Abstract
UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.
INTRODUCTION
DNA methylation mainly occurs at the fifth position of cytosine of CpG dinucleotides and is crucial for mammalian development (1). DNA methylation has been demonstrated to be a key regulatory mechanism for genomic imprinting, heterochromatin formation, X chromosome inactivation and transcriptional repression. Aberrant DNA methylation has been observed in various cancer cell types and neurological disorders (2–4). DNMT3A and DNMT3B together with a non-catalytic paralog DNMT3L establish de novo DNA methylation, whereas the maintenance DNA methyltransferase DNMT1 is responsible for propagating DNA methylation during DNA replication. Although DNA methylation has been under extensive study over the past several decades, recent advances have dramatically improved our understanding of the underlying mechanisms. For example, both DNMT1 and DNMT3A have been shown to have the auto-inhibitory effect and require other regulatory proteins to stimulate the enzymatic activities (5–8). 5-methyl cytosine can be subsequently converted to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) with Tet family enzymes (9).
Accumulating evidence has suggested that DNMT1 alone cannot maintain global DNA methylation through somatic cell divisions. Indeed, even though DNMT1 contains a PCNA-interacting peptide (PIP) box that binds replication fork associating proteins PCNA, it has been demonstrated that the interaction of DNMT1 with proliferating cell nuclear antigen (PCNA) is not sufficient for the faithful inheritance of DNA methylation during replication (10). Recently, structure-based analyses suggested that DNMT1 is a self-inhibitory enzyme (5,6,11), which required additional regulatory factors to maintain the global DNA methylation patterns. UHRF1 has emerged as one of such critical regulators. Numerous studies have demonstrated that UHRF1 plays multiple roles in maintaining DNA methylation patterns during cell division. Its SRA domain specifically recognizes hemimethylated DNA (12–17), which helps recruit DNMT1 to its targeted substrate sites. UHRF1 PHD finger and tandem Tudor domain binds favorably to histone H3R2 and H3K9me3 marks (18–20), which is important for targeting DNMT1/UHRF1 complex to the proper genomic loci, mainly at replicating heterochromatin regions (21). UHRF1 C-terminal RING domain functions as an E3 ubiquitin ligase to target histone H3 (22–24), DNMT1 binds subsequently to ubiquitinated H3 via its RFTS (replication foci targeting sequence) domain, which helps recruit DNMT1 to DNA replication sites (22). Interestingly, RFTS domain not only helps target DNMT1 to replication loci, but also exerts an auto-inhibitory effect on DNMT1 enzymatic activity (5). On the other hand, auto-inhibition of DNMT1 is relieved during DNA replication, but it remains elusive if the interaction of DNMT1 RFTS domain with ubiquitinated histone H3 has an impact on DNMT1 enzymatic activity. Although a number of studies have demonstrated UHRF1 physically associates with DNMT1 (12,13,25), the detailed molecular mechanism and biological relevance of this interaction remains unclear. A recent study has shown that DNMT1 RFTS domain binds to UHRF1 SRA domain (26), if it exists, this interaction is likely dynamic and transient as suggested previously (16). Interestingly, UHRF1 contains a N-terminal ubiquitin-like domain (UBL) but its biological function remains unknown to date.
In this study, we first verified that human DNMT1 RFTS domain directly binds to ubiquitin. We then presented the crystal structure of DNMT1 RFTS domain in complex with ubiquitin. Furthermore, we presented evidence that UHRF1 N-terminal UBL domain directly interacts with DNMT1 by nuclear magnetic resonance (NMR) titration and isothermal titration calorimetry (ITC) measurement. Interestingly we found that ubiquitin and UHRF1 UBL bind DNMT1 differently although UBL domain structurally resembles ubiquitin. Results from in vitro DNA methylation assay indicate that binding to UHRF1 UBL domain but not ubiquitin could stimulate DNMT1 enzymatic activity toward hemimethylated DNA substrate. On the other hand, binding to ubiquitinated histone H3 but not ubiquitin or histone H3 alone could alleviate DNMT1 self-inhibitory effect. At last, our cell-based data showed that introducing DNMT1 wild-type, but not the DNMT1 mutants with impaired ubiquitin binding, into DNMT1 knockout mouse embryonic stem (ES) cells can effectively restore the global DNA methylation pattern. We also showed that expression of UBL deleted or RING finger deleted UHRF1 mutant cannot restore the proper DNMT1 nuclear localization and thus cannot rescue DNA methylation defects in UHRF1 knockout mouse ES cells. Overall, our study demonstrates that the interaction between UHRF1 and DNMT1 mediated by UBL domain, as well as the interaction between DNMT1 and ubiquitinated histone H3 established by UHRF1 are required for faithful propagation of DNA methylation during DNA replication.
MATERIALS AND METHODS
Plasmid construction, protein expression and purification
Various human DNMT1 and UHRF1 fragments including DNMT1_351 (aa: 351–1616), DNMT1_621 (aa: 621–1616), RFTS domain (aa: 351–600), UHRF1 UBL domain (aa: 1–76), SRA domain (aa: 414–617), RING domain (aa: 672–793), histone H3 and H3K18CK23C (aa:1–30 with an additional tyrosine at the C-terminus), ubiquitin and its G76C mutant were cloned into either pET28a, pGEX-6P-1, a modified pRSFDuet-1 with a cleavable 6 × His-SUMO tag or pCold-GST plasmids (27). Fusion proteins were expressed in Escherichia coli strain Rosetta DE3 (Novagen) and purified via either nickel-NTA affinity column or GSTrap HP column, followed by size-exclusion chromatography. DNMT1 mutants, ubiquitin G76C mutant and UHRF1 domain deletion mutants were generated using QuikChange site-directed mutagenesis kit (Stratagene). The presence of appropriate mutations was confirmed by DNA sequencing. All mutants were purified as described for the wild-type protein. The pcDNA3.1-Myc-His tagged mouse UHRF1 and deletion mutant constructs were kindly provided by Dr Ian Marc Bonapace. Myc-DDK-tagged human DNMT1 (RC226414) and UHRF1 (RC214251) plasmids were purchased from OriGene. Flag-Myc-tagged UHRF1 wild-type and various domain deletions, DNMT1 wild-type and different mutants were reconstructed in PiggyBac vectors with CAG promoter, which are optimal for expression in ES cells.
Crystallization, data collection and structure determination
Crystals of the DNMT1 RFTS domain in complex with ubiquitin were obtained at 298K with the vapor diffusion hanging drop method by mixing 1 μl of the 1:2 molar ratio RFTS/ubiquitin complex solution with 1 μl of crystallization solution (100 mM Tris–HCl at pH 8.0, containing 200 mM sodium acetate and 25% PEG4000). All diffraction data were measured at 100K at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U. The native crystal diffracted to 2.02 Å. All data were processed with HKL-2000 (28). The crystals belong to space group P65 with unit cell dimensions of a = b = 131.76 Å, c = 61.03 Å, α = β = 90°, γ = 120°. The complex structure was solved by molecular replacement using previously determined DNMT1 structural model (PDB ID: 3AV4) and ubiquitin (PDB ID: 1UBQ) as templates. The molecular replacement solution was used subsequently for the structure refinement using Phenix and COOT (29,30). X-ray data collection and refinement statistics are listed in Table 1.
Table 1. Crystallographic data collection and refinement statistics.
| RFTS–ubiquitin complex | |
| PDB ID | 5YDR |
| Data collection | |
| Wavelength | 0.9793Å |
| Space group | P65 |
| Unit cell a, b, c (Å) | a = b = 131.76Å, c = 61.025Å |
| Resolution (Å) (highest resolution shell)a | 50.00–2.00 (2.08–2.00) |
| Observed reflections | 85 1031 |
| Unique reflections | 40 591 |
| Completeness (%) | 99.4 (98.6) |
| R merge | 0.105 (0.478) |
| I/σ | 32.2 (8.0) |
| Redundancy | 10.8 (10.4) |
| Refinement | |
| Resolution (Å) | 44.77–2.00 (2.08–2.00) |
| No. reflections | 40 583 (4016) |
| R work/Rfree (%)b | 19.9/23.3 |
| No. non-H atoms | 3341 |
| Protein | 3095 |
| Ligand | 11 |
| Water | 235 |
| B-factors (Å2) | 44.0 |
| RMSD | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.42 |
| Ramachandran favored/allowed/outliers (%) | 96.4/3.3/0.3 |
aNumbers in parentheses refer to the highest resolution shell.
b R free was calculated using 5% random data omitted from the refinement.
