Abstract
Background
It is well known that long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is closely correlated with the tumorigenesis of multiple cancers, including renal cell carcinoma (RCC). However, the potential functional mechanism is still elusive.
Material/Methods
In our present research, quantitative real-time polymerase chain reaction (qRT-PCR) was performed for the measurement of MALAT1 and miR-429. CCK-8 assay and Transwell assay were performed for the proliferation, migration, and invasion abilities of RCC cells. Dual-luciferase reporter assay was performed to validate the interaction within MALAT1 and miR-429.
Results
Data found that MALAT1 was overexpressed in RCC clinical samples and cell lines. Moreover, loss-of-functional experiments showed that MALAT1 knockdown suppress the proliferation, migration, and invasion abilities of RCC cells. RT-PCR showed that miR-429 expression was downregulated in RCC cell lines, which was negatively correlated with that of MALAT1. Bioinformatics analysis suggested that miR-429 had complementary binding sequences with MALAT1, which was confirmed by dual-luciferase reporter assay.
Conclusions
In summary, our results concluded that MALAT1 functioned as an oncogene in RCC by sponging miR-429, acting as its competing endogenous RNA (ceRNA).
MeSH Keywords: Carcinoma, Renal Cell; Nephrology; RNA, Long Noncoding
Background
Malignant kidney tumor accounts for nearly 2% of adult malignancies, among which renal cell carcinoma (RCC) accounts for approximately 85% of cases [1]. In current therapy, surgical resection is the most effective treatment because RCC patients reveal high resistance towards conventional chemotherapy and radiotherapy [2,3]. Therefore, the disease still exhibits substantial mortality due to regional or distant metastasis [4]. Previous study reported that patients with stage IV RCC have a significantly reduced 5-year survival rate (<30%) [5]. Thus, further research will be urgently required to detect the molecular mechanisms of RCC progression and to find more valid therapeutic targets of RCC.
More than 90% of the DNA sequence of the human genome is actively transcribed. However, only 2% of RNA sequence encodes proteins. The rest of the transcripts are referred to as noncoding RNAs (ncRNA), which are considered to exhibit biological functions without protein-coding capability [6]. Noncoding RNAs are classified into 2 groups according to length. Small noncoding RNAs, especially microRNAs (miRNAs), have been extensively studied for decades, and their function as oncogenes or anti-tumor genes have been widely discovered and discussed in various cancers [7]. Long noncoding RNAs (lncRNAs) have at least 200 bases and had been acknowledged as transcription noise without biological function for a long time. However, recently, increasing evidence indicates that lncRNAs take part in essential cell processes, physiologically and pathologically, including nuclear import, cell cycle control, nuclear and RNA decay, transcription, and translation [8]. These findings make lncRNAs a promising therapeutic target for cancers [9]. Thus, lncRNAs have become an emerging hotspot in cancer research [10]. Previous studies have reported on biological functions of lncRNAs in numerous cancers, including RCC [11,12].
LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was originally located on chromosome 11q13. It was reported to be overexpressed in varied cancers, including RCC [12,13], and it was also confirmed as an independent prognostic parameter for several cancers patients’ survival in previous studies [14,15]. In addition, studies revealed that lncRNA MALAT1 functioned as an oncogene in RCC development, and the underlying mechanisms were explored. Shaoan Chen et al reported that lncRNA MALAT1 could suppress proliferation and enhance apoptosis of RCC cells through upregulating linvin protein expression [16]. Research from Hiroshi Hirata’s team and Haibing Xiao’s team manifested that lncRNA MALAT1 achieved its tumor-promoting function by regulating downstream proteins expression via interfering with microRNA [17,18]. Inspired by previous studies, we designed our current study based on results of preliminary experiments, which showed that miRNA-429 was significantly downregulated in RCC. Deregulation of miR-429 is involved in suppressing different kinds of cancers, such as non-small cell lung cancer, colon cancer, and RCC [19–21].
According to our acknowledge, none of the existing articles have reported on the interaction of lncRNA MALAT1, and miR-429 of RCC tumorigenesis. Here, we constructed and conducted a study to investigate the biological effects of lncRNA MALAT1 and miR-429 on RCC tumorigenesis, as well as determine the therapeutic prospect for RCC clinic.
