Abstract
Chromatin remodelling precedes transcriptional and structural changes in heart failure. A body of work suggests roles for the developmental Wnt signalling pathway in cardiac remodelling. Hitherto, there is no evidence supporting a direct role of Wnt nuclear components in regulating chromatin landscapes in this process. We show that transcriptionally active, nuclear, phosphorylated(p)Ser675-β-catenin and TCF7L2 are upregulated in diseased murine and human cardiac ventricles. We report that inducible cardiomyocytes (CM)-specific pSer675-β-catenin accumulation mimics the disease situation by triggering TCF7L2 expression. This enhances active chromatin, characterized by increased H3K27ac and TCF7L2 occupancies to cardiac developmental and remodelling genes in vivo. Accordingly, transcriptomic analysis of β-catenin stabilized hearts shows a strong recapitulation of cardiac developmental processes like cell cycling and cytoskeletal remodelling. Mechanistically, TCF7L2 co-occupies distal genomic regions with cardiac transcription factors NKX2–5 and GATA4 in stabilized-β-catenin hearts. Validation assays revealed a previously unrecognized function of GATA4 as a cardiac repressor of the TCF7L2/β-catenin complex in vivo, thereby defining a transcriptional switch controlling disease progression. Conversely, preventing β-catenin activation post-pressure-overload results in a downregulation of these novel TCF7L2-targets and rescues cardiac function. Thus, we present a novel role for TCF7L2/β-catenin in CMs-specific chromatin modulation, which could be exploited for manipulating the ubiquitous Wnt pathway.
INTRODUCTION
Wnt signalling is evolutionarily conserved and has key roles in tissue remodelling in embryonic development and adult diseases (1–3). In the postnatal heart, activation of different components of the Wnt/β-catenin pathway was shown upon hypertrophic and ischemic stimuli in different cell types (4–7). Conversely, inhibition of Wnt signalling appears to protect the heart from ventricular remodelling (5,8–10). In the absence of β-catenin or Lymphocyte Enhancer transcription factor (Lef-1) activity, cardiomyocyte (CM) growth is impaired (11). Although, functional roles of Wnt/β-catenin signaling in the heart have been studied since about a decade, the epigenetic mechanisms and molecular networks driven by its activation remain largely unknown.
Wnt canonical signalling activates gene expression by inducing formation of complexes between transcription factors (TFs) and the co-activator β-catenin, which can be further modulated by tissue-and context-specific repressors or activators (12,13). Upon Wnt receptor activation, increased stability of β-catenin triggers target gene transcription. This is regulated by the interactions of the transcriptionally active form of β-catenin- Ser675-phosphorylated (pSer675-β-catenin) with members of the TCF/LEF family, through a displacement of repressors from the TCF/LEF complex (14–16). This leads to increased histone acetylation, resulting in chromatin remodelling and gene activation (17,18). TCF/LEF factors are essential for transducing the activation of the Wnt/β-catenin axis. Context-dependent Wnt signalling actions are further fine-tuned by recruiting cell-specific modulators to chromatin complexes (19). Transcription factor-7 like 2 (TCF7L2), one of the main transcriptional effectors of the Wnt cascade, is expressed in several tissues and was recently shown to regulate the ubiquitous Wnt target gene Myc in pathological cardiac remodelling (7). Nonetheless, TCF7L2 has been shown to have both tissue- and disease-specific roles, involving distal enhancers. Importantly, enhancers can regulate context-specific gene expression by associating to specific cardiac TFs (20–23). Overall, the genome-wide, tissue-specific regulatory complex of the ubiquitous Wnt cascade, which may help identify more selectively targetable molecules modulating disease progression in the adult heart, remains poorly understood.
In this study, we show that the very low Wnt/β-catenin activity in the healthy adult heart is increased upon pressure overload in murine and human hearts, which depends on transcriptionally active pSer675-β-catenin. Using CM-specific β-catenin stabilization, we mimic all molecular hallmarks of Wnt activation as found upon hypertrophic stimuli, which results in a hypertrophy-like phenotype and severe heart failure. We show that Wnt/β-catenin/TCF7L2 activation leads to increased genome-wide active chromatin and inducible TCF7L2 recruitment to so far unrecognized, heart-specific regulatory genomic regions, driving pathological cardiac remodelling. Conversely, β-catenin inactivation post-pressure-overload resulted in a reduced expression of these TCF7L2 target genes and prevented heart failure, confirming the validity of our findings. Most importantly, we discovered a role for the hypertrophic transcription factor GATA4 in fine-tuning Wnt/β-catenin/TCF7L2 activation, essential for adult heart homeostasis.
MATERIALS AND METHODS
Mouse models
Gain (β-cateninΔex3) and loss (β-cateninΔex2–6) of function models were achieved by mating Myh6-merCremer (24) mice with either β-cateninfloxed-ex3 (25) and β-cateninfloxed-ex2–6 (Jackson Lab). For transgenesis induction, heart-specific expression of the Cre recombinase under control of the Mhy6 promoter was activated by administration of Tamoxifen (T5648, 30 mg/kg body weight/day; Sigma–Aldrich) i.p. for 3 days. Excision of loxP-flanked exon 3 of the β-catenin coding region in αMHC-merCremer/β-cateninfloxed-ex3 resulted in a non-degradable mutant of β-catenin and in αMHC-merCremer/β-cateninfloxed-ex2–6 in non-functional β-catenin. Littermates WT at β-catenin locus and positive for Cre recombinase; and WT without Cre recombinase expression were used as controls. Genotyping primers are listed in Supplemental Table S1.
Echocardiographic analysis and disease model
Transaortic constriction (TAC) was done in 12-weeks-old mice. Pre-anesthetic and anesthetic agents are listed in Supplemental Table S2. The intervention was performed by tying a braided 5–0 polyviolene suture (Hugo Sachs Electronik) ligature around the aorta and a blunted 26-gauge needle and subsequent removal of the needle. For sham controls, the suture was not tied. To determine the level of pressure overload by aortic ligation, a high frequency Doppler probe was used to measure the ratio between blood flow velocities in right and left carotid arteries. TAC mice with blood flow gradient <60% were excluded. For echocardiography, mice were anesthetized by 2.4% isoflurane inhalation and ventricular measurements were done with a VisualSonics Vevo 2100 Imaging System equipped with a MS400, 30 MHz MicroScan transducer. The observer was unaware of the genotypes and treatments. All these procedures were performed by the SFB 1002 service unit (S01 Disease Models). All animal experiments were approved by the Niedersachsen (AZ-G 15-1840) animal review board.
Human heart samples
Left ventricular tissue was used for DNA and RNA isolation. RNA expression of foetal samples was described elsewhere (26). The investigation of human samples conforms to the principles outlined in the Declaration of Helsinki and was approved by the institutional ethics committee of the University Medical Center Goetingen (31 September 2000). DNA and RNA isolation and analyses are described in Supplemental Methods.
RNA-sequencing (RNA-seq) and data analyses
RNA-seq was performed at the Transcriptome and Genome Analysis Laboratory, University Medical Center, Goettingen, in biological triplicates. RNA was extracted, quality and integrity was assessed by Bioanalyzer (Agilent). Libraries were prepared and cDNA libraries were amplified and the size range of final cDNA libraries was determined by applying the DNA 1000 chip on the Bioanalyzer 2100 from Agilent (280 bp). cDNA libraries were sequenced using cBot and HiSeq2000 Illumina (SR; 1 × 50 bp; 51 cycles with single indexing; 6GB ca. 30–35 million reads per sample). Sequence reads were aligned to the mouse reference assembly (UCSC version mm9) using Bowtie 2.0.(27) For each gene, the number of mapped reads was counted and DESeq2 was used to analyze the differential expression (28) Gene ontology (GO) analyses were performed using default parameters and stringency in ‘ClueGO’: a Cytoscape plug-in.(29) The significant ‘GO Biological Processes’ were shown with P ≤ 0.05.
Chromatin immunoprecipitation (ChIP-seq) and data analyses
TCF7L2 and H3K27ac ChIPs in murine adult cardiac ventricular tissue were performed by 20 min crosslinking with 1.3% formaldehyde and sonicating for 45 cycles. Inputs were pre-cleared for 45 min at 4°C using protein-A-sepharose beads. For immunoprecipitation, 2 μg of anti-TCF7L2, anti-IgG (17–10109, Millipore), anti-GATA4 (sc-25310 X, SantaCruz) or anti-H3K27ac (C15410196, Diagenode) was added to the nuclear extracts and incubated O/N at 4°C. Antibodies were pulled down using protein-A-sepharose beads followed by washing and DNA extraction. For protein complex isolation, proteins were extracted from sepharose beads and supernatants were subjected to immunoblotting. ChIP-seq library preparation was performed using NEBNext Ultra DNA library prep kit for Illumina (E7370) as per manual's instructions. DNA libraries were amplified and sequenced by using the cBot and HiSeq2500 from Illumina (25–30 million reads per sample). Sequence reads were aligned to the mouse reference assembly (UCSC version mm9) using Bowtie2 (30). Peak calling was performed with Model Based Analysis of ChIPseq (MACS2) version 2.1.0.20140616.0 (31). Genes proximal to the bound chromatin regions were identified by GREAT analyses (32). Significant ‘GO Biological Processes’ were shown with P ≤ 0.05. Published/public ChIP-seq datasets were used from the following sources: TCF7L2 liver: GSE32513; GATA4, NKX2–5 and TBX3: GSM862697-(33); DNAse-seq: GSM1014166; H3K4me1: GSM769025; RNAPII: GSM918723; H3K27me3: GSM1260017; KLF15: GSM1901940 and CTCF: GSM918756.
Statistical analyses
ANOVA single factor analysis was used to calculate the P value for qPCR-based analyses. G-Power3.1 was used to determine the sample size for animal studies. For ChIP-seq and RNA-seq analyses, q-value (to call peaks) and adjusted P-value of ≤0.05 was considered for statistical significance respectively. For motif analyses, Z-score and Fisher score (negative natural logarithm of P-value) were utilized for showing significant motifs. Unpaired student's test and two-way ANOVA with Bonferroni post-test (GraphPad Prism 6.0) were used where appropriate for statistical analysis of epifluorescence measurements of calcium cycling parameters. Again, P-values <0.05 were considered statistically significant.
