Abstract
Genes that are inherently subject to strong selective constraints tend to be overretained in duplicate after polyploidy. They also continue to experience similar, but somewhat relaxed, constraints after that polyploidy event. We sought to assess for how long the influence of polyploidy is felt on these genes’ selective pressures. We analyzed two nested polyploidy events in Brassicaceae: the At-α genome duplication that is the most recent polyploidy in the model plant Arabidopsis thaliana and a more recent hexaploidy shared by the genus Brassica and its relatives. By comparing the strength and direction of the natural selection acting at the population and at the species level, we find evidence for continued intensified purifying selection acting on retained duplicates from both polyploidies even down to the present. The constraint observed in preferentially retained genes is not a result of the polyploidy event: the orthologs of such genes experience even stronger constraint in nonpolyploid outgroup genomes. In both the Arabidopsis and Brassica lineages, we further find evidence for segregating mildly deleterious variants, confirming that the population-level data uncover patterns not visible with between-species comparisons. Using the A. thaliana metabolic network, we also explored whether network position was correlated with the measured selective constraint. At both the population and species level, nodes/genes tended to show similar constraints to their neighbors. Our results paint a picture of the long-lived effects of polyploidy on plant genomes, suggesting that even yesterday’s polyploids still have distinct evolutionary trajectories.
Keywords: whole genome duplication, Arabidopsis thaliana, Brassica, dosage balance, metabolic network
Introduction
Flowering plant evolution is characterized by recurrent polyploidy events. However, the genetic redundancy that these events produce is not always long-lived. Instead, polyploid genomes are subject to diploidization, whereby homeologous sequences (which are created by genome duplication events) are then removed by unequal homologous recombination, nonhomologous recombination, and chromosome loss (aneuploidy; Soltis et al. 2015). Even when duplicated regions evade such large-scale losses, the genes they encode may also be disabled by degenerative mutations or chromosomal arrangements (dysploidy; De Storme and Mason 2014) and subsequently be lost through further genetic drift (Lynch and Conery 2000).
As an example, the Arabidopsis thaliana genome retains clear evidence of at least three ancient polyploidies in its history (Maere et al. 2005), all of which are shared with its near relative Arabidopsis lyrata, which split from A. thaliana ∼5 Ma (Van de Peer et al. 2009). The most recent of these events is termed the At-α whole-genome duplication (Blanc et al. 2003; Bowers et al. 2003) and has been dated to roughly 23 Ma (Barker et al. 2009). The majority of duplicated genes created by At-α have returned to a single-copy state, a process known as fractionation (Freeling et al. 2015). Today only ∼30% of the duplicates created by At-α survive in the A. thaliana genome (Bowers et al. 2003). At-α was an allopolyploidy (Blanc et al. 2003), meaning that the two (sub)genomes that merged to form A. thaliana’s ancestor were not identical. These initial differences appear to have driven biased fractionation, which means that retention of genes from one of the subgenomes was favored over the other (Freeling 2009; Woodhouse et al. 2014).
In addition to the At-α event, relatives of A. thaliana in the tribe Brassiceae, which includes Brassica rapa (Chinese cabbage) and Brassica oleracea (broccoli and cauliflower) share a subsequent hexaploidy, the Br-α genome triplication (Lysak et al. 2005; Arias et al. 2014; Liu et al. 2014). This event occurred through two separate hybridization steps (Tang et al. 2012) ∼16 Ma and was particularly marked by its biased pattern of resolution (Cheng et al. 2012). Thus, in B. rapa, one of the three contributing subgenomes is 70% retained, the second subgenome retains 46% of its contributed duplicates, and the most fractionated subgenome retains only 36% of its genes (Wang et al. 2011). The pattern is similar in Brassica oleracea (Liu et al. 2014; Parkin et al. 2014; Cheng et al. 2016). It is believed that the least fractionated subgenome is the one contributed by the second hybridization step in the paleopolyploidy (Tang et al. 2012), meaning that a number of duplicate losses from the other two genomes had already occurred prior to this second hybridization. In a more general sense, it has been proposed that silencing of the subgenome with the higher transposable element load through factors such as differential methylation drives the overall process of biased fractionation (Schnable et al. 2011; Garsmeur et al. 2014; Edger et al. 2017).
Besides these parent-of-origin effects, duplicate loss after polyploidy is also nonrandom with respect to gene function: genes encoding transcription factors, ribosomal proteins, and kinases have survived in duplicate after many phylogenetically independent polyploidies in organisms as diverse as amoebae, plants, vertebrates, and yeasts (Seoighe and Wolfe 1998; Blanc and Wolfe 2004; Maere et al. 2005; Aury et al. 2006; Makino and McLysaght 2010; Jiang et al. 2013; Albalat and Canestro 2016; Li et al. 2016). The force driving this convergence in the retained duplicates, known as ohnologs (Wolfe 2000), is believed to be selection to maintain dosage balance among interacting gene products (Birchler et al. 2005). In other words, because complex assembly and other macromolecular associations follow the rules of biochemical kinetics (Veitia and Birchler 2015), the duplication of only some members of such complexes will tend to drive the concentration of the functional versions of them away from their selectively optimal levels (Veitia 2002; Papp et al. 2003; Maere et al. 2005; Dopman and Hartl 2007; Wapinski et al. 2007; Coate et al. 2011; Rodgers-Melnick et al. 2012; Birchler et al. 2016). However, a duplication of the entire genome will (in general) maintain the required balance (Edger and Pires 2009; Freeling 2009; Makino and McLysaght 2010; Birchler and Veitia 2014). After such an event, selection will then tend to disfavor the loss of duplicated members of such complexes. In keeping with this idea, it is known that genes encoding highly interactive proteins that are in central network positions or genes in dense regulatory pathways are prone to be dosage-sensitive (Hakes et al. 2007; Birchler and Veitia 2012) and are overretained postpolyploidy (Freeling and Thomas 2006; Birchler and Veitia 2007; Bekaert et al. 2011; Conant 2014; Conant et al. 2014).
Such functional observations lead to the question of how natural selection acts on duplicated genes. Analyses of the pattern of selective constraint seen in duplicated and single-copy genes have shown that, despite their redundancy, duplicated genes tend to actually show relatively high selective constraint, measured as a numerically small value of the ratio of nonsynonymous to synonymous substitutions; Ka/Ks (Davis and Petrov 2004; Jordan et al. 2004). Davis and Petrov (2004) used an elegant approach of looking at the selective constraint of a duplicated gene pair’s orthologs in unduplicated outgroups to show that this difference in constraint, rather than being an effect of duplication, reflects a propensity for intrinsically more constrained genes to survive in duplicate. When this tendency is coupled to the known characteristics of surviving ohnologs, such as increased essentiality and a higher propensity for having expression phenotypes (Hakes et al. 2007; Wapinski et al. 2007), it is natural to speculate that the difference in constraint between duplicates and singletons might be partly driven by retained WGD-produced duplicates in the genomes studied. And indeed, in plants, the mean Ka/Ks ratio of duplicate genes produced by small-scale duplicates is higher than that for WGD duplicates (Carretero-Paulet and Fares 2012). In fact, a relatively clear series can be drawn: recent ohnologs have higher constraint than older ones, ohnologs are more constrained than duplicates from other mechanisms, and duplicated genes are more constrained than those without duplicates (Yang and Gaut 2011). In the same vein, Scannell and Wolfe (2008) demonstrated that the ohnologs in modern bakers’ yeast were indeed drawn from a class of genes showing higher constraint in outgroup species lacking the yeast WGD. In their analysis, they also showed that the WGD did produce a relaxation of constraint (Lynch and Conery 2000), but not enough to overcome these genes’ intrinsically higher initial constraints.
