Abstract
The expression of nitrilase in Arabidopsis during the development of the clubroot disease caused by the obligate biotroph Plasmodiophora brassicae was investigated. A time course study showed that only during the exponential growth phase of the clubs was nitrilase prominently enhanced in infected roots compared with controls. NIT1 and NIT2 are the nitrilase isoforms predominantly expressed in clubroot tissue, as shown by investigating promoter-β-glucuronidase fusions of each. Two peaks of β-glucuronidase activity were visible: an earlier peak (21 d post inoculation) consisting only of the expression of NIT1, and a second peak at about 32 d post inoculation, which predominantly consisted of NIT2 expression. Using a polyclonal antibody against nitrilase, it was shown that the protein was mainly found in infected cells containing sporulating plasmodia, whereas in cells of healthy roots and in uninfected cells of inoculated roots only a few immunosignals were detected. To determine which effect a missing nitrilase isoform might have on symptom development, the P. brassicae infection in a nitrilase mutant (nit1-3) of Arabidopsis was investigated. As a comparison, transgenic plants overexpressing NIT2 under the control of the cauliflower mosaic virus 35S promoter were studied. Root galls were smaller in nit1-3 plants compared with the wild type. The phenotype of smaller clubs in the mutant was correlated with a lower free indole-3-acetic acid content in the clubs compared with the wild type. Overexpression of nitrilase did not result in larger clubs compared with the wild type. The putative role of nitrilase and auxins during symptom development is discussed.
The infection of cruciferous hosts with the obligate biotroph Plasmodiophora brassicae Wor. leads to cell elongation and cell division in infected roots and hypocotyls, resulting in the typical hypertrophied roots (clubroot) (Ingram and Tommerup, 1972). Earlier it was speculated that growth hormones, i.e. cytokinins and auxins, are in some way involved in symptom development (Dekhuijzen and Overeem, 1971; Butcher et al., 1974). While the vegetative secondary plasmodia of the pathogen produce cytokinins (Dekhuijzen, 1981; Müller and Hilgenberg, 1986), the increase of indole-3-acetic acid (IAA) might be due to the increased synthesis and turnover of the putative host auxin precursors indole-3-acetaldoxime, indole-3-methylglucosinolate, and indole-3-acetonitrile in infected roots (Rausch et al., 1981; Searle et al., 1982; Butcher et al., 1984). Several pathways for the biosynthesis of IAA in plants have been discussed (Normanly et al., 1995). It is generally believed that in Brassicaceae the pathway involves the formation of indole-3-acetaldoxime and indole-3-acetonitrile (IAN) from Trp as a precursor (Ludwig-Müller and Hilgenberg, 1988, 1990, 1992). The turnover of indole glucosinolates may provide additional IAN (Butcher et al., 1974), in particular after tissue disruption during infection with a pathogen or after wounding. However, a pathway for the biosynthesis of IAA without involving Trp has been discovered, where not only IAA, but also high amounts of IAN have been detected (Normanly et al., 1993).
Nitrilase is discussed to play a key role for several biosynthetic pathways in Brassicaceae, because it catalyzes the last step in the Trp-dependent and probably also the Trp-independent IAA biosynthesis pathway in this plant family (Normanly et al., 1993). An increased in vivo turnover of radiolabeled IAN was correlated with a 2- to 3-fold increased extractable nitrilase activity in Brassica napus (Rausch et al., 1981, 1983). Recently, several plant nitrilase isoforms have been cloned from Arabidopsis (Bartling et al., 1992, 1994; Bartel and Fink, 1994). In particular, it was shown that a family of four closely related nitrilase isoforms, NIT1 to NIT4, were differentially expressed during plant development (Bartel and Fink, 1994). One isoform, NIT2, was specifically induced after infection with the leaf pathogen Pseudomonas syringae pv. maculicola. This result prompted us to re-address the question of whether nitrilase is induced in infected roots of club root diseased plants.
We infected Arabidopsis with P. brassicae and investigated nitrilase expression in healthy and infected roots. Recently, transgenic Arabidopsis plants carrying one of the four NIT genes in either the sense or the antisense direction were characterized to elucidate the putative role of nitrilase for IAA biosynthesis. Overexpression of nitrilase caused no changes in the phenotype of the transgenic plants, most likely due to the fact that IAN is the limiting factor in planta (Normanly et al., 1997; Grsic et al., 1998). One transgenic line overexpressing nitrilase (35SNIT2) and a mutant in the NIT1 gene (nit1-3) were used to investigate the role of nitrilase for clubroot development in vivo.
Using antibodies against nitrilase (Grsic et al., 1998) for immunolocalization, we also examined the earliest time point for nitrilase occurrence in infected cells and show that the protein is predominantly induced in cells harboring sporulating plasmodia.
MATERIALS AND METHODS
Plant Material and Infection Procedure
Arabidopsis ecotypes Nossen (No-0), Columbia (Col), Enkheim (En), Wassilewskija (WS), and Landsberg erecta (Ler), nitrilase-overexpressing line 35SNIT2, promoter-β-glucuronidase (GUS) lines D4E (35S::uidA), and NIT1-4::uidA, and nit1-3 mutants were grown on a mixture of compost:peat:sand (3:2:1) at 23°C, 60% humidity, and a light-dark regime of 16 h/8 h (28 μmol m−2 s−1) (fluorescent lights TL55 daylight and TL32 Warmton de Luxe, Philips, Eindhoven, The Netherlands). After 10 d of growth the seedlings were inoculated with a resting spore suspension of a field isolate of Plasmodiophora brassicae. To each seedling 500 μL of spore suspension (107–108 spores mL−1) were pipetted and the plants were further cultivated at 23°C, 60% humidity, and a light-dark regime of 16 h/8 h (28 μmol m−2 s−1). Roots of infected and control plants were harvested after the appropriate time period, cleaned from the soil, and used for further experiments.
Preparation of Total RNA, cDNA, and Genomic DNA
Isolation of total RNA followed the protocol of Logemann et al. (1987). First-strand cDNA was prepared from total RNA using AMV reverse transcriptase. Extraction of genomic DNA from plant material was performed according to the method of Murray and Thompson (1980). Resting spores from P. brassicae were prepared as follows for the isolation of genomic DNA. Spores were isolated from 100 g of infected Chinese cabbage root material that was homogenized in 0.1% (w/v) aq. NaCl for 5 min in a Waring blender at room temperature (neoLab, Heidelberg). The homogenate was filtered and the filtrate pressed through a 80-μm filter. The filtrate was centrifuged for 5 min at 30g, the supernatant filtered through a 20-μm filter, and the filtrate again centrifuged for 15 min at 450g. The pellet was resuspended in 1 mL of DNase buffer and incubated for 1 h at 37°C with 10 units of DNaseI. After 1 h, the reaction was stopped by the addition of 10 mg of proteinase K and further incubated at 37°C for 2 h. Following incubation, the spores were centrifuged for 10 min at 20,000g and the pellet was used for DNA extraction from the resting spores. The DNase/proteinase-treated spores were homogenized in a mortar with sea sand and liquid nitrogen to a very fine powder. The powder was then used for the extraction of genomic DNA as described for the plant material.
