Abstract
Canonical bacterial transcription activators bind to non-transcribed promoter elements to increase transcription of their target genes. Here we report crystal structures of binary complexes comprising domains of Caulobacter crescentus GcrA, a noncanonical bacterial transcription factor, that support a novel mechanism for transcription activation through the preferential binding of methylated cis-regulatory elements and the promotion of open complex formation through an interaction with region 2 of the principal σ factor, σ70. We present crystal structures of the C-terminal, σ factor-interacting domain (GcrA-SID) in complex with domain 2 of σ70 (σ702), and the N-terminal, DNA-binding domain (GcrA-DBD) in complex with methylated double-stranded DNA (dsDNA). The structures reveal interactions essential for transcription activation and DNA recognition by GcrA. These structures, along with mutational analyses, support a mechanism of transcription activation in which GcrA associates with RNA polymerase (RNAP) prior to promoter binding through GcrA-SID, arming RNAP with a flexible GcrA-DBD. The RNAP–GcrA complex then binds and activates target promoters harboring a methylated GcrA binding site either upstream or downstream of the transcription start site.
INTRODUCTION
Bacterial RNA polymerase (RNAP) is a multi-subunit (2α, β, β′, ω) enzyme responsible for decoding genetic information and transcribing DNA into RNA. RNAP core enzyme forms the holoenzyme by associating with a σ factor, which can then initiate promoter-specific transcription. The principal sigma factor, such as σ70 in Escherichia coli, contains five conserved domains (σ1, σ2, σ3, σ3/σ4 linker and σ4) and a non-conserved region (σNCR) between σ1.2 and σ2.1 (1). During transcription initiation, RNAP-σ70 holoenzyme binds the –35 and –10 promoter elements and forms the RNAP-promoter closed complex (RPc). Subsequently, σ70 unwinds ∼13 bp of promoter DNA to form the transcription bubble (2,3). The resulting RNAP-promoter open complex (RPo) is catalytically competent and capable of initiating RNA synthesis de novo (4). The transcription activity of a promoter can be modulated at each step of initiation by transcription activators (5), initiation factors (6,7), NTP concentrations (8), and small metabolites (i.e. ppGpp) (9).
Canonical bacterial transcription activators of the σ70-RNAP holoenzyme increase transcription from promoters by one of the three different mechanisms: class I activation, class II activation (10) or a conformational change of promoter DNA (5,11). During class I activation, a transcription activator binds to its cognate DNA sequence upstream of the promoter –35 element and recruits RNA polymerase via an interaction with the C-terminal domain of RNAP α subunit (12). During class II activation, a transcription activator binds to its binding site overlapping the promoter -35 element and then recruits RNAP via an interaction with domain 4 of the σ factor (σ4) and RNAP α subunit (13). In addition to these two classes of transcription activators, some proteins in the MerR family can interact with a region between the –35 and –10 elements of the promoter and alter the DNA structure to promote binding of RNA polymerase (14).
Although canonical bacterial transcription activators increase promoter activity by distinct mechanisms, they share common characteristics in terms of protein oligomerization and their DNA binding sites. Most activators consist of a DNA-binding domain and an activating domain, the latter usually responsible for receiving regulatory signals and interacting with RNAP. Activators typically dimerize through a large interface formed between two domains and then interact with their cognate DNA in a dimeric form (5), recognizing DNA sequences containing two nearly identical half-binding sites organized as direct, inverted, or palindromic repeats. Importantly, the binding sites are usually located in untranscribed promoter regions (upstream of the transcriptional start site (TSS) or upstream of –10 elements in most cases) to avoid steric clashes between RNAP and their activators.
In recent years, a few novel bacterial transcription factors have been discovered. They function as monomers and activate transcription by either interacting with σ2 (e.g. RbpA) (5) or RNAP β subunit (e.g. CarD) (6). They regulate almost all active promoters, thereby functioning as general transcription initiation factors.
GcrA is an important transcription factor in Caulobacter crescentus. It accumulates at the G1-S transition and is required for proper cell cycle progression. Loss of GcrA uncouples DNA replication and cell division, resulting in elongated cells with extra chromosomes, eventually leading to cell death in rich medium (15,16). Results from ChIP-seq and DNA microarray studies showed that GcrA directly activates expression of ∼200 genes important for cell cycle progression (15–17).
GcrA is a small protein of 173 residues comprising a N-terminal DNA binding domain (GcrA-DBD, residues 1–45), a C-terminal domain mediating interaction with σ2 of the principal sigma factor (GcrA-SID, residues 108–173), and an unstructured linker (residues 46–107) connecting the two domains. We previously showed that GcrA activates transcription of its regulon in a fundamentally different manner compared with canonical bacterial transcription factors, as (i) GcrA tightly interacts with RNAP-σ70 before RNAP binds promoters, (ii) GcrA binds σ702 not σ704; (iii) GcrA recognizes a subset of N6-adenine methylated (m6A) GANTC sites (consensus sequence YGAKTCG), which are usually methylated by the CcrM DNA methyltransferase (18) and (iv) GcrA binds to promoters harboring these sequences either up- or down-stream of transcriptional start sites to promote transcription (16). However, the structural basis and detailed molecular mechanism by which GcrA activates transcription remain elusive.
In this work, we determined the X-ray crystal structures of two binary complexes, GcrA-SID/σ702 and GcrA-DBD/m6A-DNA. The structure of GcrA-SID/σ702 reveals that GcrA-SID adopts a unique fold and forms a large interface with σ702, explaining its tight interaction with RNAP. The structures of GcrA-DBD/m6A-DNA indicate that GcrA-DBD employs two shallow pockets to read methyl groups on DNA, and conserved residues to recognize its DNA consensus sequence. This work supports a model in which GcrA activates transcription through a novel pre-recruitment pathway, with GcrA binding to RNAP before promoter recognition via its C-terminal SID. GcrA bound to σ70 equips RNAP with an extra, flexible, and sequence-specific N-terminal DBD that allows it to recognize cognate, methylated binding sites at a range of locations to promote the isomerization of RNAP at target promoters.
MATERIALS AND METHODS
Plasmids and primers
The plasmids and primers used in this study were listed in Supplemental Tables S3 and S4.
Bacterial strains and growth conditions
Caulobacter strains were grown in PYE (rich medium) at 30°C unless otherwise noted. Induction from the Pxyl and Pvan promoters was achieved by supplementing media with xylose (0.3%) or vanillate (500 μM), respectively. Antibiotics were used at the following concentrations (liquid; plates), oxytretracycline (1 μg/ml; 2 μg/ml), spectinomycin (25 μg/ml; 100 μg/ml), gentamycin (2.5 μg/ml; 1.25 μg/ml). Escherichia coli strains were grown in LB medium at 37°C unless otherwise indicated; when necessary, media were supplemented with the following antibiotic concentrations (liquid; plates): kanamycin (30 μg/ml; 50 μg/ml), oxytretracycline (12 μg/ml; 12 μg/ml), carbenicillin (50 μg/ml; 100 μg/ml).
Strain and plasmid construction
All pMT375-based plasmids were constructed using restriction sites NdeI and SacI. Point mutations in GcrA were introduced by inverted PCR using the pENTR-gcrA vector as templates. The GcrA mutant sequences were amplified by PCR and ligated into the pMT375 vector. All pKNT25 and PUT18-based plasmids containing σ70 or GcrA point mutants were generated by inverted PCR with primers listed in Supplementary Table S4 using pKNT25-σ70 and pUT18-GcrA as template respectively.
