Abstract
Large studies profiling microbial communities and their association with healthy or disease phenotypes are now commonplace. Processed data from many of these studies are publicly available but significant effort is required for users to effectively organize, explore and integrate it, limiting the utility of these rich data resources. Effective integrative and interactive visual and statistical tools to analyze many metagenomic samples can greatly increase the value of these data for researchers. We present Metaviz, a tool for interactive exploratory data analysis of annotated microbiome taxonomic community profiles derived from marker gene or whole metagenome shotgun sequencing. Metaviz is uniquely designed to address the challenge of browsing the hierarchical structure of metagenomic data features while rendering visualizations of data values that are dynamically updated in response to user navigation. We use Metaviz to provide the UMD Metagenome Browser web service, allowing users to browse and explore data for more than 7000 microbiomes from published studies. Users can also deploy Metaviz as a web service, or use it to analyze data through the metavizr package to interoperate with state-of-the-art analysis tools available through Bioconductor. Metaviz is free and open source with the code, documentation and tutorials publicly accessible.
INTRODUCTION
High-throughput sequencing of microbial communities provides a tool to characterize associations between the host microbiome and health status, to detect pathogens and to identify the interplay of an organism’s microbiome with the built environment. Recent highlights include work on the specificity of the human skin microbiome (1), diversity in the ocean microbiome (2) and cataloging the global virome (3). Effective analysis tools and appropriate statistical models for this type of data are vital to derive and communicate significant insights from these experiments. In other high-throughput sequencing assays, including those for genome, transcriptome, and epigenome, next-generation genome browsers that integrate exploratory computational and visual analysis have proven to be effective analysis tools (4,5). Exploratory analysis tools for microbiome data are scarce however, partially stemming from the challenge that microbiome features, the units of measurement and analysis, are organized in a taxonomic hierarchy. Specifically, while the linear structures of tracks and ranges used in genome browsers provide a natural scheme for navigation in genomic visualization, a hierarchical exploration technique is not readily available. In this paper, we present the Metaviz tool for effective interactive exploration, analysis and data visualization of hierarchically organized metagenomic features.
Motivation
As an illustrative use case for statistically guided interactive visualization, we consider a data analysis from the moderate to severe diarrheal (MSD) disease study among children in four countries of the developing world (6). A typical analysis for this case-control study includes statistical testing to compare taxa abundance between children with and without diarrhea to find novel associations between health and disease. The metagenomeSeq R/Bioconductor package [http://bioconductor.org/packages/release/bioc/html/metagenomeSeq.html] is a popular tool to identify differentially abundant features (7). In this paper, we target workflows after an abundance matrix has been computed. A standard workflow starts with the data analyst obtaining sequence counts indicating the abundance of annotated operational taxonomic units (OTUs) for each sample in a study with phenotypic and experimental characteristics of these samples available as metadata. The workflow proceeds by the data analyst aggregating counts to a specific level of the taxonomic hierarchy (e.g. species or genus) and obtaining differential abundance inferences by computing log fold changes and P-values for each taxa between case and control groups. She then selects features with a log fold change beyond a given threshold and P-value cutoff as differentially abundant taxa. Next, she visualizes the abundance of these filtered features across samples in a heatmap. After interpreting the plot, she may decide to change the feature selection parameters or further explore the taxonomic hierarchy, which requires another iteration of computing the feature set and visualization. In this case, each refinement of statistical analysis parameters produces another visualization with no linking between results.
Our design of the Metaviz application for interactive visualization and analysis makes this workflow much more effective: for instance, once a set of differentially abundant features is selected, the data analyst can interactively visualize abundance data for those specific features. She can then explore the hierarchy of features, aggregate counts to any level of the taxonomy and identify sub-structures that are difficult to ascertain at lower levels of the taxonomic hierarchy. Further, she may calculate differential abundance at a different level of the hierarchy then dynamically explore these inferences in the same Metaviz workspace, thus streamlining her exploration of a complex set of differential abundance results using statistical and visualization tools.
Related work
Taxonomer performs both read taxonomic assignment and visualization of results using a sunburst diagram to visualize features (8). Pathostat is a Shiny application that computes statistical metagenomic analyses, visualizes results and is integrated with different Bioconductor packages [http://bioconductor.org/packages/release/bioc/html/PathoStat.html]. Pavian is an R package that incorporates Shiny and D3.js (9) components to enable interactive analysis of results for metagenomic classification tools [https://doi.org/10.1101/084715]. Panviz is a tool for exploring annotated pan genome datasets based on D3.js libraries (10). Krona is a web-based tool for metagenomics visualization that provides a sunburst diagram to navigate the feature space (11). VAMPS is a web service that provides a JavaScript and PHP-based metagenomics visualization toolkit of datasets uploaded by researchers (12). Anvi’o is a multiomics platform that supports analysis using custom JavaScript visualizations (13). MicrobiomeDB is a web service that hosts microbiome community taxonomic profile data from open datasets and uses Shiny to visualize data (14).
Encompassing the features of these tools, Metaviz provides a comprehensive interactive visualization environment using JavaScript and D3.js for microbial marker-gene sequencing and whole metagenome shotgun sequencing data with integration to R/Bioconductor. In contrast to these tools, Metaviz uses FacetZoom, which is more suited than sunburst diagrams for browsing the hierarchical structure of metagenomic data across many samples by enabling taxonomic feature selection spanning multiple levels of a taxonomy. Further, Metaviz can analyze data from either a database or R, which makes it more efficient and scalable than Shiny-based tools which are limited by in-memory processing. Metaviz implements the WebSockets protocol directly, which allows for use of data transfer types beyond those specified in Shiny to support flexible and extensible custom JavaScript visualizations.
