Abstract
Teichoic acids (TAs) are key components of the Gram‐positive bacterial cell wall that are composed of alditol phosphate repeating units, decorated with alanine or carbohydrate appendages. Because of their microhetereogeneity, pure well‐defined TAs for biological or immunological evaluation cannot be obtained from natural sources. We present here a streamlined automated solid‐phase synthesis approach for the rapid generation of well‐defined glycosylated, glycerol‐based TA oligomers. Building on the use of a “universal” linker system and fluorous tag purification strategy, a library of glycerolphosphate pentadecamers, decorated with various carbohydrate appendages, is generated. These are used to create a structurally diverse TA‐microarray, which is used to reveal, for the first time, the binding preferences of anti‐LTA (lipoteichoic acids) antibodies at the molecular level.
Keywords: automated synthesis, glycerol phosphate, Gram-positive bacteria, microarrays, oligomers
Teichoic acids (TAs) are a diverse class of biopolymers composed of repeating alditol phosphates and carbohydrates. They are major constituents of the cell wall of most Gram‐positive bacteria and can amount up to 70 % of the dry weight of the cell wall. Two classes of TAs exist: wall teichoic acids (WTAs) that are covalently attached to the peptidoglycan, and lipoteichoic acids (LTAs) that are anchored in the cell membrane through a glycolipid. TAs are involved in determining cell shape, cell division and nutrient uptake.1 They are key virulence factors and of importance to biofilm formation and antibiotic resistance.2 TAs are targeted by the complement system (lectin ligands)3, 4 and are recognized by opsonophagocytic antibodies.5 The most common structural TA‐motif is a polymeric glycerol phosphate backbone decorated with various substituents, including d‐alanine esters and carbohydrate appendages. The TAs from the opportunistic pathogens Enterococcus faecalis, Enterococcus faecium and Staphylococcus aureus contain α‐kojibiosyl (d‐glucose‐(α‐1,2)‐glucosyl),6 α‐glucosyl5 or N‐acetyl α‐glucosamine residues,7 respectively. Positively charged glucosamine groups are also found in certain LTA types.8 Insight into how TAs interact with enzymes, receptors and antibodies may guide new antibiotic and vaccine developments and well‐defined TA fragments are indispensable tools to fuel these studies.9, 10 Because of the microheterogeneity of naturally sourced TAs, organic synthesis is the method of choice to access these compounds. We, and others, have previously reported different synthetic strategies to get access to well‐defined TA structures,11, 12, 13 and from our initial solid phase assembly campaign, in which we generated a small set of six TA‐fragments, we identified a glycerol phosphate hexamer bearing a single glucosyl substituent as a lead synthetic antigen.10 To be able to generate larger TA oligomers with a more diverse substitution pattern, we here report the development of a second generation automated solid phase approach to generate a library of TA oligomers, featuring different carbohydrate appendages at various positions on the TA chain. The method builds on the use of a “universal linker” system and the implementation of a fluorous‐tagging strategy.14 The generated TA fragments have been used to create a TA‐microarray to characterize the binding specificity of a monoclonal antibody against Staphylococcus epidermidis LTA as well as serum raised against isolated LTA from E. faecalis and serum raised against a well‐defined TA‐BSA conjugate vaccine. These studies reveal, for the first time, the recognition patterns for anti‐LTA antibodies at the molecular level, and indicate how Ab‐TA binding specificity (or lack thereof) depends on the vaccination strategy used.
