Abstract
Pathological conditions impairing functions of mitochondria often lead to compensatory upregulation of the mitochondrial DNA (mtDNA) replisome machinery, and the replicative DNA helicase appears to be a key factor in regulating mtDNA copy number. Moreover, mtDNA helicase mutations have been associated with structural rearrangements of the mitochondrial genome. To evaluate the effects of elevated levels of the mtDNA helicase on the integrity and replication of the mitochondrial genome, we overexpressed the helicase in Drosophila melanogaster Schneider cells and analyzed the mtDNA by two-dimensional neutral agarose gel electrophoresis and electron microscopy. We found that elevation of mtDNA helicase levels increases the quantity of replication intermediates and alleviates pausing at the replication slow zones. Though we did not observe a concomitant alteration in mtDNA copy number, we observed deletions specific to the segment of repeated elements in the immediate vicinity of the origin of replication, and an accumulation of species characteristic of replication fork stalling. We also found elevated levels of RNA that are retained in the replication intermediates. Together, our results suggest that upregulation of mtDNA helicase promotes the process of mtDNA replication but also results in genome destabilization.
INTRODUCTION
Replication of the mitochondrial genome is required for efficient performance of the organelle. Mitochondrial deficiencies, particularly those related to ATP production, often trigger compensatory upregulation of mitochondrial DNA (mtDNA) replication (1,2). An increase in mtDNA copy number has been reported in cases of OXPHOS limitation that result from the A3243G mutation in the tRNALeu(UUR) gene of the human mitochondrial genome (1,3,4) and inhibition of OXPHOS complex I with rotenone and complex III with antimycin A in human cultured cells results in an ∼2-fold increase in mtDNA level (5,6). A compensatory increase in mtDNA content has also been documented in muscle biopsies of Kearns-Sayre syndrome patients (7), cases of OXPHOS deficiency-related autism (8,9), and renal chromophobe carcinoma and oncocytoma associated with mutations in mtDNA (10–13).
Replication of mtDNA requires two key mitochondrial enzymes: DNA polymerase γ (Pol γ) and mtDNA helicase (14,15). Whereas upregulation of mtDNA replication requires sufficient levels of these factors, overexpression of the catalytic subunit of Pol γ in cultured human and Drosophila cells does not alter mtDNA copy number (16,17), and causes mtDNA depletion in transgenic flies (17). It has also been demonstrated that Pol γ is present at normal levels in cells lacking mtDNA, which suggests that expression of Pol γ is not responsive to variations in cellular mtDNA abundance (18). mtDNA helicase has been demonstrated to affect directly mtDNA abundance, and may be a major regulator of mtDNA copy number. Elevated expression in mouse heart and skeletal muscle increases mtDNA copy number up to 3-fold (19), concomitant with changes in mtDNA replication that manifest by generation of recombination-like mtDNA species, creating four-way junctions (20). Moreover, conditional knockout of mtDNA helicase in mouse heart and skeletal muscle results in severe and rapid mtDNA depletion (21). Studies on pathological alterations in mtDNA helicase have demonstrated a direct relationship with variations in mtDNA copy number in Drosophila Schneider cells and in fly development and physiology (22,23), and with structural rearrangements of mtDNA in human cultured cells (24).
Relevant to compensatory upregulation of mtDNA replication, overexpression of mtDNA helicase in certain pathological conditions has been reported to be beneficial. For example, an elevated level of mtDNA helicase appears to stabilize and protect mtDNA from oxidative damage induced by superoxide dismutase knockout in mouse cardiomyocytes (25), and to ameliorate cardiac fibrosis associated heart failure in a mouse model (26,27). These studies suggest that upregulation of replication induced by helicase overexpression may provide opportunities for treatment of mitochondrial deficiencies, warranting detailed evaluation of the effects of elevated levels of mtDNA helicase.
To study the effects of elevated levels of mtDNA helicase on the mitochondrial replication process in depth, we employed a combined approach of two-dimensional native agarose gel electrophoresis (2DAGE) and electron microscopy (EM) to examine replication intermediates in Drosophila melanogaster Schneider (S2) cells. We demonstrate that elevated helicase increases the abundance of replication intermediates. At the same time, the integrity of the mitochondrial genome is destabilized. We observed for the first time that excess mtDNA helicase generates mtDNA catenanes and specific deletions within the non-coding region of mtDNA in Drosophila.
MATERIALS AND METHODS
Preparation and induction of Drosophila cell lines
Drosophila Schneider S2 cells overexpressing mtDNA helicase were prepared as described previously (28). In this study, cells were treated with 0.4 mM CuSO4 only, to induce expression from the metallothionein promoter, and harvested on the fourth day after induction. Control S2 cells were cultured and treated in parallel with the helicase-overexpressing cells in all protocols. Cell pellets were frozen in liquid nitrogen and stored in –80°C prior to use.
Preparation of mitochondrial nucleic acids, enzymatic treatment, and agarose gel electrophoresis
Mitochondrial nucleic acids (mtNA) were obtained from sucrose gradient-purified mitochondria isolated from Drosophila Schneider S2 cells as described previously (29). Individual mtNA samples were isolated from pellets of ∼2 × 109 cells. Treatment of mtNA samples with restriction endonucleases ClaI and HindIII (Thermo Scientific) was performed for 4 h at 37°C using 4 units of enzyme/μg of mtNA in the manufacturer's recommended buffers (see also (29,30)). The RusA resolvase (kind gift of Dr Robert Lloyd, University of Nottingham) treatment of the HindIII-cleaved mtNA was performed on 10 μg samples in a reaction containing 0.5 μM enzyme, 25 mM Tris–HCl pH 8.0, 1 mM DTT, 0.1 mM acetylated BSA, 10% glycerol, 10 mM MgCl2, for 30 min at 37°C (see also (30)). Treatment with RNase A (Thermo Scientific) for 2DAGE and EM analyses was performed with 1 unit of enzyme per μg of mtNA in 10 mM Tris–HCl pH 8.0, 1 mM EDTA buffer for 2 h at 37°C. Treatment with RNase H1 (Thermo Scientific) was performed at 1 unit of enzyme per 1 μg of mtNA, for 30 min at 37°C in manufacturer's recommended buffer (see also (30)). In the treatment of mtNA samples with RNases A and H1 for 2DAGE, samples were first treated with RNase A, and RNase H1 was then added with the appropriate buffer for the final 30 min. After treatment, proteins were removed from mtNA samples by further digestion with Proteinase K (Thermo Scientific), which was added to reactions at a concentration of 0.5 μg per μl for 30 min at 37°C, followed by phenol–chloroform extraction and precipitation with ethanol. One-dimensional agarose gel electrophoresis was carried out by standard methods in 0.7% (Figure 3A, C andD) or 1% (Figures 3B, 4B and 6A) gels. ImageJ software was utilized for densitometric measurements. Two-dimensional agarose gel electrophoresis (2DAGE) was performed as described previously (29,30).
Figure 3.
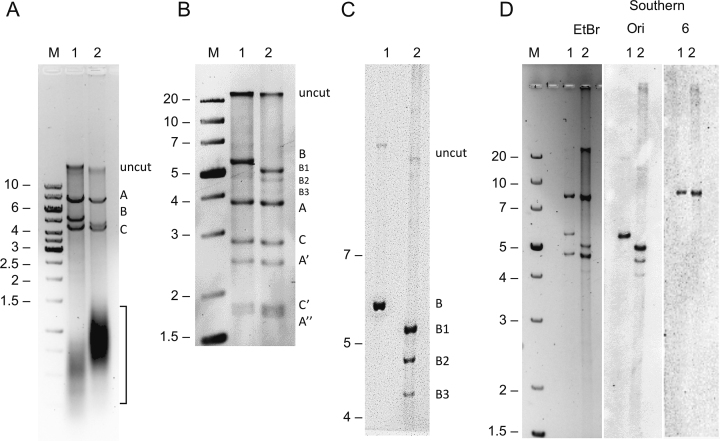
Deletions within the A+T region of mtDNA in Schneider S2 cells overexpressing mtDNA helicase. (A) Agarose gel electrophoresis of HindIII-treated mtNA (4 μg) obtained from control (lane 1) and mtDNA helicase-overexpressing (lane 2) S2 cells. The uncut material and resulting HindIII A, B and C fragments are indicated on the right. The bracket shows the distribution of RNase A-sensitive material (see Supplementary Figure S2). Fragment size is indicated in kb on the left. (B) Agarose gel electrophoresis of HindIII-SacI-treated mtNA (4 μg) obtained from control (lane 1) and mtDNA helicase-overexpressing (lane 2) S2 cells. The resulting bands are labeled relative to the HindIII fragment nomenclature on the right: uncut material and control HindIII B fragment (lane 1) remain undigested; deletion-bearing HindIII B fragments of mtDNA from helicase-overexpressing cells (lane 2) are labeled as B1, B2, B3; SacI digestion of the HindIII A fragment generates derivative fragments A, A’, A’’; SacI digestion of the HindIII C fragment generates derivative fragments C, C’. Fragment size is indicated in kb on the left. (C) Southern-blotting analysis of HindIII-treated mtNA samples obtained from control (lane 1) and mtDNA helicase-overexpressing (lane 2) S2 cells. Fragments encompassing the origin of replication were visualized by hybridization with the Ori probe (see Figure 4A, and Materials and Methods). The HindIII B fragment and its deletion-bearing derivatives B1, B2 and B3, as well as uncut material are labeled on the right. (D) Southern-blotting analysis of HindIII-treated mtNA samples obtained from control (lanes 1) and mtDNA helicase-overexpressing (lanes 2) S2 cells. Fragments were visualized by ethidium bromide staining in the agarose gel (EtBr), or by hybridization with probes specific to the replication origin site (Ori), and coding region (6, see Figure 1). M indicates a molecular weight standard with fragment sizes indicated in kb at left.