MATERIALS AND METHODS
Metaviz is a web browser-based tool for interactive exploratory microbiome data analysis. It can visualize abundance data served from an interactive R session or query data from a graph database server. Here, we present the architecture of Metaviz from the web browser application to database storage. A web browser-based application provides flexibility for users and ‘run anywhere’ functionality when deploying the tool. We built upon the D3.js project for an aesthetically pleasing and effective suite of plots and charts. The data back end serves an abundance matrix with taxonomic annotation for features, in our case OTUs, and the front end is a JavaScript application for data visualization. Given the structure of metagenomic data, the user navigation tools and the database storage are tailored to taxonomic hierarchies. We moved from a relational database model used in Epiviz (15), our previous interactive data analysis tool for functional genomic data such as gene expression and methylation data, to a graph database to manage the feature hierarchy and abundance counts. The fundamental operation enabled by this data back end is to efficiently aggregate abundance counts to a specific subset of nodes in the taxonomic hierarchy during interactive exploration.
Visualization layer
Implementing the visualization layer for this application presents several challenges for displaying, navigating and manipulating data from a feature-rich hierarchy. Design considerations for metagenomic data analysis include: (i) size of the feature space, which in datasets we visualized using Metaviz, ranges from 727 (whole metagenome shotgun data from the ‘Rampelli’ dataset in the curatedMetagenomicData R/Bioconductor Package) to 45 336 (16S OTUs in the Human Microbiome Project) features; (ii) depth of the feature hierarchy, which is a function of the annotation database and parameters used; and (iii) number of samples, with as many as 992 samples in the MSD dataset which is the largest that we host. Given these characteristics, we focused the design of Metaviz on efficient traversal of the feature space and defining feature selections across the taxonomy. In addition, we engineered the navigation tools to be applicable across datasets and persistent between user sessions for collaboration and publication of results.
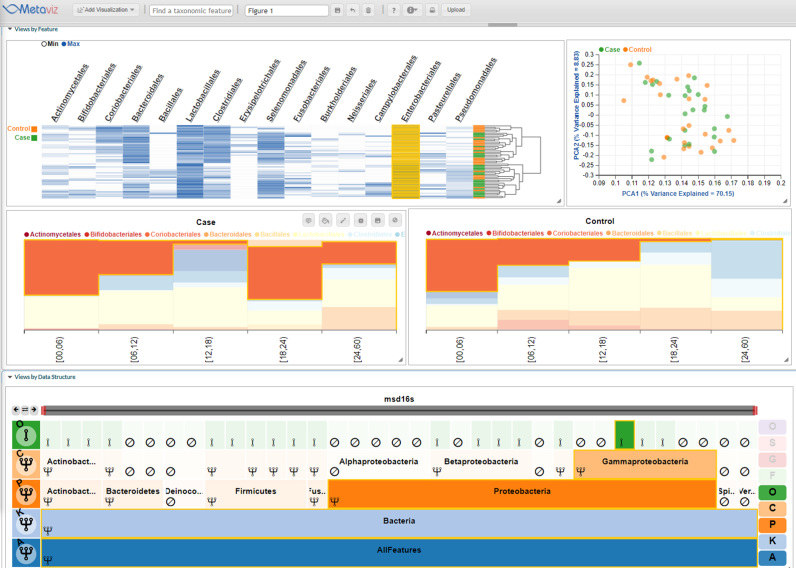
In Figure 1, we demonstrate the visualization layer of Metaviz on the MSD marker-gene survey dataset. The bottom panel is a navigation control designed to effectively explore the taxonomic feature hierarchy and aggregate count values of features to any set of taxonomic nodes. The top panel consists of a heatmap with the color intensity set as the observed count of a feature (column) in a sample (row). The rows are dynamically clustered based on Bray-Curtis distance of the count vectors for each sample and a dendrogram shows the clustering result. The top panel also includes a Principal Component Analysis (PCA) plot over all the features of the samples in the heatmap. The stacked bar plots in the second row render, for each sample (column), the proportion of counts for each microbial feature. The separate plots show case (left) or control (right) samples based on dysentery status and the columns are samples grouped by age range. This collection of charts provides multiple views of the same data and is dynamically updated upon user interaction with the navigation tool to achieve exploratory iterative visualization.
Figure 1.
Metaviz interactive visualization of childhood severe diarrhea study. A subset of 50 samples (25 case and 25 control for dysentery) from the Moderate to Severe Childhood Diarrheal Disease study (6). The FacetZoom control on the bottom panel is used for exploration of the taxonomic organization of metagenomic features. Node opacity in the FacetZoom indicates the set of taxonomic features selected across all appropriate visualizations in the Metaviz workspace. Each node can be in one of three possible states as indicated by the icon in its lower left corner: (i) aggregated, where counts of descendants of this node are aggregated and displayed in other charts, (ii), expanded, where counts for all descendants of this node are visualized in other charts or (iii) removed, where this node and all its descendants are removed from all the other charts. The left column of the FacetZoom control indicates the levels of the taxonomy and the overall selection for nodes at each taxonomic level. Hovering the mouse over FacetZoom panels highlights the corresponding features in other charts through brushing. The top left chart is a heatmap showing log-transformed counts with color intensity corresponding to the abundance of that feature (column) in that sample (row). The dynamically computed and rendered row dendrogram shows Bray-Curtis distance hierarchical clustering of samples with color indicating case/control status of each sample. The yellow highlighted column is linked between charts and FacetZoom control through brushing. The top right chart is a PCA plot over all features at the current aggregation level (order). The stacked bar plot on the left of the second row shows proportion of selected features in each case sample (columns) while the right chart shows control samples. In both, sample counts are grouped and aggregated by age range. This is available as a Metaviz workspace at http://metaviz.cbcb.umd.edu/?ws=yA4BWgUOTiq.
