Abstract
Archaea and eukaryotes have ribosomal P stalks composed of anchor protein P0 and aP1 homodimers (archaea) or P1•P2 heterodimers (eukaryotes). These P stalks recruit translational GTPases to the GTPase-associated center in ribosomes to provide energy during translation. The C-terminus of the P stalk is known to selectively recognize GTPases. Here we investigated the interaction between the P stalk and elongation factor 2 by determining the structures of Pyrococcus horikoshii EF-2 (PhoEF-2) in the Apo-form, GDP-form, GMPPCP-form (GTP-form), and GMPPCP-form bound with 11 C-terminal residues of P1 (P1C11). Helical structured P1C11 binds to a hydrophobic groove between domain G and subdomain G′ of PhoEF-2, where is completely different from that of aEF-1α in terms of both position and sequence, implying that such interaction characteristic may be requested by how GTPases perform their functions on the ribosome. Combining PhoEF-2 P1-binding assays with a structural comparison of current PhoEF-2 structures and molecular dynamics model of a P1C11-bound GDP form, the conformational changes of the P1C11-binding groove in each form suggest that in response to the translation process, the groove has three states: closed, open, and release for recruiting and releasing GTPases.
INTRODUCTION
In the living cell, the protein synthesis on ribosome requests energy, which is provided by the hydrolysis of guanosine triphosphate (GTP) by GTP binding translation factors (referred to translational GTPases) at GTPase-associated center in the large subunit of ribosome (1–4). This center contains a ribosomal protein complex known as the ‘ribosomal stalk’ (or P stalk in archaea/eukaryotes and L12 stalk in bacteria), which is highly flexible and recruits several translational GTPases to the sarcin-ricin loop (SRL), which is a GTPase binding site on the large ribosomal subunit (5–8). These GTPases include initiation factor 5B (IF5B), elongation factors (EF-2/EF-G and EF-1α/EF-Tu), and release factor 3 (RF3). This ribosomal stalk is functionally and architecturally conserved in all organisms, and is essential to promote the translational metabolic cycles. The stalk consists of an anchor protein (P0 in archaea and eukaryotes, and L10 in bacteria, these two proteins are newly named as uL10 (9)) and arm proteins (multiple copies of the aP1 homodimer in archaea, the P1•P2 heterodimer in eukaryotes, L12 homodimer in bacteria, this L12 also was newly named as bL12 (9)), which are acidic ribosomal proteins (7,10–13). The anchor proteins P0 and L10 have a similar architecture. N-domain (rRNA-anchoring domain) anchors the arm proteins at GTPase-associated center in the large subunit of ribosome, whereas C-domain is a helical spine to bind multiple copies of arm proteins. Each arm protein hands translational GTPases and recruits them to the ribosome, and it was reported that copies of arm protein involve in both efficiency and fidelity of translation (10,14). Although the rRNA-anchoring domains of P0 and L10 have similar structures, the helical spines have different conformations, which are consistent with the structural difference between aP1 (P1•P2) and L12.
In the bacteria, the structure of arm protein L12 is formed by three parts: an N-terminal domain (NTD) for dimerization and binding to helical spine of L10; a globular C-terminal domain (CTD) which is formed by three α-helices and three β-strands, and hands translational GTPases; a long ‘hinge’ region that links the NTD and CTD (15,16). Previous studies showed that the CTD of L12 (V66-V68, K70, L80 and E82) interacts with the functional insertion domain (known as subdomain G') of EF-G (E224 and E228) (17–19).
The archaeal arm proteins are three copies of aP1 homodimer, whereas the eukaryotic arm proteins are two copies of P1•P2 heterodimer (10,20). Sequence identity has shown that aP1 and P1•P2 are evolutionally related. Similar to the bacterial L12, aP1 consists of a dimerization NTD for binding to P0 helical spine, a CTD for handing translational GTPases, and a long flexible hinge between two domains (10,20). However, these domains are completely different from those of L12 in terms of both sequence and structure (21). While the CTD of L12 is composed of about 70 amino acids, the CTD of aP1 (aP1CTD) is formed by 25 amino acids that are predicted to form two α-helices (10). Such structural difference indicates that archaea/eukaryotes and bacteria will also have completely different interaction mechanisms between the stalk and translational GTPases, although the stalk proteins play the same role of recruiting GTPases to the GTPase-associated center in ribosomes (7,10). Moreover, it has been shown that the C-terminal residues L103, L106 and F107 of aP1CTD are essential to interact with GTPases and conserved through archaea and eukaryotes (21–23).
In archaea, although GTP binding domain (domain G) of translational GTPases such as aIF2γ, aIF5B, aEF-1α and aEF-2, share a similar structure, aP1CTD only recognizes aIF5B, aEF-1α and aEF-2, not aIF2γ (23). Furthermore, aEF-2 has subdomain G’ which is a functional insertion domain of the domain G, whereas aIF5B and aEF-1α do not (24,25). Previous studies showed that aP1CTD binds to a groove between domain G and III of aEF-1α (26). Taken all together, these findings indicate that aP1CTD has substrate-recognition specificity with a broad substrate-recognition ability. To recognize various translational GTPases, the aP1CTD needs to adapt to the different sequences and structures of its binding partners. Therefore, it is indispensable to investigate the details of interaction between individual translational GTPases and P1CTD by structural analysis.
To better understand the GTPase-recognition mechanism of aP1CTD, in this study, we determined the crystal structures of aEF-2 in nucleotide-free form (Apo-form), GDP-form, GMPPCP-form (GTP-form) and GMPPCP-form with 11 C-terminal residues of aP1CTD (P1C11). The structures of aEF-2 showed that the aP1C11 was located on a groove formed by domain G and subdomain G' of aEF-2. In addition, a model of aEF-2-GDP complexed with aP1C11 was built by molecular dynamics (MD) simulation based on the structures of GDP-form and GMPPCP-form in presence of aP1CTD. Combining the gel-mobility shift assay and quantitative binding assays between aP1 and aEF-2 variants with previous results of aP1 mutant analyses, we propose an aEF-2 recognition mechanism of P1 when P stalk recruits aEF-2 to the GTPase-associated center of ribosome during the elongation process.
MATERIALS AND METHODS
Construction of plasmids
The plasmid for the expression of Pyrococcus horikoshii P1 (P1) was constructed as described previously (23). The full length (735 residues) of P. horikoshii EF-2 (PhoEF-2) was expressed in Escherichia coli. The gene encoding PhoEF-2 was cloned with a C-terminal hexahistidine tag (His6) into the pET-26M vector. The plasmids of all PhoEF-2 point-mutants (Supplementary Table S1) and a truncated PhoEF-2 which was constructed by domain G, subdomain G' and domain II (amino acids: 1–389 and hereafter referred as PhoEF-2-D2) were constructed by QuikChange methods. Resulted plasmids were transformed into E. coli B834(DE3)-pRARE2. The cells were cultivated in 30 mg/ml Luria-Broth medium containing 15 μg/ml kanamycin, 34 μg/ml chloramphenicol, 1% glucose at 37°C until the optical density at 600 nm (OD600) reached 0.4–0.6. isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM to induce gene expression and cultivation was continued for 20 h at 25°C.
Preparation of proteins and a peptide
P1 was prepared as described previously (23). The harvested recombinant cells expressing PhoEF-2, its mutants or PhoEF-2-D2 were disrupted by sonication and centrifuged at 40 000g for 30 min. After heat-treatment at 70°C for 30 min following centrifugation at 10 000g for 30 min, the supernatant was subjected to 5 ml HisTrap HP (GE Healthcare) and eluted with a gradient of 50–250 mM imidazole in buffer A (20 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM β-mercaptoethanol), and then, purified using Superdex 200 pg 26/60 gel-filtration column (GE Healthcare) equilibrated with buffer B (20 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2, 1 mM DTT). The eluted fractions containing the target were mixtures of two different samples; GDP-form and Apo-form. For obtaining Apo-form, the sample was purified twice using RESOURCE Q anion-exchange column (GE Healthcare), after heat-treating at 70°C for 10 min. Firstly, the sample was injected to the column and eluted with a gradient 5–500 mM KCl in buffer C (20 mM Tris–HCl pH 8.0, 5 mM KCl, 1 mM DTT). The collected fractions were dialyzed in buffer D (20 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT), and then again applied to RESOURCE Q anion-exchange column using same protocol as described above. Purified Apo-form PhoEF-2 was confirmed by mass spectrometry analysis (23). The purification of PhoEF-2-D2 was performed by same protocol of PhoEF-2 except for the last two steps for Apo-form PhoEF-2. All purification experiments were carried at room temperature. Apo-form PhoEF-2 and PhoEF-2-D2 were concentrated to 17 and 13 mg/ml, respectively.