Material and Methods
Cell lines and clinical samples
Three cell lines were used in this research, including human RCC cell lines 786-O and ACHN, as well as human embryonic kidney (HEK) 239-E cells (Type Culture Collection of Chinese Academy of Sciences). RCC cell was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) with 10% FBS (fetal bovine serum, Gibco, Rockville, MD) and 1% penicillin-streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in humidified atmosphere with 5% CO2. HEK cell were cultured in DMEM with only 10% FBS at same thermal and gaseous conditions. All cells were used in research within 5 months. For clinical samples, we applied and obtain approval from the Research Ethics Review Committees of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. Approximately, 50 patients (38 male and 12 female) who was pathologically diagnosed as RCC were recruited in this research of Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. They were staged according to the tumor-node-metastasis classification and classified by the World Health Organization (WHO) criteria. Patient samples were obtained and storing at −80°C after patients gave their confirmation in writing.
Real-time quantitative polymerase chain reaction (PCR)
The relative expression levels of lncRNA-MALAT1 and miR-429 were determined by quantitative RT-PCR. Briefly, RNAs were obtained using the TRIzol reagent and then first-strand cDNA of RNAs were synthesized by reverse transcription using reverse transcription reagent kit (both from Invitrogen, Carlsbad, CA, USA). Real time qPCR was performed using SYBR Green PCR kit (TaKaRa, Dalian, China). Primers were all obtained from Applied Biosystems and all procedures were conducted as per the manufacturer’s instructions. Briefly, every procedure was performed under certain conditions: initial denaturation (95°C for 3.5 min), denaturation (95°C for 15 seconds), annealing (60°C for 50 seconds), elongation (72°C for 55 seconds) and the final elongation (72°C for 3 min). β-actin was tested as the internal standard. The relative gene expression levels were calculated by comparing to expression level internal standard using 2−ΔΔCT method.
Cell transfection
Transfection mimics carrying lncRNA, miRNAs, and siRNAs were used in research for regulation of target genes expression levels (Transheep, Shanghai, China), including miR-429 mimic, miR-429 siRNA mimic, MALAT1 mimic, MALAT1 siRNA mimic, and corresponding negative control mimics. Transfection was conducted using the Lipofectamine 2000 transfection reagent following manufacturers’ instructions (GenePharma, Shanghai, China).
Luciferase assays
Luciferase assay was used to clarify if there were hypothetical a binding site in 3′-UTR of MALAT1 and miR-429. The wide type (WT) or mutant type (MUT) 3′-UTR sequences of MALAT1 were cloned into pRL-CMV vectors, respectively (Promega Corporation, Madison, WI, USA) in front of the firefly luciferase gene, constructed and purified as reporter mimics. RCC 786-O cells were planted into 12-well plate (~4000 cells per well) and co-transfected with reporter mimics, miR-429 mimics or NC by Lipofectamine 2000 following manufacturers’ protocols. Additionally, every sample was also transfected with 0.05 μg pRL-CMV plasmid containing Renilla luciferase gene (Promega Corporation, Madison, WI, USA), as an internal control to observe transfection efficiency. After 48 hours, luciferase activity of firefly luciferase was measured and rectified by Renilla luciferase activity using the Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA), according to the manufacturer’s instructions.
Cell viability assay
Cells were harvested and planted into 96-well plate (~4000 cells per well) and transfected with corresponding mimics. Then we waited 48 hours before determining cell viability by a CCK-8 kit ((Beyotime Institute of Biotechnology, Jiangsu, China) following the manufacturer’s protocols.
Cell migration and invasion assay
RCC 786-O cells were co-transfected with siMALAT1 combining to miR-429, miR-inhibitor or miR-NC mimics. All cells were collected in log phase and planted into Transwell chambers (BD Biosciences, San Jose, CA, USA), which were separated as upper and lower levels both filled with RPMI-1640 medium and connected by one 8 mm pore. For invasion assay, Transwell chamber was previously coated by Matrigel membrane (BD Biosciences, San Jose, CA, USA). Afterwards, cells were incubated for 15 hours or 24 hours before migration or invasion assay. Finally, cells in lower chambers were fixed, stained, and counted under microscopic inspection (200×).
