuals per panel. The panel “Oceanian” contains the panels Papuan and Australian. The panel “EastAsian” contains the panels Cambodian, Mongola, Xibo, Daur, Hezhen, Oroqen, Naxi, Yi, Japanese, Han_NChina, Lahu, Miao, She, Han, Tujia, and Dai. The panel “NativeAmerican” contains the panels Maya, Pima, Surui, Karitiana, and Colombian. Finally, we modeled Europeans as a two-way mixture of an ancestral component related to “NativeAmerican,” and another component that split basally from the Eurasian tree and is a sister to Sardinians. This was the mixture fitted to Europeans by Lipson et al. (2013), and provides a better fit to the data than modeling Europeans merely as a sister group to East Asians and Native Americans. Though we recognize that Europeans are better modeled as a three- or four-way mixture of ancestral components (Lazaridis et al. 2014, 2016; Haak et al. 2015), it is hard to produce such a mixture without resorting to ancient DNA data (see Discussion). We tested three different versions of this graph, each containing three different sets of European populations (Figure S33C in File S1) distinguished by how much “early European farmer” (EEF) ancestry they had [based on Figure 4 of Lazaridis et al. (2014)]. “EuropeA” (low EEF) contains the following panels: Estonian, Lithuanian, Scottish, Icelandic, Norwegian, Orcadian, Czech, and English. “EuropeB” (medium EEF) contains Hungarian, Croatian, French, Basque, Spanish_North, and French_South. Finally, “EuropeC” (high EEF) contains Bulgarian, Bergamo, Tuscan, Albanian, Greek, and Spanish. The MixMapper residual norm was equal to 6.5e−07, 1.08e−06 and 1.97e−06, when fitting EuropeA, EuropeB, and EuropeC, respectively.
Trait-associated variants with significant evidence for polygenic adaptation are listed in Table 2 and the P-values of the statistic for each branch are shown in Tables S14–S16 in File S1. With these data, we are able to recapitulate the adaptive increase in height-increasing variants in Europeans we had seen before, but only observe it in populations with medium or low EEF ancestry (Figure 8 and Figures S47–S51 in File S1). This pattern is consistent with previous observations made using ancient DNA in Europeans (Mathieson et al. 2015). We also recapitulate selection patterns on variants associated with other traits, like self-reported unibrow, educational attainment, and self-reported male-pattern baldness, and observe evidence for polygenic adaptation in some additional trait-associated variants, like self-reported photic sneeze reflex (Table 2).
Table 2. Trait-associated variants with Bonferroni-corrected significant evidence of being under polygenic adaptation in the Lazaridis et al. (2014) dataset, using the statistic: where n is the number of GWAS tested, assuming a distribution.
| Graph | Trait | P | ||
|---|---|---|---|---|
| Seven-leaf graph (w/EuropeA) | Height** | |||
| Photic sneeze reflex** | 0.00087 | 0.006 | ||
| Educational attainment** | 0.001 | 0.001 | ||
| Unibrow | 0.173 | |||
| Seven-leaf graph (w/EuropeB) | Height** | |||
| Educational attainment** | 0.001 | |||
| Age at voice drop** | 0.000953 | 0.001 | ||
| Photic sneeze reflex** | 0.00086 | 0.001 | ||
| Unibrow | 0.166 | |||
| Seven-leaf graph (w/EuropeC) | Educational attainment** | |||
| Photic sneeze reflex | 0.000653 | 0.004 | 0.004 | |
| Unibrow | 0.199 |
We tested three different graphs with different sets of European panels, containing either low (EuropeA), medium (EuropeB), or high (EuropeC) EEF ancestry. We also computed P-values from 1000 samples in which we randomly switched the sign of effect size estimates, to simulate neutrality while preserving the genetic architecture of the traits (). Additionally, we computed P-values from an empirical null distribution produced using 1000 samples, each containing SNPs that were frequency-matched to the trait-associated SNPs, using their allele frequency in Europeans, to account for each GWAS’s ascertainment scheme ().
Trait-associated variants for which
Trait-associated variants for which .
Figure 8.