NMR titration
All NMR spectra were acquired at 298K on a Bruker Avance 600 MHz NMR spectrometer with cryoprobe. NMR titration was performed by recording a series of 2D 15N-HSQC spectra on uniformly 15N-labeled ubiquitin domain (about 0.1 mM) in the presence of different amounts of DNA methyltransferase (DNMT) ranging from 0 to 0.15 mM. NMR data were processed by NMRPipe and subsequently analyzed by NMRView (31,32).
Isothermal titration calorimetry
Calorimetric experiments were conducted at 25°C with a MicroCal ITC200 microcalorimeter from Microcal (GE Healthcare) following the standard procedure. Titrations were performed as follows: 50 μM ubiquitin or UHRF1 UBL solution was transferred into the sample cell, one preliminary injection of 0.5 μl of 0.5 mM RFTS sample was followed by about 36 injections of 1 μl. A 2-min delay between the injections was applied to enable the system reach equilibrium. Data were analyzed using MicroCal Origin software.
Synthesis of ubiquitinated histone analogs for in vitro DNA methylation assay
Synthesis of a dual mono-ubiquitin-linked histone H3K18CK23C (aa:1–30) was performed essentially similar as recently described (33). The purified ubiquitin G76C mutant was treated with 5 mM dithiothreitol (DTT) first, and then buffer exchanged into 50 mM sodium phosphate pH 7.5. The reduced ubiquitin mutant was mixed with a 20-fold molar excess of 5,5 -dithiobis-(2-nitrobenzoic acid) (DTNB, Sigma) and then incubated for 1 h at room temperature with vigorous shaking. The reaction solution was buffer-exchanged into the ligation buffer (20 mM Tris–HCl, pH 7.0, 50 mM NaCl and 1 mM ethylenediaminetetraacetic acid (EDTA)) using a PD-10 desalting column (GE Healthcare). The purified H3K18CK23C was also treated with 5 mM DTT first, buffer-exchanged into the ligation buffer, and mixed with a 5-fold molar excess of activated ubiquitin G76C-DTNB for 1 h. The ubiquitinated histone analog was further separated with a Resource S column.
In vitro DNA methylation assay
DNA methylation assays were performed with 50 nM DNMT1 (aa: 351–1616) or DNMT1 (aa: 621–1616) in the presence of 1 μM 30 bp hemi-methylated dsDNA (upper strand: 5′-TTGCACTCTCCTCCXGGAAGTCCCAGCTTC-3′, X = m5C) and 1 μM S-adenosyl-l-[methyl-3H]methionine (AdoMet) (specific activity 15 Ci/mmol, PerkinElmer) in methylation buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 5% glycerol, 1 mM DTT, 1 mM EDTA, 0.1 mg/ml bovine serum albumin). For comparing the effect of ubiquitin and UHRF1_UBL on DNMT1 enzymatic activity, DNMT1 proteins were pre-incubated with the indicated concentrations of ubiquitin or UHRF1_UBL on ice for 30 min. For comparing the effect of ubiquitin, histone H3 and ubiquitinated histone H3 on DNMT1 enzymatic activity, DNMT1 proteins were pre-incubated with the individual factor in methylation buffer without DTT. The reactions were carried out at 37°C and quenched at various time points from the start point to 25 min by leaving the reaction system on ice and further processed by spotting them on DE-81 membranes (GE Healthcare). The membranes were washed sequentially with 10 ml cold 0.2 M NH4HCO3, 10 ml water and 5 ml ethanol. After air-drying of the membranes, the incorporation of 3H-labeled methyl group was detected by liquid scintillation counting with a Beckman LS6500 counter.
Cell culture, generation of stable expressed ESCs and immunofluorescence analysis
Mouse WT ESCs, mUHRF1−/− ESCs, mouse J1 WT ESCs and mDNMT1−/− 36c/c ESCs were cultured on 0.1% gelatin (Sigma) in Dulbecco’s modified Eagle’s medium (high glucose, Gibco, 11 965), supplemented with 15% ES-fetal bovine serum (Gibco, 16141–079), 100 units/ml Leukemia Inhibitory Factor (Millipore, ESG1107), GlutaMAX Supplement (Life Technologies, 35 050), Sodium Pyruvate (Life Technologies, 11 360), and 2-Mercaptoethanol (Life Technologies, 31 350), maintained in a 37°C incubator with 5% CO2 and passaged every 2 days.
Plasmids including Flag-Myc-tagged DNMT1 wild-type and different mutants, Flag-Myc-tagged UHRF1 wild-type and various domain truncation mutants were transfected to UHRF1 or DNMT1 knockout ES cells using Lipofectamine 3000 (Invitrogen, L3000015) according to the manufacturer's protocol. Afterward, cells were selected using Hygromycin. Single colonies were picked to have homogeneous gene expression.
For immunofluorescence, ESCs were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at 25°C before treated with 0.1% Triton-100 (Sigma) in PBS for 10 min at 25°C. For staining of DNMT1 and UHRF1, cells were then blocked for 1 h with PBS containing 10% fetal bovine serum (FBS) at 25°C before incubation with DNMT1 antibody (1:100), UHRF1 antibody (1:100) or Flag antibody (1:200, Sigma, F1804) in PBS containing 5% FBS at 4°C overnight. For immunofluorescence analysis of 5mC content, after treatment of 0.1% Triton-100 in PBS, cells were treated with 2M HCl for 20 min at 37°C to denature the DNA. After neutralization with 0.1M Borate, pH 8.5 for 5min at 25°C twice, cells were blocked for 1 h with PBS containing 10% FBS at 25°C. Labeling was performed with 5mC (1:1000) and Flag antibody (1:500, Sigma, F7425) in PBS containing 5% FBS at 4°C overnight. After primary antibody treatment, cells were washed with 0.1% Triton-100 in PBS for three times before incubation with Alexa Fluor 568 goat anti-mouse IgG1 (1:500) and Alexa Fluor 488 donkey anti-rabbit IgG (H+L) (1:500) in PBS for 1 h at 25°C protected from light. Cells were washed with 0.1% Triton-100 in PBS and mounted using SlowFade Antifade Mountant with DAPI. Images were acquired using Carl Zeiss LSM 710 and Carl Zeiss LSM 780 confocal microscopy at 40× magnification. Images were analyzed with ImageJ. 5mC fluorescence (red channel) was quantified and normalized to DNA fluorescence (blue channel) from at least 50 cells positively stained for UHRF1 or DNMT1.
Antibodies
The following antibodies were used in this study: anti-DNMT1 (Santa Cruz, H-300, sc-20701), anti-UHRF1 (Santa Cruz, H-8, sc-373750), anti-Flag (Sigma, F1804), anti-Flag (Sigma, F7425), anti-Myc (Sigma, C3956), anti-5mC (Diagenode, 33D3, C15200081), anti-β-actin (Thermofisher, MA5–15739), Alexa Fluor 568 goat anti-mouse IgG1 (Invitrogen, A21124), Alexa Fluor 488 donkey anti-rabbit IgG (H+L) (Invitrogen, A21206).
DNA methylation quantification
The ELISA-based ‘Methylated DNA Quantification Kit (Colorimetric)’ (Abcam, ab117128) was used to quantify global DNA methylation content in mouse ES cell lines. The assay was performed in triplicates according to the manual. The 100 ng of total genomic DNA was used for 5-methyl cytosine quantification, and relative DNA methylation was quantified using the positive control provided in the kit.