RESULTS
Nuclear, phosphorylated-Ser675-β-catenin triggers Wnt transcriptional reactivation upon cardiac pressure-overload in mice and humans
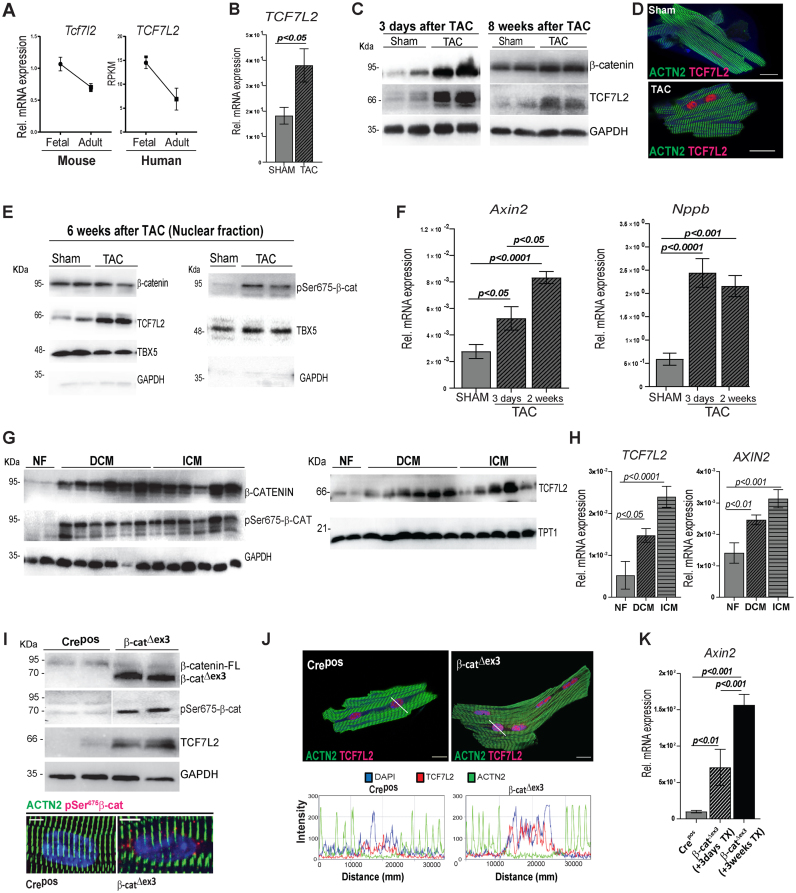
The specific contribution of TCF/LEF family members to the Wnt axis in cardiac remodelling is not well defined. We established that TCF7L2 was mainly expressed in adult ventricles and was the highest expressed TCF/LEF member with an activating function in the mouse and human left ventricle (Supplementary Figure S1A). In accordance with lowering Wnt activity during maturation, TCF7L2 decreased from foetal to adult life in mouse and human cardiac tissue (Figure 1A). However, its expression increased upon trans-aortic constriction (TAC)-induced hypertrophic remodelling (Figure 1B), indicating reactivation of a TCF7L2-dependent transcriptional program. The efficiency of TAC was confirmed by trans-aortic gradients, heart-to-body weight ratios and fractional area shortening measurements (Supplementary Figure S1B). We next examined the dynamics of protein re-expression of TCF7L2 and the association to Wnt/β-catenin activity during the course of TAC. Three days after induction of pressure overload, β-catenin and TCF7L2 proteins were significantly upregulated in left ventricular tissue, compared to sham controls. TCF7L2 remained upregulated 8 weeks post-TAC; although β-catenin levels were normalized, suggesting a sustained, TCF7L2-specific Wnt activation (Figure 1C and Supplementary Figure S1C–E). Upregulation of TCF7L2 was confirmed in isolated CMs 3 days post-TAC (Figure 1D). To test β-catenin-dependent transcriptional activation, we firstly analyzed pSer675-β-catenin, which possesses a high affinity for TCF/LEF family members for target gene regulation (34). Six weeks post-TAC, pSer675-β-catenin and TCF7L2 were more abundant in TAC nuclear fractions, while total β-catenin was not significantly changed; in line with the observation in 8 weeks post-TAC (Figure 1E and Supplementary Figure S1F). Secondly, β-catenin/TCF7L2 transcriptional activity was confirmed by upregulation of a classical Wnt target, Axin2, which increased during the course of remodelling. High levels of the Natriuretic peptide b (Nppb) post-TAC induction, confirmed pathological remodelling (Figure 1F). Furthermore, analysis of left ventricular samples from human patients with dilated (DCM) and ischemic cardiomyopathies (ICM) showed upregulation of total β-catenin, pSer675-β-catenin and TCF7L2 proteins, as well as a transcriptional activation of TCF7L2 and AXIN2, compared to non-failing (NF) heart samples (Figure 1G, H and Supplementary Figure S1G). Altogether, our results show that pSer675-β-catenin initiated and maintained a re-expression of TCF7L2, reactivating Wnt transcriptional machinery during hypertrophic remodelling in the adult mamalian heart.
Figure 1.
Nuclear phosphorylated-Ser675β-catenin triggers Wnt transcriptional reactivation upon cardiac pressure-overload in mice and humans. (A) Normalized transcript expression of murine and RPKMs of human TCF7L2 expression in foetal and adult hearts (n = 3/per group). (B) Relative transcript levels of Tcf7l2 in 8 weeks post-TAC heart tissue vs. sham control (n≥5). (C) Representative immunoblots of TCF7L2 and β-catenin expression 3 days and 8 weeks post-TAC in murine heart ventricles compared to sham (n≥4). (D) Immunofluorescence image representing increased TCF7L2 (magenta) in isolated cardiomyocytes (CM) from 3 days post-TAC murine hearts (n = 3/group). (E) Total β-catenin, TCF7L2 and pSer675-β-catenin in nuclear (TBX5-enriched) fraction, 6-weeks post-TAC. (F) Relative transcript levels of the classical Wnt target gene, Axin2, and CM hypertrophic marker, Natriuretic peptide b (Nppb) 3 days and 2 weeks post-TAC in murine ventricular tissue versus sham control (n ≥ 5). (G) Western blots showing total β-catenin, pSer675-β-catenin and TCF7L2 in cardiac ventricular biopsies from ischemic (ICM) and dilated cardiomyopathies (DCM) as compared to non-failing (NF) human hearts (NF: n = 2; DCM: n = 6; ICM: n = 6). (H) TCF7L2 and its target AXIN2 transcript levels in cardiac ventricular biopsies from DCM and ICM as compared to NF human hearts (NF: n = 7; DCM: n = 15; ICM: n = 11). (I) Immunoblot showing stabilized (70 kDa) β-catenin, pSer675-β-catenin and TCF7L2 protein in β-catΔex3 ventricles. Representative immunofluorescence images showing increased perinuclear/nuclear pSer675-β-catenin (lower panel) or (J) TCF7L2 (magenta) in isolated CM β-catΔex3 ventricles compared to Crepos along with corresponding plots below images representing fluorescence intensity profiles over the nucleus. ACTN2 is shown in green, DAPI nuclear staining in blue (n = 3/group). The profiles show DAPI (blue) signal overlapping with TCF7L2 (magenta) with a higher intensity in β-catΔex3 CM. (K) Relative transcripts of Axin2 after 3 days and 3 weeks of induction in β-catΔex3 ventricles vs. control Crepos/β-catwt (Crepos) hearts (n = 10; 3 and 5; respectively). TATA-binding protein (Tbp) (B, F) and GAPDH (H) were used for transcript normalization. GAPDH and TPT1 serve as protein loading control for whole cell lysate (C, G and I) and GAPDH for cytosolic fraction (E) and TBX5 for nuclear fraction in E. Data are mean ± SEM; t-test and ANOVA, Bonferroni's multiple comparison tests. Scale bar: I: 5 μm and D, J: 20 μm.
Wnt activation promotes heart failure by triggering developmental reprogramming in the adult heart
Next, to investigate the mechanisms triggered by β-catenin/TCF7L2-transcriptional activity, we mimicked this activation by inducing β-catenin stabilization specifically in adult CMs. Inducible stabilization was achieved by crossing a mouse possessing a Ctnnb1 (β-catenin) allele with loxP-flanked exon 3 (β-cateninfloxed-ex3) (25), with a Myh6-promoter driven tamoxifen (TX)-inducible-Cre expressing line (24). The recombined allele (β-catΔex3) produces a truncated, stabilized and GSK3-β-degradation-resistant β-catenin increasing the cytosolic and nuclear pool (Supplementary Figure S2A and B). Fourteen-weeks-old mice were induced with TX and analyzed for 3–4 weeks (Supplementary Figure S2C). To exclude effects of Cre expression and/or TX toxicity, two control groups were used for functional analyses including Myh6-merCremer/β-catwt (Crepos) and β-catflox-ex3 (Creneg), respectively. β-catΔex3 hearts showed a significant increase in stabilized β-catenin protein compared to Crepos controls. Similar to the observations in TAC-induced remodelling in mouse, TCF7L2 and pSer675-β-catenin proteins were significantly increased in β-catΔex3 cardiac tissue. Moreover, accumulation of the active pSer675-β-catenin could be visualized at the perinuclear/nuclear region in CMs, indicating Wnt activity (Figure 1I). In line with this, (i) increased TCF7L2 in β-catΔex3 CM nuclei was confirmed by confocal microscopy and fluorescence intensity profiles over the nucleus (Figure 1J) and (ii) progressively increasing TCF7L2-dependent transcriptional activation, demonstrated by Axin2 expression, was showed from 3 days to 3 weeks post-TX-induction (Figure 1K). Therefore, we analyzed the mice 3 weeks upon recombination.
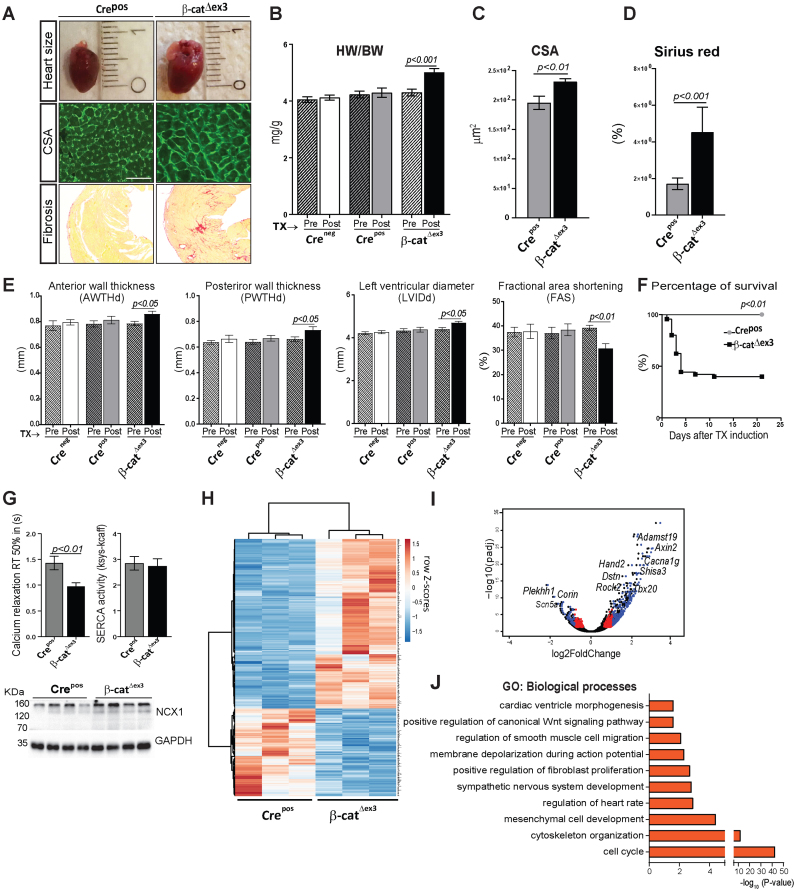
β-catΔex3 mice phenotypically resembled experimentally-induced hypertrophy with increased heart sizes, cardiac mass, myocyte cross-sectional area and fibrosis (Figure 2A–D). Maladaptive cardiac remodelling was indicated by increased anterior and posterior wall thickness and left ventricular chamber diameter along with decreased fractional area shortening and higher mortality in β-catΔex3 mice, compared to Crepos and Creneg controls (Figure 2E and F). We investigated calcium homoeostasis and observed significantly faster elimination kinetics of caffeine induced-Ca2+-transients in β-catΔex3 CMs, while SERCA2 activity remained unchanged in β-catΔex3 compared to Crepos CMs. Accordingly, expression of the Na+–Ca2+ exchanger (NCX) was higher in β-catΔex3 CMs (Figure 2G), which may be part of the genetic reprogramming in cardiac remodelling (35). Amplitude of systolic and caffeine-induced Ca2+-transients at increased half-time relaxation rate was unchanged in β-catΔex3 CMs, suggesting that NCX may contribute to the faster Ca2+-elimination in β-catΔex3 (Supplementary Figure S2D). Thus, activating β-catenin/TCF7L2-dependent transcription alone is sufficient to initiate adverse cardiac remodelling in the adult heart.