Poly-graphs for trait-associated variants that show significant evidence for polygenic adaptation in the seven-leaf admixture graph built using the Lazaridis et al. (2014) dataset and including the set of European populations with low EEF ancestry (“EuropeA”).
To check if there were systematic biases in ancestral/derived allele polarity relative to the direction of the effect size, we performed a two-tailed binomial test on each trait for which we found significant evidence of polygenic adaptation in the Lazaridis et al. (2014) or the 1000 Genomes dataset (Table S17 in File S1). We find that only schizophrenia has a significant bias, showing an excess of derived alleles with negative effect sizes (P = 0.03511), though this is not significant after Bonferroni correction. We therefore caution that the evolution of schizophrenia-associated variants may not be well-modeled by the multivariate Normal assumptions that we make to calculate the statistic or when running the MCMC.
As before, to check the robustness of our results to our modeling assumptions, we show P-values for pairwise binomial sign tests involving each of the panels in the Lazaridis et al. (2014) dataset in Tables S18–S22 in File S2. The top 10 most significant pairwise comparisons are in Tables S23–S26 in File S1, with the exception of self-reported age at voice drop, in which all comparisons had P-values due to the small number of associated SNPs. Figures S52–S55 in File S1 show, for each panel, the number of pairwise tests involving that panel that resulted in a P-value
Replication using summary statistics from the UK Biobank
Given the potential contentiousness of our educational attainment signal, we aimed to determine whether the same global patterns were observed using summary statistics from a GWAS performed on an independent cohort. For this, we resorted to the GWAS set released by the Neale laboratory (http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank) and performed on the UK Biobank cohort (Bycroft et al. 2017). Though we could not find “number of years of education” in the Neale laboratory GWAS, we found “college/university degree” and used this as a measure of educational attainment. We LD-partitioned the summary statistics in the same way as before, and then selected the SNP with the lowest P-value from each block. At we obtain a similar number of SNPs (96) as we had for the association study from Okbay et al. (2016) (86), and we find a correlation between the population genetic scores made using the data from Okbay et al. (2016) and the scores made using the UK Biobank (Figure S56 in File S1), each standardized by their respective between-population SD. At and the correlation is reduced to and respectively. Using the cutoff, we find a marginally significant overall statistic when looking at the seven populations from Figure 4 ( and ) and a significant statistic in the ancestral East Asian branch () and the terminal JPT branch (), but not in any of the other branches (). The α parameters of these two branches estimated from the PolyGraph MCMC are also positive (Figure S57 in File S1), though their magnitude is not as large as the ones obtained from the summary statistics of Okbay et al. (2016).
Discussion
We have developed a method to infer polygenic adaptation on trait-associated variants in an admixture graph, so as to be able to pinpoint where in the history of a set of populations this type of selective processes took place. Our method requires GWAS data for a particular trait, allele frequency data for a set of populations, and a precomputed admixture graph that relates these populations with each other. Importantly, the method relies on the admixture graph as an accurate description of the ancestral genome-wide relationships among the populations under study. Potential users should be careful about correctly estimating branch lengths and ghost populations that are not included in the graph but may have substantial unaccounted ancestry contributions to the populations that are included. We used MixMapper (Lipson et al. 2013) to infer the graph topology and branch lengths. Alternatively, one can also use other programs, like qpGraph (Patterson et al. 2012) or TreeMix (Pickrell and Pritchard 2012) to build graphs, though we caution that the estimated drift values of the branches in the output of these programs are scaled (in different ways) by the heterozygosity of ancestral nodes [see Supplemental Material of Lipson et al. (2013) for a way to properly obtain drift values from differences in allele frequencies between populations].
Running PolyGraph involves a two-step process, each of which is complementary to the other. The first step—the calculation of the statistic—is fast and provides a preliminary way to assess which branch in a graph has significant evidence for polygenic adaptation. However, this statistic does not model the ancestral allele frequencies at each node of the graph. The second step—the MCMC—is slower, but provides posterior distributions for selection parameters under a more parameter-rich model of population history. In our pipeline, we use the first method as a filtering step, to avoid exploring selection parameters in the MCMC for those branches that have little evidence for selection, and encourage the MCMC to be sparse in its assignments of selection in the graph. We illustrate this point in Figure S58 in File S1, where we show a side-by-side comparison of a poly-graph built using the posterior α estimates and a poly-graph built using the signed version of the statistic.