In previous studies, P1-binding affinity analyses of 10 mutants for 12 C-terminal residues (99–108aa) of P1 (1–108aa) were performed (23). The results showed that among them, F107S, L106S, and L103S lose GTPase-binding affinity. Moreover, the structure of PhoEF-1α complexed with P1 C-terminus also showed long peptide of P1C may induce effects from crystal packing. Therefore, we designed to use a short fragment of P1CTD. The peptide of 11 C-terminal amino acids of P1 (hereafter referred as P1C11), residues E98-G108 (EALAGLSALFG), was synthesized and purified by Sigma Aldrich Japan.
Crystallization and data collection
To obtain GTP-form of PhoEF-2, an analogue of GTP, GMPPCP, was added to the purified nucleotide-free PhoEF-2 in a molar ratio of 25:1 of GMPPCP:PhoEF-2 (hereafter referred to as PhoEF-2-GMPPCP) and then heat-treated at 50°C for 30 min (23). Moreover, in order to construct PhoEF-2-GMPPCP in complex with P1C11 (hereafter referred to as PhoEF-2-GMPPCP-P1C11), 0.4 mg of P1C11 powder was added to the PhoEF-2-GMPPCP samples in the ratio 20:1 of P1C11:PhoEF-2-GMPPCP, subsequently the samples were heat-treated at 50°C for 30 min and then centrifuged at 13 000g for 5 min. The PhoEF-2-D2 sample was also heat-treated at 50°C for 30 min before crystallization. The initial crystallization screening of PhoEF-2-GMPPCP, PhoEF-2-GMPPCP-P1C11, and PhoEF-2-D2 was carried out using the sitting-drop method at a small scale (96-well plate) with a PEG (polyethylene glycol) cocktail containing 10% or 20% (wt/vol) of PEG400–10 000 and 0.1 M of various buffers in a pH range of 5.0–8.0.
The initial crystals of PhoEF-2-GMPPCP appeared under several conditions containing (a) 10% PEG6000 and 0.1 M MES–NaOH pH 6.0; (b) 10% PEG3350 and 0.1 M MES–NaOH pH 6.0; (c) 10% PEG8000 and 0.1 M Citric acids pH 5.0. After optimized crystallization conditions, the diffraction-quality crystals of PhoEF-2-GMPPCP were grown up in a reservoir solution containing 6–12% PEG6000 or PEG3350 and 0.1 M MES–NaOH, pH 5.8–6.4. For the X-ray diffraction experiment, the crystals were rapidly soaked through two cryoprotectant reservoir solutions containing 5% glycerol and Paratone-N before flash-cooling under a cryostream. The diffraction data were collected at the beamline NW12A of Photon Factory (PF), Tsukuba, Japan.
The initial crystals of PhoEF-2-GMPPCP-P1C11 appeared with reservoir solution containing 10% PEG8000 or PEG10000, 0.1 M MES–NaOH pH 6.0, but most crystals were multi-crystals. For obtaining high-quality single crystals, we tried to crystallize using a salt-screening kit (NeXtal Stock Kit Salt; QIAGEN) as the additive reagents. As the result, single crystals of PhoEF-2-GMPPCP-P1C11 were grown up within one day using the micro-seeding technique, in which one drop contained a ratio 0.1:1:1 of micro-seed solution: protein: reservoir solution (10% PEG10 000, 0.1 M MES, pH 6.0, 0.5 M lithium acetate dehydrate). A diffraction data set of the PhoEF-2-GMPPCP-P1C11 crystals was collected at the beamline BL44XU of SPring-8 (Harima, Japan) (Proposal No. 2016A6611), after being soaked through cryoprotectant reservoir solutions containing 3.5–14% sucrose (3.5% stepwise) and Paratone-N.
The crystals of PhoEF-2-D2 were grown with reservoir solution containing 10% PEG1000 and 0.1 M MES–NaOH pH 6.0. After rapidly soaking the PhoEF-2-D2 crystals through a reservoir solution containing 25% glycerol, the diffraction data were collected to a resolution of 1.6 Å at the beamline BL-5A of PF.
All data sets were indexed, integrated, scaled, and merged using the XDS program package (27). The statistics for each of these are summarized in Supplementary Table S2.
Structure determination and refinement
All crystals and diffraction data were obtained as description in Supplementary information. The structure of PhoEF-2-GMPPCP was solved at 2.3 Å resolution by molecular replacement using Molrep of the CCP4 suite (28). The structure of Saccharomyces cerevisiae eEF-2 Apo-form (PDB ID: 3B78) (29) was used as a search model. The structure was refined using Phenix.refine (30) and modified manually using Coot (31). After rigid body refinement and several cycles of restrained refinement, we found the electron density maps (Fo – Fc and 2Fo – Fc) of GMPPCP in the GTP binding site. Finally, R and Rfree factors of PhoEF-2-GMPPCP structure were converged to 21.65% and 25.47%, respectively.
3.1 Å resolution structure of PhoEF-2-GMPPCP-P1C11 was determined by molecular replacement using Molrep in the CCP4 suite (28). The refined structure of PhoEF-2-GMPPCP was used as a search model. After several cycles of restrained refinement, both 2Fo – Fc and Fo – Fc map of the P1C11 were appeared, and model of P1C11 was built manually using Coot program (31). The structure of PhoEF-2-GMPPCP-P1C11 was refined using Phenix.refine (30) following manual modification using Coot (31). The R and Rfree factors were then converged to 23.02% and 28.54%, respectively.
Finally, the structure of PhoEF-2-D2 was solved by molecular replacement using Phaser of the Phenix package (32). The structure of domain G, subdomain G' and domain II of the PhoEF-2-GMPPCP-P1C11 (residues 12–386 without GMPPCP and P1C11) as a search model. Similar to structure determination of PhoEF-2-GMPPCP and PhoEF-2-GMPPCP-P1C11, the maps of GDP were appeared at GTP binding site in both 2Fo – Fc and Fo – Fc maps (hereafter referred as PhoEF-2-D2-GDP) after rigid body refinement and several cycles of restrained refinement using Phenix.refine. The structure of PhoEF-2-D2 was refined using the same method as described above. The R and Rfree factors were converged to 17.04% and 19.77%, respectively.
Summaries of refinement are presented in Supplementary Table S2. All figures of structures were drawn using PyMOL (The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC), while surface potential values were produced by APBS (33). Sequence alignments was performed by CLUSTALW (34).
Gel-mobility shift assay
Binding assay between P1 and PhoEF-2 was examined by gel-mobility shift assay using Native-PAGE for qualitative analysis under optimized condition. Each analysis was done one or two times with the condition that P1 was mixed in concentration of 100 pmol with increasing concentrations of 100–400 pmol of individual PhoEF-2 mutants (Supplementary Table S1). The binding buffer contained 20 mM Tris–HCl pH 7.5, 20 mM KCl and 10 mM MgCl2. Each mutant was mixed with GMPPCP in a molar ratio 25:1 of GMPPCP:protein and then heat-treated at 70°C for 10 min before binding assay. Each mutant with buffer D (20 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT) was then added to P1 and the binding buffer, and the mixture was incubated at 70°C for 10 min again. Following incubation, each sample was applied to Native-PAGE gel as described previously (23).
Circular dichroism (CD) spectrometry
Circular dichroism (CD) spectra were collected on a J-805 spectropolarimeter (JASCO, Japan) under an atmosphere of N2 at room temperature in a quartz cell with a path length of 1 mm. The protein samples were dialyzed in buffer E (20 mM Tris–HCl, pH 8.0, 10 mM KCl, 1 mM DTT). The concentration of PhoEF-2 and the its mutants; F226S, L214S/V216S, V198S/L214S/V216S were estimated to 0.1, 0.25, 0.2 and 0.1 mg/ml, respectively by using absorption. The CD spectra were obtained from a wavelength region of 190–300 nm by taking the average of four scans. The molar ellipticity per residues was calculated by (h) = h/(10n/c/l). Here, h is the CD signal in mdeg, n is the number of residues, c is the concentration in mol/l, and l is the length of the cuvette path (cm).