Navigation mechanism—FacetZoom
We developed Metaviz to navigate the complex hierarchical structure of microbiome feature data and perform the visualization tasks of overview, zoom and filter. We incorporate the FacetZoom (16) design, which visualizes a hierarchy using a tree structure showing a subset of levels at one time. We chose this approach to handle the limitations in the screen size and performance of rendering trees with tens of thousands of nodes. We extended the original FacetZoom design to perform interactive aggregation and removal of microbial lineages. We refer to our navigation tool, shown in the bottom panel of Figure 1, as a FacetZoom control for the rest of the manuscript.
The nodes of the FacetZoom control indicate how the abundance counts for taxonomic features are displayed in the other charts of the Metaviz workspace. Every node of the FacetZoom control can receive mouse-click input from the user. A click on a node sets that feature as the root of a dynamically rendered subtree. Each node can be in one of the three possible states as indicated by an icon in its lower left corner: (i) aggregated, where counts of descendants of this node are aggregated and displayed as a single feature in other charts, (ii) expanded, where counts for all descendants of this node are visualized as separate features in other charts or (iii) removed, where this node and all its descendants are removed completely from the other charts. The state of a node determines the state of its descendants. Node opacity in the FacetZoom control indicates the set of taxonomic units selected across all appropriate visualizations in the Metaviz workspace. Hovering the mouse over FacetZoom nodes highlights the corresponding features in other charts through brushing as shown in Figure 1. The bottom node of the FacetZoom visualization displays the taxonomic lineage of the corresponding feature at the root of the subtree currently in view.
The FacetZoom control includes a level-wise aggregation indicator panel on the left side. Each element of the indicator panel can be used to set the aggregation state of all nodes at a given depth. The letter on each element of the panel identifies the taxonomic level with ‘P’ denoting phylum and ‘O’ signifying order, for instance. The panel on the right provides a persistent global view of the hierarchy to identify where in the full taxonomy the current subtree selected by the user is located. As an example, when the FacetZoom is displaying nodes from class to genus, only these elements are highlighted in the levels indicator panel.
The bar at top of the FacetZoom sets the range of features shown in the other charts in the visualization workspace. The bar is a flexible component with arrows to control movement left or right and expansion over the full range of the current subtree. Updates to the filter bar triggers queries over the count data and those results are automatically propagated to the other charts in the workspace.
As described, the FacetZoom controls which features are included in plots and charts of count data in a Metaviz workspace. We detail our implementation of heatmaps, stacked bar plots, scatter plots, alpha diversity boxplots, PCoA plots and line plots in Supplementary Materials Section II. Among the available visualizations is a sunburst diagram, which we designed to be used in conjunction with the FacetZoom control for navigation. While the FacetZoom is used for navigation and feature selection, the sunburst provides a view of the taxonomy via a circular layout commonly used in microbiome analysis. Supplementary Figure S1 shows an example of the sunburst diagram.
Metaviz supports text-based search for quick navigation to specific taxonomic features. A user can enter the name of a taxonomic feature of interest into a search box on the toolbar. The search provides auto-complete and lists features that contain the character string in a drop-down list. Once a user selects a feature, the navigation bar in the FacetZoom control will update to encompass that feature and all linked data visualizations update as well.
Metaviz includes a dynamic boxplot, created by clicking on column labels of a heatmap, to offer details-on-demand of taxonomic feature count distributions across samples of interest. A box and whisker glyph is created for each sample group selected based on criteria defined over sample metadata. Text-search can also be used within the boxplot to select any feature in the hierarchy and display counts aggregated to that feature.
Data layer
A key difference between microbiome sequencing data and other genomic data is the hierarchical organization of its features, which drives the design of the Metaviz back end. Our data model of microbiome datasets includes the observed counts for each feature in every sample, the hierarchical taxonomic feature annotations and metadata such as phenotypic, behavioral and environmental information for each sample. A query triggered from user interaction operates over these three data types and computes aggregations on the count data to the specified hierarchy level.
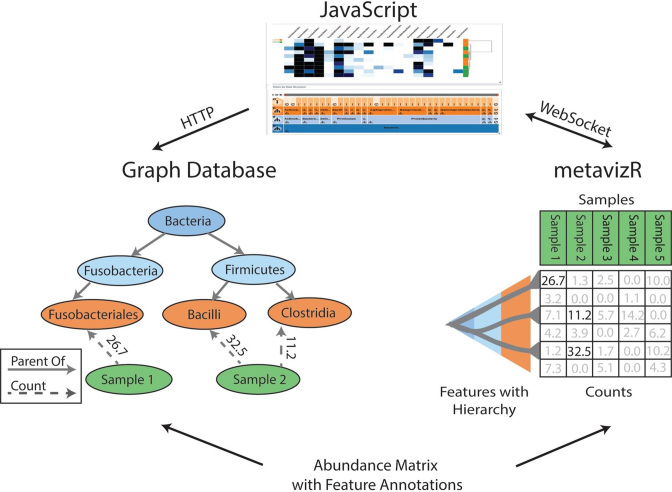
To achieve interactive visualizations with reasonable query response times, we used a graph database architecture. In a graph database, nodes and edges in a graph are objects that can be queried directly. This is a contrast to relational databases in which samples are rows and sample attributes are columns. Each table in a relational database encompasses all the required data fields for the observations in that table while keys handle relationships between tables. We use a graph database to store each taxonomic feature as a node in the graph with edges connecting nodes as specified by the taxonomic information. This system uses a natural representation of the hierarchical organization of this data while avoiding costly join operations in a relational database. We also store samples as nodes and the count value for a feature in a sample is an edge between leaf feature nodes and sample nodes. This graph database structure is shown in Figure 2.
Figure 2.