In application to human populations, we detected signals of polygenic adaptation on sets of variants that have been identified to influence height, educational attainment, and self-reported unibrow. Selection on variants associated with height in Europeans has been previously reported elsewhere (Turchin et al. 2012; Berg and Coop 2014; Mathieson et al. 2015; Robinson et al. 2015) and our results are consistent with previous findings showing that height-increasing variants are at significantly and systematically higher frequencies in northern than in southern European populations. The signal for selection affecting variants associated with self-reported unibrow is also strong, but partly driven by a few variants of large effect with large frequency differences between populations, which have documented evidence for selective sweeps in genes involved in hair, skin and eye pigmentation, and skin development (Bersaglieri et al. 2004; Voight et al. 2006; Sabeti et al. 2007; Williamson et al. 2007; Mathieson et al. 2015). Additional trait-associated variants had inconsistent evidence across datasets and graph frameworks (like schizophrenia or male-pattern baldness) and/or are driven by differences in only a few SNPs of small effect (like age at voice drop), and so we do not discuss them.
We find preliminary evidence for polygenic adaptation in East Asian populations at variants that have been associated with educational attainment in European GWAS. This result is robust to the choice of population allele frequency data we used [1000 Genomes or Lazaridis et al. (2014) panels], to the choice of GWAS summary statistics (Figure S56 in File S1), to GWAS ascertainment (Figure 6), and to our modeling assumptions, as we found a significant difference between East Asian and non-East-Asian populations even when performing a simple binomial sign test (Tables S4 and S19 in File S2 and Tables S9 and S23 in File S1). However, we caution that this pends further verification via more GWAS on the same trait. Our modeling framework suggests that, if selection truly operated on these variants, it must have done so before or early in the process of divergence among East Asian populations—at least as far back as 5 KYA (Stoneking and Delfin 2010; Fu et al. 2013; Lu et al. 2016; Wong et al. 2017)—because the signal is common to different East Asian populations (Han Chinese, Dai Chinese, Japanese, Koreans, etc.). The signal seems only very weakly present in some Siberian populations (e.g., the Even and Nganasan, and some Native American populations (e.g., the Mixe and Pima), and not present at all in other Native American populations (e.g., the Surui, Quechua, and Karitiana). This is perhaps explained by the complex demographic make-up of Siberian and Native American populations, and their divergent history from East Asians (Raghavan et al. 2015; Skoglund et al. 2015; Pugach et al. 2016).
Interpreting the educational attainment signal, and the other signals we found, requires awareness of a number of technical caveats, as well as several fundamental conceptual difficulties with the study of polygenic adaptation, some of which may ultimately prove intractable.
What is the signal of polygenic adaptation?
Before discussing these difficulties, it is worth articulating exactly what a signal of polygenic adaptation consists of. Taking the height example as a case in point, the signal is that a set of genetic variants that have been identified as associated with increased height in a European GWAS are (as a class) at higher frequency in northern Europeans today than would be expected by genetic drift alone. Though this observation is consistent with the hypothesis that natural selection has operated on these variants, it does not necessarily imply that natural selection has operated directly on “height,” nor that observed height differences between northern Europeans and other populations are necessarily genetic and due to selection.
Pleiotropy and phenotype definition
When looking at all our variant classes, we are necessarily limited by the traits that have been defined and studied by others, as we have grouped variants together based on these phenotypes. The use of previously established definitions makes it difficult to understand exactly why these variants may have been under selection in the past. This is perhaps best exemplified by the signal of polygenic adaptation for genetic variants associated with educational attainment.