Figure 2.
Wnt activation promotes heart failure by triggering developmental reprogramming in the adult heart. Representative images (A) and quantification showing increased- (B) heart-to-body weight ratios (HW/BW, n ≥ 15/group), (C) CM cross-sectional area by WGA-FITC staining (n = 3; 150 cells/mouse) and (D) fibrosis by Sirius Red staining (n = 3/group) in β-catΔex3 versus control Crepos. (E) Echocardiographic analyses showing anterior and poster wall thickness diameters (AWTHd, PWTHd), left ventricular inner diameter (LVId) and fractional area shortening (FAS), 3 weeks post-TX induction in β-catΔex3 mice compared to Crepos and Creneg/β-catwt (Creneg) controls (n≥15/group). (F) Kaplan–Meier survival curve post-TX induction, n ≥ 21. (G) Half-times of intracellular calcium relaxation (RT50%) of caffeine induced-Ca2+-transients along with SERCA2 activity in β-catΔex3 and Crepos CMs (n = 6/4–21 cells per mouse). Ventricular protein expression of Na+–Ca2+ exchanger (NCX) in β-catΔex3 and Crepos hearts, n = 4. GAPDH serves as loading control in G. (H) Heatmap representing row Z-scores of RPKM values of all 572 differentially expressed genes (DEGs) with a cut-off: log2FC ± 0.5, P < 0.05 in Crepos and β-catΔex3 cardiac ventricles. Upregulated genes and downregulated genes are depicted in red and blue respectively (n = 3/group). (I) Volcano plot depicting the DEGs. Blue: DEGs with log2FC≥0.9 and p≤0.05; red: log2FC≤0.9 and P ≤ 0.05; black: unregulated genes. (J) Gene Ontology (GO) biological processes of upregulated (P≤ 0.05) genes. Data are mean ± SEM; t-test and ANOVA, Bonferroni's multiple comparison test. Scale bar in A: 20 μm.
To explore the cellular events triggered by β-catenin/TCF7L2 activation, we performed RNA-sequencing analysis in cardiac ventricular tissue. Wnt-activated β-catΔex3 ventricles showed 376 upregulated and 196 downregulated genes compared to control Crepos (n = 3, P < 0.05 and log2FC ≥ 0.5) (Figure 2H). The most differentially regulated genes included targets of the Wnt/β-catenin-dependent (i.e. Axin2, Lef1, Cacna1g); and -independent pathways (Rock2); cardiac development (Hand2, Tbx20) and pathological genes related to heart disease condition ((Destrin (Dstn), Corin, Adamst19)) (Figure 2I). Gene ontology (GO) analysis clustered the upregulated genes into cell cycle, tissue remodelling including cytoskeleton organization, cardiac and mesenchymal development (Figure 2J). Downregulated genes included heart rate processes (Supplementary Figure S3A).
Wnt transcriptional activation results in increased CM cell cycling and cytoskeletal remodelling in the adult heart
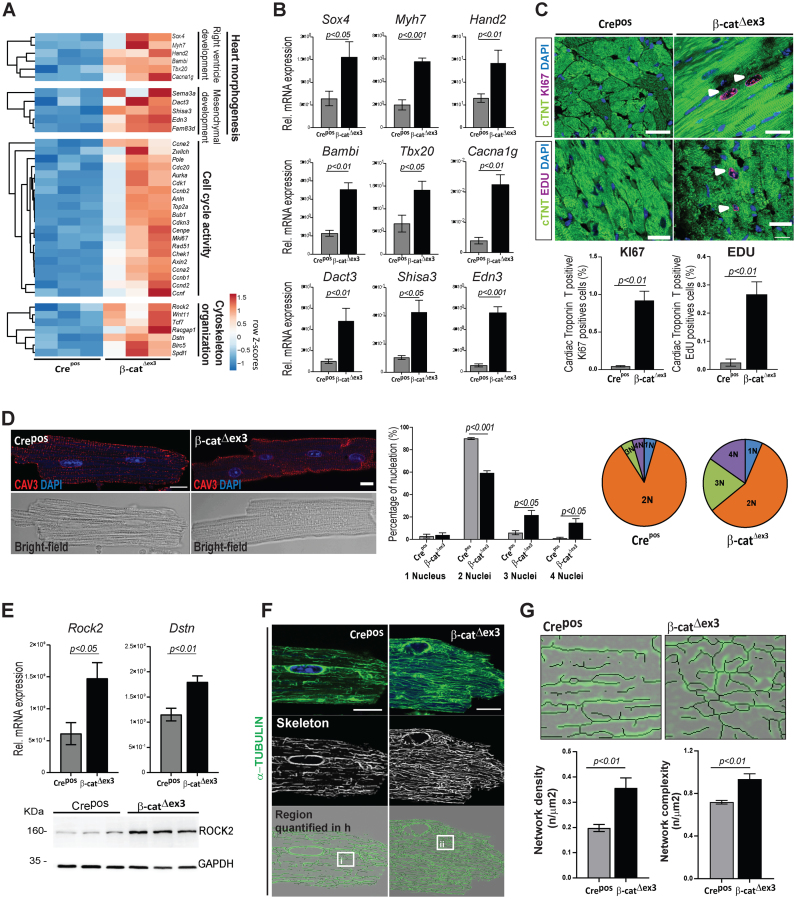
In line with the role of Wnt canonical pathway in development (36,37), β-catΔex3 hearts showed a re-expression of developmental gene clusters coordinating second heart field/right ventricle and mesenchymal/interstitial cell development, including Sox4, Tbx20, Hand2, Bambi, Edn3, Dact3 and Shisa3. Re-expression of foetal forms of calcium voltage-gated channel and myosin heavy chain (Cacna1g, Myh7) as observed in hypertrophic remodelling, was also detected in β-catΔexon3 hearts (Figure 3A and B).
Figure 3.
Wnt transcriptional activation results in increased CM cell cycling and cytoskeletal remodelling in the adult heart. (A) Heatmaps depicting row Z-scores of RPKMs of upregulated genes involved in heart morphogenesis, cell cycling and cytoskeleton organization in β-catΔex3 ventricles; with (B) corresponding qPCR validations (β-catΔex3n = 4; Crepos n = 5). (C) Confocal images of KI67 or EdU (magenta) immunostainings with quantification of double positive cTNT/KI67 and EdU cells (n = 3; >400 cells/mouse). White arrows indicate KI67 and EDU positive CM. (D) Immunostainings for Caveolin 3 (red), DAPI (blue) in single CM along with quantification of nuclei/CM (n = 3; ≥90 cells/CM per mouse). (E) qPCR validating increased cytoskeletal regulators Rock2 and Dstn; and immunoblot showing increased ROCK2 in β-catΔex3 ventricles (β-catΔex3n = 4; Creposn = 5). (F) Confocal images and (G) quantification of microtubule network density and complexity in single CMs (n = 3; 5–8 cells/mouse; i-ii represent higher magnification in F). Images are an overlay of α-TUBULIN images (green) with corresponding extracted skeletons (black). Tbp was used for normalization in B and E and GAPDH was loading control in F. Scale bar C: 20 μm, E, G: 10 μm. Confocal images were re-colored for color-safe combinations.
In line with the transcriptomic data and the role of Wnt in cell cycle regulation, we found a significantly increased expression of KI67 in cardiac troponin T (cTNT)-positive cells in myocardial tissue and isolated CMs from β-catΔex3 hearts. In vivo 5-ethynyl-2′-deoxyuridine (EdU) DNA uptake in cTNT-positive cells was increased in β-catΔex3 myocardial cells (Figure 3C and Supplementary Figure S3B). Cell cycle genes (Ccna2, Ccnb1, Ccnb2 and Ccnd2), previously implicated in hypertrophic remodelling and CM cell cycle re-entry, (38,39) were found increased in β-catΔex3 ventricles. Increased KI67 expression in cTNT-positive cells, as well as Ccna2, Ccnb1 and Ccnb2 expression was also confirmed in TAC-induced hearts (Supplementary Figure S3C). This was accompanied by a switch in CM nucleation towards 3- and 4-nuclei in β-catΔex3 hearts, with a decreased frequency of normal bi-nucleation as compared to Crepos CM (Figure 3D).
Interestingly, genes involved in cytoskeletal organization, annotating to the β-catenin-independent Wnt pathway: Wnt11, Rock2, Tcf7, Wif1 and Dstn (40), also upregulated in human cardiomyopathies, were upregulated in β-catΔex3 hearts (Figure 3E). To visualize a possible effect on cytoskeletal organization, we analyzed the CM cytoskeleton which showed an increased network density and complexity in β-catΔex3 CMs (Figure 3F and G). Adherens junctions structures showed no major alterations in β-catΔex3 CMs (Supplementary Figure S3D and E). These results suggest a cross-talk between Wnt/β-catenin-dependent and -independent signaling, which may regulate cytoskeletal rearrangements, contributing to pathological changes in adult CMs.