C. crescentus σ70 and σ702
BL21 (DE3) cells carrying pET28b-Ccσ70 or pET21b-TEV-Ccσ702(128–487) were grown to OD600 ∼0.8 at 37°C and induced with 1 mM IPTG for 18 h at 18°C. Cells were harvested by centrifugation at 8000 rpm for 5 min, and lysed in 100 ml buffer A (50 mM Tris pH8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT) at 4°C using a Avestin EmulsiFlex-C5 cell disrupter (Avestin, Inc.). The supernatant was collected by centrifugation at 16 000 rpm for 40 min, and loaded on a 20 ml column packed with 4 ml Ni-NTA resin (SMART Inc.). The resin was washed with 20 ml buffer A containing 20 mM imidazole, and eluted with 40 ml buffer A containing 250 mM imidazole. The 6xHis-tagged Ccσ702 (128–487) was cleaved by TEV protease and further loaded on a Ni-NTA column to remove 6xHis tag and other impurities. The flow-through fraction was collected and further purified by anion-exchange chromatography on a 10/100 Mono Q column (GE Healthcare, Inc.; 160 ml linear gradient of 50 mM to 1 M NaCl in 50 mM Tris–HCl, pH 8.0, 1 mM dithiothreitol and 5% glycerol). Fractions containing σ70 or σ702 were pooled, concentrated, and stored at –80°C. Yields were 8 mg/l, and purities were >95%.
C. crescentus RNAP
The endogenous C. crescentus RNAP core enzyme was purified essentially as in (16). The C. crescentus RNAP holoenzyme was reconstituted by incubating RNAP core and σ70 in a molar ratio of 1:4 and separated on a Superdex S200 column (GE healthcare) in 10 mM Tris, pH 8.0, 50 mM NaCl, 1% glycerol, 1 mM dithiothreitol. The holoenzyme was concentrated, aliquoted and stored at –80°C.
C. crescentus GcrA, GcrA-SID and GcrA-DBD
BL21 (DE3) cells carrying pET28a-TEV-gcrA(1–173), pTolo-EX5-gcrA(88–173) or pET28a-TEV-gcrA(1–45) were grown to OD600 ∼0.8 at 37°C and induced with 1 mM IPTG for 18 h at 18°C. The proteins were purified by a similar procedure as described for Ccσ702.
C. crescentus SeMet C. crescentus σ702
BL21 (DE3) cells carrying pET21b-TEV-Ccσ702 (128–487) were grown in M9 medium supplemented with 50 mg/l SeMet. The protein was expressed and purified by following the procedure for native Ccσ702 (128–487).
GcrA-SID/σ702: complex formation
CcGcrA-SID and Ccσ702 were incubated in a molar ratio of 1:4 at 4°C for 6 h, and then loaded on a Superdex S200 column in 10 mM Tris, pH 8.0, 50 mM NaCl, 1 mM dithiothreitol. The fractions of CcGcrA-SID/Ccσ702 were collected, concentrated to 8 mg/ml, and stored in –80°C.
GcrA-SID/σ702: crystallization, data collection and structure determination
The crystallization trials were performed for GcrA-SID/σ702 complex using commercial screening solutions (Emerald Biosystems, Inc.; Hampton Research, Inc.; and Qiagen, Inc.) and the sitting-drop vapor-diffusion technique (drop: 0.8 μl 8 mg/ml GcrA-SID/σ702 in 10 mM Tris–HCl, pH 8.0, 50 mM NaCl plus 0.8 μl screening solution; 22°C). Crystals appeared within 4 days in 11 conditions. All the conditions were optimized using the hanging-drop vapor-diffusion technique with larger drops (1 μl protein solution plus 1 μl reservoir solution) at 22°C. Rod-shaped crystals grown from reservoir solution A (18% PEG3350, 0.1 M sodium citrate, pH 5.5) with dimensions of 1 mm × 0.3 mm × 0.3 mm were transferred into the reservoir solution A supplemented with 7.5% butanediol and stored in LN2. The diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1, BL18U and BL19U. Data were processed using HKL2000 (19). The structure was determined by the Se-SAD method, using PHENIX AutoSol (20). The structure model was automatically built using Buccaneer (21), followed by manual model building using COOT (22) and structural refinement using PHENIX (20). The final model of GcrA-SID/σ702 was refined to Rfree and Rwork of 0.258 and 0.224 respectively and deposited in the protein data bank with accession number 5YIX.
GcrA-DBD: crystallization, data collection and crystallization
The crystallization trial of GcrA-DBD was screen as described above using a protein concentration of 10 mg/ml in the drop. Crystal grew out in many conditions. Cubic crystals of GcrA-DBD were grown to 0.3 mm × 0.4 mm × 0.5 mm in reservoir solution B (1.4 M sodium citrate, 1 M HEPES sodium, pH 7.5) after 1 day and transferred in reservoir solution B supplemented with 20% butanediol, and cooled in LN2. The structure of GcrA-DBD was solved by molecular replacement using BALBES (23). The protein model was built and refinement as described above. The final model of GcrA-DBD was refined to Rfree and Rwork of 0.211 and 0.183, respectively, and deposited into the protein data bank with accession number 5YIU.
GcrA-DBD/ m6A-DNA: complex formation and crystallization
The oligodeoxynucleotides for the crystal form 1 ( ‘+’ strand: 5′-CCTGXTTCG-3′; ‘–’ strand: 5′-CCGXATCAG-3′; X = m6A) and for the crystal form 2 (‘+’ strand: 5′-CCCTGXTTCGC-3′; ‘-’ strand: 5′- CGCGXATCAGG-3′; X = m6A) were synthesized from Sangon Biotech. The two strands were mixed at a molar ratio of 1:1 to a final concentration of 1 mM each in annealing buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl), heated to 95°C, and cooled down to 10°C with a 2°C/min rate in a thermocycler (Biorad, Inc.). GcrA-DBD/m6A-DNA complex for crystallization was prepared by mixing 100 μl 1 mM scaffold (in anneal buffer) and 500 μl 0.2 mM GcrA-DBD protein, incubating the mixture at 4°C for 6 h, and then concentrating to 10 mg/ml. Crystallization trials of the complex were screened as described above for the complex of GcrA-SID/σ702. Square bipyramid-shaped crystals of form 1 were grown from reservoir solution D (30% PEG8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M ammonium surface) and cooled in LN2 in reservoir solution D containing 20% glycerol. Cubic crystals of form 2 were grown from reservoir solution C (20% PEG8000, 0.05 M Potassium phosphate monobasic) and cooled in LN2 in reservoir solution C containing 10% butanediol.
GcrA-DBD/m6A-DNA: data collection and structure determination
The diffraction data of GcrA-DBD/m6A-DNA crystals were collected at SSRF beamline BL17U1, BL18U and BL19U, processed with HKL2000. The structure of crystal form 2 was determined by molecular replacement using GcrA-DBD as a searching model. The protein model was built using Coot, and refined using PHENIX. The nucleotides were subsequently built into the Fo – Fc difference map and further refined. The final model of GcrA-DBD/ m6A-DNA (crystal form 2) was refined to Rfree and Rwork of 0.230 and 0.205, respectively and deposited into the protein data bank with accession number 5YIW.
The structure of crystal form 1 was determined by molecular replacement using Phaser MR (24) and one molecule of GcrA-DBD from crystal form 2 as the searching template. The protein and DNA model was built and refined by a similar procedure as described above. The final model of GcrA-DBD/ m6A-DNA (crystal form 1) was refined to Rfree and Rwork of 0.242 and 0.201, respectively and deposited into the protein data bank with accession number 5YIV.
