Abstract
During translation’s elongation cycle, elongation factor G (EF-G) promotes messenger and transfer RNA translocation through the ribosome. Until now, the structures reported for EF-G–ribosome complexes have been obtained by trapping EF-G in the ribosome. These results were based on use of non-hydrolyzable guanosine 5′-triphosphate (GTP) analogs, specific inhibitors or a mutated EF-G form. Here, we present the first cryo-electron microscopy structure of EF-G bound to ribosome in the absence of an inhibitor. The structure reveals a natural conformation of EF-G·GDP in the ribosome, with a previously unseen conformation of its third domain. These data show how EF-G must affect translocation, and suggest the molecular mechanism by which fusidic acid antibiotic prevents the release of EF-G after GTP hydrolysis.
INTRODUCTION
The ribosome is a large molecular machine performing protein synthesis, ‘translation,’ in all living cells. During translation's elongation cycle, amino-acids are incorporated into a growing polypeptide chain through aminoacylated transfer RNAs (tRNAs) thanks to the decoding of mRNA codons. After each amino acid is added to the nascent peptide chain, the ribosome carries a peptidyl-tRNA at the aminoacyl A-site and a deacylated tRNA at the peptidyl P-site. In the next step, mRNA moves one codon length and the P- and A-site tRNA shift to the exit (E) and P-sites, respectively. In bacteria, this universal step of ‘translocation’ is catalyzed by elongation factor G (EF-G) (1). EF-G is a large GTPase protein made up of five distinct domains. The N-terminus (domains I and II) is separated from the C one (domains III, IV and V), and a significant hinge-like joint motion probably occurs between the two (2). EF-G is structurally similar to the ternary complex made by EF-Tu with the incoming tRNA and guanosine 5′-triphosphate (GTP), with the third EF-G domain mimicking the acceptor arm and the fourth mimicking the tRNA anticodon stem-loop (ASL) (3). Translocation is divided into several small intermediate steps that permit a gradual shifting in tRNAs positioning (4). Just after peptidyl transfer, tRNAs are in what is called the pre-translocation (PRE) state, a classic A/A and P/P state. Then a reversible 7° rotation of one ribosomal subunit relative to the other (ratcheting) allows for movement of the tRNA into the large subunit from A- to P-, and from P- to E sites, leading to the occupancy of the A/P and P/E hybrid sites. EF-G binding to a PRE-state ribosome stabilizes the rotated state and triggers GTP hydrolysis (5). An internal 18° swiveling of the head relative to the small subunit body then leads to the final post-translocation state (POST), wherein only one tRNA is bound to the P-site and the E-site tRNA is released (4,6,7). Several structures of ribosomal EF-G complexes were recently determined by X-ray or cryo-electron microscopy (cryo-EM). These yielded an accurate understanding of EF-G interactions and how they adjust with the ribosome along the translocation pathway between the PRE and POST states (3,5,8–14). So far, these detailed structures have been based on trapping EF-G in the ribosome via non-hydrolyzable GTP analogs, specific inhibitors (e.g. fusidic acid (FA), thiostrepton, viomycin and dityromycin) or a mutated form of EF-G. However, the use of these inhibitory conditions may have prevented the formation of relevant intermediate states. To explore how unobstructed EF-G acts in the ribosome, we performed experiments in the presence of natural GTP. To limit extensive release of EF-G from the ribosome after GTP hydrolysis, we increased the EF-G/ribosome molecular ratio and used a non-productive translation complex stalled on a truncated mRNA. In such a situation (15), EF-G must leave the vacant decoding site after GTP hydrolysis (16) without triggering translocation, to avoid competition with rescue systems. We identify here how EF-G interacts with stalled ribosomes. The 3.8 Å structure presented (Figure 1A and Supplementary Figures S1 and S2) reveals a hitherto unseen large motion of the EF-G’s domain III. It indicates how EFG leaves the ribosome after GTP hydrolysis and clarifies the molecular mechanism behind FA antibiotic activity.
Figure 1.