Standardized schooling, and, consequently, the concept of “educational attainment,” was only invented and implemented widely in the last few generations. It is obviously nonsensical to discuss its evolution over the past tens of thousands of years. Instead, it is likely that the set of variants for which we find evidence of selection was associated with some (unknown) phenotype(s) in the past. However, given that selection on these variants likely took place >5 KYA, it may be difficult or impossible to identify what these were. A similar problem arises when thinking about the signal of polygenic adaptation on “unibrow.” This is a self-reported phenotype, and the genetic variants that have been identified may simply be associated with pigmentation (assuming people with certain hair and/or skin pigmentation phenotypes are more likely to notice they have hair between their eyes), or alternatively with some other (unmeasured) hair-related phenotype. It is also possible that direct sexual selection for absence or presence of unibrow as an attractive facial feature in certain cultures (Vashi and Quay 2015) may be the cause of this signal. Indeed, if a selective agent is cultural, but the culture has since changed, it may be impossible to determine what actually occurred. All these variants are also likely pleiotropic (Simons et al. 2017), which makes it even harder to determine which phenotypes were truly targeted by selection.
Perhaps, one could try to find the phenotypic gradients along which selection most likely operated (Lande and Arnold 1983) by modeling the evolution of trait-associated SNPs for multiple phenotypes together. However, it is also possible that genetic correlations among traits in the present are not good proxies for genetic correlations in the past.
Relationship between polygenic scores and population mean phenotypes
Another fundamental limitation in interpreting all studies of polygenic adaptation (including this one) is that the connection between the distribution of allele frequencies today and any historical or geographic trends in phenotypes remains questionable. Indeed, though we have motivated this method as a way to identify adaptive shifts in the mean of a polygenic trait, it is a simple fact that massive changes in the mean values of many of the traits we consider have occurred by purely nongenetic environmental processes. For example, the mean height of men in the Netherlands increased from ∼166 cm in the mid-1800s to currently over 180 cm (Hatton and Bray 2010), bringing the population from around the middle of the pack among European countries to the tallest one in the world. This likely occurred for environmental reasons, such as improvements in diet and health care (Stulp et al. 2015; Tarka et al. 2015). Likewise, the average educational attainment in Iceland and North America has increased dramatically over the past century, despite a slight estimated decrease in the frequencies of genetic variants associated with an increased value of the phenotype (Beauchamp 2016; Courtiol et al. 2016; Kong et al. 2017). The somewhat paradoxical conclusion is that actual phenotypes can and do change across populations in directions that are uncorrelated to natural selection (which may in fact be a minor contributor to any such differences). It would be an understatement to say this poses challenges for the interpretation of the current study and others like it.
In fact, the trait-associated variants that we have used only explain a fraction of the narrow-sense heritability of their respective traits, even in the populations in which the association studies were performed. As we have only looked at variants that have high probability of association with a trait, this fraction is small in most cases. For example, the heritability for “educational attainment” is estimated to be ∼ and educational attainment itself is strongly determined by environmental factors (Rietveld et al. 2013). The SNPs we used in this study [themselves a subset of all SNPs tested in the original GWAS (Okbay et al. 2016)] explain only of the total variance for this particular trait. All of the aforementioned traits are likely affected by a myriad of environmental and social variables, which might contribute to determine their ultimate expression in each human individual.
Additional caveats
Beyond the above conceptual difficulties, there are a number of additional caveats with our approach to keep in mind. First, the effect sizes we have used derive from GWAS performed primarily on individuals of European ancestry. Thus, our tests can only detect if variants that have been found to be associated with a trait in European GWAS are significantly higher or lower in a particular (European or non-European) population, relative to what they should be under a pure drift model. This does not necessarily imply that populations for which we find evidence for selection have higher or lower average genetic values of such a trait than other populations. In fact, there is evidence to suggest that loci ascertained in European GWAS do not serve to make good predictors for traits in populations that are distantly related to Europeans (Martin et al. 2016). One reason for this is that many or all of the traits we are studying are likely to be influenced in non-European populations by different variants from the ones that have been discovered in European GWAS. SNPs that may be strongly associated with a trait in a particular non-European population (like an African or East Asian panel) may not have reached genome-wide significance in a European GWAS, where those SNPs may not strongly affect the trait or may be at low frequencies. It is thus possible that there are variants associated with traits like educational attainment that occur at high frequencies in East Asians, but that are missing from our analysis, or that the effect sizes in trait-associated SNPs are different in non-European populations, in such a way that the average genetic values between these populations and Europeans are not significantly different. We also do not model dominance, epistasis or gene-by-environment interactions between our trait-associated variants and the diverse environments that human populations occupy, and any of these factors may further obscure the relationship between the patterns we observe and the actual underlying genetic contribution to phenotypes in these populations.