DNA and RNA Gel-Blot Analysis
Non-radioactive DNA and RNA gel-blot analysis was performed with a biotinylated (bio-dUTP, Boehringer Mannheim/Roche, Basel) cDNA probe against NIT1 (template was cDNA prepared from total RNA of Arabidopsis leaves) prepared by PCR according to the method of Bischoff et al. (1995). Genomic DNA was digested with EcoRI, BamHI, and HindIII (Bischoff et al., 1995) and the fragments were separated on a 0.8% (w/v) agarose gel. Hybridization was performed for 16 h at 37°C and a high-stringency wash was at 48°C. Signals were detected with the Southern Light kit from Tropix (Serva, Heidelberg) for detection (Bischoff et al., 1995).
Non-radioactive northern blots were prepared according to the method of Löw and Rausch (1994). Hybridization with the biotinylated cDNA probe against NIT1 from Arabidopsis was for 16 h at 42°C, and a high-stringency wash was perfomed at 60°C. Signal development was performed by using horseradish peroxidase coupled to streptavidine and subsequent incubation with Luminol (Pierce Chemical, Rockford, IL) as a substrate according to the manufacturer's instructions. The blot was exposed for 3 min, 2 h, and overnight with x-ray film. Equal sample loading (20 μg of total RNA) was confirmed by hybridization with an 18S rRNA probe.
Western Blotting
Total proteins were isolated as described elsewhere (Zhigang et al., 1996). Denaturing SDS gel electrophoresis was carried out according to the method of Laemmli (1970) on a 12% (w/v) polyacrylamide gel. The lane with standard proteins was cut off and stained with amido black. The proteins were blotted for 1 h at 0.3 mA cm−2 onto polyvinylidene difluoride membranes (Immobilon P, Millipore, Bedford, MA) using a continuous blotting system (Grsic et al., 1998). Nitrilase was visualized on the blots using antibodies against NIT1 (Grsic et al., 1998). The primary antibodies were incubated with the membrane overnight. As a second antibody, anti-rabbit IgG coupled to horseradish peroxidase (Pierce) was used. Signal development was by incubation with Luminol (Pierce) as a substrate according to the manufacturer's instructions and subsequent exposure of the blot for 5 to 30 min with an x-ray film. Equal sample loading (100 μg of protein) was confirmed by staining the blot after development with amido black.
Immunolocalization
At different time points after inoculation, roots were washed and all soil residues were removed. The roots were fixed for 2 h in 2-(N-morpholino)-ethanesulfonic acid (MES) buffer (50 mm MES, 50 mm KCl, 2 mm MgCl2, 10 mm EGTA, and 1 mm EDTA, pH 6.7) supplemented with 4% (v/v) formaldehyde, 5% (v/v) Triton X-100, and 4% (w/v) polyethyleneglycol 8000. Afterward they were washed with MES buffer, dehydrated by an ethanol series, and embedded in methacrylate according to the method of Baskin et al. (1992). Tissue sections of 2 to 4 μm were produced by a Minot microtome (Leitz, Wetzlar, Germany) and stained according to the method of Baskin et al. (1992).
The polyclonal antiserum against nitrilase (Grsic et al., 1998), the antiserum against the Rubisco large subunit (Winter and Feierabend, 1990), and the antiserum against GUS (Molecular Probes Europe, Leiden, The Netherlands) were used as the primary antibodies, and immunosignals were obtained by fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Sigma, St.Louis). The slides were then counterstained with 2 μg mL−1 diamino-2-phenylindole (DAPI, Sigma) in phosphate-buffered saline (PBS) (pH 7.4) for 10 min, followed by a second counterstaining with ethidium bromide (10 μg mL−1) in PBS (pH 7.4) for 1 min. The preparations were analyzed with a fluorescent microscope (Orthoplan, Leitz) using different epifluorescence optics: (a) for green FITC fluorescence signals (excitation filter, 450–490 nm; dichroic mirror, 510 nm; and barrier filter, 515 nm), (b) for DAPI or propidium iodide staining (excitation filter, 340–380 nm; dichroic mirror, 400 nm; and barrier filter, 430 nm), and (c) for ethidium bromide and propidium iodide staining (excitation filter, 530–560 nm; dichroic mirror, 580 nm; and barrier filter, 580 nm). As controls, the same procedure was always performed (a) omitting the first antibody, (b) without antibody, and (c) heat-inactivated primary antibodies. For several slides in which strong signals were observed immunochemical staining after an incubation in proteinase K solution (20 μg mL−1 in PBS, pH 7.4, for 20 min) and subsequently strong washing in PBS (five times) was performed.
GUS Staining
GUS activity was determined according to the method of Jefferson (1987). Roots of control and infected plants harvested at different time points after inoculation were homogenized in 50 mmol/L Na-phosphate buffer, pH 7.4, containing 10 mmol/L NaEDTA, 0.1% (v/v) Triton X-100, 0.1% (w/v) N-lauryl-sarcosin, and 10 mmol/L dithiothreitol (DTT), and centrifuged in eppendorf tubes at 12,000g. One to 5 μL of the supernatant were mixed with 0.5 mL of reaction buffer consisting of 1 mmol/L 4-methylumbelliferyl-β-d-glucuronide and incubated at 37°C. After various time intervals (1, 5, 10, 30, and 60 min), 100 μL was taken from the assay, mixed with 1.9 mL of stop buffer (0.2 mol/L Na2CO3), and activity measured with a fluorometer (Jasco, Gross-Umstadt, Germany) with excitation at 365 nm and emission at 455 nm. Total protein content in the samples was determined with Bradford reagent (Bio-Rad Laboratories, Hercules, CA). Values are means ± se from different sets of plants of the same experiment. From each extract three determinations were performed.
Histochemical GUS staining was performed by incubating control and P. brassicae infected plants with their roots in a solution containing 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide dissolved in 0.1 m phosphate buffer, pH 7.4, containing 10 mm Na2EDTA, 0.5 mm K3(Fe[CN]6), 0.5 mm K4(Fe[CN]6), and 0.5% (w/v) Triton X-100. The incubation was stopped by placing the roots in phosphate buffer, pH 7.4, without substrate. Control roots were directly assayed by light microscopy, whereas hand sections through clubs from infected roots were performed prior to microscopic analysis.