Surface plasmon resonance
The real time measurement of the interaction between P1 and PhoEF-2-GMPPCP or PhoEF-2-GDP was performed using a BIACORE 3000 biosensor system (GE Healthcare) at 37°C. Firstly, all samples were dialyzed by buffer F (10 mM HEPES, 150 mM NaCl, 100 μM MgCl2, 0.005% polyoxyethylene sorbitan monolaurate, pH 7.4) and the buffer was used as the running buffer at the flow rate of 50 μl/min. Biotinylation using NHS-amine coupling on amino group of P1 via PEG linker was carried out using EZ-Link NHS-PEG4-Biotin (Thermo Scientific). A sensor chip CAP (GE Healthcare) was used for the immobilization of biotinylated P1. PhoEF-2-GMPPCP and PhoEF-2-GDP was injected over the immobilized P1. The binding response at each concentration was calculated by subtracting the equilibrium response measured in the control flow cell from that in the P1 flow cell. β-2-microglobulin was used as negative control. Each resonance unit was fitted to simple 1:1 Langmuir binding model (A + B ↔ AB) using least square minimization to calculate affinity constants (KD).
Molecular dynamics simulation
To investigate how P1CTD binds to the GDP-form of PhoEF-2, we performed a molecular dynamics (MD) simulation based on the structures of PhoEF-2-D2-GDP and PhoEF-2-GMPPCP-P1C11. The full-length structures of PhoEF-2-GTP-P1C11 and PhoEF-2-GDP were constructed using a strategy described previously (35,36). The missing parts were added as following: (a) the structure of the Mg2+ ion with coordinating water molecules of EF-Tu from Thermus aquaticus (PDBID: 1EFT) was used based on structural alignment with Visual Molecular Dynamics (VMD) (37,38); (b) the residues of Switch I were taken from the structure of EF-G from Thermus thermophilus (PDBID: 4V90) using the SWISS-MODEL server (39,40); (c) residues 305–310 of PhoEF-2-GMPPCP-P1C11 were taken from the PhoEF-2-D2-GDP structure; and (d) C-terminal residues (387–735) of PhoEF-2-GDP were taken from the Apo-form structure of PhoEF-2. The GMPPCP in PhoEF-2-GMPPCP-P1C11 was manually modified to GTP. Hydrogen atoms were added with the psfgen package in the NAMD software and the side chains of His were protonated at the position of ϵ-nitrogen (41). Water molecules were then added to the models using the solvate package in VMD to generate a water box of TIP3 extending 10 Å from the protein structure in every direction (38). The energy of the solvated system was minimized using an iterative process of minimizing the energy of the [water, protein]2 for each of 104 steps followed by a system minimization for 105 steps using the NAMD software (41). Following this, Na+ ions were added using the autoionize package in VMD. A final minimization of 105 steps over the entire system was then performed.
The PhoEF-2-GDP-P1C11 complex was modeled using the coordinates of P1C11 in PhoEF-2-GTP-P1C11 and PhoEF-2-GDP after 10 ns of the production time described above. To position P1C11, the structures of PhoEF-2-GDP and PhoEF-2-GTP-P1C11 were aligned, and the system was then solvated using VMD, as described previously. The added water molecules were minimized for 105 steps, following by 105 steps minimization of PhoEF-2-GDP. The PhoEF-2-GDP-P1C11 system was then minimized, ionized, equilibrated, and simulated for 50 ns, as described below.
All equilibrated MD simulations were performed using NAMD 2.9 with CHARMM 27 parameters (41,42). Each simulation used periodic boundary conditions in an NPT ensemble with a time step of 2 fs. The temperature was controlled with Langevin dynamics and the pressure was maintained at one atmosphere with a Nosé-Hoover Langevin piston. A cut-off distance of 12 Å and a switching distance of 10 Å were used. Short-range (Van der Waals) and long-range electrostatic interactions were computed every 2 fs. Each model was equilibrated at 27°C and 77°C for 150 ps. The equilibrations were then cooled to 27°C by applying the velocities of the 27°C system to the coordinates of the 77°C system, and 50-ns production runs were performed after system cooling. Snapshots of the PhoEF-2-GTP-P1C11 and PhoEF-2-GDP simulations were saved every 2 fs and were then compiled using Carma software to remove water molecules as well as movements of the protein's center of mass (43).
Building a GTPase-recruitment model
To understand the relationship between PhoEF-2 and P1 stalk at the GTPase-associated center of the ribosome, we built a GTPase-recruiting model. Because the structure of the archaeal 70S ribosome complexed with EF-2 is unavailable, we built the model in three steps. Firstly, we fitted the stalk structure of PhoP0-[P1]2[P1]2[P1]2 (PDBID: 3A1Y) (10) to the cryo-electron microscopy (cryo-EM) structure of 70S ribosome complexed with two tRNAs at the P-site and E-site from P. furiosus (PDBID: 3J2L, 3J20 and 3J21) (44). We then superposed the crystal structure of 70S ribosome with two tRNAs at the A/P-site and P/E-site and mRNA stalled EF-G from T. thermophilus (PDBID: 2WRI and 2WRJ) (45) on the cryo-EM structure of the 70S ribosome. Finally, we superposed current structure of PhoEF-2-GMPPCP-P1C11 on the structure of EF-G of the model and built the linker between P1C11 and the stalk structure of PhoP0-[P1]2[P1]2[P1]2 as previous description (31).
RESULTS
PhoEF-2 structures
The crystal structure of PhoEF-2-GMPPCP was determined at a resolution of 2.3 Å, which showed that there were two molecules in the asymmetric unit. One of these molecules was bound to GMPPCP (Figure 1A), while the other was nucleotide free (hereafter referred as PhoEF-2-Apo), despite GMPPCP being added at a high concentration for crystallization. The final model of PhoEF-2-GMPPCP contains 693 of 735 residues. Ten N-terminal residues, two C-terminal residues, and residues 51–75 and 305–310 were not able to build due to poor electron density map. The structure of PhoEF-2-Apo also had disorders, but in different regions: 11 N-terminal residues, and residues 49–75, 304–308 and 427–432. The crystal structure of PhoEF-2-D2-GDP was determined at a resolution of 1.6 Å, and there was one molecule in the asymmetric unit. The final model contains GDP molecule captured from express-host of E. coli, and residues 1–386 except for the missing residues 56–69.
Figure 1.
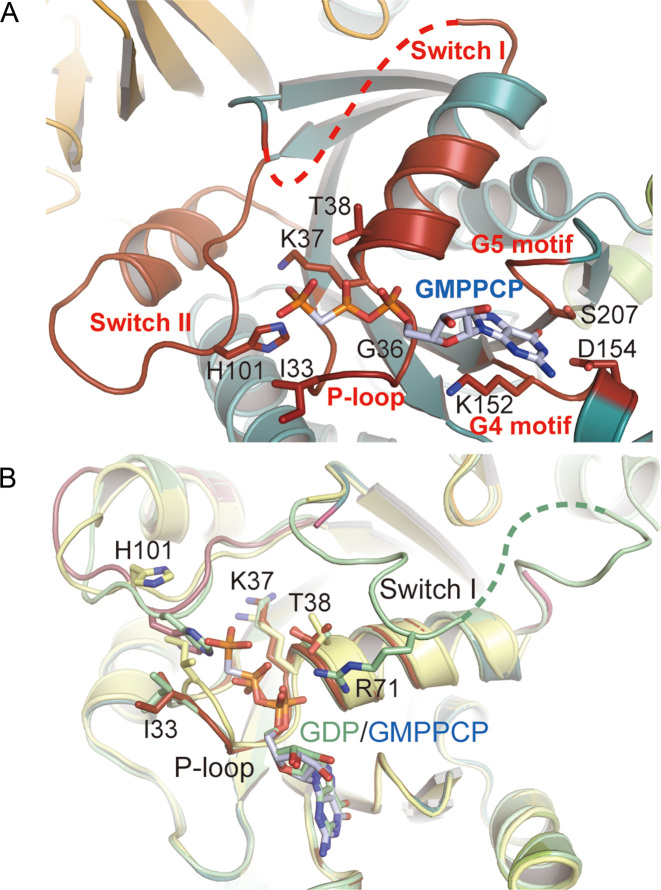
The structures of elongation factor EF-2. (A) The structure of PhoEF-2-GMPPCP. PhoEF-2 and GMPPCP are represented by ribbon and stick models, respectively. The domain G, II, III, IV, V and the subdomain G’ of PhoEF-2 are colored deep teal, orange, slate, pale pink, wheat and green, respectively. The C, N, O and P-atom of GMPPCP are colored pale blue, blue, red and orange. (B) The structures of subdomain G’ of PhoEF-2 and eukaryotic EF-2 from Saccharomyces cerevisiae (SceEF-2; PDBID: 1N0V). Both structures are shown in rainbow colors. The sequences of subdomain G’ of PhoEF-2 and SceEF-2 were compared. Completely identical amino acids are colored red, while those with conserved changes are colored green.