Second, we have assumed that all of the GWAS that we have used have properly accounted for population structure. If some of the trait-associated SNPs are, in fact, false positives caused by uncorrected structure, this could generate a false signal of polygenic adaptation. A future direction could be the incorporation of effect sizes that have been corrected for ancestry or population stratification (Robinson et al. 2015, 2017), and also effect sizes from GWAS performed on other populations (Moltke et al. 2014; Ng et al. 2014; Wen et al. 2016), in order to assess the robustness of our empirical results across variants discovered in studies involving participants of different ancestries.
Third, we made the assumption that the admixture graph for the populations that we use as input is correct. If there are additional unmodeled aspects of the history of the populations, this could induce incorrect inference about the branch on which natural selection has occurred. We also recommend that the individuals in the population panels used as leaves in the graphs have roughly similar amounts of admixture. In other words, the method works best when admixture in the population was ancient enough for the admixed ancestry to have spread uniformly among members of the admixed panel. Otherwise, an admixture graph may not be the most appropriate way to model their evolution.
Finally, we have made an explicit assumption that our model should be sparse; i.e., that polygenic adaptation is rare. If in reality adaptation is common, the PolyGraph approach will necessarily only identify selection on a small number of branches.
Future directions
A natural extension to the analyses we performed here would be to look at admixture graphs that include extinct populations or species, using ancient DNA (Slatkin and Racimo 2016). For example, present-day Europeans are known to have resulted from admixture processes involving at least four ancestral populations (Lazaridis et al. 2014, 2016), and so modeling them as a sister group to East Asians, or as a two-way mixture between a Native American-related component and a basal Eurasian component, may be overly simplistic. Incorporating ancient DNA would not require any additional theoretical work, as ancient populations can be naturally included as leaves in an admixture graph (Lazaridis et al. 2014; Mathieson et al. 2015). Care should be taken, however, in making sure that the quality of the ancient DNA data at trait-associated SNPs is accounted for while inferring the number of ancestral and derived alleles, and that there is a sufficient number of ancient individuals per population to detect polygenic adaptation. One could envision either performing pseudohaploid sampling (Patterson et al. 2012; Haak et al. 2015; Mathieson et al. 2015) or using allele frequency estimators obtained from genotype likelihoods (Nielsen et al. 2012; Korneliussen et al. 2014), while accounting for errors characteristic of ancient DNA (Jónsson et al. 2013; Soraggi et al. 2017). When working with SNP capture data (Patterson et al. 2012; Haak et al. 2015), it may be necessary to perform imputation at the GWAS SNPs, if these were not originally covered in the SNP capture array. We aim to tackle these issues in a future study.
One concern when analyzing admixture graphs is identifiability. As we mentioned before, there are multiple configurations of the α parameters that may lead to almost identical likelihoods. The use of the spike-and-slab prior and the filtering step serve to ameliorate this problem, assuming selection was sparse and only affected a few branches. An avenue of research could involve testing other types of models or constraints that may serve to better compare among different selection configurations, perhaps without having to reduce the space of possible candidate branches a priori, for example using reversible-jump MCMC for model selection (Green 1995).
In the future, it may be worth incorporating stabilizing selection into this method (Simons et al. 2017), or exploring tests of polygenic adaptation in the context of other types of demographic frameworks, like isolation-by-distance (Wright 1943) or population structure (Pritchard et al. 2000) models. For example, one could envision settings in which trait-associated variants would be best modeled as expanding or contracting over a geographically extended area over time, in a way that is not explainable by genetic drift alone.