Induced TCF7L2 and H3K27ac occupancies at disease-associated enhancers defines the cardiac epigenome upon β-catenin stabilization
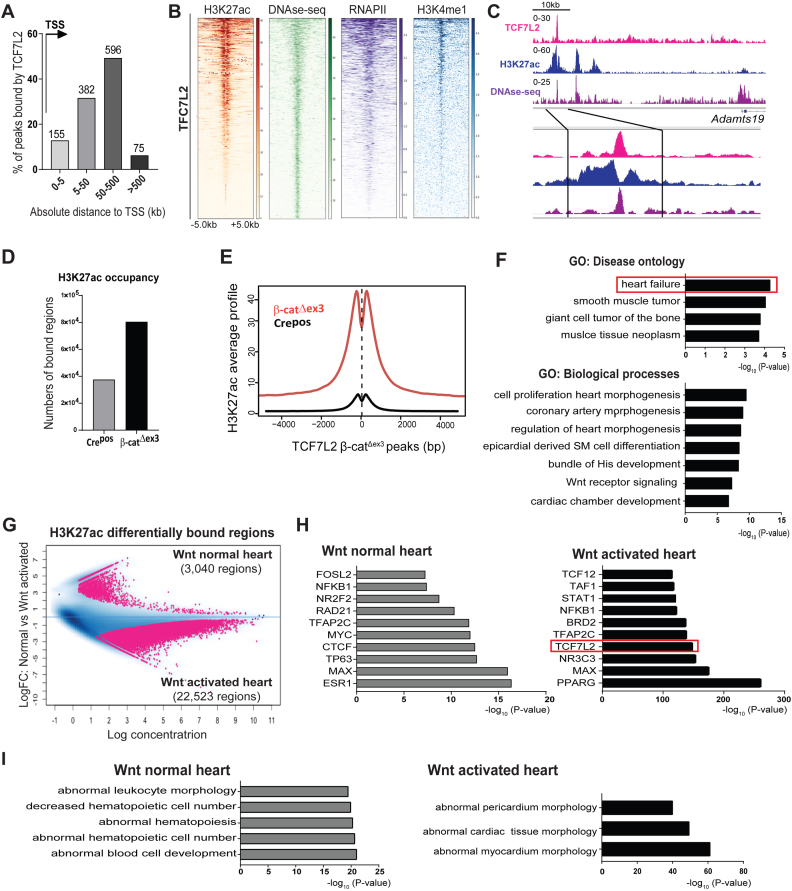
We showed that TCF7L2 was (i) the highest TCF/LEF member expressed in the mouse and human heart and (ii) re-expressed in both mouse and human cardiomyopathies. Hence, we explored the transcriptional involvement of TCF7L2 in Wnt-mediated chromatin remodelling in the adult heart. ChIP-seq analyses were performed for endogenous TCF7L2 and Histone 3 lysine 27th acetylation (H3K27ac), an established chromatin mark for active enhancers and transcriptionally active regions (41), in β-catΔex3 and Crepos ventricles. TCF7L2 occupancy was low, albeit detectable in wild-type ventricles, which correlates to the low Wnt transcriptional activity in healthy hearts (∼222 regions). Validating this ChIP, TCF7L2 occupancy was detected at known target gene promoters of Axin2 and SP5. A 20-fold enrichment of TCF7L2 occupancy on the Axin2 promoter was confirmed by qPCR (Supplementary Figure S4A and B). Upon β-catenin stabilization, genome-wide TCF7L2 binding was markedly increased, indicating an inducible occupancy, owing to its upregulation. An inducible recruitment of TCF7L2 and H3K27ac at the Tcf7l2 gene locus was observed in β-catΔex3 hearts, indicating a positive feedback regulation (Supplementary Figure S4C). We identified 1209 regions associated with 977 genes, occupied by TCF7L2 in β-catΔex3 hearts. Among these, approximately 80% of TCF7L2-bound regions were located distal to transcription start sites (TSS) (Figure 4A). TCF7L2 occupancy was closely associated with H3K27ac in β-catΔex3 hearts, and with published data for DNAse hypersensitive (DHS) regions, H3K4me1 and RNAPII binding, all marks of accessible chromatin, in the adult heart (Figure 4B and C). Global H3K27ac occupancy was markedly increased in β-catΔex3 when compared to Crepos control ventricles, notably also on TCF7L2-bound β-catΔex3 regions (Figure 4D and E). GO analysis of TCF7L2-bound regions revealed ‘heart failure’ as the most significant disease ontology along with developmental remodelling processes (Figure 4F).
Figure 4.
Induced TCF7L2 and H3K27ac recruitment to disease-associated enhancers defines the β-catΔexon3 cardiac epigenome. (A) Genomic distribution of TCF7L2 occupancy in β-catΔex3 ventricles in number of regions and as distance from TSS (arrow) in kb. (B) Heatmaps of H3K27ac in β-catΔex3 ventricles and high sensitivity DNAse-sequencing (DHS), -RNAPII, -H3K4me1 occupancy in normal adult heart, on TCF7L2 occupied regions in β-catΔex3 ventricles, aligned according to their maximum signal of TCF7L2 occupancy. Scale depicts normalized RPKM values for heatmaps. (C) Exemplary occupancy profiles of TCF7L2, H3K27ac and DHS on an identified distal enhancer of Adamts19. (D) Total number of H3K27ac bound-regions as well as (E) average signal profiles of H3K27ac on TCF7L2 occupied loci in Crepos and β-catΔex3 ventricles. (F) GO disease ontologies of TCF7L2 bound regions in β-catΔex3 ventricles. (G) Binding affinity plot of 25,563 differential ChIPseq-H3K27ac enriched regions (pink dots) in the Wnt normal (3,040 regions, log FC > +0.5, n = 3) and in β-catΔex3 (Wnt activated) (22,523 regions, log FC < –0.5, n = 2) hearts. (H) Motif analysis on differentially enriched distal H3K27ac regions (5,748 from 22,523) in ‘Wnt activated’ and control ‘Wnt normal’. TCF7L2 motif occurrence is highlighted in an orange box. (I) Disease Ontologies are for the regions in H. For heatmaps regions ± 5 kb are shown.
In order to understand the relevance of differential H3K27ac occupancy in normal and β-catΔex3 hearts, we identified 25,563 differentially-enriched genomic regions, which were specifically enriched in β-catΔex3 hearts (‘Wnt activated hearts’: 22,523 regions) or in normal hearts (‘Wnt normal hearts’: 3,040 regions) (Figure 4G). Since TCF7L2 occupied predominantly distal regions, we focused on the distally-enriched regions. Of the 22,523 regions in Wnt activated hearts, 5,748 of the β-catΔex3-specific regions were located >±5 kb from the nearest gene. We identified an extensive overlap of published TCF7L2 data (–log10P-value = 141.209) with β-catΔex3-specific distally-occupied H3K27ac regions, which was absent in the H3K27ac-bound distal regions enriched in the normal heart (Figure 4H). Functional categorization of H3K27ac-bound distal regions revealed enrichment of processes involved in hematopoietic cell biology in the normal heart, whereas Wnt activated hearts showed processes involved in abnormal cardiac muscle morphology (Figure 4I). These results indicate a particularly important chromatin-associated role of TCF7L2 in response to β-catenin stabilization in the adult heart.
TCF7L2 elicits tissue-specific gene regulation in pathological heart remodelling
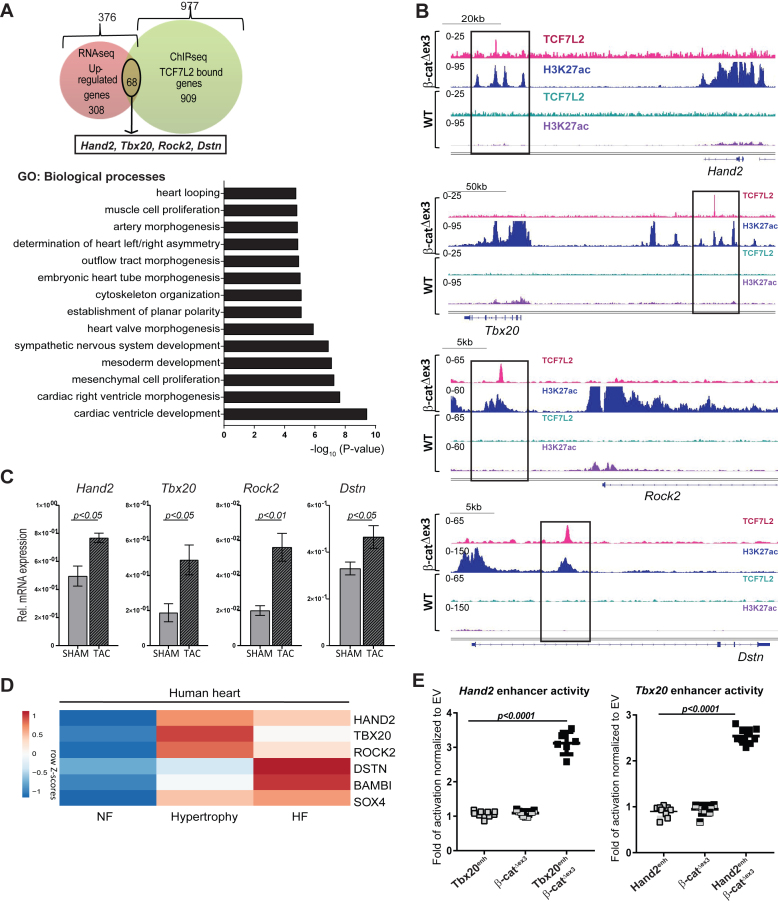
Integrative analyses of 977 TCF7L2-bound and 376 upregulated genes (RNAseq, log2FC>0.5, p<0.05) upon β-catenin stabilization, showed that TCF7L2 occupied and regulated the expression of a subset of 68 genes. Similar to the categorization of transcriptome data, these genes annotated to cardiac developmental tissue remodelling, cytoskeletal rearrangement and mesenchymal developmental processes in β-catΔex3 hearts (Figure 5A). Genes at this intersection included Hand2, Tbx20, Rock2 and Dstn. Significant occupancy of TCF7L2 and H3K27ac on the distal sites of these genes was observed in β-catΔex3, but not in normal hearts (Figure 5B). In line with previous observation (42,43), we observed their upregulation upon pathological remodelling in mouse and human hearts (Figure 5C and D). In contrast, only 8 genes showing TCF7L2 occupancy were downregulated upon β-catenin stabilization, suggesting a specific role for TCF7L2 only in pathological gene activation (Supplementary Figure S5A). Upregulated genes that did not intersect with TCF7L2 binding in β-catΔex3 hearts were enriched for DNA replication processes (Supplementary Figure S5B).
Figure 5.
TCF7L2 elicits tissue-specific gene regulation in pathological heart remodelling. (A) Venn diagram of genes bound by TCF7L2 (green, 977) and upregulated (red, 376; log2FC ≥ 0.5, P ≤ 0.05) in β-catΔex3 ventricles. GO biological processes annotation of the 68 common genes at the intersection. Hand2, Tbx20, Rock2 and Dstn are part of these common genes. (B) Occupancy profiles of TCF7L2 and H3K27ac in β-catΔex3 and Crepos ventricles on the identified putative enhancers upstream of Hand2, Tbx20, Rock2 and Dstn. (C) Hand2, Tbx20, Rock2 and Dstn relative transcript levels in ventricles upon TAC-induced hypertrophy and sham controls, n ≥ 5. (D) Heatmaps showing row Z-scores of RPKMs of dysregulated genes in human ventricular tissue from non-failing (NF), hypertrophic and failing hearts (HF). (E) Luciferase reporter assays for Hand2 and Tbx20 identified enhancers upon Wnt activation by β-catenin stabilization in HEK293 cells normalized to empty vector (Renilla luciferase was used as transfection control, n = 3/independent experiments). Tbp was normalization control in C. Data are mean ± SEM; t-test.
For functional confirmation of TCF7L2-dependency of these novel targets, we tested Hand2 and Tbx20 enhancer regions (containing 4 and 2 TCF/LEF consensus sites, respectively) upon expression of stabilized β-cateninΔex3, which mimics our β-catΔex3 mouse model in vitro. Luciferase assays showed a 3-fold (Hand2) and 2.5-fold (Tbx20) increase in luciferase activity (n = 3, Figure 5E). Altogether, these results identified novel β-catenin/TCF7L2 cardiac enhancers and highlight their significance in pathological remodelling in the adult myocardium.