Metaviz query processing and Graph DB structure. There are two deployment options, which can be used concurrently if desired. In one deployment option (left), the Metaviz JavaScript front end makes requests to a Python application querying a graph database using HTTP. In the other deployment option (right), abundance matrices are loaded into a metavizr session which uses the WebSocket protocol to communicate to the JavaScript component, allowing two-way communication between JavaScript and an interactive R session. The graph on the left shows how abundance matrices are stored in the graph database. Nodes in the graph correspond to metagenomic features or samples, edges between metagenomic features denote taxonomic relationship, edges at the leaf level of the taxonomy connect to samples and store the corresponding abundance counts. In either deployment option, aggregation queries are evaluated in response to FacetZoom control selections in the UI and require summing, for each sample, the counts for features in a selected taxonomic subtree.
Materials
We utilized several datasets during the design and testing of Metaviz. The first is the MSD dataset, gathered from a cohort of 992 children across four countries with an age range of 0–60 months. Fecal samples were gathered from subjects with diarrhea and healthy controls. Specific details for data generation, pre-processing and annotation are covered in Pop et al. (6). To study time series, we used a longitudinal Escherichia coli analysis dataset gathered from 12 participants who were challenged with E. coli and subsequently treated with antibiotics. Stool samples were gathered from participants each day starting 1 day pre-infection until 9 days post-infection. Experimental and sample details are available in Pop et al. (17). We benchmarked our system with data from the Human Microbiome Project available at the Data Analysis and Coordination website [https://www.hmpdacc.org/hmp/]. We retrieved the data as a prepared phyloseq object [http://joey711.github.io/phyloseq-demo/HMPv35.RData] and chose the subset of samples processed at the Washington University Genome Center.
RESULTS AND DISCUSSION
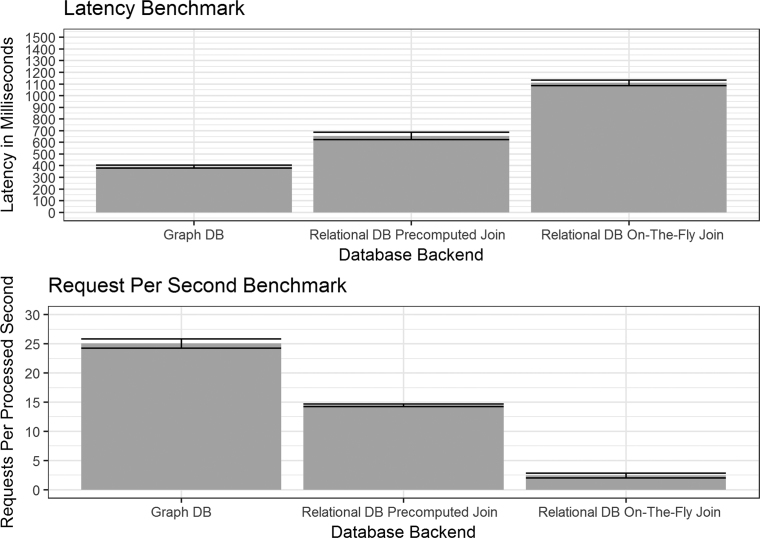
To inform the choice of database architecture, we benchmarked an implementation using a relational database against one using a graph database. The relational database uses MySQL [https://www.mysql.com/] as the database management system and PHP [http://php.net/] to handle requests from the web browser client. The graph database configuration uses Neo4j [https://neo4j.com/] and the Flask web development framework [http://flask.pocoo.org/]. In the benchmarks, we deploy our back end services on an Amazon EC2 t2.small instance and used the wrk tool [https://github.com/wg/wrk] to send HTTP requests. The testing dataset consisted of 62 samples, 973 features and 7 hierarchy levels. We observed that the graph database provides approximately five times lower latency. We also modified the relational design to pre-compute a join operation between the sample, hierarchy and count tables then store that in the database. This design decreases query response time but increases the size of the database. Compared to this implementation, our graph database implementation showed ∼50% lower latency. We present our benchmark results in Figure 3.
Figure 3.
Metaviz database architecture benchmarks. We use the wrk tool to benchmark UI requests to three database architectures for storing abundance matrices and feature hierarchies (taxonomies): (i) Graph DB, using Neo4j with a Python Flask web service, (ii) Relational DB Pre-computed Join, using a MySQL implementation with a JOIN of the three tables of features, values and samples pre-computed and stored as a table, (iii) Relational DB On-The-Fly Join, a MySQL implementation with computing a JOIN across the three tables for each query. For (ii) and (iii), a PHP application issues queries to the database in response to requests from the UI. We deployed each implementation on an Amazon EC2 t2.small instance and the dataset used across all instances consisted of 62 samples, 973 features and 7 hierarchy levels. The upper panel shows query latency including standard error across 5 days of measurements. In addition to the latency of processing each request, we also measure the number of requests per second processed providing a measure of throughput in our application. In both performance measures, we see significant benefits of a Python-Neo4j deployment compared to a PHP-MySQL stack for Metaviz tasks.
Whole metagenome shotgun sequencing data
We designed Metaviz to render community taxonomic profile data derived from whole metagenome shotgun sequencing in addition to marker gene sequencing. The results of this sequencing is often reported in relative abundance, which is converted to counts through multiplying by read depth, at inner nodes of the taxonomy instead of counts at leaf nodes only in marker gene data (18). Feature selection queries, or specification of tree cuts, at various levels of the hierarchy do not compute aggregation and instead are directly returned from the inner node counts.