Our synthetic strategy, the targeted TA fragments and the used building blocks are depicted in Scheme 1. We aimed at a library, comprising glycerol phosphates with a backbone of 15 monomers in length (Scheme 1 A) and containing either an α‐glucosyl, an α‐glucosaminyl or an α‐N‐acetyl glucosaminyl at the C‐2 of a glycerol moiety at the beginning, middle and/or end of the pentadecamers. TA fragments with a cluster of carbohydrate substituents were generated as well. In all, 16 structures (1–16) were pursued as depicted in Scheme 1 A. To assemble these TA fragments the required glycerol phosphoramidite building blocks 17–20 (Scheme 1 B) were synthesized as described previously.11 In addition we synthesised the fluorous aminospacer phosphoramidite 21[ 15] (See the Supporting Information) to give all compounds with a purification handle and, after deprotection, a ligation handle. The (commercially available) solid support used for the generation of the TA‐library bears a 3‐aminopropane‐1,2‐diol linker that is attached to the resin through a urea linkage (22, Scheme 1 C).16 This linker obviates the need for tailor‐made “pre‐loaded” resins, which require the assembly of additional building blocks. The synthesis of non‐substituted glycerol phosphate 15‐mer 1 commenced with acid‐mediated removal of the DMT‐group in 22 followed by 5‐(benzylthio)‐1‐H‐tetrazole (BTT)‐mediated condensation of phosphoramidite 17. Oxidation of the intermediate phosphite to the phosphotriester and capping of the unreacted alcohols completed the first coupling cycle. Fourteen repetitions of this cycle delivered the resin bound pentadecamer, of which the DMT group was removed by a final acid treatment. After installation of the fluorous linker, treatment with ammonia (2 m in methanol) released the pentadecamer from the resin. Prolonged treatment with aqueous ammonia then removed all remaining cyanoethyl groups from the phosphotriesters. Purification of the crude 15‐mer was readily accomplished using conventional reversed phase HPLC. Though we anticipated the need for a fluorous stationary phase to separate the target compound from the deletion sequences, the C8F17‐tail proved to be lipophilic enough to provide a similar result with a regular C18 column.
Scheme 1.
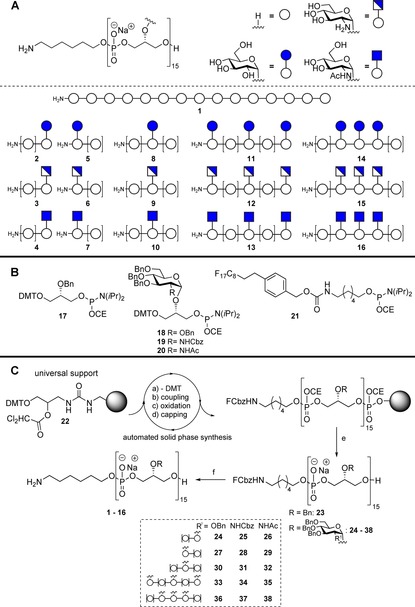
A) Target TA fragments. B) Building blocks used. C) Solid phase TA synthesis. Reagents and conditions: a) 3 % DCA, toluene; b) 17, 18, 19 or 20, 5‐(benzylthio)‐1H‐tetrazole, acetonitrile; c) I2, pyridine, H2O, acetonitrile; d) Ac2O, N‐methylimidazole, 2,6‐lutidine, acetonitrile; e) NH3, MeOH; then NH4OH, H2O. 23: 17 %, 24: 17 %, 25: 14 %,26: 7 %, 27: 10 %, 28: 15 %, 29: 8 %, 30: 16 %, 31: 16 %, 32: 7 %, 33: 5 %, 34 6 %, 35: 5 %, 36: 6 %, 37: 8 %, 38: 7 %. f) H2, Pd0, H2O. 1: 66 %, 2: 60 %, 3: 86 %, 4: 41 %, 5: 83 %, 6: 88 %, 7: 36 %, 8: 51 %, 9: 79 %, 10: 39 %, 11: 79 %, 12: 81 %, 13: 84 %, 14: 63 %, 15: 64 %, 16: 84 %.