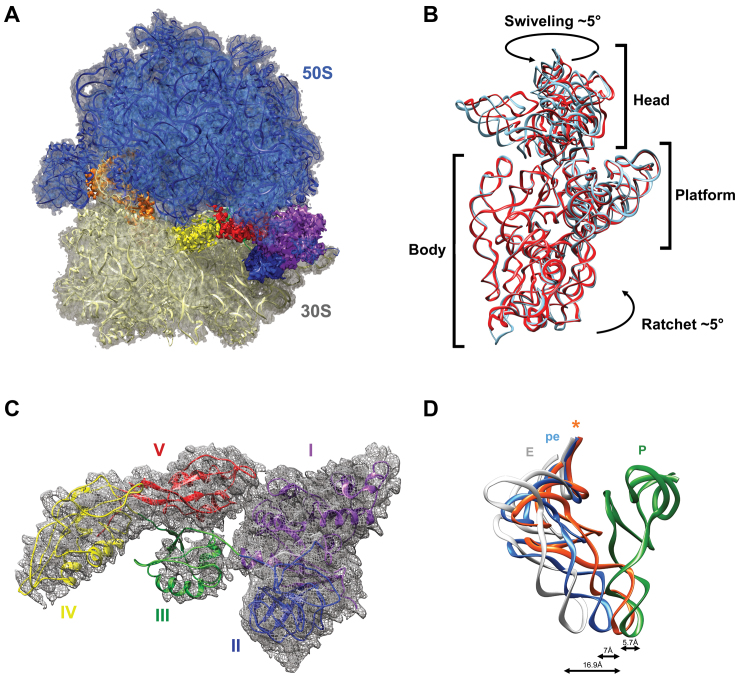
Overview of the EF-G-rotated ribosome complex and positioning of tRNA in the pe/E hybrid state. (A) Overall view of EF-G in the ribosome. The 50S subunit is blue, the 30S subunit is gold, tRNA is orange and the five (I–V) domains of EF-G are purple, blue, green, yellow and red, respectively. (B) Rotation of the 30S subunit relative to the 50S one in the EF-G complex. As compared to the non-rotated (PDB accession code: 4V51) 30S subunit (cyan), in this structure 30S (red) shows a 5° counter-clockwise rotation of the 30S body and a 5° orthogonal swiveling of the head toward the L1 stalk. The two structures are aligned on 23S rRNA. (C) Local fit of EF-G·GDP atomic model in the cryo-EM density map. The EF-G domain colors are as per (A). (D) Relative position of the current tRNA (indicated by an asterisk) with respect to P, pe and E tRNA states. The P-site is green, the pe state is blue, the current tRNA is orange and the E-site is white. The P- and E-site tRNA are from Gao et al. (8) (PDB accession code: 4V5F) while the pe state is from Zhou et al. (10) (PDB accession code: 4V9L). The distances between the current ASL and the P, pe and E ASLs are indicated. The structures were aligned using the 16S rRNA helix 44.
MATERIALS AND METHODS
Preparation of EF-G-bound ribosomes
Thermus thermophilus ribosomes were a kind gift from V. Ramakrishnan. His-tagged EF-G from T. thermophilus and His-tagged phenylalanine-tRNA synthetase (PheRS) from Escherichia coli were overexpressed in E. coli BL21 (DE3) using a T7 expression system and then isolated using Ni2+ pre-charged HiTrap chelating columns according to the manufacturer's instructions (GE Healthcare) (17). All protein concentrations were measured on a SimpliNano spectrophotometer (GE Healthcare). Phenylalanine-specific E. coli tRNA was purchased from Sigma. Aminoacylation was performed by incubating 1 μM tRNA at 37°C for 30 min in 50 mM HEPES-KOH (pH 7.5), 60 mM NH4Cl, 7 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 2 mM ATP, 30 μM phenylalanine and 1 μM PheRS. Stalled ribosomes were obtained by incubating 1 μM 70S ribosomes with 2 μM E. coli aminoacylated tRNAPhe and 2 μM mRNA with the AGGAGGUGAGGUUUU sequence containing a Shine-Dalgarno sequence and a phenylalanine codon (underlined), for 30 min at 37°C in buffer III made of 5 mM HEPES-KOH (pH 7.5), 10 mM NH4Cl, 10 mM MgOAc, 50 mM KCl, 0.1 mM EDTA and 6 mM β-mercaptoethanol. The final complexes were obtained by putting 1 μM of stalled ribosomes and 20 μM of EF-G with 2 mM GTP and mixing for 15 min at 42°C.
