Abstract
The Epstein-Barr virus (EBV) genome encodes several hundred transcripts. We have used ribosome profiling to characterize viral translation in infected cells and map new translation initiation sites. We show here that EBV transcripts are translated with highly variable efficiency, owing to variable transcription and translation rates, variable ribosome recruitment to the leader region and coverage by monosomes versus polysomes. Some transcripts were hardly translated, others mainly carried monosomes, showed ribosome accumulation in leader regions and most likely represent non-coding RNAs. A similar process was visible for a subset of lytic genes including the key transactivators BZLF1 and BRLF1 in cells infected with weakly replicating EBV strains. This suggests that ribosome trapping, particularly in the leader region, represents a new checkpoint for the repression of lytic replication. We could identify 25 upstream open reading frames (uORFs) located upstream of coding transcripts that displayed 5′ leader ribosome trapping, six of which were located in the leader region shared by many latent transcripts. These uORFs repressed viral translation and are likely to play an important role in the regulation of EBV translation.
INTRODUCTION
The Epstein-Barr virus (EBV) is a γ-herpesvirus that infects the majority of the human population and is associated with the development of ∼2% of tumors worldwide (1,2). The virus establishes lifelong latency in B lymphocytes that form the reservoir of the virus from which it can occasionally reactivate (3,4). EBV, like other herpesviruses, has a large DNA genome on which more than 70 proteins but also non-coding RNAs including miRNAs, a snoRNA and possibly long non-coding RNAs are encoded (2,5–7). The protein expression pattern of the virus appears to be tightly regulated. In infected B cells, some viral strains such as B95-8 nearly exclusively induce latency, characterized by the expression of the 8 EBV latent genes that belong to the EBNA and LMP gene families. This process results in unlimited cell proliferation and the establishment of continuously growing lymphoblastoid cell lines (LCLs) (2). In EBV-infected tumors and in specialized B cell types such as germinal center B cells, the latent protein expression pattern can be restricted to a subset of these proteins or even completely vanish (2,8). In infected epithelial cells and in B cells infected with virus strains frequently found in nasopharyngeal carcinoma such as M81, the virus undergoes lytic replication, a process that leads to the production of virus progeny and requires the sequential expression of a large number of structural proteins that build the infectious particle, as well as viral enzymes that coordinate viral DNA replication and virus assembly (9,10). High throughput sequencing technologies have recently led to the identification of several hundred new transcripts, most of which are expressed in replicating cells (11).
Although the sequence of the EBV genome has been available for >30 years, it is unclear whether all EBV proteins have been identified and how their expression is regulated (12–14). Moreover, it is still a matter of discussion whether some EBV transcripts encode proteins or are rather long non-coding RNAs (7,15). Both the proteome of EBV purified virus particles that contain large amounts of viral proteins, and the proteome of replicating cells are available, but this does not give information on the viral translation process itself (16,17). Translation ribosome profiling (TRP) identifies RNA fragments protected by the ribosome machinery after stabilization with cycloheximide (18). This approach can be refined by selectively arresting ribosomes on translation initiation sites using harringtonine (19). This strategy allows identification of new open reading frames, in particular those with non-canonical initiation sites, e.g. CUG instead of AUG. The feasibility of this approach has been amply demonstrated with cellular but also viral genomes such as HCMV or KHSV (20,21). We have applied this technology to B cells infected with weakly and strongly replicating EBV strains to generate a detailed map of the translated viral transcripts. This technology allowed us to identify new open reading frames and yielded new insights into the molecular mechanisms that condition viral protein expression.
MATERIALS AND METHODS
Ethics statement
All human primary B cells used in the experiments were isolated from anonymous buffy-coats purchased from the Blood Bank of the University of Heidelberg for which no ethical approval is required.
Cell culture
All cells used in this study were maintained in RPMI-1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Biochrom). Primary B cells were isolated from human blood buffy coats by Ficoll (GE healthcare) density gradient centrifugation and positive selection using CD19 PanB Dynabeads (Life technologies) with corresponding DETACHaBEADs (Life Technologies). Primary B cells were cultured in medium supplemented with 20% FBS until LCLs were established. HEK 293 cells are human embryonic kidney cells generated by transformation with adenovirus (ATCC: CRL-1573). The HEK293-B240 are HEK 293 cells stably transfected with the recombinant M81 BACMID and have been described previously (9). The HEK293-2089 producer cell lines have also been described previously (22).
Total RNA sequencing
Total RNA was extracted from 1 × 107 cells using Trizol (Ambion) following the manufacturer's guidelines. 10 μg of total RNA were treated with TURBO DNase (Thermo Fisher Scientific) for 15 min at 37°C. The DNase-treated RNA was re-extracted with Phenol/Chloroform. Strand-specific libraries were generated using the Agilent strand-specific RNA-Seq Library Preparation kit (Agilent). Samples were sequenced on Illumina's HiSeq 4000 sequencer.
Polysome profiling and RNA purification
Six weeks-old LCLs with a density of ∼7.5 × 105 cells/ml were split 1:3 forty eight hours prior to polysome profiling. On the day of the profiling experiment, LCLs were treated with 100μg/ml cycloheximide for 5 min to stall ribosomes on RNA. Cells were pelleted at 4°C and lysed in 250 μl ice-cold polysome lysis buffer (15 mM Tris–HCl pH 7.4; 15 mM MgCl2; 300 mM NaCl; 1% Triton X-100; 0.1% β-mercaptoethanol; 200 U/ml RNasin (Promega); 1 complete Mini Protease Inhibitor Tablet (Roche)/10 ml lysis buffer). Following 10 min incubation at 4°C, the lysate was cleared by centrifugation (10 000 rpm; 4°C; 10 min) and the supernatant was loaded onto a linear sucrose gradient ranging from 17.5–50% (w/v) sucrose (in 15 mM Tris–HCl pH 7.4; 15 mM MgCl2; 300 mM NaCl). Ultracentrifugation was carried out at 4°C at 35 000 rpm for 2.5 h in a SW60Ti rotor. Gradients were fractioned using a Teledyne Isco Foxy Jr. Gradient fractionator, which eluted the gradient into 12 fractions of 400μl volume. In parallel, the polysome profiles were recorded by measuring absorbance at 254 nm. RNA was purified from the fractions using organic solvent extraction followed by isopropanol precipitation.
Translational ribosome profiling and library generation
Translational ribosome profiling libraries were generated as described by Ingolia et al. (23) with minor modifications. Briefly, LCLs were pre-treated with cycloheximide (100 μg/ml) for 5 min at 37°C. For translation initiation mapping, samples were additionally pre-treated with harringtonine (2 μg/ml) for either 2 or 5 min. Following drug incubation, LCLs were lysed in polysome lysis buffer. Lysates were run on a linear sucrose gradient as described for polysome fractionation. Fractioned samples were treated with 600 U RNase I (Ambion) per 1 OD A260 for 15 min on a roller at room temperature. After RNAse I digestion, the samples were converted into cDNA libraries as described by Ingolia et al. (23). We used the NEBNext Indexing primers from Illumina for barcoding. Samples were sequenced on Illumina's HiSeq 2000 sequencer.
Sequence alignment
Prior to alignment, adaptor sequences were trimmed with FastX and reads mapping to rRNA sequences were removed. Alignments were carried out with STAR to the human genome issue HG-19. Unmapped reads were further aligned to the corresponding viral genomes B95-8 (accession number NC_007605.1) and M81 (accession number KF373730.1) containing the classic EBV genes using TOPHAT2. Finally, the remaining unmapped reads were aligned to the new EBV genes recently identified by O’Grady et al. (11) with TOPHAT2.
Data normalization
HTSeq-count (24) was used to quantify transcript expression levels of mapped reads from the total RNA sequencing experiments. The resulting raw reads were normalized by calculation of reads per kilobase of exon model per million mapped reads (25). The ribosome profiling-derived reads were normalized in two different ways depending on the read origin. To normalize ribosome footprints that mapped to cellular transcripts, we first calculated a scaling factor that is the ratio between the total number of reads in M81-infected LCLs and those in B95-8-infected LCLs. Absolute reads were then multiplied by this scaling factor. Footprint densities were calculated as reads per kilobase of coding DNA sequence. This method is not suitable for the low number of reads that mapped to the viral genome. Therefore, we used a modified version of the total count normalization method described by Dillies et al. (26) to compare ribosome footprints that mapped to viral genes between libraries. In this case, we divided the ribosome footprint counts of a given gene by the total number of reads that mapped to the viral genome in each of the respective library to obtain the relative read counts. We then calculated the arithmetic mean of these last two numbers that was used as a scaling factor. The relative read counts were then multiplied by this scaling factor. Analysis and figure generation were conducted using the Sashimi Plot function of the Integrative Genomics Viewer (IGV) version 2.3.25. Figures are derived from the total number of mapped reads (27) as the low coverage levels of EBV’s transcripts renders the weighted sums approach inefficient for our data (18). Read length distributions are plotted for all mapped reads. Read length determination was done using Fastq.
Metagene analysis
To analyse the aggregation of reads around start and stop codons (metagene analysis), we considered the reads that were 27 and 28 nt in length. In the case of ORFs that are located in cellular transcripts, the counts of these particular reads were normalized by transcript length. The average normalized read coverage of nucleotide positions 20 bases up- and downstream of start and stop codons was then calculated.
Identification of translation initiation sites
Harringtonine-treated libraries were used to identify translation initiation sites. The identification of novel small viral ORFs was carried out by manual curation of the alignments in IGV. Putative ORFs were scored as translated when at least two of the three harringtonine-treated libraries from a strain were positive for it. The start codon within a peak was designated as the 15th nucleotide of reads with a length of 27-28 nt and the 16th if the reads were longer. Figures showing ribosome coverage were generated using the Sashimi Plot function in IGV.
Calculation of ratios
The out:in ratios were calculated by dividing the number of RPFs mapping within 5′leaders of a transcript (out) by the number of RPFs mapping to the coding region of the transcript including the translation initiation codon (in). Read coverage was length normalized by nucleotide length of the respective feature (5′leader and coding region). 5′leader:AUG ratios were calculated by dividing the number of RPFs mapping within the 5′leaders of a transcript by the number of RPFs mapping to the start codon of a transcript (AUG) or non-canonical start codons. Read coverage was length normalized by nucleotide length of the respective feature.