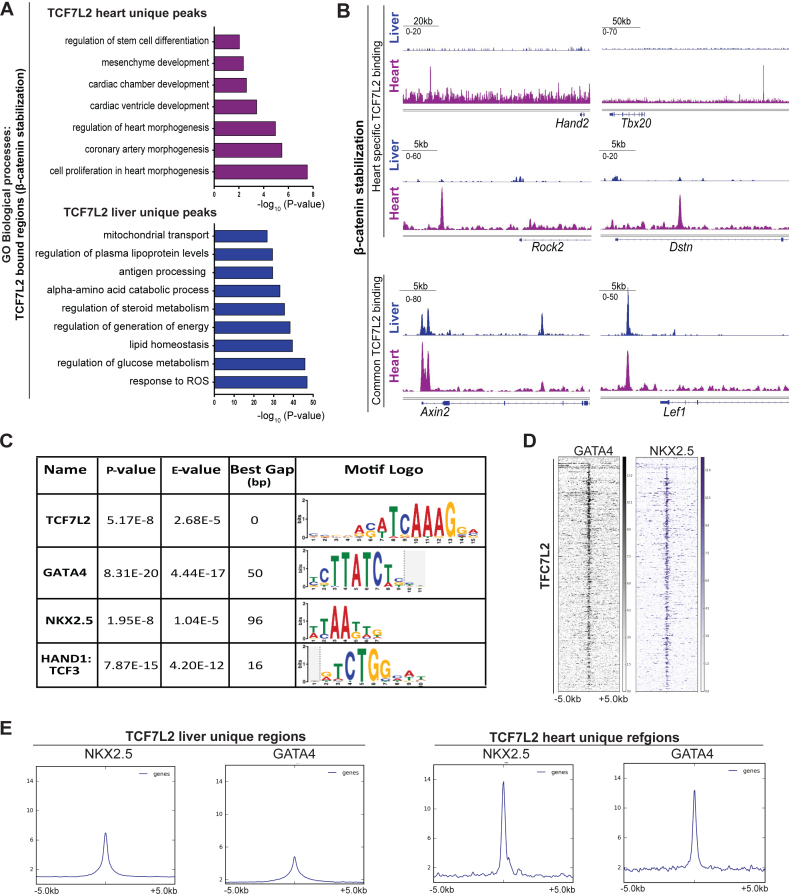
To further investigate the cardiac-specificity of TCF7L2 chromatin occupancy, we compared the binding profiles of endogenous TCF7L2 found in our CMs-β-catΔex3 model, with a published hepatocyte-specific β-catenin stabilized mouse model (44). TCF7L2-bound regions only found in β-catenin stabilized hepatocytes (compared to CMs), were associated with metabolic processes, lipid and cholesterol homeostasis. Whereas, TCF7L2-bound regions only found in β-catenin stabilized CMs were associated with cardiac developmental processes (Figure 6A). Overlapping binding regions were only found in promoter regions of classical Wnt target genes such as Axin2 and Lef1, while non-overlapping distal genomic regions belonging to Hand2, Tbx20, Rock2 and Dstn were observed only in β-catΔex3 .hearts (Figure 6B). These results strongly point to the tissue-specific functions of TCF7L2 at distal regions (23).
Figure 6.
TCF7L2 cooperates with cardiac-TFs to enable heart-specific gene regulation. (A) Comparison of TCF7L2-bound regions in in vivo models of β-catenin stabilization (β-catΔex3) in CMs and hepatocytes. GO biological processes for unique regions in CM (magenta) and hepatocytes (blue). (B) TCF7L2 occupancy profiles on identified Hand2, Tbx20, Rock2 and Dstn enhancers and common Wnt targets Axin2 and Lef1 in β-catenin stabilized CM (magenta) and hepatocytes (blue). (C) Table enlisting the transcription factors (TFs) enriched on TCF7L2-bound regions in β-catΔex3 ventricles by de-novo motif search using MEME-SpaMo. (D) Heatmap depicting the occupancy of cardiac-TFs GATA4 and NKX2.5 in the normal heart on regions occupied by TCF7L2 in β-catΔex3 hearts. Regions ± 5 kb are shown. (E) Average profiles of GATA4 and NKX2.5 occupancy on TCF7L2-bound liver and heart-specific regions. Data are mean ± SEM; t-test.
TCF7L2 cooperates with cardiac-TFs to enable heart-specific gene regulation
We next aimed to investigate the mechanisms that enabled TCF7L2 to elicit CM-specific responses. Cardiac-TFs including GATA, NKX, TBX and MEF have been shown to collaborate with other factors to direct cardiac-specific gene expression (45). De novo motif analyses revealed that TCF7L2-bound chromatin regions in β-catΔex3 hearts showed a significant enrichment for GATA4 (Gap = 50 bp, P = 8.31E–17), NKX2.5 and HAND1/TCF3 motifs (Figure 6C). This was confirmed by comparing published ChIP-seq datasets for GATA4 and NKX2.5 in the normal adult heart (46) with the TCF7L2-bound regions identified in our study (Figure 6D). Accordingly, heart-specific TCF7L2-bound regions showed clearly, a higher correlation to NKX2.5 and GATA4 occupancy as compared to their lower association with liver-specific regions (Figure 6E), further confirming its tissue-specific roles.
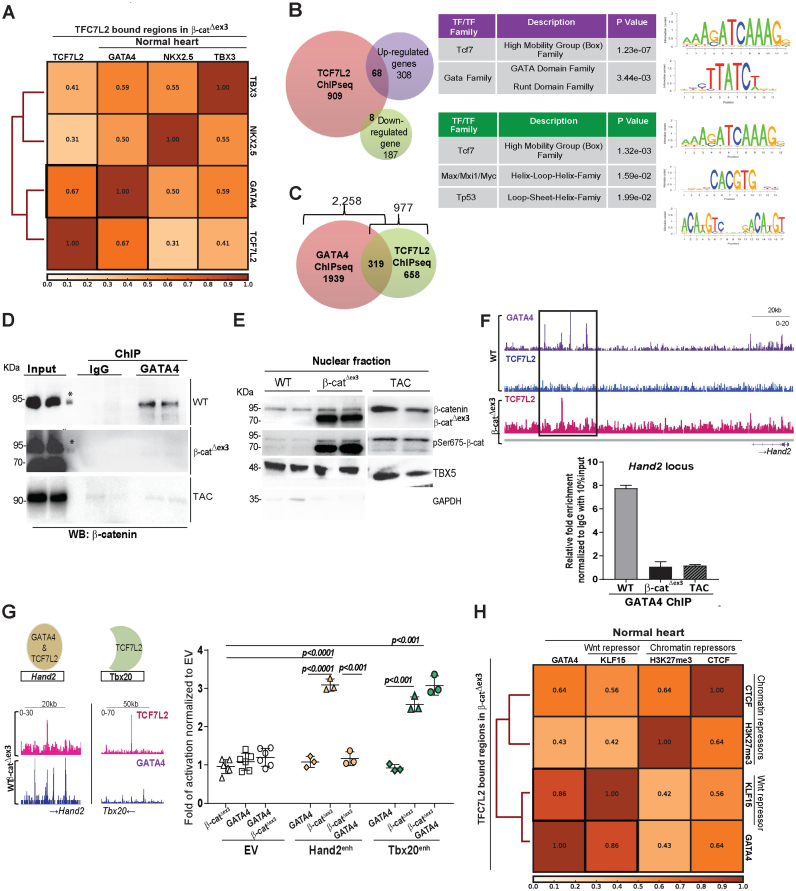
Among the analyzed cardiac-TFs, GATA4 binding in the normal heart showed the highest correlation (Spearman's correlation coefficient r = 0.67) to TCF7L2-bound regions in the β-catΔex3 hearts (Figure 7A, Supplementary Figure S6A). Next, motif analysis was performed on the genes both bound by TCF7L2, and upregulated in β-catΔex3 hearts (68 genes) showing again, a significant enrichment of the GATA motif (p value = 3.44E-3). The association with downregulated RNA-seq genes showed different motifs including TP53 (Figure 7B). Given the strong correlation of GATA4 and TCF7L2, genes co-occupied by both GATA4 and TCF7L2 in heart tissue were analyzed. Of the 2,258 GATA4-bound (46) and our 977 TCF7L2-bound genes in β-catΔex3 hearts, 1,939 were GATA4-specific, 658 were TCF7L2-specific and 319 were common targets for both GATA4 and TCF7L2. GATA4-specific target genes (Arid1a, Camk2d, Jph2, Mef2a, Myh6, Myh7, Myocd, Nppa, Nppb, Ryr2, Smad7, Tnnt2) categorized for heart and muscle development. TCF7L2-specific target genes (Axin2, Gja1, Lef1, Fzd6, Med1, Ctnnb1, Twist1, Rspo2, Sox9, Notch3, Hdac9, Fst, Klf4, Tnfrsf19) categorized for morphogenetic developmental processes. Interestingly, TCF7L2 and GATA4 common targets constituting 30% of all TCF7L2-bound genes, (Hand2, Tbx3, Tbx5, Dysf, Cited2, Smad3, Myom2, Sox4, Sox6, Tbx20, Ror2, Tead1) categorized for heart and muscle development, furthering that GATA4 confers cardiac-specificity to TCF7L2 (Figure 7C and Supplementary Figure S6B).
Figure 7.
GATA4 interacts with β-catenin and fine-tunes the molecular switch driving adult heart disease progression in vivo. (A) Spearman's correlation plot of TCF7L2 co-occupancy with GATA4, NKX2.5 and TBX3, highlighting highest correlation with GATA4 (black box). (B) Venn diagram of genes bound by TCF7L2 (orange) with upregulated (violet) or downregulated (green) genes with log2FC≥0.5, p≤0.05 in β-catΔex3 ventricles and corresponding motif enrichment of the intersections. (C) Venn diagram showing commonly bound genes (319) between TCF7L2 in β-catΔex3 hearts and GATA4 in normal hearts. (D) Immunoblot of GATA4 with β-catenin co-immunoprecipitation in WT, β-catΔex3 and 6 weeks post-TAC hearts. Input represents the total, sheared chromatin-protein complexes before immunoprecipitation, (*) protein ladder. (E) Immunoblot of total β-catenin and active pSer675 β-catenin in the nuclear fractions of control, β-catΔex3 and 6 weeks post-TAC hearts. TBX5 and GAPDH were used to detect nuclear and cytosolic enrichments respectively. (F) IGV binding profiles for TCF7L2 occupancy in β-catΔex3 hearts along with GATA4 co-occupancy in normal hearts on Hand2 enhancer locus; ChIP-qPCR for GATA4 binding on Hand2 enhancer in normal (WT), 6 weeks post-TAC (WT TAC) and β-catΔex3 hearts. Relative fold enrichment was calculated to IgG control, normalized to 10% input chromatin (n = 3 hearts/ChIP) (G) Profiles of enhancers with GATA4 and TCF7L2 overlapping occupancy (Hand2) and with only TCF7L2 occupancy (Tbx20). Luciferase reporter assay for Hand2 enhancer (-enh) and Tbx20-enh upon β-catenin stabilization, GATA4 overexpression or both normalized to empty vector (EV). (Renilla luciferase was the transfection control, n = 3/independent experiments). Data are mean ± SEM; t-test and ANOVA, Bonferroni's multiple comparison test. (H) Spearman's correlation plot depicting high correlations between GATA4 and repressive elements KLF15, H3K27me3 and CTCF in normal hearts specifically on TCF7L2-bound regions in β-catΔex3 hearts.
