Abstract
RIG-I (retinoic acid inducible gene-I) is a cytosolic innate immune protein that senses viral dsRNA with a 5′-triphosphate overhang. Upon interaction with dsRNA a de-repression of the RIG-I CARD domains takes place that ultimately leads to the production of type I interferons and pro-inflammatory cytokines. Here we investigate the RIG-I conformational rearrangement upon interaction with an activating 5′-triphosphate-10-base pair dsRNA hairpin loop (10bp) compared with a less active 5′-triphosphate-8-base pair dsRNA hairpin loop (8bp). We use size-exclusion chromatography–coupled small-angle X-ray scattering (SAXS) and limited tryptic digest experiments to show that that upon binding to 10 bp, but not 8 bp, RIG-I becomes extended and shows greater flexibility, reflecting the release of its CARDs. We also examined the effect of different ATP analogues on the conformational changes of RIG-I/dsRNA complexes. Of the analogues tested, the addition of ATP transition state analogue ADP-AlFx further assisted in the complete activation of RIG-I in complex with 10bp and also to some extent RIG-I bound to 8bp. Together these data provide solution-based evidence for the molecular mechanism of innate immune signaling by RIG-I as stimulated by short hairpin RNA and ATP.
INTRODUCTION
RIG-I (retinoic acid inducible gene-I) and other family members, MDA5 (melanoma differentiation-associated factor 5) and LGP2 (laboratory of genetics and physiology 2), serve as cytoplasmic pattern recognition receptors of viral RNA (1). They work in a coordinated manner to recognize and respond to cytosolic viral RNA products that are distinct from host RNA. RIG-I senses and interacts preferentially with short blunt ended dsRNA bearing a 5′-triphosphate (5′ppp) dsRNA (2). Such RNA products are typically formed as replication intermediates of positive strand RNA viruses such as Flavivirus, Hepacivirus and Pestivirus and replication or transcriptional intermediates of negative strand RNA viruses including Orthomyxovirus, Paramyxovirus and Rhabdovirus (3). MDA5, in contrast, detects considerably longer dsRNA regardless of the presence of 5′ phosphate end structures that can also occur as viral replication intermediates (4). MDA5 has been identified as key in the response against Picornavirus family members (5). LGP2 preferentially binds dsRNA lacking a 5′ppp (6), but differs from RIG-I and MDA5 in that it does not possess caspase activation recruitment domains (CARDs). Most recent reports propose that LGP2 plays a regulatory role in antiviral signaling via modulating the RNA interactions of MDA5 (7,8).
dsRNA binding by RIG-I or MDA5 facilitates the de-repression of their CARDs, that are then able to actively engage the CARD domain of downstream adaptor protein interferon-β promoter stimulator 1 (IPS-1, also known as VISA, MAVS and Cardiff) in a ubiquitin-stabilized manner (9–13). This results in the oligomerization of IPS-1, forming a platform for a series of signaling events that lead to the induction of type I interferons (IFNs) and other proinflammatory cytokines enabling the suppression of viral replication, clearance of virus-infected cells, and facilitation of adaptive immune responses (14,15).
The molecular basis for CARD de-repression has been a key focus of investigation for RIG-I and MDA5. Each member of the RIG-I like receptor family comprises a central DExD/H-helicase domain and a CTD (C-terminal domain) connected by a ‘pincer’ domain (Figure 1A). The helicase core, in turn, comprises two RecA-like domains HEL1 (helicase 1) and HEL2 (helicase 2) domains, with HEL2 containing an insertion domain called HEL2i. An ATP binding catalytic site exists within HEL1, as is typical of superfamily 2 helicase proteins (16). RIG-I and MDA5 (but not LGP2) also possess N-terminal tandem CARD1 and CARD2 domains.
Figure 1.
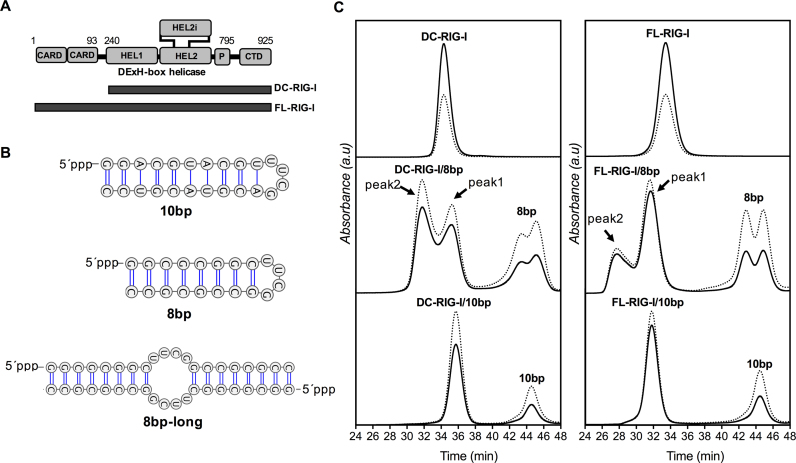
(A) Schematic representation of domain architecture of RIG-I and amino acid range of constructs used in this study (DC = deltaCARDs, FL = full-length, CTD = C-terminal domain, HEL = helicase domain, P = pincer motif), (B) Secondary structures of 10 bp and 8 bp hairpin dsRNA and the proposed structure of hybridized form of two 8mer hairpin dsRNAs (8 bp-long) [created using VARNA server (59)], (C) Size exclusion chromatography profiles of DC- and FL-RIG-I with and without dsRNAs and in the absence of any ATP analogue detected at 280 nm (solid line) and 260 nm (dotted line). Absorbance is indicated on the y-axis and elution time (in min) indicated on the x-axis.
Several crystallographically determined structures of RIG-I constructs have contributed to our understanding of the mechanism of the conformational change that occurs upon the interaction of RIG-I with viral dsRNA. Structures are available for human, duck and mouse RIG-I constructs before or after binding to different dsRNAs and with or without ATP-analogues (17–22). The only full length RIG-I structure (of duck RIG-I in the absence of RNA) shows the RIG-I HEL domains held in an ‘open conformation’ and with CARD2 interacting with the HEL2i domain, accounting for the autorepression (19). In this structure the CTD cannot be seen, likely due to flexibility. This open conformation of FL-RIG-I is also observed in the crystal structures of RIG-I lacking CARD domains (known as ‘delta-CARDs’ or DC-RIG-I) (20,23). Upon viral dsRNA exposure, the CTD of RIG-I makes high affinity specific interactions with the 5′ppp end of the dsRNA and the HEL1 and HEL2 domains form a ‘closed conformation’ around the dsRNA stem, together completely encircling the dsRNA (22,24–27). This configuration has been proposed to release the CARDs, making them available for interaction with IPS-1 (17–19). This has not been shown crystallographically, presumably owing to the flexibility of the released CARDs. SAXS data for full-length human RIG-I bound to a short dsRNA (29mer), however, has allowed a full-length RIG-I/dsRNA complex to be visualized, with extended CARDS at the end of a flexible linker (28).
The importance of RIG-I in the induction of the innate immune response and our detailed understanding of its molecular mechanism has opened up the possibility of the design of RIG-I activators that could be used therapeutically to suppress tumors by activating RIG-I mediated type I IFNs induction to promote the destruction of tumor cells (29–31). There has hence been a quest to discover a potent RIG-I activating dsRNA that can be used as an effective immunotherapeutic agonist in tumor cells (32–34). Several requirements for high affinity binding by RIG-I are known. RIG-I senses dsRNA with a 5′ppp or 5′pp with high affinity (2,35), but shows only weak affinity for dsRNA with a 5′p (35). Secondly, the minimal length of the dsRNA required for RIG-I stimulation is a consideration in agonist design. According to earlier reports, 5′ppp dsRNA of at least 20 bp was essential for optimal activation of RIG-I (36). However, in more recent attempts to determine the minimum length of dsRNA required for RIG-I activation, a 5′ppp 24-mer hairpin loop RNA containing 10 base pairs and a tetraloop (henceforth referred to as 10 bp) was shown to activate the RIG-I pathway very potently (22). Interestingly, the same study showed that a 5′ppp 20-mer hairpin loop (henceforth referred to as 8 bp) was not active. The reason for this was not clear, as both the 10 bp and the 8 bp dsRNA are able to be bound by RIG-I. A crystal structure of DC-RIG-I in complex with 8 bp and ADP shows the ability of the short dsRNA to bind to RIG-I (21). It does not appear very different from a structure solved of DC-RIG-I bound to a short hairpin dsRNA of equivalent length to 10 bp (37). It is therefore not understood why the 10 bp hairpin dsRNA is capable of activating RIG-I while the 8 bp hairpin dsRNA is not.
Another question in the field has been the role of ATP binding and ATP hydrolysis in the signaling activity of RIG-I. Being one of the members of ATP dependent helicases, the helicase domain of RIG-I has an ATP binding cleft and is able to hydrolyze ATP. It has been established that RIG-I is capable of ATP dependent unwinding of dsRNA (38) and ATP dependent translocation along dsRNA (39) that both involve ATP hydrolysis. These activities, however, are not required for the signaling activity of RIG-I, as mutant forms of RIG-I that no longer hydrolyze ATP are still able to signal (40–42). Instead, it is proposed that ATP binding by RIG-I plays an important role in helping to discriminate between pathogenic and non-pathogenic RNA. ATP is proposed to enhance the interaction of RIG-I with dsRNA and, accordingly, ATP hydrolysis serves to help disengage RIG-I from unintended RNA targets (41,42). The structural basis for this is suggested by several crystal structures of RIG-I constructs. It is evident that the RIG-I helicase core is in an open or semi-open conformation in ATP-free or ADP RIG-I/RNA complexes, with HEL2 not making close contacts with dsRNA due to being apart from the HEL1 domain (17,21,22). In the presence of the ATP analogue ADP-BeF3, the structure of DC-RIG-I bound to dsRNA adopts a ‘semi-closed’ and slightly more ordered conformation (18) and, in the presence of ADP-AlF3, the helicase domain is highly ordered and forms an even more compact closed-state conformation (19). Whether ATP binding actually assists in CARDs release has been speculated or inferred from structural, mutational and HDX-MS experiments (21,41,43) but not shown using a direct structural method.
In the current study, therefore, we address two outstanding questions for a better understanding of the requirements for CARD release by RIG-I. We use the biophysical method of small angle X-ray scattering (SAXS) to determine the effect of the short hairpin dsRNAs (10 bp and 8 bp) on the conformational rearrangement of full length RIG-I and DC-RIG-I in the presence or absence of different ATP analogues. We employ size-exclusion chromatography (SEC)-coupled SAXS to ensure the homogeneity of the RIG-I/RNA complexes. From the SAXS data we are able to report on the degree of release of the CARDs by the RIG-I/RNA complexes. The data are correlated with results of tryptic digest experiments that also reveal changes in RIG-I structure upon dsRNA and ATP analogue binding. Together these data demonstrate that the 10 bp dsRNA (but not the 8 bp dsRNA) is required for full CARDs release and that involvement of ADP-AlFx causes a further compact arrangement of the helicase domain that facilitates a more complete release of the CARDs. Therefore, our results represent the first in-solution characterization of RIG-I CARDs release by short hairpin RNAs and also demonstrate the importance of ATP binding in RIG-I activation.