Partially protected pentadecamer 23 was obtained in 17 % overall yield, based on resin loading. The synthesis of pentadecamer TAs 24–32, decorated with a single monosaccharide at three different positions in the glycerol phosphate backbone was undertaken next. After coupling glycosylated phosphoramidites 18, 19 or 20 to the resin, fourteen benzyl‐substituted glycerol phosphate units were installed and the constructs were capped with fluorous phosphoramidite 21. The isolated yields for glucose decorated 24 and N‐Cbz‐glucosamine decorated 25 (17 and 14 % respectively) were in line with the yield for undecorated oligomer 23, the N‐acetyl‐glucosamine decorated 26 was obtained in 7 % overall yield. In parallel, pentadecamers 27–29, bearing the carbohydrate appendage at the side of the spacer, were synthesized in yields ranging from 8 to 15 %. To complete the first series of compounds, the glucose‐ and glucosamine‐decorated pentadecamers, having the substituent in the middle of the glycerol phosphate chain (30, 31 and 32), were assembled (yields ranging from 7–16 %). Using the same protocol, fluorous and partially protected GTA‐fragments 33–38, bearing three monosaccharides, clustered or spread over the pentadecamer backbone, were prepared. The installation of the three carbohydrate substituted glycerol phosphate moieties proceeded with comparable efficiency as coupling of the non‐substituted glycerol phosphates (as judged from DMT count on the synthesizer) and the TA fragments bearing three carbohydrates were isolated in 5–8 %. Finally, all protecting groups (benzyl, Cbz and the fluorous Cbz) of the obtained homogeneous, partially protected and fluorous tagged pentadecamers 23–38 were removed by Pd‐catalyzed hydrogenolysis to provide the pure TA fragments (1–16) in 50–90 % yield.
Next, with the library of glycosylated TA‐pentadecamers in hand we generated TA‐microarrays to probe the interaction with anti‐LTA antibodies.17 Besides the TA library, generated here, we printed several TA‐fragments that were previously synthesized. These include a series of non‐substituted TA‐fragments of 6, 10, 14 and 20 glycerol phosphate units in length (39–42, Figure 1 A),18 TA‐hexamer 43 and kojibiosyl‐TA hexamer 44. We have previously shown that TA‐hexamer 43 can be used as a synthetic antigen in a TA‐conjugate vaccine, which can offer protection against E. faecalis in both active and passive immunization strategies.10 Notably, while kojibiose has been identified as appendage in E. faecalis TA, hexamer 44 proved to be an ineffective synthetic antigen.
Figure 1.
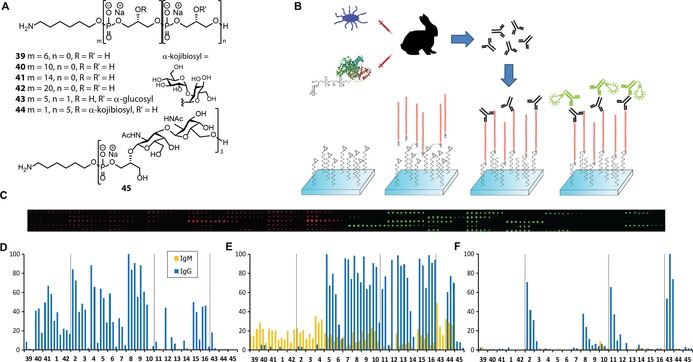
TA‐microarrays. A) Previously synthesized TA‐fragments. B) Schematic representation of teichoic acid microarray set up and sera evaluation. C) Exemplary fluorescence readout of slides after incubation with serum. D) IgM and IgG levels in the commercially available mouse anti‐ S. epidermidis monoclonal antibody (IBT Bioservices). E) IgM and IgG levels in serum obtained from rabbits immunized with LTA isolated from E. faecalis strain 12030.22 F) IgM and IgG levels in rabbit serum raised against a BSA‐43 conjugate.10 Data are represented at three concentrations (30 μm, 10 μm and 3 μm respectively, each spotted in triplicate) and are normalized to the highest intensity.