Statistical analysis
All results were extracted from 3 independent experiments and presented by mean ± standard deviation. Statistical differences were determined by the Student’s t-test, one-way ANOVA or two-way ANOVA. A P < 0.05 was considered as statistically significant.
Results
MALAT1 was upregulated in renal cell carcinoma tissues and cell lines
To detect expression level of MALAT1 in RCC patient tissues and cell lines, real time PCR (qRT-PCR) were used. Results revealed that MALAT1 expressed significantly higher in RCC tissues comparing to normal renal tissue (Figure 1A), and nevertheless, MALAT1 expression significant enhanced in RCC cells compared to HEK cells (Figure 1B). These results indicated that RCC tissues and cell lines exerted high expression lncRNA, which suggested that MALAT1 could be an oncogene.
Figure 1.
MALAT1 was upregulated in renal cell carcinoma (RCC) patient tissues and cell lines. (A) Expression of MALAT1 was significantly higher in RCC tissues than normal tissues. (B) Expression level of MALAT1 was significantly higher in RCC cell lines, 786-O and ACHN, than HEK 293-T cells. Data was presented as mean ±SD, ** P<0.01, *** P<0.001 calculated with Student’s t-test.
Downregulation of MALAT1 inhibits growing of RCC cells
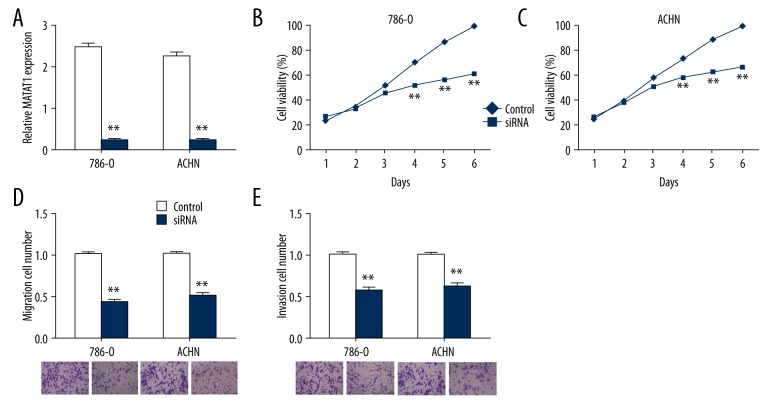
Given that qRT-PCR results revealed that MALAT1 is a potential oncogene. To figure out MALAT1 biological role in RCC, we transfected siRNA of MALAT1 into RCC cells lines 786-O and ACHN. After transfection, we detected that MALAT1 expression significantly decreased in 2 cell lines using qRT-PCR (Figure 2A). CCK-8 assay detected that knockdown of MALAT1 significantly downregulation RCC cells proliferation in a time-dependent manner (Figure 2B). Nevertheless, Transwell assay detected that migration and invasion abilities were significantly suppressed after transfection (Figure 2C, 2D). These results indicated that downregulation of MALAT1 could inhibit proliferation, migration and invasion of RCC cells.
Figure 2.
Downregulation of MALAT1 inhibited proliferation, migration, and invasion of renal cell carcinoma (RCC) cells. (A) Expression level of MALAT1 is significantly decreased after MALAT1 siRNA transfection. (B) Proliferation of RCC cells was determined at indicated time point after transfection using CCK-8 assay. (C) Migration ability of RCC cells was determined after transfection using Transwell assay. Random fields are shown below. (D) Invasion ability of RCC cells was determined after transfection using Transwell assay. Random fields are shown below. Data was presented as mean ±SD, * P<0.05, ** P<0.01, calculated with Student’s t-test.