MATERIALS AND METHODS
Protein expression and purification
Constructs of full length RIG-I (FL-RIG-I; residues 1–925) and CARDs-truncated RIG-I (DC-RIG-I; residues 239–925), with a non-cleavable C-terminal His tag and cloned in pET11, [kindly provided by Jerome Deval (Roche Palo Alto LLC, CA, USA)] (44) were transformed and expressed in Escherichia coli BL21(DE3) cells. When an OD600 of 0.6–0.8 was reached, cells were induced with 0.25 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and cultured for 16 h at 25°C overnight at 180 rpm in rich medium. Cells were collected by centrifugation at 7548 g and resuspended and lysed by an Avestin cell homogenizer in cold lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol and 1 mM DTT) also containing cOmplete EDTA-free protease inhibitor cocktail (Roche) and 1 mM PMSF (Sigma). Purification was performed using Ni-NTA agarose (QIAGEN). Nonspecific protein binding was avoided by washing with buffer containing 20 mM imidazole in addition to 50 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol and 1 mM DTT. High salt buffer (2 M NaCl and 50 mM HEPES) was then used to remove endogenous nucleic acid. An increasing step-gradient of imidazole was used for the elution of RIG-I. Eluted fractions were further purified by size exclusion chromatography using a Superdex 200 16/60 column (GE Healthcare) in 50 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol and 1 mM DTT. Final purification was achieved by applying RIG-I to a 1 ml Hi Trap Heparin HP column (GE Healthcare) with an increasing salt gradient to elute the purified protein. The proteins in high salt buffer were exchanged with buffer (25 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol and 1 mM DTT) and the purity of protein was confirmed by Coomassie Brilliant Blue staining of SDS-PAGE gels. After quantification of concentrated purified proteins at A280 using a NanoDrop (Thermo Scientific), the proteins were stored at -80°C.
Protein/RNA complex preparation and size exclusion chromatography-coupled small-angle X-ray scattering (SEC-SAXS)
5′ppp 10-base pair hairpin (10 bp) [5′ppp-GGACGUACGUUUCGACGUACGUCC] and 8-base pair hairpin (8 bp) [5′ppp-GGCGCGGCUUCGGCCGCGCC] dsRNAs were kindly provided by Dahai Luo (NTU, Singapore) [prepared as previously reported (22,43)]. RIG-I/RNA complexes at a ratio of 1:1.2 ± ATP analogues were prepared with RIG-I, at a concentration of 8 mg/ml. ATP analogues used were AMP-PNP (Calbiochem) at 0.5 mM and ADP-AlFx prepared from 2 mM ADP, 2 mM AlCl3 and 10 mM NaF, both in the samples and the SEC-SAXS elution buffer. All samples in the absence or presence of dsRNAs and ± ATP analogues were incubated on ice for 60 min and centrifuged for 5 min before performing SEC-SAXS experiments. 100 μl of each sample was subjected to SEC-SAXS using Superdex 200 10/300 column (GE Healthcare) at a flow rate of 0.4 ml/min in 25 mM HEPES pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1 mM DTT. SEC was also performed in-house under the same buffer conditions in order to obtain comparable UV traces.
SEC-SAXS data were collected at 15°C at the SAXS/WAXS beamline of the Australian Synchrotron. SAXS data were collected with 2 s exposures at 0.05 s intervals as the SEC eluent was passed through the 1 mm quartz capillary flow cell. Fractions were collected in a 96-well plate across each eluting peak. Due to dsRNA (8 bp) eluting as two peaks in SEC, fractions of RIG-I/8 bp were assessed by native and denaturing (8 M urea) PAGE (15% polyacrylamide 0.5× TBE (Tris/borate/EDTA) and stained with SYBR Gold (Invitrogen) to detect RNA. The gels were visualized using a Typhoon Trio variable mode imager (GE).
Processing of SAXS data
Australian Synchrotron beam specific software SCATTERBRAIN was utilized for SAXS data reduction (including radial integration of the SAXS images and conversion to 1-d dat files). The scattering data from samples were background-subtracted (DATOP) using scattering data taken after the flow through had cleared the column. Due to the presence of overlapping peaks in some of the SEC profiles, the recently developed software US-SOMO (Ultrascan Solution Modeler) HPLC SAS module was employed (45,46). In summary, the SAXS data were converted to the time domain, Gaussian fitting and deconvolution applied, and scattering data then back generated and used for further analyses. ATSAS programs (47) were used for further analyses which included calculation of pairwise distribution functions [P(r)], ab initio structure calculation using DAMMIF and DAMAVER (48,49) and multiphase modeling using MONSA (50). Normalized Kratky plots were calculated using the DATGNOM fitted scattering curves with a script written by MCJW. For analyses of the complexes of FL-RIG-I with 10 bp and 8 bp, the ensemble optimization method (EOM) (51) was used.
Molecular modeling against SAXS data
To assist interpretation of the SAXS data, different methods of structural modeling were used depending on the absence or presence of dsRNA and the degree of flexibility (as determined from the normalized Kratky analysis). Crystallographically determined coordinates were used for the establishment of starting models. These included coordinates from crystal structures of DC-RIG-I in complex with 8mer dsRNA (PDB ID: 4AY2) and of CARDs (PDB ID: 4P4H). Where amino acids were missing from the crystal structure flexible linkers were included. For relatively rigid molecules BUNCH (49) was used for creating a best fitting model to the SAXS data (in the absence of dsRNA), or CORAL (where dsRNA was present) (52). Alternatively, in the cases where there was evidence of flexibility, the Ensemble Optimization Method (EOM) was used to select a minimal ensemble of structures (from 10000 structures of varying conformation) that best fitted the SAXS data (51).
Limited Tryptic proteolysis of RIG-I and RIG-I/RNA complexes
Apo-FL-RIG-I and FL-RIG-I complexed with 10 bp and 8 bp dsRNA (at a 1:1.2 molar ratio) in the absence or presence of the ATP analogues were prepared as 60 μg samples and treated with 0.02 μg mass spectrometry grade Trypsin Gold (Promega) at 37°C. At equal time intervals over a time frame of 90 min, 10 μl aliquots were removed from the reaction and stopped by the addition of SDS-loading buffer and heating at 95°C for 5 min. For visualization, samples were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue.
RESULTS
Analysis of FL-RIG-I and DC-RIG-I ± dsRNA by SEC-SAXS
To investigate the conformational rearrangement of RIG-I upon interaction with short 5′ppp dsRNA and the role of ATP in RIG-I activation, we conducted SEC-coupled SAXS experiments with human FL-RIG-I (107 kDa) in the presence or absence of dsRNA hairpins and two ATP analogues. To distinguish conformational changes due to CARDs release from changes in the helicase and CTD domain arrangement, we also performed analysis of the equivalent DC-RIG-I (79 kDa) (Figure 1A). Based on a previous study determining the activation of RIG-I by hairpin dsRNA in cell-based assays (22), we used 5′ppp 10-base pair (10 bp) and 8-base pair (8 bp) hairpin loop dsRNA for the formation of RIG-I/RNA complexes ±ATP analogues (AMP-PNP and ADP-AlFx) (Figure 1B). AMP-PNP (5′-adenylyl-imidodiphosphate) is a non-cleavable stable analogue of ATP in which a nitrogen atom replaces the oxygen atom between the β and γ phosphate and the γ-phosphate has a tetrahedral geometry, mimicking the ATP-bound ground state (53). ADP-AlFx behaves as a phosphate-bound stable transition state of ATP in which AlFx mimics the γ-phosphate of ATP with planar geometry (54,55). The Al-F bond is the same length as that of P-O and fluorine can form similar H-bond interactions as oxygen, making AlFx a very good, although more electronegative, mimic of phosphate (55). Both of these analogues have been used in some of the crystal structures of RIG-I and MDA5 helicase domain and chicken LPG2 with or without dsRNA (6,19,20).
After combining 10 bp or 8 bp with the purified DC- and FL-RIG-I in a 1:1.2 ratio, RIG-I/dsRNA complexes were separated by size-exclusion chromatography (SEC) and run in-line with SAXS experiments. Figure 1C presents the elution profiles of both proteins with and without dsRNA and in the absence of ATP analogue. As a general observation, FL-RIG-I/RNA complexes eluted at a slightly earlier time than the apo-FL-RIG-I and were well separated from the dsRNA peaks. This is indicative of the complex occupying a larger volume upon complex formation. In the case of DC-RIG-I, however, DC-RIG-I/RNA complexes eluted slightly later than the apo-DC-RIG-I, consistent with a compaction of the molecular structure upon interaction with the RNA. The elution profiles of DC- and FL-RIG-I binding to 10 bp gave rise to a single peak corresponding to the complex and a second single peak corresponding to excess dsRNA. In the case of DC- and FL-RIG-I binding to 8 bp, two slightly overlapping peaks were observed for the complex as well as for the excess RNA. This is because the 8 bp existed as two species in solution, arising from self-annealing in addition to the formation of a hairpin (discussed further below). The profiles demonstrate that both species are able to bind to RIG-I (Figure 1C). For both the DC- and FL-RIG-I experiments one of the peaks (peak 1) eluted at a similar elution time as the complex formed with 10 bp, and the other (peak 2) eluted considerably earlier indicating the formation of a larger complex. This is consistent with peak 1 representing a complex with the intended 8 bp hairpin and peak 2 representing the self-annealed version of 8 bp (referred to as 8 bp-long) bearing two RIG-I molecules. The SEC profiles from SEC-SAXS of DC- and FL-RIG-I apo and with dsRNA, in the presence of AMP-PNP or ADP-AlFx were equivalent and are provided as Supplementary Figure S1.