Both of these studies note a temporal component to the selection acting on ohnologs (Scannell and Wolfe 2008; Yang and Gaut 2011) which has suggestive links to recent research outlining a functional progression in the evolution of polyploid genomes. In the earliest phases of the evolution of a polyploid species, there appears to be forces acting to favor the removal of duplicate copies of genes with functions in DNA repair or that are targeted to the organelles (Edger and Pires 2009; De Smet et al. 2013; Conant 2014). Selection for dosage balance is a critical force in the next phase, with potential functional partitioning or innovation occurring even later in time (Bekaert et al. 2011; Conant et al. 2014).
How long can dosage balance be maintained before the dosage constraints eventually change as genes diverge over time (Birchler and Veitia 2012)? How do selective patterns change after polyploidy? Are paleopolyploids still different from diploids in this regard? We hypothesized that genes retained in duplicate/triplicate remain under higher constraint even long after polyploidy. To test this hypothesis, we exploited the nested polyploid events in Arabidopsis and Brassica, using a different approach than previous work on these two polyploidies (Woodhouse et al. 2014). We chose the two nested polyploidies in the Brassicaceae family as our model system because of the availability of deep functional genomic resources and multiple sequenced genomes of A. thaliana, A. lyrata, B. rapa, and B. olecarea. Our goal here was to link the previous functional studies on the changing face of polyploidy through time with work on the selective constraint acting on the ohnologs. Hence, we asked whether the patterns of enhanced constraint among duplicates have continued to the present day by comparing the selective constraint seen between species to that seen in the circulating polymorphisms within two paleopolyploid species. We demonstrate that surviving ohnologs are under more stringent selective constraint even in present day populations, but that this pattern of constraint was nonetheless intrinsic to them prior to polyploidy.
Materials and Methods
Signatures of the At-α Duplication and Br-α Triplication
We obtained pairs of At-α duplicates using CoGe’s SynMap algorithm (Lyons, Pedersen, Kane, and Freeling 2008) and merged this list with the At-α duplicates reported by Bowers et al. (2003) using version 10.02 of the Arabidopsis thaliana Col-0 genome as our reference (The Arabidopsis Genome Initiative 2000). We excluded from our ohnolog lists any conflicts between the two lists and any “pairs” with more than two genes, resulting in 2,768 duplicated gene pairs that originated at the At-α event. Similarly, to identify regions created by the Brassica triplication, we used SynMap and GEvo in CoGe (Lyons, Pedersen, Kane, Alam, et al. 2008) to infer the orthologous relations of A. thaliana, B. rapa, and B. oleracea genes from the syntenic regions of the three genomes, requiring a syntenic score of 10 in a window of 60 genes. Versions 1.5 and 2.1 of the Brassica rapa (Wang et al. 2011) and Brassica oleracea TO1000 genomes (Liu et al. 2014; Parkin et al. 2014) were used, respectively.
Between-Species Divergences
We obtained maximum likelihood estimates of Ka (the number of nonsynonymous substitutions over the number of total possible nonsynonymous sites; Miyata and Yasunaga 1980) and Ks (the number of synonymous substitutions per synonymous site) using GenomeHistory (Conant and Wagner 2002) for three sets of homologous protein-coding genes pairs. We omitted pairs with Ks < 0.001 from our analysis. The first set of genes consists of ortholog pairs from A. thaliana Col-0 and A. lyrata v1.0 (Hu et al. 2011): we denote these data as At-Al Ka/Ks. We identified these pairs through an analysis of syntenic regions using the SynMap tool with the Last algorithm (Frith and Kawaguchi 2015) in CoGe (Lyons and Freeling 2008; Tang et al. 2011). We divided these pairs into orthologs from retained At-α duplicates and single copy syntenic orthologs and analyzed the distributions of Ka/Ks for each.
Our second set of homologous pairs is drawn from the comparison of B. rapa and B. oleracea, and we obtained genes that preserved synteny between the two species from the CoGe SynMap algorithm (Lyons, Pedersen, Kane, and Freeling 2008). Because of the WGT, a single ancestral syntenic locus is now represented by three syntenic loci in both B. rapa and B. oleracea, resulting in three sets of homology relationships for each B. rapa gene in B. oleracea. We binned the syntenic homologs and considered only loci that fell into three groups. The first group consisted of one-to-one ortholog pairs (Br-Bo 1:1), where the other two WGT-produced paralogs have been lost in both Brassica genomes. In the second group, we included only loci with all surviving genes from the WGT in both genomes, resulting in three-to-three homology relations (Br-Bo 3:3). For this second group, each B. rapa and B. oleracea orthologous triplet is equivalent to nine pairs of genes: we used a distance-based approach to resolve these three-to-three homology relationships. From each of the triplets, we took the B. rapa and B. oleracea gene pair with the closest Ka value and defined them to be the first pair of orthologs. Next, we found the B. rapa/B. oleracea pair with the closest Ka from the remaining two-to-two relations, defining them as the second orthologous pair. The remaining two genes were defined as the third pair of orthologs. For validation, we generated a list of the most plausible syntenic gene pairs from the SynMap results. We excluded one-to-many or many-to-many syntenic relations by starting from the largest synteny block containing members of the triplet and removing any other syntenic relationships for that pair of genes (which were necessarily in smaller synteny blocks). We found that 80.7% of our first pairs of orthologs (those closest in Ka) were in one-to-one synteny. The corresponding figures for the second and third pairs were 75.4% and 79.1%, respectively. Our third and final group contained B. rapa and B. oleracea genes that had each lost exactly one gene after triplication (Br-Bo 2:2). The ortholog pairs from these two-to-two homologous relationships were inferred using the approach just described. Ka/Ks was then computed for all the B. rapa and B. oleracea orthologous pairs (denoted Br-Bo Ka/Ks across all three groups).
Our final set of homolog pairs consists of A. thaliana genes with their B. rapa orthologs, subdivided into the two groups (surviving paralogs from the triplication and single-copy genes) just described (denoted At-Br Ka/Ks).