MALAT1 targeted with miR-429 at 3′-UTR
We obtained miRNA expression profiles in RCC tissue compared to normal control tissue in our earlier research (Figure 3A), in which we noticed that the miR-429 expression level was markedly low in RCC tissue, whereas it had previously been reported as a potential tumor-suppressor. Therefore, we detected the expression of miR-429 in RCC tissues and cells using qRT-PCR, which showed that miR-429 was significantly decreased in tissue and cells (Figure 3B, 3C). We performed Pearson’s correlation analysis to verify if there were correlation between expression of MALAT1 and miR-429. The results proved that MALAT1 expression was negatively correlated to miR-429 expression in RCC tissues (Figure 3D), which suggested that MALAT1 might regulate expression level of miR-429. We then determined a putative complementary region between MALAT1 and miR-429 (Figure 3E). After that, we confirmed this binding alignment between MALAT1 and miR-429 using dual-luciferase reporter assay (Figure 3F). In all, these results indicated that MALAT1 could downregulate expression of miR-429 in RCC, which may suggest a cooperating relation of their biological functions in RCC. These results suggested that the expression of MALAT1 and miR-429 was negatively related, which is highly likely presented by direct binding between MALAT1 and miR-429.
Figure 3.
MiR-429 is a direct target of MALAT1. (A) MiRNA expression profiles in renal cell carcinoma (RCC) tissue was detected by miRNA microarray. (B) Expression of miR-429 was significantly higher in RCC tissues than normal tissues. (C) Expression level of miR-429 was significantly higher in RCC cell lines, 786-O and ACHN, than HEK 293-T cells. (D) Pearson’s correlation analyzed that expression of miR-429 was negative related to expression of MALAT1 (R2=0.766). (E) A presumptive complementary alignment between MALAT1 and miR-429. (F) RCC 786-O cells were co-transfected with WT/Mut- MALAT1 vectors and miR-429/NC. Dual-luciferase reporter assay indicated that there was certain possibility of 3′-UTR of MALAT1 (WT) and miR-429 sharing a complementary binding site (firefly luciferase activity in overexpressed miR-429 786-O cells was decreased by 61% comparing to 786-O cells transfected with NC). Data was presented as mean ±SD, ** P<0.01, calculated with Student’s t-test.
Functions of MALAT1 and miR-429 on RCC cells
Considering all the results we had obtained, we decided to further explore the biological functions of MALAT1 and miR-429 at a cell level. In these experiments, we co-transfected siMALAT1 mimics combined with miR-429, miR-inhibitor or miR-NC mimics to RCC 786-O cells. This approach provided us with 3 experimental groups. For every group, CCK-8 assay, migration assay, and invasion assay were performed (Figure 4). Previous results suggested that MALAT1 exerted oncogene characteristics in RCC, while miR-429 was downregulated in RCC. Therefore, downregulation of MALAT1 could suppress renal cell carcinoma growth, co-transfection of miR-429 or miR-inhibitor might enhance or reverse siMALAT1 effects in RCC. Overall results verified this hypothesis that co-transfection of miR-inhibitor and siMALAT1 could increase cell viability and decrease migration and invasion abilities, comparing to siMALAT1+miR-NC group. However, co-transfection of miR-429 and siMALAT1 achieved the opposite results. In conclusion, these results indicated that, unlike potential oncogene MALAT1, miR-429 possesses tumor-suppressing function in RCC.
Figure 4.
Functions of MALAT1 and miR-429 on RCC 786-O cells. (A) Expression of miR-429 in RCC 786-O cells after transfections. (B) Cell viability of RCC 786-O cells determined by CCK-8 assay. (C, D) Cell migration and invasion abilities of RCC 786-O cells were determined by Transwell assays. Data was presented as mean ±SD, ** P<0.01, calculated with Student’s t-test.
Discussion
Looking back at previous research in recent decades, all experimental evidence points to the same theory that noncoding RNAs play an important role in tumorigenesis [8,22–24]. In the meanwhile, aberrant expression of lnRNA MALAT1 and miR-429 has been reported to be related to various kinds of malignancy tumors, including RCC [13,16,21]. In most articles, lncRNA MALAT1 has been shown to be an oncogene, while miR-429 has been regarded as a tumor suppressor. Underlying mechanisms of both MALAT1 and miR-429 biological functions have been explored in previous studies [17,18,25,26]. However, few studies have connected their functions in RCC.
