Abstract
Telomerase is a specialized enzyme that maintains telomere length by adding DNA repeats to chromosome ends. The catalytic protein subunit of telomerase utilizes the integral telomerase RNA to direct telomere DNA synthesis. The telomerase essential N-terminal (TEN) domain is required for enzyme function; however, the precise mechanism of the TEN domain during catalysis is not known. We report a single-molecule study of dynamic TEN-induced conformational changes in its nucleic acid substrates. The TEN domain from the yeast Candida parapsilosis (Cp) exhibits a strong binding preference for double-stranded nucleic acids, with particularly high affinity for an RNA–DNA hybrid mimicking the template–product complex. Surprisingly, the telomere DNA repeat sequence from C. parapsilosis forms a DNA hairpin that also binds CpTEN with high affinity. Mutations to several residues in a putative nucleic acid-binding patch of CpTEN significantly reduced its affinity to the RNA–DNA hybrid and telomere DNA hairpin. Substitution of comparable residues in the related Candida albicans TEN domain caused telomere maintenance defects in vivo and decreased primer extension activity in vitro. Collectively, our results support a working model in which dynamic interactions with telomere DNA and the template–product hybrid underlie the functional requirement for the TEN domain during the telomerase catalytic cycle.
INTRODUCTION
Telomeres are nucleoprotein structures that cap the ends of linear chromosomes and protect chromosomal integrity (1). Telomere DNA in most eukaryotic cells is composed of repeating units of short G-rich sequences, although some yeasts are an exception to this rule (2). Telomere erosion is an inherent consequence of the end replication problem and ultimately induces cellular senescence. To circumvent this growth limitation, rapidly dividing cells, as well as single-celled eukaryotes, express the telomerase ribonucleoprotein (RNP) to maintain telomere length through the addition of short telomere DNA repeats to chromosome ends (3). Using a template domain within its integral telomerase RNA (TR) component, telomerase is able to synthesize telomeric DNA in a processive manner (4,5). The catalytic protein subunit of the enzyme is called TERT (telomerase reverse transcriptase) and is organized into distinct domains that include: the telomerase essential N-terminal (TEN) domain, the telomerase RNA binding domain, a reverse transcriptase (RT) and the C-terminal extension (CTE) (Figure 1A) (6,7).
Figure 1.
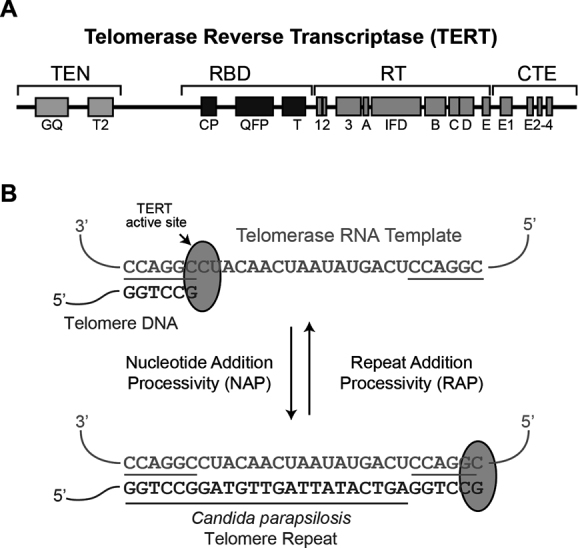
Overview of Candida parapsilosis telomere DNA synthesis. (A) Schematic diagram showing the domain organization of TERT including the TEN domain, RNA binding domain (RBD), RT and CTE. (B). Schematic of the C. parapsilosis telomerase catalytic cycle showing the RNA template, which is 29 nt in length and the 23 nt telomere DNA sequence.
A unique property of the telomerase RNP is its ability to recycle its integral RNA template during a single DNA binding event, resulting in the processive addition of multiple telomere DNA repeats (8). Upon binding to the single-stranded DNA tail of the telomere, TERT reverse transcribes the template region of telomerase RNA displaying nucleotide addition processivity (Figure 1B). To achieve repeat addition processivity (RAP), the nascent template–product hybrid must dissociate and subsequently realign with the 3′ region of the RNA template, prior to rebinding the TERT active site. In vitro cross-linking studies established that the distal 5′ end of single-stranded telomere DNA substrate associates with telomerase (9,10), and this interaction involves the TEN domain (10,11). The TEN domain has also been proposed to mediate a low affinity interaction with TR (12,13). In addition to roles in nucleic acid binding, the TEN domain contributes to telomerase recruitment in mammalian cells through interactions with the telomere binding protein TPP1 (14–17). In yeasts, the telomerase holoenzyme includes auxiliary proteins Est1 and Est3 in addition to the TERT ortholog, Est2 (18). Yeast Est3 is structurally similar to mammalian TPP1, and recent studies of the Candida parapsilosis TEN (CpTEN) domain suggest that it directly interacts with Est3 and together this heterodimeric complex binds telomere DNA (19–22).
Although the TEN domains from evolutionarily divergent TERTs display significant sequence variability, a number of highly conserved residues have been identified (Supplementary Figure S1) (17,23–25). A crystal structure of the Tetrahymena TEN domain showed that many of the conserved residues lie in solvent exposed turn regions of the structure (23). Several residues were suggested to form a DNA binding groove including the highly conserved Q168, which was required for efficient telomere DNA-TEN crosslinking (23). A second site of DNA crosslinking was mapped to W187 in Tetrahymena TEN, in a catalytically active complex (26). The results of these crosslinking studies are consistent with additional measurements of low-affinity TEN interactions using single-stranded DNA substrates (27,28). Single-molecule and direct primer extension assays showed the Tetrahymena TEN domain stabilizes the short RNA–DNA hybrid following product translocation (29). This result is in close accord with a separate study of human TEN that also implicated this domain in stabilizing short RNA–DNA hybrids in the TERT active site (30). More recently, structural studies of the TEN domain from the thermotolerant yeast, Hansenula polymorpha, mapped the surface residues required for nucleic acid binding in this organism (31). Taken together, these studies suggest TEN may directly associate with a variety of nucleic acid binding partners.
A role for the TEN domain in RAP has been demonstrated by mutagenesis and functional studies (23,24,32). Moreover, deletion of the TEN domain impairs RAP in human telomerase and adding the domain back in trans recovers partial RAP activity of a ΔTEN-TERT (30). A homology model of a yeast TEN domain was generated using the Tetrahymena structure as a scaffold, revealing several putative charged patches on the surface of the protein domain (24). Mutations to several of the positively charged residues elicited RAP defects in vitro and telomere length defects in vivo. Thus, there is substantial evidence that the TEN domain can interact with single-stranded DNA and promote telomerase activity as well as RAP during telomerase catalysis (9–11,23,26–28,32).
To explore the mechanistic details of how TEN contributes to telomerase catalysis we have used a novel single-molecule DNA binding assay to dissect the nucleic acid binding determinants for the TEN domain from the yeast CpTEN. CpTEN binding to telomere DNA can be monitored in real-time using single-molecule Förster Resonance Energy Transfer (smFRET), which reports on the distance-dependent efficiency of energy transfer between donor and acceptor dyes coupled to the DNA (33,34). Using this smFRET-based approach, we show CpTEN prefers to bind nucleic acid substrates that possess some duplex structure. Using a panel of DNA constructs, we systematically analyzed the structural properties of C. parapsilosis telomere DNA, providing evidence for the presence of a stable DNA hairpin within this sequence. Our results show that CpTEN binds to a model template–product hybrid with higher affinity than the DNA hairpin structure found within the C. parapsilosis telomere sequence. Using a homology model of the CpTEN domain as a guide, we examined the DNA binding behavior of several mutations to the TEN domain. A set of mutations intended to disrupt a positively charged patch on TEN substantially decreased the nucleic acid binding activity of CpTEN. To complement our biophysical studies we introduced similar mutations in the closely related and genetically tractable yeast, Candida albicans (Ca). Two CaTEN mutations were found to exhibit in vivo telomere length maintenance defects, as well as decreased DNA primer extension activity when assayed in vitro. Taken together, our results provide evidence for the formation of an unanticipated DNA hairpin within a yeast telomere DNA product, as well as detailed insights on the CpTEN–-nucleic acid interaction. Finally, we present a working model for how TEN binding to the various nucleic acid components in the telomerase enzyme–substrate complex may promote efficient telomere DNA synthesis.
MATERIALS AND METHODS
Protein expression and purification
The plasmid construction was described previously and the coding region includes an N-terminal 6× His tag for affinity purification, followed by a SUMO tag for increased solubility adjacent to the C. parapsilosis TEN domain followed by a FLAG tag at the C-terminus. Candida parapsilosis TEN domain was purified using a modified protocol described previously (19). In brief, the plasmid was transformed into BL21 CodonPlus RIL cells and a starter culture of 5 ml was inoculated with a single colony of cells and incubated at 37°C overnight with shaking at 220 rpm. A 1L LB broth was inoculated with the 5 ml culture and incubated with shaking at 37°C and 220 rpm until it reached an OD of 1, at which time, the flask was cooled to 18°C, and protein expression was induced by addition of 0.2 mM Isopropyl β-D-1-thiogalactopyranoside and incubation at 18°C overnight with shaking at 160 rpm.