Determination of Free IAA
Infected and control roots were harvested 5 weeks after inoculation, washed thoroughly, and dried on filter paper. Homogenization was performed in liquid nitrogen and the pulverized material was resuspended in 35% (v/v) 200 mm imidazol buffer, pH 7.0/65% (v/v) i-propanol and homogenized again. Further purification for analysis of free IAA was performed as described by Chen et al. (1988), modified according to the method of Ilic′ et al. (1996). Analysis of free IAA was performed by HPLC (Eppendorf Biotronik BT 8100, Hamburg, Germany), equipped with a 4.6 × 125 mm Lichrosorb C18, 5-μ, reverse phase column. The solvents (1% [v/v] acetic acid in water [A] and methanol [B]) were administered as a linear gradient from 30% to 60% B (0–20 min), and elution was continued for an additional 10 min with 60% B. Flow rate was 0.7 mL min−1 and detection was performed at 280 nm by co-chromatography with an authentic standard. The retention time (Rt) of IAA was 14.3 min. Values are means ± se from three independent experiments. Similar values for free IAA in Arabidopsis roots were found using gas chromatography/selected ion monitoring/mass spectrometry analysis by Ludwig-Müller et al. (1999).
RESULTS
Expression Analysis of Nitrilase in Wild-Type Arabidopsis Plants during the Development of the Clubroot Disease
Since it was shown that nitrilase activity was enhanced in Chinese cabbage during clubroot disease (Rausch et al., 1981; Grsic et al., 1999), we have investigated nitrilase expression during clubroot disease in Arabidopsis. The availability of Arabidopsis plants transformed with a promoter-GUS construct for each nitrilase isoform so far isolated from this plant (Bartel and Fink, 1994) enabled the investigation of which nitrilase isoforms are responsible for the increase in clubs compared with healthy roots. A histochemical GUS staining was performed using sections of healthy and infected roots of the respective GUS line. In control roots, only occasional GUS staining was visible in root tips of NIT1::uidA and NIT2::uidA plants (data not shown). In infected roots showing the typical clubroot symptoms, strong GUS staining was observed only in NIT1::uidA and NIT2::uidA plants. GUS activity was predominantly found in cells containing large secondary sporulating plasmodia of P. brassicae (data not shown).
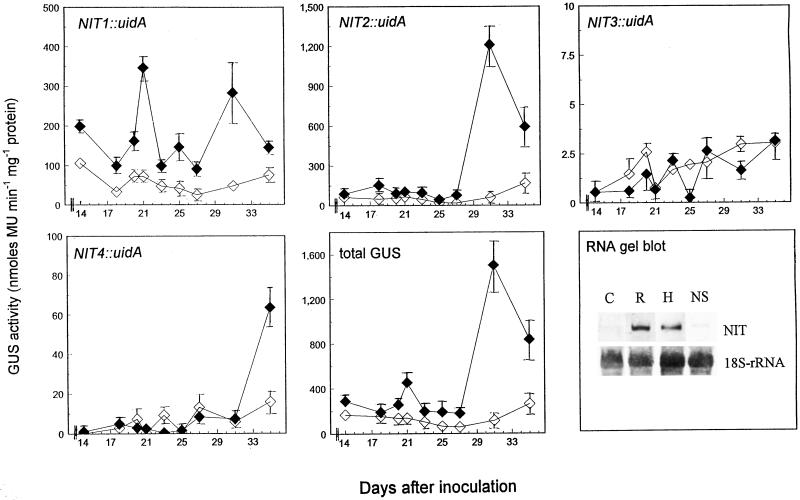
GUS activity was quantitatively measured in control and P. brassicae infected roots over a time course between 14 and 42 d post inoculation (dpi) using methylumbelliferyl-β-d-glucuronide as a substrate. It was shown that two distinct peaks of GUS activity were present throughout the development of root galls (Fig. 1). The earlier (21 dpi), smaller peak was derived from the activity of the NIT1 promoter, whereas the second, larger increase in GUS activity consisted mostly of NIT2 expression. NIT3- and NIT4-GUS-promoter fusion lines did not possess significantly higher GUS activity than the controls.
Figure 1.
Expression of different nitrilase genes during clubroot development in Arabidopsis. Control (⋄) and infected (♦) roots of transgenic Arabidopsis plants harboring the uidA gene under the control of NIT1-4 promoters were analyzed at different time points (14–35 dpi) for GUS activity. Labeling of the y-axes (GUS activity) is meant for all panels except RNA gel blot. Values are means ± se from different plants of the same experiment. Total GUS activity was calculated by addition of the activities obtained for NIT1-4::uidA plants. At 32 dpi a RNA gel-blot analysis was conducted with control roots (C), separately harvested root (R), and hypocotyl (H) clubs, as well as roots from infested soil showing no symptoms (NS). Nitrilase was detected with a NIT1-cDNA probe. Equal sample loading was confirmed by hybridizing the same blot with an 18S-rRNA probe. MU, Methylumbelliferone.
These results were confirmed by RNA gel-blot analysis of infected and control roots 32 dpi using a NIT1-cDNA probe (Fig. 1). It was shown that the increase in nitrilase mRNA was present both in hypocotyl and root clubs, two different types of infection seen during clubroot development of Arabidopsis (Ludwig-Müller, 1999). The increase in nitrilase mRNA expression was only visible in plants showing the macroscopically visible disease symptoms, but not in plants without symptoms harvested from infested soil.
Nitrilase Is Confined to Infected Root Cortex Cells during Resting Spore Formation
A polyclonal antibody raised against NIT1 from Arabidopsis overexpressed in Escherichia coli (Grsic et al., 1998) was used for detection of nitrilase in infected roots at different time points after inoculation with P. brassicae. The antiserum is specific for a 38-kD polypeptide from Arabidopsis (Grsic et al., 1998) and can detect at least NIT1 and NIT2 (both showed the same apparent molecular mass of approximately 38 kD) as shown by overexpression of these isoforms in E. coli (NIT1) and Arabidopsis (NIT2) (Grsic et al., 1998). In 32-dpi roots the pattern of protein expression follows the pattern observed in RNA gel-blot analysis and measurement of GUS activity of transgenic plants (increased protein level in infected roots compared with controls; data not shown). However, an increased amount of nitrilase protein was detectable in 42-dpi infected roots, where no differences in nitrilase mRNA between infected and control roots were visible. These results indicate that the nitrilase protein is very stable in the tissue.