Figure 4.
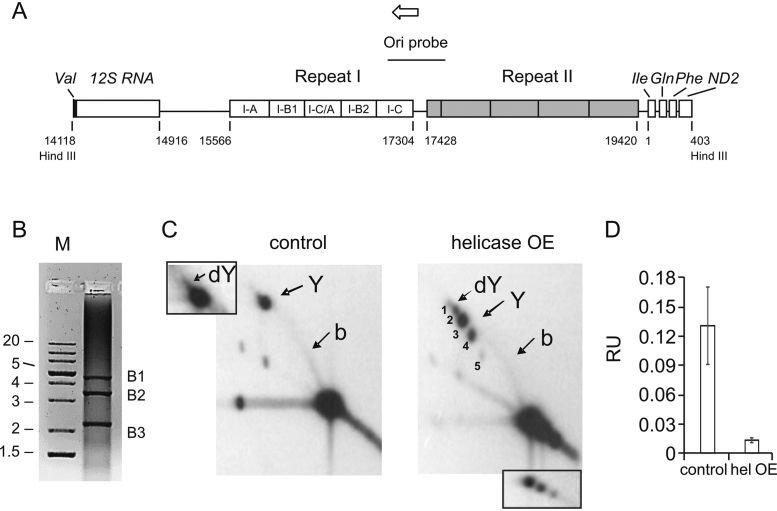
Structural rearrangements within the repeat I region of the A+T region upon mtDNA helicase overexpression. (A) Schematic representation of the A+T region (modified from Salminen et al. (36)) with regions of repeats I and II. Individual Rep1 segments are labeled: I-C 345 bp, I-B2 338 bp, I-C/A 345 bp, I-B1 338 bp, I-A 373 bp. Target site for Ori probe is indicated. Open arrow indicates the direction of replication. B, PCR analysis of the repeat 1 region of mtDNA isolated from mtDNA helicase-overexpressing cells. PCR was performed on the mtNA sample with primers flanking the repeat 1 region (see Materials and Methods). Fragments of sizes corresponding to deletions of the HindIII B1–3 fragments (indicated on the right), ∼4.5, ∼3.5 and >2 kb were amplified. M indicates a molecular weight standard with fragment sizes indicated in kb at left. (C) 2DAGE of HindIII-treated mtNA obtained from control (left panel) and mtDNA helicase-overexpressing S2 cells (right panel) hybridized with the Ori probe. Arrows indicate foci of double Y (dY) structures (labeled 1, 2 in the helicase OE image), and Y-shaped (Y) molecules resulting from progression of the replication fork from the replication origin past the HindIII restriction site (labeled 3, 4, 5 in the helicase OE image). The standard bubble arc (b) spanning from the unreplicated DNA to the Y spot results from a progressing replication fork that has not advanced past the HindIII restriction site. Inset to the control panel represents overexposed dY structures. Inset to the helicase OE panel represents lower exposure of the multiple spots of unreplicated DNA. The analysis was performed on three independent samples. (D) ImageJ quantified mean hybridization intensity (±SD) of the bubble arc derived from the Hind III B restriction fragment of mtDNA from control and mtDNA helicase-overexpressing cells that is represented in panel C. Intensity values were normalized relative to the signal intensity of unreplicated DNA (major signal at the bottom right corner) as an internal standard. Data were quantified from 2DAGE images from two independent samples.
Figure 6.
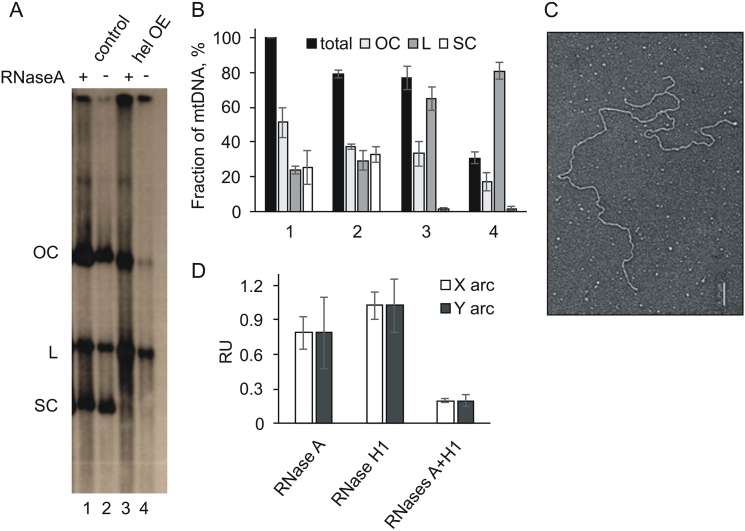
Alterations in tertiary structure and RNA incorporation into mtDNA from mtDNA helicase-overexpressing Schneider S2 cells. (A) Southern-blotting analysis of mtDNA obtained from control (lanes 1, 2) and mtDNA helicase-overexpressing (lanes 3, 4) S2 cells, treated (+, lanes 1, 3) and untreated (−, lanes 2, 4) with RNaseA. Open circular (OC), linear (L), and supercoiled (SC) forms of mtDNA were detected by hybridization with probe 6 (see Figure 1). (B) Densitometric analysis of mean hybridization intensity (±SD) of mtDNA forms from three independent Southern-blotting analyses conducted as in A. Numbering of the X axis in B corresponds to lanes 1–4 in A. Total mtDNA bars indicate the quantity of mtDNA obtained relative to that of the mtDNA from control cells that was not treated with RNaseA (taken as 100%), whereas the remaining bars within each group (OC,L,SC) represent the relative distribution of mtDNA forms within the corresponding total. (C) Representative electron micrograph of linear, branched mtNA observed frequently in undigested samples of mtDNA from helicase-overexpressing S2 cells. Scale bar 0.2 μm. (D) Relative mean hybridization intensity (+/- SD) of the Y and X arcs derived from the ClaI C fragment in 2DAGE analysis with probe 6 of mtDNA from control and helicase-overexpressing cells that was treated and untreated by RNases (see Supplementary Figure S2) was quantified by ImageJ. Intensity values were normalized relative to the signal intensity of unreplicated DNA as an internal standard.
Southern blot hybridization
Agarose gels were blotted, and radiolabeled probes were hybridized as described previously (29). The origin probe was generated by PCR using 100 ng of a gel-purified PCR product spanning nt 17061–17602 that was generated from Drosophila mtDNA as a template, using the primers stated below. The reaction mix contained Dream Taq polymerase and the manufacturer's recommended buffer, 0.8 μM [α-32P]-dTTP (Perkin-Elmer, 3000 Ci/mmol), 0.2 mM each of dATP, dCTP and dGTP, and 10 pmol per reaction of 5′-TAAATTTATTCCCCCTATTC-3′ and 5′-CATGATTTTATTATATAAATATTTTTTATAAAAATAATAC-3′ oligonucleotides as forward and reverse primers, respectively. Both for template and probe synthesis, PCR conditions were as follows: initial denaturation at 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 1 min, with final extension at 72°C for 5 min. Probes were purified by gel filtration on Sephadex G-50 spin columns (Roche). Radiolabeled probe 6 hybridizes to nt 6801–7378, and was generated as described previously (30).