To assess if the selective constraint of surviving polyploid-produced duplicates differed from that of the single copy genes, we took the (nonzero) Ka/Ks values for the two groups and analyzed their distribution. We first fit separate lognormal distributions to the Ka/Ks values from the At-α duplicates and the single copy genes, computing the likelihood of observing the set of Ka/Ks values for each (Lduplicate and Lsingle-copy, respectively). We then fit a single log-normal distribution to the pooled Ka/Ks values from both, yielding the likelihood Lcombined. We then tested the hypothesis of a difference in these distributions using a likelihood ratio test, comparing D, twice the natural log of the ratio of (Lduplicate×Lsingle-copy) over Lcombined, to a chi-square distribution with 2 degrees of freedom (Wilks 1938):
| (1) |
We performed the same analysis with the triplicated genes and single-copy genes from the comparison of B. rapa and B. oleracea.
Within-Species Variation
To quantify the actions of selection at the population level, we estimated the number of circulating synonymous and nonsynonymous polymorphisms using SNP data. For A. thaliana, we obtained polymorphism data in variant call format (VCF) from 1,135 natural inbred lines curated by the 1001 Genomes Project (Alonso-Blanco et al. 2016). SNPs in protein-coding genes were annotated as synonymous or nonsynonymous polymorphisms using SnpEff (Cingolani et al. 2012).
The B. rapa SNPs were called from the transcriptomes of 126 accessions (Qi et al. 2017), and also annotated with SnpEff. Low quality SNPs were removed with vcffilter (https://github.com/vcflib/vcflib; last accessed March 21, 2018) and VCFtools (Danecek et al. 2011): only SNPs identified in regions with read depth >10 and root mean square mapping quality >30 were used for subsequent analyses.
To normalize the number of observed SNPs per gene, we counted the number of nonsynonymous positions in each protein-coding gene showing polymorphism and divided that number by the total number of nonsynonymous changes possible in the gene. We denote this ratio as pN. Similarly, pS is the number of polymorphic synonymous positions divided by the number of possible synonymous sites (McDonald and Kreitman 1991). We then calculated the ratio of pN/pS for all possible protein-coding genes in A. thaliana and B. rapa. We removed genes with pS = 0 and pN ≤ 1; for those with pS = 0 and pN > 1, pN/pS was set to 1. We fit the distributions of pN/pS to lognormal curves as above.
Identifying Changes in Selective Constraints Prior to and Postpolyploidy
Using the syntenic orthology relations inferred for A. thaliana versus A. lyrata, A. thaliana versus B. rapa, and B. rapa versus B. oleracea that we obtained from SynMap (Lyons, Pedersen, Kane, and Freeling 2008), we linked the orthologous genes of these four species together. All four species share the At-α duplication, and the two Brassica species share the Br-α triplication. We calculated Ka/Ks for the comparison of A. thaliana and A. lyrata orthologs (At-Al), A. thaliana and B. rapa orthologs (At-Br), and B. rapa and B. oleracea orthologs (Br-Bo), respectively. At the within-species level, we calculated pN/pS for A. thaliana genes using resequencing data from 1,135 accessions (Alonso-Blanco et al. 2016) and for B. rapa from transcriptomic data on 126 accessions (Qi et al. 2017).
We partitioned each group of selective constraints into four subgroups, groups where: 1) both the Arabidopsis and Brassica lost the extra copies and returned to singleton state after the At-α duplication and the Br-α triplication, respectively, 2), Arabidopsis retained both At-α duplicates, but Brassica lost the triplicated copies returned to singleton state, 3) Arabidopsis lost the duplicated copy, but Brassica retained all three copies after the WGT, 4) Arabidopsis retained At-α duplicates and Brassica retained Br-α triplets.
We created notched boxplots for the selective constraints across the four different subgroups in R. By comparing the selective constraints in these subgroups, we could observe how selective constraints shifted after At-α WGD, before Br-α WGT, and after Br-α. We could also assess whether these changes were still preserved among populations of extant species. Nonparametric multiple comparison tests using Kruskal–Wallis tests as a pairwise basis (Siegel and Castellan 1988) were performed using the kruskalmc function in the pgirmess package in R (https://github.com/pgiraudoux/pgirmess; last accessed March 21, 2018).
To again examine the differences in functions between genes retained after polyploidy versus single copy genes, we used the gene list analysis tool from the PANTHER classification system (Mi et al. 2017) and performed overrepresentation tests of molecular function and biological process Gene Ontology (GO) terms using a list of A. thaliana genes that are orthologous to surviving triplicates in B. rapa, compared against another A. thaliana gene list that contains genes in one-to-one orthology relationships with single-copy (with respect to Br-α) genes in B. rapa.
Metabolic Network Analysis
We employed an updated version of the Arabidopsis thaliana metabolic network (de Oliveira Dal’Molin et al. 2010) that we have previously described as AraGEM v1.2 (Bekaert et al. 2012). Each node in this network represents a biochemical reaction (with associated enzyme-coding genes), and edges connect pairs of nodes with shared metabolites. We estimated the selective constraint for each node in the network by taking the average Ka/Ks or pN/pS of all the genes mapped to that reaction. To observe how the selective constraint changes from the population level to species level, we only included A. thaliana genes that both have At-Al Ka/Ks values and within-species pN/pS values. We also inferred a draft-quality B. rapa metabolic network by mapping the reactions catalyzed by A. thaliana genes onto their corresponding orthologs in B. rapa. We further refined the inferred B. rapa network by limiting it to the subset of B. rapa genes that have small nonsynonymous distances to their A. thaliana orthologs (i.e., having At-Br Ka values below the 75% percentile for the full set of genes in the network). The assumption here is that such orthologs are even more likely to retain the enzymatic function of their A. thaliana counterparts. The metabolic networks were visualized using Gephi v0.9.1 (Bastian et al. 2009) with the layout algorithm Force Atlas.
We computed three measurements of importance for the nodes in the network. The first was the node degree, that is, the number of edges connected to that node (Hakimi 1962). The next was the clustering coefficient, defined as the ratio of the number of observed connections among each triplet of nodes to the maximum number of such connections possible (Watts and Strogatz 1998). Third and finally, we computed each node’s betweenness centrality, which is the number of the network’s shortest paths passing through that node (Brandes 2001). For each statistic, we calculated the Spearman’s correlation coefficient between the nodes’ mean selective constraints and the statistic in question.
We also conducted an analysis of the similarity of the selective constraint of adjacent nodes, defining the weight of the edge connecting two nodes as the absolute value of the difference in the constraint values of those two nodes (e.g., pN/pS or Ka/Ks). To assess if adjacent nodes were more similar in constraint than expected, we generated 10,000 random networks with identical structure but randomized assignments of constraints to nodes. We compared the average and sum of the edge weights for these random networks to those of the real networks.