Bisulfide sequencing
Genomic DNA was extracted and purified with PureLink Genomic DNA mini kit (Invitrogen). For bisulfite conversion, 1 μg genomic DNA was treated and recovered using EpiTect Bisulfite kit following manufacturer's manual. The IAP and LINE1 regions were amplified from bisulfite converted genomic DNA via polymerase chain reaction using following primer pairs.
IAP F: ATTTTGTTGATTAAATAAATTATTATTGGG
IAP R: TAAAACATATCCTCTAATCATTTCTACTCA.
LINE1 F: TTATTTTGATAGTAGAGTT
LINE1 R: CAAACCAAACTCCTAACAA.
IAP and LINE1 amplicons were ligated into pGEM-T vector (Promega) for cloning. Eight colonies were sequenced for each assay and the sequencing data were analyzed by QUMA (http://quma.cdb.riken.jp/).
RESULTS
Ubiquitin and UHRF1 UBL domain directly interact with DNMT1
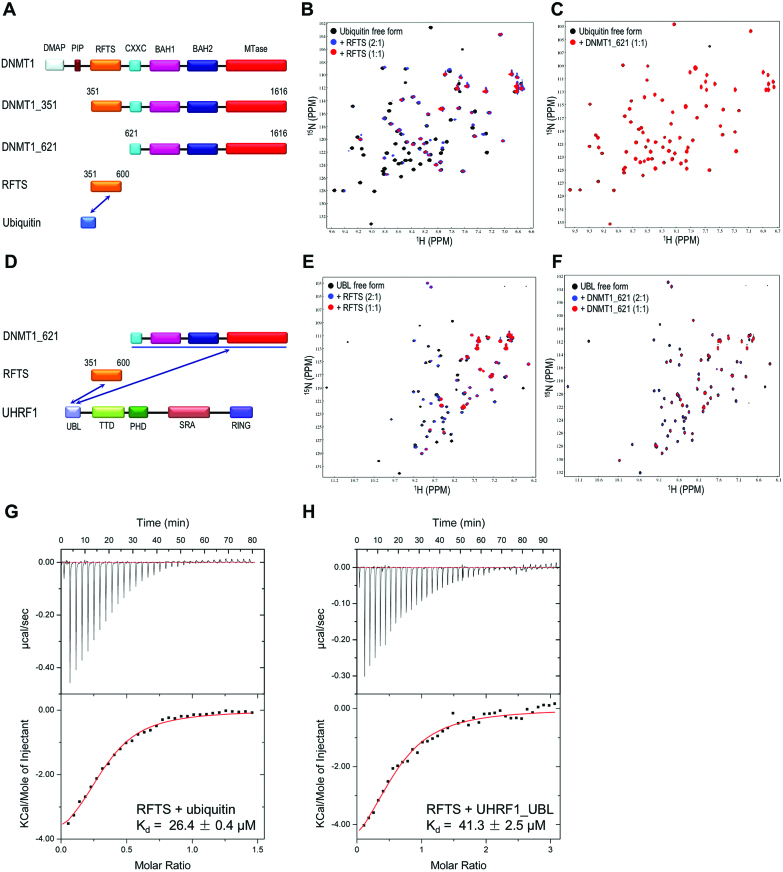
Recent studies have suggested DNMT1 binds to ubiquitinated H3 that helps the localization of DNMT1 to replication sites for efficient DNA methylation maintenance (22,23). Here we generated a number of human DNMT1 fragments and applied NMR titration to assess the interaction between DNMT1 and ubiquitin (Figure 1A). As shown in superimposed 2D 15N-HSQC spectra (Figure 1B and Supplementary Figure S1A), a number of well-resolved cross peaks showed progressive line broadening eventually disappeared when titrating DNMT1_351 (aa: 351–1616) or RFTS domain (aa: 351–600) into 15N-labeled ubiquitin solution. However, adding DNMT1_621 (aa: 621–1616) into 15N-labeled ubiquitin solution does not exhibit any obvious line broadening or chemical shift perturbation (Figure 1C), indicating DNMT1 interacts directly with ubiquitin through its RFTS domain. This conclusion was further confirmed by reciprocal NMR titration of ubiquitin into 15N-labeled RFTS solution as a number of cross peaks disappeared owning to the serious line broadening effect (Supplementary Figure S1B). This result was also verified by ITC titration showing the DNMT1 RFTS domain binds to ubiquitin with apparent Kd of 26.0 μM (Figure 1G).
Figure 1.
Molecular determinants of DNMT1 binding to ubiquitin and UBL domain of UHRF1. (A) Domain organization of DNMT1. Three DNMT1 fragments were prepared for ubiquitin binding analysis. (B and C) 15N HSQC spectra show that ubiquitin binds to DNMT1 RFTS domain (aa: 351–600) but not DNMT1_621 (aa: 621–1616), as illustrated by strong selective line broadening effect induced by addition of DNMT1 RFTS domain but not DNMT1_621. (D) Domain organization of DNMT1 fragments that were prepared for UHRF1 UBL binding. (E and F) NMR titration indicates UHRF1 UBL binds to DNMT1 RFTS domain as well as DNMT1_621, as illustrated by strong line broadening of certain resonances induced by the binding of DNMT1 RFTS (E) and DNMT1_621 fragment (F). (G and H) ITC confirms the molecular interaction of DNMT1 RFTS domain with ubiquitin and UHRF1 UBL domain. The top panel shows experimental ITC curve of titrating DNMT1 RFTS domain into ubiquitin (G) and UHRF1 UBL (H) respectively. The lower panel shows fitted curves of calorimetric titrations. The measured dissociation constant (Kd) is shown.
Previous studies have indicated that UHRF1 forms a complex with DNMT1 (12,13,25,34,35), but results from domain mapping experiments by different studies are inconsistent. Given that UHRF1 N-terminal UBL domain structurally resembles ubiquitin, as structure superimposition gives a RMSD of 0.52 Å (Supplementary Figure S1C), we therefore tested if UHRF1 N-terminal UBL domain interacts with DNMT1. Overlay of 2D 15N-HSQC spectra of UHRF1 UBL domain in free form and in mixture with DNMT1_351 (aa: 351–1616) or RFTS domain reveals a number of UBL amide resonances were broadened beyond detection or underwent significant chemical shift perturbations (Figure 1E and Supplementary Figure S1D), the reciprocal NMR titration of UBL to 15N-labeled RFTS domain produced the similar result (Supplementary Figure S1E), suggesting UHRF1 UBL domain binds to DNMT1 RFTS domain as well. These NMR titration results are in good agreement with Kd of 41.3 μM derived from ITC titration (Figure 1H). Strikingly, titrating DNMT1_621 (aa: 621–1616) into 15N-labeled UBL domain also caused obvious line broadening effect (Figure 1F), as many cross peaks intensities are greatly attenuated. While under the same molar ratio, the line broadening effect caused by DNMT1_351 is much more significant when compared with the addition of DNMT1_621, indicating UHRF1 UBL domain interacts with multiple regions of DNMT1.