Electron microscopy
Samples of purified mtDNA in 0.25 M ammonium acetate pH 7.5, isolated from control and helicase overexpressing Schneider cells, were spread on a film of denatured cytochrome C protein formed on an air-buffer interface and picked up with parlodion-coated copper grids followed by dehydration and rotary metal shadow casting with 80% platinum–20% palladium as previously described (31,32). At least 100 images for each study group were captured using a Gatan Orius CCD camera (Pleasanton, CA, USA) attached to an FEI Tecnai T12 TEM/STEM instrument (Hillsboro, OR, USA) operated at 40 kV. Contour lengths of the DNAs were measured using Gatan Digital Micrograph software. Statistical analysis was performed using the non-parametric Mann–Whitney U-test and data are presented as mean values. P-value <0.05 was considered statistically significant. The micrographs for publication were adjusted for brightness and contrast using Adobe Photoshop and shown in reverse contrast.
Amplification of the Repeat 1 segment of the A+T region of mtDNA
The fragment of the mtDNA A+T region containing the Repeat 1 region was amplified using 100 ng of mtNA obtained from the helicase overexpressing cells and Dream Taq polymerase (Thermo Scientific), in a reaction mix containing indicated buffer, 0.4 mM dNTPs, and 10 pmol per reaction of primers spanning in wild-type mtDNA nt 12841–17602: forward: 5′-AAACCAACCTGGCTTACACC-3′, reverse: 5′-GATTTTATTATATAAATATTTTTTATAAAAATAATAC-3′. PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 90 s, 47°C for 90 s, 72°C for 3.5 min, with final extension at 72°C for 10 min.
RESULTS
Elevated levels of mtDNA helicase alleviate pausing of the replication fork and increase the abundance of replication intermediates
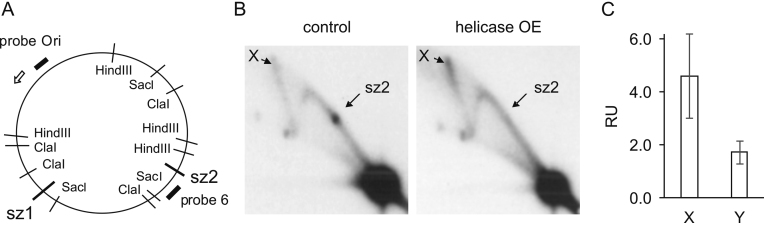
We established stable lines of D. melanogaster Schneider cells expressing wild-type mtDNA helicase (28). Immunoblot analysis of whole cell extracts indicated 0.3 μg of the recombinant mtDNA helicase per 106 overexpressing cells after 4 days of induction (Materials and Methods), whereas the endogenous helicase was undetectable in the control cells grown in parallel (data not shown). Based on earlier studies, we estimate that this represents a 7–20-fold increased expression relative to control cells (22,28). In order to evaluate the effects of elevated mtDNA helicase levels on the mtDNA replication process, we analyzed replication intermediates (RIs) in samples of total mitochondrial nucleic acid (mtNA) obtained from sucrose gradient-purified mitochondria of the control and mtDNA helicase-overexpressing cells, by 2DAGE as described previously (Materials and Methods, (29,30)). First, we cleaved the samples with ClaI restriction endonuclease (Figure 1A), and probed the resulting fragment bearing the previously-identified replication pause site that defines replication slow zone 2 (sz2) (29) (i.e. using probe 6 (30)). Notably, we observed that the signal from this prominent replication pause site is depleted strongly in samples from cells overexpressing the helicase (Figure 1B), indicating that mtDNA helicase alleviates pausing of the replication machinery. Furthermore, we observed an increase in the intensity of the RI signal in the mtDNA helicase-overexpressing samples; we detected a ∼1.6-fold increase in the signal of Y-shaped structures generated by duplex replication forks (Y arc), and a >4.5-fold increase in the signal of cruciform structures (X arc, Figure 1C). The increase in cruciform structures may relate to replication, repair and/ or recombination events. We reported previously that overexpression of mtDNA helicase in D. melanogaster Schneider cells increases mtDNA copy number ∼1.2-fold (28). We evaluated this in the current study using qPCR on total cellular DNA extracts, but observed no significant changes in the mtDNA copy number in helicase-overexpressing cells as compared to the control cells (data not shown). This discrepancy likely results from the fact that mtDNA helicase overexpression was induced for 14 days in the previous study as compared to four days in this study. Similarly, studies on human cell lines overexpressing mtDNA helicase for three days also showed unaltered mtDNA copy number (33,34). The substantial increase in the signal of RIs in the absence of mtDNA copy number changes suggests that overexpression of mtDNA helicase promotes increased initiation without complete replication of mtDNA molecules. This possibility is explored in more detail in later sections.
Figure 1.
Analysis of mtDNA replication intermediates from Schneider S2 cells overexpressing mtDNA helicase. (A) Schematic map of D. melanogaster mtDNA indicating the positions of hybridization probes as black, bold lines, replication slow zones sz1 and sz2 (29), and restriction endonuclease sites for ClaI, HindIII and SacI. Open arrow indicates the direction of replication. (B) 2DAGE of ClaI-treated mtNA isolated from control S2 cells (left panel) or mtDNA helicase-overexpressing S2 cells (right panel) hybridized with probe 6 (see Materials and Methods for details). Arrows indicate discrete spots on standard Y arcs that represent the major replication pause site at sz2 (see (30)). (C) ImageJ quantified mean hybridization intensity (±SD) of the X and Y arcs derived from the ClaI C restriction fragment of mtDNA from mtDNA helicase-overexpressing cells, relative to that from control DNA. Intensity values were normalized relative to the signal intensity of unreplicated DNA (major signal at the bottom right corner) as an internal standard. Data were quantified from 2DAGE images from three independent samples.
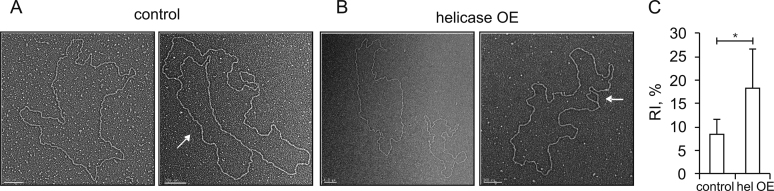
Electron microscopy identifies circular molecules bearing deletions in mtDNA from helicase- overexpressing cells
We applied electron microscopy (EM) to examine the effects of elevated levels of mtDNA helicase using a complementary method. We utilized the same samples as for 2DAGE, but first removed RNA by RNaseA digestion to reduce its interference in imaging (see Materials and Methods). As a control to test whether removal of free RNA affects the RI profiles, we analyzed these samples by 2DAGE, but found no differences from those containing total mtNA (see Figure 6D). In our EM analyses, we observed circular molecules bearing theta replication intermediate structures of various sizes (Figure 2A and B, right panels), comprising ∼8% of total circular molecules in the control samples versus ∼18% in samples from mtDNA helicase-overexpressing cells (Figure 2C). This increase of >2 fold in RIs is consistent with that inferred from 2DAGE analyses (Figure 1B and C). Next, we measured the contour length of circular mtDNA molecules from the control and helicase-overexpressing cell lines. Measuring the length of 336 control molecules, we observed a mean value of 6.1 μm (±0.3 μm), which appeared to fall within a single distribution that is consistent with the theoretical size (19517 bp) of the D. melanogaster mitochondrial genome. In striking contrast, the distribution of the same number of molecules of mtDNA from the helicase-overexpressing cells had a peak at a smaller size (mean of 5.6 ± 0.1 μm) together with a clear tailing towards shorter species. Indeed, only 27 of the 336 molecules examined fell within a secondary distribution with a mean value of 6.1 μm, similar to the control group. In contrast, 229 had a peak at a smaller size of 5.6 μm and 80 were distributed between 5 and 0.3 μm (Figure 2B). The latter could result from large deletions, albeit their relative abundance must be too low to detect either by ethidium bromide staining or Southern blotting with a probe containing the coding region sequence (see below).
Figure 2.
Electron microscopy of mtDNA replication intermediates isolated from control and mtDNA helicase-overexpressing Schneider S2 cells. (A) Representative electron microscopic images of circular (full-length (∼6 μm), left panel) and theta replication intermediate (right panel, indicated by the white arrow) mtDNA molecules obtained from control S2 cells. Scale bars 200 nm. (B) Representative electron microscopic images of circular (left panel; full-length (∼6 μm, upper molecule) and deleted (∼5 μm, lower molecule)) and theta replication intermediate (right panel, indicated by the white arrow; deleted, (∼5.6 μm)) mtDNA molecules obtained from mtDNA helicase-overexpressing S2 cells. Scale bars 200 nm. (C) EM quantification of mtDNA theta replication intermediates obtained from control and helicase-overexpressing S2 cells expressed as percentage of total monomeric mtDNA circles. The analysis was performed in three independent experiments with duplicate samples in each group. 3730 and 459 molecules were scored in control and helicase-overexpressing mtDNA samples, respectively. Statistical analysis was performed using the non-parametric Mann–Whitney U-Test and the data are presented as mean values. A P-value <0.05 was considered statistically significant.