Lastly, we note that, despite some clear methodological and conceptual differences, our method bears a close relationship to a number of methods for inferring changes in the rate of phenotypic evolution on species phylogenies over macroevolutionary timescales. Our use of the Normal model of drift as an approximation to the Wright-Fisher diffusion is closely analogous to the use of Brownian motion models in some phylogenetic methods (Eastman et al. 2011; Venditti et al. 2011; Revell et al. 2012; Rabosky et al. 2013; Jhwueng and O’Meara 2015). It may also be worth exploring the relationship between Ornstein-Uhlenbeck models for phenotypic evolution on phylogenies (Uyeda and Harmon 2014; Khabbazian et al. 2016) and the aforementioned hypothetical extension of our method to include stabilizing selection, as the two processes are closely related (Lande 1976; Simons et al. 2017).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300148/-/DC1.
Acknowledgments
We thank Claude Bhérer, Guy Sella, Molly Przeworski, Graham Coop, Joshua Schraiber, Simon Myers, Benjamin Peter, and members of the Sella and Przeworski laboratories for helpful advice and discussions. We also thank an anonymous reviewer for valuable feedback, and Thomas Mailund for assistance with running the admixturegraph R package as well as helpful comments on the manuscript. Finally, we thank Mark Lipson for help with running MixMapper. This work was supported by National Institute of General Medical Science (NIGMS) grant 1R01GM121372-01 to J.K.P.
Note added in proof: See Novembre and Barton 2018 (pp. 1351–1355) in this issue for a related work.
Footnotes
Communicating editor: J. Novembre
Literature Cited
- Auton A., Abecasis G. R., Altshuler D. M., Durbin R. M., Bentley D. R., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., de Vladar H. P., 2009. Statistical mechanics and the evolution of polygenic quantitative traits. Genetics 181: 997–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp J. P., 2016. Genetic evidence for natural selection in humans in the contemporary United States. Proc. Natl. Acad. Sci. USA 113: 7774–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. J., Coop G., 2014. A population genetic signal of polygenic adaptation. PLoS Genet. 10: e1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. J., Zhang X., Coop G., 2017. Polygenic adaptation has impacted multiple anthropometric traits. bioRxiv. https://doi.org/10.1101/167551. [Google Scholar]
- Berisa T., Pickrell J. K., 2016. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics 32: 283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T., Sabeti P. C., Patterson N., Vanderploeg T., Schaffner S. F., et al. , 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74: 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft, C., C. Freeman, D. Petkova, G. Band, L. T. Elliott et al., 2017 Genome wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. DOI: 10.1101/166298. [Google Scholar]
- Cavalli-Sforza L. L., Barrai I., Edwards A., 1964. Analysis of human evolution under random genetic drift. Cold Spring Harb. Symp. Quant. Biol. 29: 9–20. [DOI] [PubMed] [Google Scholar]
- Chen H., Patterson N., Reich D., 2010. Population differentiation as a test for selective sweeps. Genome Res. 20: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Witonsky D., Di Rienzo A., Pritchard J. K., 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtiol A., Tropf F. C., Mills M. C., 2016. When genes and environment disagree: making sense of trends in recent human evolution. Proc. Natl. Acad. Sci. USA 113: 7693–7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Burgess Publishing Co., Minneapolis. [Google Scholar]
- Csardi G., Nepusz T., 2006. The igraph software package for complex network research. Int. J. Complex Sys. 2006: 1695. [Google Scholar]
- Das S., Forer L., Schönherr S., Sidore C., Locke A. E., et al. , 2016. Next-generation genotype imputation service and methods. Nat. Genet. 48: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O., Zagury J.-F., Marchini J., 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10: 5–6. [DOI] [PubMed] [Google Scholar]
- Eastman J. M., Alfaro M. E., Joyce P., Hipp A. L., Harmon L. J., 2011. A novel comparative method for identifying shifts in the rate of character evolution on trees. Evolution 65: 3578–3589. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1973. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25: 471. [PMC free article] [PubMed] [Google Scholar]
- Field Y., Boyle E. A., Telis N., Gao Z., Gaulton K. J., et al. , 2016. Detection of human adaptation during the past 2000 years. Science 354: 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Meyer M., Gao X., Stenzel U., Burbano H. A., et al. , 2013. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. USA 110: 2223–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., et al. , 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349: 1343–1347. [DOI] [PubMed] [Google Scholar]