Crystal structure of DNMT1 RFTS domain in complex with ubiquitin
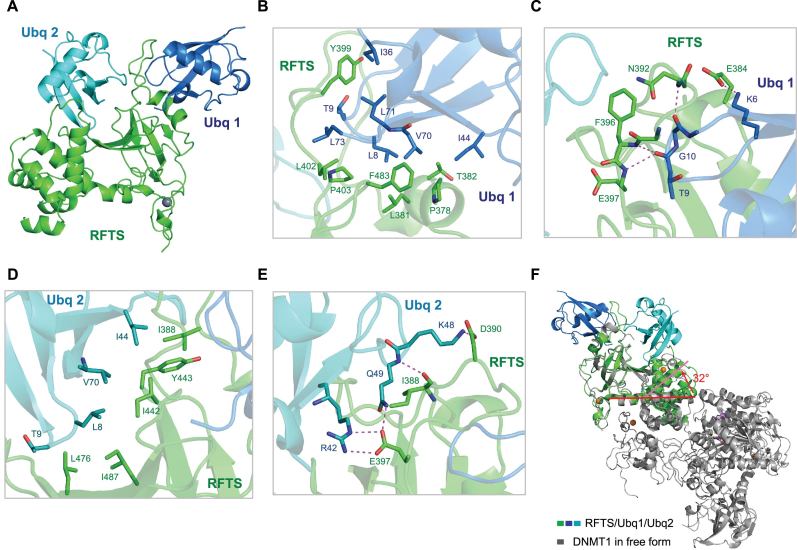
To further explore the detailed molecular mechanism of ubiquitin binding by DNMT1 RFTS domain, we determined the three-dimensional crystal structure of human DNMT1 RFTS domain in complex with ubiquitin at 2.0 Å resolution in a P65 lattice containing one complex per asymmetric unit (Figure 2 and crystallographic statistics is given in Table 1). Ubiquitin and DNMT1 RFTS domain are present at 2:1 stoichiometry in an asymmetric unit, which is consistent with sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis of the protein crystal (Supplementary Figure S2A). As shown in Figure 2A, RFTS domain mainly consists of a zinc-binding motif, an antiparallel seven-stranded β-barrel followed by an α-helical bundle. The long loop encompassing residues 386–404, which is invisible in previous reported structures due to the high flexibility (5,7), protrudes from the β-barrel and is sandwiched by two ubiquitin molecules (Supplementary Figure S2B). Two ubiquitin chains, though sharing almost identical conformations, bind RFTS in a very different fashion and bury the surface area of 1790 Å2 and 1520 Å2 respectively (Supplementary Figure S2C–E). The contact surface for each ubiquitin is relatively large compared to other ubiquitin binding domains, agreeing well with relatively high affinity of RFTS binding to ubiquitin. Ubiquitin 1 sits on one end of the β-barrel whereas ubiquitin 2 lies outside of the β-barrel. We noted that ubiquitin 2 molecule has higher B factors and less well-defined electron density maps compared to that of ubiquitin molecule 1, indicating ubiquitin 2 has the higher flexibility.
Figure 2.
Crystal structure of DNMT1 RFTS domain in complex with ubiquitin. (A) 1:2 DNMT1 RFTS domain-ubiquitin contents of the asymmetric unit are shown as ribbon diagrams. DNMT1 RFTS domain is colored in green, two ubiquitin molecules are colored in marine and cyan respectively. (B and C) Close-up view of the interactions between the RFTS domain and ubiquitin 1 (Ubq1). (D and E) Detailed view of the interaction network of the RFTS domain with ubiquitin 2 (Ubq2). Residues that form the binding interface are depicted as stick models and labeled. Hydrogen bonds and salt bridges are shown in magenta dashes. (F) DNMT1 RFTS domain undergoes conformational changes upon ubiquitin binding. The α-helix (aa: 493–518) of RFTS domain adopts as a straight α-helical conformation in DNMT1 (PDB ID: 4WXX) but bends about 32° in the middle of α-helix (aa: 493–518) at the position of Met502 in RFTS/ubiquitin complex. Ubiquitin binding caused the bending of RFTS α-helix (aa: 493–518) that connects the preceding β-barrel (aa: 400–490) to the α-helical bundle, this eventually leads to large changes in the relative orientation of the β-barrel with respect to α-helical bundle.
An extensive network of hydrophobic interactions and hydrogen bonding contribute to the RFTS and ubiquitin complex formation. The complex structure has clearly displayed a new ubiquitin-binding mode. On the RFTS–ubiquitin 1 interface (Figure 2B), Pro378, Leu381 and Thr382 of the RFTS α-helix encompassing residue 377–382, as well as side chains of Pro403 and Phe483 interact with the canonical hydrophobic Ile44 patch that is mainly comprised of residues Leu8, Ile44 and Val70. Additionally, residues Tyr399 inserts in a hydrophobic core formed by the side chains of Thr9, Ile36 and Leu71 of ubiquitin 1, whereas Leu402 makes hydrophobic contacts with Leu73. Ubiquitin 1 hydrophobic Ile44 patch and Ile36 patch (composed of Ile36, Leu71 and Leu73) are both involved in RFTS binding, which is rarely observed in the ubiquitin binding. Furthermore, the β1–β2 loop of ubiquitin is deeply inserted into the long loop (Leu386-Gln404) and stabilizes this acidic loop through the hydrogen bonding and complementary electrostatic interactions (Figure 2C). For example, Glu384 side chain carboxyl group forms a salt bridge with lysine 6 side chain ϵ-amino group. The amides of Asn392 and Phe396 form hydrogen bonds with main chain carbonyl groups of Gly10 and Thr9 respectively. At the RFTS–ubiquitin 2 interface (Figure 2D), Ile388, Ile442, Tyr443, Leu476 and Ile487 of RFTS domain engage in hydrophobic interactions with Ile44 patch of ubiquitin 2. In addition, Ubiquitin 2 Lys48 side chain points to a concave cleft mainly formed by Glu384 and Asp390 (Figure 2E). Gln49 main chain amide group forms a hydrogen bond with backbone carbonyl group of Ile388, at the same time Gln49 side chain amide group donates a hydrogen bond to side chain carboxyl group of Glu397. Moreover, Ubiquitin 2 Arg42 side chain guanidinium group forms salt bridges with side chain carboxyl group of Glu397.
Structural comparison of human DNMT1 (aa: 351–1600; PDB ID: 4WXX) with our RFTS–ubiquitin complex revealed that two major segments of RFTS domain including seven-stranded β-barrel and the α-helical bundle in both structures are similar (36). Structure superimposition of the β-barrel (aa: 400–490) of human DNMT1 RFTS domain (aa: 351–1600; PDB ID: 4WXX, color in gray) with that of RFTS–ubiquitin complex (color in green/marine/cyan) gives an RMSD of 0.55 Å (Figure 2F), while superimposing the corresponding α-helical bundle of RFTS domain (aa: 493–589) of free and ubiquitin bound form gives an RMSD of 0.95 Å (Supplementary Figure S3). However, the long helix (aa: 493–518) that connects the β-barrel and the followed α-helical bundle has an unusual structural dynamic feature. It adopts as a straight α-helix conformation in DNMT1 free form structure (PDB ID: 4WXX) but bends about 32° in the middle of α-helix at the position of Met502 in in our structure (Figure 2F), which cause the orientation of the β-barrel relative to the followed α-helical bundle significantly different in two structures. We reasoned that the ubiquitin binding contributes to the severe bending of the long helix (aa: 493–518) and the change in the relative orientation of two RFTS subdomains. Another obvious conformational difference is found at C-terminal region of RFTS in which the α-helical region (Leu592-Gln598) of RFTS–ubiquitin complex and human DNMT1 differed in the orientation. Interestingly, the corresponding region in human DNMT1 (PDB ID: 4WXX) exists as a random coil (36). Structural comparison also highlighted that the loop Leu386-Gln404 are disordered and thus invisible in the apo state, but are stabilized upon ubiquitin binding and well defined in our complex structure.