The difference in the mean sizes of the two peaks (0.5 μm) corresponds to a deletion of approximately 1600 bp in the helicase-overexpressing line. Importantly, we observed that circular molecules putatively bearing such deletions do contain replication bubbles (Figure 2B, right panel). This observation implies the retention of the replication origin in the larger classes of deleted molecules, as indicated by the Southern blotting analysis described below. Taken together, the EM observations indicate that mtDNA molecules carrying deletions can remain circular and undergo replication.
Elevated levels of mtDNA helicase promote rearrangement of the A+T region
To examine further the effect of elevated levels of mtDNA helicase on the organization of the mtDNA, we treated mtNA samples with restriction endonuclease HindIII (Figure 1A). As described previously, the HindIII fragment B (HindIII B) encompasses the entire A+T region that contains the origin of replication (35). We observed that the HindIII B fragment of mtDNA obtained from the helicase-overexpressing cells migrates faster than that from the control cells cultured in parallel, whereas no change in the sizes of the A or C fragments was apparent (Figure 3A). Notably, despite the fact that equivalent amounts of total mtNA samples were analyzed, mtDNA in the samples from helicase-overexpressing cells was underrepresented (∼2.5 fold less), whereas there was a substantial increase in the quantity of heterogeneous material migrating with apparent molecular weight of ∼1 kb (Figure 3A). (That this material may represent RNA is suggested by subsequent analyses using RNases A and H1, see Figure 6D). To examine possible small size differences in the HindIII A and C fragments, we treated the HindIII-digested samples with SacI restriction endonuclease. SacI cleaves within the HindIII A and C fragments of mtDNA, but not the HindIII B fragment (Figure 1A). As for the HindIII digestion alone, SacI–HindIII digestion of the helicase-overexpressing sample showed no apparent differences as compared to the digestion pattern of the control cell sample run in parallel (Figure 3B). In addition, we analyzed the HindIII-cleaved mtNA samples by Southern blotting using a probe specific to the HindIII A fragment comprising a major portion of the coding region, including replication slow zone 2 (29). Slow zone 2 represents a major site of replication fork stalling, double-strand breaks, and possibly deletions (Figure 3D, see also Figure 1A and B, (30)). Nonetheless, we did not find any size alterations of the HindIII A fragment. This identifies the HindIII B fragment as the major site bearing size alterations that result from helicase overexpression. In addition to fragments consistent with the predicted restriction pattern for control mtDNA, we observed two additional bands in the helicase-overexpressing sample that migrate faster than the HindIII B fragment that were absent in the control sample (Figure 3B). To determine if these represent additional size variants of the HindIII B fragment, we analyzed the HindIII-cleaved mtNA samples by Southern blotting, for which we designed a probe specific for the A+T region that encompasses the site of the putative origin of replication (Figure 3C, see Figures 1A and 4A, and Materials and Methods). The analysis confirmed that the additional bands hybridized to the origin site-containing probe and are thus also HindIII B derivatives (Figure 3C). We estimate the sizes of the three HindIII B fragments (relative to a molecular weight standard run in parallel) in the helicase-overexpressing sample to be 5430 bp for HindIII B1, 4730 bp for HindIII B2, and 4080 bp for HindIII B3, in each case ± 90 bp. Given that the size of the Hind III B control mtDNA fragment is 5802 bp, the analysis indicates that helicase overexpression results in deletions of ∼370 bp (HindIII B1), ∼1070 bp (HindIII B2), and ∼1720 bp (HindIII B3) within the A+T region. The sizes estimated for the HindIII B1–3 deletions are consistent with the range of deletion sizes observed by EM.
Next, we analyzed by densitometry the molar fraction of the HindIII B fragments relative to the similarly migrating HindIII-SacI A fragment (Figure 3B). The molar fraction of the HindIII B fragment to the HindIII-SacI A fragment in the control sample was 1 (∼8 fmol for each). However, the molar fraction of these fragments in the helicase-overexpressing sample was 0.5 for the HindIII B1, 0.15 for the HindIII B2, and 0.05 for the HindIII B3 fragment, which together comprise ∼0.7 of the molar fraction of the HindIII-SacI fragment. This suggests that ∼30% of the remaining HindIII B fragment molecules bearing deletions are not detectable as discrete bands by ethidium bromide staining. Because in our Southern-blotting analyses we did not detect any HindIII B derivatives other than the B1–3 species in the helicase-overexpressing sample (Figures 3C, see also 4B), the remaining putative deletion molecules have likely lost that part of the A+T region that is complementary to the probe (see Figures 1A and 4A). Alternatively, they may be extremely heterogeneous.
The major classes of mtDNA deletions generated by elevated levels of mtDNA helicase map to the repeat I region of A+T region
D. melanogaster exhibits a complex organization of the A+T region of its mtDNA (Figure 4A). The A+T region leftward from the replication origin towards the HindIII restriction site and adjacent to the 12S rRNA gene bears five repeated sequence elements, designated as repeat I, which vary in size from 338 to 373 bp (Rep1, Figure 4A, (35)). We demonstrated recently that the number of repeated elements in this region varies from three to six among wild-type strains of D. melanogaster (36). Hence, we reasoned that this region may represent a hot spot for deletions resulting from overexpression of the mtDNA helicase. Notably, the sizes of the three major deletions we observed match those of the Rep1 elements; deletions in the HindIII B1 (Δ∼370 bp), B2 (Δ∼1070), and B3 (Δ∼1720) fragments correspond approximately to the loss of one (up to 373 bp), three (up to 1063 bp) and all five (1739 bp) Rep1 elements, respectively.
To test whether the HindIII B fragment deletions in the helicase-overexpressing mtNA samples map to the Rep1 region, we analyzed the region by PCR using primers covering nts 12841 (16S rRNA gene) -17602 (origin probe rev primer) downstream of the Rep1 region through the replication origin (Figure 4A, Materials and Methods). Strikingly, the PCR reaction on the mtNA from the helicase-overexpressing cells yielded three distinct products of ∼4.5 (Δ∼300 bp), ∼3.5 (Δ∼1 kb), and >2 kb (Δ∼ 2 kb); the deletions implied by these products correspond in size to those of the HindIII B1–3 fragment deletions (Figure 4B). This identifies the repeat I region as the specific site of the helicase-generated deletions.
Salminen et al. demonstrated that the number of Rep1 elements correlates positively with mtDNA copy number and developmental time, which implies a relationship between them and the replication process (36). To investigate how deletions within the Rep1 region affect replication initiation, we analyzed RIs within the HindIII B fragments by 2DAGE, using the probe specific for the replication origin (ori probe, see Figure 4A). We observed that mtNA from control cells yields three prominent signals corresponding with the HindIII B fragment (Figure 4C). The first represents molecules in which the replication fork has progressed unidirectionally from the replication origin through the Rep1 region and past the first-encountered HindIII restriction site (29), which are observed as a focus of Y-shaped molecules of uniform length that we have designated as the ‘Y spot’. Replication forks that did not yet progress beyond the HindIII restriction site yielded the second prominent signal of a standard bubble arc (Figure 4C, left panel). Notably, the intensity of the bubble arc in the helicase-overexpressing samples was ∼10-fold lower than in the control cells (Figure 4D). The third signal represents double Y (dY) structures generated as the replication fork approaches the origin/ terminus, prior to termination of replication within the A+T region (Figure 4C, left panel inset). The dY signal is relatively faint and is obscured by the prominent Y spot. The minimal separation of the dY molecules from the Y spot is consistent with the near proximity of the termination site and the HindIII B restriction site, at this level of resolution of high-molecular weight species. In contrast, the helicase-overexpressing sample gave rise to five distinct foci of uneven intensity and migration (Figure 4C, right panel). Foci 1 and 2 likely represent termination intermediates. The variable positions of the Y spots indicated as 3, 4 and 5 (Figure 4C, right panel) likely result from deletions within the Rep1 region of the HindIII B fragment, which is evident as three corresponding signals within the unreplicated DNA signal (Figure 4C, right panel inset). The observation of multiple foci generated in the helicase-overexpressing sample indicates that molecules bearing deletions can still be replicated, a finding that is consistent with our EM observations.