GATA4 interacts with β-catenin and fine-tunes the molecular switch driving adult heart disease progression in vivo
To get an insight into the mechanism of regulation of GATA4 on Wnt signalling, we tested the potential physical interaction of GATA4 with nuclear Wnt components in the healthy and Wnt-activated (genetically and induced by pressure-overload) adult hearts. GATA4 was immunoprecipitated, followed by chromatin-associated protein complex isolation and immunoblotting in the normal, β-catΔex3 and in 6 weeks post-TAC ventricular tissue. The suitability of the anti-GATA4 antibody was validated both by ChIP-qPCR on a known target gene Natriuretic peptide a (Nppa) and by protein detection (Supplementary Figure S6C). We observed that GATA4 and β-catenin interacted in the normal, but not in the β-catΔex3 or TAC ventricular tissue (Figure 7D). Although β-catenin was clearly present in the nucleus (Supplementary Figure S6D), it was transcriptionally inactive in the healthy heart, based on low pSer675 β-catenin levels. However, pSer675 β-catenin was found abundantly enriched in the nuclei of β-catΔex3 and in TAC tissue (Figure 7E), suggesting that GATA4 could bind only to transcriptionally inactive β-catenin, having a direct role on Wnt gene regulation.
To further this hypothesis, we selected and analyzed enhancers showing GATA4 and TCF7L2 co-occupancy such as Hand2 and Tbx3. They showed TCF7L2 occupancy upon Wnt activation but not in the healthy heart, whereas GATA4 occupancy was observed in the healthy heart. Consistently, ChIP-qPCR analyses revealed enriched GATA4 binding to the enhancer regions of Hand2 and Tbx3 in the healthy heart, which was significantly decreased upon Wnt activation in β-catΔex3 and TAC hearts (Figure 7F, Supplementary Figure S6E). We next investigated the functional role of GATA4 in this context. For this purpose, we analyzed the regulation of two TCF7L2-targets: Hand2, displaying co-occupancy of GATA4 (in WT healthy hearts) and TCF7L2 (in β-catΔex3 hearts) and Tbx20, displaying TCF7L2, but not GATA4 occupancy. By performing luciferase assays, we showed that GATA4 co-expression prevented β-catΔex3-mediated Hand2, but not Tbx20-enhancer activation (Figure 7G). This supports a repressive role of GATA4 on a subset of Wnt target genes in the normal adult heart.
Next, we analyzed the correlation of GATA4 with transcriptional repressors like Krüppel-like Factor 15 (KLF15), a nuclear Wnt repressor interacting with β-catenin and TCF7L2 in cardiac tissue (47). Using published KLF15 ChIP-seq data from the adult heart (33), we observed that TCF7L2-bound regions in β-catΔex3 hearts displayed a remarkable overlap with KLF15 and GATA4 co-occupancy (Spearman's correlation coefficient r = 0.86) in the normal heart. This co-occupancy was further correlated to H3K27me3 and CTCF, known players associated with chromatin insulating activity (48,49) (Figure 7H). Altogether, these results showed a fine-tuning effect of GATA4 on Wnt-dependent cardiac genomic regions (Figure 8G).
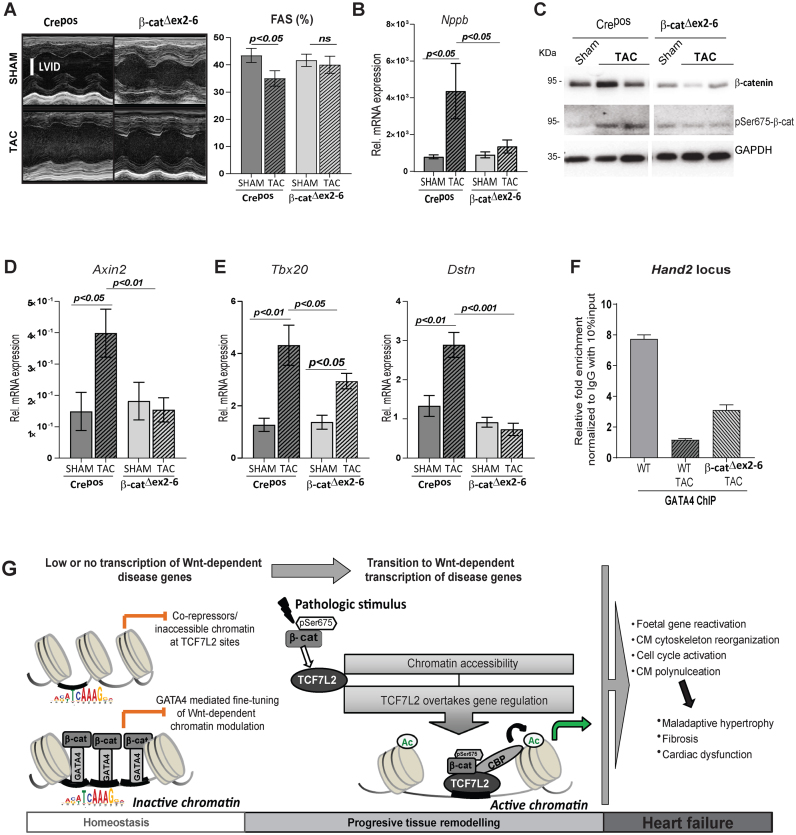
Figure 8.
β-catenin loss of function in CM rescues Wnt-dependent pathological gene regulation in vivo. (A) Representative examples of M-mode echocardiograms, and quantification of fractional shortening (FAS) by echocardiographic analysis of 3 weeks TAC-induced Crepos control and β-catΔex2–6 mice, n ≥ 7. (B) Relative transcript levels of hypertrophic marker Nppb in β-catΔex2–6 and controls Crepos; sham and TAC (n≥8/per group). (C) Immunoblot depicting total β-catenin and pSer675-β-catenin upon TAC in β-catenin loss of function (β-catΔex2–6). GAPDH was protein-loading control (n = 2/group). Relative transcript levels of (D) classical Wnt target Axin2 and (E) newly identified cardiac Wnt targets Tbx20 and Dstn, 6 weeks post-TAC in β-catΔex2–6 and controls, n≥7. Data are mean ± SEM; ANOVA, Bonferroni's multiple comparison test. Tbp was used for transcript normalization. (F) ChIP-qPCR for GATA4 binding on Hand2 enhancer in normal (WT), 6 weeks post-TAC (WT TAC) and β-catΔex2–6 TAC hearts. Relative fold enrichment was calculated to IgG control, normalized to 10% input chromatin (n = 3 hearts/ChIP). (G) Schematic representation of the findings of this study. In the healthy adult heart, β-catenin/TCF7L2-dependent loci are inactive, inaccessible or bound by transcriptional repressors, resulting in low transcription. On a subset of these loci, GATA4 binds to transcriptionally inactive β-catenin, fine-tuning Wnt-dependent transcription. This chromatin state guaranties normal homeostasis in the adult heart. Pathological stimuli leading to active pSer675-β-catenin accumulation activates Wnt signalling and the epigenetic state switches on to ‘active’, replaced by transcriptionally active pSer675-β-catenin bound to TCF7L2, leading to enriched H3K27ac occupancy and a high Wnt transcriptional activity. This results in the expression of disease-associated genes leading to adverse remodelling and heart failure.
β-Catenin loss of function in CM rescues Wnt-dependent pathological gene regulation in vivo
As a proof of concept, we analyzed murine hearts with CMs specific inducible β-catenin loss of function (β-catΔex2–6) with a confirmed, reduced total β-catenin expression subjected to TAC-induced hypertrophy (Supplementary Figure S7A and B). The efficiency of TAC was confirmed by a homogenous trans-aortic gradient indicating similar induced pressure overload among the different groups (Supplementary Figure S7C). Supporting a beneficial effect of blocking Wnt– transcriptional activation, reduction of fractional area shortening and upregulation of the hypertrophic marker Nppb was prevented in β-catΔex2–6 upon TAC (Figure 8A, B). Upregulation of pSer675-β-catenin protein expression was attenuated in β-catΔex2–6 upon 6-weeks post-TAC (Figure 8C, Supplementary Figure S7D). Accordingly, Wnt-mediated transcriptional activation was significantly decreased, as demonstrated by lower Axin2 expression in β-catΔex2–6 sham and TAC mice, whereas Axin2 activation was corroborated upon TAC in Crepos ventricles (Figure 8D). In line, with the low activity of Wnt in the adult heart, β-catenin loss of function did not significantly affect the expression of novel disease genes at baseline (Supplementary Figure S7E). Finally, activation of the identified novel Wnt associated disease-enhancers Tbx20, Dstn, Hand2 and Rock2 was decreased 6-weeks after TAC in β-catΔex2–6 hearts (Figure 8E and Supplementary Figure S7F). Interestingly, ChIP-qPCR analyses showed that GATA4 binding to the enhancer regions of Hand2 and Tbx3 was partially restored in β-catΔex2–6 hearts upon TAC (Figure 8F and Supplementary Figure S7G), indicating that GATA4’s occupancy at TCF7L2 disease-target genes is associated to improved cardiac performance, reinstating its protective role at these loci.
DISCUSSION
Activation of the Wnt/β-catenin signalling occurs upon heart remodelling (5,7,8) by mechanisms which are so far not well understood. In this study, (i) we elucidated the mechanisms by which Wnt signalling activity regulates chromatin remodelling and (ii) further identified the cellular processes mediated by this activation in CMs of the adult mammalian heart. We established the relevance of TCF7L2 as a transducer of global Wnt/β-catenin-dependent transcription, controlling this process. In the adult mouse and human heart, we showed that TCF7L2 is the mainly expressed TCF/LEF family member, with known transcriptional activating functions, confirming its relevance in Wnt-mediated functions in this tissue, as also reported elsewhere (7). In accordance with decreasing Wnt activity during heart maturation, TCF7L2 expression decreases from foetal to adulthood and is elevated upon pathological remodelling in the mouse and human heart ventricles. These findings suggest TCF7L2 involvement in inducing foetal gene reprogramming in the stressed adult heart. Despite low Wnt activity, we detected β-catenin and TCF7L2 in the nuclei of CMs in the healthy adult heart. Our data suggest that inactivity of the pathway could be explained by low levels of pSer675 β-catenin in the healthy heart, in contrast to its abundance in hypertrophic murine and human hearts. Increased pSer675-β-catenin subsequently triggered TCF7L2 activation and its recruitment to the chromatin. Further amplifying the signalling, there exists a positive feedback regulation on TCF7L2 expression following β-catenin/TCF7L2 activation. Thus, pSer675-β-catenin initiates and sustains pathological heart remodelling (scheme, Figure 8G). Importantly, this regulation is evolutionarily conserved in the human heart, allowing us to confidently address the relevance of Wnt/TCF7L2-mediated transcription in adult tissue remodelling in the murine heart model.