Similar to eEF-2 and EF-G, the overall structure of PhoEF-2 consists of five structural domains and a functional subdomain (Figure 1A): domain G (residues 1–213 and 245–259), subdomain G’ (residues 214–245), domain II (residues 260–386), domain III (residues 387–466), domain IV (residues 467–624 and 701–735) and domain V (residues 625–700). However, the subdomain G’ differs between eEF-2 and PhoEF-2 in both size and structure, consisting of 109 residues with six α-helices and one β-sheet (composed of four β-strands) in eEF-2, and only 31 residues with a helical structure (three α-helices) in PhoEF-2 (Figure 1B).
A structural comparison among PhoEF-2, eEF-2 (PDBID: 1N0V) (46), and EF-G (PDBID: 2XEX) (47–50) showed that although the folding of the individual domains excepting for subdomain G’, is similar between these three proteins, the orientation of each domain is different, indicating highly flexibility between the domains (Supplementary Figure S1). The large rotation of domains III-V of EF-G has previously been demonstrated by comparing ribosome bound and unbound forms (51). Likewise, the domains III-V in PhoEF-2-Apo and PhoEF-2-GMPPCP exhibited different orientation even in ribosome unbound state, with the orientations of the decoding tips in domain IV showing particularly large differences. When domain G was superposed, the shift and rotation angle between two tips of PhoEF-2-Apo and PhoEF-2-GMPPCP were 11.8 Å and 14.9°, respectively (Supplementary Figure S2A), and domain III also exhibited orientation differences of 4.5 Å and 5.7°, respectively (Supplementary Figure S2A). By contrast, the relative position of domains III-V hardly changed, and the contact surface between domain G and V was retained (Supplementary Figure S2B).
The domain G of PhoEF-2 contains five GTP binding motifs, P-loop (A30-N43), Switch I (A48-N76), Switch II (D97-G117), G4 (N151-D154), and G5 (S207-Y209) (Figure 2A). All those motifs of sequences except G5 are well-conserved in other translational GTPases (Supplementary Figure S3) (52,53). The Switch I region is very flexible and could not be visualized in previous studies. In this study, we were able to build 12 residues of this region in PhoEF-2-D2-GDP structure. As shown in Figure 2B, the α-phosphate of GDP is recognized not only by P-loop, but also by the side chain of R71 of the Switch I. This R71 is highly conserved in all GTPases (R65 in S. cerevisiae eEF-2 and R59 in E. coli EF-G) and it is known that the E. coli mutant of R59A deceases GTPase activity on the ribosome and causes defect in promoting translocation (54–56).
Figure 2.
Detailed views of the GTP binding site in different forms. EF-2 and GMPPCP, GDP, and the side chains of residues which involved in GMPPCP or GDP binding are represented by ribbon and stick models, respectively. The O, N atoms are colored by red and blue, respectively. (A) Structure of GTP-binding site of PhoEF-2-GMPPCP. P-loop, the Switch I and Switch II region, and the G4 and G5 motif (including C atoms of side chains) are colored by red. The C-atoms of GMPPCP are colored by pale blue. (B) Structural comparison of the GTP binding sites of PhoEF-2-Apo (pale yellow), PhoEF-2-D2-GDP (pale green) and protein of PhoEF-2-GMPPCP (red) by superposing domain G. The C atom of GMPPCP and GDP is colored pale blue and pale green, respectively. The residues changed conformation are labeled.
Interaction between PhoEF-2 and P1CTD
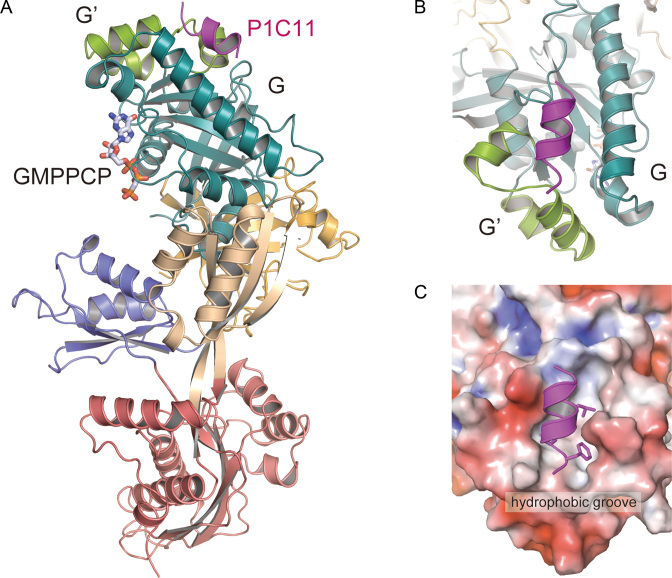
To reveal the mechanism by which P1 recognizes PhoEF-2, we determined the crystal structure of PhoEF-2-GMPPCP in complex with the P1C11 (PhoEF-2-GMPPCP-P1C11) at 3.1 Å resolution. Both molecules (molecule A and B) in the asymmetric unit bound GMPPCP and P1C11. The final model of PhoEF-2-GMPPCP-P1C11 complex contains residues 1–735 of PhoEF-2 and 98–108 of P1C11. The disordered region in molecule A is 11 N-terminal residues, 51–75 and 304–308 residues of PhoEF-2 and two N-terminal residues of P1C11, while that of the molecule B is 10 N-terminal residues, 51–76, 305–310 residues and two C-terminal residues of PhoEF-2 and one N-terminal residue of P1C11.