UORF conservation analysis
Genome sequences of different EBV strains were downloaded from the NCBI database and genomic regions of interest were aligned against each other using the MacVector software version 15.1.1.
Immunostaining
LCLs were stained for BMRF1 (Clone MAB8186) or BZLF1 (Clone BZ.1) after fixation with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. Permeabilization of PFA-fixed samples was performed by immersion in PBS/0.5% Triton X-100 for 2 min. For gp350 (Clone 72A1) and BMRF2 (Rabbit polyclonal) stainings, LCLs were fixed in acetone for 20 min. Primary antibody incubation was carried out in a humidity chamber at 37°C for 30 min, followed by three times washing in PBS. The slides were incubated with a secondary antibody conjugated to Cy-3 (Dianova) for another 30 min, again followed by washing in PBS. Nuclei were counterstained with DAPI for 2min and washed in PBS three times. We used 90% glycerol in PBS for sample embedding. All antibodies were diluted in 10% heat-inactivated goat serum/PBS.
Real-time quantitative PCR
Total RNA was extracted with Trizol (Ambion) from LCLs. Reverse transcription of 400ng total RNA was performed using AMV reverse transcriptase (Roche) and random hexamers (Invitrogen) according to the manufacturer's protocol. SYBR green RT-qPCR analysis was run with the following cycling parameters: 10 min at 95°C for initial polymerase activation followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The Taqman RT-qPCR experiments were performed using the thermal cycling protocol on the ABI StepOnePlus Real Time PCR System (Applied Biosystems). All samples were run in duplicates and unless stated otherwise the human GAPDH gene was used for normalization among samples. The primer and probe sequences used for transcript detection are listed in Supplementary Table S4.
Western blotting
Cell pellets were lysed in RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; protease inhibitor cocktail (1:1000; Sigma)) with subsequent sonication. Protein concentration was determined by a Bradford assay. 50 μg of protein were denatured in Laemmli buffer for 5 min at 95°C and separated on 10% SDS-polyacrylamide gels. Proteins were blotted onto nitrocellulose membranes (Hybond C, Amersham) by wet transfer. Blotted membranes were blocked in 3% milk in PBS/0.1% Tween-20) for 30 min. Primary antibodies were incubated for at least 1 h at room temperature or overnight at 4°C. The antibodies used in this study were directed against BZLF1 (Clone BZ.1), HA-tag (Clone C29F4) and actin (Clone ACTN05C4). Secondary antibodies conjugated to HRP were purchased from Promega. ECL detection (PerkinElmer) was used to visualize protein bands.
Luciferase assays
Selected 5′leader sequences up to the translation initiation codon were synthesized and cloned into a pEX-A2 plasmid. The sequences were excised and cloned into the Hind III/PvuII(Bsp120I) (see Supplementary Table S5 for details on individual constructs) restriction site preceding the Firefly luciferase gene in the pGL4.5 vector. HEK 293 cells were seeded at a cell density of 1.2 × 103 cells per well in a 24-well plate. The following day, the cells were transfected with 200 ng of each construct and 200 ng of the pRL Renilla control vector (Promega) using Metafectene (Biontex). After 24 h, the cells were washed with PBS and lysed in 100 μl Passive Lysis Buffer (Promega) for 15 min at room temperature with gentle shaking. Renilla and Firefly luciferase signals were measured using the Dual-Luciferase Assay System Kit (Promega) according to manufacturer's instructions. Renilla luciferase signals were used for normalization of the Firefly luciferase signal. The Perkin Elmer Wallac 1420 multilabel counter was used to record the signals.
Cloning and expressing the HA-tagged expression constructs
Selected viral open reading frames along with their 5′leader sequence were amplified by PCR from a DNA library generated with the M81 EBV DNA. The primers used in these experiments are listed in the Supplementary Table S6. The HA-tag was included in the sequence of the reverse primer used to amplify the viral ORF. Constructs lacking the uORFs were generated in parallel by PCR mutagenesis. The PCR products were cloned into the pcDNA3.1(+) expression vector and the constructs were validated by sequencing. HEK 293 cells were seeded at a cell density of 2.5 × 103 cells per well on a 6-well plate. The following day, the cells were transfected with 1μg of each construct using Metafectene (Biontex). 500 ng of the pEGFPC1 plasmid was co-transfected to control for transfection efficiency. After 24 hours, the cells were washed with PBS and lysed in RIPA buffer. The samples were processed as described in the western blotting section.
Statistical analysis
For all experiments, statistical analysis was performed using GraphPad Prism version 6.0.c (GraphPad Software, La Jolla, CA, USA). For all data obtained, we used the unpaired t-test to infer significance.
RESULTS
Overview of the EBV ribosome profile
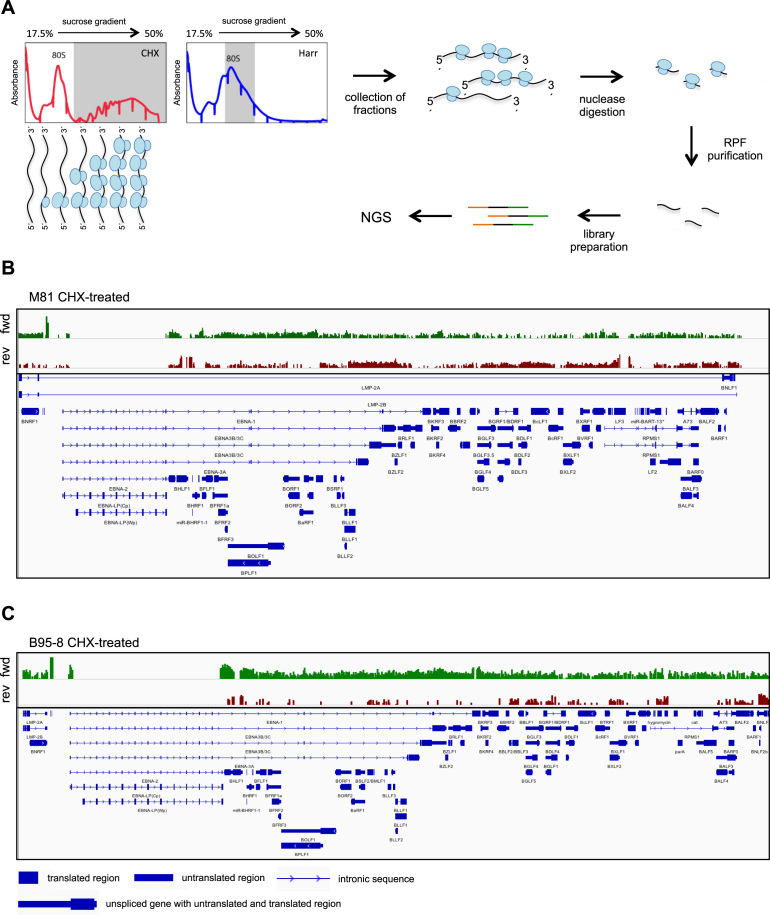
We wished to characterize viral translation and to map all translated viral open reading frames (ORF) in B cells infected with B95-8 or M81 using a TRP approach. To that end, we generated four libraries with ribosome-protected RNA fragments (RPF) purified from the polysomes of cycloheximide-treated EBV-infected B cells (sample 1) to exclude non-translating RNAs coupled to 80S ribosomes. We also purified RPFs from monosomes isolated from infected cells treated with harringtonine (Figure 1A, Supplementary Figure S1A). B cells infected with M81 support viral lytic replication and thus express EBV lytic genes (Supplementary Figure S1B). The generated libraries were subjected to high throughput sequencing, and the sequence was aligned to the viral genomes, allowing identification of ORFs (Figure 1B and C for an overview, Supplementary Table S1). We identified a large number of translated genes on both strands of the genome, although the number of reads varied substantially between them (Figure 1B and C). Cells infected with M81 showed many more signals on the leftward EBV genome strand, where many lytic genes are encoded, than cells infected with B95-8. Alternative splicing of the EBNA genes generates very large introns that contain exons used in other coding transcripts. Thus, reads that map to these exons falsely appear to be located in introns in the figure (Figure 1B and C).
Figure 1.
M81-infected LCLs show evidence for global bidirectional translation of the viral genome. (A) Ribosome profiling of EBV-infected LCLs. The schematic shows representative polysome profiles obtained after sucrose density centrifugation of RNA-protein complexes extracted from LCLs treated with either cycloheximide (CHX) (left) or harringtonine (Harr) (right). The fractions of the gradient collected for library generation are shaded in grey. After collection of the fractions, the RNAs are subjected to nuclease digestion and the ribosome-protected fragments (RPF) are used to generate sequencing libraries. NGS: next generation sequencing. (B) The upper panel shows an overview of M81-specific ribosome-protected transcripts after cycloheximide treatment. Reads that map to rightward oriented transcripts are shown in green, those that map to the leftward transcripts are shown in red. The lower panel shows the main EBV transcripts. (C) Same as in (B) but with cells infected with B95–8 and treated with cycloheximide. Please also see Supplementary Figures S1–S3.