Metavizr
Metaviz expands the analysis that can be performed from Bioconductor through the metavizr package [https://bioconductor.org/packages/release/bioc/html/metavizr.html]. Interactive visualization of microbiome statistical analysis results allows a user to explore the data at various levels of detail and report those findings in an accessible, aesthetically pleasing interface. Metavizr uses the metagenomeSeq R/Bioconductor package to load the feature, count and sample data into a data object. Metavizr communicates with a Metaviz web browser application instance using a WebSocket connection. A FacetZoom control along with data charts and plots can be added to the Metaviz workspace interactively from the R session. A user can specify taxonomic features for visualization from the results of statistical testing as discussed in the Motivation section. Metaviz can be used with other R/Bioconductor packages beyond metagenomeSeq for analysis. As an example, we use the vegan CRAN package to compute alpha diversity [https://cran.r-project.org/web/packages/vegan/index.html] for microbial community-level analysis. Github Gists can be used through metavizr to modify any plot or chart display setting using JavaScript in addition to customization facilities provided directly by the metavizr package itself. We provide a guide to using Github Gists in the Supplementary Materials—Tutorial. Finally, a persistent workspace identifier can be used to reproduce the visual analysis of a collaborator after metavizr loads the dataset. To measure the performance of metavizr, we benchmarked the memory usage and run-time of aggregation operations using a subset of the Human Microbiome Project dataset, which we describe in the Materials and Methods section. We ran the benchmark on an AWS ec2 t2.large instance to simulate the configurations of a typical laptop used for analysis using R/Bioconductor. We present the performance results in Supplementary Figure S2. We found metavizr to provide suitably responsive behavior for datasets up to 1000 samples and 25 000 features and recommend switching to the graph database back end for larger datasets.
UMD metagenome browser
We loaded samples from a variety of marker gene and whole metagenome shotgun sequencing studies into the UMD Metagenome Browser—a Metaviz instance hosted by the University of Maryland Center for Bioinformatics and Computational Biology at http://metaviz.cbcb.umd.edu. The whole metagenome shotgun data are from the R/Bioconductor package curatedMetagenomicData [https://bioconductor.org/packages/release/data/experiment/html/curatedMetagenomicData.html] which provide curated data from metagenomic studies for dozens of diseases across multiple body sites (19). A total of 7115 samples from published studies are available from the UMD Metagenome Browser. Figure 4 lists the datasets, sample sites and descriptions of the available metadata. With the UMD Metagenome Browser, an analyst can choose from the datasets available to complete a study and share results through a persistent Metaviz workspace. New datasets can be added to the UMD Metagenome Browser by loading the abundance matrix, sample metadata and taxonomic hierarchy into the database hosted at UMD. We plan to continuously load new datasets and encourage users to contact us with datasets they would like to host publicly in the UMD Metagenome Browser and we will load those into the database.
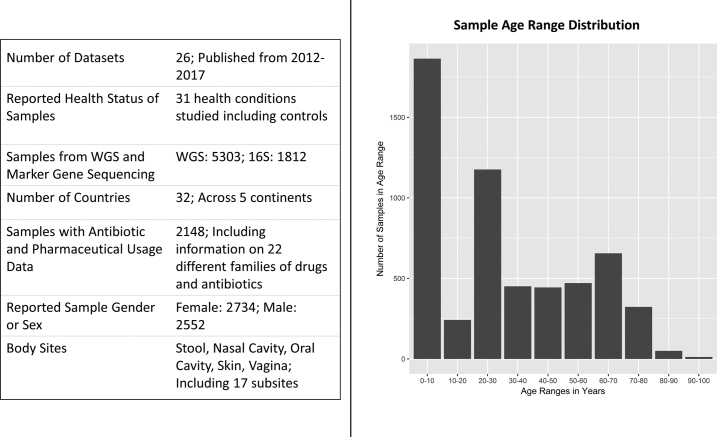
Figure 4.
UMD Metagenome Browser Sample Summary. The publicly available Metaviz instance at http://metaviz.cbcb.umd.edu hosts data from several published studies which were generated using marker gene survey and whole metagenome shotgun sequencing. A total of 26 datasets with 7115 samples from published studies across 31 health conditions and 32 countries are available. Host age ranges from 0 months to subjects over 90 years old. Among the metadata available is reported gender or sex of subject, antibiotic or pharmaceutical usage data, and time course measurements. The number and types of datasets are being updated continuously.
Deployment
We support two other deployment mechanisms of Metaviz for users to interactively visualize an abundance matrix with hierarchical feature annotations depending on analysis needs. For interactive joint exploratory statistical and visual analysis, data analysts can load the abundance matrices into a Metaviz instance through metavizr. Also, we provide Docker [http://www.docker.com] scripts so users can build and deploy containers of the database, load the abundance matrix to the database, and host the web browser application as an independent Metaviz instance [https://github.com/epiviz/metaviz-docker].
Use cases
We employ Metaviz to perform exploratory visual analysis on two published microbiome datasets. The exploratory analysis is coupled with statistical testing methods available from the metagenomeSeq R/Bioconductor package. We envision Metaviz being used along with a statistical testing framework to identify the significance of analysis results. We note statistically significant and non-significant results of our visual exploration in use cases that follow.
Use case 1: exploration of MSD childhood diarrhea study in developing countries
To demonstrate the analysis utility of Metaviz we report on a new analysis of the MSD dataset. To visualize and explore samples, we examined the data from each of the four countries in the study separately and aggregated taxonomic features to the order level. In this analysis, we set case status as those with dysentery and control status as those without blood in stool, meaning that samples with diarrhea and healthy samples are in the control group for dysentery. We chose this analysis to expand upon the work from the author’s original investigation, which studied healthy versus diarrhea and dysenteric versus non-dysenteric diarrhea (6). This analysis is exemplary of case-control studies commonly employed in microbiome data investigation. For our exploration, we used three visualizations, a heatmap, a dynamic boxplot and two stacked bar plots to identify differences in the microbial communities in case and control across age ranges by country. We created boxplots for details-on-demand of specific taxonomic features based on visual analysis of the heatmap. In the heatmap, row colors were set by dysentery status and each stacked bar plot consisted of the case and control samples for dysentery of each country. We also grouped the samples in the stacked bar plot by age range.