Cryo-electron microscopy
After dilution of 150 nM ribosomes in buffer III, 2 μl of each complex was applied to glow-discharged holey carbon grids (Quantifoil R2/2) and flash-frozen in liquid ethane using a Vitrobot Mark III (FEI). The grids were then transferred to a CS-corrected Titan Krios electron microscope (FEI), operated at 300 kV. Using FEI EPU software, images were automatically recorded on a Falcon II direct electron detector. This was set to a nominal magnification of 59 000 X, corresponding to a calibrated pixel size of 1, 162 Å/pixel and with a defocus range of −1.5 to −3.0 μm. The images were acquired in movie mode at 7 frames per exposure (1 s). The original image stacks were corrected for drift and beam-induced motion and summed using the optical flow approach (18) accessed through Scipion (19). The contrast transfer function parameters and defocus values for each micrograph were determined using CTFFIND 4 (20). EMAN2 (21) e2boxer software was used for semi-automatic particle picking, and all subsequent steps used RELION (version 1.4) (22). A total of 96 509 particles were extracted from 949 micrographs. Particles were split into height classes during the first round of 3D classification, with a map of the mature 70S ribosome (low-pass filtered to 60 Å) as the initial model. Based on the presence of EF-G, three classes were combined for a total of 61 404 particles, reflecting the rather high occupancy of EF-G on stalled ribosomes. To boost the resolution, Relion’s particle polishing procedure was applied. After 3D auto-refinement and post-processing, this yielded a 3.7 Å map at a gold-standard Fourier shell correlation (FSC) of 0.143 with fuzzy densities for EF-G domains III and IV. To further improve the quality of the map, the theoretical pixel size was re-assessed by calculating the cross-correlation between our map and an X-ray structure of the ribosome with EF-G trapped in the post-translocational state (PDB accession code: 4V5F). This led to a readjustment of the pixel size from 1.162 to 1.103 Å/pixel. The defocus values for each micrograph were re-calculated, and the 3D auto-refinement and post processing were re-performed on the 61 404-particle subset, leading to an improved density map with a resolution of 3.5 Å. To get around the structural heterogeneity, we then performed a focused classification on EF-G, subtracting the signal from the rest of the complex (Supplementary Figure S4) (23). The best 30, 000 particles produced a density map with better definition of EF-G domains III and IV. The refined maps were corrected for the detector’s modulation transfer function, and the high spatial frequencies were boosted using B-factor sharpening (24), resulting in a 3.8 Å density map. The local resolution map was estimated using ResMap (Supplementary Figure S1) (25). The initial modeling template used was the crystal structure of the T. thermophilus EF-G-ribosome complex in a PRE ribosome (PDB accession code: 4V90).The models of the 16S, 23S and 5S rRNA, tRNA, mRNA and the ribosomal proteins were docked separately on the density map by hand using UCSF Chimera (26). We then improved the atomic model by real-space refinement in PHENIX (27) against the density map, with secondary structure and geometry constraints applied. Alternating rounds of manual model adjustment using Coot (28) and model real space refinement using PHENIX (29) were then carried out. Finally the RNA atomic model was adjusted using ERRASER (30) and a final real space refinement was performed. To avoid overfitting, different density map weights were tested during the refinement procedure and a .75 coefficient was chosen to balance good geometry and small differences between FSCwork and FSCtest and good fits to the density. Cross-validation against overfitting was performed as previously described (31,32). To do this, the positions of the atoms in the atomic model were first randomly shifted by 0.5Å. The model was then refined against a map recreated from half of the data produced by RELION during the final high-resolution structural refinement iteration. The absence of overfitting was verified by comparing the FSC of our model and the two half maps. The statistics of the final model were evaluated using MolProbity (33).