Results
Arabidopsis Genes Retained after At-α Polyploidy Are under Stronger Selective Constraint
We compared the between-species selective constraint, measured with the ratio of Ka/Ks, for genes retained in duplicate and those returned to single-copy after polyploidy. Ka/Ks values <1.0 suggest purifying selection against amino acid substitutions and values >1.0 are indicative of directional selection (Li 1997). We made similar comparisons using the within-species constraints estimated with the ratio pN/pS (Materials and Methods), a ratio that reports the proportion of all nonsynonymous and synonymous sites with circulating polymorphisms in a population. Thus, we calculated five groups of selective constraints: three at the species level: At-Al (A. thaliana to A. lyrata) Ka/Ks, At-Br (A. thaliana to B. rapa) Ka/Ks, and Br-Bo (B. rapa to B. oleracea) Ka/Ks; and the other two at population level: pN/pS for A. thaliana and for B. rapa. We expected that the ratio pN/pS would exceed Ka/Ks due to the circulation of low frequency, mildly deleterious polymorphisms in populations, some of which are eventually purged over the longer times represented by the between-species comparisons.
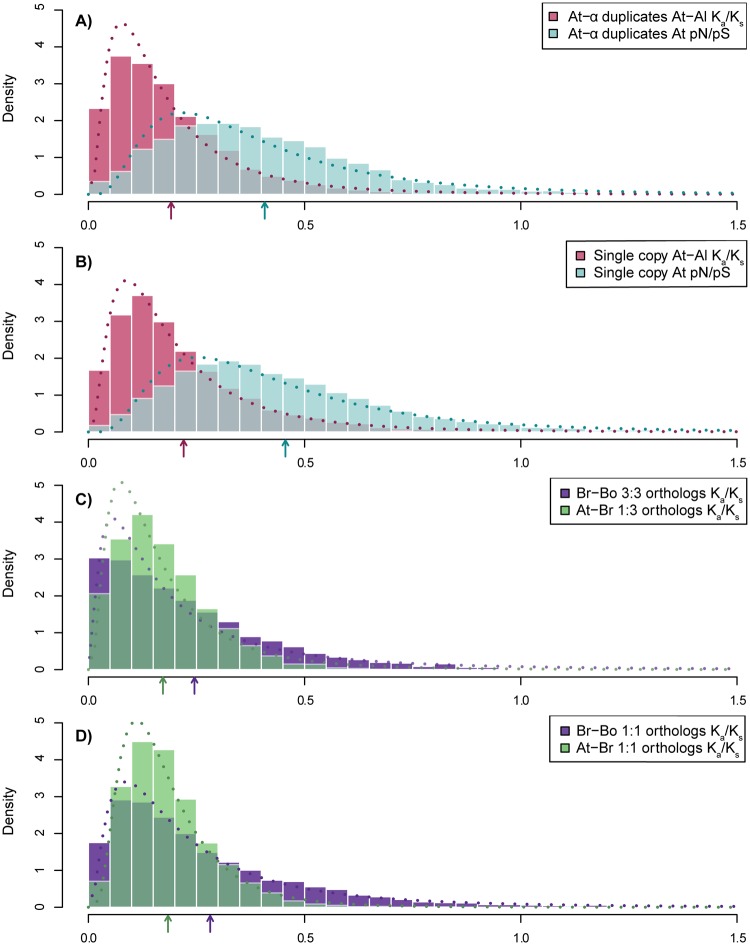
Figure 1 shows the density distributions of Ka/Ks and pN/pS for retained At-α duplicates and Br-α triplicates and for genes that returned to single copy after these polyploidy events. We fit the selective constraints to lognormal distributions and performed likelihood ratio tests to compare each pair of distributions. The distributions of selective constraint for retained duplicates/triplicates and for single copy genes are significantly different, both for the within-species population data and for between-species comparisons, with the duplicates having higher constraint in all cases (see also table 1).
Fig. 1.
—Distributions of measures of selective constraints. (A and B) Selective constraints of Arabidopsis thaliana genes that retained both At-α duplicates, and genes that returned to single copy after At-α. (A) Red: distribution of At-Al Ka/Ks for A. thaliana genes that survived At-α, blue: distribution of within-species pN/pS for corresponding retained At-α duplicates that have orthologs in A. lyrata; (B) Red: distribution of Ka/Ks for At-Al 1:1 orthologs, blue: distribution of pN/pS for the corresponding A. thaliana genes. Dotted lines: fitted lognormal density distribution curves. The distributions of Ka/Ks from retained At-α duplicates and from single-copy genes are significantly different (P < 0.00001), as are the two distributions for pN/pS (P < 0.00001). (C and D): Selective constraints of Brassica rapa genes that survived the Br-α triplication, and those of the single-copy genes, as well as the selective constraints of their A. thaliana orthologs. (C) Purple: distribution of Ka/Ks for Br-Bo orthologs that preserved three copies in both B. rapa and B. oleracea, green: distribution of Ka/Ks for At-Br orthologs where A. thaliana genes are orthologous to the same B. rapa gene set (retained triplets). (D) Purple: distribution of Ka/Ks for single copy Br-Bo orthologs, green: distribution of Ka/Ks for single copy At-Br orthologs. Dotted lines are as for (A) and (B). The distributions of Ka/Ks for Br-Bo 3:3 and Br-Bo 1:1 orthologs are significantly different (P < 0.00001); the distributions of Ka/Ks for At-Br 1:3 and At-Br 1:1 orthologs are also significantly different (P < 0.00001). Arrows mark the average selective constraint of each distribution. See also table 1.
Table 1.
Average Selective Constraints
| Selective Constraints | Single-Copy Orthologs |
Retained Duplicates/Triplicates |
% Differencec | ||
|---|---|---|---|---|---|
| Sample Sizea | Mean Valueb | Sample Sizea | Mean Valueb | ||
| At vs. Al Ka/Ksd | 11,966 | 0.2203 | 4,261 | 0.1914 | −13.13 |
| At vs. Br Ka/Kse | 5,367 | 0.1843 | 5,069 | 0.1724 | −6.42 |
| Br vs. Bo Ka/Ksf | 7,604 | 0.2814 | 5,680 | 0.2454 | −12.79 |
| At 1135 ecotypes pN/pSg | 14,293 | 0.4557 | 4,839 | 0.4078 | −10.50 |
| Br 126 accessions pN/pSh | 1,316 | 0.1269 | 1,031 | 0.1285 | 1.19 |
Note.—See also figure 1.
Sample size for the calculation of mean selective constraint (b): for Ka/Ks this value corresponds to the number of orthologous pairs; for pN/pS to the number of genes.
Mean value of the measure of selective constraint in question (i.e., Ka/Ks or pN/pS, left).
The difference as a percentage of the selective constraints of single copy genes.
The average Ka/Ks computed between Arabidopsis thaliana and A. lyrata.
The average Ka/Ks computed between A. thaliana and Brassica rapa.
The average Ka/Ks computed between B. rapa and B. oleracea.