Cells were harvested by centrifugation at 4000 rpm, resuspended in 30 ml lysis buffer per liter of cells (50 mM Tris pH 7.4, 50 mM NaCl) containing protease inhibitors (0.3 ml 100 mM Phenylmethylsulfonyl fluoride and one tablet of cOmplete mini-EDTA free protease inhibitor, Roche). Cell lysis was carried out by passing cells through a cell disruptor for three passes. Post lysis, cells were treated with Turbo DNAse (40 µl per 1 l of cells) for 30 min at 4°C to degrade any DNA bound to the protein. After DNAse treatment, lysate salt concentration was brought up to 500 mM NaCl along with the addition of 10 mM Imidazole and incubated with 2.5 ml Ni Sepharose 6 Fast Flow (GE) resin in a rotating 50 ml falcon for 30 min to bind the His-tagged protein to the resin. The resin was washed with 20 ml of wash buffer (50 mM Tris pH 7.4, 250 mM NaCl, 100 mM Imidazole), and protein was eluted with 300 mM Imidazole in 50 mM Tris pH 7.4, 500 mM NaCl.
Nickel column purified protein was sized on an S200 column in sizing buffer (50 mM Tris pH 7.4, 250 mM NaCl) and sized CpTEN–SUMO fractions were pooled together. To the pooled fractions, Ulp1 protease was added at a ratio of 1:40 (Ulp1: Protein w/w), incubated for 1 h at 4°C on a rotator, to cleave the SUMO tag. After cleavage, TEN domain was purified away from the SUMO and 6×His tags by passing the cleavage products on a 1 ml HiTrap His column (GE Healthcare) and collecting the flow through. Concentrated TEN domain was stored in 50 mM Tris pH 7.4, 250 mM NaCl, 10% v/v glycerol after flash freezing in liquid nitrogen.
Single molecule experiments
DNA oligonucleotides were purchased from Integrated DNA Technologies. Cy3 and Cy5 handles were both labeled with the respective Cy dye as described previously (35). Ultra-stable dyes, directly coupled to triplet state quenchers, were purchased from Lumidyne Inc. See Supplementary Table S1 for the complete sequences of oligonucleotides used in the study. The various nucleic acid constructs were annealed to a Cy5 handle containing a biotin and a Cy3 labeled handle at the other end by heating the oligonucleotides in a ratio of 1:2:2 Cy5 handle: DNA of interest: Cy3 handle at 95°C for 5 min in the presence of buffer (10 mM Tris pH 7.4, 50 mM NaCl) and slowly cooling to room temperature over several hours. To prepare surface with single DNA molecules, chambers were first flushed with 50 μL of 10 mg/ml bovine serum albumin (BSA) and allowed to incubate at room temperature for 20 min. The channels were then washed with 100 μl of T50 buffer (10 mM Tris pH 7.4, 50 mM NaCl) and incubated with 60 μl of 0.2 mg/ml streptavidin (Molecule Probes) in T50 for 10 min. Excess streptavidin was washed with 2 × 100 μl of T50 buffer. Labeled DNA constructs were flowed into the chamber at concentrations of 8–15 pM in T50 to obtain a suitable density for single molecule experiments and incubated for 10 min. Unbound DNA molecules were washed out with 2 × 100 μL of T50 buffer. TEN protein in imaging buffer (10 mM Tris pH 7.4, 50 mM K-Acetate, 0.1 mg/ml BSA, 0.1 mg/ml tRNA, 0.8% w/v glucose, 0.2 mg/ml catalase, 1 mg/ml glucose oxidase, 10% glycerol) at varying concentrations (0, 10, 25, 50, 100, 200, 400, 800 and 2000 nM) was flowed into individual chambers and slides were visualized under the microscope after incubation for 5 min to allow the binding to reach a steady state equilibrium. Details of slide preparation, microscope setup, data acquisition, analysis, HaMMY and dwell time analysis have been described in previously (29,35,36).
RNA–DNA hybrid molecule preparation
DNA oligonucleotides were ordered from IDT and RNA oligonucleotides, with 5′ Phosphate were ordered from DharmaCon. The molecules were constructed via a two part splinted ligation. DNA splint (0.4 nmol), RNA oligonucleotide (1.2 nmol) and 5′ Phosphate DNA oligonucleotide (1.2 nmol) were mixed together in a 200 μl of 50 mM Tris buffer pH 7.4. The nucleic acid mixture was heated to 75°C for 3 min and slow cooled to room temperature over several hours. After 3–4 h, 200 μl of ligation mixture containing 10 mM MgCl2, 10 mM Dithiothreitol, 1 mM adenosine triphosphate, 3200U of RNAsin Plus (Promega N2615) and 8000U of T4 DNA Ligase (NEB M0202L) in 50 mM Tris buffer pH 7.4 was added to the annealed oligonucleotide mixture and ligation was carried out at 37°C overnight. DNA from the ligation sample was extracted by ethanol precipitation after being cleaned by phenol–chloroform extraction and purified on an 8% polyacrylamide denaturing gel by gel electrophoresis. The ligated DNA was identified by UV shadowing, gel band was excised and DNA was extracted by vigorous shaking in TE buffer for 48 h at room temperature. DNA cleaned by phenol–chloroform extraction was ethanol precipitated and reconstituted in 15 μl of nuclease free water. A second ligation was carried to ligate the 5′ DNA tail to the RNA–DNA hybrid generated by the first ligation step. The RNA was ordered with a 5′ Phosphate to enable its ligation to the DNA tail. The second ligation was carried out in a manner similar to the first step with different concentrations of the oligonucleotides: 0.2 nmol DNA, 0.08 nmol RNA–DNA hybrid from the first ligation step, 0.2 nmol DNA splint. In the first ligation step, the DNA splint was limiting, whereas in the second ligation step, the RNA–DNA hybrid from the first ligation step was limiting. Sequences of the RNA–DNA hybrid molecules and the oligonucleotides used to construct the molecules are listed in Supplementary Table S1. DNA oligonucleotides were ordered for use as in vitro transcription templates by T7 RNA polymerase to generate the TEL46–RNA hairpin construct. Following in vitro transcription, DNA template oligonucleotides were digested with Turbo DNase I (Ambion) and polyacrylamide gel electrophoresis purified prior to use in smFRET experiments.
Candida albicans telomerase mutants
The functions of C. albicans EST2/TERT TEN domain mutants were investigated by analyzing the telomeres and telomerase activities tert-ΔΔ strains harboring a re-integrated copy of wild type or mutant TERT. To facilitate the analysis, the re-integrated alleles were each fused to a FLAG-TAP affinity tag (designated TERT-TAG). The construction of the integrating plasmid carrying TERT-TAG (named pBS-CaEST2-TAG-URA) has been described previously (37). The G78A and HRK149AAA mutations were introduced into the integrating plasmid using the Quik-Change protocol, and following cleavage by Bst EII, the resulting plasmids were transformed into a tert-ΔΔ strain. Proper integration of the tagged TERT alleles at the native chromosomal locus was confirmed by southern analysis.
Telomere length analysis
Chromosomal DNAs were isolated from 4 ml of saturated C. albicans culture by smash and grab, digested with AluI and NlaIII, fractionated in 0.9% agarose gels and the telomere restriction fragments detected by southern as previously described (Singh et al., (40)).
Fluorescence anisotropy measurements
Two-hundred nM 5′ fluorescein labeled TEL23 oligo was incubated on ice with increasing concentrations of CpTEN for 20 min in 20 μl binding buffer (20 mM Tris–HCl, pH8.0, 50 mM KCl, 10 mM DTT, 0.5 mM MgCl2 and 0.1% NP-40). Fluorescence polarization (mP) values (triplicates for each protein concentration) were obtained at 23°C with a SpectraMax M5 (Molecular Devices), and then converted to anisotropy (mA). The binding data were plotted using Prism 5.0 for Windows, GraphPad Software and fitted with a standard binding expression.
Telomerase activity analysis
Whole cell extracts of C. albicans were prepared and the binding of tagged telomerase to IgG-Sepharose beads were carried out as previously described (37). The primer extension assays were performed using either of two 13-nt primers and labeled P32-TTP only (38). Previous studies show that yeast telomerase activities are more easily measured by using selected nucleotides and limiting the extent of primer extension, because this method enables greater enzyme turnover and concentrates all the signals in just a few bands (39,40).
Homology model construction
Model structure of the C. parapsilosis TEN domain was obtained through the threading approach available in the I-TASSER structure prediction software (41). The crystal structure of Tetrahymena thermophila TEN domain (PDB ID: 2B2A) and the solution structure of the TEN domain of Ogatea polymorpha (PDB ID: 5LGF) was used as template for generating the model structure. Although the sequence identity is 13.3%, the global fold similarity of the model to Tetrahymena TEN is 56.9%.