Bacterial two-hybrid assays
The T18/T25 (cAMP-based) bacterial two-hybrid assay was performed as described previously (25). Cells were grown in M63 minimal media supplemented with maltose (0.2%), IPTG (1 mM), carbenicillin, and kanamycin. Saturated overnight cultures were then spotted onto MacConkey agar (40 g/l) plates supplemented with maltose (1%), IPTG (1 mM), and appropriate antibiotics. Plates were incubated at 30°C and pictures taken two days post-incubation.
Open complex formation assay
Reactions were performed at 30°C. Reaction mixes were made in which an individual time point reaction was 15 μl including 8 μl Caulobacter RNAP holoenzyme diluted in transcription buffer (40 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 0.014% Tween-20, 50 μg/ml BSA (Ambion) and 1 mM DTT) and 2 μl GcrA diluted in GcrA storage buffer. A 10 min incubation at 30°C was performed prior to addition of 5 μl of labeled DNA (0.1–0.25 nM) diluted in (10 mM Tris–HCl, pH 8.5) and pre-incubated at 30°C for 10 min. Aliquots (14 μl) were removed at the indicated times and challenged with 2 μl heparin (50 μg/ml final) for 20 s, bound to prewashed nitrocellulose filters (Millipore) and immediately washed with 4 × 1 ml of wash buffer (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA). Filters were then incubated in 4 ml of scintillation liquid (National Diagnostics) and counts were read on a scintillation counter Tri-Carb 2910 TR (Perkin Elmer).
In vitro transcription assay
Templates for in vitro transcription assay were amplified by PCR (see Supplementary Table S3 for primer sequences) and methylated as described in (16). Reactions mixtures (20 μl) containing 40 nM Caulobacter RNAP holoenzyme, 250 nM GcrA or GcrA mutants, 100 μM GTP, 100 μM ATP, 100 μM CTP and 100 μM [α-32P]UTP (0.89 Bq/fmol) were pre-incubated at 30°C for 15 min in transcription buffer (40 mM Tris–HCl, pH 8.0, 75 mM NaCl, 5 mM MgCl2, 12.5 % glycerol, 2.5 mM DTT). The reactions were initiated by addition of promoter DNA (40 nM final), incubated at 30°C for 20 min, and terminated by addition of 5 μl stop buffer (8 M urea, 20 mM EDTA, 0.025% xylene cyanol, and 0.025% bromophenol blue). The reaction products were heated at 95°C for 2 min, transferred to ice for 5 min, electrophoresed on 15% TBE-Urea polyacrylamide gels, and analyzed by phosphorimaging.
Western blot assay
The western blot assays were performed as described in (16).
RESULTS
Structure of C. crescentus GcrA-SID complexed with region σ702
To provide insight into the mechanism of transcription activation by GcrA, we determined the X-ray crystal structure of the C. crescentus GcrA-SID/σ702 complex to 2.3 Å resolution. We purified GcrA-SID (residues 88–173) and σ702 (residues 128–487) separately (Figure 1A), mixed them together, and then isolated the complex by size exclusion chromatography (Supplemental Figure S1). We obtained crystals of the complex, collected a native dataset at 2.3 Å and a Se-Met dataset at 3.0 Å (Supplementary Table S1), and then solved the structure by experimental phasing from anomalous signals of Se atoms (Supplemental Figure S2A).
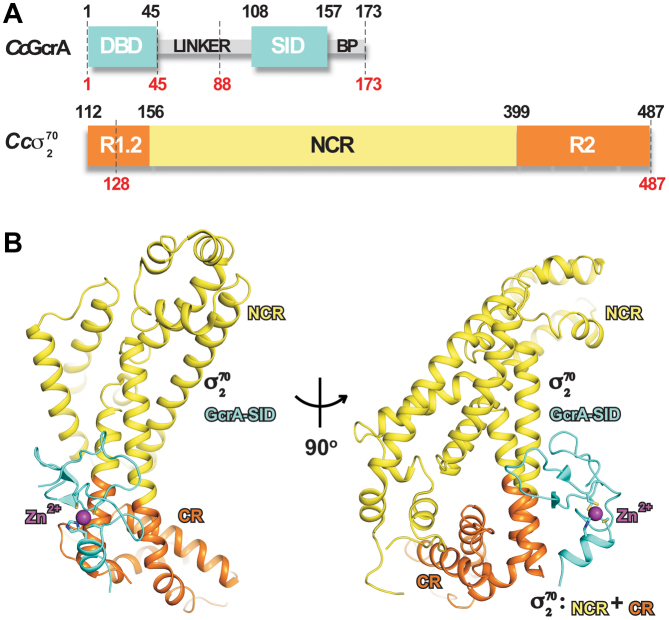
Figure 1.
The crystal structure of GcrA-SID/σ702. (A) The schematic of GcrA and σ702. The regions used for crystallization are indicated by dashes. (B) Front and side views of the crystal structure of GcrA-SID/σ702. GcrA-DBD and GcrA-SID, cyan; the internal linker and C-terminal basic patch (BP), grey; the conserved regions of σ702 (region 1.2 and 2), orange; the non-conserved region of σ702, yellow. The zinc atom is shown as sphere and colored in purple. The zinc coordination residues are showed as sticks.
The crystal structure contains one molecule of GcrA-SID and one molecule of σ702 in an asymmetric unit (Figure 1B), suggesting that GcrA-SID interacts in a 1:1 molar ratio with σ702. The electron density map reveals clear density for 48 residues (residues 105–152) of GcrA-SID (Supplemental Figure S2A), and 330 residues (residues 128–198 and 229–487) of σ702. The 17 residues (residues 88–104) of the linker region and the C-terminal basic patch (residues 157–173) of GcrA appear disordered in the structure (Figure 1A) although they were included during crystallization, consistent with the structural prediction that these two regions of GcrA are unstructured. Residues 199–228 of σ702 are not visible in the structure either. This region, which corresponds to the acidic loop in E. coli σ70, contains multiple acidic residues and is also disordered in the crystal structure (Supplemental Figure S2C and D) (26).
The GcrA-SID comprises mainly loops and a very short helix at the C-terminus. The loops are intertwined and stabilized by a CCCH zinc finger motif (C-X14-C-X9-C-X2-H) (Figure 1B and Supplemental Figure S2B). A structural similarity search on the Dali server (27) found no similar fold in bacterial proteins, and only one poor match with a human ubiquitin–protein ligase (Z score of 2.6; PDB: 4LJP), suggesting that GcrA-SID adopts a novel fold for CCCH type zinc-finger domains. The zinc-coordinating residues C118, C133, C143 and H146 are well conserved in GcrA homologs and essential for its interaction with σ70 (Figure 2C and D). GcrA-SID mutants bearing alanine substitution of any of these residues failed to interact with σ702 (Figure 2C) (16), probably due to misfolding of the protein without a correct zinc finger.