RESULTS AND DISCUSSION
Our single-particle cryo-EM reconstruction clearly shows extra densities for P-site tRNA and EF-G (Figure 1A). Compared to the classic ribosomal state (34), the 30S body is rotated slightly counterclockwise by ∼5° with respect to the 50S subunit (Figure 1B). This ratchet movement is lower than the 7° usually observed in PRE state ribosomes, but similar to that previously observed for complexes in intermediate POST states. This suggests a back-ratcheting of the 30S body after GTP hydrolysis but before complete translocation (6,10,35). The 30S head is also swiveled, but by only about 5° (Figure 1B). This rather short swiveling may reflect the start of the partial back-swiveling movement of the head as tRNA transits from the hybrid P/E state to the classic E/E one. As a result of 30S ratcheting, the P-site tRNA moves to an intermediate hybrid state between the ribosomal P- and E-sites. To better identify the position of the ASL with respect to these sites, we first aligned the small subunit’s helix 44 to compare our structure with tRNA bound in intermediate PRE or POST states (8,10) (Figure 1C). This showed that in our structure the ASL is positioned in a chimeric ‘pe’ state, meaning that it interacts with parts of the 30S P-site head and E-site platform. More precisely, the ASL is positioned 5.7 Å downstream from a canonical P-site ASL and 16.9 Å upstream from a canonical E-site one (8). This disposition resembles the structure recently described by Zhou et al. (10) in the presence of FA or GDPNP non-hydrolyzable analog of GTP. On the other hand, the acceptor stem has reached the E-site of the 50S subunit, explaining the absence of density accounting for the amino acid side chain on the tRNA. Concomitantly, the L1 stalk has moved inward into an intermediate conformation to interact with the tRNA’s elbow (Supplementary Figure S3). These interactions allow L1 to chaperone the p/E tRNA as it translocates to the E/E conformation (11). Since the structure contains only a single tRNA, it is not a real translocation intermediate. However, our data provides insights into a late translocation step. The ribosome present a small 5° intersubunit rotation and a 5° swiveling of the 30S head. The deacylated tRNA is trapped in a late intermediate pe/E translocation state, just prior to EF-G·GDP dissociation.
In our structure, the EF-G density is well-defined and all five domains were easily attributed. Domains I, II and V are stable and well-resolved, allowing for an accurate atomic reconstruction (Figure 1d and Supplementary Figure S1). On the other hand, domains III and IV are highly flexible and required the use of masked classification with residual signal subtraction in order to unambiguously attribute their electron densities (Supplementary Figure S4). The overall conformations and positions of domains I and II are very similar to that of previously-reported structures, in both PRE and POST states. Domain I is the GTPase active site of EF-G and contains switches I and II, two mobile elements that are essential for EF-G’s unidirectional cycle during protein synthesis (16,36). In our map, switch I (amino acids 38–64) is disordered (Supplementary Figure S5), a feature which is characteristic of EF-G after GTP hydrolysis into guanosine diphosphate (GDP) (3). Accordingly, the distinct density within the EF-G catalytic center correctly accommodates a GDP nucleotide molecule, but even at a high threshold it is too small for GTP (Supplementary Figure S5). The complex therefore looks like EF-G after GTP hydrolysis, including an intersubunit rotation and 30S subunit head swiveling, as well as having pe/E tRNA positioned close to the L1 stalk.