We sequenced four additional libraries from cells obtained from a second, independent, blood sample (sample 2) that we infected with B95-8 or M81 and treated with harringtonine for 2 or 5 min (Supplementary Table S1). Finally, we sequenced one harringtonine-treated, lytically-induced 293 cell line that carries the B95-8 genome (Supplementary Table S1). In parallel, we generated four transcriptomes from blood samples 1 and 2 infected each with B95-8 or M81 (Supplementary Table S1). We performed multiple quality controls on the thirteen sequenced libraries. We assessed the reproducibility of the results obtained in terms of read identity between the transcriptomes generated from two blood samples that we considered as biological replicates (Supplementary Figure S2A and B). We used the same approach for the TRP generated after treatment with harringtonine for 2 min (Supplementary Figure S2C and D). This analysis showed a R2 Pearson correlation coefficient ranging between 0.77 and 0.96 as previously observed (28). Analysis of the length distribution of the reads showed a predominance of 29 to 30 nt-long fragments in cells infected with M81, and 28 to 29 nt-long fragments in cells infected with B95-8 (Supplementary Figure S2E and F). These results are consistent with previous reports on ribosome profiling (19). The RPFs were centered on the initiation codon in B95-8 and M81-infected cells treated with harringtonine (Supplementary Figure S2G and H). The distribution was more complex in cells treated with cycloheximide, with reads increasingly accumulating after the initiation codon and decreasing slowly after the stop codon, as previously described for RNAs purified from polysomes (Supplementary Figure S2G and H) (29). We analyzed the triplet periodicity of the reads (28 to 29 nt RPF for B95-8 and 29 to 30 nt for M81) to identify the base position within codons on which ribosomes are located after harringtonine or cycloheximide treatment and indeed found a relative base predominance among the codons (Supplementary Figure S3A and B). The predominance was obvious for samples treated with harringtonine but less clearly defined for cycloheximide-treated samples (See discussion). Finally, we analyzed the distribution of the reads within the various components of the gene unit and found a dominance of reads in the coding regions (Supplementary Figure S3C–F). Cells treated with harringtonine showed an accumulation on the translation initiation codon, relative to those treated with cycloheximide (Supplementary Figure S3C–F).
Ribosome profiling of M81-infected cells
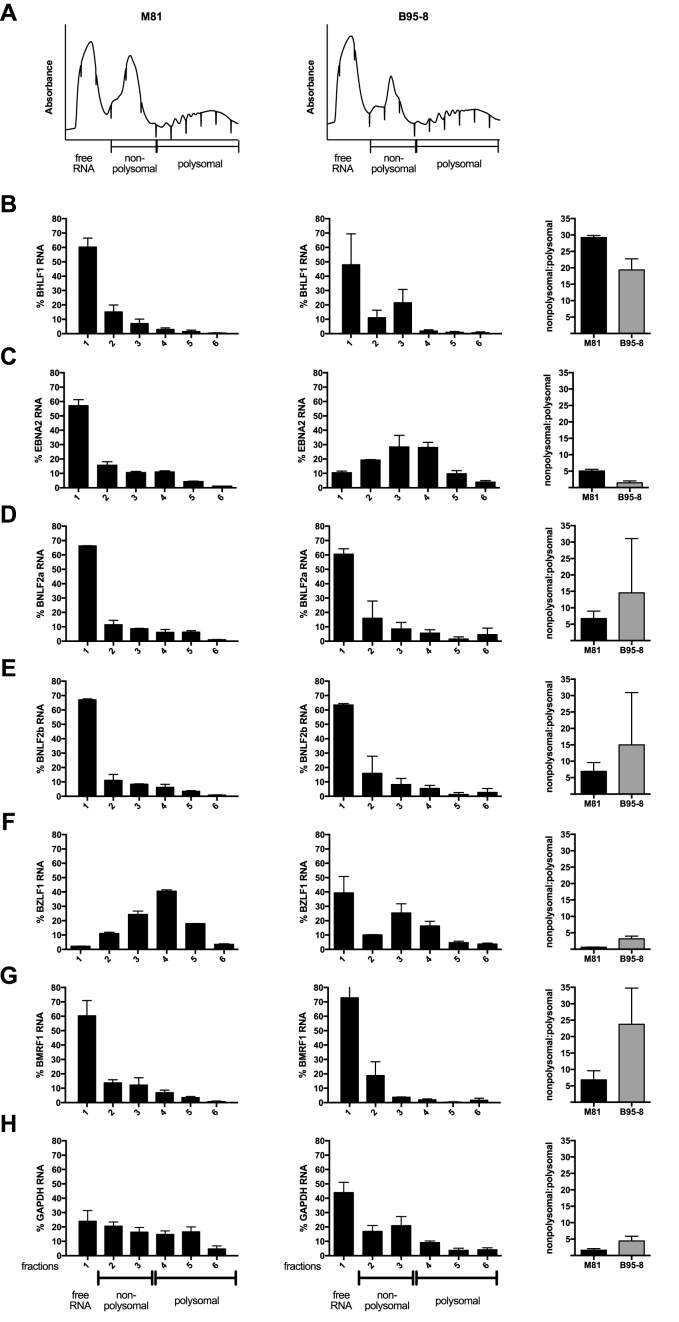
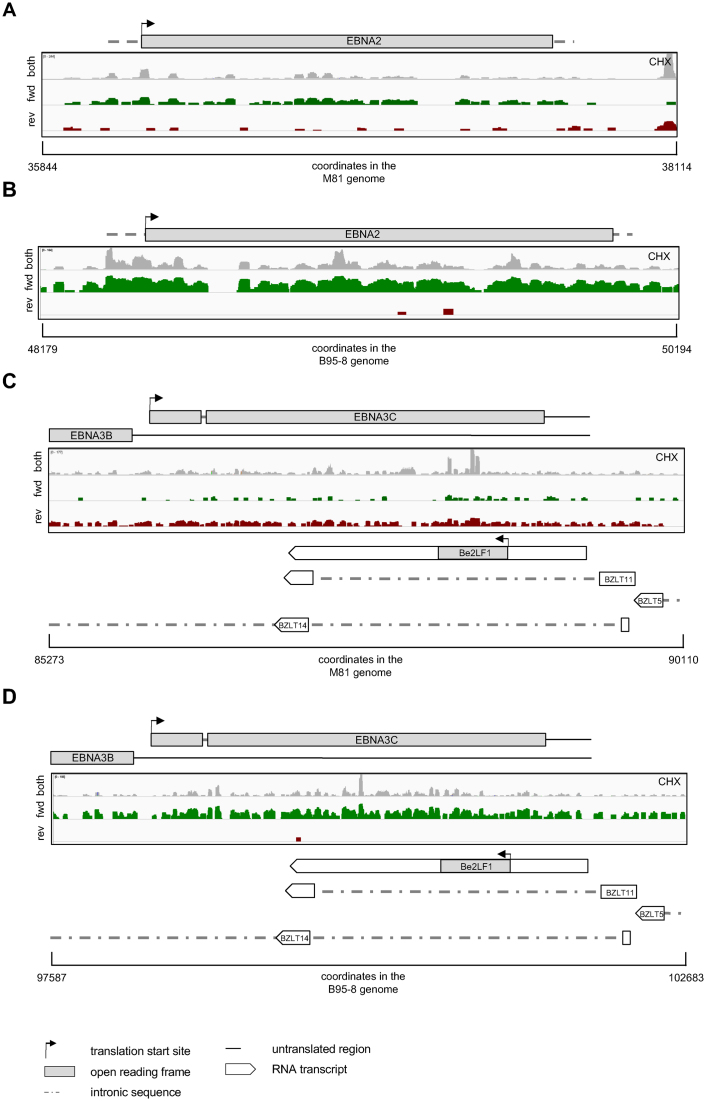
Detailed analysis of the TRP of M81-infected cells treated with cycloheximide showed a great heterogeneity in viral translation that ranged from 0.02 to 2.5 RPFs per nucleotide (Supplementary Figure S4A). Some poorly or controversially characterized EBV transcripts such as the BXRF1, BVLF1 and BTRF1 hardly showed any ORF ribosome recruitment (Supplementary Figure S4A). Similar remarks apply to the A73, the RPMS1, and the LF1 ORFs that are located within the BART transcripts (Supplementary Figure S4A). There were more ribosome footprints in the LF2 ORF (Supplementary Figure S4A). In contrast, the BHLF1 and LF3 transcripts that are generated by gene homologs displayed extremely abundant coding DNA sequence (CDS) ribosome footprints (Supplementary Figure S4A). Therefore, we purified the different types of transcripts on a sucrose gradient to isolate free RNA, transcripts covered by monosomes and transcripts covered by increasing numbers of polysomes. Quantification of the BHLF1 transcripts present in the different sucrose fractions using qPCR showed that the BHLF1 mRNA is mainly present as free RNA or associated with monosomal ribosomes, rather than with actively translating polysomes (Figure 2A and B). Interestingly, BHLF1 was previously recognized as an actively transcribed gene for which evidence of translation was lacking (30–33). E. Flemington and colleagues recently described a new group of lytic transcripts (11) that were generally transcribed at much lower rates than the usual lytic transcripts in our samples (Supplementary Figure S4B). Because the large majority of these new lytic transcripts are included within or overlap with previously characterized lytic transcripts, the precise quantification of the TRP reads that map them in cells treated with cycloheximide is generally impossible. However, the number of reads covering the transcripts that are antisense to EBNA2 and EBNA3A, 3B and 3C can be unequivocally determined in M81-infected cells (Figure 3, Supplementary Figure S5). The EBNA2 antisense transcript was hardly covered by RPFs, an observation in line with a previous report that this transcript is probably non-coding, as it is mainly located in the nucleus of infected cells (34). In contrast, the transcripts antisense to each of the EBNA3 genes all carried numerous ribosomes, suggesting active translation (Figure 3, Supplementary Figure S5). We also observed reads in some of the introns of these antisense genes, although they were generally less abundant than those located on the spliced transcripts. This could indicate some degree of intron retention, as has been previously described for EBNA3 mRNA (Figure 3) (35). In rare instances, we could not ascribe the antisense ribosome reads to any known transcripts (Supplementary Figure S5A). This could indicate the presence of so far unidentified transcripts in this region. Importantly, the analysis of the TRP in the same cells treated with harringtonine could not identify any read accumulation around putative initiation codons. This suggests that these antisense transcripts are not actively translated, at least not at rates seen with the ‘classical’ EBV transcripts.
Figure 2.
Distribution of EBV transcripts in various ribosome fractions. The figure shows the distribution of viral transcripts within monosomal and polysomal ribosome fractions after gradient separation. Viral transcripts devoid of ribosomes are given as ‘free RNA’. (A) The figure shows representative polysome absorbance profiles for M81-infected LCLs (left) and B95–8-infected LCLs (right) after gradient fractionation. We quantified the viral transcripts present in the different fractions by RT-qPCR, and give their proportion in these fractions relative to the total viral RNA. The graph of bars gives the ratio between transcripts covered by monosomal ribosomes and those covered by polysomal ribosomes. The indicated values represent the mean ± standard deviation of at least two independent experiments. (B–H). Distribution of BHLF1 (B), EBNA2 (C), BNLF2a (D), BNLF2b (E), BZLF1 (F), BMRF1 (G) and GAPDH (H) RNAs within the gradient.