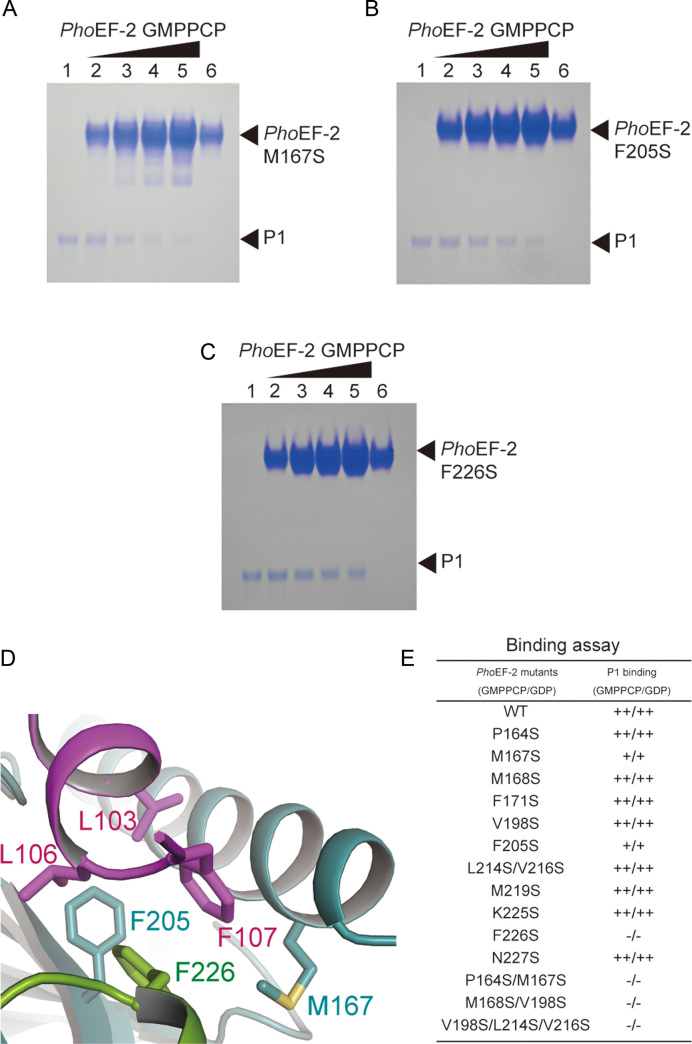
The complex structure of PhoEF-2-GMPPCP-P1C11 revealed that P1C11 formed an α-helix (Figure 3A), and bound hydrophobically to a groove between the domain G and the subdomain G’ of PhoEF-2 (Figure 3B and C). Such binding manner was completely different from that of PhoEF-1α and also bacterial EF-G (17,18) in terms of both binding-position and recognizing-sequence (31). The domain G of PhoEF-2 is involved mainly in the formation of the P1C11-binding groove, whereas the subdomain G’ seems to play a direct interacted role, as indicated by the gel-mobility shift assay described below (Supplementary Figure S4A and Figure 4). In the PhoEF-2-GMPPCP-P1C11 structure, three C-terminal residues, L103, L106, and F107 of P1C11, which are crucial for binding to GTPases (23), were located on the same side of the α-helix and interacted hydrophobically with the groove between domain G and subdomain G' of PhoEF-2. Furthermore, G102 of P1C11 also interacted with PhoEF-2, and the mutant G102S completely disrupted the binding to PhoEF-2 as well as to L103, L106, and F107 of P1 (Supplementary Figures S4A and S5). The P1C11-binding hydrophobic groove was formed by the residues P164, M167, M168, F171, V198, and F205 in domain G, and L214, V216, M219, K225, F226, and N227 in subdomain G’ of PhoEF-2. (Supplementary Figures S4A and S4B). Among these residues, the side chain of F226 in subdomain G' exhibited π-stacking and hydrophobic interactions with L106 and F107 of P1C11 at distances of approximately 3.7 and 3.8 Å, respectively (Figure 4D and Supplementary Figure S4B). The structure also revealed that M167 of domain G, contacted to F107 of P1C11 with a distance of 3.3 Å (Figure 4D and Supplementary Figure S4B). Similarly, F205 in domain G interacted hydrophobically with P1C11 L103 and L106 at distances of approximately 3.4 and 3.6 Å, respectively (Figure 4D and Supplementary Figure S4B).
Figure 3.
Structure of PhoEF-2-GMPPCP in complex with 11 C-terminal residues of P1 (PhoEF-2-GMPPCP-P1C11). (A) Overall structure of PhoEF-2-GMPPCP-P1C11. The PhoEF-2 and GMPPCP are shown same as in Figure 1A. P1C11 is represented by a ribbon model (magenta). (B) Diagram illustrating how P1C11 was bound to a groove between subdomain G′ and domain G. (C) The electrostatic molecular surface of P1-binding groove of PhoEF-2. Positive, negative, and neutral electrostatic surface potentials are shown in blue, red, and white, respectively. Residues, G102, L103, L106, and F107 of P1C11, which are critical for binding PhoEF-2 (23), are represented by stick models.
Figure 4.
P1-binding analysis of PhoEF-2 using point mutants. (A–C) are results for GMPPCP-bound PhoEF-2 mutants M167S, F205S, and F226S, respectively. The homodimer of P1 (100 pmol) was incubated without PhoEF-2 mutants (lane 1), or with 100 pmol (lane 2), 200 pmol (lane 3), 300 pmol (lane 4) or 400 pmol (lane 5) of the PhoEF-2 mutants in 10 μl of solution at 70°C. Each PhoEF-2 mutant (100 pmol) was also incubated without P1 (lane 6). (D) Detailed view of the interaction between M167, F205, and F226 of PhoEF-2 and L103, L106, and F107 of P1C11. The side chains of these residues are represented by stick models. (E) Comparison of the P1-binding ability of GMPPCP/GDP-bound PhoEF-2 mutants. The binding ability of each mutant is displayed as ++ (comparable to WT), + (less closely than WT) or – (undetectable).
To confirm the structural data of these interactions, we performed mutation analysis using a gel-mobility shift assay for 12 residues of PhoEF-2 (P164, M167, M168, F171, V198, and F205 in domain G; L214, V216, M219, K225, F226, and N227 in subdomain G’). Circular dichroism spectroscopy was used to confirm that there were no significant conformational changes between the wild-type and mutants (Supplementary Figure S6). F226S completely disrupted P1-binding (Figure 4C, E and Supplementary Figure S7C), highlighting the crucial role of F226 in P1-binding. Furthermore, the PhoEF-2 mutants M167S and F205S partially affected P1-binding affinity (Figure 4A, B, E and Supplementary Figure S7C), suggesting that M167 and F205 not only play a role in hydrophobic groove formation but also exhibit an important interaction with P1. Although the residues P164, M168, F171, and V198 in domain G, and L214, V216, M219, K225, and N227 in subdomain G' are involved in P1C11-binding hydrophobic groove formation, no effects were detected for the point mutations P164S, M168S, F171S, V198S, M219S, K225S, and N227S, and even the double mutation L214S/V216S (Figure 4E, Supplementary Figure S7 and Supplementary Table S1). However, the interaction between P1 and PhoEF-2 mutants of plural amino acid substitutions P164S/M167S, M168S/V198S, or V198S/L214S/V216S were entirely disrupted (Figure 4E, Supplementary Figures S7B and S7D). This result indicates that these hydrophobic groove formation residues work together to bind P1.
Conformational changes of P1C11-bound PhoEF-2
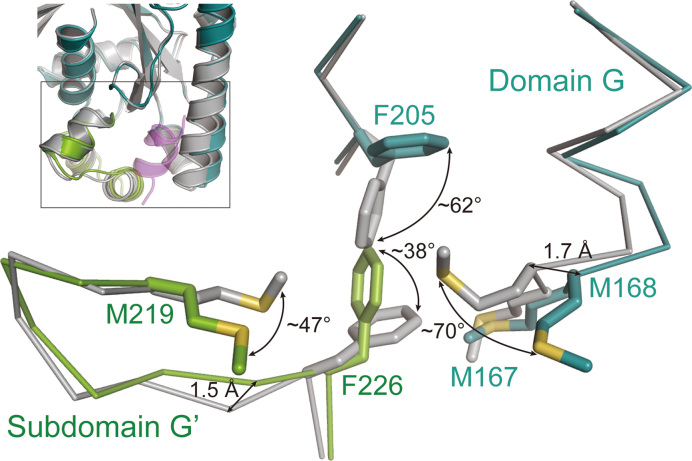
As described above, the folds of each domain of PhoEF-2 between its different forms were almost same, although their orientations were different. However, the P1-binding residues M167, F205, and F226 of PhoEF-2 exhibited conformational changes when P1C11 was bound (Figure 5). Compared with PhoEF-2-GMPPCP, the side chains of F205 and F226 in PhoEF-2-GMPPCP-P1C11 maintained the similar hydrophobic interaction to those of PhoEF-2-GMPPCP, but rotated by approximately ∼62° and ∼38°, respectively, for interacting with L106 and F107 of P1C1 (Figures 4D and 5). Furthermore, although M168 in domain G and M219 in subdomain G' of PhoEF-2 did not interact with P1C11 (Figure 4E, Supplementary Figures S4A, S4B and S7A), their side chains rotated by approximately 70° and 47°, respectively, when bound to P1C11, suggesting these two amino acids worked as a gate for P1C11 (Figure 5). In addition, some main chains surrounding the P1C11-binding groove of PhoEF-2 also showed small conformational changes associated with P1C11 binding. Helix P164-Y185 in domain G and region T222-K235 in subdomain G’ shifted by a maximum distance of 1.7 Å and 1.5 Å, respectively (Figure 5). The conformational change of M167 appeared to correspond with that of F205 and F226 of subdomain G’ (Figure 5).
Figure 5.
Structural comparison of P1-binding region by superposing domain G between PhoEF-2-GMPPCP and PhoEF-2-GMPPCP-P1C11. PhoEF-2-GMPPCP is colored by gray and PhoEF-2-GMPPCP-P1C11 is shown same as in Figure 3A. In closed-up view, PhoEF-2 structures and side chains of residues that have conformational changes by binding to P1 are represented by line and stick models, respectively.