- George E. I., McCulloch R. E., 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88: 881–889. [Google Scholar]
- Green P. J., 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82: 711–732. [Google Scholar]
- Günther T., Coop G., 2013. Robust identification of local adaptation from allele frequencies. Genetics 195: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., et al. , 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton T. J., Bray B. E., 2010. Long run trends in the heights of European men, 19th–20th centuries. Econ. Hum. Biol. 8: 405–413. [DOI] [PubMed] [Google Scholar]
- Jhwueng D.-C., O’Meara B., 2015. Trait evolution on phylogenetic networks. bioRxiv. https://doi.org/10.1101/023986. [Google Scholar]
- Jónsson H., Ginolhac A., Schubert M., Johnson P. L., Orlando L., 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29: 1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabbazian M., Kriebel R., Rohe K., Ané C., 2016. Fast and accurate detection of evolutionary shifts in Ornstein–Uhlenbeck models. Methods Ecol. Evol. 7: 811–824. [Google Scholar]
- Kong A., Frigge M. L., Thorleifsson G., Stefansson H., Young A. I., et al. , 2017. Selection against variants in the genome associated with educational attainment. Proc. Natl. Acad. Sci. USA 114: E727–E732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. Angsd: analysis of next generation sequencing data. BMC Bioinformatics 15: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30: 314–334. [DOI] [PubMed] [Google Scholar]
- Lande R., Arnold S. J., 1983. The measurement of selection on correlated characters. Evolution 37: 1210–1226. [DOI] [PubMed] [Google Scholar]
- Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., et al. , 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N., et al. , 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppälä K., Nielsen S. V., Mailund T., 2017. admixturegraph: an R package for admixture graph manipulation and fitting. Bioinformatics 33: 1738–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M., Loh P.-R., Levin A., Reich D., Patterson N., et al. , 2013. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol. Biol. Evol. 30: 1788–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lou H., Yuan K., Wang X., Wang Y., et al. , 2016. Ancestral origins and genetic history of Tibetan highlanders. Am. J. Hum. Genet. 99: 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. R., Gignoux C. R., Walters R. K., Wojcik G. L., Neale B. M., et al. , 2016. Human demographic history impacts genetic risk prediction across diverse populations. bioRxiv. https://doi.org/10.1101/070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., McVean G., 2013. Estimating selection coefficients in spatially structured populations from time series data of allele frequencies. Genetics 193: 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., et al. , 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. J., Beauchamp J. J., 1988. Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83: 1023–1032. [Google Scholar]
- Moltke I., Grarup N., Jørgensen M. E., Bjerregaard P., Treebak J. T., et al. , 2014. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512: 190–193. [DOI] [PubMed] [Google Scholar]
- Ng M. C., Shriner D., Chen B. H., Li J., Chen W.-M., et al. , 2014. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 10: e1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson G., Smith A. V., Jónsson F., Gústafsson Ó., Stefánsson K., et al. , 2002. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J. R. Stat. Soc. Series B Stat. Methodol. 64: 695–715. [Google Scholar]
- Nielsen R., Korneliussen T., Albrechtsen A., Li Y., Wang J., 2012. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One 7: e37558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A., Beauchamp J. P., Fontana M. A., Lee J. J., Pers T. H., et al. , 2016. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovaskainen O., Karhunen M., Zheng C., Arias J. M. C., Merilä J., 2011. A new method to uncover signatures of divergent and stabilizing selection in quantitative traits. Genetics 189: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., et al. , 2012. Ancient admixture in human history. Genetics 192: 1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., 2014. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94: 559–573 (erratum: Am. J. Hum. Genet. 95: 126). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Pritchard J. K., 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8: e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Berisa T., Liu J. Z., Ségurel L., Tung J. Y., et al. , 2016. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Di Rienzo A., 2010. Adaptation—not by sweeps alone. Nat. Rev. Genet. 11: 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I., Matveev R., Spitsyn V., Makarov S., Novgorodov I., et al. , 2016. The complex admixture history and recent southern origins of Siberian populations. Mol. Biol. Evol. 33: 1777–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky D. L., Santini F., Eastman J., Smith S. A., Sidlauskas B., et al. , 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4: 1958. [DOI] [PubMed] [Google Scholar]