Figure 3.
Association of transcripts antisense to EBNA2 and EBNA3C with ribosomes. In each of these figures, we indicated the region of the viral genome to which the ribosome-protected reads map as well as their coordinates in their respective genomes. The grey reads show the total ribosome-protected fragments (RPFs) obtained from infected B cells that were treated with cycloheximide (CHX). The rightward reads among those are shown in green, the leftward reads in red. (A) The figure shows reads that mapped to the viral segment that contains the EBNA2 gene in cells infected with M81. (B) Same as in (A) but with cells infected with B95–8. (C) The figure shows reads that mapped to the viral segment that contains the EBNA3C gene in cells infected with M81. (D) Same as in (C) with cells infected with B95–8. Please also see Supplementary Figure S5.
The data provided by the TRPs will allow detailed characterization on the regulation of the translation of any single EBV gene or group of genes that frequently proved to be complex. The BMRF1 and BMRF2 transcripts that extensively overlap with each other and with BMRT3 and BMRT4 transcripts provide one example. BMRF1 and BMRF2 showed a similar number of reads in the TRP of cells treated with cycloheximide (Supplementary Figure S6A). However, the BMRF2 transcript recruited a much lower number of ribosomes than BMRF1 in cells treated with harringtonine (Supplementary Figure S6A). We could confirm by immunostaining that the BMRF2 protein is indeed expressed at clearly lower levels than BMRF1, a feature previously noted in the proteome of replicating cells (17) (Supplementary Figure S6B). Closer examination of the BMRF2 ribosome profile revealed an accumulation of reads in the last third of the transcript. This suggests the existence of ribosome pausing that would be congruent with the reduced BMRF2 protein expression.
Viral genes are translated with variable efficiency
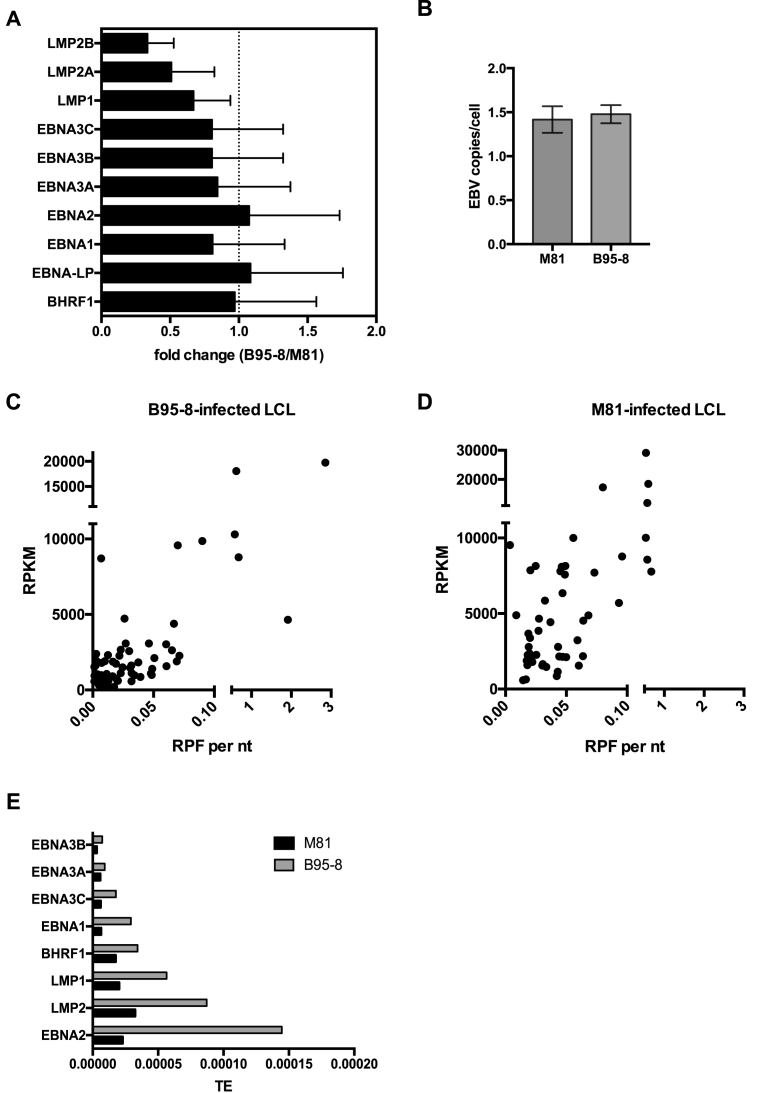
We determined the translation efficiency of the various viral genes by building the ratio between ribosome reads and transcript reads. M81 and B95-8 infected cells expressed EBV-specific transcripts within a very wide range (233 RPKM for BCRF1 or viral IL10 to 29 144 RPKM for BKRF4 in M81-infected cells) (Supplementary Figures S4B and S7B). Many lytic genes were transcribed at higher levels than some latent genes in M81-infected cells. Taking into account that only a minority of M81-infected cells undergoes lytic replication, the transcription level of many lytic genes appears to be several orders of magnitude higher than those of latent genes (Supplementary Figure S4B). Cells infected with B95-8 showed a reduced transcription of lytic genes relative to M81 that was marked for some genes such as those located on the A segment of the EBV genome, but was much milder for other genes such as BZLF2, BNLF2a, BNLF2b or BRLF1 that reached 10–30% of the M81 levels (Supplementary Figures S7B and S8A). We also found a mildly reduced latent gene transcription in B95-8 transformed cells, relative to cells infected with M81 (Figure 4A). This was not due to differences in EBV copy numbers in the different types of infected cells (Figure 4B). For each gene, we then plotted the transcription level against the abundance of ribosome-protected transcripts (Figure 4C and D, Supplementary Figures S4A, B, S7A, B). This analysis showed a broad range in translation efficiency over two orders of magnitude (Figure 4C and D, Supplementary Figures S4C and S7C). Within the latent gene family, the EBNA3 genes were less efficiently translated than EBNA2 or the LMP genes (Figure 4E). There was also a difference between cells infected with B95-8 or with M81, the translation efficiency of latent genes being generally higher in cells infected with the former virus. We used polysome profiling and qPCR to confirm these data. While only little free EBNA2 mRNA was visible in cells infected with B95-8, there was a five-fold higher proportion of polysomes detected on the translated mRNA, relative to M81-infected cells (Figure 2C).
Figure 4.
Viral gene transcription and efficiency of translation in infected B cells. (A) The graph of bars shows a comparative analysis of latent gene transcription in cells infected with either M81 or B95–8. It gives the mean relative fold change of latent gene expression levels with standard deviation in B95–8-infected LCLs relative to M81-infected LCLs. (B) Genomic DNA was extracted from four independently established LCLs at 10 days post infection and the EBV copy number per cell was determined by qPCR. (C) and (D) The dot plots show the efficiency of translation in cells infected with B95–8 (C) or M81 (D). Each dot corresponds to one viral gene, whose y coordinate gives the transcription rate as reads per kilobase per million mapped reads (RPKM) and the x coordinate the number of RNAse-resistant ribosome protected footprint (RPF) reads per nucleotide (nt). (E) The graph shows a comparative analysis of the latent gene translation efficiency (TE) in cells infected with B95–8 or M81, displayed as the ratio between translation given as RPF and total transcription given as RPKM. Please also see Supplementary Figures S4, S7 and S8.
We compared the transcription, translation and translation efficiency of different families of viral lytic transcripts (Supplementary Figure S9A–C). We found that immediate early and early transcripts were, on average, generated at a slightly lower rate than late transcripts. However, the differences between the early and late transcript groups disappeared in the ribosome profile. We also assessed the transcription and RPF rates of viral gene groups dedicated to different viral functions and found that these rates were highest for the glycoprotein group (Supplementary Figure S9D–F). We finally compared genes whose expression is dependent on BGLF3 with those that is not and found that the former are expressed on average 6.9 times more and are covered by ribosomes on average 7.5 times more than the second (36) (Supplementary Figure S9G–I).
BKRF3 shows alternative translation initiation sites
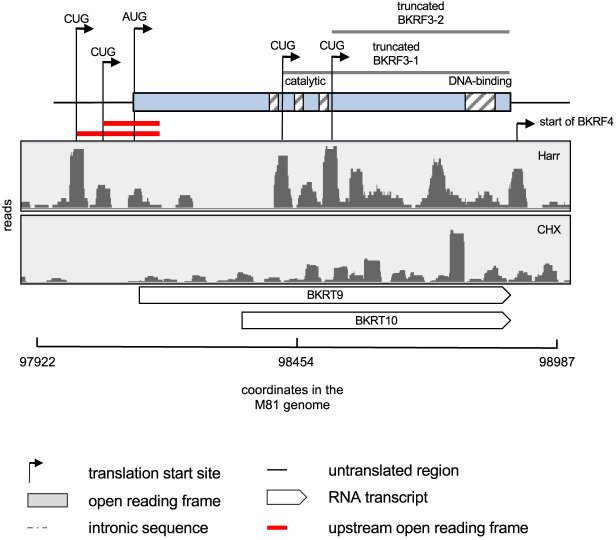
We could map new alternative translation initiation sites located upstream or downstream of previously identified initiation codons, thereby extending or truncating the main protein product, up to 101 amino acids in the case of the BKRF3 protein (Table 1). These alternative initiation codons were either AUGs or CUGs (Table 1 and Figure 5). The analysis of the BKRF3 gene was particularly interesting as it exemplifies the complexity of translation in the EBV genome (Figure 5). This gene overlaps with several newly identified lytic transcripts, including BKRT9 and BKRT10 that are contained within the BKRF3 locus, and is readily followed by the BKRF4 gene. We could identify two new possible initiation codons for these genes, located 3′ of the annotated BKRF3 AUG. More work will be needed to learn whether they correspond to translation products from the distinct BKRF3, BKRT9 or the BKRT10 transcripts, or are instead all translated from the BKRF3 transcript.