The structure of PhoEF-2-GMPPCP-P1C11 shows that the P1C11-binding groove looks to be a long distance from the GTP binding site (∼20 Å). However, the P1-binding groove connects to parts of GTP binding motifs, G4 and G5 (Supplementary Figure S8). The side chain of V153 in G4 motif forms a hydrophobic core with P1C11-bound residues M167, F205, and F226, and moreover F205 is involved in G5 motif (Supplementary Figure S8). Such structural relationships indicate that conformational changes of the P1-binding groove may be affected by nucleotide binding. Considering the results of previous gel-mobility shift assay and present binding assay that P1 also binds to GDP-form of PhoEF-2 with same level of GTP-form, we tried to build a docking model of GDP-form PhoEF-2 in presence of P1C11 (hereafter referred as PhoEF-2-GDP-P1C11) by MD simulation based on crystal structures solved in this study (Supplementary Figures S9A and S9B). This PhoEF-2-GDP-P1C11 model can be hypothesized to be a state that PhoEF-2 probably dissociates with P1 from the ribosome after the GTP hydrolysis.
In the PhoEF-2-GDP-P1C11 MD-simulated model, the structure of the P1C11 C-terminal was similar to that of PhoEF-2-GMPPCP-P1C11, forming an α-helix and displaying a similar binding in terms of both positions and interactions with PhoEF-2, but with slight conformational changes (Supplementary Figure S9C). Compared with the GDP-form, conformational changes of M168, M219, and F226 seemed to be according to interaction with F107 of P1C11, whereas two residues, M167 and F205 maintained almost similar positions in the PhoEF-2-GDP structure (Supplementary Figure S9D).
Thus, the P1C11-binding groove structure in the GDP-form of PhoEF-2 is almost similar to that of the GTP-form, suggesting that P1 could bind to PhoEF-2 GDP-form at a level comparable with that of the GTP-form. The slight conformational change seems to be favorable towards the next step that is P1-dissociation. To confirm such a nucleotide-independent binding manner, we performed all P1-binding assay experiments using both GTP- and GDP-forms of PhoEF-2, and the results demonstrated no differences in the P1-binding affinity of the two forms as previously reported (23). We further confirmed the quantitative P1-binding affinity of PhoEF-2-GMPPCP and PhoEF-2-GDP using surface plasmon resonance. The KD value of the GDP-form was comparable with that of the GTP-form, although the koff values could not be measured (Supplementary Figure S10). The results showed that nucleotide-independent P1-binding affinity is extremely weak (only detected μM level), and dissociation of P1 from EF-2 is exceedingly fast.
DISCUSSION
The crystal structures determined in the present study shed light on the conformational details of PhoEF-2 in its different forms (Apo, GTP, and GDP), as well as the interaction between PhoEF-2 and P1CTD. Among five GTP binding motifs, Switch I always retained its flexibility, and P-loop and Switch II changed their conformation, responding to the binding-state, whereas the G4 and the G5 motifs exhibited similar conformations in the different forms. In this study, it was clearly shown that the α-phosphate of GDP was recognized not only by P-loop, but also by the side chain of the residue R71 of Switch I (Figure 2B), which is conserved in all organisms, and involved in increasing the GTPase activity on the ribosome and promoting translocation (55,56).
Recognition of recruitment partners by P1CTD
Comparing to the structure of PhoEF-1α-GDP-P1CTD (PDBID: 3WY9) (26), the P1-binding regions of PhoEF-2 and PhoEF-1α are completely different in both of their position and sequence, although the α-helix of P1CTD is uniform (Supplementary Figure S11). The P1-binding features of PhoEF-2 are (i) only hydrophobic residues interact each other; (ii) the most hydrophobic residues in groove work together except F226; (iii) GTP-binding site is close to P1-binding groove (Supplementary Figure S8). Combining with the results of previous research that the conserved C-terminal residues of aP1 recognized directly individual translational GTPases aEF-2, aEF-1α and aIF5B (23), it can be considered that responding to the diversity of binding partners, the P stalk does not recognize the specific sequences and positions, but may recognize the hydrophobic groove of its binding partners by hydrophobic interactions with G102, L103, L106, and F107 of aP1. Based on this proposal, we discuss the P1CTD-bind behavior of other translational GTPases by superposing their domain G upon that of PhoEF-2-GMPPCP-P1C11. As a P stalk binding translational GTPase, aIF5B shares domain G with aEF-2 and aEF-1α, but does not have subdomain G’ (24,57). By superposing domain G, we found a hydrophobic groove (helices α7–α9) in the opposite side of the GTP binding site of ApeIF5B (PDBID: 5FG3) domain G (Supplementary Figure S12), which may relate to interact with aP1CTD.
Because previous studies that aP1CTD bound to both eEF-2 and aEF-2 (58), we compared two structures of SceEF-2 (PDBID: 1N0V) and PhoEF-2-GMPCP-P1C11 (Supplementary Figure S13A). Although the P1C11-binding site of PhoEF-2 was arranged inside of subdomain G', we found a hydrophobic groove (α10-α12) on SceEF-2 subdomain G' which may be involved in the interaction with P1•P2 (Supplementary Figure S13B). The large subdomain G’ of eEF-2 may be evolved from its ancestor for substituting a part of domain G to form P1•P2 binding groove.
Considering the diversity of recruitment partners, it is indispensable to confirm the prediction of P1-binding grooves of aIF5B, eEF-2 by the structural comparison with PhoEF-2-GMPPCP-P1C11 as described above. Therefore, the structural analysis of P stalk bound other GTPases is imperative future for full understand the diversified interactions between translational GTPases and P stalk
Additionally, it was recently reported that eukaryotic P stalk also recruits of eukaryote-specific RIPs (ribosome inactivating protein) to SRL, similar to translational GTPase (59,60). Eukaryotic RIPs depurinate a conserved adenine residue at SRL of 28S rRNA to inactivate ribosome (60). In this study, we showed that P stalk does not recognize the specific sequences and positions for diversity of binding partners (translational GTPases) with week binding affinity. In contrast, in eukaryotes, RIPs are specifically recognized by P stalk (59), such as RTA with the KD approximately 200-fold stronger than that of PhoEF-2 (61,62). Considering the characteristics of partner-binding and their functions, it was indicated that P stalk possesses completely different recognition manner for translational GTPases and RIPs, which is that, P stalk possesses a partner-function-dependent recognition mechanism.