Elevated levels of mtDNA helicase promote formation of cruciform-like structures within the A+T region, and excessive catenation of mtDNA molecules
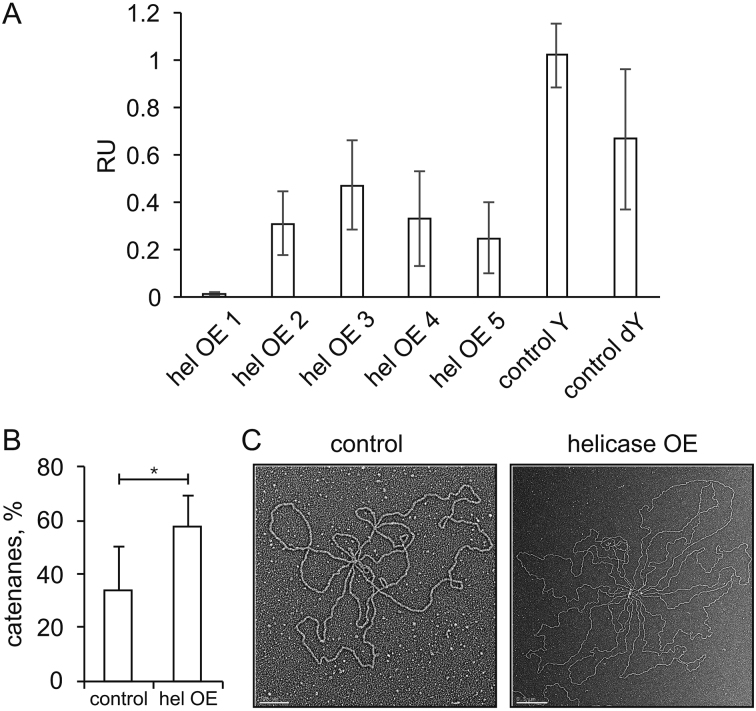
Because overexpression of the replicative helicase enhanced substantially the quantity of cruciform-like RIs (Figure 1C), we tested the sensitivity of the helicase overexpression-generated RIs within the HindIII B fragment to the cruciform-cleaving enzyme, RusA. 2DAGE analysis of the treated versus untreated samples showed a >2-fold decrease in signal of all of the Y-spots in the helicase-overexpressing sample, and a decrease in the abundance of the dY RIs, whereas it had no effect on the corresponding RIs of the control sample (Figure 5A, see also Supplementary Figure S1).
Figure 5.
Putative replication-stalling intermediates and catenanes in mtDNA helicase-overexpressing Schneider S2 cells. (A) Densitometric analysis of the sensitivity of dY- and Y-spot replication intermediates (see Figure 4C) to the cruciform-structure resolvase RusA. HindIII-digested mtNA samples obtained from control and mtDNA helicase-overexpressing S2 cells were treated with RusA (see Materials and Methods). The treated and untreated samples were analyzed together by 2DAGE and hybridization with the Ori probe (Supplementary Figure S1). The relative mean hybridization intensity (±SD) of dY and Y spots derived from mtDNA that was treated and untreated by RusA from control and helicase-overexpressing cells (where hel OE 1, 2…5 designate the species labeled 1,2….5 in Figure 4C) was quantified with Image-J. Intensity values were normalized relative to the signal intensity of unreplicated DNA as an internal standard. (B) Quantitation of catenated mtDNA obtained from control and helicase-overexpressing S2 cells, expressed as a percentage of total open and supercoiled monomeric mtDNA circles, based on visualization by EM. The analysis was performed in three independent experiments with duplicate samples in each group. Statistical analysis was performed using the non-parametric Mann–Whitney U-Test and the data were presented as mean values. A P-value ≤0.01 was considered statistically significant. (C) Representative electron micrographs of catenated mtDNA from control (left panel) and helicase-overexpressing (right panel) S2 cells. Molecules were considered as catenated if multiple mtDNA genomes were found joined together. Scale bars are 0.2 and 0.5 μm for the images of control and helicase-overexpressing samples, respectively.
Furthermore, earlier studies reported that a significant fraction of D. melanogaster mtDNA remains physically interlinked, and that catenation is related to the mtDNA replication process (37). We evaluated the effect of helicase overexpression on catenation of mtDNA by EM. We observed that ∼30% of circular mtDNA molecules from control cells form catenanes comprising two to five genome monomers. Helicase overexpression results in an ∼2-fold increase in these apparently catenated molecules, with up to ten genomic monomers per catenane (Figure 5B and C).
Elevated levels of mtDNA helicase promote incorporation of RNA into mtDNA
To assess further the impact of elevated levels of mtDNA helicase on structural aspects of the mitochondrial genome, we used a Southern-blotting analysis of the undigested mtNA samples (Figure 6A, B). We observed that a majority of mtDNA molecules extracted from control S2 cells consist of approximately equal fractions of open circular, supercoiled and linear forms (Figure 6A and B, lane 2). Notably, removal of free RNA by RNaseA treatment increases the overall quantity of mtDNA that migrates through the gel during electrophoresis (compare ± RNase lanes in Figure 6A and B). Moreover, removal of RNA shifts modestly the proportions of mtDNA forms, such that open circular forms comprise ∼50%, and linear and supercoiled forms each comprise ∼25%. This corroborates earlier studies by Joers et al. (30) in which the authors suggested that a significant fraction of D. melanogaster mtDNA is complexed with RNA, forming structures that are too large to enter the electrophoretic gel. This effect appears to be exacerbated in samples obtained from the mtDNA helicase-overexpressing cells, as only ∼30% of the total mtDNA entered the gel in the absence of RNaseA treatment as compared to ∼70% in the case of control samples (Figure 6A and B, lanes 3 and 4). These data show that elevated levels of helicase promote complexing of mtDNA molecules with RNA, which is also corroborated indirectly by the appearance of an apparent excess of RNA (Figure 6D) relative to mtDNA that we observed in helicase-overexpressing samples (see Figure 2A). In contrast to the control sample in the absence of RNase digestion, ∼80% of mtDNA that entered the gel is linear, and the remainder is open circular (Figure 6A and B, lanes 3 and 4). Treatment with RNaseA shifted this only slightly to ∼65% linear molecules and ∼30% open circular molecules. The large increase in the fraction of linear forms implies strongly that overexpression of the helicase results in double-strand breakage. Notably, the supercoiled form was undetectable in both RNase-treated and untreated samples (Figure 6A and B, lanes 3 and 4), implying that it may have been damaged as a result of excessive unwinding. Indeed, we observed by electron microscopy multiple nicked or broken molecules (Figure 6C), which were much more abundant in all of the helicase-overexpressing samples analyzed.
To investigate the effect of RNA incorporation into mtDNA molecules on the replication process, we analyzed RNaseA- (specific for free RNA) and RNaseH1- (RNA–DNA hybrid specific) treated samples by 2DAGE. Although as mentioned earlier we did not observe any effects of removing free RNA on the RI profile (Figure 6D, Supplementary Figure S2), we found that combined RNase A and H1 treatment of the helicase-overexpressing samples reduced dramatically (>5-fold) the relative signal of RIs (Figure 6D), with the appearance of a faint sub-Y arc in the 2DAGE image (Supplementary Figure S2). The loss of the signal indicates a strong contribution (or retention) of RNA in RIs, with the sub-Y arc representing extensive ssDNA patches resulting from removal of the annealed RNA.
DISCUSSION
Mitochondrial replisome components are often upregulated under mitochondrial stress conditions, which can be interpreted as a compensatory response to reduced mitochondrial function. mtDNA helicase has been proposed as a major regulator of mtDNA copy number, as its overexpression and knockdown result, respectively, in a substantial increase and decrease of mtDNA copy number across various taxa (19,22). Notably, elevated levels of helicase are associated with various pathological conditions. For example, treatment of colon cancer cells with doxorubicin (a widely-used anticancer drug) results in an ∼4-fold increase in mtDNA helicase level (38), and increased expression of mtDNA helicase has been observed in a mouse model of dilated cardiomyopathy (21). Our 2DAGE and EM analyses showed that overexpression of the helicase in Schneider cells results in a net increase in the abundance of replication intermediates representing replication forks progressing through the coding region (as detected by hybridization to probe 6, see Figure 1A). Further, it alleviated programmed pausing at the mTTF binding site designated as sz2 (see Figure 1B). By contrast, in the non-coding (A+T) region, our analysis showed an ∼10-fold decrease in the relative abundance of the bubble arc (as detected by hybridization to the ori probe, see Figure 4D). Taken together, accumulation of RIs within the coding region and their apparent decrease in the vicinity of the replication origin would be consistent with the hypothesis that overexpression of helicase gives rise to increased unwinding that enhances fork assembly and/ or movement within the ori region but as a result of the generation of more complex replicating structures and stalling intermediates, fork movement is reduced as it progresses around the molecule. An increase in the fraction of replication intermediates in the absence of changes in mtDNA copy number has been also reported in other studies on mtDNA maintenance, and is considered to be a hallmark of replication-fork stalling (39,40).