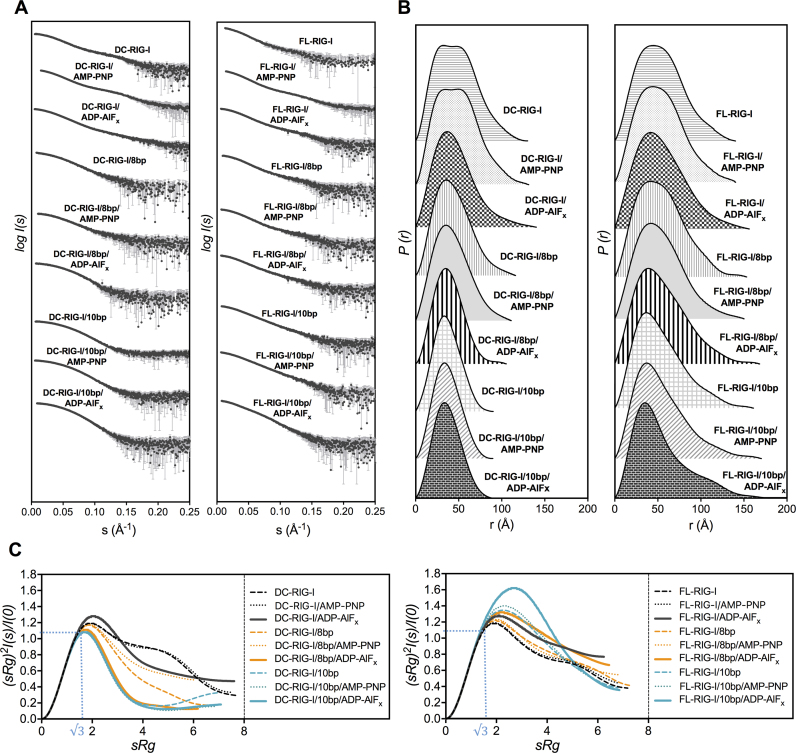
Due to the presence of overlapping peaks and/or shoulders in some of the SEC-SAXS profiles, the Ultrascan solution modeler (US-SOMO) HPLC-SAS module was used to deconvolute each of the overlapping peaks (45,46). US-SOMO is based on Gaussian decomposition of non-baseline-separated SEC-SAXS peaks by converting background subtracted raw SAXS scattering intensity [I(q) represented as a function of scattering vector q] into a scattering intensity represented as a function of elution time [I(t)]. This allows for the deconvolution of more than one species co-eluting as overlapping peaks. SAXS data [as I(q) versus q] is then back generated from each Gaussian decomposed peak and used for further analyses. Supplementary Figure S2 shows the Gaussian decomposition of the baseline subtracted SAXS data. Figure 2A, B and C represents the experimental scattering curves (obtained from Gaussian decomposed peaks for each sample); the pairwise distribution function (P(r)) profiles and normalized Kratky plots, respectively, and Supplementary Table S1 reports the data collection and SAXS invariants and other parameters.
Figure 2.
SAXS analyses of apo-RIG-I and RIG-I:RNA complexes ± ATP analogues. (A) Experimental scattering profiles of DC- and FL-RIG-I, with or without 10 bp or 8 bp and ATP analogues, obtained by plotting the log of scattering intensity [log I(s)] as a function of forward scattering vector in the units of Å−1 and represented by black spheres along with gray error bars showing the mean ± SD, (B) pairwise distribution [P(r)] profiles obtained from the scattering data by plotting P(r) as a function of r in the units of Å, (C) normalized Kratky profiles of DC- and FL-RIG-I apo and with dsRNAs in the absence or presence of any ATP analogues. Note: SAXS data for apo-RIG-I constructs in the presence of AMP-PNP are reproduced from (28) with permission for comparative purposes.
DC- and FL-RIG-I molecular conformation in the absence of dsRNA
SAXS data acquired for DC- and FL-RIG-I apo proteins ±ATP analogues were used for analyzing the conformation of the molecule in the absence of dsRNA. SAXS data for DC- and FL-RIG-I in the presence of AMP-PNP have been described previously (28). These data indicated that DC-RIG-I/AMP-PNP adopts a bilobular shape in which the CTD resides alongside the helicase domain as a distinct lobe. The FL-RIG-I/AMP-PNP complex appeared in an open conformation with the CARDs positioned against the helicase domain and the CTD positioned along the other side of helicase domain (28). These data, including scattering curves, P(r) and normalized Kratky plots are shown again (with permission) for comparison with other data sets (Figure 2).
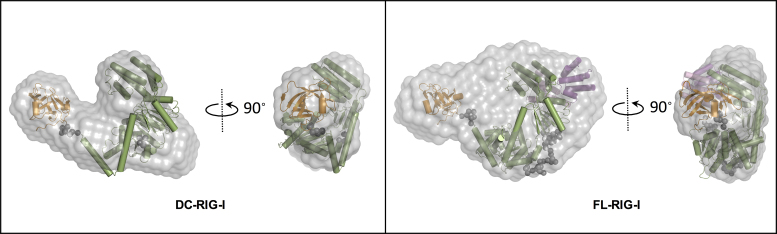
The scattering intensities and the P(r) plots (Figure 2A and B) for DC-RIG-I in the absence of ATP analogue showed no detectable difference from those collected in the presence of AMP-PNP. As can be seen from the scattering profiles and the P(r) plots (Figure 2A and B), the same bilobar shape is assumed by DC-RIG-I. The radius of gyration (Rg) of 36.7 Å determined by Guinier approximation, and Rmax = 130 Å are close to the values previously reported for DC-RIG-I/AMP-PNP (28). The DC-RIG-I normalized Kratky profile shows a non-parabolic curve with a peak maximum slightly >1.1 at about √3 sRg, indicating that this molecule possesses only a limited amount of flexibility (56). A single best-fitting model of DC-RIG-I in the absence of ATP analogue was generated using the program BUNCH (49) with protein domain coordinates from the crystal structure of DC-RIG-I in complex with 8 bp (PDB ID: 4AY2) and movement between the HEL1 and HEL2 domains allowed. Figure 3 shows this model superposed on an ab initio shape reconstruction independently calculated using DAMMIF (48). This shows visually that the molecular model produced by BUNCH is in agreement with the DAMMIF-derived envelope. The predicted scattering curve derived from the model fits well with the experimental scattering data with a χ2 value = 0.68 (Supplementary Figure S3 and Table S1). The generated model is similar to our previously reported model, analyzed in the presence of AMP-PNP (28).
Figure 3.
SAXS derived models of DC- and FL-RIG-I in the absence of any ATP analogue. DAMMIF (48,49) was used to calculate the ab initio shape reconstructions represented as grey envelopes. BUNCH (49) derived models of DC- and FL-RIG-I are superposed on ab initio shapes. The CTD is coloured orange, the helicase core is coloured green and the linker regions are shown as gray spheres. In the case of FL-RIG-I, the two CARDs are shown in purple and pink. Fits are shown in Supplementary Information Figure S3.
Likewise, the SAXS data indicate that the shape of FL-RIG-I in the absence of ATP analogue closely resembles that acquired for FL-RIG-I/AMP-PNP. The P(r) profile, Rg value of 43.4 Å, Rmax value of 140 Å and normalized Kratky profile of FL-RIG-I are similar to those of FL-RIG-I/AMP-PNP, consistent with the same open conformation of the helicase domain and the CTD and CARDs arranged alongside (28). A single best-fitting structural model of FL-RIG-I in the absence of any ATP analogue was generated using BUNCH using the protein domain coordinates from DC-RIG-I (PDB ID: 4AY2) and the CARDs (PDB ID: 4P4H) with flexibility allowed between CARDs, helicase and CTD domains. The resulting model showed good agreement with a DAMMIF generated ab initio shape reconstruction, and again showed a similar arrangement of RIG-I subdomains as determined for its AMP-PNP bound equivalent (Figure 3). The predicted scattering curve from this model was in close agreement with raw scattering data (χ2 value = 0.22) (Supplementary Figure S3 and Table S1).
DC- and FL-RIG-I molecular conformation in the presence of ADP-AlFx
In contrast to the previous data, SAXS data collected for DC-RIG-I/ADP-AlFx indicated a compaction of the helicase domain. In the presence of ADP-AlFx, DC-RIG-I exhibits a similar Rg of 35.1 Å but an extended Rmax of 140 Å compared to DC-RIG-I in the absence of ATP analogue or presence of AMP-PNP. The P(r) profile shifts toward a more bell-shaped curve at low r values indicating a shift toward a more compact or globular domain structure, but, with a slightly extended tail at high r value. The normalized Kratky plot of DC-RIG-I in the presence of ADP-AlFx appears as a partially parabolic curve with a slightly higher peak maxima at √3 sRg, indicating some parts of the molecule may be flexible in the presence of ADP-AlFx but with the bulk in a rigid state.
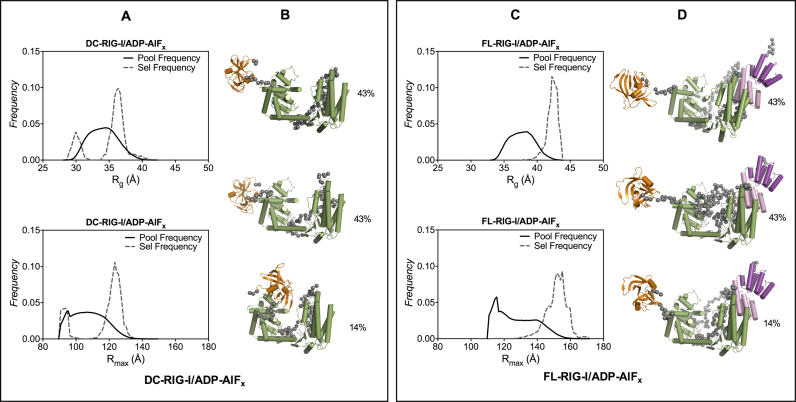
It was not possible to generate a model that gave an acceptable fit to the data for DC-RIG-I/ADP-AlFx using rigid body modeling, likely due to flexible features within DC-RIG-I/ADP-AlFx. Instead, EOM (the Ensemble Optimization Method) was employed to provide a set of structural models that, together, can be used to describe flexible proteins in solution as better reflects molecules in equilibrium between different forms (57). The helicase domain and CTD coordinates from the structure of DC-RIG-I in complex with 8 bp (PDB ID: 4AY2) were set as a rigid bodies linked by a flexible linker to create an ensemble of 10000 randomly generated structures. From these, structures are selected that, together, provided a best fit against the SAXS data. Figure 4A represents the Rg and Rmax distributions of the ensemble of randomly generated model structures (shown by solid line) and the selected structures (shown by dashed line). From these populations the minimum number structures that fit well with the experimental scattering data were selected. In case of DC-RIG-I/ADP-AlFx, three structures were selected from the ensemble that, together, provide agreement with the experimental scattering data (χ2 value = 0.42) (Supplementary Figure S4A and Table S1). While the predicted scattering for the individual conformers do not match the experimental scattering data, when combined they are a good match (Supplementary Figure S4B). The structures include both longer and more compact forms of DC-RIG-I indicating that DC-RIG-I/ADP-AlFx may exist in equilibrium between fairly closed structures and ones in which the CTD extends away from the helicase domain (Supplementary Figure S4B).
Figure 4.
Ensemble optimization method analysis of SAXS data for DC- and FL-RIG-I in the presence of ADP-AlFx. (A and C) Rg and Rmax distributions of best fitting ensembles (dashed line) from EOM (57) selected structures of DC- and FL-RIG-I in the presence of ADP-AlFx from a pool of 10 000 conformers (solid line); (B and D) EOM selected representative models of DC- and FL-RIG-I that, together, best predict the scattering data (with their proportion contribution indicated as percentages). Fits are shown in Supplementary Information Figure S4.