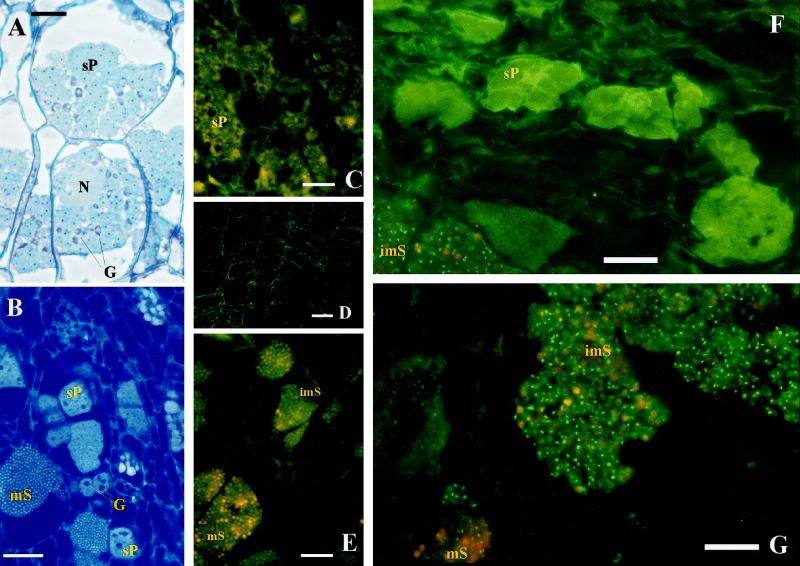
Since we have found that nitrilase mRNA and protein was enhanced in clubs compared with controls (Fig. 1), the question was raised as to whether nitrilase is expressed only in certain cells and therefore an increase is not detectable at earlier stages of development. A typical cortical root infection shows secondary plasmodia (Fig. 2A), which are stained by toluidine blue and basic fuchsin, and also sporangia with resting spores (Fig. 2B), whose nuclei can be seen after staining with DAPI and ethidium bromide. The multinucleate plasmodia are also clearly visible using this staining technique. It should be noted that cortical root cells that contain plasmodia or resting spores are enlarged compared with uninfected cells.
Figure 2.
Immunolocalization of nitrilase in longitudinal sections of infected roots of Arabidopsis using a specific antibody against NIT1. Secondary plasmodia (sP), immature spores (imS), and mature (mS) resting spores of P. brassicae as well as nuclei (N) and starch granules (G) of the cortical host plant cells are labeled. Bars indicate 20 μm. A and B, Light microscopic pictures of large secondary plasmodia (23 dpi) stained with toluidine blue and basic fuchsin (A) and plasmodia and resting spores (29 dpi) stained with ethidium bromide and DAPI using epifluorescence optics specific for DAPI (B). Using different staining techniques, different parts of the host cell and the pathogen can be distinguished. In A the plasmodia and the enlarged host cells are clearly visible using toluidine blue and basic fuchsin staining, while in B the multinucleate plasmodia and the nuclei of the resting spores are stained with DAPI and ethidium bromide. Immunosignals of nitrilase in infected cells at 21 dpi (F) and 35 dpi (G) using FITC-specific epifluorescence optics, and the corresponding controls only using the secondary antibody for staining at 21 dpi (C) and 35 dpi (E). Immunolocalization of nitrilase in non-infected roots of a nitrilase-overproducing line (35SNIT2) 28 d after germination is also shown (D).
The antibody against NIT1 was used for immunolocalization of this protein during clubroot development in Arabidopsis. In uninfected roots almost no immunosignal could be detected. To demonstrate that the antibody recognizes nitrilase within the root cells, transgenic Arabidopsis plants that overexpress NIT2 constitutively and show a strong immunosignal in western-blot analysis with the anti-NIT1 antibody were used (Grsic et al., 1998). Within this line a prominent immunosignal was detected near the cell wall of uninfected roots (Fig. 2D), demonstrating the location of this isoform on the plasma membrane, as previously shown by Bartling et al. (1994).
In P. brassicae-infected roots, immunosignals at two different developmental stages of the pathogen could be detected using the NIT1 antibody. Weak signals associated with large secondary plasmodia were observed at 21 to 30 dpi (Fig. 2F; Table I). Strong signals associated with sporulating plasmodia (immature resting spores) could be detected after 29 dpi (Fig. 2G; Table I). The amount of the latter signal increased during club development parallel with this developmental stage of the pathogen, and peaked between 35 and 42 dpi (Table I), confirming the western analysis results (data not shown). During this phase of club development, plasmodia, developing sporangia, and resting spores are visible at the same time, as demonstrated in a cross-section of clubs 35 dpi (Fig. 2B). At later time points (45 dpi and older roots) the signal density decreased again. No signal was detected when (a) no antibody (data not shown), (b) only second antibody (Fig. 2, C and E), (c) heat-inactivated primary antibody (data not shown), or (d) antibody against Rubisco as the primary antibody (data not shown) were used.
Table I.
Summary of the microscopic analysis of 11- to 44-dpi plants
| Immunohistochemical Signal | Time
|
||||||
|---|---|---|---|---|---|---|---|
| 11 dpia | 21 dpib | 29 dpib | 35 dpic | 38 dpic | 42 dpic | 44 dpid | |
| Nitrilase in ecotype Nossen | − | + | + | +++ | ++++ | ++++ | ++ |
| GUS in line Nit1∷uidA | − | + | + | ++++ | NDe | ++ | ND |
| GUS in line Nit2∷uidA | − | − | + | ++++ | ND | ++ | ND |
The symbols indicate detectable (+) and not detectable (−) specific FITC signals. The number of signs are a semiquantitative indicator of the estimated amount of signals within the slides at this stage.
Secondary plasmodia could be detected in few cells.
Secondary plasmodia of different developmental stage could be observed. Metaphase plates of pathogen could be viewed. The formation of spores could be rarely detected.
Secondary plasmodia of all different developmental stages could be observed. The spore maturation has been finished in some cells whereas in other cells it is still in progress.
Most cells are overcrowded with mature spores. Younger plasmodia could be rarely observed.
ND, Not determined.
After proteinase K treatment, the signals also vanished (data not shown), although in samples with a high density of immunosignals an incubation time of 2 h was necessary to eliminate the signals completely. Interestingly, it was shown that nitrilase was located mainly in cells where sporulating plasmodia were present (Table I; Fig. 2, F and G). Cells containing large secondary plasmodia (Fig. 2F) and mature resting spores (mS; Fig. 2G) showed only weak nitrilase immunosignals. Thus, it could be demonstrated that nitrilase expression during clubroot is confined to certain stages of pathogen development, where cell enlargement occurs, as detected by light microscopy (Fig. 2A). In healthy cells, only occasionally was a single membrane-bound signal observed.