In the array, we also included E. faecium WTA fragment 45 that features a di‐N‐acetyl galactosamine‐glycerolphosphate backbone,19 as a negative control. All TA‐fragments were printed in varying concentrations (3, 10 and 30 μm) on epoxide functionalized microarray glass slides.20 With the generated microarrays, we probed the binding of a commercially available (IBT Bioservices) mouse anti S. epidermidis monoclonal antibody (used at a 1:6000 dilution), serum obtained from rabbits immunized with LTA isolated from E. faecalis strain 1203021 (used at a 1:500 dilution), and rabbit serum raised against the previously reported BSA‐TA 43 conjugate10 (used at a 1:500 dilution) (Figure 1 B). The fluorescence intensities (Figure 1 C) were quantified and are graphed in Figures 1 D, E and F. Figure 1 D depicts the interaction of the monoclonal antibodies against S. epidermidis LTA with the immobilized TAs. It becomes immediately apparent that various TA stretches are recognized. There is no selective recognition for either of the carbohydrate appendages and it seems that the monoclonal antibody mainly recognizes the glycerol phosphate backbone. The appendage of multiple carbohydrate substituents (fragments 11–16) or larger carbohydrates (as in kojibiosyl TA 44) seems to perturb binding of the antibody. The fact that undecorated or minimally decorated TA stretches are the main interaction partners for the monoclonal antibody, accounts for the fact that this antibody is cross reactive to various Gram‐positive bacteria, which share the glycerolphosphate backbone as a common epitope.5, 9b We next analyzed the serum, obtained from rabbits immunized with LTA isolated from E. faecalis strain 12030. Here, large differences are observed in the binding of IgM and IgG antibodies. IgM antibodies in the serum bound both the glycosylated and non‐glycosylated TAs, again indicating the glycerolphosphate backbone as the prime recognition target. Surprisingly, the IgG antibodies only bound glycosylated fragments. No binding of IgG antibodies to the “terminally“ glycosylated fragments 2–4 and 43 was detected, rather a preference for internally functionalized fragments (as present in TA fragments 8–16) was observed. This lack of selective recognition of one glycosylated TA over another, can explain the cross‐reactivity of serum, raised against E. faecalis 12030, with respect to other Gram‐positive bacteria such as S. aureus and E. faecium. Previously, the cross‐reactivity of the serum was attributed to recognition of the non‐decorated glycerolphosphate backbone by IgG antibodies, which clearly is not observed for the IgG antibodies probed with these arrays.22 Lastly, we investigated the serum raised against a BSA–TA 43 conjugate (Figure 1 F). This microarray shows that the serum mainly contains TA‐recognizing antibodies of the IgG‐type, and only very low levels of IgM antibodies, confirming the effectiveness of the used vaccine modality. Besides conjugate‐component TA 43, also TA 2, essentially an elongated analogue of TA 43, and compounds 8 and 11, harboring the full structure of 43 embedded in or attached to extra glycerol phosphate residues, are recognized, showing the high specificity of the serum for TA stretches with a terminal glucose appendage. The selective recognition observed with this array highlights the power of the analytical potential of the here presented arrays and shows, for the first time, the binding specificity of antibodies at the molecular level. In addition, they indicate that it is possible to generate antibodies against a well‐defined TA epitope, that do not (or only minimally) cross react with other (non‐substituted) TA‐fragments. Where it could be expected that the glycerolphosphate backbone represents the immunodominant epitope in LTA, it is clear that TA sequence specific antibodies can be raised if the appropriate conjugate is used for vaccination.
In summary, an efficient automated solid phase synthesis for the rapid generation of a library of TA oligomers, decorated with a diverse pattern of carbohydrate substituents, is described. The methodology allows for the generation of long glycerol phosphate TA stretches, with carbohydrate appendages positioned at will along on the backbone. The assembled TAs were used to generate TA‐microarrays, which have been employed to evaluate the binding preferences of monoclonal antibodies, as well as antibodies in serum, raised against different TA preparations. The arrays have revealed the difference in binding selectivity (or lack thereof) of antibodies, of varying origin. The arrays have shown that very selective antibodies can be generated to recognize specific and well‐defined TA epitopes, with minimal binding to the common glycerol phosphate backbone. This in turn implies that it should be possible to generate antibodies that selectively recognize bacterium specific TA stretches for vaccination strategies targeting a specific Gram‐positive bacterium. Clearly, the use of TAs isolated from bacterial sources for vaccine purposes can only lead to non‐selective serum. The arrays will be valuable tools to probe other biomolecules, such as host immune system lectins or bacteriophage proteins, for binding and they can be used to investigate enzymes involved in the biosynthesis and diversification of TAs.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank the Netherlands Organization for Scientific Research (NWO, ECHO, grant number 711.011.014) and the EU (Marie‐Curie ITN GLYCOVAX, grant number 675671) for financial support.