- Racimo F., 2016. Testing for ancient selection using cross-population allele frequency differentiation. Genetics 202: 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M., Steinrücken M., Harris K., Schiffels S., Rasmussen S., et al. , 2015. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349: aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell L. J., Mahler D. L., Peres-Neto P. R., Redelings B. D., 2012. A new phylogenetic method for identifying exceptional phenotypic diversification. Evolution 66: 135–146. [DOI] [PubMed] [Google Scholar]
- Rietveld C. A., Medland S. E., Derringer J., Yang J., Esko T., et al. , 2013. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340: 1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. R., Hemani G., Medina-Gomez C., Mezzavilla M., Esko T., et al. , 2015. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 47: 1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. R., Kleinman A., Graff M., Vinkhuyzen A. A., Couper D., et al. 2017. Genetic evidence of assortative mating in humans. Nature Human Behaviour 1: 0016. [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Varilly P., Fry B., Lohmueller J., Hostetter E., et al. , 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y. B., Bullaughey K., Hudson R. R., Sella G., 2017. A model for the genetic architecture of quantitative traits under stabilizing selection. arXiv. https://doi.org/10.1101/219238.
- Skoglund P., Mallick S., Bortolini M. C., Chennagiri N., Hünemeier T., et al. , 2015. Genetic evidence for two founding populations of the Americas. Nature 525: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Racimo F., 2016. Ancient DNA and human history. Proc. Natl. Acad. Sci. USA 113: 6380–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soraggi S., Wiuf C., Albrechtsen A., 2017. Improved D-statistic for low-coverage data. bioRxiv. https://doi.org/10.1101/146043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M., Delfin F., 2010. The human genetic history of East Asia: weaving a complex tapestry. Curr. Biol. 20: R188–R193. [DOI] [PubMed] [Google Scholar]
- Stulp G., Barrett L., Tropf F. C., Mills M., 2015. Does natural selection favour taller stature among the tallest people on Earth? Proc. Biol. Sci. 282: 20150211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarka M., Bolstad G. H., Wacker S., Räsänen K., Hansen T. F., et al. , 2015. Did natural selection make the Dutch taller? A cautionary note on the importance of quantification in understanding evolution. Evolution 69: 3204–3206. [DOI] [PubMed] [Google Scholar]
- Turchin M. C., Chiang C. W., Palmer C. D., Sankararaman S., Reich D., et al. , 2012. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 44: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda J. C., Harmon L. J., 2014. A novel Bayesian method for inferring and interpreting the dynamics of adaptive landscapes from phylogenetic comparative data. Syst. Biol. 63: 902–918. [DOI] [PubMed] [Google Scholar]
- Vashi N. A., Quay E. R., 2015. Subjective aspects of beauty, pp. 63–81 in Beauty and Body Dysmorphic Disorder. Springer, Berlin. [Google Scholar]
- Venditti C., Meade A., Pagel M., 2011. Multiple routes to mammalian diversity. Nature 479: 393–396. [DOI] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72 (erratum: PLoS Biol. 4: e154); [corrigenda: PLoS Biol. 5: e147 (2007)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Kato N., Hwang J.-Y., Guo X., Tabara Y., et al. , 2016. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci. Rep. 6: 17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. H., Hubisz M. J., Clark A. G., Payseur B. A., Bustamante C. D., et al. , 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. H., Khrunin A., Nichols L., Pushkarev D., Khokhrin D., et al. , 2017. Reconstructing genetic history of Siberian and Northeastern European populations. Genome Res. 27: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1943. Isolation by distance. Genetics 28: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1949. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X. P., et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.