RESULTS
Candida parapsilosis TEN domain dynamically binds and remodels telomere DNA
The crystal structure of the Tetrahymena TEN domain revealed the presence of a DNA binding groove (23). Mutagenesis studies, together with single-stranded DNA cross-linking experiments, further support the notion that TEN binds directly to DNA (10,11,23,25,26,42). A recent study showed that the bacterially expressed and purified TEN domain from C. parapsilosis, was able to crosslink to C. parapsilosis telomere DNA, indicating a direct TEN–telomere DNA interaction (19). However, the few reported assays to measure TEN–telomere binding in the absence of covalent crosslinking have suggested the isolated TEN domain possesses a low affinity for DNA (27,28,31), despite being essential for RAP during telomerase catalysis (23,24,32,42). Single-molecule FRET (smFRET) is a powerful technique to probe dynamics of protein–nucleic acid interactions and has been employed previously to investigate TEN domain stabilization of short RNA–DNA hybrids and to monitor human telomere DNA dynamics during the telomerase catalytic cycle (29,36). Therefore, we used bacterially expressed and purified CpTEN to investigate the mechanistic details of TEN–telomere DNA interactions.
Using an oligonucleotide annealing approach, we introduced Cy3 and Cy5 FRET labels, as well as a single terminal biotin moiety, into short DNA duplexes flanking a 46 nt segment of DNA comprising two repeat sequences of C. parapsilosis telomere DNA (TEL46) (Figure 2A). The labeled DNA constructs were surface immobilized onto a microscope slide via a biotin–streptavidin interaction and varying amounts of CpTEN were introduced into the sample chamber. DNA molecules were imaged using a custom-built total internal reflection fluorescence microscope. The background corrected intensities of the donor and acceptor dyes for each DNA molecule were used to calculate the FRET ratio, defined as the acceptor intensity divided by the sum of the donor plus acceptor intensities. Unless specified otherwise, all smFRET measurements were carried out at room temperature in CpTEN binding buffer including 50 mM K+ (see ‘Materials and Methods’ section for details). In the absence of any CpTEN protein, the TEL46 DNA exhibits a stable FRET signal of ∼0.49 (Figure 2B). Interestingly, addition of CpTEN to the TEL46 DNA molecules induced dynamic transitions to a higher FRET state at ∼0.8 (Figure 2B). We note that attempts to monitor CpTEN–DNA binding interactions by electrophoretic mobility shift assays were not successful (data not shown), consistent with the protein–DNA complex being highly dynamic. The frequency of the FRET transitions to the high FRET state increased with elevated CpTEN concentrations (Figure 2B). Based upon this observation, we assign the high FRET state to the CpTEN ‘bound’ state of the DNA and the low FRET state to the ‘unbound’ DNA conformation. To further substantiate these FRET state assignments, we analyzed the kinetics of the CpTEN–DNA interactions. Hidden Markov modeling of individual smFRET trajectories was performed to generate idealized FRET trajectories (Supplementary Figure S2) (43), which were then used to analyze the dwell time distributions for both the low and high FRET states. The dwell time distributions were well-fit by single-exponential decay functions, consistent with a stochastic two-state process underlying the FRET dynamics (Supplementary Figure S3). The inverse of the mean lifetime for the low FRET state is the observed ‘on’ rate constant (kon) and the inverse of the mean lifetime for the high FRET state is the observed ‘off’ rate constant (koff). For a simple bimolecular binding reaction given by the following expression:
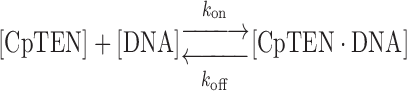 |
kon is expected to increase linearly with increasing CpTEN concentrations and koff is expected to remain constant. The observed kon values for CpTEN binding increased linearly as a function of [CpTEN], while koff was far less sensitive to changes in protein concentration (Figure 2C). The slight negative slope we observe for koff as a function of [CpTEN] is likely due to the limited time resolution of our measurements, which results in an underestimate of koff at high [CpTEN]. The equilibrium dissociation constant (KD), which is a measure of the binding affinity, is the [CpTEN] at which the rate of binding is equal to the rate of dissociation from DNA. By fitting kon and koff values measured in our CpTEN titration experiment to linear functions, we obtained KD = 244 nM for CpTEN binding to the TEL46 telomere DNA substrate.
Figure 2.
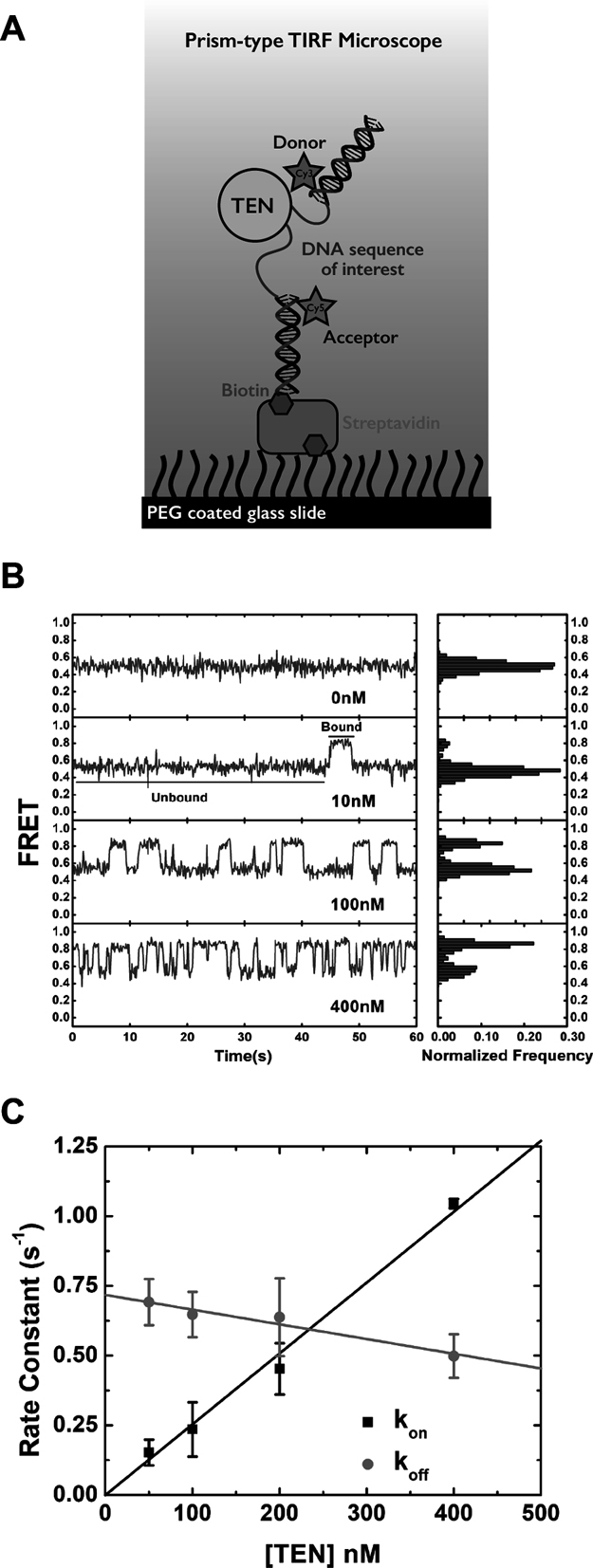
Dynamic interactions between the CpTEN domain and Candida parapsilosis telomere DNA. (A) Experimental set up showing a DNA bound to a quartz surface via biotin–streptavidin–biotin linkage. DNA is labeled with Cy3 and Cy5, one at each end of the molecule of interest through complementary DNA handles. One of the handles is labeled with a biotin to attach it to the surface. FRET before and after addition of purified CpTEN domain is monitored. (B) Representative FRET traces at different [CpTEN], showing transient changes from a no protein FRET efficiency of ∼0.5 to a protein bound FRET value of ∼0.8 on addition of CpTEN. Frequency of the transitions from low to high FRET increases with increasing [CpTEN]. Histogram created by binning FRET efficiency at each time point over a time of 60s shows FRET changing from a single peak in the absence of any [TEN] to a bimodal distribution on addition of [CpTEN]. (C) Kinetic rate constants, kon (squares) and koff (circles), are plotted at each concentration of CpTEN measured as an average value from three separate experiments analyzing N > 300 molecules. Error bars represent the standard deviation.
The dissociation constant may also be estimated for a binding reaction at equilibrium by analyzing the fractional occupancy of the DNA in the unbound (low FRET) and the bound (high FRET) states (Figure 3A). FRET histograms, generated from data collected on TEL46 DNA molecules in the presence of increasing amounts of CpTEN protein, show a clear bimodal distribution corresponding to the ‘unbound’ and ‘bound’ TEL46 DNA states. Fitting these distributions with Gaussian functions provides a measure of the fraction of DNA in the CpTEN bound conformation, which was then plotted and fit with a standard binding equation. The Kd measured in this way (209 ± 13 nM) is in good quantitative agreement with the kinetics analysis and the Hill factor of ∼1 indicates that CpTEN binds DNA in a non-cooperative manner (Figure 3B and Supplementary Table S2). To determine the specificity of the observed CpTEN–DNA interaction, we next conducted our smFRET experiment with a non-telomeric PolyT46 construct and did not observe any changes in signal upon addition of CpTEN (Supplementary Figure S4). The PolyT46 experiment not only provides an important negative control for the sequence of the DNA substrate, but also demonstrates the presence of the smFRET dyes is not sufficient to support the high-affinity CpTEN–DNA binding interaction detected in our assay. Thus, the binding activity of CpTEN appears to be specific to telomere DNA and exhibits a highly dynamic binding/dissociation equilibrium. Furthermore, the transition to a higher FRET state upon CpTEN binding indicates the protein domain remodels the telomere substrate into a significantly more compact conformation.