Prior to our current research, we found that there were potential complementary sequences between MALAT1 and miR-429 in starBase (http://starbase.sysu.edu.cn) [27]. Therefore, we determined MALAT1 expression level in RCC tissues and cell lines by qRT-PCR. MALAT1 was overexpressed in tissue samples and cell lines, which was consistent with previous results. Next, we confirmed the oncogenic function of MALAT1 in RCC by knockdown MALAT1 in RCC cell lines. Our overall results showed that downregulation of MALAT1 significantly suppressed cell viability, migration, and invasion ability. Research from Tripathi et al suggested that MALAT1 enhance tumorigenesis by upregulating oncogenic transcription factor BMYB [28]. Hirata et al reported that MALAT1 facilitates oncogenesis of RCC by binding Ezh2 and interference with miR-205 [17]. Chen et al found in their study that MALAT1 promoted RCC cells proliferation and metastasis by increasing livin expression [12].
We have obtained miRNA expression profiles of RCC tissue compared to normal tissue in previous research. From these profiles, we noticed that miR-429 expression was remarkable low. In this study, we confirmed that expression of miR-429 in RCC tissues and cell lines was significant lower than normal controls. This finding coincided with previous results. Machackova and Wu both reported that miR-429 was downregulated in RCC cell lines, acting as a tumor suppressor [21,26]. We found that miR-429 expression level was negatively related to MALAT1 expression level using Pearson’s correlation, which predicted that potential binding sites existing within miR-429 and 3′-UTR of MALAT1. We further clarified the presumptive connection between MALAT1 and miR-429 using dual-luciferase reporter assay, from which we learned that luciferase activity was significantly decreased when RCC cells were co-transfected with MALAT1-WT and miR-429 mimics. By verifying this connection, we learned that there was a structural foundation for cooperative biological functions of MALAT1 and miR-429 in RCC.
To support our theory, we further investigated miR-429 tumor suppressor functions on the basis of downregulation of MALAT1 by introducing siMALAT1 mimics to 786-O cells. Our overall results indicated that transfection of miR-429 could enhance tumor-suppressive effects of siMALAT1, while transfection of miR-inhibitor could reverse siMALAT1 tumor-suppressive impacts on RCC cells proliferation, migration, and invasion ability. All those findings indicate that MALAT1 and miR-429 have a synergistic relationship in regulating tumorigenesis of RCC. Similar conclusions have been supported by previous research in RCC and other kinds of malignancies. For instance, Haibing et al reported that MALAT1 stimulated proliferation and metastasis of clear cell kidney carcinoma by sponging miR-200s in order to regulate expression of ZEB2 [18]. In the Tan study, they found that MALAT1 achieved its oncogenic function in human hilar cholangiocarcinoma via interacting with miR-204 to modulate CXCR4 expression [29]. To our acknowledge, this is the first study to illustrate the synergistic relationship between MALAT1 and miR-429 in proliferation and metastasis of RCC. Although, we could not provide details regarding underlying mechanism of this regulation pathway in this study, we did connect 2 significant factors to presents a novel entry point for future research to explore pathogenesis and clinical therapy of RCC.
The competing endogenous RNA (ceRNA) theory is the most canonic theory for the lncRNA on pathological physiology, including RCC [30]. For instance, lncRNA DLX6-AS1 is upregulated in RCC tumor tissues compared with normal kidney tissues, which promotes RCC progression via acting as ceRNA of miR-26a to regulates PTEN protein [31]. LncRNA NEAT1 is upregulated in RCC tissue and the high NEAT1 expression is associated with tumor progression and poor survival in RCC patients by acting as a competitive sponge for miR-34a, which prevents inhibition of c-Met [32]. In this study, we found that MALAT1 acted as a ceRNA for miR-429, providing a new insight for RCC.