Table 1. Viral genes with alternative translation initiation sites.
| Gene | Start codon | Gene coordinate in M81 | Location relative to the annotated start codon [aa] |
|---|---|---|---|
| BFRF3 | CUG | 49317 | −14 |
| BLRF2 | AUG | 76828 | +17 |
| BFRF1a | AUG | 46526 | +51 |
| BKRF2 | CUG | 97899 | +93 |
| BKRF2 | CUG | 98052 | +111 |
| BKRF3 | CUG | 98318 | +101 |
| BKRF3 | CUG | 98570 | +151 |
| BALF1 | AUG | 164920 | +39 |
aa: amino acids.
Figure 5.
BKRF3 illustrates the complexity of viral translation. Ribosome profile of the genome region encoding BKRF3 in M81-infected LCLs. The hatched boxes within the BKRF3 coding region highlight functional domains of the protein. The two upstream open reading frames (uORFs) located upstream of BKRF3 are highlighted in red. The figure shows two CUG codons within the annotated ORF that could initiate the translation of truncated BKRF3 protein isoforms. We also give the location of two RNA transcripts identified by Grady et al. (11) that are located in the BKRF3 region.
EBV latent transcripts show 5’leader ribosome recruitment
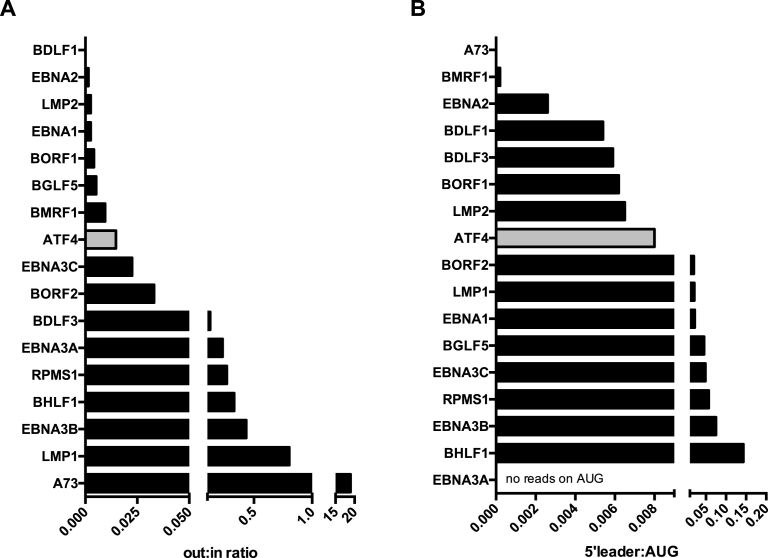
We used normalized libraries to quantify the ribosome footprints in the transcript leader region, around the initiation codon, and within the coding region of every large ORF. Analysis of cycloheximide-treated LCLs generated with either type of virus showed that all latent genes carry ribosomes in their long leader region (Supplementary Figure S10A). We calculated the ratio between the number of reads located in the leader region of the gene and the number of reads located in the CDS region (out:in ratio) to gather information on the existence of leader regulation (Figure 6A, Supplementary Figure S10B). We also investigated ATF4, a cellular gene subjected to 5′leader regulation (37). This analysis revealed that LMP1, EBNA3A, EBNA3B were subjected to leader region regulation at a higher rate than ATF4 (Figure 6A). It is important to note that in the latent genes part of the 5′leader sequences are shared. Thus, it is not always possible to unequivocally ascribe reads to the leader region of a given latent gene. While some degree of leader region regulation was visible for the EBNA3C and EBNA1 transcripts, this was hardly the case for EBNA2 and LMP2. We then used the information provided by the harringtonine libraries to quantify the ratio between reads located in the leader region and those located on the initiation codon (5′leader:AUG ratio) (Figure 6B, Supplementary Figure S10C). This analysis confirmed that LMP1 and the EBNA3 transcripts have a large proportion of reads located upstream of the initiation translation codon, consistent with an active negative regulation. It was not possible to analyze the regulation of BHRF1 in M81-infected cells, as this gene is transcribed from different promoters during latency or lytic replication and the ribosome profiling does not distinguish CDS-reading ribosomes on the same transcript produced by different promoters.
Figure 6.
Ribosome recruitment to the leader regions in EBV genes. (A) The graph shows the out:in ratios of selected EBV and cellular transcripts in cells infected with M81 and treated with cycloheximide. These ratios are obtained by dividing the number of RNAse-resistant ribosome protected footprints (RPF) that mapped to the 5′ leader region (out) by the number of reads that mapped to the open reading frame of the same gene (in). (B) The figure depicts the ratios between reads that mapped to the 5′ leader and those that mapped to the translation initiation codon of the same gene, or 5′ leader:AUG ratios in cells treated with harringtonine. More examples are given in Supplementary Figures S10–S14.
EBV lytic transcripts show 5’leader ribosome recruitment
Ten lytic genes had a substantial number of reads in their 5′leader region after treatment with cycloheximide, and showed out:in ratios comparable to those recorded with EBNA3A or B, e.g. for BALF1 and BALF4 (Supplementary Figure S11). However, the lytic genes frequently overlap and render unambiguous mapping difficult in many transcripts. This problem disappears after treatment with harringtonine. In that case, approximately half of the lytic transcripts recruited ribosomes to the 5′leader (Supplementary Figure S12A). Some genes such as BKRF3, BFRF3, BFLF2 or BcLF1 showed a high 5′leader:AUG ratio, comparable to what was found for EBNA3B, others such as BALF1 or BBRF1 showed lower ratios, similar to those found with EBNA2 (Supplementary Figure S12B).
The BHLF1 and LF3 transcripts carried ribosomes both in their CDS and in their leader region or around their initiation AUG in cells transformed by M81 or B95-8 (Supplementary Figure S13). Consequently, the 5′leader:AUG ratio was high, in particular in cells infected with B95-8. These data suggested that translation is actively repressed in this gene. A73 had abundant footprints in its leader region. Together with the paucity of reads in the CDS region, this suggests that these genes are not substantially translated and that translation of A73 is actively repressed (Supplementary Figure S14).
Comparison of ribosome profiles of B95-8 and M81-infected cells
We compared the TRPs obtained with M81 and B95-8-infected cells and noted lower numbers of RPFs specific to the EBV latent genes in cells infected with the former virus (Supplementary Figure S7D). As expected, lytic genes were much more actively translated in M81-infected cells, although here again some of these transcripts showed strong ribosome coverage after infection with B95-8 (Supplementary Figures S7A and S8B). The BNLF2a and BNLF2b transcripts in particular had RPF levels that are in the range of latent transcripts in cells infected with B95-8. This suggests that they are translated in infected cells independently of their latent or lytic status. We assessed BNLF2a and BNLF2b transcription with qPCR and also included cells from the same blood sample that were infected with the non-replicating ΔZR M81 mutant in the analysis (Supplementary Figure S7E). The latter cells also showed transcription of BNLF2a and BNLF2b, albeit reduced in intensity. We conclude that these genes are transcribed during latent infection but transcription increases upon induction of lytic replication. Quantification by qPCR of the fractions generated by a new round of transcript profiling showed a similar profile in M81 and B95-8 infected cells, with a large proportion of free BNLF2a and 2b RNA and an average ratio of monosomal versus polysomal ribosomes in M81-infected cells (Figure 2D and E). This ratio was twice as high in B95-8 infected cells, suggesting a lower efficiency of translation in these latently infected cells. Altogether these results fit with the previous suggestion that BNLF2a is a latent gene, although the low efficiency of translation requires further characterization of protein production using specific antibodies (38). Other lytic genes showed a reduced but significant ribosome coverage, most notably BZLF1 in B95-8 infected cells. The transcript profile coupled to qPCR showed very little free BZLF1 RNA and a high proportion of polysomal ribosomes in M81-infected cells (Figure 2F). In contrast, cells infected with B95-8 BZLF1 showed abundant free RNA and a higher proportion of monosomal ribosomes. This suggests the existence of repressive mechanisms in non-permissive cells that are located downstream of transcription and ribosome recruitment. Similar observations were made with BMRF1 (Figure 2G). Using qPCR, we found that this gene was expressed in cells infected by B95-8 at half the levels seen in M81-infected cells (Supplementary Figure S6C). However, these levels were higher than those recorded in cells infected with the replication-deficient M81ΔZR virus, that were themselves higher than in cells infected with a BMRF1 knockout virus, suggesting that this lytic gene is actively transcribed in cells infected with B95-8, to some extent independently of the BZLF1 and BRLF1 proteins. Immunostains for BMRF1 revealed only very rare cells positive for the protein, relative to M81-infected counterparts (Supplementary Figure S6D). The TRP from harringtonine-treated cells confirmed that approximately half of the lytic genes are hardly translated in B95-8 infected cells relative to M81 (Supplementary Figure S8C). However, many other transcripts in B95-8 infected cells, including BZLF1, BRLF1 and BMRF1, carried ribosomes on their start codons at rates 30–120% of those recorded after infection with M81 (Supplementary Figure S8C). The newly identified family of lytic transcripts was hardly translated in cells infected with B95-8 (Figure 3D).