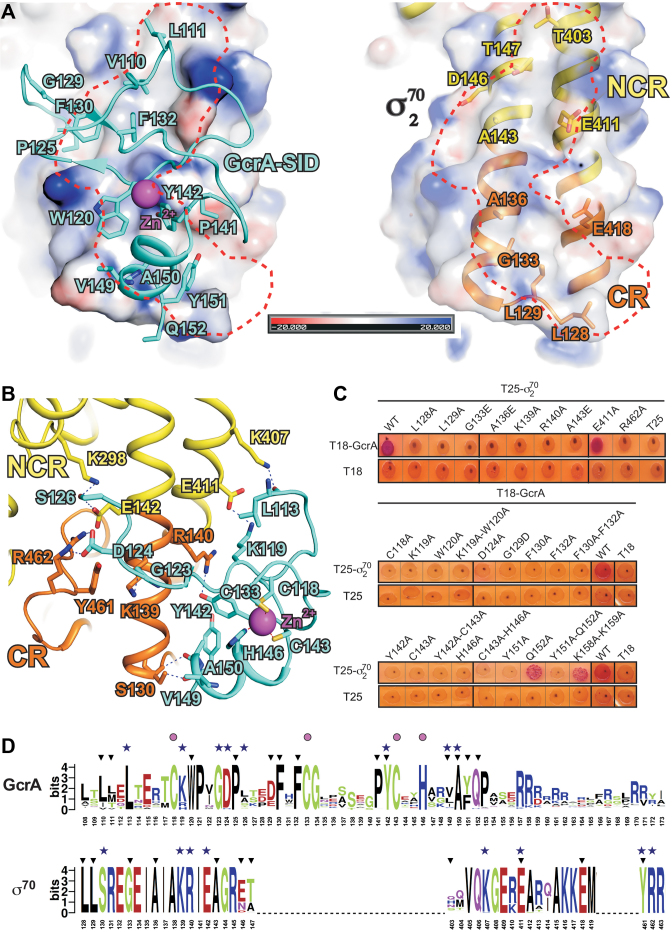
Figure 2.
The interactions between GcrA-SID and σ702. (A) Electrostatic potential surface representation of σ702 and the hydrophobic interactions on the interface. The electrostatic surface of σ702 was generated using the APBS tools in Pymol, with partial charges determined by the PDB2PQR server. Left, a shallow hydrophobic groove (red dashes) on the surface of σ702 and the residues of GcrA inserting into the groove; right, the residues of σ702 creating the hydrophobic groove. Surface with partial negative charges, red; surface with partial positive charges, blue; carbon, oxygen, and nitrogen atoms of GcrA-SID, cyan, red and blue; carbon, oxygen, and nitrogen atoms of σ702, yellow, red, and blue; H-bonds, blue dashes. (B) The polar interactions between GcrA-SID and σ702. Colors are as described above. (C) Bacterial-two hybrid for the interactions between WT and derivatives of GcrA-SID or σ702. (D) Alignments of ∼1000 Sequences of GcrA-SID and σ702 homologs. The sequences were extracted from UniProt Database by BLAST. The alignment was performed by Clustal Omega and the sequence logos were generated on the WebLogo server (http://weblogo.berkeley.edu/logo.cgi) (38). Black filled triangles, residues involved in hydrophobic interactions; blue filled stars, residues forming H-bonds; purple filled circles, residues coordinating the zinc atom.
C. crescentus σ702 is composed mainly of α-helices, similar to the reported crystal structures of σ70 from other bacterial species (26,28,29). The conserved regions of σ702 (σR1.2, σR2.1-σR2.4) form a structural unit, which are present in all principal σ factors, whereas the non-conserved region (NCR) folds into an independent unit, and connects to the conserved domain (CR) through a coiled-coil bridge.
The molecular interface of GcrA-SID and σ702
The GcrA-SID contacts σ702 through a large interface of ∼820 Å2, consisting mainly of hydrophobic interactions, as well as a few H-bond contacts. GcrA-SID inserts several hydrophobic residues (L110, L113, W120, P125, F130, F132, Y142, V149, A150 and Y151; Figure 2A) into a shallow groove created by residues (L128, L129, G133, A136, A143, D146, T147, T403, E411 and E418) on the coiled-coil bridging the CR and NCR of σ702 (Figure 2A). GcrA-SID also makes a few H-bond interactions with σ702 (Figure 2B) with residues L113, K119, G123, D124, S126, Y142, V149 and A150 of GcrA-SID interacting with residues S130, K139, R140, E142, K298, K407, E411, Y461 and R462 of σ702.
To determine which residues are essential for the GcrA-SID/σ702 interaction, we performed bacterial-two hybrid analyses with individual alanine-substitutions of interfacial residues in both GcrA-SID and σ70. In this system, GcrA-SID and σ70 are fused to the T18 and T25 fragments of adenylate cyclase respectively, with an interaction leading to the production of cAMP, which activates a reporter gene that produces red colonies. The results show that substitutions in several GcrA residues (W120A, G129D, F130A, F132A, Y142A and Y151A) contributing to the hydrophobic interface between the two proteins disrupted the interaction (Figure 2C). As the residues mutated are solvent-exposed, the diminished interaction likely results from a direct disruption of the interface, although we cannot formally rule out global unfolding of the mutant proteins in this assay. Similarly, mutating most of the σ702 residues forming the hydrophobic surface that interacts with GcrA-SID (L128A, L129A, G133E, A136E and A143R) also diminished the interaction measured by bacterial two-hybrid (Figure 2C). Alanine mutations of most polar residues (K119A, Y142A, and D124A of GcrA; K139A, R140A, R462A of σ70) forming H-bonds and salt bridges also impaired complex formation.
An alignment of ∼1000 GcrA homologs suggests that GcrA-SID is a generally well conserved domain and that most residues responsible for interacting with σ702 are highly conserved (Figure 2D), implying that GcrA probably contacts the principal σ factor in those organisms in a similar manner as GcrA in C. crescentus. We also aligned ∼1000 homologs of the principal σ factor from the same group of bacteria. Not surprisingly given the importance of the principal σ factor, this alignment indicated extremely high sequence identity (Figure 2D). Notably the residues of C. crescentus σ702 responsible for contacting GcrA are almost identical in the aligned σ70 sequences.
The crystal structure explains why GcrA interacts specifically with σ70 and not with other σ factors (16). The GcrA-contacting surface of σ70 is located on the surface of the coiled-coil bridge, which is composed of residues from both the CR (σR1.2 and σR2.1; orange) and the NCR (Figure 2B, Supplemental Figure S3A-B; yellow). Of the 19 predicted σ factors in C. crescentus, only σ70 and σ32 contain σR1.2 and σNCR (the other σ factors include 16 ECF σ factors and 1 σ54 factor). A protein sequence alignment of σ70 and σ32 reveals several residues essential for contacting GcrA in σ70 that are not conserved in σ32 (Supplemental Figure S3B), which explains the inability of GcrA to bind σ32 and further confirms that residues from both the conserved and non-conserved regions of σ702 are critical for GcrA binding.
Structures of GcrA-DBD and GcrA-DBD bound to methylated DNA
To understand the structural basis for GcrA's DNA binding specificity to N6-adenine methylated sites with a consensus of YGAKTCG (16), we determined a crystal structure of GcrA-DBD alone and two crystal structures of GcrA-DBD bound to its methylated DNA consensus sequence (Supplementary Table S2). The structure of GcrA-DBD alone has one molecule in the asymmetric unit, folding into a compact, three-helix bundle (Figure 3A). A search for homologous structures surprisingly found σ4 of bacterial principal σ factors, the domain that normally binds the -35 promoter element, among the most-related structures (Z score of 5.4). A sequence alignment and three-dimensional structure superimposition of GcrA-DBD and T. aquaticus σ704 revealed 23.8% sequence identity and ∼1.33 RMSD (Cα atoms) between the two domains (Supplemental Figure S4H–G), respectively, suggesting that the two domains might interact with DNA in a similar manner.