For visual inspection of differential abundance, we ordered each heatmap by dysentery status so that all case and control samples are grouped together. We looked at the heatmap and removed features with low abundance using the FacetZoom control. We then examined each column individually, identifying the number of samples with a feature present and the distribution of samples with high or low intensity. For features of interest, we then created a boxplot by clicking the column label in the heatmap. The boxplot shows the counts aggregated to that feature for case and control dysentery groups. Using these two visualizations of count data, we called the feature as more abundant in case samples, more abundant in control samples, or as no difference in abundance across groups. When we identified a difference in abundance for a feature, we used the FacetZoom to aggregate counts to the next level lower in the hierarchy, restrict the heatmap to show only children of that feature and updated the boxplot to identify differences in abundance at that level of the hierarchy. We performed this systematic approach to inspect each feature from the order level to the species level. We compared the results of visual analysis by computing the log fold change using metagenomeSeq and report those features detected through our visualization process and list the results of statistical testing. When using metagenomeSeq, counts were normalized using cumulative sum scaling (with P = 0.75) and binary dysentery status as the variable of interest in the fitFeatureModel method for differential abundance. The threshold for differential abundance was an absolute log fold change of at least 1 and an adjusted P-value < 0.1 when comparing samples using dysentery status.
Supplementary Figures S3 and S4 show our visual analysis for Bangladesh samples. From the heatmap and boxplot analysis of these samples, the following taxa appear more abundant in the samples with dysentery than the control samples: Actinomycetales, Enterobacteriales, Lactobacillales, Pasteurellales, Pseudomonadales, Micrococcaceae, Enterobacteriaceae, Carnobacteriaceae, Streptococcaceae, Pasteurellaceae, Moraxellaceae, Rothia, Escherichia, Shigella, Granulicatella, Streptococcus, Haemophilus, Acinetobacter, E. coli, Escherichia sp. oral clone 3RH-30, Granulicatella adiacens, Streptococcus equinus, Streptococcus mitis, Streptococcus parasanguinis, Streptococcus salivarius, Haemophilus parainfluenzae and Acinetobacter sp. SF6. Correspondingly, the following taxa appear more abundant in the control samples as compared to the case samples: Coriobacteriales, Bacteroidales, Clostridiales, Coriobacteriaceae, Bacteroidaceae, Porphyromonadaceae, Clostridiaceae, Eubacteriaceae, Lachnospiraceae, Ruminococcaceae, Collinsella, Bacteroides, Clostridium, Eubacterium, Dorea, Faecalibacterium, Ruminococcus, Collinsella sp. CB20, Bacteroides fragilis, Faecalibacterium prausnitzii, Faecalibacterium sp. DJF_VR20 and Ruminococcus gnavus. Examining the stacked bar plots at the order level, Clostridiales exhibits low proportion in the case samples at 0–6 and 6–12 months, a lower level compared to control samples at 12–18 months and then a similar proportion in both groups for 18–24 and 24–60 months. With the control samples, Bacteroidales shows a greater proportion at all intervals after 0–6 months.
Using metagenomeSeq, we find the following taxa to have significant difference in abundance for Bangladesh samples: Enterobacteriales (log fold change = 1.38, adjusted P-value = 1.46E-04), Pasteurellales (2.47, 4.16E-12), Coriobacteriales (−1.38, 9.88E-04), Bacteroidales (−1.19, 7.56E-04), Clostridiales (−1.09, 6.45E-04), Enterobacteriaceae (1.37, 2.26E-04), Carnobacteriaceae (1.52, 3.23E-05), Streptococcaceae (1.41, 5.00E-05), Pasteurellaceae (2.46, 1.43E-11), Coriobacteriaceae (−1.37, 1.95E-03), Bacteroidaceae (−1.09, 1.16E-02), Ruminococcaceae (−1.09, 3.17E-03), Escherichia (1.33, 6.50E-04), Granulicatella (1.51, 8.29E-05), Streptococcus (1.33, 2.91E-04), Haemophilus (2.42, 6.12E-11), Collinsella (−1.48, 3.89E-03), Bacteroides (−1.08, 2.27E-02), Ruminococcus (−1.18, 3.89E-03), E. coli (1.33, 1.71E-03), G. adiacens (1.51, 1.92E-03), S. mitis (1.16, 1.50E-02), S. parasanguinis (1.07, 1.71E-03), S. salivarius (1.02, 2.11E-02), H. parainfluenzae (2.26, 3.04E-07), Collinsella sp. CB20 (−1.26, 3.68E-02) and R. gnavus (−1.18, 3.48E-02). We present the results for visual analysis and metagenomeSeq differential abundance calculation for each country in Supplementary Tables S1–4 and in Section III of Supplementary Materials.
The previously published analysis of dysenteric versus non-dysenteric diarrhea grouped samples from all countries and identified OTUs associated with dysenteric stool, including those from the following taxa: Haemophilus, Streptococcus, Granulicatella, E. coli and Enterobacter cancerogenus (6). While using the heatmap, boxplot and FacetZoom control to explore each country we observed greater abundance in case samples for Haemophilus in Bangladesh, The Gambia, Mali and Kenya; S. salivarius in Bangladesh; Granulicatella in Bangladesh and The Gambia; E. coli in Bangladesh and The Gambia; and E. cancerogenus in Kenya. Examining results across all countries, three taxa showed greater abundance among case samples through visual inspection and were statistically significant using metagenomeSeq: Pasteurellales, Pasteurellaceae and Haemophilus.
Features that showed statistically significant difference in abundance in more than one country but not all are Enterobacteriales and Enterobacteriaceae in Bangladesh and The Gambia. Some features with differential abundance in only one country include Coriobacteriales, Bacteroidales, Coriobacteriaceae, Collinsella, Ruminococcus, Collinsella sp. CB20, R. gnavus and S. parasanguinis in Bangladesh. A literature examination revealed that R. gnavus has been identified as present in patients with Crohn’s Disease that relapsed 6 months after surgical treatment (20). Streptococcus parasanguinis has been identified as having higher relative abundance in cancers of the gastric body in patients without Helicobacter pylori infection (21). In samples from The Gambia, Actinomycetales is more abundant in case than control which is notable given that Tropheryma whipplei is the only identified enteric pathogen in the order (22,23) and that was not identified as differentially abundant by either visual analysis or statistical testing. It is important to note that for The Gambia, Kenya and Mali, samples without dysentery outweighed those with dysentery.