The EBV genome encodes upstream open reading frames
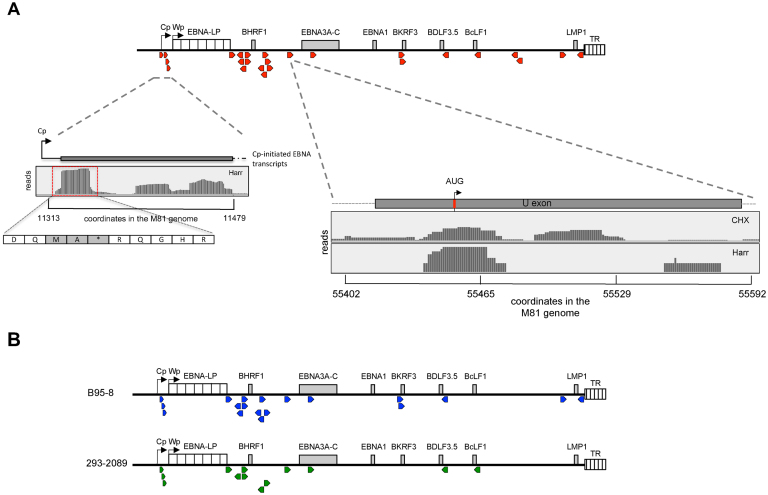
The libraries generated from infected cells treated with harringtonine revealed the existence of 25 short open reading frames with a size ranging from 1 to 74 amino acids (Figure 7, Table 2). These ORFs were located upstream of well-characterized genes and thus represent upstream open reading frames (uORFs). We investigated the parameters that have been found to influence the strength of the uORF, including their Kozak sequence, the cap to first uORF distance and the distance from last uORF to the main ORF (39–41) (Supplementary Table S2). The sequences of the four shortest uORFs were perfectly conserved in all 115 EBV strains for which this sequence was available (Supplementary Table S3). This also applied to the initiation codon of 17 of the 21 remaining uORFs. Four uORFs showed mutations of their initiation codon in very rare strains. In two of these polymorphic uORFs, the mutation generated another non-canonical translation start site, whilst in the remaining two it destroyed the uORF (BKRF3 uORF2, BFRF3 uORF1). However, these two genes possess an additional uORF that is conserved in all strains. We also found polymorphisms within some of the uORFs, none of which interrupted the oligopeptide translation (Supplementary Table S3). The uORFs located upstream of LMP1 and BHLF1 uORF1 showed the highest degree of polymorphisms among the EBV strains (22 strains out of 121 and 26 out of 53, respectively) (Supplementary Figure S15A).
Figure 7.
The EBV genome contains multiple upstream open reading frames. (A) The schematic shows the distribution of upstream open reading frames (uORFs) in the M81 EBV genome (Red arrowheads). The left close-up shows the uORFs behind the Cp/Wp promoter after treatment with harringtonine (Harr). The right close-up underneath shows the reads that mapped to the U exon after ribosome profiling of M81-infected LCLs treated with harringtonine or cycloheximide (CHX). (B) The schematic shows the distribution of upstream open reading frames (uORFs) identified in the ribosome profiling libraries derived from LCLs transformed with the B95–8 EBV strain (Blue arrowheads) or from HEK 293 cells that carry the B95–8 BACmid that had been induced into lytic replication (green arrowheads).
Table 2. Identification of uORFs in the EBV genome.
| Proposed nomenclature | Length of the encoded peptide [aa] | Gene coordinates in M81 | Potentially regulated genes | Start codon |
|---|---|---|---|---|
| Cp uORF1 | 1 | 11140–11145 | EBNA-LP & other EBNAs (Cp) | AUG |
| Cp uORF2 | 2 | 11229–11237 | EBNA-LP & other EBNAs (Cp) | AUG |
| Cp uORF3 | 1 | 11395–11400 | EBNA-LP & other EBNAs (Cp) | AUG |
| Cp uORF4 | 1 | 11341–11346 | EBNA-LP & other EBNAs (Cp) | AUG |
| BHLF1 uORF1 | 35 | 40361–40254 | BHLF1 | CUC |
| BHLF1 uORF2 | 20 | 40518–40456 | BHLF1 | CUG |
| BHLF1 uORF3 | 7 | 40563–40456 | BHLF1 | UUG |
| Y2 uORF | 7 | 35565–35588 | EBNA2; BHRF1; EBNA1; EBNA3A-C | AUG |
| BHRF1 uORF1 | 14 | 41658–41702 | BHRF1 | CUG |
| BHRF1 uORF2 | 74 | 41726–41914 | BHRF1 | CUG |
| BFLF2 uORF1 | 12 | 44838–44800 | BFLF2 | CUG |
| BFLF2 uORF2 | 30 | 44885–44793 | BFLF2 | AUG |
| BFRF1 uORF | 10 | 46698–46730 | BFRF1 | UUG |
| BFRF3 uORF1 | 25 | 49246–49323 | BFRF3 | ACG |
| BFRF3 uORF2 | 41 | 49310–49435 | BFRF3 | UUG |
| U uORF | 6 | 55343–55363 | EBNA1; EBNA3A-C | AUG |
| BORF2 uORF | 17 | 64134–64187 | BORF2 | GUG |
| BKRF3 uORF1 | 45 | 97899–98036 | BKRF3 | CUG |
| BKRF3 uORF2 | 27 | 97953–98036 | BKRF3 | CUG |
| BDLF3.5 uORF | 8 | 116982–116956 | BDLF3.5 | AUG |
| BcLF1 uORF | 11 | 125269–125234 | BcLF1 | CUC |
| BXLF2 uORF1 | 24 | 130846–130772 | BXLF2 | UUG |
| BXLF2 uORF2 | 30 | 130803–130711 | BXLF2 | AUG |
| LMP2A uORF | 20 | 165917–165979 | LMP2A | UUG |
| LMP1 uORF | 15 | 168919–168872 | LMP1 | CUG |
uORF: upstream open reading frame.
aa: amino acids.
Four uORFs were located between the Cp and the Wp promoters that drive the expression of the EBNA genes and of BHRF1, an antiapoptotic protein (Figure 7A, Supplementary Figure S15B). The short but strongly translated 7 aa long (MKTKSQA) Y2uORF straddles the EBNA-LP stop codon (Table 2). This ORF begins in the region that encodes the DE repeats, but is translated in another reading frame than EBNA-LP. Remarkably, several uORFs were found in the leader region of the same transcript. Five uORFs are present in all Cp-driven EBNA mRNAs and on the mRNA that encodes a latent form of BHRF1 (Supplementary Figure S15B). The EBNA1 and the EBNA3 transcripts additionally contained the uORF located in the U exon, another non-coding exon located in their common transcript leader region (42). Interestingly, this uORF is very close to a putative IRES (43) (Supplementary Figure S15C). Thus, the Cp-driven EBNA and the latent BHRF1 mRNAs contain five to six uORFs in their leader region. We also found that the leader region of the BHLF1 transcript contained three uORFs, whilst the leader region of the lytic BHRF1, of BFRF3 and of BKRF3 contained 2 uORFs each. In the latter two cases, some of the uORFs overlapped with the initiation codon of the main ORF but were located in a different reading frame. The uORFs located upstream of the EBNA transcripts began with an AUG. With the exception of BDLF3.5 uORF and BFLF2 uORF2, all other uORFs used a non-canonical initiation codon. Comparison between the different libraries revealed some variation. B cells infected with M81 expressed the highest number of uORFs, B cells infected with B95-8 did not express six of these uORFs and infected 293–2089 cells lacked the expression of twelve of them (Figure 7B). UORFs have been implicated in active retention of ribosomes in the leader region of downstream genes. Therefore, we tested whether the presence of uORF correlated with the intensity of ribosome recruitment in this region and found that this is indeed the case (Supplementary Figure S12).
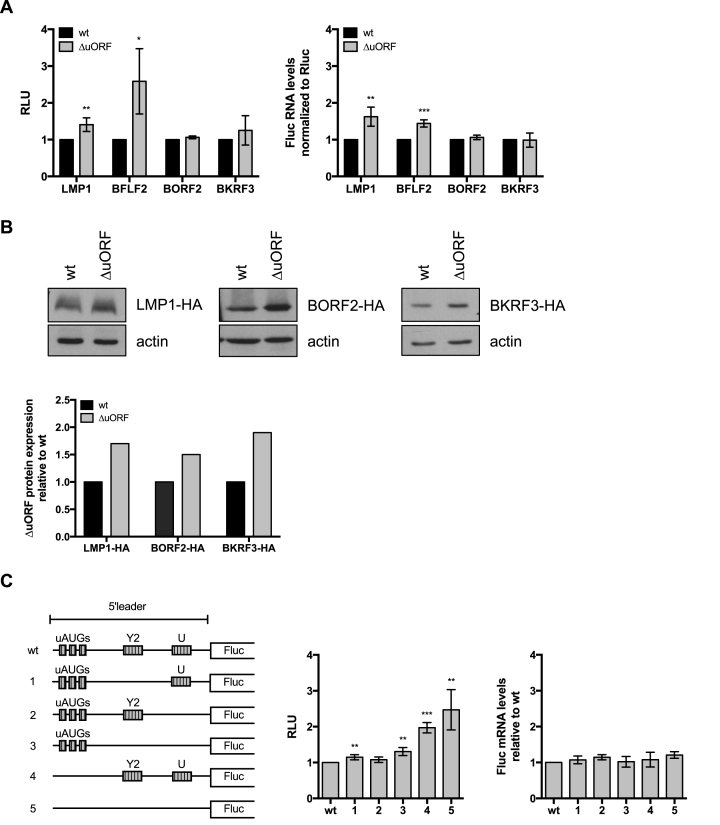
We cloned 4 EBV (LMP1, BFLF2, BORF2, BKRF3) wild type or mutated uORFs with an inactive initiation codon 5′ of the Firefly luciferase ORF. We transfected these constructs in 293 cells and performed luciferase assays with their protein extracts. The Renilla luciferase was co-transfected in these experiments and served as a transfection control. These experiments showed that the luciferase activity was higher after transfection with two of the constructs carrying mutated uORFs (Figure 8A). For LMP1 and BFLF2, luciferase RNA levels were higher in the constructs carrying the inactive uORF. These assays showed that for LMP1 and BFLF2, both the RNA levels and the luciferase activity were increased. There was also a similar but very weak effect with BORF2 and BKRF3. We therefore fused the BORF2, BKRF3 and LMP1 genes with a human influenza hemagglutinin (HA) tag. These genes were preceded by their respective uORFs, either in their wild type or in their mutated versions. Transfection of these constructs in 293 cells showed that BORF2, BKRF3 and LMP1 are expressed at higher levels in the absence of a functional uORF (Figure 8B). We then investigated the EBNA3 5′ leader region using luciferase constructs (Figure 8C). Inactivation of 3 uAUGs doubled the luciferase activity without affecting the luciferase RNA levels and this effect was slightly potentiated by the inactivation of the Y2 and U uORFs. Constructs lacking a functional Y2 uORF, a functional U uORF or both also showed a mild increase in luciferase activity. We conclude from this set of experiments, that all tested EBV uORFs repress gene translation, although the intensity of the effects varied between the uORFs.