D. van der Es, F. Berni, W. F. J. Hogendorf, N. Meeuwenoord, D. Laverde, A. van Diepen, H. S. Overkleeft, D. V. Filippov, C. H. Hokke, J. Huebner, G. A. van der Marel, J. D. C. Codée, Chem. Eur. J. 2018, 24, 4014.
References
- 1.
- 1a. Neuhaus F. C., Baddiley J., Microbiol. Mol. Biol. Rev. 2003, 67, 686–723; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Brown S., Santa Maria J. P., Walker S., Annu. Rev. Microbiol. 2013, 67, 313–336; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Percy M. G., Gründling A., Annu. Rev. Microbiol. 2014, 68, 81–100; [DOI] [PubMed] [Google Scholar]
- 1d. Schneewind O., Missiakas D., J. Bacteriol. 2014, 196, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Weidenmaier C., Peschel A., Nat. Rev. Microbiol. 2008, 6, 276–287; [DOI] [PubMed] [Google Scholar]
- 2b. Rockel C., Hartung T., Front. Pharmacol. 2012, 3, 1–19; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Lynch N. J., Roscher S., Hartung T., Morath S., Matsushita M., Maennel D. N., Kuraya M., Fujita T., Schwaeble W. J., J. Immunol. 2004, 172, 1198–1202. [DOI] [PubMed] [Google Scholar]
- 3. Kurokawa K., Takahashi K., Lee B. L., Immunobiology 2016, 221, 1091–1101. [DOI] [PubMed] [Google Scholar]
- 4. Jung D.-J., An J.-H., Kurokawa K., Jung Y.-C., Kim M.-J., Aoyagi Y., Matsushita M., Takahashi S., Lee H.-S., Takahashi K., Lee B. L., J. Immunol. 2012, 189, 4951–4959. [DOI] [PubMed] [Google Scholar]
- 5. Kodali S., Vinogradov E., Lin F., Khoury N., Hao L., Pavliak V., Jones C. H., Laverde D., Huebner J., Jansen K. U., Anderson A. S., Donald R. G. K., J. Biol. Chem. 2015, 290, 19512–19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huebner J., Wang Y., Krueger W. A., Madoff L. C., Marrirosian G., Boisot S., Goldmann D. A., Kasper D. L., Tzianabos A. O., Pier G. B., Infect. Immun. 1999, 67, 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanderson A. R., Strominger J. L., Nathenson S. G., Sanderson R., J. Biol. Chem. 1962, 237, 3603–3613. [PubMed] [Google Scholar]
- 8. Cot M., Ray A., Gilleron M., Vercellone A., Larrouy-Maumus G., Armau E., Gauthier S., Tiraby G., Puzo G., Nigou J., PLoS One 2011, 6, e26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Martin C. E., Broecker F., Eller S., Oberli M. A., Anish C., Pereira C. L., Seeberger P. H., Chem. Commun. 2013, 49, 7159–7161; [DOI] [PubMed] [Google Scholar]
- 9b. Chen Q., Dintaman J., Lees A., Sen G., Schwartz D., Shirtliff M. E., Park S., Lee J. C., Mond J. J., Snapper C. M., Infect. Immun. 2013, 81, 2554–2561; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Zhou Z., Ding W., Li C., Wu Z., J. Carbohydr. Chem. 2017, 36, 205–219. [Google Scholar]
- 10. Laverde D., Wobser D., Romero-Saavedra F., Hogendorf W., van der Marel G., Berthold M., Kropec A., Codee J., Huebner J., PLoS One 2014, 9, e110953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. Hogendorf W. F. J., Kropec A., Filippov D. V., Overkleeft H. S., Huebner J., van der Marel G. A., Codée J. D. C., Carbohydr. Res. 2012, 356, 142–151; [DOI] [PubMed] [Google Scholar]
- 11b. Hogendorf W. F. J., Lameijer L. N., Beenakker T. J. M., Overkleeft H. S., Filippov D. V., Codée J. D. C., Van der Marel G. A., Org. Lett. 2012, 14, 848–851. [DOI] [PubMed] [Google Scholar]
- 12.