The average pN/pS for A. thaliana genes with an ortholog in A. lyrata. About 1,610 genes with pS = 0 and pN ≤ 1 were removed. pN/pS values for 150 genes with pS = 0 and pN > 1 were set to 1. The total number of genes after filtering was 25,806. Only genes with orthologs in A. lyrata were included in the analysis (as noted in a).
The average pN/pS for B. rapa genes with orthologs in B. oleracea. About 266 genes with pS = 0 and pN ≤ 1 were removed. pN/pS values for 129 genes with pS = 0 and pN > 1 were set to 1. The total number of genes after filtering was 5,128. Only genes with orthologs in B. oleracea were included in the analysis (as noted in a).
We also note that the separation between values of pN/pS and Ka/Ks was smaller for genes surviving in multiple copies post-WGD/WGT relative to those returned to single copy (fig. 1A and B). It takes more time for purifying selection to act on mildly deleterious polymorphisms than on more strongly deleterious ones (Hartl and Clark 1997). Hence, this observation might suggest stronger selection acting on these retained paralogs, such that, even at the population level, there is relatively fast-acting evolutionary pressure to remove deleterious variants (Cao et al. 2011). For both the within-population and the between-species comparisons of A. thaliana and A. lyrata, constraint also decreases as one moves from surviving At- α duplicates found in both genomes to At-α duplicates specific to A. thaliana to single-copy genes (supplementary fig. S1A, Supplementary Material online).
Similar patterns are seen when comparing the triplicated and single-copy genes between B. rapa and B. oleracea and for the comparison with their corresponding A. thaliana orthologs (fig. 1C and D;table 1), with both triplicated Brassica genes and their A. thaliana orthologs being more constrained. For completeness, we also considered the case of Br-Bo 2:2 pairs (i.e., both genomes retained exactly two syntenic copies after triplication; supplementary fig. S1B, Supplementary Material online). As expected, the average selective constraint of the At-Br 1:2 orthologs falls between the At-Br 1:1 and At-Br 1:3 cases. An apparently similar trend was seen in the B. rapa population data, but we lacked the statistical power to detect differences in pN/pS between the two groups when using only 126 transcriptomes for our SNP detection.
Non-WGT Orthologs of Retained Brassica Triplets Are under Strong Purifying Selection
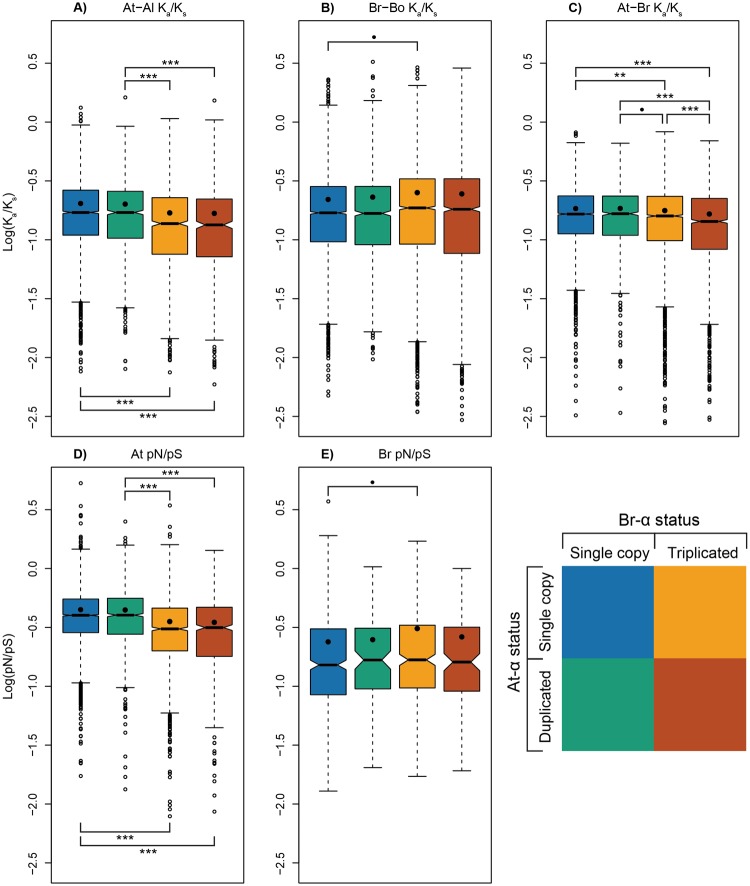
Since the Br-α triplication is specific to genus Brassica, we used the Arabidopsis genomes to estimate the patterns of selective constraint that may have acted on the Brassica genomes prior to that triplication. In particular, we can partly assess whether the ohnologs’ reduction in constraint is driven by their intrinsic properties or the polyploidy event itself. Figure 2 shows the selective constraints for the At-Al-Br-Bo syntenic orthologs in the cases where duplicates or triplicates are either lost or preserved (i.e., the four subgroups described in the Materials and Methods). The estimated constraint of the triplicated Brassica genes absent the triplication, inferred using the constraint seen between these genes’ orthologs in A. thaliana and A. lyrata (which both lack the Brassica hexaploidy), was smaller than for the corresponding single-copy genes by ∼17% (supplementary table S1, Supplementary Material online). This difference was seen regardless of whether the At-α duplicates were retained (green/red boxplots in fig. 2A and D, P < 0.0001) or lost (blue/yellow boxplots in fig. 2A and D, P < 0.0001). The WGT apparently ameliorated this difference in constraint: when comparing Ka/Ks in B. rapa and B. oleracea orthologs that are also in synteny with A. thaliana, we observed a 12% increase in the average selective constraint in the triplicated orthologs relative to the single copy pairs (fig. 2B and supplementary table S1, Supplementary Material online). This relaxation in constraint among these triplicates may be due to the redundancy they introduce (Conant and Wolfe 2008). However, when taking all Br-Bo orthologs into consideration, regardless of whether there is syntenic orthology in A. thaliana, we observed a ∼13% reduction in the Ka/Ks of the Br-Bo 3:3 genes relative to the Br-Bo 1:1 orthologs (table 1 and supplementary fig. S1B, Supplementary Material online). This apparent inconsistency is caused by the presence of a group of fast-evolving single copy Brassica genes that lack synteny with A. thaliana and that increase the average Ka/Ks value for the total set of single-copy Brassica genes.
Fig. 2.
—Notched box plots of log selective constraints among Al-At-Br-Bo syntenic orthologs. Colors indicate the loss/retention state of Arabidopsis orthologs after At-α WGD, and Brassica orthologs after Br-α WGT. “Lost”: genes returned to singleton state after polyploidy, “retained”: duplicated/triplicated copies are preserved in the genome. Subplots are boxplots of (A) log(Ka/Ks) for A. thaliana versus A. lyrata, (B) log(Ka/Ks) for B. rapa versus B. oleracea, (C) log(Ka/Ks) for A. thaliana versus B. rapa; and (D) log(pN/pS) for 1,135 A. thaliana ecotypes, (E) log(pN/pS) for 126 B. rapa accessions. The notches are 95% confidence intervals of the medians. Kruskal–Wallis multiple comparison tests were performed to evaluate significant differences across medians, P values: ***P < 0.0001, **P < 0.001, *P < 0.01, •P < 0.05. The black dots represent the log(mean) selective constraints. See also supplementary table S1, Supplementary Material online.