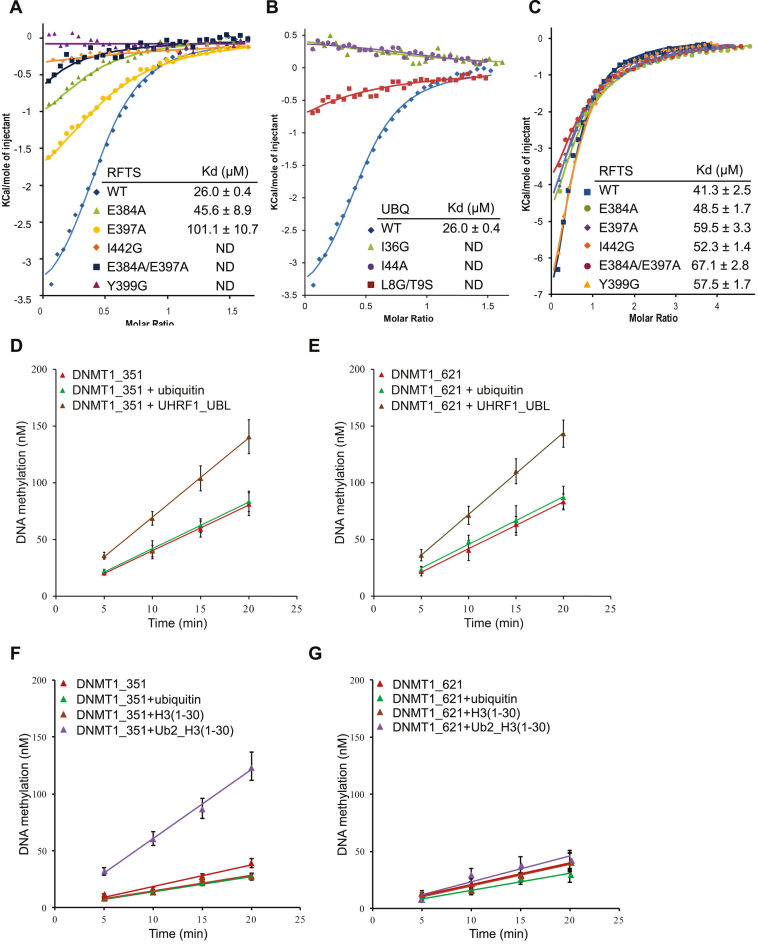
To analyze the significance of specific interactions identified in the RFTS–ubiquitin complex, we made a number of amino-acid substitutions including E384A, E397A, Y399G, I442G and E384A/E397A mutations on RFTS and I44A, I36G and L8G/T9S mutations on ubiquitin. Single mutation of I44A or double mutation of L8G/T9S on ubiquitin abolishes the binding to RFTS domain (Figure 3A). This is not surprising since I44 hydrophobic patches of both ubiquitin molecules are involved in RFTS binding. But unexpectedly, a single mutation of ubiquitin Ile36 with glycine also blocks the binding of RFTS even though only ubiquitin 1 Ile36 hydrophobic patch participates in the interaction of RFTS. Consistently, we observed that a single mutation on RFTS Y399G disrupts the ubiquitin binding though residue Tyr399 has extensive interactions with the Ile36 patch of ubiquitin 1 but not ubiquitin 2 in our complex structure. On the other hand, mutation on RFTS I442G abolishes the ubiquitin binding though I442 only participate in the recognition of ubiquitin 2 but not ubiquitin 1 (Figure 3B), implying that the binding of two ubiquitin molecules to RFTS is interdependent. Mutating the charged residue Glu384 or Glu397 of RFTS domain to alanine reduced the ubiquitin binding about 2-fold, but the double mutant E384A/E397A seriously compromised ubiquitin binding. Overall, ITC measurement showed that mutations of critical residues in either ubiquitin or RFTS compromise RFTS–ubiquitin binding, thus confirmed our structural findings. It is worth noting that these RFTS mutations have little impact on the binding to UHRF1 UBL domain (Figure 3C), suggesting these RFTS residues directly involved in ubiquitin binding are not required for UHRF1 UBL binding. Altogether, our results demonstrate that ubiquitin binding is defined by key interactions with residues of RFTS within the loops Leu386-Gln404, Lys440-Val453, Thr467-Leu476 and Ser480-Glu485 and the helical region of Glu377-Glu384. A recently study has identified a ubiquitin-interacting motif in RFTS but most of residues defined this ‘UIM motif’ do not directly participate in ubiquitin recognition (23).
Figure 3.
Experimental measurement of DNMT1 RFTS domain binding to ubiquitin or UHRF1 UBL domain. (A) ITC fitting curves for binding of DNMT1 RFTS domain and its mutants to ubiquitin, the insert lists the calculated dissociation constant (Kd). ND, not detectable. (B) ITC fitting curves for binding of DNMT1 RFTS domain to ubiquitin wild-type and mutants, along with the calculated Kd. (C) ITC fitting curves for binding of DNMT1 RFTS domain and its mutants with UHRF1 UBL domain with dissociation constant (Kd) values indicated. (D and E) In vitro DNA methylation reactions were performed as a function of time and the concentration of ubiquitin or UHRF1 UBL. (D) DNMT1_351 (aa: 351–1616) or (E) DNMT1_621 (aa: 621–1616) was used as the enzyme, and a 30-bp long hemimethylated DNA fragment as the substrate. (F and G) In vitro DNA methylation reactions were performed as a function of time and the concentration of ubiquitin, histone H3 or H3ub2. (F) DNMT1_351 (aa: 351–1616) or (G) DNMT1_621 (aa: 621–1616) was used as the enzyme.
Binding of UHRF1 UBL domain, or a two mono-ubiquitinated histone H3 but not ubiquitin or histone H3 can stimulate DNMT1 enzymatic activity
Several recent studies have revealed that RFTS domain of DNMT1 regulates its enzymatic activity through an auto-inhibition mechanism (5,7,37), as explained by the structural analyses showing the RFTS domain of DNMT1 occupies its own catalytic pocket and prevents substrate DNA binding. Interestingly, a previous study demonstrated that SRA domain of UHRF1 interacts with DNMT1 RFTS domain and relieves its inhibitory effect to a certain degree (35). Since we found that ubiquitin and UBL domain of UHRF1 can interact with RFTS domain, we attempted to test whether these interactions could have an impact on DNMT1 enzymatic activity. We conducted in vitro DNA methylation assays to evaluate DNMT1_351 (aa: 351–1616) and DNMT1_621 (aa: 621–1616) enzymatic activities in the presence and absence of UHRF1 UBL domain by employing a 30-bp long hemimethylated DNA as the substrate. As shown in Figure 3D and E, pre-incubation of either DNMT1_351 or DNMT1_621 with UHRF1 UBL domain resulted in a 2-fold increase of DNMT1 activity. In contrast, no stimulation was observed when either DNMT1_351 or DNMT1_621 was pre-incubated with ubiquitin. Interestingly, about 3-fold increase of methylation activity was observed when pre-incubating DNMT1_351 with a two mono-ubiquitin-linked histone H3K18CK23C peptide (Figure 3F), this is in line with a recent report suggesting ubiquitinated histone H3 but not ubiquitin or histone H3 alone could stimulate DNMT1 enzymatic activity (33). However, this dual mono-ubiquitin-linked histone H3 peptide does not exert obvious stimulatory effect on DNMT1_621, as shown in Figure 3G. Taken together, the results from in vitro DNA methylation assay indicate that binding of the two mono-ubiquitinated histone H3 but not ubiquitin or histone H3 alone to DNMT1 RFTS could alleviate its self-inhibitory effect. On the other hand, though UHRF1 UBL can interact with both RFTS and DNMT_621, the binding of UBL domain of UHRF1 to DNMT1_621 fragment is essential for its stimulatory effect on DNMT1 enzymatic activity.