Figure 3.
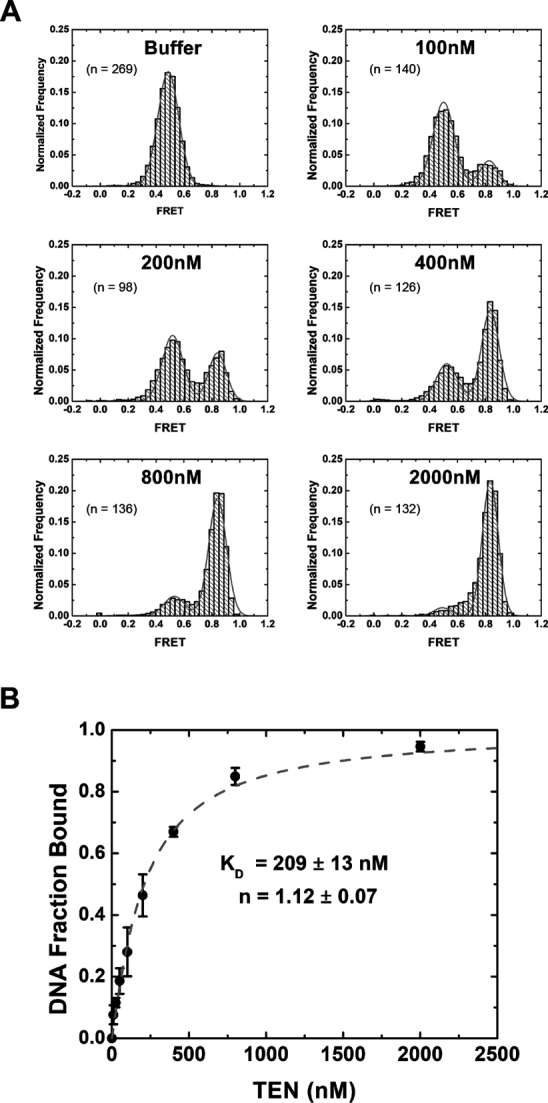
Analysis of CpTEN–DNA interactions from equilibrium smFRET histograms. (A) Representative FRET histograms measured for each indicated concentration of CpTEN. Data are normalized to the total number of molecules analyzed for each experiment. Results showing a bimodal distribution of FRET between ∼0.45 and ∼0.8 were fit to a Gaussian distribution and area under the two peaks was used to calculate the fraction of DNA in a bound conformation at each [CpTEN]. (B) Fraction of DNA in bound conformation as a function of [CpTEN] is plotted with standard deviations calculated from three separate experiments. Fitting the data to a standard binding curve yielded a dissociation constant Kd = 209 ± 13 nM and a Hill coefficient of 1.12 ± 0.07.
Structural analysis of C. parapsilosis telomere DNA
In the course of our studies of CpTEN binding to telomere DNA substrates described above, we noted that the FRET distribution observed for the TEL46 and PolyT46 DNAs, both of which are 46 nt in length, varied considerably (compare Figure 3A with Supplementary Figure S4). The PolyT46 molecule is expected to be largely unstructured which is consistent with the low FRET value that we observe (44). In contrast, the significantly higher FRET value we measured for TEL46 suggests the telomere DNA substrate might adopt a long-lived structured conformation. It is well-established that the G-rich short telomere DNA repeats in many vertebrates, ciliates, plants, and some yeasts form stable G-quadruplex structures in vitro and possibly in vivo (45,46). However the long, regular repeats in C. parapsilosis telomere DNA appear to lack any G-quadruplex forming potential (47). Therefore, we used secondary structure calculations to analyze what, if any, structures were predicted to form in telomere DNA repeats from several budding yeast species (Supplementary Figure S5) (48). Interestingly, a DNA hairpin structure was predicted to form in several yeast telomere sequences provided there was greater than one complete telomere DNA repeat included in the calculation. Estimates for the length of the single-stranded DNA tail at the end of yeast telomeres range from ∼20–200 nt (49–51); thus, it is conceivable that the predicted DNA hairpins may indeed form in vivo. Of the structures examined, the C. parapsilosis telomere DNA is predicted to form the most stable hairpin (Figure 4A and Supplementary Figure S5), and we next set out to analyze this putative DNA structure experimentally.
Figure 4.
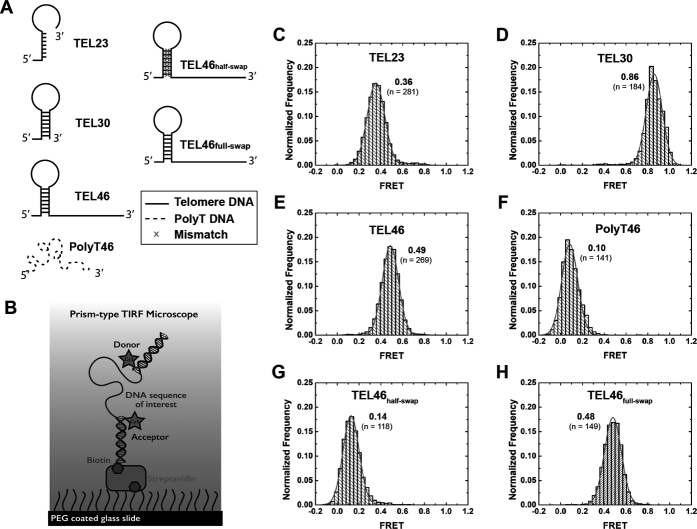
Candida parapsilosis telomere DNA forms a hairpin structure. (A) Schematic diagrams of the various DNA sequences studied by smFRET. (B) Schematic of the smFRET experimental set up to study DNA conformational changes in the absence of CpTEN protein. Representative smFRET histograms for (C) TEL23, containing a single telomere sequence; (D) TEL30, composed of one complete telomere DNA sequence and 7 nt of the next repeat; (E) TEL46 composed of two repeating units of the telomere DNA; (F) PolyT46, a 46 nt long homopolymeric DNA composed of dTTP; (G) TEL46HalfSwap, for which one half of the hairpin forming sequence is swapped with its complementary sequence, destabilizing the hairpin; (H) TEL46FullSwap, in which the two arms of hairpin forming sequences were swapped, preserving the hairpin structure. All histograms are fit with single Gaussian distribution functions.
To analyze the structural properties of C. parapsilosis telomere DNA sequences by smFRET, we labeled the DNA constructs with Cy3 and Cy5 dyes as described earlier (Figure 4B). A single C. parapsilosis telomere DNA repeat of 23 bases (TEL23) exhibited a FRET signal whose value was centered at ∼0.36 (Figure 4C). Surprisingly, a DNA construct composed of a 30 base sequence (TEL30), including one complete telomere repeat plus an additional 7 nt, exhibited a significantly higher FRET peak centered at ∼0.86 (Figure 4D), while a DNA construct composed of two complete 23 base telomere DNA repeats (TEL46) yielded a FRET distribution centered at ∼0.49 (Figure 4E). In the absence of any secondary structure, single-stranded DNA behaves as a freely jointed chain (52), whose end-to-end distance is expected to increase monotonically as a function of increased DNA length. Thus, the observation of a non-monotonic change in the measured FRET value with progressively longer C. parapsilosis telomere DNA substrates supports the hypothesis that part of the DNA forms a stable structure. Consistent with this notion, a PolyT46 DNA, which is expected to be unstructured, exhibited a FRET distribution centered at ∼0.10 (Figure 4F), in stark contrast to the TEL46 DNA. To further investigate the nature of the C. parapsilosis telomere DNA structure, we made mutations to the TEL46 sequence, in which one half of the predicted stem sequence was substituted with its complementary sequence to disrupt the predicted DNA hairpin structure. The FRET distribution for this TEL46half-swap DNA was centered at a value of ∼0.14 (Figure 4G), close to the value obtained with the unstructured PolyT46 DNA construct. Finally, a last TEL46 mutant was analyzed in which the sequences from both arms of the stem–loop were swapped, a change that is predicted to preserve the DNA hairpin structure. Indeed, the FRET distribution for the TEL46full-swap construct recovered a FRET value of ∼0.48 (Figure 4H), very similar to the original wild-type TEL46 DNA sequence. Taken together, the data collected on the various C. parapsilosis telomere DNA sequences strongly support the conclusion for persistent structure being present in DNA substrates >30 nt in length, likely reflecting the propensity of the DNA to fold into a hairpin structure.