In the presence of ADP-AlFx, the SAXS data for FL-RIG-I follows a similar trend. The data show a similar Rg value of 41.9 Å and longer Rmax of 156 Å compared to those of FL-RIG-I with AMP-PNP or without any ATP analogue. Differences can be seen more clearly in the P(r) profile, which shows a narrower bell shape centered at a lower r value and a slight tail at higher r value. Moreover, the normalized Kratky profile of FL-RIG-I/ADP-AlFx again has a higher peak maxima at √3 sRg compared to FL-RIG-I with no analogue or with AMP-PNP, which is indicative of a changed conformation and flexibility of FL-RIG-I when ADP-AlFx is present. As was conducted for DC-RIG-I/ADP-AlFx, EOM was used for structural modeling of FL-RIG-I/ADP-AlFx using the protein coordinates from the structures of DC-RIG-I (PDB ID: 4AY2) and the CARDs (PDB ID: 4P4H), with the helicase and CARDs positioned by analogy to their juxtaposition in the structure of duck RIG-I (19) and appropriate length flexible linkers. The structures that together were in agreement with the scattering data were selected from the larger Rg and longer Rmax ends of the ensemble (Figure 4C). From these populations the minimum number structures that fit well with the experimental scattering data were selected. The predicted scattering curves derived from these three structures matched well with the experimental scattering data (χ2 value = 0.25) (Supplementary Figure S4 and Table S1). These structures show that the data are consistent with helicase and CARDs of FL-RIG-I/ADP-AlFx in a fairly compact form but with the CTD extended away and showing some flexibility.
DC-RIG-I conformation upon binding 10 bp dsRNA
As 10 bp was reported to be the minimum length of hairpin dsRNA able to effectively activate RIG-I (22), we were interested to investigate the conformational change in RIG-I upon binding to 10 bp ±ATP analogues. It was anticipated that 10 bp binding to FL-RIG-I would provide direct biophysical evidence of CARDs release when examined in comparison to DC-RIG-I. From the SAXS data of the DC-RIG-I/10 bp complex in the absence of any ATP analogue, we observed an Rg of 28.2 Å and Rmax of 90 Å with the P(r) profile appearing as a unimodal curve (Figure 2A and B). In the presence of AMP-PNP and ADP-AlFx, the DC-RIG-I/10 bp complex also gave rise to a similar Rg of 28.2 Å and Rmax values of 90 Å and 89 Å respectively and a bell-shaped P(r) curve (Figure 2A and B). When compared with apo-DC-RIG-I ±ATP-analogues, the P(r) of DC-RIG-I/10 bp complex appears significantly more compact. Furthermore, the normalized Kratky plots of DC-RIG-I/10 bp in the presence of ATP analogues were similar with bell shaped curves with maxima of 1.1 at >√3 sRg and approaching zero at higher sRg. This reflects an ordered and compact conformation of DC-RIG-I after binding with 10 bp.
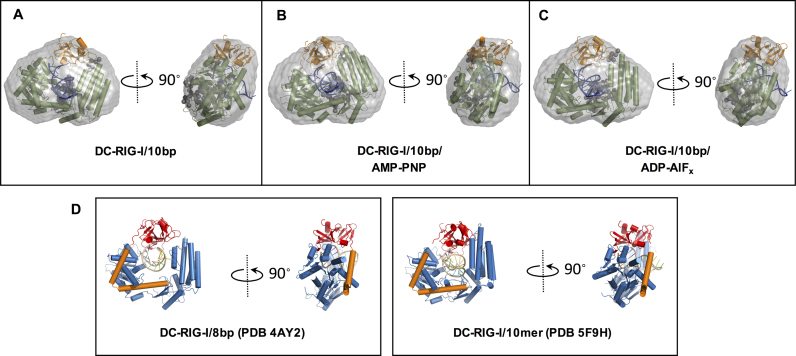
To obtain structural models of DC-RIG-I/10 bp ±ATP analogues, CORAL (COmplexes with RAndom Loops) was used, which is based on rigid body modeling of complexes with flexible created missing loops against the SAXS data and allows for the inclusion of coordinates for RNA. By using CORAL, the optimal configuration of available high resolution models of domains can be found (52). For this purpose, the crystallographically determined structure of DC-RIG-I/8 bp complex (PDB ID: 4AY2) was used as a starting point, however, the 8-base pair dsRNA in the crystal structure was replaced by a 10-base pair dsRNA generated by MC-Sym pipeline (58). In the case of DC-RIG-I/10 bp/ADP-AlFx, a best fitting model was generated when all domains were kept rigid against the SAXS data, but for DC-RIG-I/10 bp or DC-RIG-I/10 bp/AMP-PNP a best fit was obtained when the CTD domain was allowed to move relative to the helicase domain. The models generated are shown superposed within the ab initio shape reconstruction of DC-RIG-I/10 bp ±ATP analogues calculated using DAMMIF (22) (Figure 5A–C). Also shown in Figure 5 are the crystallographically derived structures of DC-RIG-I bound to 8 bp (4AY2) and a 10mer dsRNA hairpin (5F9H). It can be seen that the compact arrangement of DC-RIG-I around the dsRNA as shown by SAXS is consistent with these crystallographically derived models. The resulting predicted scattering curves from these models fit well with the experimental scattering data of DC-RIG-I/10 bp (χ2 value = 0.52), DC-RIG-I/10 bp/AMP-PNP (χ2 value = 0.4) and DC- RIG-I/10 bp/ADP-AlFx (χ2 value = 0.57) (Supplementary Figure S3 and Table S1). These data demonstrate that 10 bp binding results in the compact rearrangement of helicase and CTD domains ±ATP analogues. Further, the data suggest that the orientation of helicase subdomains is most compact when ADP-AlFx is present in addition to 10 bp binding.
Figure 5.
Rigid-body models of DC-RIG-I bound to 10 bp derived from SAXS data using CORAL (49). (A–C) CORAL-derived models of DC-RIG-I in complex with 10 bp in the absence of ATP analogue and the presence of AMP-PNP or ADP-AlFx which are superposed on ab initio reconstructions (represented as grey envelopes). The CTD is colored orange, the helicase core is colored green and the linker regions are shown as gray spheres. Fits are shown in Supplementary Information Figure S3. (D) shows the crystallographically derived structures of DC-RIG-I bound to 8 bp (PDB ID: 4AY2) and to a 10mer dsRNA hairpin (PDB ID: 5F9H) for comparison. In these structures the CTD is colored red, the helicase core is colored blue, pincer domain orange and RNA yellow.
FL-RIG-I conformation upon binding 10 bp dsRNA
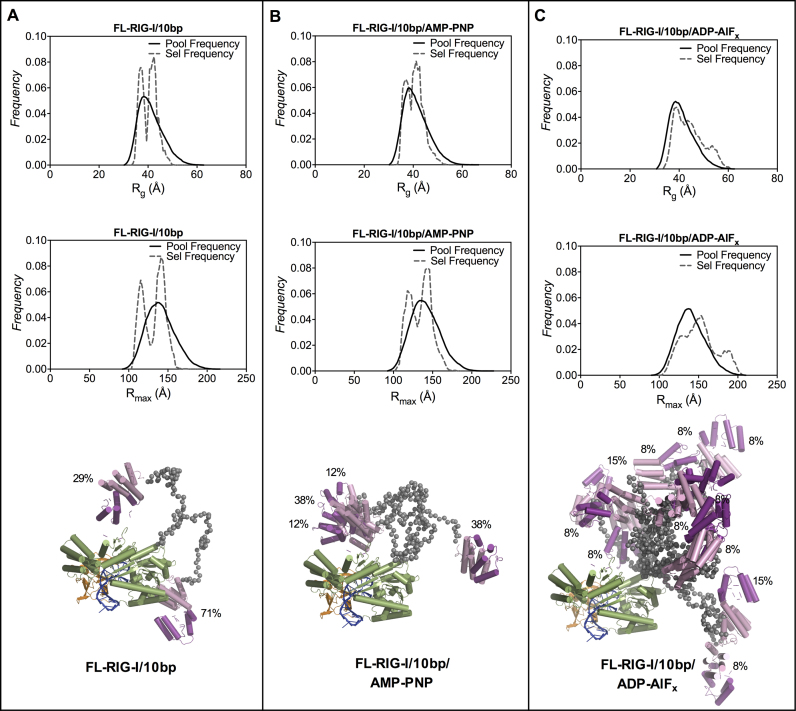
In contrast to the compact structure of the DC-RIG-I/10 bp complex, the SAXS data for FL-RIG-I/10 bp complex indicate a significantly elongated structure consistent with the release of CARDs. FL-RIG-I/10 bp complex with no ATP analogue gave rise to an Rg of 40.8 Å and Rmax of 161 Å, and, the complex with AMP-PNP resulted in an Rg of 42.0 Å and Rmax of 170 Å. However, when ADP-AlFx, was present, the SAXS data indicated an Rg of 40.4 Å and a slightly higher Rmax value of 183 Å. The P(r) profiles of the FL-RIG-I/10 bp complexes show a narrowing of the bell shaped peak at lower r values, as also seen for DC-RIG-I, overlaid with a more pronounced shoulder at higher r value. This is consistent with an asymmetric extended and flexible molecule with domains tethered by long disordered linker regions (56). This effect is most pronounced for the FL-RIG-I/10 bp complex in the presence of ADP-AlFx in which the bell shaped peak at the higher r(q) value is narrowest, the shoulder is most defined and the Rmax is longest. Similarly, while the normalized Kratky profiles for FL-RIG-I/10 bp ± AMP-PNP show some increased flexibility of the molecule with a peak maxima of about 1.3 at >√3 sRg, in the presence of ADP-AlFx the normalized Kratky plot shows a significantly higher peak maxima that is shifted toward a value of 1.6 at ∼2.7 sRg. This indicates a significant change in the flexibility upon complex formation with 10 bp. Together this indicates that, particularly in the presence of ADP-AlFx, the FL-RIG-I/10 bp complex becomes an elongated molecule with the dsRNA-engaged CTD and helicase domain being more compact and CARDs completely released from the rest of molecule.
The SAXS data collected for FL-RIG-I/10 bp complexes were analyzed using EOM (57) as a way of depicting the range of conformations that may be adopted by FL-RIG-I/10 bp complexes. EOM was conducted using models created from CORAL derived output structures of DC-RIG-I/10 bp complexes and CARD domains (PDB ID: 4P4H) treated as rigid bodies and connected by appropriate length flexible linkers. Figure 6 (A, B, C) presents the Rg and Rmax distributions of the ensemble of randomly generated model structures (shown by solid lines) and the selected structures (shown by dashed line) from within the ensemble best matching the experimental scattering data. In case of the FL-RIG-I/10 bp complex without any analogue or with AMP-PNP, the selected structures resulted into two populations with average Rg values of 40.6 Å and 40.9 Å and Rmax values of 133 Å and 135 Å respectively. From these populations the minimum number structures that fit well with the experimental scattering data were selected (χ2 value = 0.69 and 0.72 for FL-RIG-I/10 bp and FL-RIG-I/10 bp/AMP-PNP) (Supplementary Figure S4). These selected structures (Figure 6A and B) indicate an equilibrium between structures with CARDs away from the rest of the molecule (corresponding to larger Rg and Rmax values) and structures with CARDs closer to rest of the molecule (corresponding to smaller Rg and Rmax values). The overall similar selection of structures in terms of Rg and Rmax, for FL-RIG-I/10 bp complexes both without ATP analogue and with AMP-PNP, indicates that the addition of AMP-PNP does not considerably affect the conformational rearrangement of RIG-I, consistent with the P(r) and Kratky analysis.