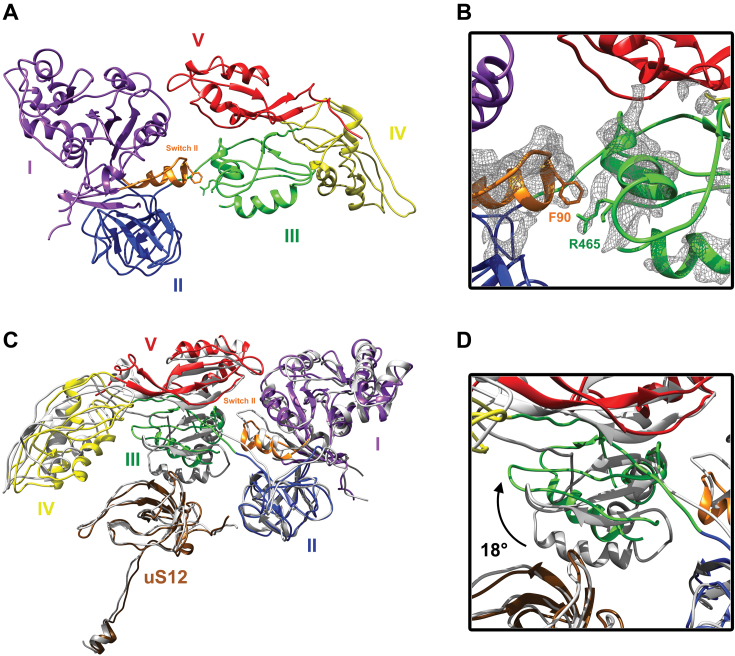
The switch II loop (amino acids 84–107) is one of the most stable EF-G elements in our complex (Supplementary Figure S6). The loop has moved away from the catalytic site to join domain III, and this conformation differs from all of the known structures of EF-G ribosomal complexes (Figure 2 and Supplementary Figure S6). Due to a large motion of the third domain (see below), the Phe90 found in the tip of switch II is positioned to interact with Arg465 or the neighboring residues in the B3 helix (the resolution of the map does not allow unambiguous side chain modeling for this part of domain III). The switch II shift is similar to that encountered in the crystal structure of the free EF-G·GDP (PDB accession code: 2BM0), confirming that the factor is frozen in a post-GTP hydrolysis conformation. However, the interaction with the B3 helix is not seen in the free EF-G·GDP in solution (Supplementary Figure S7) as the domain III, IV and V are folded on the domain I and II.
Figure 2.
Interactions between EF-G switch II and domain III, and movement of this domain away from ribosomal protein uS12. (A) Overall view of EF-G, with the five (I–V) domains colored in purple, blue, green, yellow and red, respectively, and the switch II in orange. (B) Focus on the interaction between switch II (orange) and domain III (green). The local mesh volume shows the interaction between phenylalanine 90 (F90) and arginine 465 (R465) and its neighboring residues. (C) Superposition of the structure of EF-G in a classic 70S ribosome translocation intermediate state trapped with FA (gray: PDB accession code: 4V9L) and our EF-G model. Ribosomal protein uS12 is brown. The two EF-G structures are aligned on EF-G domains I and II. (D) Details of the domain III movement as it relates to ribosomal protein uS12. The third domain undergoes an 18° movement (black arrow) toward domain V, leading to a loss of interaction with uS12.
Domain III is critical for EF-G activity, as its deletion impairs GTP hydrolysis and translocation (5,37). Among the EF-G domains in our structure, it is the least well-defined, suggesting its dynamic nature (Supplementary Figures S1 and 2). In comparison with the PRE state, domain III has moved toward domain V in a large motion of about 18 Å. This moves domain III away from uS12, the only ribosomal protein located near the decoding site (Figure 2 and Supplementary Figure S6). It also allows new interactions to occur between domain III helix B3 (arginine 465 and neighboring residues) and switch II phenylalanine 90 (Figure 2). These data suggest that domain III is not part of a rigid body composed of domains III–V, but is instead subjected to large rotations during translocation (Supplementary Movies S1 and 2). While this motion has never been described in structural studies, it has been suggested as a result of chemical crosslinking and single-molecule polarized fluorescence microscopy examinations of normal translocation (i.e. without blockers that might freeze the EF-G conformation) (38,39). Our study shows the movement observed after GTP hydrolysis and before EF-G release, confirming that domain III can cause the subsequent movement of mRNA and tRNA (5). Unlike domain III, domain V does not undergo a large motion, although it is slightly pushed toward protein L11.