Candida parapsilosis TEN domain preferentially binds to structured nucleic acids
Having discovered an unanticipated DNA structure within the C. parapsilosis telomere repeat, we next designed a set of DNA substrates with which to probe the role of DNA structure in mediating CpTEN–DNA interactions (Figure 5A). As described above, using the smFRET assay to monitor CpTEN-induced DNA conformational changes yielded a Kd = 209 ± 13 nM for the CpTEN–TEL46 interaction (Figures 3B and 5B, closed circles). In contrast, a DNA composed of a single C. parapsilosis telomere repeat sequence of 23 bases (TEL23), which is unable to form a hairpin structure, bound with ∼4-fold lower affinity with a measured Kd = 838 ± 54 nM (Figure 5B, open squares). Disrupting the putative DNA hairpin by deleting the first 6 bases on the 5′ end of the DNA substrate (Δ6-TEL46) also significantly reduced the measured binding affinity of CpTEN to a Kd = 1012 ± 49 nM (Figure 5B, open circles). An even more dramatic decrease in binding affinity of ∼10-fold (Kd = 2040 ± 127 nM) was observed for a 46 base C. parapsilosis telomere DNA substrate harboring five strategically chosen mutations (intended to disrupt the putative hairpin but maintain the length of the DNA (Figure 5B, open triangles). Collectively, these data point to the ability of CpTEN to specifically recognize structured DNA substrates. This result is consistent with independent measurement of CpTEN binding to an unstructured template assayed by fluorescence anisotropy, which yielded a Kd ∼4 uM (Supplementary Figure S6). To examine whether a DNA hairpin is both necessary and sufficient for high-affinity CpTEN interaction, we next made a DNA construct that comprised a PolyT DNA cassette into which the native C. parapsilosis hairpin sequence was inserted (PolyTHairpin). Strikingly, the PolyTHairpin DNA bound CpTEN with a Kd = 194 ± 48 nM (Figure 5B, closed squares), comparable to the native TEL46 telomere DNA sequence. In a separate experiment, we introduced a unique helical segment with a non-telomere sequence into the PolyT cassette and CpTEN bound this substrate with an estimated dissociation constant of ∼176 nM (Supplementary Figure S7). Taken together with the observation that a completely PolyT46 sequence shows no detectable CpTEN binding (Supplementary Figure S4), the results obtained with the PolyT-cassette substrates are consistent with a model wherein DNA structure, and not sequence, underlies the high-affinity CpTEN interaction.
Figure 5.
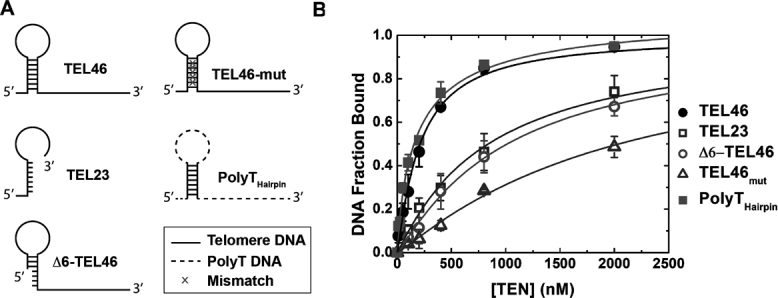
CpTEN–DNA interaction is structure dependent. (A) Schematic diagrams of the various hairpin structures and disruptions to the hairpin that were studied by smFRET. (B) Binding isotherm for CpTEN binding to the Candida parapsilosis DNA constructs and its variants. TEL46 (closed circles); TEL23 (open squares); Δ6-TEL46 (open circles); TEL46-mut (open triangles); and PolyThairpin (closed squares). Plotted data represent average values derived from three separate experiments with n > 300 molecules. Error bars represent the standard deviation.
Recently reported biochemical and biophysical experiments suggested the TEN domain might directly interact with and stabilize the template–product hybrid during the telomerase catalytic cycle (29,30); however, these prior studies did not directly probe TEN–nucleic acid interactions. Therefore, we designed a model RNA–DNA hybrid to mimic the template–product duplex expected to occur within the C. parapsilosis telomerase active site (Figure 6A). The RNA–DNA hybrid construct consisted of a 7 nt 5′ DNA tail, an 8 bp RNA–DNA duplex closed off by an RNA tetraloop, followed by a 10 base 3′ RNA tail (Figure 6A). The initial FRET distribution observed for the RNA–DNA hybrid construct was centered at a value of ∼0.4, and upon addition of CpTEN, transient excursions to a higher FRET value of ∼0.8 were observed (Supplementary Figure S8). Kinetics analysis was consistent with prior experiments performed with the DNA hairpin substrates, wherein the low- and high-FRET states represent the ‘unbound’ and ‘bound’ conformations, respectively (Supplementary Figure S8). Using the fractional occupancy of the RNA–DNA hybrid in the CpTEN bound and unbound states, we determined the apparent dissociation constant for this interaction to be 83 ± 9 nM with a Hill coefficient of 0.89 (Figure 6B, open circles). Thus, the RNA–DNA hybrid binds to CpTEN domain with an ∼3 fold higher affinity as compared with the DNA hairpin present in the TEL46 telomere DNA (Figure 6B, compare open and closed circles). To explore whether the polarity of the phosphodiester backbone within the RNA–DNA hybrid impacts CpTEN binding, we constructed a ‘reverse-polarity’ RNA–DNA hybrid (Figure 6A). For this substrate the RNA and DNA segments were exchanged, resulting in an RNA–DNA hybrid with single-stranded tails having the opposite polarity as that expected to occur in the telomerase active site. Surprisingly, the reverse-polarity RNA–DNA hybrid substrate bound equally well to CpTEN with an apparent Kd = 70 ± 6 nM (Figure 6B, diamonds).
Figure 6.
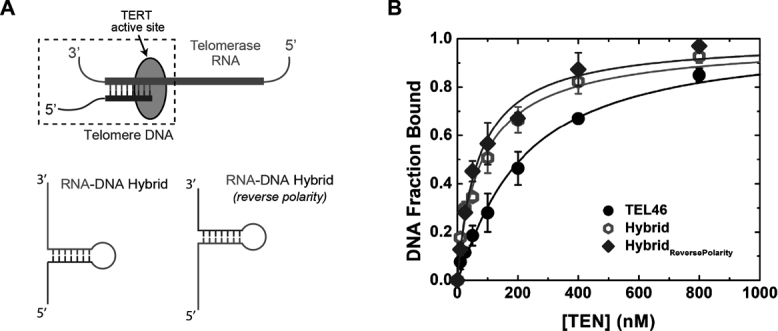
CpTEN binds to an RNA–DNA hybrid with high affinity. (A) Schematic of a RNA–DNA hybrid in the active site of telomerase enzyme. RNA is represented in red whereas telomere DNA is shown in blue. RNA–DNA hybrid constructs used in FRET assays with the correct polarity, reverse polarity and a DNA hairpin molecule. (B) Binding isotherm for CpTEN binding to TEL46 (DNA Hairpin, closed circles), RNA–DNA hybrid (open circles) and RNA–DNA hybridReversePolarity (closed diamonds). Data points for the isotherm represent average values derived from three separate experiments and n > 300 molecules. Error bars represent the standard deviation.
The higher binding affinity exhibited by CpTen for the RNA–DNA hybrid could be derived from the expected A-form helical properties of a heteroduplex, from the presence of 2′ hydroxyl groups in the in RNA segment, or both. To explore these possibilities, we next set out to investigate whether the CpTEN domain binds to an RNA duplex. We designed an RNA construct with the same sequence as the two repeat C. parapsilosis telomere DNA, TEL46–RNA, which is also expected to form a hairpin structure. Interestingly, TEL46–RNA showed even tighter binding than either TEL46 or the RNA–DNA hybrid (Supplementary Figure S9). The starting FRET of TEL46–RNA was ∼0.65, a higher value than either TEL46 or RNA–DNA hybrid. The higher starting FRET observed for TEL46–RNA may reflect a subtle structural difference between the A-form helix formed by this construct and the RNA–DNA hybrid. Upon addition of CpTEN, the FRET peak shifted to a higher value and this shift was dependent on the concentration of CpTEN added, similar to observations with TEL46 and RNA–DNA hybrid (Supplementary Figure S8). We estimated the dissociation constant for the CpTEN–TEL46–RNA interaction to be ∼29 nM with a Hill coefficient of 0.98. Collectively, these results demonstrate that CpTEN dynamically binds a broad range of duplex nucleic acid substrates, with an apparent structural preference for nucleic acids that can adopt an A-form helix and/or have affinity for 2′-OH groups. Furthermore, the capacity of CpTEN to bind and compact the single-stranded regions flanking the duplex is not dependent upon the nucleotide sequence or phosphodiester backbone polarity.