More detailed analyses of mtDNA molecules using both 2DAGE and EM demonstrated that elevated levels of mtDNA helicase promote deletions that occur specifically within the Rep1 region of the non-coding segment. Intriguingly, the organization of the affected region is variable among Drosophila species and has been correlated recently with mtDNA copy number and developmental time in D. melanogaster (36). Furthermore, mtDNA copy number has been correlated directly with longevity and aging (41), which together suggest a possible relationship between organization of mitochondrial genome organization and aging.
Drosophila melanogaster mtDNA replication is unidirectional (29), and formation and expansion of the replication bubble during initiation and the early stages of elongation of the replication process occur within the Rep1 region (Figure 4A). The Rep1 region has a GC content as low as 5.8% among Drosophila species, facilitating duplex melting during replication initiation. This unusually high A+T content could facilitate excessive unwinding by the helicase under overexpression conditions. The Rep1 region comprises many shorter DNA sequence repeats even within the individual repeated Rep1 elements (35), likely making it highly susceptible to the formation of multiple secondary structures. Stalling of the replication fork at sites of secondary structure formation often generates deletions due to DNA polymerase slippage (42,43), when DNA synthesis is reinitiated by recapture of the free 3′-OH end to bypass the sequence of the looped out secondary structure. Although each of the Rep1 repeats contains multiple palindromic motifs that could promote DNA polymerase slippage, it appears that Rep1 deletions likely occur in a specific manner. The estimated sizes of the HindIII B1–3 fragments obtained from the mtDNA helicase overexpressing samples indicate that either 1, 3 or 5 Rep1 elements are deleted, rather than each consecutively (see Figure 3C and related text). To investigate this, we subjected the Rep1 region sequence to computational analysis for the potential to form secondary structures (see Materials and Methods). We found that a putative hairpin formed by the entire Rep1 region may involve pairing of repeated elements from opposite ends, while the middle (third) repeat undergoes internal base pairing; that is, repeat 1-A pairs with 1-C, 1-B pairs with 1-B2, and 1-C/A pairs internally (Supplementary Figure S3). Each of the Rep1 elements contains a conserved palindromic sequence of a repeated TA stretch spanning nts 65 to 84, and a stretch of TTTTTTTTnAAAAAAAAA (where n indicates a T in all the Rep1 elements but I-C/A, in which it is A) ∼90 nt downstream, which facilitate hairpin structure formation. The I-C/A segment contains a second copy of the conserved palindromic sequence at its opposite end (nts 312–332) that enables internal pairing. Given this scenario, it is likely that deletions are generated by slippage of Pol γ over the 1-C/A hairpin, resulting in a single deletion to yield the HindIII B1 fragment, a 1-B:1-B2 together with a 1-C/A-generated hairpin resulting in a triple Rep1 element deletion to yield the HindIII B2 fragment, or a 1-A:1-C (+ 1-B:1-B2, and intrinsic 1-C/A)-generated hairpin resulting in deletion of the entire Rep1 region to yield the HindIII B3 fragment. This proposed mechanism is consistent with the sizes of prominent deletions observed upon mtDNA helicase overexpression (Figure 3C and related text). Notably, cases of large deletions within the human mitochondrial genome have also been proposed to result from polymerase slippage, though this mechanism has been debated (44–47).
Interestingly, we also observed that helicase overexpression results in accumulation of X-shaped RIs (Figure 1C), which have been attributed to recombination events (30). Homologous recombination serves to repair double-strand breaks in mtDNA, and deletions are often generated in the process (24,48). Homologous recombination-generated size variations of the D. melanogaster mitochondrial genome have been documented recently, and the authors of the study speculated that homologous recombination explains the diversity of A+T region size among fly mitochondrial genomes (48). Hence, deletions within the Rep1 region could result from homologous recombination instead of replication slippage. Consistent with this possibility, we show that deleted molecules can remain circular (see Figure 2B) and be replicated (see Figure 4C and related text). Because the repeated elements within the Rep1 repeat region share high sequence identity, they may serve as hot spots for homologous recombination, though the precise mechanism of recombination-related deletions remains to be investigated.
We also investigated whether deletions can occur outside of the A+T region by probing the coding region in the vicinity of slow zone 2 (see Figure 1A), which is a region of prominent replication fork stalling (see Figure 1B, control panel), double-strand breaks and possible deletions (30). However, we detected no size alterations within the coding region (see Figure 3A and D). This implies that most aberrant replication intermediates resulting from elevated helicase levels, at least those within the coding region, can be processed efficiently by reinitiation or turnover mechanisms, avoiding the production of rearranged mitochondrial genomes.
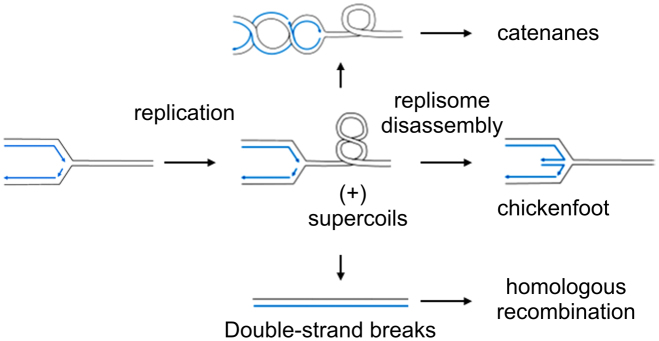
Frequent and extensive deletions, together with the observed increase in RIs in the absence of changes in mtDNA copy number, would suggest that increased helicase levels may result in replication fork stalling. Consistent with that, we also observed a large increase in the fraction of linear mtDNA in the helicase-overexpressing samples. Double-strand breaks are a prominent consequence of replication fork stalling, and have been associated with mtDNA helicase dysfunction in mammals (24). Though the direct cause of replication stalling under our experimental conditions remains to be elucidated, one possibility is that enhanced helicase activity results initially in unhampered, rapid replication fork movement coupled with helix unwinding, leading to an increase in positive supercoiling of the parental duplex ahead of the replication fork. If unresolved, the mechanical strain imposed may result in replisome stalling and disassembly (49). Superhelical strain can be relieved by regression of the replication fork and generation of duplex cruciform-like, ‘chicken-foot’ structures (Figure 7), which are RusA-sensitive (30). Indeed, we observed a signature of chicken-foot structures within the A+T region consistent with the HindIII B fragment deletions (i.e., between the replication origin and first HindIII restriction site (see Figure 5A and Supplementary Figure S1, and related text)). Interestingly, the appearance of RusA-sensitive chicken-foot structures was documented in D. melanogaster upon knockdown of mitochondrial transcription termination factor (mTTF) (30). Superhelical strain may be also relieved by inter-winding of the daughter duplexes behind the fork (Figure 7), which may give rise to catenanes (49), which we also observed (see Figure 5 B, C and related text). Superhelical strain can ultimately cause double-strand breaks. Consistent with that, we observed a depletion in the supercoiled form and an increase in the fraction of linear form mtDNA from helicase-overexpressing cells (see Figure 6 A and B, and related text). Together, the appearance of putative chicken-foot structures, an increase in the fraction of catenated molecules, and a substantial increase in double-strand breaks are consistent with a conclusion that elevated levels of the mtDNA helicase result in replication fork stalling.
Figure 7.
Schematic representation of replication fork-imposing strain in the DNA template. Supercoiling ahead of the fork may result in regression of the replication fork and the appearance of ‘chicken-foot’ structures, or inter-coiling of daughter duplexes behind the fork, which may lead to catenation of daughter molecules (image modified from Postow et al. (49)). Replication fork stalling may ultimately result in double-strand breaks, which can be repaired in mitochondria by homologous recombination (24). Parental DNA strands are shown in black and daughter DNA strands are in blue.