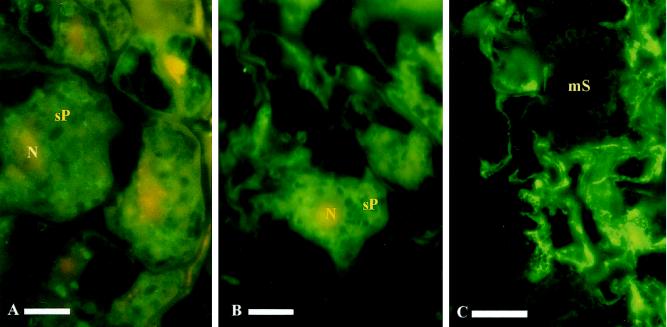
Using transgenic lines carrying the nitrilase-promotor uidA-gene fusion constructs, an antibody against GUS was used to localize expression of GUS in infected roots. Studying the lines NIT1::uidA and NIT2::uidA a similar pattern as described for nitrilase was observed (Table I). In large secondary plasmodia, 21- and 29-dpi weak signals could be detected (Fig. 3, A and B). However, GUS-derived immunosignals associated with sporulating plasmodia decreased more rapidly than signals obtained with NIT1 antibody (Table I). Infected roots from a plant line constitutively expressing the uidA gene (D4E; 35S::uidA) showed no signals associated with the pathogen after incubation with a GUS antibody (Fig. 3C), thus demonstrating that the immunosignals obtained with anti-GUS-antibodies (Fig. 3, A and B) in the lines Nit1::uidA and Nit2::uidA are specific.
Figure 3.
Immunolocalization of GUS in longitudinal sections of infected roots of Arabidopsis using a monospecific antibody against GUS. Secondary plasmodia (sP) and mature resting spores (mS) of P. brassicae as well as nuclei (N) of the cortical plant host cells are marked. GUS signals in NIT1::uidA lines 21 dpi (A) and 29 dpi (B) and in infected roots of a GUS-overproducing line (35S::uidA) 28 dpi (C). Bars indicate 10 μm.
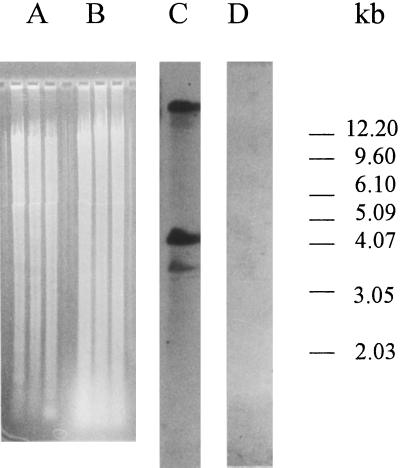
The immunosignal obtained with antibodies against nitrilase and GUS seemed to be associated with the immature resting spores. Therefore, we investigated whether a nitrilase sequence similar to the plant sequence is present in P. brassicae. Genomic DNA was isolated from Arabidopsis and resting spores of P. brassicae, and it was possible to amplify a approximately 600-bp fragment by PCR using the primers for nitrilase amplification published by Bischoff et al. (1995) from Arabidopsis DNA, but not from P. brassicae DNA (data not shown). After digestion of the genomic DNA with EcoRI, strong bands could be detected with Arabidopsis genomic DNA (Fig. 4, lane C), but not with P. brassicae genomic DNA (Fig. 4, lane D) using a homologous NIT1-cDNA probe. The same results were obtained after digestion with HindIII and BamHI (data not shown). In Figure 4, lanes A and B, the restricted genomic DNA of Arabidopsis and P. brassicae, respectively, are shown (from left BamHI, EcoRI, and HindIII digest). The blot was probed with moderate stringency (see “Materials and Methods”) to allow detection of putative P. brassicae nitrilase sequences with low homology to Arabidopsis nitrilase sequences. It is thus very likely that P. brassicae has no nitrilase sequence with homology to plant nitrilase. Therefore, the nitrilase signals detected with the anti-NIT1 antibody must be derived from the plant protein(s).
Figure 4.
Detection of nitrilase from genomic DNA of Arabidopsis and P. brassicae by DNA gel-blot analysis. Lanes A and B show the digested genomic DNA of Arabidopsis and P. brassicae, respectively, prior to blotting after separation on a 0.8% (w/v) agarose gel. From left to right: BamHI, EcoRI, and HindIII digest. Southern blot from genomic DNA of Arabidopsis (lane C) and P. brassicae (lane D) digested with EcoRI was performed with a NIT1-cDNA probe. No signals are visible in the lane with P. brassicae DNA.
The Role of Nitrilase for the Clubroot Disease in Vivo
Since only NIT1 and NIT2 were notably increased in Arabidopsis roots after infection with P. brassicae, only plants affected in either NIT1 or NIT2 were used to further study the role of nitrilase for clubroot development. The transgenic plants of Arabidopsis overexpressing one nitrilase isoform (35SNIT2) have been characterized elsewhere (Normanly et al., 1997; Grsic et al., 1998). They show no distinct phenotype, but a strong overexpression of mRNA, protein, and an increased enzyme activity (Grsic et al., 1998). Additionally, a mutant in the NIT1 gene (nit1-3) was used for this study. The nit1-3 mutant has a mutation that prematurely terminates the polypeptide (Normanly et al., 1997).
For the determination of fresh weight and free IAA in 35SNIT2 and nit1-3 plants, only roots from infested soil, which showed visible clubroot symptoms, were used. The clubs on nit1-3 plants were smaller than the clubs of wild-type and 35SNIT2 plants, as shown by determination of the ratio in fresh weight of control and infected roots (Table II). Determination of the infection rate showed that nit1-3 plants also had a reduced infection rate compared with the respective wild type. Overexpression of nitrilase did not result in larger clubs, as demonstrated by the ratio of the fresh weight of infected to control roots of 35SNIT2 plants (Table II).
Table II.