Our study shows that β-catenin stabilization in the adult heart not only results in CM hypertrophy, but also in CM de-differentiation, a feature of pathological remodelling, leading to severe heart failure. As a part of the de-differentiation phenotype, β-catΔex3 hearts showed upregulation of genes participating in heart morphogenesis, more confined to second heart field development and expansion, in line with the role of Wnt during embryogenesis in these processes (36,37). In heart development, initial activation of Wnt/β-catenin signalling (canonical) is followed by an activation of the Wnt/β-catenin-independent (non-canonical) pathway, which represses the canonical signalling and regulates cell polarity (36). In line with this, β-catenin stabilization in adult CM was followed by an upregulation of ROCK2, a Wnt/planar cell polarity (PCP) non-canonical component and key regulator of the cytoskeleton. Additionally, we identified Destrin, an actin dynamizing factor previously involved in rearrangements of cytoskeletal filaments, human cardiomyopathies and heart failure (40) as a novel Wnt target in cardiac tissue. This regulation was reflected by a major reorganization of CM cytoskeleton as indicated by a denser and interlinked cytoskeleton in β-catenin stabilized hearts. Disorganization of cytoskeleton is a feature of hypertrophic and failing CM, which contributes to alterations in intracellular signalling and CM function, (50) and may explain the increased mortality in β-catΔex3 mice. Thus, our study showed a so far unappreciated cross-talk between components of Wnt signalling branches and uncovered a direct TCF7L2 regulation of Rock2 and the novel Wnt target Dstn, driving cytoskeleton reorganization in pathological cardiac remodelling.
In line with activation of cell cycle genes, increased CM cycling (as indicated by KI67 expression and EdU incorporation) was observed in β-catΔex3 similar to TAC hearts, the latter supporting previous studies (51). Increased cell cycle in β-catΔex3 CM resulted in multi-nucleated cells, suggesting endo-reduplication, rather than newly formed myocytes. Inactivation of GSK3β, which allows for β-catenin accumulation led to increased cycling in adult CM in vitro, which previously allowed for the conclusion that Wnt signalling pathway is a potential target for stimulating cardiac regeneration (52). Of note, one-day postnatal murine CM, which maintains a high regenerative potential (53), showed Wnt signalling enriched gene networks after ischemic heart injury (54). Since Wnt signalling becomes inactivated in the postnatal heart during later stages, it is tempting to speculate that reactivation of the signalling will confer regenerative capacity to the adult heart (54). However, our results show that sustained CM cycling and reprogramming stimulated by Wnt activation, leads to organ functional decay and not to tissue regeneration. Similarly, persistent activation of the Hippo-pathway leads to cell cycle reactivation in adult CM accompanied by loss of cardiac function (55). Thus, activation of the Wnt pathway in the adult heart triggers a program of CM dedifferentiation and remodelling. This may implicate an initial protective mechanism of the stressed heart to preserve CM function and structure, which fails eventually upon sustained activation of the pathway. This may be explained by a low developmentally permissive transcriptional state of the adult CM in comparison to the early postnatal stages (54).
The ability of the Wnt pathway to elicit a large variety of transcriptional responses requires that β-catenin and TCFs distinguish between genes to be activated and genes to remain silent in a specific context. We explored the so far uninvestigated chromatin landscape modifications (23) upon increased, transcriptionally active β-catenin and TCF7L2 in the adult heart. This activation resulted in increased active chromatin, both genome-wide and at TCF7L2-bound regions as indicated by an enhanced H3K27ac genomic recruitment in β-catΔex3 hearts. We detected stronger H3K27ac global signal intensities and also an increase in TCF7L2 binding sites, compared to normal adult heart. Consistent with our transcriptomic data, TCF7L2 occupied active enhancers of genes involved in heart failure and cardiac developmental remodelling. Furthermore, distal regions differentially enriched for H3K27ac in β-catΔex3 were associated with TCF7L2 binding. This is in line with stronger TCF7L2 recruitment to distal regions upon Wnt stimulation (23), possibly by increased DNA-binding affinities of TCF7L2 bound to the transcriptionally active pSer675-β-catenin. TCF7L2 binding could thus create a local chromatin environment that allow for enhanceosome formation, promoting activation of normally silenced genes (23). Increased TCF7L2 occupancy was mostly associated with upregulated genes, only a few genes showing TCF7L2 occupancy were downregulated upon Wnt activation. This implies that TCF7L2 is indeed, a part of an ‘activated’ disease program and plays little or no role in the regulation of ‘protective genes’ which are downregulated. TCF7L2 specifically gets activated in disease and seems to occupy a low, albeit an extremely specific set of genes driving tissue remodelling. Additionally, we cannot exclude the contribution of other downstream Wnt effectors such as LEF members.
Although most Wnt targets are tissue and context-specific, a surprisingly short list of confirmed specific targets is known (56). A previous study associated TCF7L2 activation with the ubiquitous Wnt target c-Myc in the heart (7). Using integrative, unbiased genome-wide analyses, we identified and validated novel TCF7L2 cardiac-specific target genes involved in tissue remodelling- such as Hand2, Tbx20, Rock2 and Dstn (40,42,43,57). β-catenin stabilization induced TCF7L2 recruitment to distal regions of these novel targets in the heart, but not in liver; whereas recruitment of TCF7L2 to proximal regions of ubiquitously described Wnt targets Axin2 and Lef1 was induced in both tissues. This demonstrated the cardiac-specific association of TCF7L2 to the distal regulatory regions of these genes.
A striking finding of our present study is the repressive function of GATA4 on disease-associated-enhancers co-occupied by GATA4 and TCF7L2. We uncovered a chromatin-bound Wnt nuclear complex including GATA4 and β-catenin in the healthy adult heart. Cooperation of GATA3 was shown to repress TCF7L2-mediated transcription in cancer cells (21), suggesting a context-specific cooperation of members of the GATA family with TCF7L2. Moreover, GATA1 has been involved in context-dependent regulation at the chromatin level, indicating that the GATA factors are cell- and context specific cofactors controlling homeostasis and disease (58,59). We unearthed a GATA4-β-catenin interaction and observed GATA4 occupancy on TCF7L2-disease-enhancers, which are normally lowly, expressed in the healthy adult heart. However, upon pathological remodelling, characterized by Wnt activation, GATA4 and β-catenin binding was abrogated, and GATA4 occupancy on TCF7L2-target genes was reduced, superseded by TCF7L2. Our findings support that tissue-specific Wnt-independent TFs, such as GATA4, collaborate with β-catenin to modulate chromatin activity (23). This highlights the role of the inactive nuclear β-catenin in the healthy heart, where quiescent Wnt/β-catenin-dependent transcription maintains homeostatic gene expression. Accordingly, GATA4 is necessary for maintenance of adult heart homeostasis, since its loss of function resulted in hypertrophic remodelling (60,61). This suggests that cooperation of GATA4 and Wnt signalling may contribute to the inhibition of pathological cascades in the normal heart and also confers cardiac specificity to Wnt target genes. In line with a repressive role of GATA4 on Wnt targets, we showed that on TCF7L2-bound β-catΔex3 regions, GATA4 and KLF15, a previously identified Wnt nuclear inhibitor, not only showed a remarkable correlation with each other, but also with known repressive loci occupied by H3K27me3 and CTCF in the normal heart. This, in addition to the luciferase assay and immunoprecipitation results, encouraged us to suggest the repressive role of GATA4 in Wnt-mediated transcription in the normal adult heart. Accordingly, a dynamic, context-specific GATA4 occupancy in heart homeostasis and disease has been demonstrated (62).
Upon pathological conditions, this balance is impaired by aberrant activation of pSer675-β-catenin, followed by TCF7L2 positive feedback activation, inducing a shift towards active chromatin. This is done by displacement of repressors at the Wnt complex, including GATA4. Consequently, a de-repression of normally silenced genomic regions, leading to CM de-differentiation, occurs. This results in pathological heart remodelling and culminates in heart failure (scheme, Figure 8G).
To further validate the implications of the Wnt signal activation on these identified disease targets, we used a CM-β-catenin loss of function in vivo model. In line with previous finding, β-catenin loss of function in CM resulted in preserved cardiac function upon TAC (9,11,63,64). Here, we showed that upon β-catenin loss of function, reduced levels of pSer675-β-catenin under pathological stimulus prevented Wnt nuclear feedback activation and disease-specific upregulation of our identified novel Wnt disease targets. This strongly supports their dependency on Wnt/TCF7L2 activation during pathological remodelling and their influence on functional deterioration. Moreover, GATA4 binding to TCF7L2-disease-enhancers was partially restored. Our findings indicate that the feedback mechanism on Wnt activation is an important trigger for the progression of pathological remodelling, which is attenuated in β-catΔex2–6 TAC hearts. Importantly, GATA4 occupancy on TCF7L2-disease-enhancers is positively correlated to heart homoeostasis.
Overall, our study presents the first genome-wide occupancy mapping of heart-specific targets of TCF7L2 in vivo. This enabled us to provide a novel mechanistic insight, showing a nuclear, cardiac-specific regulatory complex driving tissue and context-specific Wnt-dependent transcriptional switch at the transition to disease. This is mediated by a context and tissue-specific epigenetic function of β-catenin/TCF7L2, which cooperate with specific co-regulators to activate reprogramming and tissue remodelling. More broadly, the identification of TCF7L2-specific regulation may provide us with a framework to identify novel therapeutic avenues to reduce the increased activity of the ubiquitously expressed Wnt pathway in a tissue-specific manner, for preventing heart damage and heart failure progression. Approaches stabilizing GATA4/β-catenin repressive functions may help maintain cardiac homeostasis and prevent adverse remodelling.
DATA AVAILABILITY
Sequencing data files have been deposited in NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97763) under accession GSE97763. ChIP-seq datasets: GSE97761, RNA-seq datasets: GSE97762.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ines Mueller and Daniela Wolter for superb technical assistance; Collaborative Research Center (CRC) (Sonderforschungsbereiche (SFB)) 1002 service units (S01 Disease Models for echocardiography measurements and analysis; S02 High resolution fluorescence microscopy for advancing cell staining and cytoskeleton analysis and INF Information infrastructure for providing platform structure) and Dr. Gabriela Salinas (Head of Transcriptome and Genome Analysis Laboratory, University of Goettingen) for advices on RNAseq. The authors thank Prof. W.-H. Zimmermann (WHZ) for revising the article and providing helpful advice. The founders had no role in study design, data collection, analysis and/or decision to publish.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [ZE900-3 to L.C.Z.]; CRC 1002 [Project C07 to L.C.Z. and A11 to T.H.F.]; German Center for Cardiovascular Disease (DZHK). Funding for open access charge: Internal institutional funding (UMG).
Conflict of interest statement. None declared.