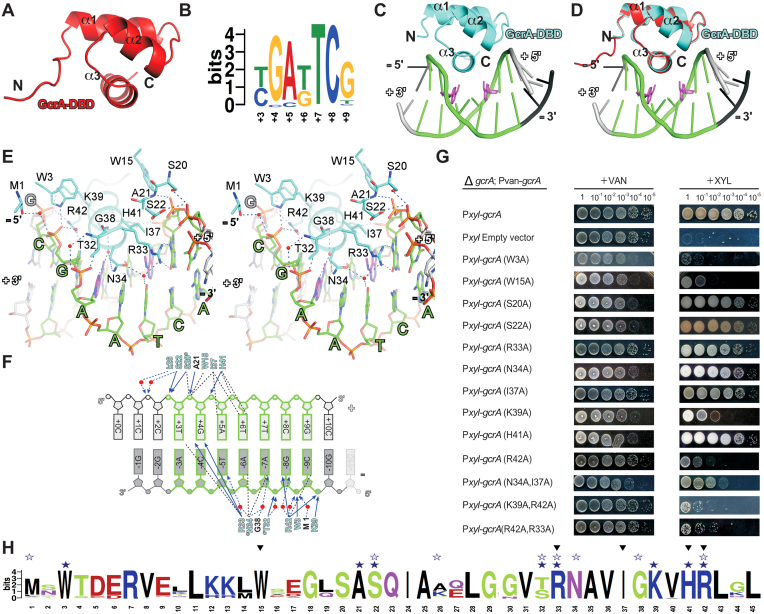
Figure 3.
The sequence-specific recognition of its cognate DNA by GcrA-DBD. (A) The crystal structure of GcrA-DBD. (B) The sequence logo of GcrA consensus sequences adapted from (16). (C) The crystal structure of GcrA-DBD in complex with m6A-DNA. GcrA, cyan; ‘+’ strand of DNA, gray; ‘–’ strand of DNA, black; the consensus motif, green; methylated adenines, purple. (D) Superimposition of GcrA-DBD/m6A–DNA binary complex structure and GcrA-DBD apo structure (red). Colors are as above. (E) A stereo presentation of the interactions between GcrA-DBD and DNA. Carbon, oxygen, nitrogen and phosphorus atoms of consensus DNA motif, green, red, blue and orange; water molecules, red spheres. The other colors are the same as above. (F) The summary of interactions between DNA and GcrA-DBD. Residues contacting DNA with main-chain atoms, black letters; residues contacting DNA with side-chain atoms, cyan letters; direct H-bonds, blue solid arrows; water-mediated H-bonds, dashed arrows; Van der Waals interactions and stacking interactions, black dashed lines; water molecules, red spheres. (G) The in vivo complementation experiments showing the effect on C. crescentus growth of GcrA mutants. C. crescentus cells (ΔgcrA; Pvan-gcrA; Pxyl-gcrA-mutant) growing on vanillate plates (inducing expression of wild-type GcrA) serve as controls and C. crescentus cells (ΔgcrA; Pvan-gcrA; Pxyl-gcrA-mutant) growing on xylose plates (inducing expression of GcrA mutants) serve as experiment groups. (H) Alignment of GcrA-DBD sequence from the ∼1000 GcrA homologs. Black filled triangles, residues involved in hydrophobic interactions; blue filled stars, residues forming direct H-bonds; blue open stars, residues forming water-mediated H-bonds.
We also obtained crystal structures of GcrA-DBD bound to methylated DNA fragments (10 and 11 bp long) containing the sequence TGATTCG (hereafter referred to as m6A-DNA; Figure 3B) in two different crystal forms (Supplementary Table S2 and Figure S4A and B). The first crystal form contains four GcrA-DBD molecules and four dsDNA molecules in an asymmetric unit, in which each GcrA-DBD molecule binds to one dsDNA molecule in an essentially identical manner (Supplemental Figure S4A). The second crystal form contains three GcrA-DBD molecules and one dsDNA molecule in an asymmetric unit, in which two of the three GcrA-DBD molecules (molecule 1 and 3 in Supplemental Figure S4B) bind to opposite surfaces of the DNA. Importantly, the GcrA–DNA interactions in the first crystal form are also present in the second crystal form (the interaction between molecule 1 of GcrA-DBD and DNA; Supplemental Figure S4C). The fact that we observed the same interactions between GcrA-DBD and DNA in both crystal forms suggests a physiological relevance of this interaction. These results also suggest that GcrA-DBD binds to DNA as a monomer, although the full-length GcrA could bind differently, possibly making two different types of contact, as seen in the second crystal form.
Determinants of DNA sequence recognition by GcrA-DBD
The structure of GcrA-DBD bound to m6A-DNA from crystal form 2 was refined at 1.55 Å and is discussed hereafter because of its higher resolution (Supplementary Table S1). The 2Fo – Fc map contoured at 2σ shows clear, sharp densities for all nucleotides and for the methyl groups on m6A (Supplemental Figure S4D and E).
GcrA-DBD binds to DNA without any conformational change of the protein (∼0.24 Å RMSD of GcrA-DBD Cα atoms between the GcrA-DBD apo structure and the GcrA-DBD/m6A–DNA structure; Figure 3D). The helix-turn-helix of GcrA-DBD (α2 and α3; Figure 3C) binds to the major groove of dsDNA and recognizes the identity of the nucleotides through a few direct H-bonds, stacking interactions, and various Van der Waals interactions with the nucleotide bases. In the structure, bases of nucleotides at all seven (TGATTCG) positions of its consensus sequence (Figure 3B) make direct contact with GcrA-DBD (Figure 3E and F), explaining GcrA’s preference for a subset of GANTC methylation sites: the +3T stacks on the guanidine moiety of R33; the +4G makes two direct H-bonds to R33 with its O6 and N7 atoms; the +5A contacts I37 through Van der Waals interactions and the –5T makes water-mediated H-bonds with R33 and N34; the +6T and –6A make multiple Van der Waals interactions with I37, H41, and N34; the –7A contacts N34 and G38 through Van der Waals interactions and two water-mediated H-bonds; the –8G makes one direct H-bond and one water-mediated H-bond with R42; and the –9C contacts R42 through Van der Waals interactions. Besides the interactions with exposed bases in the major groove, several residues (M1, W3, W15, S20, A21, S22, T32, K39, H41 and R42; Figure 3E) of GcrA also stabilize the dsDNA binding through multiple direct or water-mediated H-bonds to backbone phosphates (Figure 3E and F).
To validate the residues identified from this structure as critical to DNA binding by GcrA, we performed complementation experiments in which we expressed GcrA bearing single or double alanine substitutions in a strain in which the wild-type copy of GcrA can be depleted (16). The results show that alanine substitutions of base-contacting residues (R42A, N34A/I37A and R42A/R33A) or backbone-contacting residues (K39A and K39A/R42A) resulted in impairment or loss of GcrA function while not affecting protein stability (Figure 3G and Supplemental Figure S4I), suggesting an essential role for DNA binding by GcrA in C. crescentus. The contribution of W3 and W15 is uncertain, as the W3A and W15A mutants were destabilized in vivo (Supplemental Figure S4I).