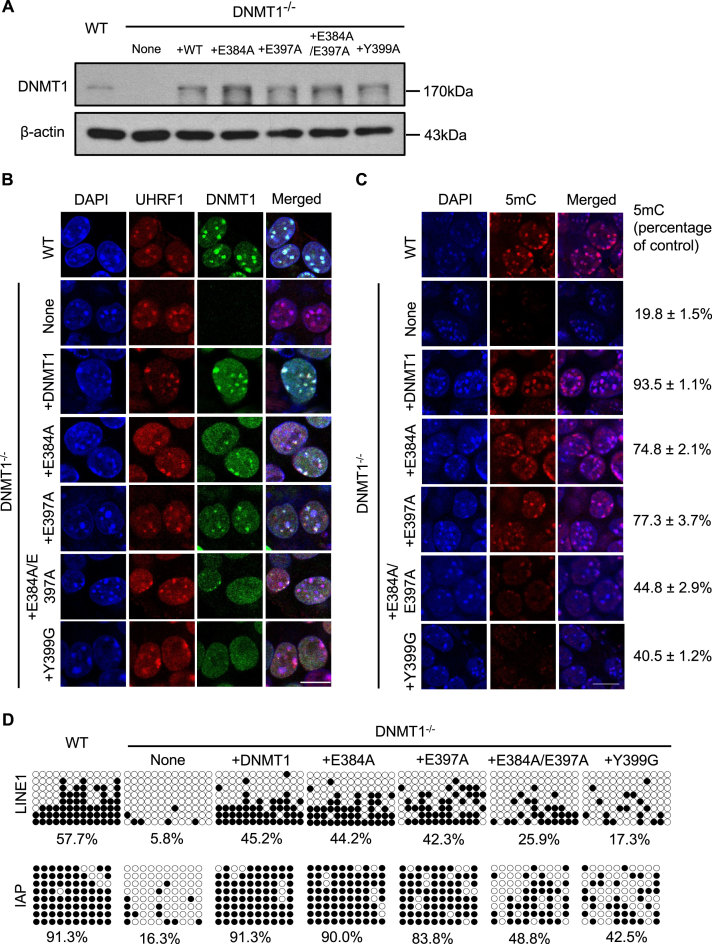
Expression of DNMT1 mutants with impaired binding to ubiquitin is unable to restore global DNA methylation in DNMT1−/− ES cells
Considering that DNMT1 RFTS domain physically interacts with ubiquitin, and binding to ubiquitinated histone H3 stimulates DNMT1 enzymatic activity in vitro, we further assessed the impact of the interaction between DNMT1 and ubiquitinated histone H3 on global DNA methylation patterns in mouse ES cells. We first established DNMT1 knockout mouse ES cell lines stably expressing DNMT1 wild-type, E384A, E397A or Y399A single point mutant, or E384A/E397A double mutant. The DNMT1−/− ES cells that stably expressed exogenous DNMT1 or mutants at a level similar to endogenous DNMT1 in J1 ES cells are further selected for the subsequent immunofluorescence analysis (Figure 4A). As shown in Figure 4B, DNMT1 wild-type, E384A or E397A single point mutant is co-localized with UHRF1 with clearly visible nuclear foci in DNMT1−/− ES cells, whereas DNMT1 Y399G single mutant or E384A/E397A double point mutant did not form clear nuclear foci and exhibited much dimmer signal dispersed throughout the nucleus. We also assessed global DNA methylation by immunostaining using anti-5-methylcytosine antibodies (Figure 4C). Global DNA methylation level in DNMT1−/− ES cells is substantially lower than that of the wild-type cells. Stable expression of fl-DNMT1 in DNMT1−/− ES cells restored the DNA methylation level comparable to that of wild-type ES cells, whereas expression of DNMT1 E384A or E397A single point mutant restored DNA methylation level to ∼75% of that of wild-type ES cells. In contrast, stable expression of DNMT1 Y399G single mutant or E384A/E397A double point mutant only recovered the 5-methyl cytosine level to about 40–45% of that of wild-type cells. The observation is consistent with the results from ELISA-based methylated DNA quantification assay (Supplementary Figure S4A), that DNMT1−/− ES cells stably expressing DNMT1 Y399G single mutant or E384A/E397A double point mutant have a significant decrease of global DNA cytosine methylation compared to wild-type mouse ES cells or DNMT1−/- ES cells stably expressing fl-DNMT1. We also examined the effect of mutations on DNA methylation status of the regulatory regions in retrotransposons using bisulfite sequencing analysis. A dramatic decrease of DNA methylation in regulatory region of LINE-1 (long interspersed nuclear element-1) and IAP (intracisternal A particle) retrotransposons in DNMT1 knockout ES cells was observed (Figure 4D). Stable expression of fl-DNMT1 effectively restored the DNA methylation level, but expression of DNMT1 Y399G or E384A/E397A mutant only partially rescued the DNA methylation defect observed in DNMT1−/− ES cells. Our results indicated that disruption of the interaction between DNMT1 and ubiquitinated histone H3 could affect the nuclear localization of DNMT1 and the maintenance of DNA methylation.
Figure 4.
Assessing nuclear localization and DNA methylation status in DNMT1−/− mouse embryonic stem cells stably expressing DNMT1 wild-type or mutants. (A) Flag-Myc-tagged wild-type DNMT1, DNMT1 E384A, E397A, E384A/E397A and Y399A mutants were stably expressed in DNMT1−/− mouse ESCs, as analyzed by western blot using antibodies that recognize DNMT1. The expression level of these exogenous proteins similar to endogenous DNMT1 was selected for the subsequent study. β-actin was selected as a loading control. (B) Immunofluorescence analysis of DNMT1 focal staining pattern in DNMT1 wild-type or knockout mouse ES cells or DNMT1−/− ES cells stably expressing DNMT1 wild-type, E384A or E397A or Y339G single point mutant, or E384A/E397A double point mutant respectively. Scale bars, 10 μm. (C) Immunostaining using an antibody against 5mC in control and DNMT1−/− mouse ES cells after genetic complementation with DNMT1 wild-type or various mutants. 5mC fluorescence signals from ∼50 cells were quantified and normalized against the wild-type cells, the mean value with a standard error has been provided. (D) The DNA methylation status of LINE1 and IAP was analyzed by bisulfite sequencing in control, DNMT1−/− ESCs and DNMT1−/− ESCs stably expressing DNMT1 wild-type, E384A, E397A, E384A/E397A and Y399A mutants. The percentage of 5mC was calculated and shown.
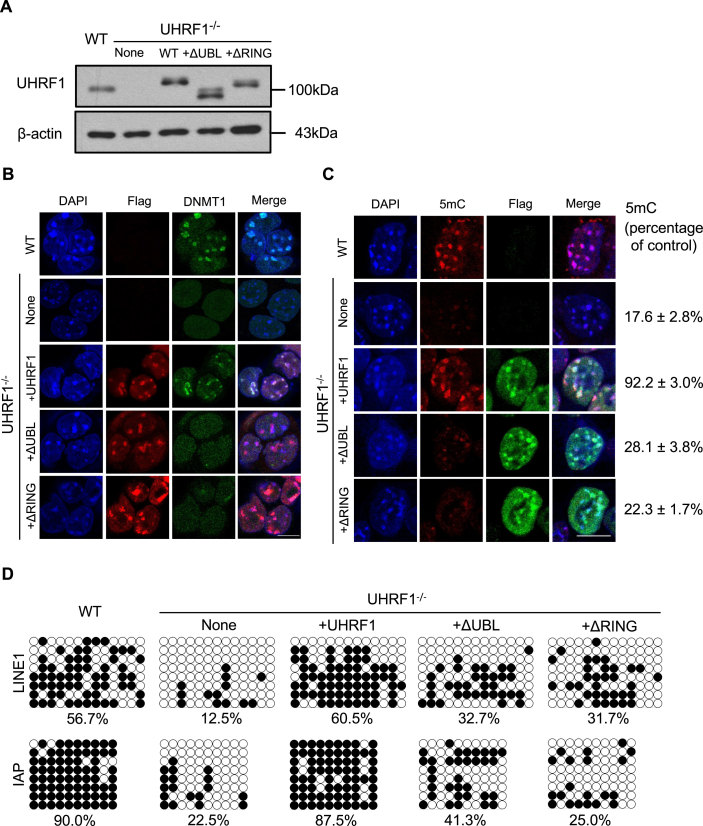
UBL and RING domains of UHRF1 are required for proper DNMT1 nuclear localization and the maintenance of DNA methylation
Given that UHRF1 UBL domain is directly involved in UHRF1–DNMT1 interaction, we therefore performed immunofluorescence assay to analyze whether UBL domain is required for proper nuclear localization of DNMT1. We obtained stable UHRF1−/− ES cell lines expressing either UHRF1 wild-type, UBL-deleted or RING-deleted UHRF1 at a level comparable to that of endogenous UHRF1 in mouse ES cells (Figure 5A). Similar to previous observation (12,13,21), DNMT1 has a diffuse nuclear localization pattern in UHRF1−/− ES cells (Figure 5B). Expression of full length UHRF1 restored DNMT1 nuclear foci, two proteins are mainly co-localized at heterochromatin region. In contrast, introducing either UBL-deleted or RING-deleted form of UHRF1 into UHRF1−/− ES cells failed to restore DNMT1 localization at heterochromatin regions, DNMT1 exhibited a diffuse nuclear localization pattern although both UHRF1 domain-deleted mutants show clear heterochromatin localization patterns. Taken together, these results highlighted not only the interaction between DNMT1 and UHRF1, but also the interaction between DNMT1 and ubiquitinated histone H3 established by UHRF1 are critical for proper DNMT1 nuclear localization. It is worth noting that RFTS-mediated ubiquitinated histone H3 recognition and UHRF1 binding actually provides a mechanistic explanation for the previous observation showing DNMT1 RFTS domain is critical for the association of DNMT1 with late replication heterochromatin region (38).
