Abstract
The most abundant protein of resting rhizomes of Calystegia sepium (L.) R.Br. (hedge bindweed) has been isolated and its corresponding cDNA cloned. The native protein consists of a single polypeptide of 212 amino acid residues and occurs as a mixture of glycosylated and unglycosylated isoforms. Both forms are derived from the same preproprotein containing a signal peptide and a C-terminal propeptide. Analysis of the deduced amino acid sequence indicated that the C. sepium protein shows high sequence identity and structural similarity with plant RNases. However, no RNase activity could be detected in highly purified preparations of the protein. This apparent lack of activity results most probably from the replacement of a conserved His residue, which is essential for the catalytic activity of plant RNases. Our findings not only demonstrate the occurrence of a catalytically inactive variant of an S-like RNase, but also provide further evidence that genes encoding storage proteins may have evolved from genes encoding enzymes or other biologically active proteins.
Many plants accumulate large quantities of presumed storage proteins in various vegetative storage organs. These proteins play a primary role in nitrogen storage and distribution, and make an important contribution to the survival of the plant in its natural environment (Staswick, 1994). According to currently accepted ideas, vegetative storage organs such as bulbs, tubers, corms, rhizomes, and bark act as sinks for soluble nitrogen compounds (mainly amino acids) generated from the leaf proteins when the plant enters a senescing phase. After transport through the phloem into the storage organs, the amino acids are incorporated into (storage) proteins in the storage parenchyma cells. These cells are capable of accumulating large quantities of proteins and store away a corresponding amount of nitrogen in a biologically harmless form. When the plant resumes growth after a resting or dormancy period, the vegetative storage organs become a source of nitrogen. Environmental and/or endogenous stimuli induce a regulated degradation of the storage proteins, resulting in a massive release of amino acids that are subsequently transported to the new shoots to satisfy the high nitrogen demand of the rapidly growing tissues. Since a rapid growth after a period of dormancy is often an absolute prerequisite for biannual or perennial plants to successfully compete for light and nutrients, the survival of these plants in their normal habitat is certainly favored by the ample availability of ready-to-use nitrogen compounds. It is evident, therefore, that vegetative storage proteins (VSPs), even in the absence of a biological activity, are essential for the plant.
Although VSPs have received less attention than their functional counterparts from seeds, the available data leave no doubt that they are widespread among higher plants and form a heterogeneous group of proteins. An extended list of storage proteins has been identified, indeed, in various typical vegetative storage tissues of plant species from different taxonomic groups. Classical examples are the tuber storage proteins from potato (Solanum tuberosum) and sweet potato (Ipomoea batatas), which are commonly known as patatin and sporamin, respectively (Mignery et al., 1984; Maeshima et al., 1985). VSPs have also been found in the bark of deciduous trees such as poplar (Populus deltoides) (Coleman et al., 1991), elderberry (Sambucus nigra) (Van Damme et al., 1997b), and several legume trees. Some of these bark proteins have been identified on the basis of their biological activity. For example, elderberry bark accumulates almost exclusively lectins and ribosome-inactivating proteins (Van Damme et al., 1997b).
The most abundant storage proteins in the bark of the legume trees Sophora japonica (Japanese pagoda tree), Cladrastis lutea (yellow wood), Robinia pseudoacacia (black locust), and Maackia amurensis are genuine lectins (Hankins et al., 1988; Van Damme et al., 1995a, 1995b, 1997a, 1997c). In addition to bark, storage-protein-like lectins have been identified in bulbs of Allium sativum (garlic) (Van Damme et al., 1992) and Allium ursinum (ramsons) (Van Damme et al., 1993), Tulipa sp. (tulip) (Van Damme et al., 1996b), Amaryllidaceae species such as snowdrop (Galanthus nivalis) and daffodils (Narcissus sp.) (Van Damme et al., 1988), and in rhizomes of ground elder (Aegopodium podagraria) (Peumans et al., 1985). VSPs are not confined to typical storage organs but are also found in non-storage tissues. The best-studied examples of this group are the so-called soybean VSPs called VSPα and VSPβ, which under certain conditions accumulate in large quantities in leaves, seed pods, and hypocotyls, when these tissues act as or are forced to act as a nitrogen sink (Staswick, 1989a, 1989b).