Determinants of m6A recognition by GcrA-DBD
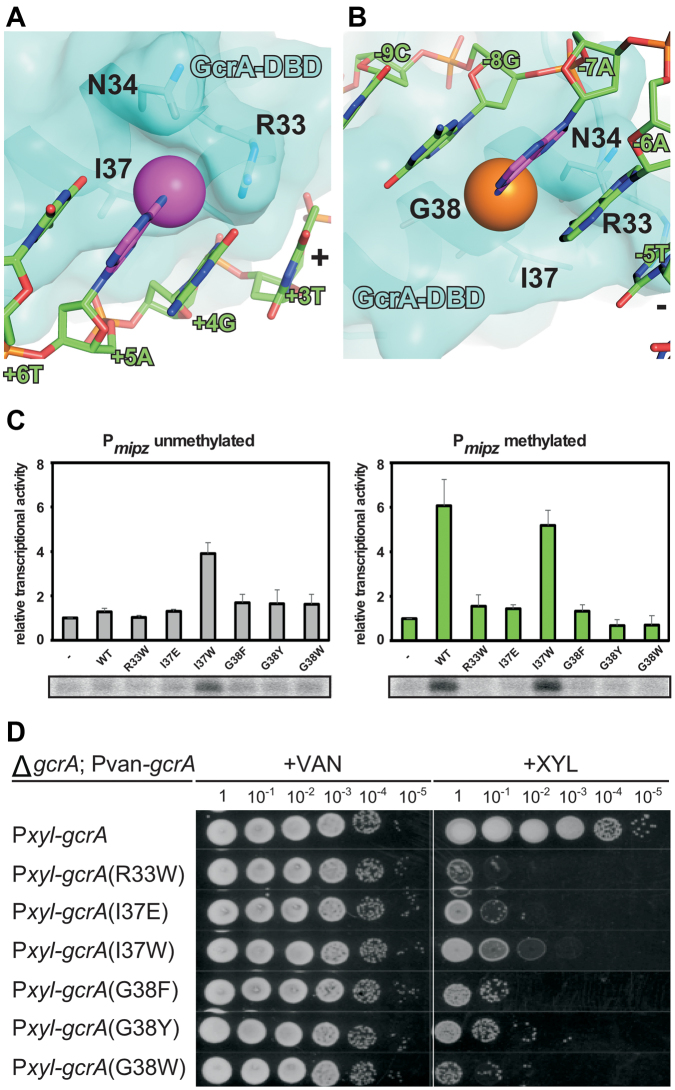
The crystal structure of GcrA-DBD/m6A-DNA also reveals the structural basis for m6A recognition. We discovered two pockets on the GcrA-DBD surface involved in binding methyl groups on both strands of the DNA. The first pocket holding the methyl group on the ‘+’ strand involves I37 on the bottom and N34 and R33 on the sides (Figure 4A), whereas the second shallow pocket holding the methyl group on the ‘–’ strand involves G38 on the bottom and N34 on the side (Figure 4B). To explore how GcrA-DBD binds to non-methylated DNA, we obtained a crystal structure of GcrA-DBD with non-methylated dsDNA at 1.60 Å (Supplementary Table S2). The structure shows that GcrA makes essentially the same interactions with non-methylated DNA as in GcrA-DBD/m6A-DNA except that the two protein pockets for methyl groups are empty (Supplemental Figure S4K–M).
Figure 4.
The methyl-group specific recognition by GcrA-DBD. Surface presentation of GcrA-DBD shows the pockets for methyl groups on ‘+’ (A) or ‘–’ (B) strand of m6A-dsDNA. The colors are as in previous figures. (C) The in vitro transcription experiments showing the effect on transcription activation of GcrA mutants. (D) The in vivo complementation experiments showing the effect on C. crescentus growth of GcrA mutants.
To validate the role of these residues in methyl group recognition and to explore the importance of such recognition for GcrA function, we generated substitutions in the residues forming the methyl-group pockets that should disrupt methyl group recognition. I37E was designed to eliminate the hydrophobic property of the first pocket and R33W, I37W, G38Y and G38W were designed to fill the pockets while not affecting interactions with other atoms of the nucleotide bases. Most mutants showed severely impaired trans-activation activity on methylated promoter DNA in an in vitro transcription assay (Figure 4C). Interestingly, the I37W derivative of GcrA showed substantial trans-activation activity on both methylated and unmethylated promoter DNA. All of the mutants severely impaired viability while not affecting the stability of GcrA, suggesting that R33, I37 and G38 are important for the recognition of m6A DNA (Figure 4C and Supplemental Figure S4J). The fact that the over-active I37W derivative of GcrA also caused reduced cell viability highlights the importance of finely-regulated GcrA activity by DNA methylation in C. crescentus. The alignment of ∼1000 GcrA homologs shows that residues responsible for interacting with DNA are generally highly conserved (Figure 3H). These results also support the notion suggested previously that m6A modifications of chromosomal DNA in C. crescentus is important for cell cycle regulation (30), and that GcrA functions as an m6A reader to activate certain methylated promoters (16).
Transactivation mechanism of GcrA
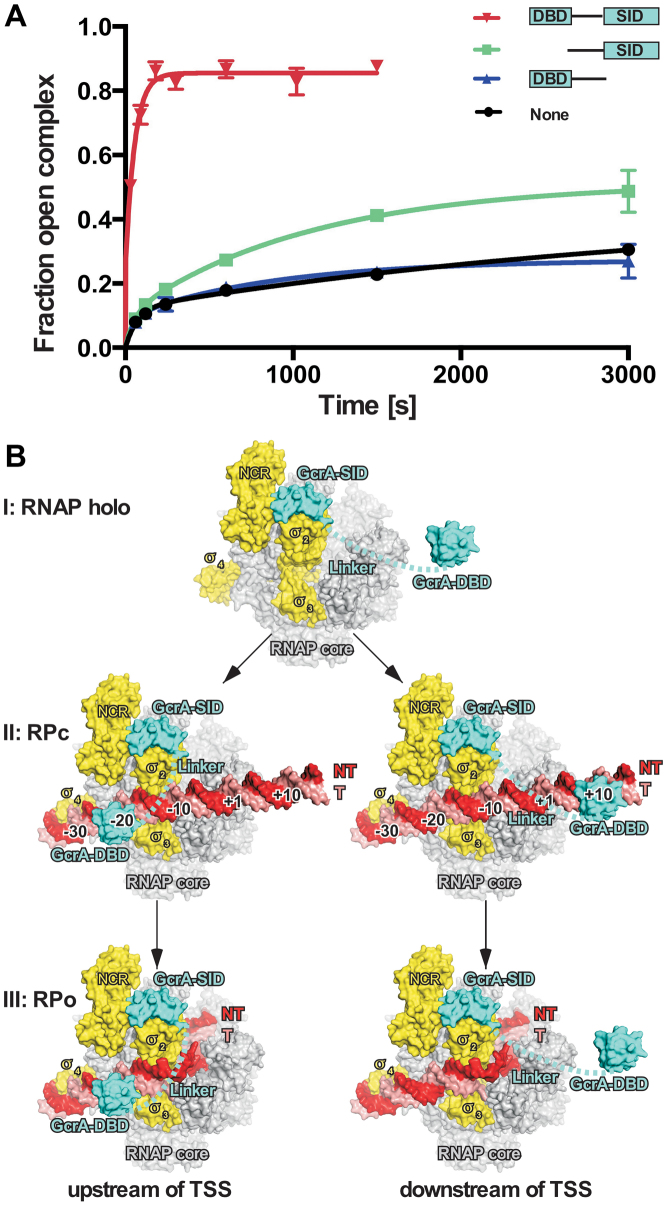
Our structures show that GcrA interacts with σ70-RNAP holoenzyme through its C-terminal SID and binds DNA through its N-terminal DBD. To explore the contribution of the individual domains to transcription activation by GcrA, we measured RPo formation kinetics in the presence of GcrA truncations lacking the DNA binding domain (GcrA-ΔDBD) or the σ70 interacting domain (GcrA-ΔSID). As reported previously, wild-type GcrA strongly increases the rate of promoter isomerization (16) whereas GcrA-ΔSID (residues 1–107) completely loses the ability to promote open complex formation (Figure 5A). Interestingly, GcrA-ΔDBD (residues 46–173) shows a slightly increased rate of promoter melting (Figure 5A), suggesting that the GcrA-SID and domain linker alone are capable of facilitating promoter melting although with relatively low activity.
Figure 5.