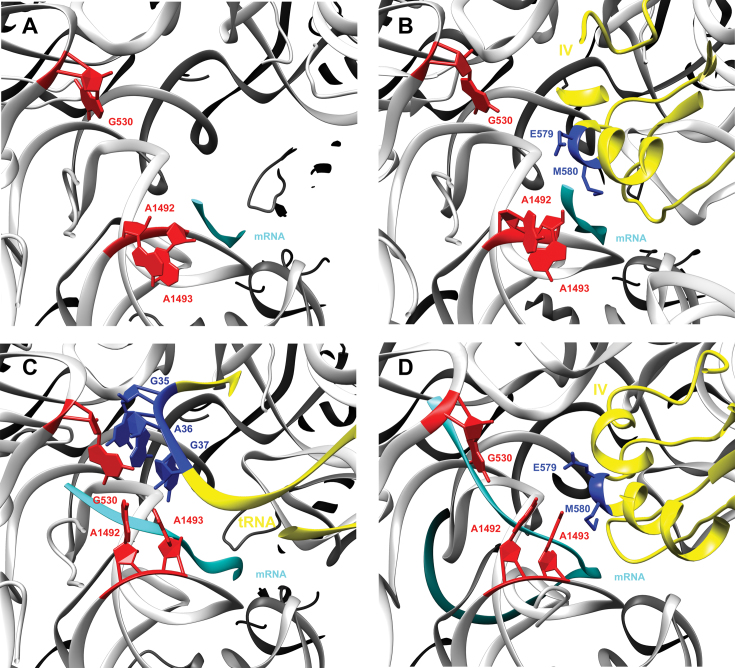
The fourth domain establishes close contact with the decoding site on the small 30S subunit thus is critical for translocation catalysis. The decoding site cannot simultaneously bind tRNA and a translation factor, so obviously EF-G, and particularly domain IV, undergoes structural rearrangements during translocation (14,40). Despite the absence of a codon, this domain fully occupies the decoding center in a similar location as in ribosome with elongation factor G trapped in the post translocational state (8), with loop I at the tip of domain IV extending toward the P-site to make contact with pe/E tRNA. Within helix 44, there is no deep remodeling of nucleotides G530, A1492 and A1493, as occurs during ribosomal tRNA selection (41). Thus this domain cannot directly interact with these nucleotides (Figure 3) as it does during canonical translocation (13). This implies that EF-G detects the presence or absence of codon-anticodon interactions within the A-site by sampling the positions of these nucleotides. Our results also confirm that the molecular clamp formed by these bases is necessary for triggering translocation, but not for EF-G binding and GTP hydrolysis (42), which are instead found to be due to EF-G flanking restrictions imposed by the III and V domains in the rotated ribosome (5).
Figure 3.
Comparison of the presented structure’s decoding center with the structures of empty 30S, tRNA-EF-Tu and PRE state EF-G. Detailed interactions of EF-G domain IV with the universally conserved G530, A1492 and A1493 16S rRNA bases (red) and mRNA (cyan). (A) The empty 30S subunit, without tRNA in its A-site (PDB accession code: 1J5E). (B) The presented structure showing E579 and M580 (blue) at the tip of EF-G domain IV (yellow). (C) The canonical tRNA (yellow with anticodon in blue) that decodes mRNA in the A-site (PDB accession code: 4V87). (D) The EF-G (yellow and E579 and M580 in blue) in a classic 70S ribosome translocation intermediate state trapped with FA (PDB accession code: 4V9L).
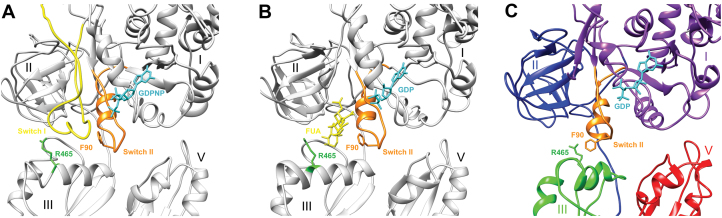
According to various structures bound to FA, the antibiotic bonds occur between domains II and III, preventing conformational changes in EF-G that are required for its release from the ribosome after GTP hydrolysis (8,10,12). In our map, obtained without FA, domains I and III are linked through a hitherto unseen interaction between helix B3 (arginine 465 and neighboring residues) and phenylalanine 90 (Figures 2 and 4). We can therefore assume that after GTP hydrolysis, switch I disorganization causes the release of domain III, which then changes conformation. This movement allows new interactions to occur between domain III and switch II, triggering EF-G·GDP release from the ribosome (Supplementary movies S1 and 2). However, after GTP hydrolysis, FA replaces the switch I position on domain III and prevents binding of R465-F90 amino acids. As a consequence, EF-G preserves a ‘GTP-like’ conformation and stays within the ribosome. This is in line with previous work showing that FA overlaps residues in the conserved core of switch loop I (10), and that the antibiotic interacts with both arginine 465 and phenylalanine 90 (8). Consequently, mutation of these residues is known to confer FA resistance (43), and most FA-resistant EF-G mutations have altered switch II or domain III properties including these two amino acids (43,44).