Many, but not all, VSPs exhibit a well-defined biological activity. For example, patatin is considered to be a lipid acyl hydrolase (Andrews et al., 1988), whereas sporamin belongs to the superfamily of trypsin-inhibitors (Yeh et al., 1997). As already mentioned above, several bark and bulb storage proteins are lectins or ribosome-inactivating proteins. Similarly, the soybean VSPs have a low acid phosphatase activity (De Wald et al., 1992). Other VSPs, such as the bark proteins from apple and poplar, exhibit no (known) enzymatic or other biological activity. For some of the biologically active VSPs it is believed that, through their carbohydrate-binding, ribosome-inactivating, or trypsin-inhibiting activity, they acquired in addition to their storage function a role in plant defense (Peumans and Van Damme, 1995; Yeh et al., 1997). To further corroborate this presumed defensive role it is important to search for novel types of VSPs.
In this report we describe the isolation and cloning of the major storage protein from rhizomes of Calystegia sepium (hedge bindweed). This protein closely resembles plant RNases with respect to its amino acid sequence and structure, but is completely devoid of RNase activity because one of the His residues which is essential for enzymatic activity is replaced by a Lys. Our work on the C. sepium RNase-related protein (CalsepRRP) not only demonstrates for the first time the occurrence of an enzymatically inactive S-like RNase homolog, but also enables the purification of large quantities of this protein for comparative biochemical and structural studies.
MATERIALS AND METHODS
Plant Material
Rhizomes of Calystegia sepium (L.) R.Br. (hedge bindweed) were collected in Leuven, Belgium in December and stored at −20°C. Whole rhizomes were used for the isolation of the RNase-related protein.
Isolation of CalsepRRP
CalsepRRP was isolated from resting rhizomes of C. sepium by classical protein purification techniques. Frozen rhizomes (200 g) were broken into small pieces, immersed in 10 volumes (v/w) of a solution of 0.1% (w/v) ascorbic acid (adjusted to pH 6.0), and homogenized in a blender. The homogenate was squeezed through a double layer of cheesecloth and centrifuged at 8,000g for 10 min. The supernatant was decanted, adjusted to pH 8.7 with 1 m NaOH, re-centrifuged at 8,000g for 10 min, and filtered through filter paper. Subsequently, the crude extract was applied onto a column (5 × 5 cm; 100-mL bed volume) of Q Fast Flow (Pharmacia, Uppsala) equilibrated with 20 mm Tris-HCl (pH 8.7). After loading the extract, the column was washed with 1 L of the same Tris buffer and eluted with 0.1 m Na-OAc (pH 5.0). This partially purified protein fraction was diluted with 4 volumes of distilled water, the pH raised to 8.7 with 1 m NaOH, and loaded on a second column (5 cm × 2.5 cm; 25-mL bed volume) of Q Fast Flow equilibrated with 20 mm Tris-HCl (pH 8.7).
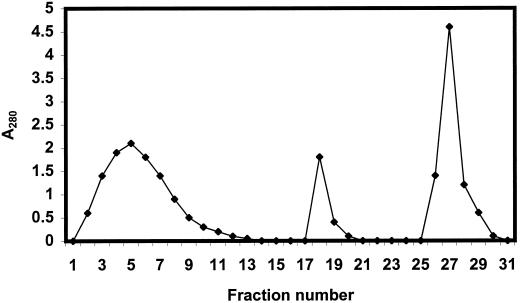
After washing the column until the A280 fell below 0.01, the bound proteins were eluted with 0.3 m NaCl in Tris buffer. The resulting concentrated protein solution (25 mL) was loaded onto a column (40 × 5 cm; 800-mL bed volume) of Sephacryl 100 equilibrated with phosphate buffered saline (PBS) (1.5 mm KH2PO4, 10 mm Na2HPO4, 3 mm KCl, and 140 mm NaCl, pH 7.4). Under these conditions, the proteins eluted in two well-resolved peaks with an apparent molecular mass around 200 and 30 kD, respectively. The proteins present in the second peak were dialyzed against Tris buffer and applied onto a small column (5 × 1.5 cm; bed volume) of Q Fast Flow equilibrated with Tris buffer. Immediately after loading, the column was eluted with 0.3 m NaCl in Tris buffer, yielding a small volume of concentrated protein. Five-milliliter aliquots of this concentrated protein solution were loaded onto a column (70 × 2.6 cm; 350-mL bed volume) of Sephacryl 100, and chromatographed using PBS as a running buffer. Peak fractions were pooled and used as a source of total CalsepRRP.