For those retained Br-α triplets in the B. rapa and B. oleracea (yellow and red boxplots in fig. 2), the stronger selective constraints associated with being members of a surviving At-α duplicate pair can be observed both before (fig. 2A and C) and after triplication (fig. 2B). This pattern also extends to the population level (fig. 2D and E). However, among the Brassica genes that returned to singleton state (blue and green boxplots in fig. 2), those genes deriving from surviving Arabidopsis ohnologs show a relaxation in constraint in the Brassica lineage (supplementary table S1, Supplementary Material online).
Biased fractionation allows researchers to detect three distinct subgenomes in B. rapa: the least-fractionated subgenome (LF), and the two more-fractionated subgenomes (MF1 and MF2; Wang et al. 2011; Cheng et al. 2012). It is natural to ask if our conclusions regarding selective constraint are sensitive to this fractionation pattern. As shown in supplementary figure S2, Supplementary Material online, the At-Br Ka/Ks estimates are consistent across three subgenomes. Brassica rapa and B. oleracea orthologs from the LF subgenome are slightly more constrained when compared with genes from the other two subgenomes for both the Br-Bo 1:1 and Br-Bo 3:3 cases. Thus, biased fractionation will not significantly affect our overall conclusions regarding the constraints observed for retained ohnologs and single-copy genes, but it is intriguing that genes from the more retained genome are also apparently more constrained.
Selective Constraints Are Correlated with Clustering Coefficients in the Metabolic Network
We reduced the full Arabidopsis metabolic network to include only nodes representing reactions where the enzyme-coding genes involved have A. lyrata orthologs. This simplified network contains 1,068 nodes (reactions) and 14,864 edges (representing shared metabolites between the reactions of the connected nodes). Our draft version of the inferred Brassica metabolic network contains 949 nodes and 11,499 edges. When we required that the nodes in question had estimated values for both Ka/Ks and pN/pS in B. rapa, the resulting network contains 595 nodes and 5,064 edges.
Table 2 shows the Spearman’s rank-order correlation coefficient (ρ) between a number of network statistics and the selective constraints Ka/Ks and pN/pS, respectively. Selective constraints and clustering coefficients were significantly positively correlated in both the Arabidopsis and Brassica metabolic networks, and the correlation was stronger for the between-species comparisons. This observation also holds for a further reduced subset of Brassica network, where only highly conserved B. rapa genes were included (Materials and Methods; values in parentheses in table 2). Clustering coefficient is defined as the ratio of the number of observed connections between a node’s neighbors to the maximum number of possible connections (Watts and Strogatz 1998). Only Ka/Ks for the At-Al orthology comparisons was significantly correlated with node degree, in contrast to other studies reporting either no association or the expected weak negative association (Fraser et al. 2002; Bloom and Adami 2003; Jordan et al. 2003; Hahn et al. 2004). The selective constraints showed no significant correlations with betweenness centrality (the number of shortest paths passing through the node) in the Arabidopsis network, which differs from the pattern seen in protein-interaction networks (Hahn and Kern 2005). There was a significant negative correlation between selection and betweenness centrality in the Brassica network. In general, all of the correlations observed, significant or otherwise, were numerically small, suggesting that metabolic network structure is probably not a major driver of constraint in these taxa.
Table 2.
Spearman’s Correlations of Selective Constraints and Network Statistics
| Gene Setsa | Ka/Ks or pNpS | Number of Nodesb | Number of Edgesc | Selection and Node Degreed |
Selection and Clustering Coefficiente |
Selection and Betweenness Centralityf |
|||
|---|---|---|---|---|---|---|---|---|---|
| Spearman’s Correlation | P Value | Spearman’s Correlation | P Value | Spearman’s Correlation | P Value | ||||
| At-Al ∩ At | At-Al Ka/Ksg | 1,068 | 14,864 | 0.0856 | 0.002* | 0.1459 | 0.000* | 0.0067 | 0.443 |
| At pN/pSh | 1,068 | 14,864 | 0.0249 | 0.208 | 0.0882 | 0.001* | −0.0353 | 0.128 | |
| Br-Bo | Br-Bo Ka/Ksi | 949 (865) | 11,499 (9467) | −0.0358 | 0.120 | 0.1175 | 0.000* | −0.1295 | 0.000* |
| −0.0472 | 0.083 | 0.0975 | 0.002* | −0.1002 | 0.002* | ||||
| Br-Bo ∩ Br | Br-Bo Ka/Ksj | 595 (548) | 5,064 (4455) | 0.0030 | 0.503 | 0.0479 | 0.104 | −0.1010 | 0.003* |
| 0.0149 | 0.392 | 0.0986 | 0.008* | −0.0910 | 0.010* | ||||
| Br pN/pSk | 595 (548) | 5,064 (4455) | 0.0879 | 0.014* | −0.0126 | 0.397 | 0.0284 | 0.254 | |
| 0.1216 | 0.001* | 0.0183 | 0.343 | 0.0757 | 0.040* | ||||
P value < 0.05.
Metabolic networks defined from three gene sets: 1) the Arabidopsis network with genes appearing in both the Ka/Ks and the pN/pS analyses, 2) the full Brassica network and 3) the reduced Brassica network with genes that appeared in both the Ka/Ks and the pN/pS analyses.
Number of nodes in the metabolic network. Each node is a biochemical reaction.
Number of edges in the metabolic network. An edge connects two nodes in the network if the reactions for those nodes share a metabolite.
Node degree is the number of edges connected to a node.
Clustering coefficient is defined as the ratio of existing links connecting a node’s neighbors to each other over the maximum possible number of such links.
Betweenness centrality is the number of the network’s shortest paths that pass through a node.
The Ka/Ks for each node, calculated by taking the average of Ka/Ks of enzyme-coding genes corresponding to the reaction of the node, computed between A. thaliana and A. lyrata.
The average pN/pS for each node calculated in a similar way, for A. thaliana genes with an ortholog in A. lyrata.
The average Ka/Ks for each node, computed between B. rapa and B. oleracea. Results for a subset of B. rapa genes for which At-Br Ka < 0.1127 are shown in parentheses.
The average Ka/Ks for each node, computed between B. rapa and B. oleracea, for an intersection set of genes that appeared in both the Ka/Ks and the pN/pS analyses. Results for a subset of B. rapa genes for which At-Br Ka < 0.1127 are shown in parentheses.
The average pN/pS for each node, for B. rapa genes with orthologs in B. oleracea. Results for a subset of B. rapa genes for which At-Br Ka < 0.1127 are shown in parentheses.