Figure 5.
Both UHRF1 UBL and RING finger are critical for DNMT1 proper nuclear localization and maintenance DNA methylation in mouse embryonic stem cells. (A) UHRF1−/− mouse ES cell lines stably expressing Flag-Myc-tagged UHRF1 wild-type or various mutants were analyzed by western blot using antibodies that recognize UHRF1. Flag-Myc-tagged UHRF1 or mutants expressed at a level similar to endogenous UHRF1 were selected for the study. β-actin was selected as a loading control. (B) Immunofluorescence analysis of DNMT1 focal staining pattern in UHRF1 wild-type, UHRF1 knockout mouse ES cells and UHRF1−/− ES cells transfected with UHRF1-ΔUBL or UHRF1-ΔRING truncated mutant. Exogenous expression of UHRF1 and mutants was detected by Flag antibodies. Scale bars, 10 μm. (C) Immunostaining using an antibody against 5mC in control and UHRF1−/− mouse ES cells after genetic complementation with wild-type or UHRF1-ΔUBL or UHRF1-ΔRING. The 5mC levels relative to wild-type ESCs were shown. Error bars represent ± s.e.m. (D) The DNA methylation status of LINE1 and IAP was analyzed by bisulfite sequencing in wild-type ESCs (as control), UHRF1−/− ESCs and UHRF1−/- ESCs stably expressing UHRF1 wild-type, or UHRF1-ΔUBL or UHRF1-ΔRING mutants. The percentage of 5mC was calculated and shown.
The above observation also prompted us to investigate UHRF1 UBL or RING deleted mutants in DNA methylation by assessing their ability to rescue DNA methylation defects in UHRF1−/− ES cells. As expected, immunofluorescence staining for 5-methyl cytosine revealed that global DNA methylation in UHRF1−/− ES cells is greatly reduced compared with that in wild-type cells (Figure 5C). Expression of Flag-Myc-tagged UHRF1 in UHRF1−/− ES cells restored the level of DNA methylation to ∼80% of that measured in the wild-type ES cells, whereas expression of either UBL or RING deleted UHRF1 mutants only recovered a small portion of DNA methylation but the level is still far lower than that measured in wild-type UHRF1 ES cells (Figure 5C). This observation is in line with ELISA-based relative quantification of global DNA cytosine methylation shown in Supplementary Figure S4B and recent reports (22,39). In addition, bisulfite sequencing analysis also revealed that fl-UHRF1, but not UHRF1 UBL or RING deleted UHRF1 mutant, could restore DNA methylation pattern in the regulatory regions of IAP and LINE1 retrotransposons (Figure 5D). These results strongly indicate that interactions of DNMT1 with both UHRF1 and ubiquitinated histone H3 are crucial for maintaining DNA methylation in ES cells.
DISCUSSION
Ubiquitination, the covalent attachment of mono-ubiquitin or linkage-specific poly-ubiquitin to the target proteins, modulates a wide range of important cellular processes by regulating protein activity and interactions (40). Ubiquitinated proteins are recognized by distinct ubiquitin-binding domains found in a variety of proteins (41). Mono- or poly-ubiquitin chains serve as the versatile cellular signaling messengers to be interpreted by these ubiquitin-binding proteins and thus regulate the downstream signaling. A number of studies have shown histones can be subject to ubiquitination and involve in transcriptional regulation, DNA damage response and cell cycle regulation (42,43). For example, histone H2B ubiquitination plays a role in transcriptional regulation and DNA replication (44–46), H2A ubiquitination is required for damaged DNA repair (47–49). UHRF1 has recently been reported to be an E3 ubiquitin ligase that targets multiple lysines on histone H3 including Lys23, Lys18 and Lys14 (22–24), ubiquitinated histone H3 can be further recognized by RFTS domain of DNMT1, thus facilitates localization of DNMT1 to replication foci and promotes the faithful maintenance of DNA replication. In current study, we provide a detailed molecular basis of DNMT1 mediated ubiquitinated histone binding at replication foci by solving the high-resolution crystal structure of DNMT1 RFTS domain in complex with ubiquitin. Interestingly, one RFTS domain can bind two ubiquitin molecules simultaneously. Ile44-centered and Ile36-centered hydrophobic patches of ubiquitin 1, as well as Ile44-centered hydrophobic patch of ubiquitin 2 are involved in the RFTS interactions whereas electrostatic interactions, hydrogen bonding and salt bridges also contribute complementary to the complex formation. This mode of ubiquitin binding is quite different from most of other ubiquitin recognition patterns (40). Ishiyama et al have just reported the structure of RFTS in complex with histone H3 linked two mono-ubiquitin molecules and observed very similar interaction network between RFTS and two ubiquitin molecules (33). Intriguingly, though the conjugated histone H3 peptide contributes to RFTS binding, it has no visible impact on the interaction network between RFTS and ubiquitin.
Notably, we also found that DNMT1 RFTS domain can interact with UHRF1 N-terminal UBL domain thus mediates the interaction between UHRF1 and DNMT1. Unlike ubiquitin, UHRF1 UBL domain binds to both DNMT1 RFTS domain and DNMT1_621 (aa: 621–1616). The results from in vitro DNA methylation assay suggest that the binding of ubiquitinated histone H3 to RFTS domain of DNMT1 alleviates DNMT1 auto-inhibitory effect. Strikingly, binding of ubiquitin or histone H3 alone failed to show the stimulatory effect on DNMT1 enzymatic activity. On the other hand, the N-terminal UBL domain of UHRF1, which structurally resembles ubiquitin, not only binds to DNMT1 but also stimulates its methyltransferase activity. Considering ubiquitinated histone H3 is enriched at replication foci, we postulate that ubiquitinated histone H3 binding to DNMT1 not only helps accumulate DNMT1 at replication foci, but also alleviates DNMT1 auto-inhibitory effect. Similarly, the binding of UHRF1 UBL to DNMT1_621 as well as RFTS domain not only helps target DNMT1 to the hemimethylated CpG substrate sites, but also enhances DNMT1 enzymatic activity. Intriguingly, our data suggest the stimulatory mechanisms by UHRF1 UBL and ubiquitinated histone H3 are different. UHRF1 UBL domain stimulates DNMT1 enzymatic activity mainly through the interaction with the DNMT_621 fragment, whereas ubiquitinated histone H3 binding to DNMT1 RFTS domain may facilitate RFTS domain to dissociate from catalytic domain which in turn promotes the access of substrate DNA. UBL domains have been found in many proteins, they typically share very similar three-dimensional structure but not necessary sequence similarity to ubiquitin. The sequence variation of UBL domains provides the opportunity to expand the selectivity of ubiquitin binding domain interactions considering ubiquitin is evolutionarily highly conserved in primary sequence across the eukaryotic species (50). Indeed, a number of ubiquitin binding domains have been found to bind to UBL domain too, but with different binding affinities in many occasions (51,52). Mutations on RFTS I442G or E384A/E397A abolished ubiquitin interaction but had no effect on UHRF1 UBL binding, indicating the interaction network of RFTS–ubiquitin complex is distinct from that of RFTS-UBL. Nonetheless, future structural analysis is required to unravel the detailed molecular basis of UHRF1 UBL binding to RFTS domain. Moreover, the quantitative ITC measurement indicate that ubiquitin binds more strongly to RFTS domain compared to UHRF1 UBL domain, whereas UBL domain, but not ubiquitin, has the capability to bind to DNMT1_621 (aa: 621–1616).