An electrostatic patch within the TEN domain mediates nucleic acid binding and telomerase function in vitro and in vivo
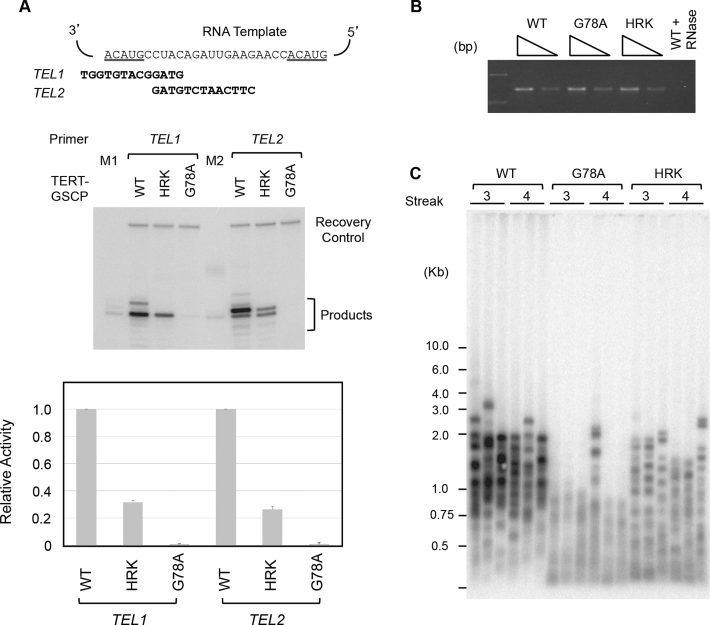
Alignment of TEN domain sequences from a broad set of organisms reveals limited sequence identity and conservation (Supplementary Figure S1). Nevertheless, there are several highly conserved amino acids, which are anticipated to be important for TEN domain folding and/or function. Indeed mutations to several of these conserved residues within the TEN domain have been reported to elicit functional defects in a variety of model systems (10,23–26,28,32). To explore the possible role(s) of several of these conserved residues in mediating nucleic acid binding, we made a series of CpTEN mutants and attempted to express and purify the proteins from bacteria. In many cases, mutation of conserved amino acids (i.e. N72A, L123A/L124A, Q139A and G142A) resulted in proteins that were insoluble, likely driven by protein misfolding, precluding further biophysical characterization (data not shown). However, we successfully generated soluble purified proteins for two mutant CpTEN constructs. In the first, a highly conserved glycine (G77) was substituted with an alanine. The G77 is predicted to reside in a solvent exposed cleft of the CpTEN based on a homology model generated using the Tetrahymena and O. polymorpha TEN domains as reference structures (23,53) (Supplementary Figure S7). The CpTENG77A was tested for both DNA hairpin and RNA–DNA hybrid binding using the smFRET assay (Figure 7A and B, open circles). In both cases, the CpTENG77A bound to nucleic acids with similar affinity to the wild type CpTEN protein. Interestingly, when the corresponding mutation (G78A) was introduced into C. albicans, a substantial loss of telomerase primer extension activity was observed in vitro (Figure 8A and B), and a telomere shortening phenotype was observed in vivo (Figure 8C). Thus, while mutation to G77 (G78 in C. albicans) clearly elicits a telomere maintenance defect, the mechanism underlying this phenotype is likely due to disruption of some aspect of telomerase function other than nucleic acid binding (see ‘Discussion’ section for details).
Figure 7.
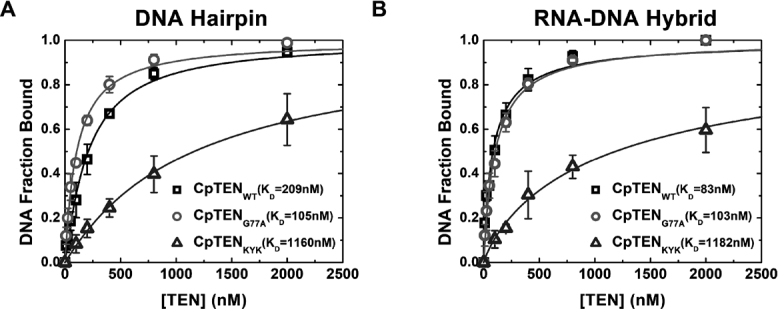
Binding affinities of CpTEN domain mutants for different structured nucleic acids. CpTEN binding data collected with for the DNA Hairpin (A) and RN–DNA hybrid (B) with wild-type CpTEN (open squares), CpTENKYK (open triangles) and CpTENG77A (open circles).
Figure 8.
Analysis of Candida albicans TEN domain mutants. (A) Primer extension assays of tagged C. albicans telomerase with wild-type or mutated TEN domain. The telomerase RNA template region and the two primer sequences are displayed at the top and a set of representative assays shown at the bottom. M1 and M2 are primer+1 size markers prepared by labeling the TEL1 and TEL2 primer with P32-dTTP and terminal transferase. The relative activities were quantified and plotted at the bottom. Data (averages ±S.D.) are from three independent sets of assays. (B) The levels of TER1 RNA in the IgG-Sepharose beads used for primer extension assays were measured by RT-PCR. Two different amounts of RNA, corresponding to 1/270 and 1/810 of the sample used for primer extension, were tested. (C) The lengths of telomeres in strains carrying wild-type or mutated TERT were analyzed ∼125 or 150 generations (streak 3 or streak 4) after the tagged alleles were introduced into a TERT null strain.
A second feature that emerges from the TEN sequence alignments is the prevalence of polar residues at the C-terminal end of the TEN domain near the region of the polypeptide that forms a linker to the TERT RNA binding domain (Supplementary Figures S1 and 10). Previous structural modeling and functional work in Saccharomyces cerevisiae suggested these polar residues may comprise a patch required for TEN domain function, perhaps by mediating nucleic acid interactions (24). To test this hypothesis, we made a triple mutant in this region of CpTEN, substituting two positively charged lysines for negatively charged glutamate residues and one tyrosine for an alanine (K144E/Y147A/K151E, hereafter referred to as CpTENKYK). When assayed by smFRET, CpTENKYK exhibited a severely compromised binding affinity for both the DNA hairpin and RNA–DNA hybrid (Figure 7A and B, open triangles; Supplementary Table S3). Moreover, when mutations were introduced to a similar region of the C. albicans TEN domain (H149A/R150A/K151A hereafter referred to ask HRK), a reduction in telomerase primer extension activity (Figure 8A and B) and a corresponding telomere length defect was also observed (Figure 8C). These experiments support a model in which a positively charged patch located at the C-terminus of the TEN domain is required for nucleic acid binding in order to support normal levels of DNA primer extension in vitro and telomere length maintenance in vivo.
The phenotype of the CpTENKYK mutant, which disrupts a C-terminal patch of positively charged residues, suggested that CpTEN–nucleic acid binding dynamics may be sensitive to changes in ionic strength. To test this hypothesis, we analyzed the binding of TEL46 to CpTEN in the presence of 50, 75, 100 and 150 mM K+, while keeping all other reaction conditions constant. Representative FRET traces at the four different salt conditions measured in the presence of 200 nM CpTEN are shown in Supplementary Figure S11. Dynamic transitions between the low- and high-FRET states are observed at 50, 75 and 100 mM K+. Interestingly, both the low→high and high→low FRET transitions become more frequent at increasing ionic strengths. At the highest salt concentration tested (150 mM K+), we were no longer able to resolve distinct FRET transitions due to limited time resolution of the measurement; however an increase in the observed FRET value is still detected, likely the result of rapid CpTEN binding dynamics that are time-averaged in our experiments. Estimates for the dissociation constants for binding at 75 and 100 mM K+ yielded values of ∼120 and ∼156 nM, respectively (Supplementary Figures S12-13 and Table S4). These estimates are similar to the measured KD of ∼209 nM for TEL46 binding to CpTEN at 50 mM K+, suggesting changes in ionic strength influence the rates of both CpTEN binding and dissociation. This result may reflect a stabilization of DNA structure by increased ionic strength together with destabilization of the electrostatic interactions between CpTEN and DNA. Importantly, these experiments at higher ionic strengths support the conclusion that the CpTEN–nucleic acid interaction we have characterized in vitro is likely to hold under typical physiological conditions.
DISCUSSION
The TERT–TEN domain contributes to telomerase function in several distinct ways. TEN has long been suggested to mediate binding interactions with telomere DNA substrates (9–11,23–25,27,28,32,42,54), and more recently, to stabilize short template–product hybrids in the RT active site (29,30). In addition to these nucleic acid binding activities, TEN also governs the cell-cycle dependent recruitment of telomerase to telomeres via interactions with telomere-associated proteins (14–17,55,56). However, while this body of evidence implicates the TEN domain as a central regulator of telomerase catalysis and recruitment, the precise mechanism of how the TEN domain contributes to telomerase function remains to be determined. Many previous studies of TEN–nucleic acid interactions relied on UV crosslinking using single-stranded telomere DNA (9–11,23,26), and the few reported studies designed to detect DNA interactions with a purified TEN domain in the absence of covalent crosslinking revealed only weak binding with its cognate single-stranded telomere DNA (27,28,31).