The mechanism of transcription activation by GcrA. (A) The kinetics of open complex formation at the methylated mipZ promoter by C. crescentus RNAP holoenzyme in the presence of wild type GcrA or truncated GcrAs. (B) Three-step model of transcription activation by GcrA on two different types of promoters (left, promoters with GcrA binding sites at upstream of TSS; right, promoters with GcrA binding sites at downstream of TSS). GcrA, cyan; promoter DNA nontemplate strand, red; promoter template strand, salmon; σ70, yellow; RNAP core enzyme, gray. Error bars represent SDs. n = 2.
We previously showed that GcrA colocalizes genome-wide with RNAP holoenzyme containing σ70 at virtually all σ70-dependent promoter regions but only activates transcription of ∼200 genes (16). Those findings indicated that GcrA activates transcription through a pre-recruitment pathway in vivo, in which GcrA interacts first with RNAP and then scans the promoter DNA together with RNAP. Based on our structural and biochemical data, we propose that GcrA activates transcription through a three-step mechanism: (i) GcrA forms a tight complex with RNAP through its C-terminal SID prior to the recruitment of RNAP to promoters (the top of Figure 5B); (ii) GcrA-RNAP then preferentially binds to promoter DNA containing a GcrA-binding site (GcrA-DBD is able to reach out to its binding sites located at various positions upstream or downstream of the TSS due to its flexible, unstructured linker) and increases the rate of RPc formation (the middle of Figure 5B); (iii) GcrA facilitates promoter melting and thus increases the rate of RPo formation (the bottom of Figure 5B).
DISCUSSION
We have studied the molecular mechanism of a non-canonical transcription activator GcrA from C. crescentus using structural, biochemical, and genetic approaches. Structure prediction had suggested that GcrA adopts a unique domain architecture composed of two domains (the N-terminal DBD and C-terminal SID) connected by a long unstructured linker. We determined a crystal structure of GcrA-SID in complex with σ702, and two crystal structures of GcrA-DBD in complex with dsDNA containing the methylated (m6A) consensus sequence. The structures reveal that both GcrA-SID/σ702 and GcrA-DBD/m6A–DNA form binary complexes in a 1:1 molar ratio, suggesting that GcrA likely functions as a monomer to activate transcription. A previous report measured the average radius of GcrA by SAXS and suggested that GcrA might exist as a dimer in solution (17). We do not fully exclude the possibility that GcrA can dimerize alone, but our data suggest that GcrA likely functions in a monomeric form to associate with RNAP and promoter DNA.
The distribution of GcrA-binding sites on promoter DNA is distinct from the promoter-locations of both class I and class II activator binding sites. The results presented here and previously suggest that GcrA is able to increase transcription from promoters with GcrA-binding sites located upstream (peaked at –20 and –30) or downstream (peaked at +8 and +18) of TSS (16). The binding sites of class I transcription activators are typically located upstream of the –35 element and interspaced by 10 bp (one helix turn) to ensure that the activator binds on the same surface of promoter DNA for optimal interaction with the CTD of the α subunit (5,11), whereas the binding sites of class II transcription activators usually overlap the –35 element to promote an interaction with σ4 (5,11,13). The distribution of GcrA-binding sites contrasts with both classes of transcription activators. Additionally, GcrA is particularly unusual in that very few transcription factors bind downstream of the TSS to activate transcription, supporting the notion that GcrA activates transcription in a novel manner.
Our models of GcrA-RPc complexes provide an explanation for the unique distribution of GcrA-binding sites (Figure 5). In these models, GcrA-SID associates with σ702, GcrA-DBD binds to promoter DNA, and the flexible linker connects the SID and DBD of GcrA. We found that GcrA-DBD is able to fit into the major groove of the DNA centered at –20 ± 1 and –30 ± 2 without introducing steric clashes with RNAP (Supplemental Figure S5A and B). The limited linker length of GcrA (∼64 residues; ∼180 Å in a fully-stretched state) and the presence of σ704 bound at the –35 element prevent the GcrA-DBD from reaching out to promoter DNA upstream of –35 elements. We also modeled the GcrA-DBD onto several positions downstream of the TSS from +5 to +30, and found that GcrA has less restriction when binding to DNA downstream of the TSS (Supplemental Figure S5C), as the dsDNA has not been loaded into RNAP and is largely solvent-exposed in RPc. The maximum length of the linker (∼180 Å) in a fully stretched conformation allows the GcrA-DBD to extend to ∼+50, in agreement with the previous observation that GcrA-binding sites can be found at locations up to +40 for some GcrA-activated promoters (16). Further efforts are being made to obtain the structure of a functional complex comprising GcrA, RNAP holoenzyme, and promoter DNA, which would provide more insight into the mechanisms by which GcrA affects transcription.
The unique distribution of GcrA-binding sites suggests that GcrA activates transcription in a fundamentally different manner than canonical transcription activators. Transcription activation typically occurs at two steps of transcription initiation: (i) RNAP binds to promoter dsDNA and forms the RNAP-promoter closed complex (RPc); and (ii) RNAP bends dsDNA around the –10 and +3 positions, unwinds ∼13 bp dsDNA, anchors non-template ssDNA onto surfaces of the σ and β subunits, and loads the template ssDNA into its active center to form the open complex (RPo) (4,31). As shown previously (16), GcrA activates transcription by increasing the rates of both steps. GcrA binds much more tightly to RNAP than to DNA (16), indicative of a pre-recruitment mechanism, in which GcrA first associates with RNAP, and then with promoter DNA. After engaging with RNAP, the GcrA-RNAP complex has higher affinity for promoters harboring GcrA-binding sites and increases the rate of RPc formation at these promoters. Our previous data showed that GcrA also accelerated RPo formation, the second step of transcription initiation (16). We propose that GcrA increases the rate of promoter isomerization by stabilizing the RPo complex, and we infer that residues of GcrA-SID and the linker (unresolved in our crystal structures) are probably involved in such interactions (Figure 5A); future work is necessary to dissect how GcrA-SID interacts with and affects promoter melting during the isomerization step of transcription initiation. Several positively charged residues from the internal linker of GcrA might stabilize the phosphate backbone of upstream fork junction in a similar way as with M. tuberculosis RbpA (7). Moreover the C-terminal basic patch (resides 157–173; Figure 1A and Supplemental Figure S5D) of GcrA, mainly comprising positively charged residues, is physically nearby and might directly contact and stabilize the nontemplate ssDNA of the –10 element (Supplemental Figure S5D). GcrA-DBD probably remains associated with DNA during the isomerization steps of transcription initiation if the binding site of GcrA is located upstream of the –10 element (the bottom left of Figure 5B), but has to fall off if the GcrA-DBD binds to regions downstream of TSS (the bottom right of Figure 5B). GcrA has a much weaker affinity for DNA compared to its affinity for σ70, so it is likely that GcrA still associates with RNAP and facilitates promoter isomerization when the GcrA-DBD dissociates from DNA.
The domain σ4 typically serves as a hub for anchoring various bacterial transcription factors and phage proteins to activate transcription but few transcription activators interact with domain σ2. Here we showed that GcrA uses a new structural fold to anchor itself onto σ2 and promote transcription in a new way. RbpA from Mycobacterium tuberculosis (7,32) and GrgA from Chlamydia trachomatis (33) are the only two other proteins reported to interact with σ2. RbpA clamps onto σ2 through two short helices in the C-terminal domain, binds to the RNAP β' zipper domain through its core domain, and stabilizes the backbone of promoter DNA upstream of -10 elements through three positively charged residues of the linker region. It is intriguing that RbpA and GcrA contact the same region on the coiled-coil bridge connecting NCR and CR of σ2 (Supplemental Figure S2E), although the interface of RbpA/σ2 is much smaller. However, RbpA functions as a general initiation factor to activate most primary σ factor-regulated genes in a nonspecific manner by totally different mechanisms than GcrA. GrgA was also reported to interact with both the NCR of σ2 and promoter DNA, but the detailed structural basis of this interaction and the activation mechanism are unknown.