GTPase recruitment process
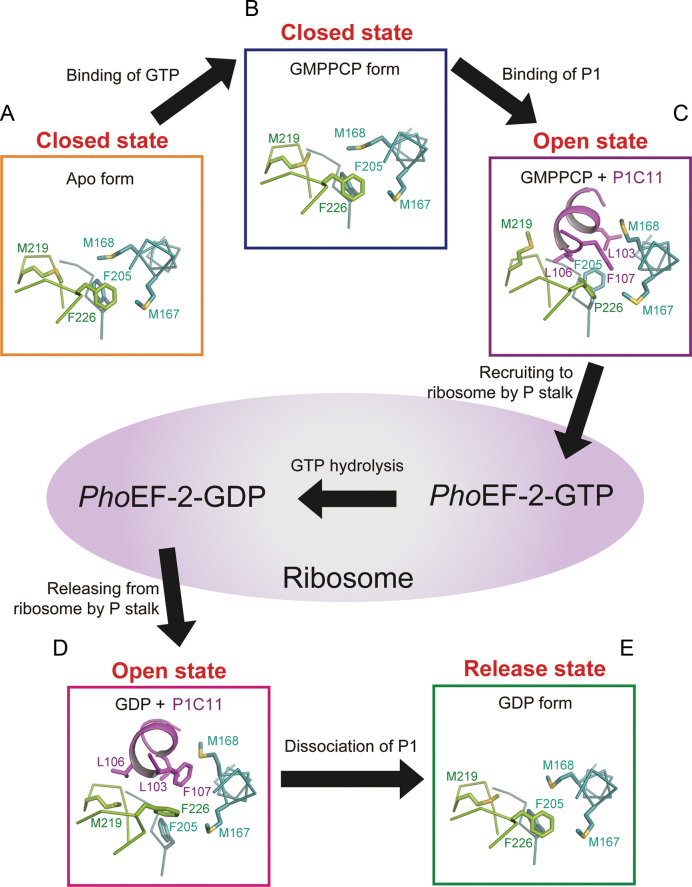
The most remarkable findings in the present study are conformational changes of PhoEF-2 around P1C11-binding groove not only in response to the presence or absence of P1C11 but also in the Apo, GMPPCP, and GDP-forms. During the elongation cycle, PhoEF-2 works through five stages: Apo, GTP-bound, GTP/P1-bound, GDP/P1-bound, and GDP-bound. In this study, we obtained the crystal structures of four forms among them, and remained GDP/P1-bound form was calculated by MD simulation (Supplementary Figure S9A). By structural comparison, the conformational changes were found between different forms. Taken all together, our studies suggest that the PhoEF-2 responses to the processes of recruiting and releasing GTPases through three states: ‘closed’, ‘open’, and ‘release’ of P1-binding groove (Figure 6). Firstly, the P1-binding groove of PhoEF-2-Apo is in a closed state (Figure 6A). When GMPPCP binds to PhoEF-2, PhoEF-2 keeps the closed state, although the side chain of M219 changes the conformation to make a space in P1-binding groove (Figure 6B). The side chains of M167, M168, F205, M219, and F226 then rotate to enter an open state for receiving and binding to P1, following which PhoEF-2-GMPPCP is recruited to the ribosome (Figure 6C). After GTP hydrolysis in the ribosome, the P1-binding groove maintains the open state with a slight conformational change towards the release state for P1 dissociation (Figure 6D). The similar conformation in both the GTP- and GDP-forms of PhoEF-2 are considered to reflect the nucleotide-independent binding manner, and the balance of the binding and recruiting GTP-forms could be adjusted according to the GTPase-GTP concentrations in the cell. When PhoEF-2-GDP dissociates from the ribosome and P1, the groove becomes to a release state (Figure 6E). In this state, despite the side chains of five residues (M167, M168, F205, M219, and F226) go back to the conformations of the closed state, but the main chain of the α-helix (P164-E169) changes to intermediate conformation between the open and the closed states (Figure 6E). Finally, after GDP dissociates from PhoEF-2, the conformation of the groove returns to the closed state (Figure 6A). Comparing to PhoEF-2, the GTP binding site is further away from P1-binding region in PhoEF-1α than that in PhoEF-2 (Supplementary Figure S8), implying that rather than GTP or GDP-binding, the tRNA-binding may affect the P1CTD-binding for PhoEF-1α.
Figure 6.
The responses of P1-binding groove of PhoEF-2 during recruitment process by P1 stalk. The domain G and subdomain G’ of PhoEF-2, and P1C11 are shown in colors same as in Figure 3A. P1C11, PhoEF-2 and side chains of residues are represented by ribbon, line and stick models, respectively. (A) The ‘closed’ state of P1-binding groove in Apo form of PhoEF-2. (B) GMPPCP bound PhoEF-2 with ‘closed’ state of P1-binding groove. (C) The ‘open’ state of the P1-binding groove of GMPPCP bound PhoEF-2. When P1C11 binds to PhoEF-2-GMPPCP, the side chains of M168 and M219 rotate to an ‘open’ state for receiving P1C11, following which PhoEF-2-GMPPCP is recruited to the ribosome. (D) The ‘open’ state of the P1-binding groove in GDP-bound PhoEF-2. After GTP hydrolysis in the ribosome, the P1-binding groove maintains the ‘open’ state with slight conformational changes. (E) The ‘release’ state of the P1-binding groove. When PhoEF-2-GDP dissociated from ribosome and P1 stalk, the groove changes its conformation to a ‘release’ state. Once GDP has dissociated from PhoEF-2, the P1-binding groove of PhoEF-2-Apo returns to the ‘closed’ state (A).
In the present study, we revealed how PhoEF-2 is recognized by P1CTD. Furthermore, the structure of the ribosomal P stalk and elongation factor complex on ribosome is indispensable to address how the stalk complex recruits/releases translation factors to/from the ribosome SRL during protein synthesis. Previous functional and structural analyses have shown EF-G or eEF-2 on the ribosome after recruiting (39,45,63,64), therefore we are able to build a GTPase-recruitment model of ribosome with stalk P0-[P1]2[P1]2[P1]2 and PhoEF-2-GMPPCP-P1C11 complex providing an understanding of the positional relations among PhoEF-2, P1CTD and the ribosome after the PhoEF-2 is delivered into the ribosome (Supplementary Figure S14). This model shows that P1CTD binding region of PhoEF-2 protrudes from the ribosome. Because P1CTD-binding region of PhoEF-2 is located outside of the ribosome, the interaction does not affect the GTP hydrolysis of PhoEF-2 in the ribosome. Such arrangement indicates that P1-binding manner on PhoEF-2 is advantage efficiently for the recruiting translational GTPase. Moreover, P1 binds to different positions of GTPases may be requested by how GTPases perform their functions in individual position and orientation on the ribosome.
DATA AVAILABILITY
Atomic coordinates and structure factors have been deposited to RCSB Protein Data Bank with the accession numbers 5H7J for the PhoEF-2-GMPPCP/PhoEF-2-Apo, 5H7K for the PhoEF-2-D2-GDP and 5H7L for PhoEF-2-GMPPCP-P1C11 complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Naoko Nomura (Hokkaido University) for sample preparation, and Dr Yoshikazu Tanaka for many discussions. We also thank the beamline staffs of the PF, PF-AR and SPring-8 for their technical assistance with data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KAKENHI Grant-in-Aid for Scientific Research (B) [17H03637 to M.Y., 24370073 to T.U.]; Challenging Exploratory Research [26650013 to M.Y.]; Innovative Areas [17H05656 to M.Y.] from MEXT, Japan. Funding for open access charge: MEXT, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta. 1978; 505:95–127. [DOI] [PubMed] [Google Scholar]
- 2. Bourne H.R., Sanders D.A., McCormick F.. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991; 349:117–127. [DOI] [PubMed] [Google Scholar]
- 3. Liljas A., al-Karadaghi S.. Structural aspects of protein synthesis. Nat. Struct. Biol. 1997; 4:767–771. [DOI] [PubMed] [Google Scholar]
- 4. Wilson D.N., Nierhaus K.H.. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005; 40:243–267. [DOI] [PubMed] [Google Scholar]
- 5. Kavran J.M., Steitz T.A.. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 2007; 371:1047–1059. [DOI] [PubMed] [Google Scholar]
- 6. Wahl M.C., Möller W.. Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Peptide Sci. 2002; 3:93–106. [DOI] [PubMed] [Google Scholar]
- 7. Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J.M., Tonevitsky A.G., Stark H., Rodnina M.V., Wahl M.C.. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005; 121:991–1004. [DOI] [PubMed] [Google Scholar]
- 8. Mohr D., Wintermeyer W., Rodnina M.V.. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002; 41:12520–12528. [DOI] [PubMed] [Google Scholar]
- 9. Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M. et al. . ScienceDirect A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014; 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naganuma T., Nomura N., Yao M., Mochizuki M., Uchiumi T., Tanaka I.. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J. Biol. Chem. 2010; 285:4747–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lalioti V.S., Pérez-Fernández J., Remacha M., Ballesta J.P.G.. Characterization of interaction sites in the Saccharomyces cerevisiae ribosomal stalk components. Mol. Microbiol. 2002; 46:719–729. [DOI] [PubMed] [Google Scholar]
- 12. Hagiya A., Naganuma T., Maki Y., Ohta J., Tohkairin Y., Shimizu T., Nomura T., Hachimori A., Uchiumi T.. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J. Biol. Chem. 2005; 280:39193–39199. [DOI] [PubMed] [Google Scholar]
- 13. Krokowski D., Boguszewska A., Abramczyk D., Liljas A., Tchórzewski M., Grankowski N.. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006; 60:386–400. [DOI] [PubMed] [Google Scholar]
- 14. Wawiórka L., Molestak E., Szajwaj M., Michalec-Wawiórka B., Mołoń M., Borkiewicz L., Grela P., Boguszewska A., Tchórzewski M.. Multiplication of ribosomal P-stalk proteins contributes to the fidelity of translation. Mol. Cell. Biol. 2017; 37:e00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bocharov E.V., Sobol A.G., Pavlov K.V., Korzhnev D.M., Jaravine V.A., Gudkov A.T., Arseniev A.S.. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 2004; 279:17697–17706. [DOI] [PubMed] [Google Scholar]
- 16. Leijonmarck M., Liljas A.. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A. J. Mol. Biol. 1987; 195:555–579. [DOI] [PubMed] [Google Scholar]