Recent UV crosslinking experiments showed that a purified TEN domain from the budding yeast CpTEN is competent to form a covalent complex with its cognate telomere DNA (19). To directly assay the binding characteristics of CpTEN without need for crosslinking, we employed a single-molecule assay that utilizes FRET to report on CpTEN-induced conformational changes in nucleic acid substrates. The smFRET assay permitted us to directly detect and quantitatively characterize high-affinity, yet dynamic, CpTEN–nucleic acid interactions. Our results indicate CpTEN binds preferentially to double-stranded nucleic acids, including DNA hairpins and RNA–DNA hybrids. Furthermore, while CpTEN binding involves recognition of duplex nucleic acids, the smFRET assay also reveals that the single-stranded DNA flanking a helix adopts a compacted conformation upon protein binding, implying some protein–DNA contacts near the ds-ssDNA junction. Using mutagenesis studies, we find that an electrostatic patch expected to reside on the surface of CpTEN is required for binding to both a DNA hairpin and the RNA–DNA hybrid. Mutation of the corresponding residues in the related and genetically tractable organism C. albicans elicited a telomere shortening phenotype in vivo and reduced telomerase primer extension activity in vitro. Taken together, our studies reveal that CpTEN is capable of forming multiple dynamic interactions with various nucleic acid binding substrates, a finding that has implications for how this essential domain contributes to processive telomere DNA synthesis.
The short G-rich DNA repeat sequences found at many telomeres are often associated with the formation of G-quadruplexes (2,45); however, the C. parapsilosis telomere sequence does not meet the criteria required for G-quadruplex formation (47). Nevertheless, FRET measurements performed on the 46-nucleotide long telomere DNA substrate (TEL46) and a control PolyT46 molecule suggested the C. parapsilosis telomere DNA has the capacity to form a stable structure. Indeed, secondary structure prediction calculations also showed that the C. parapsilosis telomere DNA as well as sequences from several related yeast organisms have the potential to fold into a hairpin structure (Supplementary Figure S2) (48). We experimentally tested this structure prediction with a series of smFRET measurements taken on C. parapsilosis telomere DNAs of varying length, together with several control sequences. These experiments provide strong support for the formation of a DNA hairpin structure in C. parapsilosis telomere DNAs that are longer than a single telomere repeat. In the future, it will be interesting to further characterize the proposed DNA hairpin structure at atomic resolution by nuclear magnetic resonance (NMR) or X-ray crystallography. Prior analysis of telomeres from several yeasts suggests that the G-strand overhangs range anywhere from ∼20–200 nt (49–51), raising the possibility that such a structure may indeed form and play a functional role at telomeres in vivo.
The discovery of an unanticipated DNA structure in a C. parapsilosis telomere substrate that binds CpTEN with high affinity prompted further investigation into the nucleic acid preferences of the CpTEN domain. Our results show that CpTEN binds a DNA hairpin with a significantly higher affinity than unstructured DNA. Surprisingly, the presence of the C. parapsilosis telomere hairpin (as well as a non-telomere DNA hairpin) within an otherwise completely PolyT backbone supports a high-affinity interaction with CpTEN. This result suggests CpTEN binds nucleic acids in a structure-specific and largely sequence-independent manner. Previously published reports of a weak interaction with single-stranded DNA are consistent with our observations of lower-affinity binding to the unstructured TEL23, Δ6-TEL46 and TEL46-mut substrates measured by smFRET and fluorescence anisotropy (27,28). Several biochemical and single-molecule experiments suggested the TEN domain stabilizes a short RNA–DNA hybrid in the active site during processive telomere repeat addition (29,30,57). Our result that directly demonstrates the CpTEN domain binds to a model template–product hybrid with high affinity is in close accordance with these prior reports. Interestingly, a model RNA duplex also binds the CpTEN domain with very high affinity, suggesting CpTEN harbors a preference for binding to RNA substrates. In contrast to earlier studies using covalent crosslinking methods to trap TEN–nucleic acid complexes (9–11,19,23,26), results from our single-molecule binding assay illustrate the high-affinity CpTEN interaction with its nucleic acid substrates is characterized by an exceedingly dynamic binding/dissociation equilibrium. This result is reminiscent of single-molecule studies of the single-stranded DNA binding protein from Escherichia coli, which also described a highly dynamic protein–DNA interaction for a protein that binds DNA with nanomolar affinity (58,59).
In a recently published study by Petrova et al., a high-resolution structure of the TEN domain from the thermotolerant yeast, H. polymorpha (Hp), was reported (31). Despite the lack of obvious sequence conservation with other TEN domains, the overall topology of the HpTEN domain was found to be very similar to the previously reported T. thermophila TEN domain structure (23). NMR studies of HpTEN demonstrated the ability of the isolated protein domain to interact with a variety of nucleic acid substrates, with an apparent preference for single-stranded RNA and RNA–DNA hybrids. These results are in very good agreement with our results on the CpTEN domain, although the binding affinities we have measured are several orders of magnitude stronger than those reported for HpTEN. The difference in the observed strength of binding may reflect the unique catalytic requirements for the TEN domain in yeasts with long, regular, telomere DNA repeats such as those found in C. parapsilosis. In addition, HpTEN did not bind DNA substrates with high-affinity, whereas CpTEN bound to a DNA hairpin very well in our assay. This finding may relate the the propensity of C. parapsilosis telomere repeats to form a DNA hairin, a property that is not obviously present in the telomere DNA sequence of H. polymorpha. Most importantly, this recent work using a complementary set of structure probing techniques helps to support the conclusion that the TEN domain is a versatile nucleic acid binding scaffold that has adapted its specific functions to the needs of divergent organisms that rely upon telomerase catalysis for survival.
Mutagenesis studies on the TEN domain have implicated a number of residues in telomerase function. In the present study, we characterized the nucleic acid binding properties of two CpTEN mutants, G77A and K144E/Y147A/K151E. When these same mutations were made to the corresponding residues in C. albicans, we observed a telomere shortening phenotype in vivo and compromised primer extension activity in vitro. This result is consistent with the finding that a similar mutation (G85A) showed defects in telomere maintenance in S. cerevisiae (25). In the case of the K144E/Y147A/K151E mutant, we observed a substantial defect in the ability to bind both DNA hairpin and RNA–DNA hybrid substrates, suggesting this positively charged surface is required for high-affinity CpTEN binding to nucleic acids. Interestingly, a separate study implicated this region of the TEN domain in binding Est3 (60), raising the intriguing possibility that Est3 can influence the nucleic acid-binding activity of TEN. In contrast, the G77A mutant showed no reduction in nucleic acid binding affinity as measured in the smFRET assay. This result suggests the mechanism underlying the phenotypes observed for this mutation are not related to the nucleic acid binding properties of the TEN domain. Instead, we suggest the glycine mutation disrupts an essential protein–protein interaction made by the TEN domain. Indeed, mutation of the corresponding residue, G100, in human TEN was shown to disrupt interactions with the TPP1 protein, which mediates TEN-dependent recruitment to telomeres and processivity enhancement by the POT1/TPP1 complex (16,17). Interestingly, a crystal structure of the Est3 protein from S. cerevisiae reveals close structural homology to TPP1 (21). Therefore, it is likely that mutation of the highly conserved glycine residue in CaTEN disrupts a protein-binding interface between the TEN domain and Est3, leading to defects in telomerase and telomere maintenance. Future experiments will be required to directly test this hypothesis.
We propose a working model in which dynamic binding of TEN to double-stranded nucleic acids promotes telomerase-catalyzed DNA synthesis and RAP (Figure 9). In the model, TEN binding to a DNA hairpin in the 3′ single-stranded DNA tail contributes to telomerase recruitment to telomeres. Once bound to telomere DNA, the dynamic binding behavior of the TEN domain facilitates formation and stabilization of the short RNA–DNA hybrid in the telomerase active site, consistent with findings from studies of Tetrahymena and human telomerase (29,30). In the case of yeasts, which often have particularly long RNA templates, it is expected that rearrangements during the synthesis of a single telomere repeat are required. For example, as repeat synthesis progresses, the RNA template must be drawn into the active site, while at the same time a growing RNA–DNA hybrid will be formed. We speculate that at some stage in this process a strained conformation occurs, caused by crowding effects of the growing RNA–DNA hybrid and downstream RNA template. In C. parapsilosis, gradual melting of the template–product hybrid would coincide with the propensity of the nascent DNA to form a DNA hairpin and would serve to relieve the strained conformation. Dynamic TEN binding to the DNA hairpin and/or the RNA–DNA hybrid could in principle stabilize this rearrangement. Related models for gradual melting of the RNA–DNA hybrid during repeat synthesis, as well as for structure formation in the nascent DNA chain have been proposed in other telomerase systems (61–63). More recently a hairpin model for telomerase catalysis was proposed, wherein formation of non-canonical telomere DNA hairpins may interact with telomerase protein domains, the CTE for example, to promote telomere RAP (64). The related model we propose here for the specialized case of long telomere DNA sequences in yeasts posits that multiple TEN-mediated interactions with nucleic acid structural intermediates are necessary for the completion of the telomere repeat synthesis reaction and to promote the required conformational changes for RAP.
Figure 9.