Figure 4.
Overview of the structural mechanism of FA. (A) Structure of EF-G in a 70S ribosome translocation intermediate state trapped with GDPNP (PDB accession code: 4V9K), showing the switch I loop (yellow), the switch II (orange) and GDPNP (cyan). (B) In the 70S ribosome translocation intermediate state trapped with FA (PDB accession code: 4V9L), switch I is disordered and replaced by FA (yellow) and GDPNP is replaced by GDP (cyan). (C) Position modifications of domain III and switch II in the current structure without GDPNP or FA. GDP is cyan, switch II is orange and EF-G domain I, II, III and V are in purple, blue, green and red respectively. The key residues phenylalanine 90 (F90) and arginine 465 (R465) are always in orange and green, respectively.
CONCLUSION
Together, our data have provided an atomic model of EF-G·GDP bound to a rotated ribosomal state. The tRNA is in a chimeric intermediate pe/E state between the P/E and E/E ones, close to the L1 stalk of the large ribosomal subunit. Although our complex is not a real translocation intermediate because of a single deacylated tRNA it provides insights into the late translocation steps, just prior to EF-G·GDP recycling. The absence of translocation blockers allows for the first visualization of a large motion of EF-G’s domain III, a shift which leads to the movement of the switch II loop from the catalytic site to join domain III. This conformational change occurs before the complete translocation of the tRNA from the P- to the E-site, and could therefore act as the facilitator for that final movement. The interaction between Phe90 with Arg465 or the neighboring residues in the domain III rationalizes the effect of FA in stabilizing EF-G binding to the ribosome. It explains how the antibiotic FA stops this conformational change and thus prevents EF-G release after GTP hydrolysis.
DATA AVAILABILITY
The maps were deposited with the EMDB under the accession code EMD-3852. Atomic coordinates were deposited with the Protein Data Bank under the accession code 5OT7.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the use of electron microscopes from the Microscopy Rennes Imaging Center (MRic) facilities, from Instruct at IGBMC-CERBM Strasbourg and the Netherlands Centre for Electron Nanoscopy (NeCEN) (Leiden University). We thank Juliana Berland for insightful comments on the manuscript. Computer time was provided by GENCI (IDRIS and CINES, #i2015037391).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Netherlands Organization for Scientific Research (NOW); European Regional Development Fund of the European Commission; French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05]; Agence Nationale de la Recherche [ANR-14-ASTR-0001]. Funding for open access charge: University of Rennes 1.
Conflict of interest statement. None declared.
REFERENCES
- 1. Achenbach J., Nierhaus K.H.. The mechanics of ribosomal translocation. Biochimie. 2015; 114:80–89. [DOI] [PubMed] [Google Scholar]
- 2. Connell S.R., Takemoto C., Wilson D.N., Wang H., Murayama K., Terada T., Shirouzu M., Rost M., Schüler M., Giesebrecht J. et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell. 2007; 25:751–764. [DOI] [PubMed] [Google Scholar]
- 3. Pulk A., Cate J.H.D.. Control of ribosomal subunit rotation by elongation factor G. Science. 2013; 340:1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holtkamp W., Wintermeyer W., Rodnina M.V.. Synchronous tRNA movements during translocation on the ribosome are orchestrated by elongation factor G and GTP hydrolysis. Bioessays. 2014; 36:908–918. [DOI] [PubMed] [Google Scholar]
- 5. Li W., Liu Z., Koripella R.K., Langlois R., Sanyal S., Frank J.. Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G. Sci. Adv. 2015; 1:e1500169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ratje A.H., Loerke J., Mikolajka A., Brünner M., Hildebrand P.W., Starosta A.L., Dönhöfer A., Connell S.R., Fucini P., Mielke T. et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010; 468:713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. [DOI] [PubMed] [Google Scholar]
- 8. Gao Y.-G., Selmer M., Dunham C.M., Weixlbaumer A., Kelley A.C., Ramakrishnan V.. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009; 326:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin J., Gagnon M.G., Bulkley D., Steitz T.A.. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015; 160:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou J., Lancaster L., Donohue J.P., Noller H.F.. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013; 340:1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tourigny D.S., Fernández I.S., Kelley A.C., Ramakrishnan V.. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013; 340:1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramrath D.J.F., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H.F., Spahn C.M.T.. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Feng S., Kumar V., Ero R., Gao Y.-G.. Structure of EF-G-ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol. 2013; 20:1077–1084. [DOI] [PubMed] [Google Scholar]