Figure 6.
Ensemble optimization method of analysis of SAXS data for FL-RIG-I/10 bp ± ATP analogues. Shown are the Rg and Rmax distribution of best fitting ensembles (dashed line) of EOM (57) selected structures of FL-RIG-I complexed with 10 bp in the absence of any ATP-analogue (A) or the presence of AMP-PNP (B) or ADP-AlFx (C) from a pool of 10000 conformers (solid line). The EOM selected best representative structures of FL-RIG-I/10 bp ± ATP analogues that, together, best predict the scattering data, are shown superposed (with their proportion contribution indicated as percentages). The coloring scheme is as used in previous figures, with the addition of blue used to depict the dsRNA. Fits are shown in Supplementary Information Figure S4.
In contrast, in the case of FL-RIG-I/10 bp complex with ADP-AlFx, the selected structures showed a different profile, with a selection of more structures with higher Rg and longer Rmax (Figure 6C). The scattering data derived from the set of selected structures matched the experimental scattering data well (χ2 value = 0.28) (Supplementary Figure S4), with an average Rg of 44.6 and Rmax of 152. Some of the selected structures also show an Rg of 54.0 Å and an Rmax of 185 Å, which shows a greater flexibility and elongation of the molecule. The selected wider range of structures, including elongated species, reflects a shift in equilibrium toward a greater extent of elongation of the FL-RIG-I/10 bp in the presence of ADP-AlFx compared to FL-RIG-I/10 bp with no analogue or AMP-PNP bound.
DC-RIG-I conformation upon binding with hairpin 8 bp dsRNA
In contrast to the potent activation of RIG-I by 10 bp dsRNA, it is reported that the 8 bp dsRNA is less effective in activating RIG-I (22). Intriguingly, a crystal structure of DC-RIG-I in complex with this 8 bp dsRNA indicates that RIG-I is able to engage this 8 bp species (21). Its lack of activity is therefore unexplained and unverified by biophysical means. We therefore undertook SEC-SAXS to answer the question of whether the 8 bp dsRNA is able to effect CARD release. As RIG-I/8 bp complexes eluted as two peaks in the SEC profiles, we interpreted the latter eluting peak to represent the 1:1 protein/dsRNA complex and refer to it as peak1 (the validation of this is discussed further below). The SAXS data from peak1 of DC-RIG-I/8 bp complex without any ATP analogue and with AMP-PNP, indicated an Rg value of 33.7 Å and 32.0 Å and Rmax of 116 Å and 111 Å respectively. The P(r) profiles in both cases were similar and appeared as unimodal curves but were distinctly broad compared to DC-RIG-I/10 bp complexes, showing that 8 bp is less able to make a compact arrangement of the helicase domain of RIG-I (Figure 2B). In the presence of ADP-AlFx, DC-RIG-I/8 bp complex gave rise to a slightly lower Rg of 29.6 Å and Rmax of 105 Å, and with a P(r) profile showing a bell shaped curve more similar to the DC-RIG-I/10 bp complexes. In addition, the normalized Kratky plots of DC-RIG-I/8 bp in the absence of ATP analogues or the presence of AMP-PNP were not a perfect bell shaped curve like those of DC-RIG-I/10 bp complexes, also reflecting a less compact molecular shape (Figure 2C). In contrast, in the presence of ADP-AlFx, the Kratky curve was shifted toward a more bell shaped curve which approached zero at high sRg in a manner similar to those of DC-RIG-I/10 bp complexes. This reflects a more ordered and compact conformation of DC-RIG-I/8 bp only after the addition of ADP-AlFx.
To model DC-RIG-I/8 bp/ADP-AlFx, CORAL was used with all domains of the starting structure (PDBID: 4AY2) kept rigid but loops between structured domains mobile. The model generated is shown superposed within the ab initio shape reconstruction of DC-RIG-I/8 bp/ADP-AlFx calculated using DAMMIF (48) (Figure 7A). The resulting predicted scattering curve from the molecular model fitted the experimental scattering data with a χ2 value of 0.27 (Supplementary Figure S3 and Table S1). These data suggest that DC-RIG-I in complex with 8 bp in the presence of ADP-AlFx adopts a compact conformational rearrangement of its helicase and CTD domains. In contrast, a model that gave an acceptable fit to the data could not be generated by CORAL to match the raw scattering data of DC-RIG-I/8 bp without ATP analogue or with AMP-PNP, and therefore, EOM was used for structural modeling of these complexes. To generate a pool of models the structure of DC-RIG-I (PDB ID: 4AY2) was used as a starting point and CTD bound to 8 bp allowed to move relative to the rest of the molecule. In each case, two main populations of structures were selected from across the ensemble with Rg values ranging from 27 Å to 43 Å and Rmax values from 80 Å to 130 Å (Figure 7B and C). From these populations the minimum number structures that fit well with the experimental scattering data were selected. The predicted scattering curves derived from these selected structures matched with the experimental scattering data with χ2 values of 0.36 and 0.24 for DC-RIG-I/8 bp and DC-RIG-I/8 bp/AMP-PNP respectively) (Supplementary Figure S4 and Table S1). The range of selected structures are consistent with an equilibrium between fully engaged and partially open DC-RIG-I molecules interacting with the 8 bp dsRNA. Overall, this indicates that 8 bp is not sufficient for complete engagement by DC-RIG-I except in the presence of ADP-AlFx.
Figure 7.
SAXS derived models of DC-RIG-I in complex with 8 bp ± ATP analogues. (A) CORAL-derived model of DC-RIG-I in complex with 8 bp in the presence of ADP-AlFx superposed on ab initio reconstruction (represented as a grey envelope). (B and C) Ensemble optimization method of analysis of SAXS data for DC-RIG-I/8 bp complexes ± AMP-PNP. Rg and Rmax distributions of best fitting ensembles (dashed line) of EOM (57) selected structures of DC-RIG-I/8 bp ± AMP-PNP from a pool of 10000 conformers (solid line) are shown. EOM derived representative selected models of DC-RIG-I in complex with 8 bp ± AMP-PNP are shown (with their proportion contribution indicated as percentages). The coloring scheme is as used in previous figures. Fits are shown in Supplementary Information Figure S4.
FL-RIG-I conformation upon binding with hairpin 8 bp dsRNA
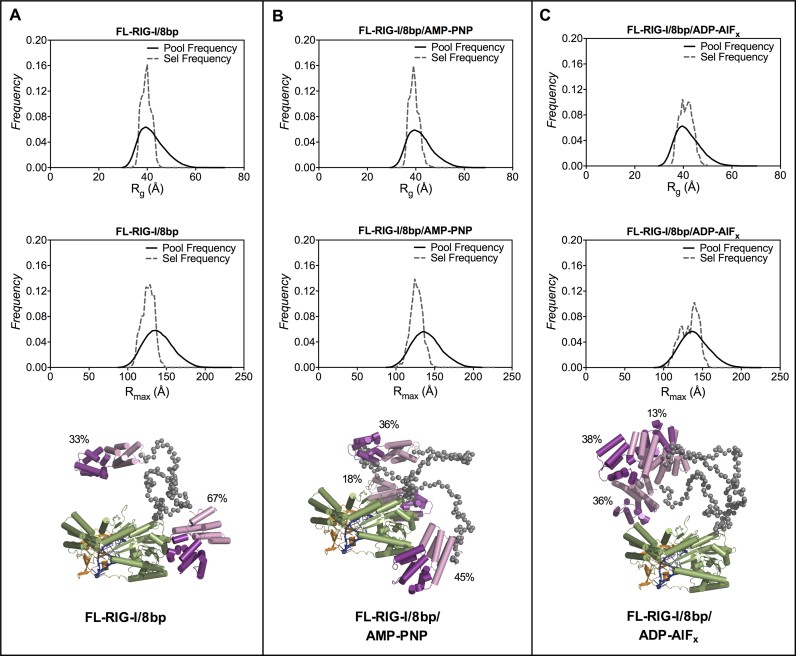
Analysis of the SAXS data for peak1 of FL-RIG-I/8 bp is consistent with a limited degree of CARD release. The SAXS data corresponding to peak1 of the FL-RIG-I/8 bp complex gave rise to an Rg of 42.6 Å and an Rmax of 153 Å, with the P(r) profile showing a wider bell-shaped curve than that obtained for FL-RIG-I/10 bp. This indicates that 8 bp binding does not cause the same degree of shape change as observed with 10 bp dsRNA. In the presence of AMP-PNP the Rg was 41.1 Å and Rmax was 150 Å. The P(r) was similar to that of FL-RIG-I/8 bp in the absence of ATP analogue complex except for a slight narrowing of the curve at lower r value (Figure 2B). In the presence of ADP-AlFx, however, while the Rg remains a similar value of 42.4 Å the Rmax is increased to 168 Å, which shows some elongation of FL-RIG-I has taken place. The P(r) profile, however, showed a broad curve that was less defined than that of FL-RIG-I/10 bp complex with ADP-AlFx. Accordingly, the normalized Kratky profiles of FL-RIG-I/8 bp complexes without ATP analogue and with AMP-PNP show non-parabolic curves with peak maxima of 1.2 at slightly greater than √3 sRg, indicating some degree of flexibility. In contrast, in the presence of ADP-AlFx, the Kratky profile showed a greater increase in peak maxima, indicative of a greater degree of flexibility, though not the same degree of flexibility as seen for FL-RIG-I/10 bp/ADP-AlFx. This overall suggests that the addition of ADP-AlFx can affect the structure of FL-RIG-I bound with 8 bp, but not to the extent seen for FL-RIG-I bound to 10 bp.