The decrease in site-specific pausing in replication slow zones that we observed in mtDNA from helicase-overexpressing cells was also observed upon mTTF knockdown, resulting in the generation of abnormal RIs containing extensive tracts of incorporated RNA (30). Overexpression of the mtDNA helicase results in an ∼4-fold increase in apparent RNA levels and the formation of high molecular weight RNA-mtDNA complexes that are unable to enter the gel during agarose gel electrophoresis (See Figure 6A, B). Notably, we found that the RIs of helicase-overexpressing samples remain sensitive to combined treatment with RNases A and H1, although not to either of these individually (see Figure 6D, and Supplementary Figure S2), whereas the mTTF knockdown-generated RIs were RNase H1 sensitive. Moreover, in contrast to the RIs generated under conditions of mTTF knockdown, the treatment of mtNA obtained from the helicase-overexpressing mitochondria with RNases did not yield an abundant sub-Y arc, but rather decreased the overall signal of RIs (see Figure 6D) to yield a faint sub-Y arc (see Supplementary Figure S2). This suggests that the resulting RNA-containing RIs are even more extensive than those generated by mTTF knockdown, most likely with fragile sites or nicks on both strands. The minimal alteration in migration of the Y arc in material from control cells upon RNase H1 treatment (30), implies that tracts of incorporated RNA are usually short and/ or transient. The disturbances produced by mTTF knockdown (30) and helicase overexpression are qualitatively similar, but the latter appear to be more dramatic. Their similar effects may suggest that mtDNA helicase and mTTF function in concert. Both manipulations led to the generation of aberrant RIs containing extensive RNA tracts, whilst MTERF1, a mammalian homologue of mTTF, has been shown previously to function as a directional contrahelicase (50) against TWINKLE, the mammalian orthologue of the Drosophila mtDNA helicase. mTTF may thus interact with mtDNA helicase to limit the progression of replication forks into the heavily and oppositely transcribed regions lying beyond its binding sites. Either mTTF knockdown or helicase overexpression overcomes this regulation, leading to the generation of aberrant RIs containing incorporated RNA, and impairing genome stability. Intriguingly, the MTERF1 binding site in humans is a hotspot for genomic rearrangements (51), and the mTTF binding sites in Drosophila have been associated with inter-mtDNA recombination (48).
The physiological relevance of the increased level of RNA and the mechanism of its incorporation into mtDNA replication intermediates upon mtDNA helicase overexpression warrants further investigation. We speculate that it may reflect a compensatory effect to increase the level of mitochondrial respiratory proteins, and/ or to protect long stretches of ssDNA resulting from increased helicase activity. The latter is reminiscent of the RITOLS model of replication reported in vertebrates (52,53) and interestingly, human mtDNA helicase has been speculated to promote RNA incorporation in the RITOLS replication mode (54). To our knowledge, our study reports the first direct evidence linking mtDNA helicase and incorporation of RNA into mitochondrial replication intermediates. At the same time, we note that DNA strand breakage is elevated upon helicase overexpression in our system. A possible direct interaction of mtDNA helicase and RNA (or the transcriptional machinery) should also be investigated.
Earlier studies have demonstrated that helicase overexpression increases mtDNA copy number (19,22) and provides beneficial effects in various pathological conditions (25–27,55). The latter may relate directly to the complex structure of human mtDNA found in heart muscle, in which a substantial fraction is found as catenated multimers that accumulate with age (56). mtDNA catenanes are postulated to be maintained stably to assure genome function at a high level. The same study demonstrated that although murine heart catenanes are not generally found, helicase overexpression promotes their formation. In this study, we observed that overexpression of helicase results in at least a 2-fold increase in the fraction of catenated molecules, as well as the number of mtDNA molecules per catenane (see Figure 5B). Notably, our earlier studies on helicase overexpression demonstrated that upon exposure to high helicase levels much longer than that applied in this study (14 versus 4 days, respectively), mtDNA copy number increased 1.2-fold (28). We suggest that this may be due to an accumulation of catenated molecules, similar to that observed in the human heart. The observation of catenanes in the absence of mtDNA copy number changes may be suggestive of early, direct effects of excess mtDNA helicase, whereas increased mtDNA copy number may represent an indirect, cumulative effect. Indeed, excess mtDNA helicase introduces strain to the template DNA, which is likely to promote the formation of catenanes (Figure 7). Interestingly, a strand exchange activity of mtDNA helicase has been documented recently, which may contribute to their formation (57). Overall, this implies that the formation of catenanes is a mechanism for stabilization of the mtDNA genome to assure its function in a stressful environment. The endogenous mechanism regulating mtDNA helicase is as yet unknown. An understanding of this mechanism might pave the way for the development of therapeutic strategies to stabilize mtDNA in human diseases.
In sum, our current study indicates that elevated levels of mtDNA helicase serve to enhance mtDNA replication, albeit to the extent of destabilizing the structural integrity of the genome. It seems likely that the activity of mtDNA helicase is regulated in vivo and / or coordinated by additional factors. For example, formation of catenanes and retention of RNA in replication intermediates may serve to stabilize the genome in cases of aberrant mtDNA activity.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Päivi Lillsunde for her contributions in developing the protocols for preparation of mtDNA samples for 2DAGE and EM analyses of replication intermediates, and for developing the general approach for mtDNA analysis by 2DAGE used in this study. We also thank Merja H. Jokela for preparation of the wild-type and helicase-overexpressing cells, and for immunoblot analyses of overexpression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM45295 to L.S.K., GM31819 and ES013773 to J.D.G]; Academy of Finland [139587, 256615, 272376 to H.T.J.]; University of Tampere [to G.L.C in part]. Funding for open access charge: Internal funds from Michigan State University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Monnot S., Samuels D.C., Hesters L., Frydman N., Gigarel N., Burlet P., Kerbrat V., Lamazou F., Frydman R., Benachi A. et al. . Mutation dependance of the mitochondrial DNA copy number in the first stages of human embryogenesis. Hum. Mol. Genet. 2013; 22:1867–1872. [DOI] [PubMed] [Google Scholar]
- 2. Lee H.C., Wei Y.H.. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. In. J. Biochem. Cell Biol. 2005; 37:822–834. [DOI] [PubMed] [Google Scholar]
- 3. Moraes C.T., Ciacci F., Silvestri G., Shanske S., Sciacco M., Hirano M., Schon E.A., Bonilla E., DiMauro S.. Atypical clinical presentations associated with the MELAS mutation at position 3243 of human mitochondrial DNA. Neuromusc. Disord.: NMD. 1993; 3:43–50. [DOI] [PubMed] [Google Scholar]
- 4. Liu H., Ma Y., Fang F., Zhang Y., Zou L., Yang Y., Zhu S., Wang S., Zheng X., Pei P. et al. . Wild-type mitochondrial DNA copy number in urinary cells as a useful marker for diagnosing severity of the mitochondrial diseases. PLoS One. 2013; 8:e67146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miyako K., Kai Y., Irie T., Takeshige K., Kang D.. The content of intracellular mitochondrial DNA is decreased by 1-methyl-4-phenylpyridinium ion (MPP+). J. Biol. Chem. 1997; 272:9605–9608. [DOI] [PubMed] [Google Scholar]
- 6. Lee H.C., Yin P.H., Lu C.Y., Chi C.W., Wei Y.H.. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem. J. 2000; 348:425–432. [PMC free article] [PubMed] [Google Scholar]
- 7. Bonod-Bidaud C., Giraud S., Mandon G., Mousson B., Stepien G.. Quantification of OXPHOS gene transcripts during muscle cell differentiation in patients with mitochondrial myopathies. Exp. Cell Res. 1999; 246:91–97. [DOI] [PubMed] [Google Scholar]
- 8. Gu F., Chauhan V., Kaur K., Brown W.T., LaFauci G., Wegiel J., Chauhan A.. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl. Psychiatry. 2013; 3:e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giulivi C., Zhang Y.F., Omanska-Klusek A., Ross-Inta C., Wong S., Hertz-Picciotto I., Tassone F., Pessah I.N.. Mitochondrial dysfunction in autism. JAMA. 2010; 304:2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H., Buhay C., Kang H., Kim S.C., Fahey C.C. et al. . The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014; 26:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reznik E., Miller M.L., Senbabaoglu Y., Riaz N., Sarungbam J., Tickoo S.K., Al-Ahmadie H.A., Lee W., Seshan V.E., Hakimi A.A. et al. . Mitochondrial DNA copy number variation across human cancers. eLife. 2016; 5:e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mayr J.A., Meierhofer D., Zimmermann F., Feichtinger R., Kogler C., Ratschek M., Schmeller N., Sperl W., Kofler B.. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res. 2008; 14:2270–2275. [DOI] [PubMed] [Google Scholar]