Conclusions
Overall, both lncRNA MALAT1 and miR-429 play important parts in tumorigenesis of RCC. Our current study reveals that MALAT1 is an oncogene, functioning by sponging miR-429 or acting as its ceRNA. Therefore, MALAT1 is a pivotal factor in RCC and may have potential to be a promising therapeutic target in the future.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Jonasch EGJ, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljungberg BBK, Canfield S, Dabestani S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913–24. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Escudier BSC, Porta C, Gore M. Treatment selection in metastatic renal cell carcinoma: expert consensus. Nat Rev Clin Oncol. 2012;9:327–33. doi: 10.1038/nrclinonc.2012.59. [DOI] [PubMed] [Google Scholar]
- 4.Ljungberg B, Campbell SC, Choi HY. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–21. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Atkins MB, Ernstoff MS, Figlin RA, et al. Innovations and challenges in renal cell carcinoma: Summary statement from the Second Cambridge Conference. Clin Cancer Res. 2007;13:667–70. doi: 10.1158/1078-0432.CCR-06-2231. [DOI] [PubMed] [Google Scholar]
- 6.Djebali SDC, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Research. 2006;66:7390–94. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni W, Song E, Gong M, et al. Downregulation of lncRNA SDPR-AS is associated with poor prognosis in renal cell carcinoma. Onco Targets Ther. 2017;10:3039–47. doi: 10.2147/OTT.S137641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Ma P, Zhao Y, et al. Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: Role of the MALAT-1-Livin protein interaction. J Physiol Sci. 2016;7:77–85. doi: 10.1007/s12576-016-0486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–39. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi XSM, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: Systematic review and meta-analysis. Br Med J Open. 2015;5:965–75. doi: 10.1136/bmjopen-2015-008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Ma P, Zhao Y. Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: Role of the MALAT-1-Livin protein interaction. J Physiol Sci. 2017;67(5):577–85. doi: 10.1007/s12576-016-0486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–37. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao H, Tang K, Liu P. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–15. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon KA, Yoon H, Park S, et al. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2012;144:794–807. doi: 10.1016/j.jtcvs.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Tian X, Wei Z, Wang J, et al. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling. Oncol Rep. 2015;34:707–14. doi: 10.3892/or.2015.4039. [DOI] [PubMed] [Google Scholar]
- 21.Machackova T, Mlcochova H, Stanik M. MiR-429 is linked to metastasis and poor prognosis in renal cell carcinoma by affecting epithelial-mesenchymal transition. Tumor Biol. 2016;85:1–6. doi: 10.1007/s13277-016-5310-9. [DOI] [PubMed] [Google Scholar]
- 22.Huarte M. Large non-coding RNAs: Missing links in cancer? Hum Mol Genet. 2010;19:152–58. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 24.Lin YL, Wang YP, Li HZ, Zhang X. Aberrant promoter methylation of PCDH17 (Protocadherin 17) in serum and its clinical significance in renal cell carcinoma. Med Sci Monit. 2017;23:3318–23. doi: 10.12659/MSM.902077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W, Xu H, Zhu D, et al. MiR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2012;69:723–31. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Niu X, Pan H, et al. Tumor-suppressing effects of microRNA-429 in human renal cell carcinoma via the downregulation of Sp1. Oncol Lett. 2016;12:2906–11. doi: 10.3892/ol.2016.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JH, Liu S, Zhou H, et al. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:3368–76. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan X, Zhiguo H, Li X. Long non-coding RNA MALAT1 interacted with miR-204 to modulates human hilar cholangiocarcinoma proliferation, migration and invasion by targeting CXCR4. J Cell Biochem. 2017;76:122–43. doi: 10.1002/jcb.25862. [DOI] [PubMed] [Google Scholar]
- 30.Xie W, Wang L, Sheng H, et al. Metformin induces growth inhibition and cell cycle arrest by upregulating microRNA34a in renal cancer cells. Med Sci Monit. 2017;23:29–37. doi: 10.12659/MSM.898710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng X, Hu Z, Ke X, et al. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16:2212–19. doi: 10.1080/15384101.2017.1361072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, Chen N, Gong Y, et al. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8:62927–38. doi: 10.18632/oncotarget.17757. [DOI] [PMC free article] [PubMed] [Google Scholar]