Figure 8.
Influence of the 5′leader on downstream translation of the main open reading frame. (A) Four viral 5′leader sequences containing some of the identified upstream open reading frames (uORFs) and mutants thereof lacking a functional translation initiation codon were cloned upstream of the Firefly luciferase reporter gene. Dual-reporter luciferase assays were performed by transient transfection of the Firefly luciferase constructs together with a Renilla luciferase expression plasmid into HEK293 cells. The relative light units (RLU) between the luminescence levels generated by expression of the mutated uORF and the wild type uORFs are given in a graph of bars that represent the mean of at least three independent experiments with error bars representing the standard deviation. Asterisks indicate statistical significance determined by unpaired t-test (*P< 0.05, **P< 0.05 and ****P< 0.0001). The relative luciferase mRNA levels of the wild type and ΔuORF luciferase reporter constructs were measured by RT-qPCR. As for the luciferase assays, the Renilla luciferase was used as an internal control. (B) Three EBV open reading frames together with their 5′leader containing the putative uORFs were HA-tagged and cloned into an expression vector. A mutant version of the expression constructs that carried mutated uORF start codons was generated in parallel. The constructs were transiently transfected into HEK293 cells and subjected to a western blot analysis with antibodies specific to the HA-tag. Actin was used as loading control. Representative western blots are shown for each expression construct. The relative intensity of the bands was quantified, normalized by the actin signals and is shown in the graph of bars below. (C) The 5′leader of the EBNA3A mRNA with all five identified uORFs was cloned upstream of the Firefly luciferase gene. A schematic overview of the wild type and the different mutant constructs lacking various uORF translation initiation codons is shown. Transient transfection experiments were performed as described in (A). The relative light units (RLU) between the luminescence levels generated by expression of the mutated uORF and the wild type uORFs are given in a graph of bars that represent the mean of at least three independent experiments with error bars representing the standard deviation. Asterisks indicate statistical significance determined by unpaired t-test (*P< 0.05, **P< 0.05 and ****P< 0.0001). The relative luciferase mRNA levels of the wild type and ΔuORF luciferase reporter constructs were measured by RT-qPCR. As in the luciferase assays the Renilla luciferase was used as an internal control.
DISCUSSION
In this paper, we used translational ribosome profiling to obtain a detailed and comprehensive map of the open reading frames encoded by EBV. We did not detect any new large open reading frame, suggesting that the EBV large size proteome is essentially complete, but we obtained evidence that several of the viral ORFs are not translated in infected B cells. The central information is that the EBV genome encodes 25 new small uORFs that coincide with 5′ leader ribosome recruitment. We found evidence that at least nine of these uORFs are functional and down regulate downstream protein expression. The construction of viral recombinants with mutations in the uORFs will help defining their precise function in the context of viral infection. Moreover, we identified alternative translation initiation sites in a few viral genes, some of which non-canonical, that possibly endow the different viral isoforms they produce with variable properties. Last, we found evidence of low-level extensive monosomal ribosome coverage of some EBV lytic transcripts that does not equate with translation but points to a new layer of translation regulation that could be important to repress virus replication in infected cells.
The analysis of the EBV TRP is difficult because of the extensive overlaps between viral genes. This approach was facilitated by the harringtonine-treatment of the samples that allowed clear-cut identification of the translation initiation sites. The results obtained after cycloheximide-treatment were more difficult to interpret and the analysis of the triplet periodicity, a quality control typically performed in TRP assays showed only a limited predominance of one reading frame. This could reflect a poor quality of the library or be the result of viral infection (44). However, it should be noted that libraries treated with harringtonine, that showed perfect evidence of triplet periodicity, were also treated with cycloheximide. Thus, an inappropriate cycloheximide treatment should also have deleterious effects on the harringtonine-treated libraries. Moreover, the size of the reads obtained in the TRP that directly reflects the protection of the RNA by ribosomes during RNAse treatment showed the expected distribution. Finally, all results obtained with the cycloheximide-treated cells, except the translation efficiency, were backed by other experiments, either by the results obtained with cells treated with harringtonine or by independent ribosome profiles coupled to qPCR analyses. It should be noted that multiple abnormalities of the translation machinery have been identified in virus-infected cells, including leaky scanning and ribosome frame-shifting (44). Thus, it is possible that EBV infection has a general, so far unrecognized, effect on translation that affects the accuracy of ribosome reading.
The LF1, LF2, LF3, BHLF1, RPMS1 and A73 transcripts had in common that their protein product is difficult to detect (15,45–48). However, they showed highly variable features. While LF1, LF2, LF3, RPMS1 and A73 are transcribed approximately at the same level in M81-infected cells, BHLF1 had much higher transcription rates, as previously reported (49–51). BHLF1, and to a lesser extent LF3, also showed a much higher ribosome coverage in the CDS than the other members of this group. All transcripts except LF2 showed ribosome recruitment in their leader region that led to a high to very high out:in ratio. LF2 also had more ribosome coverage in the CDS region. Both features fit with the detection of the LF2 protein in infected cells (52). In LF1, A73 and RPMS1, the combination of low CDS ribosome coverage and substantial ribosome recruitment in the leader region suggested that these genes are subjected to an active repression of translation. Although these features also apply to LF3 and to BHLF1 whose leader region contains multiple uORFs, ribosome coverage in their CDS region was substantial and could in principle lead to protein translation. However, these transcripts were preferentially associated with monosomal ribosomes and rarely with polysomes, which could explain the previously reported absence of protein synthesis. (30,32,33). There is one report of BHLF1 protein expression in induced EBV-infected cells (46). Interestingly, BHLF1 protein expression in that case included abundant degradation products. Protein degradation could be caused by a nonsense-mediated mRNA decay response that has previously been reported for long non-coding RNAs (53). This response is frequently triggered by uORFs (54). Whether or not the uORFs located in the BHLF1 leader are responsible for the ribosome recruitment in these regions needs to be investigated in more detail. Altogether, our results fit with the suggestion that BHLF1 is a long non-coding RNA, as previously suggested by its role in the regulation of the origin of lytic replication (7). Similar remarks probably apply to LF3, a BHLF1 homolog, although we could not identify any uORF in the leader region of this gene (49,50,55). The recently identified class of lytic transcripts was significantly transcribed and translated in M81-infected cells only (11). However, the absence of translation initiation in M81-infected cells treated with harringtonine suggests that they represent non-coding RNAs, a result that fits with the previous report that they only encode short ORFs (11).
Similar though distinct observations were made in cells infected with the weakly replicating virus B95-8. We found that a significant proportion of the EBV lytic genes, including the master transactivators BZLF1 and BRLF1 showed decreased, but nevertheless significant CDS ribosome coverage, mainly with monosomal ribosomes. The observation that in harringtonine-treated cells ribosomes accumulate in the leader region of these genes and on their translation initiation codon confirms that these lytic transcripts recruit ribosomes, but it is clear that it does not lead to protein production. This suggests the existence of inhibitory mechanisms of viral translation that are relieved in cells such as M81, thereby allowing recruitment of polysomes to the CDS and efficient translation.
The identification of a large number of uORFs in the EBV genome suggests the existence of a new layer of regulation in EBV protein expression. The high degree of conservation of these elements across a large number of EBV strains suggests that they are important for viral functions. Multiple parameters modulate the inhibitory effects of uORFs, including the number of uORFs within the leader region, the strength of the Kozak sequence, the cap to first uORF distance and the distance from the last uORF to the main ORF (reviewed in: (41)). When comparing the characteristics of the EBV uORFs to those of well-characterized cellular uORFs that repress translation, it is clear that the regulatory sequences associated with the EBNA genes and with BHLF1 display features consistent with such a function. We performed luciferase and translation assays that confirmed that at least some of the uORFs are functional and repressive, although their effects were generally weak. This might explain why some genes such as the EBNA genes carry several of them. It is also possible that the full effects of the uORFs will only become visible in B cells infected with a complete virus and undergoing lytic replication. The results of the TRP assays that showed frequent ribosome trapping in the leader region of the genes preceded by the uORF are also concordant with a repressive role of these genetic elements. However, the number of genes showing evidence of ribosome trapping in the leader region exceeded the number of those equipped with uORFs. Other factors such as a high GC content might also influence ribosome distribution in these RNA domains (56). UORFs have previously been identified in mammalian but also viral genomes such as Ebola, KHSV and HCMV (20,21,57). Ribosome profiles obtained from various eukaryotic cell lines have revealed that up to 65% of cellular transcripts contain uORFs (19,58). UORFs can serve multiple functions but generally negatively impact protein expression, up to 80%, by recruiting ribosomes that upon dissociation from the mRNA less frequently reach the start codon of the downstream ORF (59–62). Translation is blocked particularly efficiently when the initiation site from the downstream mRNA and the uORF overlap (58). Transcripts such as BKRF3 and BFRF3 carried uORFs that extend downstream of the AUG of the main ORF, albeit in a different frame, and are very likely to negatively interfere with the translation of their downstream gene, as previously demonstrated for ATF4 (63). We also found evidence for alternative translation starts within the BKRF3 gene. UORFs have previously been shown to drive the preferential expression of particular protein isoforms and this might also be the case for BKRF3 (64,65). We also noted that the BFRF1 and BFLF2 genes both harbor a uORF in their leader regions. This might facilitate a synchronized and equivalent expression of these 2 proteins that interact together to facilitate virus egress across the nuclear membrane (66).
In summary, TRP of EBV-infected cells revealed that the translation of viral genes is highly complex and suggest an important role for ribosome recruitment to the transcripts leader regions. Further work will reveal the role played by the uORFs during viral infection. UORFs rarely permanently repress translation, although this might be the case for non-coding RNAs, as we found for the BHLF1 gene, but rather become activated under stress circumstances. It will be important to delineate these circumstances in the context of viral infection and how they relate to viral functions.