Role of nitrilase in vivo in the development of clubroot disease
| Feature | Arabidopsis Line
|
|||
|---|---|---|---|---|
| No-0 | 35SNIT2 | Col | nit1-3 | |
| Infection rate (%) | 70.3 ± 3.8 | 77.3 ± 6.5 | 70.0 ± 5.1 | 56.3 ± 3.4 |
| Ratio inf∶con roots 35 dpi | 8.36 ± 0.29 | 8.00 ± 0.22 | 5.28 ± 0.13 | 2.31 ± 0.05 |
| Free IAA control roots (ng g−1 fresh wt) | 106.3 ± 5.1 | 227.9 ± 40.0 | 150.9 ± 10.5 | 147.8 ± 40.1 |
| Free IAA infected roots (ng g−1 fresh wt) | 199.8 ± 20.8 | 376.9 ± 28.0 | 263.9 ± 2.0 | 161.1 ± 19.4 |
| Free IAA ratio inf∶con | 1.88 | 1.65 | 1.75 | 1.08 |
For the determination of the infection rate, at least 50 plants were analyzed per line and the experiments were performed three times. Values are means ± se from these three independent experiments. The ratio of infected (inf) to control (con) roots was calculated from the fresh weight of control and infected roots showing the typical clubroot symptoms. Roots from infested soil without symptoms were not included in the measurements. IAA analyses were performed three times with independently cultivated plants 35 dpi, and mean values ± se are given.
The free IAA concentration in P. brassicae-infected roots of wild-type and transgenic plants was determined 32 dpi in clubs and control roots of ecotype Nossen and 35SNIT2 and ecotype Columbia and nit1-3 plants (Table II). Healthy roots of 35SNIT2 plants showed a 2-fold increase in free IAA concentration compared with the wild type, whereas the nit1-3 mutant had the same IAA levels as wild-type roots. In infected roots the free IAA content was higher in 35SNIT2 plants compared with the controls, as it was also seen in both wild types, whereas the nit1-3 mutant showed similar auxin concentrations in infected roots compared with controls. The free IAA content in clubbed roots compared with controls was increased by about 180%, 165%, 175%, and 108% in ecotype Nossen, 35SNIT2, ecotype Columbia, and nit1-3 roots, respectively.
DISCUSSION
The development of clubroot disease symptoms is correlated with an increase of auxin (Ludwig-Müller et al., 1993, 1996) and cytokinin (Dekhuijzen and Overeem, 1971), thus resulting in increased cell division and cell elongation. The high IAA content was attributed to the conversion of indole glucosinolates to IAN by the enzyme myrosinase, which is compartmented against its substrate in healthy tissue, and further conversion by nitrilase to IAA (Butcher et al., 1974). Several components of IAA synthesis via the indole glucosinolate pathway are induced during clubroot in Chinese cabbage (Brassica rapa subsp. pekinensis). The activity of the enzyme, which converts Trp to indole-3-acetaldoxime, presumably the first step in indole glucosinolate biosynthesis (Ludwig-Müller and Hilgenberg, 1988), was enhanced in infected roots (Ludwig-Müller et al., 1997), presumably leading to the elevated glucosinolate levels found in clubs (Ludwig-Müller et al., 1993, 1997). The increase in myrosinase expression was also demonstrated in infected roots of Chinese cabbage (Grsic et al., 1999). The activity of nitrilase was enhanced in hypertrophied roots of Brassica napus (substrate 3-cyanopyridine; Rausch et al., 1981) and B. rapa (substrate IAN; Grsic et al., 1999), and in vivo studies showed elevated IAN levels in infected roots after feeding of labeled Trp (Rausch et al., 1983).
The expression of nitrilase during clubroot disease in Arabidopsis was investigated by using transgenic plants carrying promoter-reporter gene fusions for each nitrilase isoform, as well as by RNA gel-blot analysis during symptom development. While no differences between nitrilase expression in younger infected and control roots were found in RNA gel-blot analysis (data not shown), in 32 dpi roots, corresponding to the exponential growth phase of clubs, the expression of nitrilase was prominently enhanced in infected roots (Fig. 1). This has been confirmed by western-blot analysis using polyclonal antibodies against the 38-kD protein from Arabidopsis. At later stages no differences were visible on the mRNA level, but the amount of nitrilase protein was still higher, indicating that it is more stable than nitrilase mRNA (data not shown). In a previous report it was shown that nitrilase expression was not enhanced in Chinese cabbage clubs compared with controls (Bischoff et al., 1995), although nitrilase activity was enhanced in clubs compared with healthy roots (Grsic et al., 1999). One explanation of these differences between Chinese cabbage and Arabidopsis might be that the ratio of infected cells containing nitrilase to non-invaded cells in Chinese cabbage is smaller than in Arabidopsis, and, therefore, no differences in NIT mRNA are visible. Alternatively, in Chinese cabbage a novel nitrilase isoform is induced during clubroot that does not cross-hybridize to NIT1 of Arabidopsis.
These results, however, do not show which nitrilase isoforms are involved in symptom development. Arabidopsis plants transformed with promoter-GUS-fusions for each NIT gene indicated expression of NIT1 and NIT2 in control roots, which were both enhanced in infected roots (Fig. 1). Two GUS activity peaks were observed, one at 21 dpi and a second one at 32 dpi. While the first peak was only the result of NIT1 expression, the second peak was predominantly due to expression of NIT2. The increase in NIT expression 21 dpi can be correlated with the growth of plasmodia, the increase 32 dpi with sporulating plasmodia and development of sporangia (data not shown). Histochemical GUS staining in infected roots of NIT1::uidA and NIT2::uidA plants 32 dpi showed a correlation between GUS expression and sporangia-containing cells (data not shown), confirming the results of the immunolocalization of nitrilase protein. Our results support the data on increased NIT2 expression after pathogen infiltration in leaves (Bartel and Fink, 1994) and point to an additional role for NIT1 in the development of the hypertrophy.
Althought we made every effort to synchronize the different experiments (measurement of GUS activity, determination of GUS expression via antibodies, and RNA-blot analysis), slight variations in temperature and light in the greenhouse may have led to differences in the velocity of club development, which may account for the differences observed for expression of NIT1::uidA in two different experiments. While GUS activity was measured every 3rd d, thus making it possible to detect variations within few days, the experiments for immunolocalization were carried out at prominent time points during club development and may thus have missed a peak in NIT1 expression (see Fig. 1 versus Table I).
Rausch et al. (1983) proposed that only very small amounts of auxins are needed for club formation, and, therefore, enhancement of auxins in a few cells would be sufficient to induce symptom development. Therefore, the cellular localization of nitrilase in clubs using polyclonal antibodies against nitrilase was investigated. Only occasional nitrilase signals occurred in control roots and non-invaded cells of clubs (data not shown), but at certain stages of P. brassicae development, high amounts of nitrilase signals were observed, which correlated tightly with sporulating plasmodia. Only weak signals were found in cells harboring secondary plasmodia, and no signals in cells with mature resting spores. A time course study showed that cells containing strong nitrilase signals were first visible 29 dpi, with the number of these cells peaking 35 to 42 dpi and then declining again. This confirms the western analysis results, which showed increased nitrilase protein 35 and 42 dpi, although NIT mRNA was only increased 32 dpi and then declined again.