- 12a. Hogendorf W. F. J., Meeuwenoord N., Overkleeft H. S., Filippov D. V., Laverde D., Kropec A., Huebner J., Van der Marel G. A., Codée J. D. C., Chem. Commun. 2011, 47, 8961–8963; [DOI] [PubMed] [Google Scholar]
- 12b.For the first synthesis of a TA-fragment see: van Boeckel C. A. A., Visser G. M., Hermans J. P. G., van Boom J. H., Tetrahedron Lett. 1981, 22, 4743–4746; [Google Scholar]
- 12c.For the first automated synthesis of a TA-fragment see: Westerduin P., Veeneman G. H., Pennings Y., van der Marel G. A., van Boom J. H., Tetrahedron Lett. 1987, 28, 1557–1560; [Google Scholar]
- 12d.For the first synthesis of a LTA-fragment see: Fukase K., Matsumoto T., Ito N., Yoshimura T., Kotani S., Kusumoto S., Bull. Chem. Soc. Jpn. 1992, 65, 2643–2654; [Google Scholar]
- 12e.For a pivotal example of the synthesis of a carbohydrate-containing TA-fragment see: Pedersen C. M., Figueroa-Perez I., Lindner B., Ulmer A. J., Zähringer U., Schmidt R. R., Angew. Chem. Int. Ed. 2010, 49, 2585–2590; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 2639–2644. [Google Scholar]
- 13.For a review on all current synthetic methods towards teichoic acids see: van der Es D., Hogendorf W. F. J., Overkleeft H. S., van der Marel G. A., Codée J. D. C., Chem. Soc. Rev. 2017, 46, 1464–1482. [DOI] [PubMed] [Google Scholar]
- 14. Zhang W., Chem. Rev. 2009, 109, 749–795. [DOI] [PubMed] [Google Scholar]
- 15. Filippov D. V., van Zoelen D. J., Oldfield S. P., van der Marel G. A., Overkleeft H. S., Drijfhout J. W., van Boom J. H., Tetrahedron Lett. 2002, 43, 7809–7812. [Google Scholar]
- 16. Yagodkin A., Weisel J., Azhayev A., Nucleosides, Nucleosides Nucleotides Nucleic Acids 2011, 30, 475–489. [DOI] [PubMed] [Google Scholar]
- 17. Park S., Gildersleeve J. C., Blixt O., Shin I., Chem. Soc. Rev. 2013, 42, 4310–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Theilacker C., Kropec A., Hammer F., Sava I., Wobser D., Sakinc T., Codée J. D. C., Hogendorf W. F. J., van der Marel G. A., Huebner J., J. Infect. Dis. 2012, 205, 1076–1085. [DOI] [PubMed] [Google Scholar]
- 19. van der Es D., Groenia N. A., Laverde D., Overkleeft H. S., Huebner J., van der Marel G. A., Codée J. D. C., Bioorg. Med. Chem. 2016, 24, 3893–3907. [DOI] [PubMed] [Google Scholar]
- 20.For the used protocol see: van Diepen A., Smit C. H., van Egmond L., Kabatereine N. B., Pinot de Moira A., Dunne D. W., Hokke C. H., PLoS Neglected Trop. Dis. 2012, 6, e1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huebner J., Quaas A., Krueger W. A., Goldmann D. A., Pier G. B., Infect. Immun. 2000, 68, 4631–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The non-selective binding of the IgM antibodies may also contribute to the cross reactivity of the serum.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