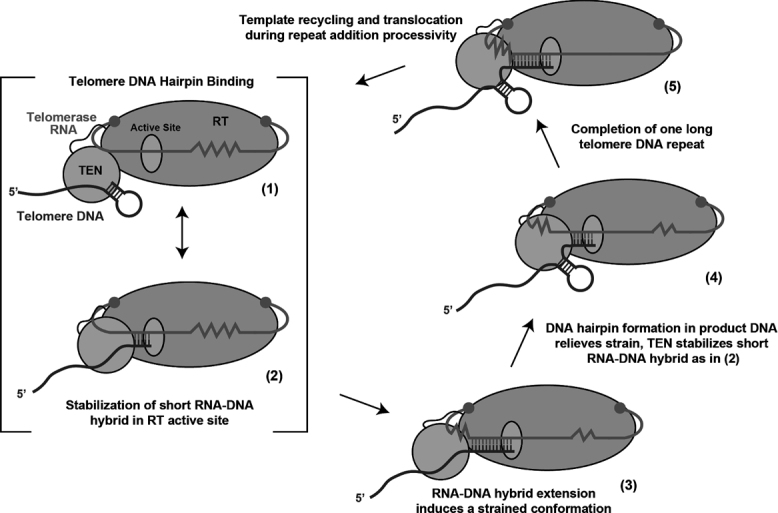
Model of CpTEN domain function during telomerase catalysis. (1) CpTEN domain binds to telomere DNA, perhaps through interactions with Candida parapsilosis DNA hairpin structure. (2) The 3′ end of the DNA primer forms a short RNA–DNA hybrid in the active site to prime telomere repeat synthesis. (3). In yeasts with long telomere DNA repeats, the growing template–product hybrid and compressed downstream RNA template may induce a strained conformation. (4) The strained conformation can be relieved by partial melting of the nascent RNA–DNA hybrid, which may be energetically aided by formation of DNA hairpin structure in telomere product. The TEN domain can in principle serve to stabilize both the short RNA–DNA hybrid and/or the telomere DNA hairpin. (5) Completion of a single telomere DNA repeat, which must be followed by the necessary rearrangements to realign the new DNA 3′-end with the downstream region of the RNA template to promote RAP.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences (NIGMS) [GM107287 to M.D.S., N.L; GM096850 to M.D.S.]. Funding for open access charge: NIGMS [GM107287].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Podlevsky J.D., Bley C.J., Omana R.V., Qi X., Chen J.J.. The telomerase database. Nucleic Acids Res. 2008; 36:D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blackburn E.H., Collins K.. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2011; 3:a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greider C.W., Blackburn E.H.. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989; 337:331–337. [DOI] [PubMed] [Google Scholar]
- 5. Greider C.W. Telomerase is processive. Mol. Cell. Biol. 1991; 11:4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan H., Wang Y., Feigon J.. Progress in human and tetrahymena telomerase structure determination. Annu. Rev. Biophys. 2017; 46:199–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R.. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997; 276:561–567. [DOI] [PubMed] [Google Scholar]
- 8. Wu R.A., Upton H.E., Vogan J.M., Collins K.. Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 2017; 86:439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hammond P.W., Lively T.N., Cech T.R.. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 1997; 17:296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lue N.F. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005; 280:26586–26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wyatt H.D., Lobb D.A., Beattie T.L.. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell. Biol. 2007; 27:3226–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moriarty T.J., Marie-Egyptienne D.T., Autexier C.. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 2004; 24:3720–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O’Connor C.M., Lai C.K., Collins K.. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005; 280:17533–17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armbruster B.N., Etheridge K.T., Broccoli D., Counter C.M.. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol. Cell. Biol. 2003; 23:3237–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Armbruster B.N., Linardic C.M., Veldman T., Bansal N.P., Downie D.L., Counter C.M.. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 2004; 24:3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt J.C., Dalby A.B., Cech T.R.. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife. 2014; 3, doi:10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaug A.J., Podell E.R., Nandakumar J., Cech T.R.. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010; 24:613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hug N., Lingner J.. Telomere length homeostasis. Chromosoma. 2006; 115:413–425. [DOI] [PubMed] [Google Scholar]
- 19. Yen W.F., Chico L., Lei M., Lue N.F.. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20370–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J., Mandell E.K., Tucey T.M., Morris D.K., Lundblad V.. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat. Struct. Mol. Biol. 2008; 15:990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao T., Lubin J.W., Armstrong G.S., Tucey T.M., Lundblad V., Wuttke D.S.. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu E.Y., Wang F., Lei M., Lue N.F.. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 2008; 15:985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacobs S.A., Podell E.R., Cech T.R.. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Structu. Mol. Biol. 2006; 13:218–225. [DOI] [PubMed] [Google Scholar]
- 24. Lue N.F., Li Z.. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res. 2007; 35:5213–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia J., Peng Y., Mian I.S., Lue N.F.. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 2000; 20:5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romi E., Baran N., Gantman M., Shmoish M., Min B., Collins K., Manor H.. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8791–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finger S.N., Bryan T.M.. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008; 36:1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sealey D.C., Zheng L., Taboski M.A., Cruickshank J., Ikura M., Harrington L.A.. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010; 38:2019–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akiyama B.M., Parks J.W., Stone M.D.. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015; 43:5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu R.A., Collins K.. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014; 33:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrova O.A., Mantsyzov A.B., Rodina E.V., Efimov S.V., Hackenberg C., Hakanpaa J., Klochkov V.V., Lebedev A.A., Chugunova A.A., Malyavko A.N. et al. . Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wyatt H.D., Tsang A.R., Lobb D.A., Beattie T.L.. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One. 2009; 4:e7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ha T., Enderle T., Ogletree D.F., Chemla D.S., Selvin P.R., Weiss S.. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stryer L., Haugland R.P.. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. U.S.A. 1967; 58:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long X., Parks J.W., Bagshaw C.R., Stone M.D.. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013; 41:2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parks J.W., Stone M.D.. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat. Commun. 2014; 5:4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu M., McEachern M.J., Dandjinou A.T., Tzfati Y., Orr E., Blackburn E.H., Lue N.F.. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:11682–11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh S.M., Lue N.F.. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lingner J., Cech T.R., Hughes T.R., Lundblad V.. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11190–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh S.M., Steinberg-Neifach O., Mian I.S., Lue N.F.. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell. 2002; 1:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008; 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaug A.J., Podell E.R., Cech T.R.. Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 2008; 15:870–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McKinney S.A., Joo C., Ha T.. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006; 91:1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murphy M.C., Rasnik I., Cheng W., Lohman T.M., Ha T.. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004; 86:2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Galati A., Micheli E., Cacchione S.. Chromatin structure in telomere dynamics. Front. Oncol. 2013; 3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lipps H.J., Rhodes D.. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009; 19:414–422. [DOI] [PubMed] [Google Scholar]
- 47. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fridholm H., Astromskas E., Cohn M.. Telomerase-dependent generation of 70-nt-long telomeric single-stranded 3′ overhangs in yeast. Nucleic Acids Res. 2013; 41:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Larrivee M., LeBel C., Wellinger R.J.. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004; 18:1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Steinberg-Neifach O., Lue N.F.. Modulation of telomere terminal structure by telomerase components in Candida albicans. Nucleic Acids Res. 2006; 34:2710–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith S.B., Cui Y., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 53. Polshakov V.I., Petrova O.A., Parfenova Y.Y., Efimov S.V., Klochkov V.V., Zvereva M.I., Dontsova O.A.. NMR assignments of the N-terminal domain of Ogataea polymorpha telomerase reverse transcriptase. Biomol. NMR Assign. 2016; 10:183–187. [DOI] [PubMed] [Google Scholar]
- 54. Steczkiewicz K., Zimmermann M.T., Kurcinski M., Lewis B.A., Dobbs D., Kloczkowski A., Jernigan R.L., Kolinski A., Ginalski K.. Human telomerase model shows the role of the TEN domain in advancing the double helix for the next polymerization step. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9443–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nandakumar J., Cech T.R.. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013; 14:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sexton A.N., Regalado S.G., Lai C.S., Cost G.J., O’Neil C.M., Urnov F.D., Gregory P.D., Jaenisch R., Collins K., Hockemeyer D.. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014; 28:1885–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Karademir Andersson A., Gustafsson C., Krishnankutty R., Cohn M.. Multiple DNA interactions contribute to the initiation of telomerase elongation. J. Mol. Biol. 2017; 429:2109–2123. [DOI] [PubMed] [Google Scholar]
- 58. Roy R., Kozlov A.G., Lohman T.M., Ha T.. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009; 461:1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou R., Ha T.. Single-molecule analysis of SSB dynamics on single-stranded DNA. Methods Mol. Biol. 2012; 922:85–100. [DOI] [PubMed] [Google Scholar]
- 60. Tucey T.M., Lundblad V.. Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 2014; 28:2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gavory G., Farrow M., Balasubramanian S.. Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vitro. Nucleic Acids Res. 2002; 30:4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gilley D., Blackburn E.H.. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol. 1996; 16:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jarstfer M.B., Cech T.R.. Effects of nucleotide analogues on Euplotes aediculatus telomerase processivity: evidence for product-assisted translocation. Biochemistry. 2002; 41:151–161. [DOI] [PubMed] [Google Scholar]
- 64. Yang W., Lee Y.S.. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat. Struct. Mol. Biol. 2015; 22:844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.