REFERENCES
- 1. van Amerongen R., Nusse R.. Towards an integrated view of Wnt signaling in development. Development. 2009; 136:3205–3214. [DOI] [PubMed] [Google Scholar]
- 2. Ozhan G., Weidinger G.. Wnt/beta-catenin signaling in heart regeneration. Cell Regeneration. 2015; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gessert S., Kuhl M.. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 2010; 107:186–199. [DOI] [PubMed] [Google Scholar]
- 4. Hermans K.C., Blankesteijn W.M.. Wnt signaling in cardiac disease. Comprehensive Physiol. 2015; 5:1183–1209. [DOI] [PubMed] [Google Scholar]
- 5. van de Schans V.A., Smits J.F., Blankesteijn W.M.. The Wnt/frizzled pathway in cardiovascular development and disease: friend or foe. Eur. J. Pharmacol. 2008; 585:338–345. [DOI] [PubMed] [Google Scholar]
- 6. Nakagawa A., Naito A.T., Sumida T., Nomura S., Shibamoto M., Higo T., Okada K., Sakai T., Hashimoto A., Kuramoto Y. et al. . Activation of endothelial beta-catenin signaling induces heart failure. Scientific Rep. 2016; 6:25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hou N., Ye B., Li X., Margulies K.B., Xu H., Wang X., Li F.. Transcription factor 7-like 2 mediates canonical Wnt/beta-catenin signaling and c-Myc upregulation in heart failure. Circ. Heart Failure. 2016; 9:e003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dawson K., Aflaki M., Nattel S.. Role of the Wnt-Frizzled system in cardiac pathophysiology: a rapidly developing, poorly understood area with enormous potential. J. Physiol. 2013; 591:1409–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van de Schans V.A., van den Borne S.W., Strzelecka A.E., Janssen B.J., van der Velden J.L., Langen R.C., Wynshaw-Boris A., Smits J.F., Blankesteijn W.M.. Interruption of Wnt signaling attenuates the onset of pressure overload-induced cardiac hypertrophy. Hypertension. 2007; 49:473–480. [DOI] [PubMed] [Google Scholar]
- 10. Xiang F.L., Fang M., Yutzey K.E.. Loss of beta-catenin in resident cardiac fibroblasts attenuates fibrosis induced by pressure overload in mice. Nat. Commun. 2017; 8:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen X., Shevtsov S.P., Hsich E., Cui L., Haq S., Aronovitz M., Kerkelä R., Molkentin J.D., Liao R., Salomon R.N. et al. . The β-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Mol. Cell. Biol. 2006; 26:4462–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hatzis P., van der Flier L.G., van Driel M.A., Guryev V., Nielsen F., Denissov S., Nijman I.J., Koster J., Santo E.E., Welboren W. et al. . Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008; 28:2732–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cadigan K.M. Wnt-beta-catenin signaling. Curr. Biol.: CB. 2008; 18:R943–R947. [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Shao Y., Tong Y., Shen T., Zhang J., Li Y., Gu H., Li F.. Nucleo-cytoplasmic shuttling of PAK4 modulates beta-catenin intracellular translocation and signaling. Biochim. Biophys. Acta. 2012; 1823:465–475. [DOI] [PubMed] [Google Scholar]
- 15. Hino S., Tanji C., Nakayama K.I., Kikuchi A.. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005; 25:9063–9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon M.D., Nusse R.. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 2006; 281:22429–22433. [DOI] [PubMed] [Google Scholar]
- 17. van Veelen W., Le N.H., Helvensteijn W., Blonden L., Theeuwes M., Bakker E.R., Franken P.F., van Gurp L., Meijlink F., van der Valk M.A. et al. . Beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011; 60:1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nusse R. Wnt signaling in disease and in development. Cell Res. 2005; 15:28–32. [DOI] [PubMed] [Google Scholar]
- 19. Xin N., Benchabane H., Tian A., Nguyen K., Klofas L., Ahmed Y.. Erect wing facilitates context-dependent Wnt/wingless signaling by recruiting the cell-specific Armadillo-TCF adaptor earthbound to chromatin. Development. 2011; 138:4955–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Papait R., Cattaneo P., Kunderfranco P., Greco C., Carullo P., Guffanti A., Vigano V., Stirparo G.G., Latronico M.V., Hasenfuss G. et al. . Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20164–20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frietze S., Wang R., Yao L., Tak Y.G., Ye Z., Gaddis M., Witt H., Farnham P.J., Jin V.X.. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012; 13:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Norton L., Chen X., Fourcaudot M., Acharya N.K., DeFronzo R.A., Heikkinen S.. The mechanisms of genome-wide target gene regulation by TCF7L2 in liver cells. Nucleic Acids Res. 2014; 42:13646–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mosimann C., Hausmann G., Basler K.. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 2009; 10:276–286. [DOI] [PubMed] [Google Scholar]
- 24. Sohal D.S., Nghiem M., Crackower M.A., Witt S.A., Kimball T.R., Tymitz K.M., Penninger J.M., Molkentin J.D.. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001; 89:20–25. [DOI] [PubMed] [Google Scholar]
- 25. Harada N., Tamai Y., Ishikawa T.-o., Sauer B., Takaku K., Oshima M., Taketo M.M.. Intestinal polyposis in mice with a dominant stable mutation of the beta -catenin gene. EMBO J. 1999; 18:5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tiburcy M., Hudson J.E., Balfanz P., Schlick S., Meyer T., Chang Liao M.L., Levent E., Raad F., Zeidler S., Wingender E. et al. . Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation. 2017; 135:1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pages F., Trajanoski Z., Galon J.. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010; doi:10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L., Prosdocimo D.A., Bai X., Fu C., Zhang R., Campbell F., Liao X., Coller J., Jain M.K.. KLF15 establishes the landscape of diurnal expression in the heart. Cell Rep. 2015; 13:2368–2375. [DOI] [PubMed] [Google Scholar]
- 34. Valenta T., Hausmann G., Basler K.. The many faces and functions of beta-catenin. EMBO J. 2012; 31:2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sipido K.R., Volders P.G., Vos M.A., Verdonck F.. Altered Na/Ca exchange activity in cardiac hypertrophy and heart failure: a new target for therapy. Cardiovasc. Res. 2002; 53:782–805. [DOI] [PubMed] [Google Scholar]
- 36. Cohen E.D., Tian Y., Morrisey E.E.. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008; 135:789–798. [DOI] [PubMed] [Google Scholar]
- 37. Ai D., Fu X., Wang J., Lu M.F., Chen L., Baldini A., Klein W.H., Martin J.F.. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Senyo S.E., Lee R.T., Kuhn B.. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014; 13:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahuja P., Sdek P., MacLellan W.R.. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol. Rev. 2007; 87:521–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kubin T., Poling J., Kostin S., Gajawada P., Hein S., Rees W., Wietelmann A., Tanaka M., Lorchner H., Schimanski S. et al. . Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell. 2011; 9:420–432. [DOI] [PubMed] [Google Scholar]
- 41. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. . Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dong M., Liao J.K., Fang F., Lee A.P., Yan B.P., Liu M., Yu C.M.. Increased Rho kinase activity in congestive heart failure. Eur. J. Heart Failure. 2012; 14:965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dirkx E., da Costa Martins P.A., De Windt L.J.. Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta. 2013; 1832:2414–2424. [DOI] [PubMed] [Google Scholar]
- 44. Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P.D., Godard C., Moldes M., Burnol A.F., Dubuquoy C. et al. . T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014; 59:2344–2357. [DOI] [PubMed] [Google Scholar]
- 45. He A., Kong S.W., Ma Q., Pu W.T.. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van den Boogaard M., Wong L.Y., Tessadori F., Bakker M.L., Dreizehnter L.K., Wakker V., Bezzina C.R., ’t Hoen P.A., Bakkers J., Barnett P. et al. . Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Invest. 2012; 122:2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noack C., Zafiriou M.P., Schaeffer H.J., Renger A., Pavlova E., Dietz R., Zimmermann W.H., Bergmann M.W., Zelarayan L.C.. Krueppel-like factor 15 regulates Wnt/beta-catenin transcription and controls cardiac progenitor cell fate in the postnatal heart. EMBO Mol. Med. 2012; 4:992–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hosogane M., Funayama R., Shirota M., Nakayama K.. Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell reports. 2016; 16:696–706. [DOI] [PubMed] [Google Scholar]
- 49. Lutz M., Burke L.J., Barreto G., Goeman F., Greb H., Arnold R., Schultheiss H., Brehm A., Kouzarides T., Lobanenkov V. et al. . Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000; 28:1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sequeira V., Nijenkamp L.L., Regan J.A., van der Velden J.. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim. Biophys. Acta. 2014; 1838:700–722. [DOI] [PubMed] [Google Scholar]
- 51. Liu Z., Yue S., Chen X., Kubin T., Braun T.. Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ. Res. 2010; 106:1498–1506. [DOI] [PubMed] [Google Scholar]
- 52. Tseng A.S., Engel F.B., Keating M.T.. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem. Biol. 2006; 13:957–963. [DOI] [PubMed] [Google Scholar]
- 53. Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A.. Transient regenerative potential of the neonatal mouse heart. Science. 2011; 331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quaife-Ryan G.A., Sim C.B., Ziemann M., Kaspi A., Rafehi H., Ramialison M., El-Osta A., Hudson J.E., Porrello E.R.. Multi-cellular transcriptional analysis of mammalian heart regeneration. Circulation. 2017; 136:1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tian Y., Liu Y., Wang T., Zhou N., Kong J., Chen L., Snitow M., Morley M., Li D., Petrenko N. et al. . A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015; 7:279ra238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parker D.S., Ni Y.Y., Chang J.L., Li J., Cadigan K.M.. Wingless signaling induces widespread chromatin remodeling of target loci. Mol. Cell. Biol. 2008; 28:1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mittal A., Sharma R., Prasad R., Bahl A., Khullar M.. Role of cardiac TBX20 in dilated cardiomyopathy. Mol. Cell. Biochem. 2016; 414:129–136. [DOI] [PubMed] [Google Scholar]
- 58. Bresnick E.H., Katsumura R.K., Lee H.Y., Johnson K.D., Perkin A.S.. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012; 40:5819–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bresnick K.R.K.A.E.H. The GATA factor revolution in hematology. Blood. 2017; 129:2092–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oka T., Maillet M., Watt A.J., Schwartz R.J., Aronow B.J., Duncan S.A., Molkentin J.D.. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 2006; 98:837–845. [DOI] [PubMed] [Google Scholar]
- 61. Bisping E., Ikeda S., Kong S.W., Tarnavski O., Bodyak N., McMullen J.R., Rajagopal S., Son J.K., Ma Q., Springer Z. et al. . Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. He A., Gu F., Hu Y., Ma Q., Ye L.Y., Akiyama J.A., Visel A., Pennacchio L.A., Pu W.T.. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat. Commun. 2014; 5:4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haq S., Michael A., Andreucci M., Bhattacharya K., Dotto P., Walters B., Woodgett J., Kilter H., Force T.. Stabilization of beta -catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4610–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zelarayan L.C., Noack C., Sekkali B., Kmecova J., Gehrke C., Renger A., Zafiriou M.P., van der Nagel R., Dietz R., de Windt L.J. et al. . Beta-Catenin downregulation attenuates ischemic cardiac remodeling through enhanced resident precursor cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19762–19767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data files have been deposited in NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97763) under accession GSE97763. ChIP-seq datasets: GSE97761, RNA-seq datasets: GSE97762.