Since the nitrilase signals in immunolocalization were always associated with developing spores, we investigated the possibility that P. brassicae might contain a nitrilase similar to plant nitrilase. Several lines of evidence suggest that the nitrilase detected during club development is of plant and not fungal origin. First, Southern analysis with a plant-specific NIT1 cDNA probe (Bischoff et al., 1995) revealed that no nitrilase sequence similar to plant nitrilase is present in P. brassicae (Fig. 4). Similarities between NIT1 and the other nitrilase genes NIT2, NIT3, and NIT4 sequences were 95%, 87%, and 69%, respectively, according to sequence alignment on the nucleotide level. The NIT1 cDNA probe used in this study clearly cross-hybridizes with NIT1-3, but not very strongly with NIT4 (N. Kasperczyk and J. Ludwig-Müller, unpublished results). Therefore, it is highly unlikely that an antibody against the plant protein derived from this DNA would recognize a distantly related putative nitrilase from P. brassicae. However, it may be still formally possible that a nitrilase with different primary amino acid sequence could be structurally similar enough to cross-react with the NIT1 antibody. Since transgenic Arabidopsis plants overexpressing NIT4 (the least homologous Arabidopsis nitrilase) do not show an increased signal with the NIT1 antibody compared with wild type (S. Grsic-Rausch and J. Ludwig-Müller, unpublished results), this is highly unlikely. Second, isolated resting spores from P. brassicae did not show immunosignals with the NIT1 antibody. Third, in NIT::uidA lines, the immunosignal obtained with an antibody against GUS is located in the same cells as is the signal obtained with the NIT1 antibody. Measurement of GUS activity in clubs of untransformed plants have shown that P. brassicae does not possess GUS activity; therefore, the immunosignal obtained with the GUS antibodies must also derive from the plant promoter.
It was hypothesized that the increase in IAA is derived from the decompartmentalization of host cells by P. brassicae (Butcher et al., 1974), and that, subsequently, indole glucosinolates stored in the vacuole (Helmlinger et al., 1983) can be converted by myrosinase to IAN, which is then converted to IAA by nitrilase. As can be seen in Figure 2, the host cells are still intact during the development of secondary plasmodia, whereas sporangia-containing cells are enlarged compared with uninfected cells, and the compartmentation is disrupted. This correlates with an increase in nitrilase signals. At later stages of sporangia development, the host cell might be completely destroyed, so that no protein synthesis can take place, which is reflected in reduced nitrilase signals. Since nitrilase can be induced by its own substrate (Allard, 1995; Grsic et al., 1998), one possible regulation mechanism for increased nitrilase expression could be the increased amounts of IAN synthesized by myrosinase from indole glucosinolates. However, other regulatory mechanisms cannot be ruled out.
It was also interesting to investigate the effect of P. brassicae infection on plants missing one nitrilase isoform to determine the role of this enzyme for club development and to see whether one isoform can substitute for another. We therefore used transgenic plants that overexpress one nitrilase isoform (35SNIT2; Normanly et al., 1997; Grsic et al., 1998) and the nit1-3 mutant (Normanly et al., 1997) to address this question. Interestingly, it was found in this study that roots of healthy 35SNIT2 plants showed a 2-fold increase in free IAA content compared with wild type. This is in contrast to previous findings from our laboratory (Grsic et al., 1998) and from investigations of Normanly et al. (1997), where no differences in free IAA were found between 35SNIT2 plants and wild type. However, in both investigations other plant parts have been used (5-d-old seedlings and green parts of mature plants; Grsic et al., 1998; 7-d-old seedlings; Normanly et al., 1997). Overexpression of nitrilase does not result in more severe symptoms because nitrilase activity was already induced above a sufficient level. However, the club size and infection rate were reduced in nit1-3 mutants, showing that loss of one NIT gene results in a less severe phenotype of club formation, although it was previously shown that one NIT isoform can substitute for another (Normanly et al., 1997).
In conclusion, our results have provided further evidence of a role for nitrilase in IAA biosynthesis during clubroot disease, because: (a) symptom development is negatively influenced in Arabidopsis nitrilase mutant plants (nit1-3), resulting in a less severe phenotype of clubroot and a reduced infection rate; (b) nitrilase expression is increased in clubbed roots approximately 32 dpi, as shown by northern analysis; nitrilase protein was also enhanced at a later time point, as shown by western analysis and immunolocalization; (c) nitrilase was specifically located in infected cells harboring sporulating plasmodia. In cells containing secondary plasmodia, only weak signals were observed, and in cells containing older sporangia, no nitrilase signals were observed, indicating a role of nitrilase in cell elongation during sporulation of the pathogen. The mechanism that leads to the induction of nitrilase during clubroot formation (signals from the pathogen or from the host plant) has yet to be investigated.
ACKNOWLEDGMENTS
The transgenic lines (35SNIT2 and NIT-GUS-fusions) and nit1-3 mutant were a generous gift from Dr. Bonnie Bartel, Rice University, Houston, TX. The transgenic line 35S::uidA was kindly supplied by Prof. Wolf Frommer, University of Tübingen, Germany. The anti-Rubisco antibody was a gift from Prof. J. Feierabend, Botanisches Institut, J.W. Goethe-Universität, Frankfurt. We would like to thank Kerstin Pieper, Birgit Schubert, and Annette Nöh for technical assistance.
Footnotes
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (grant no. Lu 500/2–2 to J.L.-M.).