We also identified protein pockets in GcrA that probably read the m6A modification in GcrA-binding sites (Figure 4A and B). Disrupting the pockets by either introducing bulkier side chains or eliminating the hydrophobic property of the pocket severely inhibited C. crescentus cell growth (Figure 4D). The results support the observation that GcrA preferentially activates methylated promoters compared with unmethylated ones (16,17), and are also consistent with the finding that removing either the m6A writer–CcrM (a methyl transferase in C. crescentus responsible for methylation of GANTC sequence) (34), or the m6A reader–GcrA, causes defects in cell cycle progression (16). The recognition of m6A by GcrA is somewhat similar to other DNA m6A reader proteins, such as the methylation-specific restriction endonuclease R.DpnI (35), the oriC binding protein SeqA (36), and the DNA mismatch repair protein MutH (37), all of which use hydrophobic pockets to accommodate the methyl group of m6A in dsDNA.
In summary, we have presented the structural basis for transcription activation by the non-canonical transcription activator GcrA. Our structures reveal that GcrA binds σ702 through its C-terminal domain, and reads the DNA and its methylation states through its N-terminal domain. These interactions are not shared with virtually any other transcription activator characterized to date and account for the unique mechanism of transcription activation by GcrA. GcrA is extremely well-conserved among α-proteobacteria, suggesting a wide distribution of this unique transcription activation mechanism.
DATA AVAILABILITY
The atomic coordinates and structure factors of CcGcrA-DBD, CcGcrA-DBD/m6A–DNA binary complex (crystal form 1), CcGcrA-DBD/m6A-DNA binary complex (crystal form 2), CcGcrA-SID/σ702 binary complex, and CcGcrA-DBD/DNA binary complex have been deposited into the Protein Data Bank with accession code 5YIU, 5YIV, 5YIW, 5YIX and 5Z7I respectively (http://wwpdb.org/).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at beamline BL18U1/BL19U1 of National Center for Protein Science Shanghai (NCPSS), and at beamline BL17U1 of Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank Prof. Zhaocai Zhou for generous gift of plasmid pET28a-TEV and Tolo Biotechnology for generous gift of pTolo-EX vectors. We thank B. Perchuk for assistance with the bacterial two-hybrid and growth assays. We thank Richard H. Ebright for many informative discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
National Natural Science Foundation of China [31670067 to Y.Z.]; Chinese Thousand Talents Program; Leading Science Key Research Program from Chinese Academy of Sciences [QYZDB-SSW-SMC005 to Y.Z.]; Chinese Academy of Sciences pilot B cultivation project [XDPB0402 to Y.Z.]; Howard Hughes Medical Institute International Predoctoral Fellowship (to D.L.H.); National Institutes of Health [R01GM082899 to M.T.L.], who is also an Investigator at the Howard Hughes Medical Institute. Funding for open access charge: National Natural Science Foundation of China [31670067 to Y.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Feklistov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. 2015; 4:e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saecker R.M., Record M.T. Jr, Dehaseth P.L.. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011; 412:754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 6. Bae B., Chen J., Davis E., Leon K., Darst S.A., Campbell E.A.. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. eLife. 2015; 4:e08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S., Darst S.A., Campbell E.A.. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. eLife. 2017; 6:e22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaal T., Bartlett M.S., Ross W., Turnbough C.L. Jr, Gourse R.L.. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997; 278:2092–2097. [DOI] [PubMed] [Google Scholar]
- 9. Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu. Rev. Microbiol. 1975; 29:301–318. [DOI] [PubMed] [Google Scholar]
- 10. Busby S., Ebright R.H.. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 1999; 293:199–213. [DOI] [PubMed] [Google Scholar]
- 11. Decker K.B., Hinton D.M.. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013; 67:113–139. [DOI] [PubMed] [Google Scholar]
- 12. Hudson B.P., Quispe J., Lara-Gonzalez S., Kim Y., Berman H.M., Arnold E., Ebright R.H., Lawson C.L.. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19830–19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Philips S.J., Canalizo-Hernandez M., Yildirim I., Schatz G.C., Mondragon A., O’Halloran T.V.. TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015; 349:877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holtzendorff J., Hung D., Brende P., Reisenauer A., Viollier P.H., McAdams H.H., Shapiro L.. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004; 304:983–987. [DOI] [PubMed] [Google Scholar]
- 16. Haakonsen D.L., Yuan A.H., Laub M.T.. The bacterial cell cycle regulator GcrA is a sigma70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev. 2015; 29:2272–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fioravanti A., Fumeaux C., Mohapatra S.S., Bompard C., Brilli M., Frandi A., Castric V., Villeret V., Viollier P.H., Biondi E.G.. DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genet. 2013; 9:e1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohapatra S.S., Fioravanti A., Biondi E.G.. DNA methylation in Caulobacter and other Alphaproteobacteria during cell cycle progression. Trends Microbiol. 2014; 22:528–535. [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W.. [20]Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 20. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cowtan K. Completion of autobuilt protein models using a database of protein fragments. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 23. Long F., Vagin A.A., Young P., Murshudov G.N.. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008; 64:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karimova G., Pidoux J., Ullmann A., Ladant D.. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murakami K.S., Masuda S., Darst S.A.. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002; 296:1280–1284. [DOI] [PubMed] [Google Scholar]
- 27. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin W., Mandal S., Degen D., Liu Y., Ebright Y.W., Li S., Feng Y., Zhang Y., Jiang Y., Liu S. et al. . Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Mol. Cell. 2017; 66:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murakami K.S. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J. Biol. Chem. 2013; 288:9126–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohapatra S.S., Fioravanti A., Biondi E.G.. DNA methylation in Caulobacter and other Alphaproteobacteria during cell cycle progression. Trends Microbiol. 2014; 22:528–535. [DOI] [PubMed] [Google Scholar]
- 31. Feklistov A., Bae B., Hauver J., Lass-Napiorkowska A., Kalesse M., Glaus F., Altmann K.H., Heyduk T., Landick R., Darst S.A.. RNA polymerase motions during promoter melting. Science. 2017; 356:863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D., Darst S.A., Campbell E.A., Paget M.S.. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7171–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bao X., Nickels B.E., Fan H.. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of sigma66. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16870–16875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzalez D., Kozdon J.B., McAdams H.H., Shapiro L., Collier J.. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 2014; 42:3720–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mierzejewska K., Siwek W., Czapinska H., Kaus-Drobek M., Radlinska M., Skowronek K., Bujnicki J.M., Dadlez M., Bochtler M.. Structural basis of the methylation specificity of R.DpnI. Nucleic Acids Res. 2014; 42:8745–8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chung Y.S., Brendler T., Austin S., Guarne A.. Structural insights into the cooperative binding of SeqA to a tandem GATC repeat. Nucleic Acids Res. 2009; 37:3143–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J.Y., Chang J., Joseph N., Ghirlando R., Rao D.N., Yang W.. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005; 20:155–166. [DOI] [PubMed] [Google Scholar]
- 38. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of CcGcrA-DBD, CcGcrA-DBD/m6A–DNA binary complex (crystal form 1), CcGcrA-DBD/m6A-DNA binary complex (crystal form 2), CcGcrA-SID/σ702 binary complex, and CcGcrA-DBD/DNA binary complex have been deposited into the Protein Data Bank with accession code 5YIU, 5YIV, 5YIW, 5YIX and 5Z7I respectively (http://wwpdb.org/).