- 17. Nechifor R., Murataliev M., Wilson K.S.. Functional Interactions between the G' subdomain of bacterial translation factor EF-G and ribosomal protein L7/L12. J. Biol. Chem. 2007; 282:36998–37005. [DOI] [PubMed] [Google Scholar]
- 18. Nechifor R., Wilson K.S.. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J. Mol. Biol. 2007; 368:1412–1425. [DOI] [PubMed] [Google Scholar]
- 19. Helgstrand M., Mandava C.S., Mulder F.A.A., Liljas A., Sanyal S., Akke M.. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007; 365:468–479. [DOI] [PubMed] [Google Scholar]
- 20. Lee K.M., Yu C.W.H., Chiu T.Y.H., Sze K.H., Shaw P.C., Wong K.B.. Solution structure of the dimerization domain of the eukaryotic stalk P1/P2 complex reveals the structural organization of eukaryotic stalk complex. Nucleic Acids Res. 2012; 40:3172–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grela P., Bernadó P., Svergun D., Kwiatowski J., Abramczyk D., Grankowski N., Tchórzewski M.. Structural relationships among the ribosomal stalk proteins from the three domains of life. J. Mol. Evol. 2008; 67:154–167. [DOI] [PubMed] [Google Scholar]
- 22. Lee K.M., Yusa K., Chu L.O., Yu C.W.H., Oono M., Miyoshi T., Ito K., Shaw P.C., Wong K.B., Uchiumi T.. Solution structure of human P1*P2 heterodimer provides insights into the role of eukaryotic stalk in recruiting the ribosome-inactivating protein trichosanthin to the ribosome. Nucleic Acids Res. 2013; 41:8776–8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nomura N., Honda T., Baba K., Naganuma T., Tanzawa T., Arisaka F., Noda M., Uchiyama S., Tanaka I., Yao M. et al. . Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3748–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roll-Mecak A., Cao C., Dever T.E., Burley S.K.. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000; 103:781–792. [DOI] [PubMed] [Google Scholar]
- 25. Vitagliano L., Masullo M., Sica F., Zagari A., Bocchini V.. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with GDP reveals novel features in nucleotide binding and exchange. EMBO J. 2001; 20:5305–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito K., Honda T., Suzuki T., Miyoshi T., Murakami R., Yao M., Uchiumi T.. Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1α. Nucleic Acids Res. 2014; 42:14042–14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 29. Jørgensen R., Wang Y., Visschedyk D., Merrill A.R.. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008; 9:802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J Appl Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A.. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wieden H.-J., Mercier E., Gray J., Steed B., Yawney D.. A combined molecular dynamics and rapid kinetics approach to identify conserved three-dimensional communication networks in elongation factor Tu. Biophys. J. 2010; 99:3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mercier E., Girodat D., Wieden H.-J.. A conserved P-loop anchor limits the structural dynamics that mediate nucleotide dissociation in EF-Tu. Sci Rep. 2015; 5:7677–7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kjeldgaard M., Nissen P., Thirup S., Nyborg J.. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993; 1:35–50. [DOI] [PubMed] [Google Scholar]
- 38. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 39. Chen Y., Feng S., Kumar V., Ero R., Gao Y.-G.. Structure of EF-G-ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol. 2013; 20:1077–1084. [DOI] [PubMed] [Google Scholar]
- 40. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al. . SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K.. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Foloppe N., MacKerell A.D. Jr. All‐atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000; 21:86–104. [Google Scholar]
- 43. Glykos N.M. Software news and updates. Carma: a molecular dynamics analysis program. J. Comput. Chem. 2006; 27:1765–1768. [DOI] [PubMed] [Google Scholar]
- 44. Armache J.-P., Anger A.M., Márquez V., Franckenberg S., Fröhlich T., Villa E., Berninghausen O., Thomm M., Arnold G.J., Beckmann R. et al. . Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 2013; 41:1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao Y.-G., Selmer M., Dunham C.M., Weixlbaumer A., Kelley A.C., Ramakrishnan V.. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009; 326:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jørgensen R., Ortiz P.A., Carr-Schmid A., Nissen P., Kinzy T.G., Andersen G.R.. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 2003; 10:379–385. [DOI] [PubMed] [Google Scholar]
- 47. AEvarsson A., Brazhnikov E., Garber M., Zheltonosova J., Chirgadze Y., al-Karadaghi S., Svensson L.A., Liljas A.. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994; 13:3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Czworkowski J., Wang J., Steitz T.A., Moore P.B.. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994; 13:3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansson S., Singh R., Gudkov A.T., Liljas A., Logan D.T.. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 2005; 579:4492–4497. [DOI] [PubMed] [Google Scholar]
- 50. Chen Y., Koripella R.K., Sanyal S., Selmer M.. Staphylococcus aureus elongation factor G–structure and analysis of a target for fusidic acid. FEBS J. 2010; 277:3789–3803. [DOI] [PubMed] [Google Scholar]
- 51. Li W., Trabuco L.G., Schulten K., Frank J.. Molecular dynamics of EF-G during translocation. Proteins. 2011; 79:1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dever T.E., Glynias M.J., Merrick W.C.. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:1814–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kjeldgaard M., Nyborg J., Clark B.F.. The GTP binding motif: variations on a theme. FASEB J. 1996; 10:1347–1368. [PubMed] [Google Scholar]
- 54. al-Karadaghi S., AEvarsson A., Garber M., Zheltonosova J., Liljas A.. The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure. 1996; 4:555–565. [DOI] [PubMed] [Google Scholar]
- 55. Mohr D., Wintermeyer W., Rodnina M.V.. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 2000; 19:3458–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bartish G., Nygård O.. Importance of individual amino acids in the Switch I region in eEF2 studied by functional complementation in S. cerevisiae. Biochimie. 2008; 90:736–748. [DOI] [PubMed] [Google Scholar]
- 57. Murakami R., Miyoshi T., Uchiumi T., Ito K.. Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix. Proteins. 2016; 84:712–717. [DOI] [PubMed] [Google Scholar]
- 58. Nomura T., Nakano K., Maki Y., Naganuma T., Nakashima T., Tanaka I., Kimura M., Hachimori A., Uchiumi T.. In vitro reconstitution of the GTPase-associated centre of the archaebacterial ribosome: the functional features observed in a hybrid form with Escherichia coli 50S subunits. Biochem. J. 2006; 396:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choi A., Wong E., Lee K.-M., Wong K.-B.. Structures of eukaryotic ribosomal stalk proteins and its complex with Trichosanthin, and their implications in recruiting ribosome-inactivating proteins to the ribosomes. Toxins. 2015; 7:638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Endo Y., Mitsui K., Motizuki M., Tsurugi K.. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987; 262:5908–5912. [PubMed] [Google Scholar]
- 61. Li X.P., Grela P., Krokowski D., Tchórzewski M., Tumer N.E.. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin A chain to the ribosome for depurination. J. Biol. Chem. 2010; 285:41463–41471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grela P.X.A., Li X.P., Horbowicz P., ska M.D.X.W.X., rzewski M.T.X., Tumer N.E.. Human ribosomal P1-P2 heterodimer represents an optimal docking site for ricin A chain with a prominent role for P1 C-terminus. Sci. Rep. 2017; 7:5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tourigny D.S., Fernández I.S., Kelley A.C., Ramakrishnan V.. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013; 340:1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Imai H., Miyoshi T., Murakami R., Ito K., Ishino Y., Uchiumi T.. Functional role of the C-terminal tail of the archaeal ribosomal stalk in recruitment of two elongation factors to the sarcin/ricin loop of 23S rRNA. Genes Cells. 2015; 20:613–624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited to RCSB Protein Data Bank with the accession numbers 5H7J for the PhoEF-2-GMPPCP/PhoEF-2-Apo, 5H7K for the PhoEF-2-D2-GDP and 5H7L for PhoEF-2-GMPPCP-P1C11 complex.