The SAXS data collected for FL-RIG-I/8 bp complexes were analyzed using EOM with an ensemble of structures generated from DC-RIG-I (PDB ID: 4AY2) linked to CARDs (PDB ID: 4P4H) by a flexible linker. In the case of FL-RIG-I/8 bp complexes without any analogue or with AMP-PNP the structures were selected from the population with lower Rg and Rmax values, indicating a lesser tendency toward CARD extension compared to RIG-I bound to the 10 bp RNA (Figure 8A and B). In the case of FL-RIG-I/8 bp complex with ADP-AlFx, the structures were selected from a broader range of the ensemble (Figure 8C), some with greater Rg and Rmax values than selected for no analogue or AMP-PNP counterparts. This reflects the contribution of ADP-AlFx to the conformational rearrangement of the molecule. However, the selected structures did not include those with the highest values of Rg and Rmax as seen in the EOM analysis of RIG-I binding to 10 bp. From these populations, the minimum number structures that fit well with the experimental scattering data were selected and are also shown in Figure 8. The predicted scattering curves derived from these selected structures matched with the experimental scattering data with χ2 values of 0.50, 0.59 and 0.48 for FL-RIG-I/8 bp with no ligand, with AMP-PNP and with ADP-AlFx respectively) (Supplementary Figure S4 and Table S1). Together this is consistent with the 8 bp RNA inducing some degree of RIG-I CARD release, particularly in the presence of ADP-AlFx, but not to the extent seen upon the interaction of RIG-I with the 10 bp species.
Figure 8.
Ensemble optimization method analysis of SAXS data of FL-RIG-I/8 bp ± ATP analogues. Rg and Rmax distribution of best fitting ensembles (dashed line) of EOM (57) selected structures of FL-RIG-I complexed with 8 bp in the absence of any ATP-analogue (A) or the presence of AMP-PNP (B) or ADP-AlFx (C) from a pool of 10 000 conformers (solid line). EOM selected best representative structures of FL-RIG-I/8 bp ± ATP analogues are shown superposed (with their proportion contribution indicated as percentages). The linker regions between CARDs and rest of molecule are shown by gray spheres. The coloring scheme is as used in previous figures. Fits are shown in Supplementary Information Figure S4.
8 bp-long binds two DC-RIG-I molecules
Upon determining that 8 pb eluted from the SEC as two peaks (Figure 1C), we proposed that the first eluting peak might be a fusion of two ssRNA molecules forming a longer palindromic duplex dsRNA (8 bp-long) (Figure 1B). This hybridized 8 bp-long dsRNA would contain two 8-base-paired regions and may include a bulge (caused by non-complimentary tetraloops) and a 5′ppp at each terminus (Figure 1B). Such a structure would allow for the binding of two RIG-I molecules. In order to characterize the two species, we collected the SEC fractions from peak 1 and peak 2 of ATP-analogue free RIG-I/8 bp complex as well as those from free RNA and subjected them to denaturing and native PAGE and staining with SYBR Gold (Supplementary Figure S5). Peaks 1 and 2 were shown to be equivalent in denaturing PAGE, migrating in between self-annealing 24mer and 10mer control RNA. The native PAGE showed peak 2 of free RNA migrating close to the 24mer self-annealed RNA control, consistent with it being a self-annealed species of a similar size. The same gel showed peak 1 of free RNA migrating close to the self-annealing 10mer consistent with it being a monomeric species. In the presence of protein, peak 1, but not peak 2, showed migration to the lower MW position, again consistent with the monomeric species being present. Migration was also seen to the higher MW position in these lanes possibly due to peak overlap. Overall, the native and denaturing acrylamide gel electrophoresis are consistent with the two species observed in the SEC profiles being derived from chemically homogenous 8 bp dsRNA.
Since it is possible that any 8 bp preparation may contain a proportion of self-annealed duplex dsRNA, it was of interest to analyze the SAXS data correlating with peak2 to ascertain whether this species induces CARD release upon binding by RIG-I. Supplementary Figure S6A shows experimental scattering curves (obtained from Gaussian decomposed peaks for each sample) of DC-RIG-I and FL-RIG-I with 8 bp-long ±ATP analogues. The SAXS data correlating to peak2 of DC-RIG-I/8 bp-long complexes without any ATP analogue, with AMP-PNP and ADP-AlFx resulted in Rg values of 40.5, 37.7 and 39.2 Å respectively which are higher than those derived for respective 8 bp complexes analyzed from peak 1 (Supplementary Table S1). The Rmax values of 150, 154 and 160 Å are also greater for DC-RIG-I/8 bp-long without ATP analogue, with AMP-PNP or ADP-AlFx respectively. Furthermore, the molecular weights estimated from the Perod volumes corresponded very well with the molecular weight expected for a 2:1 complex formation between DC-RIG:I with 8 bp-long, further verifying the 2:1 protein:RNA nature of this species (Supplementary Table S1). The P(r) profiles in all three cases were similar unimodal curves but show a longer tail at high r value and a larger Rmax value compared to DC-RIG-I/8 bp complexes (Supplementary Figure S6B). In the case of DC-RIG-I/8 bp-long, no significant differences were observed in the presence of ADP-AlFx, compared to the presence of AMP-PNP or without any analogue.
The SAXS data for the DC-RIG-I/8 bp-long complexes are thus entirely consistent with 8 bp-long existing as a self-annealed species able to be bound by two RIG-I molecules. In order to visualize this, we undertook structural modeling by creating a longer in silico dsRNA (8 bp-long), the length of which was based on the nonlinear fusion of two 8 bp hairpin dsRNAs at an angle of almost 120°, using MC-Sym pipeline (58). Four SAXS data sets were used to calculate a multiphase ab initio SAXS reconstruction for DC-RIG-I in complex with 8 bp-long in a 2:1 ratio in the presence of ADP-AlFx using MONSA (50) (Supplementary Figure S7A) including DC-RIG-I/8 bp-long/ADP-AlFx (peak 2), a set derived from the 8 bp-long model and two sets calculated from DC-RIG-I protein only structure coordinates (PDB ID: 4AY2). The 8 bp-long is shown in the middle of the molecular envelope, with two helicase domains wrapping around on both sides. By using SASREF (49), a molecular model of two molecules of DC-RIG-I bound to 8 bp-long was generated by using two copies of DC-RIG-I crystal structure bound to 8 bp dsRNA (PDB ID: 4AY2) as rigid bodies connected by a 25-Å linker (Supplementary Figure S7B). SAXS data from peak2 of the DC-RIG-I/8 bp/ADP-AlFx fitted well with the SASREF derived model with a nonlinear orientation of the two rigid bodies (χ2 value = 0.26) (Supplementary Figure S7). The arrangement of helicase domains in this model was consistent with the MONSA molecular envelope. Altogether this is consistent with two DC-RIG-I molecules interacting with each terminus of 8 bp-long created by the fusion of two 8 bp dsRNA molecules.
8 bp-long has a similar effect as 8 bp on FL-RIG-I
With the knowledge that the 8 bp-long species is able to bind to two DC-RIG-I molecules, it was of interest to determine what effect it could have on FL-RIG-I CARDs release. The SAXS data from peak 2 of FL-RIG-I/8 bp-long complexes without any ATP analogue, with AMP-PNP and with ADP-AlFx resulted in Rg values of 57.1, 57.8 and 53.2 Å respectively which are higher than respective complexes of FL-RIG-I with 8 bp as analyzed from peak 1 (Supplementary Table S1). The Rmax values of FL-RIG-I in complex with 8 bp-long without ATP analogue, with AMP-PNP and ADP-AlFx were 246, 246 and 251 Å, respectively, which are also greater than obtained for corresponding experiments with 8 bp and indicate the relatively longer sizes of the complexes formed (Supplementary Table S1). The SAXS derived molecular weight values were also consistent with larger 2:1 protein:RNA complexes having formed (Supplementary Table S1). The P(r) profiles in all three cases of FL-RIG-I/8 bp-long (Supplementary Figure S6B) showed broader curves and longer tails and overall represent much bigger molecules compared to FL-RIG-I/8 bp complexes. Again, no difference in the presence of ADP-AlFx was observed. Together, these data are consistent with two RIG-I molecules binding to the 8 bp-long dsRNA, and some degree of CARD release occurring, as similarly observed for 8 bp binding to FL-RIG-I.
Limited proteolysis of RIG-I and RIG-I/dsRNA complexes using trypsin
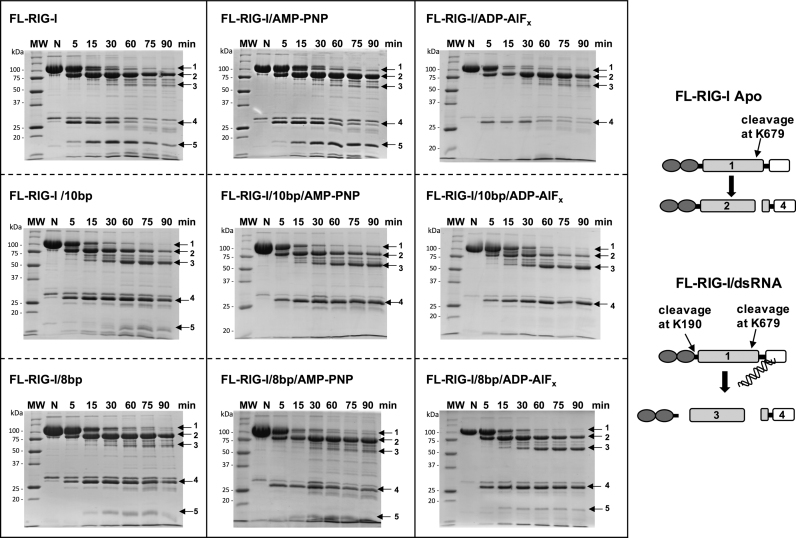
To further validate the release of the CARDs by interaction of FL-RIG-I with 10 bp, versus 8 bp dsRNA, the protein/RNA complexes were tested for their susceptibility to cleavage by trypsin at K190, which becomes accessible when the CARDs are released. RIG-I bound to 10 bp and 8 bp dsRNA, ±ATP analogues were subjected to limited tryptic digestion (Figure 9). We have previously determined that 4 main diagnostic bands arise in the tryptic digest pattern of RIG-I (28). While band 1 corresponds to the uncleaved FL-RIG-I (band 1; ∼107 kDa), band 2 corresponds to the N-terminal portion of FL-RIG-I cleaved at K679 (band 2; ∼90 kDa) and band 4 is the remaining C-terminal portion (band 4; ∼28 kDa). Band 3 (∼56 kDa) arises upon further cleavage at K190 of N-terminal portion of FL-RIG-1 cleaved at K679 and is thus diagnostic of CARDs release. As a comparison, apo-FL-RIG-I in the absence or the presence of ATP analogues was also subjected to tryptic digestion.
Figure 9.
The effect of 10 bp versus 8 bp on the tryptic digest patterns of FL-RIG-I ± ATP analogues. Arrows 1, 2, 3, 4 and 5 indicate the fragments generated by tryptic digestion and identified by mass spectrometry. In the schematic representation, the accessible cleavage sites for trypsin are shown. resulting in the observed cleavage products.
In the case of apo-FL-RIG-I ±ATP analogues, band 2 is very prominent, but band 3 is faint, indicating limited susceptibility to tryptic digestion at K190. Furthermore, an additional band (band 5 ∼17 kDa) is observed for apo-FL-RIG-I in the absence of ATP analogue and in the presence of AMP-PNP, but not when ADP-AlFx is added to apo-FL-RIG-I. Band 5 was identified by N-terminal sequencing to be a cleavage product of the C-terminal region of the protein (giving rise to band 4). This could reflect a changed conformation of the RIG-I in the presence of ADP-AlFx.