Use case 2: analysis of longitudinal metagenomic studies
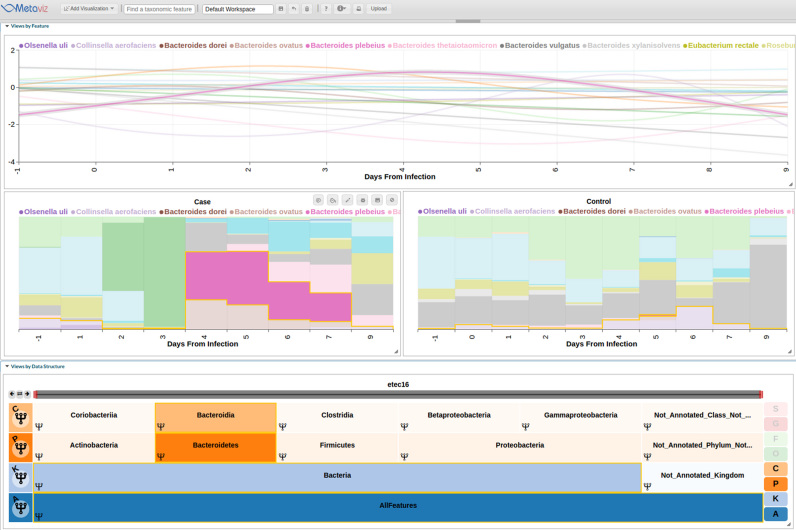
Another use case of Metaviz is the analysis of longitudinal metagenomic datasets. We followed the analysis using smoothing spline ANOVA as described in Paulson et al. [https://doi.org/10.1101/099457] for a longitudinal dataset characterizing host response to a challenge with enterotoxigenic E. coli (17). The metagenomeSeq R/Bioconductor package provides the fitMultipleTimeSeries function for fitting a smoothing spline and performing SS-ANOVA testing. Using fitMultipleTimeSeries, the formula considered if diarrhea developed at any day as well when antibiotics were given to the individual. To visualize the results, we use a line plot with time points on the X-axis, log fold change on the Y-axis and each line representing a taxonomic feature. The FacetZoom is linked to the line plot and the path through the hierarchy is highlighted when hovering over a given line. We also created a stacked line plot of counts aggregated to the species level for those species that were found to be differentially abundant for an interval of at least 2 days using the SS-ANOVA model. Figure 5 shows the Metaviz workspace for this analysis with the spline plot on the top, one sample with diarrhea on the left and one sample without diarrhea at any day on the right. We chose one pair because antibiotics were administered on different days across samples therefore averaging counts across case and control groups is not representative of response for the treatment applied. Each column in the stacked line plots represent the measurement taken at the day since infection. Antibiotics were administered at days 3 through 5 for the case sample and days 4 through 6 for the control sample. Examining the stacked plots Bacteriodes plebeius shows high proportion in the case sample on the day after antibiotics are administered then a decrease 2 days after the treatment was completed to a similar level as in the control sample. This procedure can be generalized to time series analysis of microbiome data when investigating differential abundance across time points. Also, the smoothing parameter for the spline can be updated through the user interface and this process is detailed in Supplementary Materials—Tutorial.
Figure 5.
Interactive visualization of smoothing spline differential analysis of longitudinal study. We use Metaviz to explore a longitudinal analysis of the dataset from an enterotoxigenic Escherichia coli study (17). Count data were aggregated to the species level and a smoothing-spline ANOVA model was fit using the fitTimeSeries function of the metagenomeSeq R/Bioconductor package. Features with a statistically significant interval of 2 days or longer as estimated by the smoothing spline model at any time point were selected for visualization. The line plot is linked via brushing with the FacetZoom control and a stacked plot showing feature count proportions for a sample that developed diarrhea and a sample with no diarrhea.
CONCLUSION
In this paper, we presented the design and performance of Metaviz, a web browser-based interactive visualization and statistical analysis tool for microbiome data. We described design decisions for operating over abundance matrices with tens of thousands of features, thousands of samples and complex feature hierarchies. We use a graph database for storing community abundance profile matrices as the features have a hierarchy derived from taxonomic databases. We also developed the metavizr R/Bioconductor package providing tight integration of the Metaviz interactive visualization tool and computational and statistical analyses using R/Bioconductor packages. We used Metaviz to analyze existing datasets and our results highlight the power of interactive visualization coupled with complementary statistical analysis to examine microbiome data. A major contribution of this work is the navigation utility that adapts information visualization techniques to effectively explore and manipulate the rich feature hierarchy of metagenomic datasets. Another significant contribution is the UMD Metagenome Browser web service available to host abundance matrices that allows researchers to explore and share results. We expect that Metaviz will prove useful for researchers in analyzing microbiome sequencing studies as genome browsers have for genomic data.
An avenue for continued research in this area is robust visualization of whole metagenome shotgun sequencing data. This will involve both navigation of the feature taxonomy tree as well as exploration of specific genes for each bacterial feature. This will be a useful visualization as strain level analysis of metagenomic datasets will likely be essential for research and clinical applications. Also, functional annotations could be incorporated to explore associations with host health status. These features could be examined alongside metabolome data to inspect interactions and identify the associations between microbiome community abundances and host cellular processes.