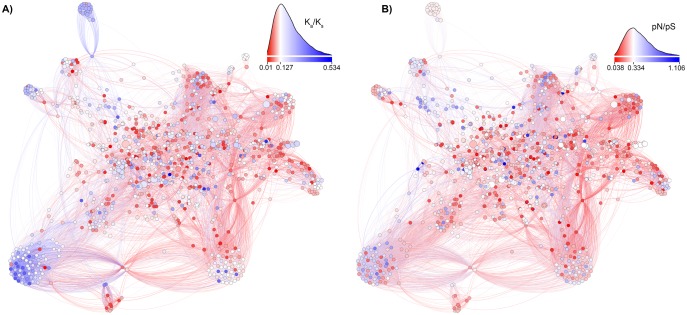
Figure 3 shows the selective constraint both between species (At-Al Ka/Ks) and within species (At pN/pS) for nodes in the Arabidopsis metabolic network. Nodes where the Ka/Ks and pN/pS are below the mean for all nodes are colored in red, and those that are less constrained than average are in blue. We observed that visually tight clusters of nodes in this diagram appear to be less constrained, leading to our analyses testing whether neighboring nodes shared similar constraints (next section).
Fig. 3.
—The distributions of selective constraints in the Arabidopsis thaliana metabolic network. Nodes represent biochemical reactions and are colored with the average selective constraints of genes encoding enzymes for each reaction. The diameter of each node is in proportion to the number of genes for the node. Edges connect two nodes if the two reactions share compounds. (A) Nodes are colored by At-Al Ka/Ks, with red indicating a Ka/Ks is below the network mean, and blue above that mean. The histogram shows the density distribution of At-Al Ka/Ks. (B) Nodes are colored by At pN/pS, with red indicating below-average constraints, and blue above. The histogram shows the density distribution of At pN/pS.
Adjacent Enzymes Share Similar Selective Constraints
We defined the weight of an edge in our network as the absolute value of the difference between the mean selective constraints (pN/pS or Ka/Ks) of its incident nodes. Supplementary table S2, Supplementary Material online, shows the sum and average edge weights of the real network and of randomized networks. At both species level and population level in A. thaliana, the sum of edge weights of the real metabolic networks is smaller than those of randomized networks, suggesting that genes under similar selective pressure tend to cluster in the network. A similar trend was seen for the comparisons between B. rapa and B. oleracea, but no significant difference was observed between the real and randomized networks for the comparison of the B. rapa populations, likely due to small sample sizes.
Discussion
As reported by others (Jordan et al. 2004; Scannell and Wolfe 2008; Yang and Gaut 2011), it is clear that polyploidy-produced duplicates are under stronger constraint than their single-copy counterparts. In a sense, this result is also not surprising, as it parallels the known functional biases of the retained ohnologs: ohnologs tend to fall into functional groups such as regulation of transcription, intracellular signal transduction and formation of multisubunit complexes (Warren et al. 2010; Wang et al. 2011; supplementary tables S3 and S4, Supplementary Material online). Such ohnologs rely on kinetic and stoichiometric balance (Birchler et al. 2016), and dosage perturbations in them can only be tolerated in a narrow range. As a result, mutations that could alter dosage are strongly selected against (Birchler and Veitia 2014; Pires and Conant 2016). It is possible that other types of mutations are equally selected against, accounting for the globally reduced selective constraints. On the other hand, there is also an interesting relationship between gene expression and selective constraint, with high gene expression strongly predicting high constraint (Drummond et al. 2005, 2006). Patterns of these kinds may drive our observations of the relaxation in selection on the Brassica triplicates (where genes from more fractionated subgenomes show relaxed constraint relative to the less fractionated subgenome), given that genes in the less fractioned subgenome also tend to show higher expression levels (Woodhouse et al. 2014).
In yeast, Scannell and Wolfe (2008) showed that a) ohnologs had intrinsically higher constraint, that b) WGD relaxed this constraint somewhat, and c) that even a relatively long time after WGD, the constraint on the ohnologs had not fully returned to the pre-WGD level. Hence, it is natural to ask two related questions: 1) can we detect both the intrinsically higher constraint and its postpolyploidy relaxation in plants as well? and 2) is the increased constraint on ohnologs acting even today for paleopolyploid species?
In answer to question #1, we have clearly shown that genes retained in duplicate/triplicate are intrinsically more constrained. When compared with the subset of Brassica single-copy genes with syntenic orthologs in A. thaliana, triplicated Brassica genes actually show less constraint than do those single copy genes. When all Brassica genes are considered, regardless of their status in Arabidopsis, the triplicated genes are on average more constrained than the single-copy genes, but still show relaxation in constraint relative to what would be predicted based on their constraint in unduplicated outgroup genomes, again arguing that intrinsic constraint did relax after the triplication. Nevertheless, speaking to question #2, we see that the balance of high intrinsic constraint relaxed by polyploidy described in question #1 is a long-lasting one: the patterns of constraints on surviving ohnologs from both At-α and Br-α are mimicked at the population level. As these population data are as near as we can come to “selection at this instant”, it appears that whatever the postpolyploidy evolution of ohnologs, they have not specialized or diverged enough to lose the characteristics that marked them as ohnologs. Our findings of the long-lived effects of polyploidy (and nested polyploidy) on a gene’s selective constraint also complement the findings of Woodhouse et al. (2014), who found that the expression differences between ohnologs due to allopolyploidy can persist even through subsequent polyploidy events.
We also see some association between selective constraint and position in the metabolic network, although these associations are weak, as has been seen in other network analyses (Bloom and Adami 2003; Jordan et al. 2003; Hahn et al. 2004; Vitkup et al. 2006). Selective constraint is correlated with clustering coefficient in the metabolic network. Higher clustering in the network means that more neighbors interact with each other, that is, there are more alternative paths connecting two reactions. Selection pressure might be relaxed in the highly clustered regions in the network because of this increased redundancy, but be relatively more stringent in less clustered parts. The fact that genes with similar selective constraints are also more likely to be neighbors in the network may describe a similar phenomenon.
We have previously suggested that postpolyploidy evolution might be seen as proceeding in phases (Bekaert et al. 2011; Conant 2014; Conant et al. 2014). Our results here suggest, however, that such phases should not be taken too literally, as the ohnologs created by polyploidy are distinct in their character at their origin and retain much of this distinctiveness, at least in their sequence evolution, long after the polyploidy events. Such a result suggests again how polyploidy continues to shape the evolution of its possessors long afterward.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This project was supported by the National Science Foundation-IOS-1339156 (Y.H., E.L., J.C.P., and G.C.C.) and NSF-DBI-1358997 (J.R., B.N., and G.C.C.). We would like to thank M. Barker, H. An, and X. Qi for the B. rapa transcriptome data. We also appreciate the helpful suggestions from J. R. Ash, H. Truong, R. Patil, and T. Beissinger.