In case of the RIG-I/10 bp complexes, band 3 is prominent in the absence or presence of any ATP analogues. This indicates that 10 bp dsRNA is sufficient to induce a conformational rearrangement of RIG-I that avails the cleavage site K190 to trypsin. A progressively diminished band 2 (region of RIG-I including CARDs) was observed in cases of RIG-I/10 bp complexes, especially in the presence of ADP-AlFx, consistent with the cleavage of CARDs by trypsin. In contrast, with 8 bp bound RIG-I in the absence of ATP analogue and the presence of AMP-PNP, band 3 is faint, similar to apo-FL-RIG-I. Band 3 is more prominent, however, when ADP-AlFx is in the presence of the RIG-I/8 bp complex.
Overall, the tryptic digest patterns correlate with the SAXS data analysis, suggesting that the RIG-I CARDs are most completely released in the presence of 10 bp ±ATP analogue, but especially in the presence ADP-AlFx. 8 bp dsRNA, however, appears unable to facilitate CARDs release except upon the addition of ADP-AlFx. This suggests that for the complete activation of RIG-I, 10 bp is more effective that 8 bp, ATP binding also has a role in the activation mechanism of RIG-I and their combined action leads to the most complete extension of the CARD domains.
DISCUSSION
RIG-I plays a frontline role in the detection of viral dsRNA in the cytosol, and initiation of the innate immune response. It is activated through recognition and binding of 5′ppp-dsRNA and ATP that together act to trigger the release of the tandem CARDs, allowing them to interact with a downstream partner IPS-1. Crystal structures have revealed the details of molecular recognition of 5′ppp-dsRNA end structures and shown that the closing of the helicase and CTD domains around the dsRNA is likely to displace the CARDs from their auto-repressed state. It has also been shown through crystallographic studies that the binding of ATP analogues, in particular ADP-AlF3, accentuates the formation of this closed conformation (17–27).
What has not been shown through crystallography, however, is the nature of the full-length dynamic system before and after dsRNA and ATP binding. Neither the CTD of the apo-FL-RIG-I nor the CARDs of the FL-RIG-I/dsRNA have been visualized in crystal structures, presumably due to their flexibility. Nor has the effect of ATP binding been directly shown to assist CARD release. Thus, we have utilized SAXS to provide valuable overall shape information for FL-RIG-I before and after dsRNA and ATP analogue binding, including the characterization of released CARDs. Since SAXS reflects the dynamic ensemble of molecular shape in solution it is able to reveal shifts in dynamic equilibria. It therefore complements the crystallographic and functional studies by allowing the dynamics of RIG-I in solution to be revealed, helping to complete the model of the molecular mechanism of RIG-I activation. It should be noted that SAXS does not provide detailed structural information—thus all molecular models produced in this study are for the purpose of visualizing structures or sets of structures that are consistent with the overall shape information from the SAXS experiment.
Here, we have applied SAXS to address whether the 10 bp 5′ppp-hairpin loop is truly more effective than the 8 bp equivalent species in causing RIG-I CARD release. Cellular assays have shown that 10 bp activates the RIG-I pathway whereas 8 bp is far less effective (22). Structural studies using X-ray crystallography, however, show that DC-RIG-I can bind to either the 8 bp (21) or 10 base pair hairpin RNA (37) and thus do not reveal the basis for this difference in activity. Understanding whether 10 bp is truly the shortest effective RNA activator of RIG-I discovered to date is important for its potential development as a therapeutic acting through the RIG-I pathway. In addition, since ATP has been shown to act alongside the dsRNA in the activation of RIG-I, our experiments were conducted in the absence and presence of ATP analogue AMP-PNP and transition-state analogue ADP-AlFx.
The SAXS data were collected as the RIG-I or RIG-I complexes eluted from a size exclusion column (SEC-SAXS). This ensured the data were being collected for stoichiometric complexes and with no interference from aggregation. The quality of the data was further improved using a Gaussian decomposition method that allows the contributions to the scattering, from partially overlapped peaks, to be distinguished and separated from one another. This was particularly important for studies of the 8 bp dsRNA which existed as two species – the intended hairpin (8 bp) and a self-annealed duplex (8 bp-long). Together these methods allowed for the analysis of data of higher quality than could have been collected using a static sample SAXS approach.
In parallel with SAXS studies of the FL-RIG-I, it was also important to collect comparative data for DC-RIG-I. This is because the activation of RIG-I involves the compaction of the helicase-CTD domains simultaneous with the elongation of the CARDs. Thus, the overall increase of the maximal length (Rmax) of the RIG-I as it engages with dsRNA is not very striking. But when viewed side by side with the SAXS profiles for DC-RIG-I, the profiles for FL-RIG-I clearly indicate the extent to which the CARDs are released. The comparative set of P(r) and normalized Kratky profiles shown in Figure 2 reveal the progressive compaction of the DC-RIG-I from the most extended open conformation of the apo-DC-RIG-I to a more compact structure formed upon binding 8 bp, and finally to the very compact structure formed in complex with 10 bp. Thus, a difference in the degree of engagement of 8 bp compared to 10 bp is clear. Further to this, the addition of the ATP analogue also has an effect. Even in the absence of dsRNA, ADP-AlFx is able to stabilize a compacted form of DC-RIG-I. In the presence of dsRNA, AMP-PNP is able to slightly stabilize the compact form, and ADP-AlFx even more so.
With the effect of dsRNA and ATP analogues on DC-RIG-I revealed, the FL-RIG-I SAXS profiles become highly informative. Although the Rmax for FL-RIG-I is not greatly increased upon binding dsRNA compared to apo-FL-RIG-I, the P(r) profiles show a change in shape reflecting the simultaneous compacting of the DC-RIG-I and extension of the CARDs. The binding of 8 bp can thus be seen to result in some CARD release, but binding of 10 bp clearly effects an even greater degree of CARD release. In addition, particularly in the case of 10 bp, the addition of ADP-AlFx results in the most dramatic change of shape of FL-RIG-I that we interpret as full CARD release. These data are also fitted by molecular models that allow the forms of RIG-I to be visualized. Here EOM methods were used to generate ensembles of molecular structures that reflect the dynamic nature of the molecular structure of FL-RIG-I. These show that CARD release likely exists in dynamic equilibrium between forms of RIG-I in which the CARDs have only a limited degree of freedom and forms where the CARDs are completely released. These models depict that when 10 bp and AMP-AlFx are present, this equilibrium is shifted to the most extreme degree of CARDs release. This was also supported by tryptic digest experiments in which cleavage in the CARDs linker region could be monitored. While RIG-I bound to 8 bp showed some susceptibility to cleavage at this site (mainly in the presence of ADP-AlFx), RIG-I bound to 10 bp showed the greatest susceptibility (particularly in the presence of ADP-AlFx).
The current studies also revealed the presence of a duplex form of 8 bp in the preparation. It was considered as to whether this unintended species could contribute to the lack of activity reported for 8 bp. However, our SAXS experiments showed that this species (8 bp-long) was able to be bound by two RIG-I molecules, as would be expected for a duplex with two 5′ppp-extensions of 8 bp each, and trigger a degree of CARDs release much like 8 bp. Thus, it is unlikely that the presence of this species, if it was present in biological assays, would be the cause of the reduced RIG-I activity observed for 8 bp.
Together the data support our understanding that 10 bp is a strong activator of RIG-I by very effectively initiating CARDs release. It is also clear that while 8 bp can effect partial release of the CARDs it is not to the extent seen for 10 bp, which may be critical for the initiation of the signaling pathway in cells. Why this is the case is not revealed from crystallographic studies that show an apparently tight complex of DC-RIG-I bound to 8 bp (21). The current data suggest that the differences between the 10 bp and 8 bp complexes with RIG-I lie in the dynamic equilibrium of the interactions that are not revealed using crystallography. The addition of the two extra pairs of bases in the 10 bp binding tunnel potentially provides for the extra stability of the closed form of the complex in which the CARDs are fully released, leading to full RIG-I activation.
It is also clear that the ATP transition-state analogue ADP-AlFx is extremely effective at acting alongside the dsRNA to form a tight and stable closed form of RIG-I around the dsRNA. This is consistent with observations from crystallographic studies in which this ATP analogue is co-crystallized with duck RIG-I helicase domain in the presence of a dsRNA 19mer and shown to interact with both the HEL1 and HEL2 domains that come together in the closed conformation (19). In particular glutamine and arginine sidechains emanating from HEL2 formed hydrogen bond interactions with the AlF3 moity that mimics the gamma phosphate. Interestingly, they also co-crystallized the same structure with AMP-PNP, but do not report whether this structure differs from that of RIG-I helicase domain bound to ADP-AlFx. However, our observations are consistent with a hydrodynamic radius study of RIG-I bound to dsRNA which demonstrated that ADP-AlFx effected the greatest shift in sedimentation coefficient compared to other ATP analogues, also interpreted as the formation of a fully closed complex (21). Together this supports the model of ATP binding assisting RIG-I CARDs release through stabilizing the closed conformation around the dsRNA (41,42). As the helicase domain becomes tightly compacted, the CARDs become displaced resulting in RIG-I’s activation. The transition-state analogue ADP-AlFx was clearly more effective than the ground state analogue AMP-PNP in achieving this - either through its ability to stabilize a bone fide transition state of the helicase domain, or through its stronger electostatic interactions with HEL1 and HEL2.
In conclusion, the model of RIG-I activation is supported by the SAXS data, allowing this flexible system to be better understood. We have also shown that the 10 bp dsRNA species is shown to be more effective than 8 bp in triggering CARDs release in vitro, likely underlying its superior potency in cellular assays. This understanding is anticipated to assist in the development of molecules that activate RIG-I that can be used therapeutically in the future.