LITERATURE CITED
- Allard R. The functional analysis of plant nitrilases in transgenic plants. PhD thesis. Germany: Göttingen; 1995. . ISBN 3–89588–172–4. [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Mithöfer A, Weiler EW. Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone indole-3-acetic acid. Eur J Biochem. 1992;205:417–424. doi: 10.1111/j.1432-1033.1992.tb16795.x. [DOI] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler EW. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc Natl Acad Sci USA. 1994;91:6021–6025. doi: 10.1073/pnas.91.13.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Busby CH, Fowke LC, Sammut M, Gabler F. Improvements in immunostaining samples embedded in methacrylate: localisation of microtubules and other antigens throughout developing plants of diverse taxa. Planta. 1992;187:405–413. doi: 10.1007/BF00195665. [DOI] [PubMed] [Google Scholar]
- Bischoff M, Löw R, Grsic S, Rausch T, Hilgenberg W, Ludwig-Müller J. Infection with the obligate biotroph Plasmodiophora brassicae, the causal agent of the clubroot disease, does not affect expression of NIT1/2-related nitrilases in roots of Chinese cabbage. J Plant Physiol. 1995;147:341–345. [Google Scholar]
- Butcher DN, Chamberlain K, Rausch T, Searle LM. Biochemical Aspects of Synthetic and Naturally Occurring Plant Growth Regulators. British Plant Growth Regulator Group, Monograph 11: 91–101. 1984. Changes in indole metabolism during the development of clubroot symptoms in Brassica. [Google Scholar]
- Butcher DN, El-Tigani S, Ingram DS. The role of indole glucosinolates in the clubroot disease of the Cruciferae. Physiol Plant Pathol. 1974;4:127–141. [Google Scholar]
- Chen K-H, Miller AN, Patterson GW, Cohen JD. A rapid and simple procedure for purification of indole-3-acetic acid prior to GC-SIM-MS analysis. Plant Physiol. 1988;86:822–825. doi: 10.1104/pp.86.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekhuijzen HM. The occurrence of free and bound cytokinins in plasmodia of Plasmodiophora brassicae isolated from tissue cultures of clubroots. Plant Cell Rep. 1981;1:18–20. doi: 10.1007/BF00267649. [DOI] [PubMed] [Google Scholar]
- Dekhuijzen HM, Overeem JC. The role of cytokinins in clubroot formation. Physiol Plant Pathol. 1971;1:151–161. [Google Scholar]
- Grsic S, Kirchheim B, Pieper K, Fritsch M, Hilgenberg W, Ludwig-Müller J. Auxin biosynthesis in clubroot diseased Chinese cabbage plants and induction by jasmonic acid. Physiol Plant. 1999;105:521–531. [Google Scholar]
- Grsic S, Sauerteig S, Neuhaus K, Albrecht M, Rossiter J, Ludwig-Müller J. Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in sense or antisense direction. J Plant Physiol. 1998;153:446–456. [Google Scholar]
- Helmlinger J, Rausch T, Hilgenberg W. Localization of newly synthesized indole-3-methylglucosinolate (=glucobrassicin) in vacuoles from horseradish (Armoracia rusticana) Physiol Plant. 1983;58:302–310. [Google Scholar]
- Ilic′ N, Normanly J, Cohen JD. Quantification of free plus conjugated indoleacetic acid in Arabidopsis requires correction for nonenzymatic conversion of indolic nitriles. Plant Physiol. 1996;111:781–788. doi: 10.1104/pp.111.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DS, Tommerup IC. The life history of Plasmodiophora brassicae Woron. Proc R Soc Lond B. 1972;180:103–112. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Löw R, Rausch T. Sensitive nonradioactive northern blots using alkaline transfer of total RNA and PCR-amplified biotinylated probes. Biotechniques. 1994;17:1026–1030. [PubMed] [Google Scholar]
- Ludwig-Müller J. Plasmodiophora brassicae, the causal agent of clubroot disease: a review on molecular and biochemical events in pathogenesis. Z Pflanzenkr Pflanzenschutz. 1999;106:109–127. [Google Scholar]
- Ludwig-Müller J, Bendel U, Thermann P, Ruppel M, Epstein E, Hilgenberg W. Concentrations of indole-3-acetic acid in plants of tolerant and susceptible varieties of Chinese cabbage infected with Plasmodiophora brassicae Woron. New Phytol. 1993;125:763–769. doi: 10.1111/j.1469-8137.1993.tb03926.x. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Epstein E, Hilgenberg W. Auxin-conjugate hydrolysis in Chinese cabbage: characterization of an amidohydrolase and its role during the clubroot disease. Physiol Plant. 1996;97:627–634. [Google Scholar]
- Ludwig-Müller J, Hilgenberg W. A plasma membrane-bound enzyme oxidizes l-tryptophan to indole-3-acetaldoxime. Physiol Plant. 1988;74:240–250. [Google Scholar]
- Ludwig-Müller J, Hilgenberg W. Conversion of indole-3-acetaldoxime to indole-3-acetonitrile by plasma membranes from Chinese cabbage. Physiol Plant. 1990;79:311–318. [Google Scholar]
- Ludwig-Müller J, Hilgenberg W. Tryptophan oxidizing enzyme and basic peroxidase isoenzymes in Arabidopsis thaliana (L.) Heynh.: are they identical? Plant Cell Physiol. 1992;33:1115–1125. [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana L. glucosinolate mutants and the development of the clubroot disease. Planta. 1999;208:409–419. doi: 10.1007/s004250050576. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Schubert B, Pieper K, Ihmig S, Hilgenberg W. Glucosinolate content in susceptible and tolerant Chinese cabbage varieties during the development of the clubroot disease. Phytochemistry. 1997;44:407–414. [Google Scholar]
- Müller P, Hilgenberg W. Isomers of zeatin and zeatin riboside in clubroot tissue: evidence for trans-zeatin biosynthesis by Plasmodiophora brassicae. Physiol Plant. 1986;66:245–250. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA. 1993;90:10355–10374. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Slovin JP, Cohen JD. Rethinking auxin biosynthesis and metabolism. Plant Physiol. 1995;107:323–329. doi: 10.1104/pp.107.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Butcher DN, Hilgenberg W. Nitrilase activity in clubroot diseased plants. Physiol Plant. 1981;52:467–470. [Google Scholar]
- Rausch T, Butcher DN, Hilgenberg W. Indole-3-methylglucosinolate biosynthesis and metabolism in clubroot diseased plants. Physiol Plant. 1983;58:93–100. [Google Scholar]
- Searle LM, Chamberlain K, Rausch T, Butcher DN. The conversion of 3-indolemethylglucosinolate to 3-indoleacetonitrile by myrosinase and its relevance to the clubroot disease of the Cruciferae. J Exp Bot. 1982;33:935–942. [Google Scholar]
- Winter U, Feierabend J. Multiple coordinate controls contribute to a balanced expression of ribulose-1,5-bisphosphate carboxylase/oxygenase subunits in rye leaves. Eur J Biochem. 1990;187:445–453. doi: 10.1111/j.1432-1033.1990.tb15324.x. [DOI] [PubMed] [Google Scholar]
- Zhigang A, Löw R, Rausch T, Lüttge U, Ratajczak R. The 32 kDa tonoplast polypeptide Di associated with the V-type H+-ATPase of Mesembryanthemum crystallinum L. in the CAM state: a proteolytically processed subunit B? FEBS Lett. 1996;389:314–318. doi: 10.1016/0014-5793(96)00556-x. [DOI] [PubMed] [Google Scholar]