- 13. Simonnet H., Demont J., Pfeiffer K., Guenaneche L., Bouvier R., Brandt U., Schagger H., Godinot C.. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis. 2003; 24:1461–1466. [DOI] [PubMed] [Google Scholar]
- 14. Kaguni L.S. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004; 73:293–320. [DOI] [PubMed] [Google Scholar]
- 15. Ciesielski G.L., Oliveira M.T., Kaguni L.S.. Animal Mitochondrial DNA Replication. Enzymes. 2016; 39:255–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spelbrink J.N., Toivonen J.M., Hakkaart G.A., Kurkela J.M., Cooper H.M., Lehtinen S.K., Lecrenier N., Back J.W., Speijer D., Foury F. et al. . In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000; 275:24818–24828. [DOI] [PubMed] [Google Scholar]
- 17. Lefai E., Calleja M., Ruiz de Mena I., Lagina A.T. 3rd, Kaguni L.S., Garesse R.. Overexpression of the catalytic subunit of DNA polymerase gamma results in depletion of mitochondrial DNA in Drosophila melanogaster. Mol. Gen. Genet.: MGG. 2000; 264:37–46. [DOI] [PubMed] [Google Scholar]
- 18. Davis A.F., Ropp P.A., Clayton D.A., Copeland W.C.. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res. 1996; 24:2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A.. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004; 13:3219–3227. [DOI] [PubMed] [Google Scholar]
- 20. Pohjoismaki J.L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P.J., Griffith J.D., Holt I.J. et al. . Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 2009; 284:21446–21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milenkovic D., Matic S., Kuhl I., Ruzzenente B., Freyer C., Jemt E., Park C.B., Falkenberg M., Larsson N.G.. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 2013; 22:1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsushima Y., Kaguni L.S.. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J. Biol. Chem. 2007; 282:9436–9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez-Martinez A., Calleja M., Peralta S., Matsushima Y., Hernandez-Sierra R., Whitworth A.J., Kaguni L.S., Garesse R.. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS One. 2012; 7:e43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pohjoismaki J.L., Goffart S., Spelbrink J.N.. Replication stalling by catalytically impaired Twinkle induces mitochondrial DNA rearrangements in cultured cells. Mitochondrion. 2011; 11:630–634. [DOI] [PubMed] [Google Scholar]
- 25. Pohjoismaki J.L., Williams S.L., Boettger T., Goffart S., Kim J., Suomalainen A., Moraes C.T., Braun T.. Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19408–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka A., Ide T., Fujino T., Onitsuka K., Ikeda M., Takehara T., Hata Y., Ylikallio E., Tyynismaa H., Suomalainen A. et al. . The overexpression of Twinkle helicase ameliorates the progression of cardiac fibrosis and heart failure in pressure overload model in mice. PLoS One. 2013; 8:e67642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikeda M., Ide T., Fujino T., Arai S., Saku K., Kakino T., Tyynismaa H., Yamasaki T., Yamada K., Kang D. et al. . Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS One. 2015; 10:e0119687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsushima Y., Kaguni L.S.. Functional importance of the conserved N-terminal domain of the mitochondrial replicative DNA helicase. Biochim. Biophys. Acta. 2009; 1787:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joers P., Jacobs H.T.. Analysis of replication intermediates indicates that Drosophila melanogaster mitochondrial DNA replicates by a strand-coupled theta mechanism. PLoS One. 2013; 8:e53249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joers P., Lewis S.C., Fukuoh A., Parhiala M., Ellila S., Holt I.J., Jacobs H.T.. Mitochondrial transcription terminator family members mTTF and mTerf5 have opposing roles in coordination of mtDNA synthesis. PLoS Genet. 2013; 9:e1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Griffith J.D., Christiansen G.. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 1978; 7:19–35. [DOI] [PubMed] [Google Scholar]
- 32. Thresher R., Griffith J.. Electron microscopic visualization of DNA and DNA-protein complexes as adjunct to biochemical studies. Methods Enzymol. 1992; 211:481–490. [DOI] [PubMed] [Google Scholar]
- 33. Wanrooij S., Goffart S., Pohjoismaki J.L., Yasukawa T., Spelbrink J.N.. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007; 35:3238–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rajala N., Gerhold J.M., Martinsson P., Klymov A., Spelbrink J.N.. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 2014; 42:952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis D.L., Farr C.L., Farquhar A.L., Kaguni L.S.. Sequence, organization, and evolution of the A+T region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 1994; 11:523–538. [DOI] [PubMed] [Google Scholar]
- 36. Salminen T.S., Oliveira M.T., Cannino G., Lillsunde P., Jacobs H.T., Kaguni L.S.. Mitochondrial genotype modulates mtDNA copy number and organismal phenotype in Drosophila. Mitochondrion. 2017; 34:75–83. [DOI] [PubMed] [Google Scholar]
- 37. Rubenstein J.L., Brutlag D., Clayton D.A.. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977; 12:471–482. [DOI] [PubMed] [Google Scholar]
- 38. Yadav N., Pliss A., Kuzmin A., Rapali P., Sun L., Prasad P., Chandra D.. Transformations of the macromolecular landscape at mitochondria during DNA-damage-induced apoptotic cell death. Cell Death Dis. 2014; 5:e1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Torregrosa-Munumer R., Goffart S., Haikonen J.A., Pohjoismaki J.L.. Low doses of ultraviolet radiation and oxidative damage induce dramatic accumulation of mitochondrial DNA replication intermediates, fork regression, and replication initiation shift. Mol. Biol. Cell. 2015; 26:4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torregrosa-Munumer R., Forslund J.M.E., Goffart S., Pfeiffer A., Stojkovic G., Carvalho G., Al-Furoukh N., Blanco L., Wanrooij S., Pohjoismaki J.L.O.. PrimPol is required for replication reinitiation after mtDNA damage. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:11398–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camus M.F., Wolf J.B., Morrow E.H., Dowling D.K.. Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr. Biol.: CB. 2015; 25:2717–2722. [DOI] [PubMed] [Google Scholar]
- 42. Pinder D.J., Blake C.E., Lindsey J.C., Leach D.R.. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 1998; 28:719–727. [DOI] [PubMed] [Google Scholar]
- 43. Viguera E., Canceill D., Ehrlich S.D.. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001; 20:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shoffner J.M., Lott M.T., Voljavec A.S., Soueidan S.A., Costigan D.A., Wallace D.C.. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:7952–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Srivastava S., Moraes C.T.. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 2005; 14:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W., Wanrooij S., Spelbrink J.N., Lightowlers R.N., Turnbull D.M.. What causes mitochondrial DNA deletions in human cells. Nat. Genet. 2008; 40:275–279. [DOI] [PubMed] [Google Scholar]
- 47. Phillips A.F., Millet A.R., Tigano M., Dubois S.M., Crimmins H., Babin L., Charpentier M., Piganeau M., Brunet E., Sfeir A.. Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol. Cell. 2017; 65:527–538. [DOI] [PubMed] [Google Scholar]
- 48. Ma H., O’Farrell P.H.. Selections that isolate recombinant mitochondrial genomes in animals. eLife. 2015; 4, doi:10.7554/eLife.07247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Postow L., Crisona N.J., Peter B.J., Hardy C.D., Cozzarelli N.R.. Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shi Y., Posse V., Zhu X., Hyvarinen A.K., Jacobs H.T., Falkenberg M., Gustafsson C.M.. Mitochondrial transcription termination factor 1 directs polar replication fork pausing. Nucleic Acids Res. 2016; 44:5732–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kajander O.A., Rovio A.T., Majamaa K., Poulton J., Spelbrink J.N., Holt I.J., Karhunen P.J., Jacobs H.T.. Human mtDNA sublimons resemble rearranged mitochondrial genoms found in pathological states. Hum. Mol. Genet. 2000; 9:2821–2835. [DOI] [PubMed] [Google Scholar]
- 52. Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J.. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006; 25:5358–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reyes A., Kazak L., Wood S.R., Yasukawa T., Jacobs H.T., Holt I.J.. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013; 41:5837–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Holt I.J., Jacobs H.T.. Unique features of DNA replication in mitochondria: a functional and evolutionary perspective. BioEssays. 2014; 36:1024–1031. [DOI] [PubMed] [Google Scholar]
- 55. Inoue T., Ikeda M., Ide T., Fujino T., Matsuo Y., Arai S., Saku K., Sunagawa K.. Twinkle overexpression prevents cardiac rupture after myocardial infarction by alleviating impaired mitochondrial biogenesis. Am. J. Physiol. Heart Circ. Physiol. 2016; 311:H509–H519. [DOI] [PubMed] [Google Scholar]
- 56. Pohjoismaki J.L., Goffart S., Taylor R.W., Turnbull D.M., Suomalainen A., Jacobs H.T., Karhunen P.J.. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One. 2010; 5:e10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sen D., Patel G., Patel S.S.. Homologous DNA strand exchange activity of the human mitochondrial DNA helicase TWINKLE. Nucleic Acids Res. 2016; 44:4200–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.