DATA AVAILABILITY
SAXS data (and models) have been deposited in the SASDB under the accession numbers:
SASDB78: Full-length RIG-I
SASDB88: Full-length RIG-I plus ADP-AlFx
SASDB98: Full-length RIG-I plus bound 10mer hairpin dsRNA
SASDBA8: Full-length RIG-I plus 10mer hairpin dsRNA /AMP-PNP
SASDBB8: Full-length RIG-I plus 10mer hairpin dsRNA and ADP-AlFx
SASDBD8: Full-length RIG-I plus 8mer hairpin dsRNA (SEC-peak1)
SASDBE8: Full-length RIG-I 8mer hairpin dsRNA/AMP-PNP (SEC-peak1)
SASDBF8: Full-length RIG-I 8mer hairpin dsRNA/ADP-AlFx (SEC-peak1)
SASDBG8: Delta-CARDs RIG-I
SASDBH8: Delta-CARDs RIG-I plus ADP-AlFx
SASDBJ8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA
SASDBK8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA and AMP-PNP
SASDBL8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA and ADP-AlFx
SASDBM8: Delta-CARDs RIG-I plus 8mer hairpin dsRNA (SEC-peak1)
SASDBN8: Delta-CARDs plus 8mer hairpin dsRNA/AMP-PNP (SEC-peak1)
SASDBP8: Delta-CARDs plus 8mer hairpin dsRNA/ADP-AlFx (SEC-peak1)
SASDBQ8: Full-length RIG-I plus 8mer hairpin dsRNA (SEC-peak2)
SASDBR8: Full-length RIG-I 8mer hairpin dsRNA/AMP-PNP (SEC-peak2)
SASDBS8: Full-length RIG-I 8mer hairpin dsRNA/ADP-AlFx (SEC-peak2)
SASDBT8: Delta-CARDs RIG-I plus 8mer hairpin dsRNA (SEC-peak2)
SASDBU8: Delta-CARDs plus 8mer hairpin dsRNA/AMP-PNP (SEC-peak2)
SASDBV8: Delta-CARDs plus 8mer hairpin dsRNA/ADP-AlFx (SEC-peak2)
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Associate Professor Dahai Luo and Ms Hui Yee Yong for the 10bp and 8bp dsRNA and their discussion of this work. We also wish to thank all the beamline staff at the SAXS/WAXS and MX2 beamlines at the Australian Synchrotron, Victoria, Australia. In particular, Dr Nigel Kirby and Dr Tim Ryan provided exceptional assistance with data collection. We would also like to thank Emre Brookes for his guidance on the Gaussian Deconvolution of SAXS data.
Author contributions: J.W. and M.W. designed the experiments. N.S. prepared the proteins and performed the SEC-SAXS experiments under the guidance of S.B., M.W. and N.S. analyzed the SAXS data. N.S. prepared the figures. N.S. and J.W. wrote the manuscript with input from all others.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council of Australia [APP1044390 to J.A.W., M.C.J.W.]; National Health and Medical Research Senior Research Fellowship [FP1079611 to M.C.J.W.]. Funding for open access charge: Monash University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Yoneyama M., Onomoto K., Jogi M., Akaboshi T., Fujita T.. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015; 32:48–53. [DOI] [PubMed] [Google Scholar]
- 2. Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G. et al. . Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009; 31:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwasaki A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012; 66:177–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S.. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008; 205:1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. et al. . Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006; 441:101–105. [DOI] [PubMed] [Google Scholar]
- 6. Uchikawa E., Lethier M., Malet H., Brunel J., Gerlier D., Cusack S.. Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5. Mol. Cell. 2016; 62:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez K.R., Bruns A.M., Horvath C.M.. MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J. Virol. 2014; 88:8194–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu Z., Zhang X., Wang G., Zheng H.. The laboratory of genetics and physiology 2: emerging insights into the controversial functions of this RIG-I-like receptor. BioMed Res. Int. 2014; 2014:960190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S.. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005; 6:981–988. [DOI] [PubMed] [Google Scholar]
- 10. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J.. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005; 437:1167–1172. [DOI] [PubMed] [Google Scholar]
- 11. Peisley A., Wu B., Xu H., Chen Z.J., Hur S.. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014; 509:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B.. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005; 19:727–740. [DOI] [PubMed] [Google Scholar]
- 13. Seth R.B., Sun L., Ea C.K., Chen Z.J.. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682. [DOI] [PubMed] [Google Scholar]
- 14. Akira S., Uematsu S., Takeuchi O.. Pathogen recognition and innate immunity. Cell. 2006; 124:783–801. [DOI] [PubMed] [Google Scholar]
- 15. Takeuchi O., Akira S.. Pattern recognition receptors and inflammation. Cell. 2010; 140:805–820. [DOI] [PubMed] [Google Scholar]
- 16. Cordin O., Banroques J., Tanner N.K., Linder P.. The DEAD-box protein family of RNA helicases. Gene. 2006; 367:17–37. [DOI] [PubMed] [Google Scholar]
- 17. Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M.. Structural insights into RNA recognition by RIG-I. Cell. 2011; 147:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M. Jr, Patel S.S., Marcotrigiano J.. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011; 479:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S.. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011; 147:423–435. [DOI] [PubMed] [Google Scholar]
- 20. Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S., Carell T., Hopfner K.P.. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011; 12:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo D., Kohlway A., Vela A., Pyle A.M.. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure. 2012; 20:1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M.. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013; 14:772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deimling T., Cui S., Lammens K., Hopfner K.P., Witte G.. Crystal and solution structure of the human RIG-I SF2 domain. Acta Crystallogr. F: Struct. Biol. Commun. 2014; 70:1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P.. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010; 18:1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. et al. . Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010; 17:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P.. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011; 39:1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S.. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013; 152:276–289. [DOI] [PubMed] [Google Scholar]
- 28. Beckham S.A., Brouwer J., Roth A., Wang D., Sadler A.J., John M., Jahn-Hofmann K., Williams B.R., Wilce J.A., Wilce M.C.. Conformational rearrangements of RIG-I receptor on formation of a multiprotein:dsRNA assembly. Nucleic Acids Res. 2013; 41:3436–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poeck H., Besch R., Maihoefer C., Renn M., Tormo D., Morskaya S.S., Kirschnek S., Gaffal E., Landsberg J., Hellmuth J. et al. . 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 2008; 14:1256–1263. [DOI] [PubMed] [Google Scholar]
- 30. Hou J., Zhou Y., Zheng Y., Fan J., Zhou W., Ng I.O., Sun H., Qin L., Qiu S., Lee J.M. et al. . Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell. 2014; 25:49–63. [DOI] [PubMed] [Google Scholar]
- 31. Kubler K., Gehrke N., Riemann S., Bohnert V., Zillinger T., Hartmann E., Polcher M., Rudlowski C., Kuhn W., Hartmann G. et al. . Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Res. 2010; 70:5293–5304. [DOI] [PubMed] [Google Scholar]
- 32. Zitvogel L., Kroemer G.. Anticancer immunochemotherapy using adjuvants with direct cytotoxic effects. J. Clin. Invest. 2009; 119:2127–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Besch R., Poeck H., Hohenauer T., Senft D., Hacker G., Berking C., Hornung V., Endres S., Ruzicka T., Rothenfusser S. et al. . Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 2009; 119:2399–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Palchetti S., Starace D., De Cesaris P., Filippini A., Ziparo E., Riccioli A.. Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J. Biol. Chem. 2015; 290:5470–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J. et al. . Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014; 514:372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlee M., Hartmann G.. The chase for the RIG-I ligand–recent advances. Mol. Ther. 2010; 18:1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J.. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M. Jr, Inagaki F., Fujita T.. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008; 29:428–440. [DOI] [PubMed] [Google Scholar]
- 39. Myong S., Cui S., Cornish P.V., Kirchhofer A., Gack M.U., Jung J.U., Hopfner K.P., Ha T.. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009; 323:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bamming D., Horvath C.M.. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009; 284:9700–9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rawling D.C., Fitzgerald M.E., Pyle A.M.. Establishing the role of ATP for the function of the RIG-I innate immune sensor. eLife. 2015; 4, doi:10.7554/eLife.09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Louber J., Brunel J., Uchikawa E., Cusack S., Gerlier D.. Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5. BMC Biol. 2015; 13:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zheng J., Yong H.Y., Panutdaporn N., Liu C., Tang K., Luo D.. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015; 43:1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gee P., Chua P.K., Gevorkyan J., Klumpp K., Najera I., Swinney D.C., Deval J.. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 2008; 283:9488–9496. [DOI] [PubMed] [Google Scholar]
- 45. Brookes E., Demeler B., Rosano C., Rocco M.. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J.: EBJ. 2010; 39:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brookes E., Perez J., Cardinali B., Profumo A., Vachette P., Rocco M.. Fibrinogen species as resolved by HPLC-SAXS data processing within the UltraScan Solution Modeler (US-SOMO) enhanced SAS module. J. Appl. Crystallogr. 2013; 46:1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M. et al. . ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Franke D., Svergun D.I.. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009; 42:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petoukhov M.V., Svergun D.I.. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005; 89:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bernado P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I.. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007; 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 52. Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bagshaw C. ATP analogues at a glance. J. Cell Sci. 2001; 114:459–460. [DOI] [PubMed] [Google Scholar]
- 54. Wittinghofer A. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr. Biol.: CB. 1997; 7:R682–685. [DOI] [PubMed] [Google Scholar]
- 55. Chabre M. Aluminofluoride and beryllofluoride complexes: a new phosphate analogs in enzymology. Trends Biochem. Sci. 1990; 15:6–10. [DOI] [PubMed] [Google Scholar]
- 56. Receveur-Brechot V., Durand D.. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012; 13:55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bernadó P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I.. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007; 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 58. Parisien M., Major F.. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008; 452:51–55. [DOI] [PubMed] [Google Scholar]
- 59. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAXS data (and models) have been deposited in the SASDB under the accession numbers:
SASDB78: Full-length RIG-I
SASDB88: Full-length RIG-I plus ADP-AlFx
SASDB98: Full-length RIG-I plus bound 10mer hairpin dsRNA
SASDBA8: Full-length RIG-I plus 10mer hairpin dsRNA /AMP-PNP
SASDBB8: Full-length RIG-I plus 10mer hairpin dsRNA and ADP-AlFx
SASDBD8: Full-length RIG-I plus 8mer hairpin dsRNA (SEC-peak1)
SASDBE8: Full-length RIG-I 8mer hairpin dsRNA/AMP-PNP (SEC-peak1)
SASDBF8: Full-length RIG-I 8mer hairpin dsRNA/ADP-AlFx (SEC-peak1)
SASDBG8: Delta-CARDs RIG-I
SASDBH8: Delta-CARDs RIG-I plus ADP-AlFx
SASDBJ8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA
SASDBK8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA and AMP-PNP
SASDBL8: Delta-CARDs RIG-I plus 10mer hairpin dsRNA and ADP-AlFx
SASDBM8: Delta-CARDs RIG-I plus 8mer hairpin dsRNA (SEC-peak1)
SASDBN8: Delta-CARDs plus 8mer hairpin dsRNA/AMP-PNP (SEC-peak1)
SASDBP8: Delta-CARDs plus 8mer hairpin dsRNA/ADP-AlFx (SEC-peak1)
SASDBQ8: Full-length RIG-I plus 8mer hairpin dsRNA (SEC-peak2)
SASDBR8: Full-length RIG-I 8mer hairpin dsRNA/AMP-PNP (SEC-peak2)
SASDBS8: Full-length RIG-I 8mer hairpin dsRNA/ADP-AlFx (SEC-peak2)
SASDBT8: Delta-CARDs RIG-I plus 8mer hairpin dsRNA (SEC-peak2)
SASDBU8: Delta-CARDs plus 8mer hairpin dsRNA/AMP-PNP (SEC-peak2)
SASDBV8: Delta-CARDs plus 8mer hairpin dsRNA/ADP-AlFx (SEC-peak2)