DATA AVAILABILITY
All sequencing data are available under the following NCBI accession number: GSE81802.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the High Throughput Sequencing unit of the Genomics & Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing excellent sequencing services. We are grateful to Helge Lips and Helmut Bannert for expert technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Cancer Research Center [F100]; Institut national de la santé et de la recherche médicale [INSERM]; DKFZ PhD stipend (to M.B.); Jose Carreras charity (to M.H.T.). Funding for open access charge: German Cancer Research Center [F100].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009; 392:1–10. [DOI] [PubMed] [Google Scholar]
- 2. Rickinson A.B., Kieff E.. Knipe DM, Howley PM. Fields Virology. 2007; Philadelphia: Lippincott Williams & Wilkins; 2603–2654. [Google Scholar]
- 3. Laichalk L.L., Thorley-Lawson D.A.. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 2005; 79:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyashita E.M., Yang B., Babcock G.J., Thorley-Lawson D.A.. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 1997; 71:4882–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutzinger R., Feederle R., Mrazek J., Schiefermeier N., Balwierz P.J., Zavolan M., Polacek N., Delecluse H.J., Huttenhofer A.. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009; 5:e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfeffer S., Zavolan M., Grasser F.A., Chien M., Russo J.J., Ju J., John B., Enright A.J., Marks D., Sander C. et al. Identification of virus-encoded microRNAs. Science. 2004; 304:734–736. [DOI] [PubMed] [Google Scholar]
- 7. Rennekamp A.J., Lieberman P.M.. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 2011; 85:2837–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babcock G.J., Hochberg D., Thorley-Lawson A.D.. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000; 13:497–506. [DOI] [PubMed] [Google Scholar]
- 9. Tsai M.H., Raykova A., Klinke O., Bernhardt K., Gartner K., Leung C.S., Geletneky K., Sertel S., Munz C., Feederle R. et al. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep. 2013; 5:458–470. [DOI] [PubMed] [Google Scholar]
- 10. Temple R.M., Zhu J., Budgeon L., Christensen N.D., Meyers C., Sample C.E.. Efficient replication of Epstein-Barr virus in stratified epithelium in vitro. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16544–16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Grady T., Wang X., Honer Zu Bentrup K., Baddoo M., Concha M., Flemington E.K.. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 44. 2016; e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baer R., Bankier A.T., Biggin M.D., Deininger P.L., Farrell P.J., Gibson T.J., Hatfull G., Hudson G.S., Satchwell S.C., Seguin C. et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984; 310:207–211. [DOI] [PubMed] [Google Scholar]
- 13. de Jesus O., Smith P.R., Spender L.C., Elgueta Karstegl C., Niller H.H., Huang D., Farrell P.J.. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 2003; 84:1443–1450. [DOI] [PubMed] [Google Scholar]
- 14. Dolan A., Addison C., Gatherer D., Davison A.J., McGeoch D.J.. The genome of Epstein-Barr virus type 2 strain AG876. Virology. 2006; 350:164–170. [DOI] [PubMed] [Google Scholar]
- 15. Lin Z., Wang X., Strong M.J., Concha M., Baddoo M., Xu G., Baribault C., Fewell C., Hulme W., Hedges D. et al. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J. Virol. 2013; 87:1172–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johannsen E., Luftig M., Chase M.R., Weicksel S., Cahir-McFarland E., Illanes D., Sarracino D., Kieff E.. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16286–16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ersing I., Nobre L., Wang L.W., Soday L., Ma Y., Paulo J.A., Narita Y., Ashbaugh C.W., Jiang C., Grayson N.E. et al. A temporal proteomic map of Epstein-Barr virus lytic replication in B cells. Cell Rep. 2017; 19:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A.S., Bellare P., Holdorf M., Weissman J.S., Ganem D.. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014; 10:e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stern-Ginossar N., Weisburd B., Michalski A., Le V.T., Hein M.Y., Huang S.X., Ma M., Shen B., Qian S.B., Hengel H. et al. Decoding human cytomegalovirus. Science. 2012; 338:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delecluse H.J., Hilsendegen T., Pich D., Zeidler R., Hammerschmidt W.. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 26. Dillies M.A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., Keime C., Marot G., Castel D., Estelle J. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinformatics. 2013; 14:671–683. [DOI] [PubMed] [Google Scholar]
- 27. Irigoyen N., Firth A.E., Jones J.D., Chung B.Y., Siddell S.G., Brierley I.. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016; 12:e1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cenik C., Cenik E.S., Byeon G.W., Grubert F., Candille S.I., Spacek D., Alsallakh B., Tilgner H., Araya C.L., Tang H. et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015; 25:1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P.. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife. 2014; 3:e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hummel M., Kieff E.. Epstein-Barr virus RNA. VIII. Viral RNA in permissively infected B95-8 cells. J. Virol. 1982; 43:262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeang K.T., Hayward S.D.. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J. Virol. 1983; 48:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laux G., Freese U.K., Bornkamm G.W.. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J. Virol. 1985; 56:987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfitzner A.J., Tsai E.C., Strominger J.L., Speck S.H.. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J. Virol. 1987; 61:2902–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O’Grady T., Cao S., Strong M.J., Concha M., Wang X., Splinter Bondurant S., Adams M., Baddoo M., Srivastav S.K., Lin Z. et al. Global bidirectional transcription of the Epstein-Barr virus genome during reactivation. J. Virol. 2014; 88:1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kienzle N., Young D.B., Liaskou D., Buck M., Greco S., Sculley T.B.. Intron retention may regulate expression of Epstein-Barr virus nuclear antigen 3 family genes. J. Virol. 1999; 73:1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McKenzie J., Lopez-Giraldez F., Delecluse H.J., Walsh A., El-Guindy A.. The Epstein-Barr virus immunoevasins BCRF1 and BPLF1 are expressed by a mechanism independent of the canonical late pre-initiation complex. PLoS Pathog. 2016; 12:e1006008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vattem K.M., Wek R.C.. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strong M.J., Laskow T., Nakhoul H., Blanchard E., Liu Y., Wang X., Baddoo M., Lin Z., Yin Q., Flemington E.K.. Latent expression of the Epstein-Barr Virus (EBV)-encoded major histocompatibility complex class I TAP inhibitor, BNLF2a, in EBV-positive gastric carcinomas. J. Virol. 2015; 89:10110–10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987; 7:3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999; 234:187–208. [DOI] [PubMed] [Google Scholar]
- 41. Barbosa C., Peixeiro I., Romao L.. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013; 9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sawada K., Yamamoto M., Tabata T., Smith M., Tanaka A., Nonoyama M.. Expression of EBNA-3 family in fresh B lymphocytes infected with Epstein-Barr virus. Virology. 1989; 168:22–30. [DOI] [PubMed] [Google Scholar]
- 43. Isaksson A., Berggren M., Ricksten A.. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene. 2003; 22:572–581. [DOI] [PubMed] [Google Scholar]
- 44. Reineke L.C., Lloyd R.E.. Animal virus schemes for translation dominance. Curr. Opin. Virol. 2011; 1:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Al-Mozaini M., Bodelon G., Karstegl C.E., Jin B., Al-Ahdal M., Farrell P.J.. Epstein-Barr virus BART gene expression. J. Gen. Virol. 2009; 90:307–316. [DOI] [PubMed] [Google Scholar]
- 46. Nuebling C.M., Mueller-Lantzsch N.. Identification and characterization of an Epstein-Barr virus early antigen that is encoded by the NotI repeats. J. Virol. 1989; 63:4609–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith P.R., de Jesus O., Turner D., Hollyoake M., Karstegl C.E., Griffin B.E., Karran L., Wang Y., Hayward S.D., Farrell P.J.. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 2000; 74:3082–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xue S.A., Lu Q.L., Poulsom R., Karran L., Jones M.D., Griffin B.E.. Expression of two related viral early genes in Epstein-Barr virus-associated tumors. J. Virol. 2000; 74:2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dambaugh T.R., Kieff E.. Identification and nucleotide sequences of two similar tandem direct repeats in Epstein-Barr virus DNA. J. Virol. 1982; 44:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gao Y., Smith P.R., Karran L., Lu Q.L., Griffin B.E.. Induction of an exceptionally high-level, nontranslated, Epstein-Barr virus-encoded polyadenylated transcript in the Burkitt's lymphoma line Daudi. J. Virol. 1997; 71:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hudewentz J., Delius H., Freese U.K., Zimber U., Bornkamm G.W.. Two distant regions of the Epstein-Barr virus genome with sequence homologies have the same orientation and involve small tandem repeats. EMBO J. 1982; 1:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Calderwood M.A., Holthaus A.M., Johannsen E.. The Epstein-Barr virus LF2 protein inhibits viral replication. J. Virol. 2008; 82:8509–8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tani H., Torimura M., Akimitsu N.. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013; 8:e55684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Muhlemann O.. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011; 17:2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Parker B.D., Bankier A., Satchwell S., Barrell B., Farrell P.J.. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990; 179:339–346. [DOI] [PubMed] [Google Scholar]
- 56. Babendure J.R., Babendure J.L., Ding J.H., Tsien R.Y.. Control of mammalian translation by mRNA structure near caps. RNA. 2006; 12:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shabman R.S., Hoenen T., Groseth A., Jabado O., Binning J.M., Amarasinghe G.K., Feldmann H., Basler C.F.. An upstream open reading frame modulates ebola virus polymerase translation and virus replication. PLoS Pathog. 2013; 9:e1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee S., Liu B., Lee S., Huang S.X., Shen B., Qian S.B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ghilardi N., Wiestner A., Skoda R.C.. Thrombopoietin production is inhibited by a translational mechanism. Blood. 1998; 92:4023–4030. [PubMed] [Google Scholar]
- 60. Hughes T.A., Brady H.J.. Expression of axin2 is regulated by the alternative 5′-untranslated regions of its mRNA. J. Biol. Chem. 2005; 280:8581–8588. [DOI] [PubMed] [Google Scholar]
- 61. Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lu P.D., Harding H.P., Ron D.. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004; 167:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kochetov A.V., Ahmad S., Ivanisenko V., Volkova O.A., Kolchanov N.A., Sarai A.. uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 2008; 582:1293–1297. [DOI] [PubMed] [Google Scholar]
- 65. Pelechano V., Wei W., Steinmetz L.M.. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013; 497:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gonnella R., Farina A., Santarelli R., Raffa S., Feederle R., Bei R., Granato M., Modesti A., Frati L., Delecluse H.J. et al. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 2005; 79:3713–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available under the following NCBI accession number: GSE81802.