- 14. Brilot A.F., Korostelev A.A., Ermolenko D.N., Grigorieff N.. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giudice E., Macé K., Gillet R.. Trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 2014; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ticu C., Nechifor R., Nguyen B., Desrosiers M., Wilson K.S.. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009; 28:2053–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weis F., Bron P., Giudice E., Rolland J.-P., Thomas D., Felden B., Gillet R.. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO J. 2010; 29:3810–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abrishami V., Vargas J., Li X., Cheng Y., Marabini R., Sorzano C.Ó.S., Carazo J.M.. Alignment of direct detection device micrographs using a robust Optical Flow approach. J. Struct. Biol. 2015; 189:163–176. [DOI] [PubMed] [Google Scholar]
- 19. de la Rosa-Trevín J.M., Quintana A., Del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016; 195:93–99. [DOI] [PubMed] [Google Scholar]
- 20. Rohou A., Grigorieff N.. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J.. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- 22. Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai X., Rajendra E., Yang G., Shi Y., Scheres S.H.. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015; 4:e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H.W., Henderson R.. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013; 135:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kucukelbir A., Sigworth F.J., Tagare H.D.. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 27. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chou F.C., Sripakdeevong P., Dibrov S.M., Hermann T., Das R.. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods. 2012; 10:74–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amunts A., Brown A., Bai X., Llácer J.L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S.H.W., Ramakrishnan V.. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014; 343:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernández I.S., Bai X.-C., Murshudov G., Scheres S.H.W., Ramakrishnan V.. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014; 157:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Selmer M., Dunham C.M., Murphy F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 35. Yamamoto H., Qin Y., Achenbach J., Li C., Kijek J., Spahn C.M.T., Nierhaus K.H.. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 2014; 12:89–100. [DOI] [PubMed] [Google Scholar]
- 36. Vetter I.R., Wittinghofer A.. The guanine nucleotide-binding switch in three dimensions. Science. 2001; 294:1299–1304. [DOI] [PubMed] [Google Scholar]
- 37. Martemyanov K.A., Gudkov A.T.. Domain III of elongation factor G from Thermus thermophilus is essential for induction of GTP hydrolysis on the ribosome. J. Biol. Chem. 2000; 275:35820–35824. [DOI] [PubMed] [Google Scholar]
- 38. Chen C., Cui X., Beausang J.F., Zhang H., Farrell I., Cooperman B.S., Goldman Y.E.. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7515–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nechifor R., Wilson K.S.. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J. Mol. Biol. 2007; 368:1412–1425. [DOI] [PubMed] [Google Scholar]
- 40. Salsi E., Farah E., Dann J., Ermolenko D.N.. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15060–15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogle J.M., Brodersen D.E., Clemons W.M., Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 42. Garcia-Ortega L., Stephen J., Joseph S.. Precise alignment of peptidyl tRNA by the decoding center is essential for EF-G-dependent translocation. Mol. Cell. 2008; 32:292–299. [DOI] [PubMed] [Google Scholar]
- 43. Laurberg M., Kristensen O., Martemyanov K., Gudkov A.T., Nagaev I., Hughes D., Liljas A.. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 2000; 303:593–603. [DOI] [PubMed] [Google Scholar]
- 44. Chen Y., Koripella R.K., Sanyal S., Selmer M.. Staphylococcus aureus elongation factor G–structure and analysis of a target for fusidic acid. FEBS J. 2010; 277:3789–3803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The maps were deposited with the EMDB under the accession code EMD-3852. Atomic coordinates were deposited with the Protein Data Bank under the accession code 5OT7.