Literature Cited
- Albalat R, Canestro C.. 2016. Evolution by gene loss. Nat Rev Genet. 177:379–391. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, et al. , 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias T, Beilstein MA, Tang M, McKain MR, Pires JC.. 2014. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am J Bot. 1011:86–91. [DOI] [PubMed] [Google Scholar]
- Aury JM, et al. , 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 4447116:171–178. [DOI] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME.. 2009. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 10:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M.. 2009. Gephi: An open source software for exploring and manipulating networks. Third International AAAI Conference on Weblogs and Social Media; San Jose, CA: AAAI Publications. p. 361–362.
- Bekaert M, Edger PP, Hudson CM, Pires JC, Conant GC.. 2012. Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytol. 1962:596–605. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC.. 2011. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative followed by absolute dosage constraints. Plant Cell 235:1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Johnson AF, Veitia RA.. 2016. Kinetics genetics: incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 245:128–134. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA.. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 214:219–226. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2007. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 192:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 10937:14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2014. The gene balance hypothesis: dosage effects in plants. Plant epigenetics and epigenomics: methods and protocols. Totowa (NJ: ): Humana Press; p. 25–32. [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH.. 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 132:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH.. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 167:1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Adami C.. 2003. Apparent dependence of protein evolutionary rate on number of interactions is linked to biases in protein-protein interactions data sets. BMC Evol Biol. 3:21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH.. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 4226930:433–438. [DOI] [PubMed] [Google Scholar]
- Brandes U. 2001. A faster algorithm for betweenness centrality. J Math Sociol. 252:163–177. [Google Scholar]
- Cao J, et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 4310:956–963. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Fares MA.. 2012. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications. Mol Biol Evol. 2911:3541–3551. [DOI] [PubMed] [Google Scholar]
- Cheng F, et al. , 2012. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 75:e36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, et al. , 2016. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet. 4810:1218–1224. [DOI] [PubMed] [Google Scholar]
- Cingolani P, et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 62:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate JE, Schlueter JA, Whaley AM, Doyle JJ.. 2011. Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 1554:2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC. 2014. Comparative genomics as a time machine: how relative gene dosage and metabolic requirements shaped the time-dependent resolution of yeast polyploidy. Mol Biol Evol. 3112:3184–3193. [DOI] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC.. 2014. Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol. 19:91–98. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wagner A.. 2002. GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res. 3015:3378–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 912:938–950. [DOI] [PubMed] [Google Scholar]
- Danecek P, et al. , 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Petrov DA.. 2004. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 23:e55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK.. 2010. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 1522:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, et al. , 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci U S A. 1108:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Mason A.. 2014. Plant speciation through chromosome instability and ploidy change: cellular mechanisms, molecular factors and evolutionary relevance. Curr Plant Biol. 1:10–33. [Google Scholar]
- Dopman EB, Hartl DL.. 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 10450:19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 10240:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO.. 2006. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 232:327–337. [DOI] [PubMed] [Google Scholar]
- Edger PP, et al. , 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 299:2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC.. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 175:699–717. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW.. 2002. Evolutionary rate in the protein interaction network. Science 2965568:750–752. [DOI] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60:433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M, Scanlon MJ, Fowler JE.. 2015. Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr Opin Genet Dev. 35:110–118. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC.. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 167:805–814. [DOI] [PubMed] [Google Scholar]
- Frith MC, Kawaguchi R.. 2015. Split-alignment of genomes finds orthologies more accurately. Genome Biol. 16:106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur O, et al. , 2014. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 312:448–454. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Conant GC, Wagner A.. 2004. Molecular evolution in large genetic networks: connectivity does not equal constraint. J Mol Evol. 582:203–211. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Kern AD.. 2005. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol. 224:803–806. [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL.. 2007. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 810:R209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi SL. 1962. On realizability of a set of integers as degrees of the vertices of a linear graph. I. J Soc Ind Appl Math. 103:496–506. [Google Scholar]
- Hartl DL, Clark AG.. 1997. Principles of population genetics. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Hu TT, et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 435:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WK, Liu YL, Xia EH, Gao LZ.. 2013. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 1614:1844–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV.. 2003. No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol Biol. 3:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV.. 2004. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 4:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. 1997. Molecular evolution. Sunderland: Sinauer Associates Incorporated. [Google Scholar]
- Li Z, et al. , 2016. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28:326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. , 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 5:3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 2905494:1151–1155. [DOI] [PubMed] [Google Scholar]
- Lyons E, Freeling M.. 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 534:661–673. [DOI] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Alam M et al. , 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: coGe with rosids. Plant Physiol. 148:1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M.. 2008. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the Rosids. Trop Plant Biol. 1:181–190. [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I.. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 154:516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, et al. , 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 10215:5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A.. 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 10720:9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 3516328:652–654. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. , 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Yasunaga T.. 1980. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J Mol Evol. 161:23–36. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD.. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 4246945:194–197. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, et al. , 2014. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 156:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires JC, Conant GC.. 2016. Robust yet fragile: expression noise, protein misfolding, and gene dosage in the evolution of genomes. Annu Rev Genet. 50:113–131. [DOI] [PubMed] [Google Scholar]
- Qi X, et al. , 2017. Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol Ecol. 2613:3373–3388. [DOI] [PubMed] [Google Scholar]
- Rodgers-Melnick E, et al. , 2012. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 221:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Wolfe KH.. 2008. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 18:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M.. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 10810:4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH.. 1998. Extent of genomic rearrangement after genome duplication in yeast. Proc Natl Acad Sci U S A. 958:4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel S, Castellan N.. 1988. Nonparametric statistics for the behavioral sciences. 2nd ed New York: McGraw-Hill Int; p. 213–214. [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE.. 2015. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 35:119–125. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. , 2011. Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics 12:102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. , 2012. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 1904:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 4086814:796–815. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 1010:725–732. [DOI] [PubMed] [Google Scholar]
- Veitia RA. 2002. Exploring the etiology of haploinsufficiency. BioEssays 242:175–184. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Birchler JA.. 2015. Models of buffering of dosage imbalances in protein complexes. Biol Direct 10:42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitkup D, Kharchenko P, Wagner A.. 2006. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 75:R39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. , 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 4310:1035–1039. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A.. 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 4497158:54–61. [DOI] [PubMed] [Google Scholar]
- Warren AS, Anandakrishnan R, Zhang L.. 2010. Functional bias in molecular evolution rate of Arabidopsis thaliana. BMC Evol Biol. 10:125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH.. 1998. Collective dynamics of ‘small-world’ networks. Nature 3936684:440–442. [DOI] [PubMed] [Google Scholar]
- Wilks SS. 1938. The large-sample distribution of the likelihood ratio for testing composite hypotheses. Ann Math Statist. 91:60–62. [Google Scholar]
- Wolfe KH. 2000. Robustness: it’s not where you think it is. Nat Genet. 251:3–4. [DOI] [PubMed] [Google Scholar]
- Woodhouse MR, et al. , 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A. 11114:5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Gaut BS.. 2011. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 288:2359–2369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.