DATA AVAILABILITY
The msd16s and etec16s datasets analyzed during this study are openly available as Bioconductor data packages available at [http://bioconductor.org/packages/release/data/experiment/html/msd16s.html] and [http://bioconductor.org/packages/release/data/experiment/html/etec16s.html], respectively. The scripts for analysis performed in this manuscript and for generating Figure 5 are available at [https://github.com/jmwagner/MetavizManuscriptScripts]. Metaviz code, documentation and tutorials are available through [http://www.metaviz.org]. Metaviz docker scripts are available at [https://github.com/epiviz/metaviz-docker]. Metavizr is available for download through Bioconductor with the project page at [http://bioconductor.org/packages/3.5/bioc/html/metavizr.html].
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Todd Treangen and Mihai Pop for providing feedback on the Metaviz application; and O. Colin Stine for helpful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was partially supported by National Institutes of Health (NIH) [RO1GM114267 to J.W., J.K., H.C.B., in part; U54DK102556 to J.W., V.F., A.M., H.C.B.]; US National Science Foundation Graduate Research Fellowship [DGE0750616 to J.N.P.]. Funding for open access charge: NIH [RO1GM114267].
Conflict of interest statement. None declared.
REFERENCES
- 1. Oh J., Byrd A.L., Deming C., Conlan S., Kong H.H., Segre J.A., Barnabas B., Blakesley R., Bouffard G., Brooks S. et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014; 514:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A. et al. Structure and function of the global ocean microbiome. Science. 2015; 348:1261359. [DOI] [PubMed] [Google Scholar]
- 3. Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C.. Uncovering Earth's virome. Nature. 2016; 536:425–430. [DOI] [PubMed] [Google Scholar]
- 4. Chelaru F., Smith L., Goldstein N., Bravo H.C.. Epiviz: interactive visual analytics for functional genomics data. Nat. Methods. 2014; 11:938–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pop M., Walker A.W., Paulson J., Lindsay B., Antonio M., Hossain M.A., Oundo J., Tamboura B., Mai V., Astrovskaya I. et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014; 15:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paulson J.N., Stine O.C., Bravo H.C., Pop M.. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013; 10:1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flygare S., Simmon K., Miller C., Qiao Y., Kennedy B., Di Sera T., Graf E.H., Tardif K.D., Kapusta A., Rynearson S. et al. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016; 17:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bostock M., Ogievetsky V., Heer J.. D³: Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011; 17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 10. Pedersen T.L., Nookaew I., Wayne Ussery D., Månsson M.. PanViz: interactive visualization of the structure of functionally annotated pangenomes. Bioinformatics. 2017; 33:1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ondov B.D., Bergman N.H., Phillippy A.M.. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011; 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huse S.M., Welch D.B.M., Voorhis A., Shipunova A., Morrison H.G., Eren A.M., Sogin M.L.. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics. 2014; 15:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eren A.M., Esen Ö.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L., Delmont T.O.. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015; 3:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliveira F.S., Brestelli J., Cade S., Zheng J., Iodice J., Fischer S., Aurrecoechea C., Kissinger J.C., Brunk B.P., Stoeckert C.J. Jr et al. MicrobiomeDB: a systems biology platform for integrating, mining and analyzing microbiome experiments. Nucleic Acids Res. 2017; 46:D684–D691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chelaru F., Bravo H.C.. Epiviz: a view inside the design of an integrated visual analysis software for genomics. BMC Bioinformatics. 2015; 16(Suppl. 11):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dachselt R., Frisch M., Weiland M.. Czerwinski M, Lund A, Tan D. FacetZoom: a continuous multi-scale widget for navigating hierarchical metadata. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. 2008; NY: CHI ’08. ACM; 1353–1356. [Google Scholar]
- 17. Pop M., Paulson J.N., Chakraborty S., Astrovskaya I., Lindsay B.R., Li S., Bravo H.C., Harro C., Parkhill J., Walker A.W. et al. Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genomics. 2016; 17:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C.. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Meth. 2012; 9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pasolli E., Schiffer L., Manghi P., Renson A., Obenchain V., Truong D.T., Beghini F., Malik F., Ramos M., Dowd J.B. et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods. 2017; 14:1023–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mondot S., Lepage P., Seksik P., Allez M., Tréton X., Bouhnik Y., Colombel J.F., Leclerc M., Pochart P., Doré J. et al. Structural robustness of the gut mucosal microbiota is associated with Crohn's disease remission after surgery. Gut. 2016; 65:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sohn S.-H., Kim N., Jo H.J., Kim J., Park J.H., Nam R.H., Seok Y.-J., Kim Y.-R., Lee D.H.. Analysis of gastric body microbiota by pyrosequencing: possible role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. J. Cancer Prev. 2017; 22:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fenollar F., Minodier P., Boutin A., Laporte R., Brémond V., Noël G., Miramont S., Richet H., Benkouiten S., Lagier J.C. et al. Tropheryma whipplei associated with diarrhoea in young children. Clin. Microbiol. Infect. 2016; 22:869–874. [DOI] [PubMed] [Google Scholar]
- 23. Keller P.M., Rampini S.K., Büchler A.C., Eich G., Wanner R.M., Speck R.F., Böttger E.C., Bloemberg G.V.. Recognition of potentially novel human disease-associated pathogens by implementation of systematic 16S rRNA gene sequencing in the diagnostic laboratory. J. Clin. Microbiol. 2010; 48:3397–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The msd16s and etec16s datasets analyzed during this study are openly available as Bioconductor data packages available at [http://bioconductor.org/packages/release/data/experiment/html/msd16s.html] and [http://bioconductor.org/packages/release/data/experiment/html/etec16s.html], respectively. The scripts for analysis performed in this manuscript and for generating Figure 5 are available at [https://github.com/jmwagner/MetavizManuscriptScripts]. Metaviz code, documentation and tutorials are available through [http://www.metaviz.org]. Metaviz docker scripts are available at [https://github.com/epiviz/metaviz-docker]. Metavizr is available for download through Bioconductor with the project page at [http://bioconductor.org/packages/3.5/bioc/html/metavizr.html].