Previous studies have demonstrated that UHRF1 binds to hemimethylated DNA and methylated histone H3K9, which are critical for targeting DNMT1 to proper genomic foci for the faithful epigenetic inheritance of DNA methylation. In this study, we performed structure guided genetic complementation experiments to show the binding of DNMT1 to ubiquitinated histone H3, as well as the interaction between DNMT1 and UHRF1 UBL domain plays equally important role in efficient DNA methylation maintenance. It is conceivable that all modular domains of UHRF1 participate and act in concert with DNMT1 to maintain DNA methylation pattern during DNA replication.
DATA AVAILABILITY
The coordinate and structure factor for the reported crystal structure has been deposited in the Protein Data Bank with the following accession code: 5YDR.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to acknowledge the use of the Shanghai synchrotron radiation facility (beamline BL17U) for X-ray data collection, and NMR facility at the Hong Kong polytechnic University and Dr Kenneth Yan for NMR data collection. We thank Dr Xiaochun Yu at Michigan University for providing UHRF1 wild-type and knockout mouse embryonic stem cells, and Dr TaiPing Chen at MD Anderson Cancer Center for providing DNMT1 wild-type and knockout mouse embryonic stem cells, and Dr Ian Marc Bonapace for providing the pcDNA3.1-Myc-His tagged mouse UHRF1 and deletion mutants constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hong Kong Research Grants Council, General Research Fund [17127715, 17117514, 776313 to C.Q.; 17123816, HKU2/CRF/13G to Z.Z.]. Funding for open access charge: Hong Kong Research Grants Council, General Research Fund [17127715, 17117514, 776313 to C.Q.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 2. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007; 8:286–298. [DOI] [PubMed] [Google Scholar]
- 3. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klein C.J., Botuyan M.V., Wu Y., Ward C.J., Nicholson G.A., Hammans S., Hojo K., Yamanishi H., Karpf A.R., Wallace D.C. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 2011; 43:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S.. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9055–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song J., Rechkoblit O., Bestor T.H., Patel D.J.. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011; 331:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Syeda F., Fagan R.L., Wean M., Avvakumov G.V., Walker J.R., Xue S., Dhe-Paganon S., Brenner C.. The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J. Biol. Chem. 2011; 286:15344–15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo X., Wang L., Li J., Ding Z., Xiao J., Yin X., He S., Shi P., Dong L., Li G. et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015; 517:640–644. [DOI] [PubMed] [Google Scholar]
- 9. Wu H., Zhang Y.. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014; 156:45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spada F., Haemmer A., Kuch D., Rothbauer U., Schermelleh L., Kremmer E., Carell T., Langst G., Leonhardt H.. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 2007; 176:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song J., Teplova M., Ishibe-Murakami S., Patel D.J.. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012; 335:709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E.. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007; 317:1760–1764. [DOI] [PubMed] [Google Scholar]
- 13. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007; 450:908–912. [DOI] [PubMed] [Google Scholar]
- 14. Hashimoto H., Horton J.R., Zhang X., Bostick M., Jacobsen S.E., Cheng X.. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008; 455:826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S.. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008; 455:822–825. [DOI] [PubMed] [Google Scholar]
- 16. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M.. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008; 455:818–821. [DOI] [PubMed] [Google Scholar]
- 17. Qian C., Li S., Jakoncic J., Zeng L., Walsh M.J., Zhou M.M.. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 2008; 283:34490–34494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie S., Jakoncic J., Qian C.. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 2012; 415:318–328. [DOI] [PubMed] [Google Scholar]
- 19. Arita K., Isogai S., Oda T., Unoki M., Sugita K., Sekiyama N., Kuwata K., Hamamoto R., Tochio H., Sato M. et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng J., Yang Y., Fang J., Xiao J., Zhu T., Chen F., Wang P., Li Z., Yang H., Xu Y.. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 2013; 288:1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J.. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013; 4:1563. [DOI] [PubMed] [Google Scholar]
- 22. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F. et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013; 502:249–253. [DOI] [PubMed] [Google Scholar]
- 23. Qin W., Wolf P., Liu N., Link S., Smets M., La Mastra F., Forne I., Pichler G., Horl D., Fellinger K. et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015; 25:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harrison J.S., Cornett E.M., Goldfarb D., DaRosa P.A., Li Z.M., Yan F., Dickson B.M., Guo A.H., Cantu D.V., Kaustov L. et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. Elife. 2016; 5:e17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Felle M., Joppien S., Nemeth A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Langst G.. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011; 39:8355–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berkyurek A.C., Suetake I., Arita K., Takeshita K., Nakagawa A., Shirakawa M., Tajima S.. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J. Biol. Chem. 2014; 289:379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayashi K., Kojima C.. pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expr. Purif. 2008; 62:120–127. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski T., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 30. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 32. Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004; 278:313–352. [DOI] [PubMed] [Google Scholar]
- 33. Ishiyama S., Nishiyama A., Saeki Y., Moritsugu K., Morimoto D., Yamaguchi L., Arai N., Matsumura R., Kawakami T., Mishima Y. et al. Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell. 2017; 68:350–360. [DOI] [PubMed] [Google Scholar]
- 34. Achour M., Jacq X., Ronde P., Alhosin M., Charlot C., Chataigneau T., Jeanblanc M., Macaluso M., Giordano A., Hughes A.D. et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008; 27:2187–2197. [DOI] [PubMed] [Google Scholar]
- 35. Bashtrykov P., Jankevicius G., Jurkowska R.Z., Ragozin S., Jeltsch A.. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. The Journal of biological chemistry. 2014; 289:4106–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z.M., Liu S., Lin K., Luo Y., Perry J.J., Wang Y., Song J.. Crystal Structure of Human DNA Methyltransferase 1. J Mol Biol. 2015; 427:2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bashtrykov P., Jankevicius G., Smarandache A., Jurkowska R.Z., Ragozin S., Jeltsch A.. Specificity of Dnmt1 for methylation of hemimethylated CpG sites resides in its catalytic domain. Chem Biol. 2012; 19:572–578. [DOI] [PubMed] [Google Scholar]
- 38. Leonhardt H., Page A.W., Weier H.U., Bestor T.H.. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992; 71:865–873. [DOI] [PubMed] [Google Scholar]
- 39. Smets M., Link S., Wolf P., Schneider K., Solis V., Ryan J., Meilinger D., Qin W., Leonhardt H.. DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation. Human molecular genetics. 2017; 26:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Komander D., Rape M.. The ubiquitin code. Annu Rev Biochem. 2012; 81:203–229. [DOI] [PubMed] [Google Scholar]
- 41. Dikic I., Wakatsuki S., Walters K.J.. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009; 10:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weake V.M., Workman J.L.. Histone ubiquitination: triggering gene activity. Mol Cell. 2008; 29:653–663. [DOI] [PubMed] [Google Scholar]
- 43. Jackson S.P., Durocher D.. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013; 49:795–807. [DOI] [PubMed] [Google Scholar]
- 44. Kim J., Hake S.B., Roeder R.G.. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005; 20:759–770. [DOI] [PubMed] [Google Scholar]
- 45. Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D.. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005; 20:601–611. [DOI] [PubMed] [Google Scholar]
- 46. Trujillo K.M., Osley M.A.. A role for H2B ubiquitylation in DNA replication. Mol Cell. 2012; 48:734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J.. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. [DOI] [PubMed] [Google Scholar]
- 48. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S. et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012; 150:697–709. [DOI] [PubMed] [Google Scholar]
- 49. Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K.. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 50. Winget J.M., Mayor T.. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010; 38:627–635. [DOI] [PubMed] [Google Scholar]
- 51. Zhang D., Chen T., Ziv I., Rosenzweig R., Matiuhin Y., Bronner V., Glickman M.H., Fushman D.. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009; 36:1018–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raasi S., Varadan R., Fushman D., Pickart C.M.. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005; 12:708–714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinate and structure factor for the reported crystal structure has been deposited in the Protein Data Bank with the following accession code: 5YDR.