The total preparation of CalsepRRP was further subjected to affinity chromatography on immobilized concanavalin A (ConA) to separate the glycosylated and un-glycosylated isoforms, as described in the legend to Figure 2. The CalsepRRP fractions obtained by affinity chromatography on immobilized ConA were subsequently purified by ion-exchange chromatography using a FPLC system (Pharmacia). Samples containing 2 mg of protein were loaded on a Mono Q column (HR5/5, Pharmacia) equilibrated with Tris buffer. After washing the column with 4 mL of buffer, proteins were eluted with a linear gradient (56 mL) of increasing NaCl concentration (0–0.5 m in the same buffer) at a flow rate of 2 mL/min. Peak fractions were collected manually and used for further analyses.
Figure 2.
Affinity chromatography of CalsepRRP on ConA-Sepharose. Fifty milligrams of total CalsepRRP dissolved in 20 mL of PBS was applied onto a column (2.6 × 10 cm; 50-mL bed volume) of ConA-Sepharose 4B pre-equilibrated with PBS. After loading the proteins, the column was washed with PBS until the A280 fell below 0.01. Bound glycoproteins were desorbed by consecutive elution of the column with 0.2 m α-methylmannoside in PBS and 20 mm Tris-HCl (pH 10.0), respectively. The flow rate was 2 mL/min; the fraction size was 5 mL.
Analytical Methods
Proteins were analyzed by SDS-PAGE using 12.5% to 25% (w/v) acrylamide gradient gels as described by Laemmli (1970). Proteins separated by SDS-PAGE and electroblotted on an Immobilon P membrane were stained for carbohydrate using periodic acid Schiff's reagent. Total neutral sugar was determined by the phenol/H2SO4 method (Dubois et al., 1956) with d-Glc as standard. Polypeptides separated by SDS-PAGE and electroblotted on a PVDF membrane were sequenced on an protein sequencer (model 477A, Perkin-Elmer/Applied Biosystems, Foster City, CA) interfaced with an on-line analyzer (model 120A, Perkin-Elmer/Applied Biosystems).
Analytical gel filtration of the purified proteins was performed on a Superose 12 column (Pharmacia) using PBS as running buffer. Molecular mass reference markers were catalase (240 kD), aldolase (160 kD), bovine serum albumin (67 kD), ovalbumin (45 kD), chymotrypsinogen (25 kD), and cytochrome c (12.5 kD).
Electrospray spectra were obtained with a tandem quadruple mass spectrometer (Quattro-II, Micromass, Manchester, UK). The electrospray carrier solvent was water:acetonitrile (50:50, v/v), and was applied at a flow rate of 30 μL/min. The capillary voltage was 90 V, and the source temperature 80°C. Data were acquired in the multichannel mode by averaging five scans and scanning the mass range from 800 to 2,000 D at a rate of 4 s/scan. Data processing was performed with Masslynx software (Micromass, Manchester, UK).
RNase Activity
RNase activity was detected by an electrophoretic method. Total RNA (25S + 18S rRNA) from young elderberry (Sambucus nigra) leaves (isolated as described by Van Damme and Peumans, 1993) was used as a substrate. One microgram of RNA was incubated in 10 μL of 25 mm Tris-HCl (pH 7.4) containing 25 mm KCl and 5 mm MgCl2 in the presence of different concentrations of purified CalsepRRP at 30°C for 15 min. The reaction mixture was then analyzed in a 1.2% (w/v) agarose gel. Gels were stained with ethidium bromide (0.5 μg/mL) for 30 min, then destained in 0.5 m NH4Ac prior to photodocumentation using a short-wavelength UV lamp. A crude extract of tobacco (Nicotiana tabacum cv Samsun) styles was used as a positive control for plant RNase. The extract was prepared by grinding 0.1 g of styles in 1 mL of Tris buffer in a mortar and pestle, and centrifuging the homogenate at 13,000g for 10 min.
RNase activity was also assayed by the perchloric acid precipitation method described by Brown and Ho (1986) using yeast extract as a substrate. For this test an extract from tobacco styles was also used as a positive control.
RNA Isolation, Construction, and Screening of cDNA Library
Rhizomes destined for the isolation of RNA were collected in early October because at that time the plants accumulate storage carbohydrates and proteins in their underground storage organs. Apexes (top centimeter) were excised and stored at −80°C until use. Total cellular RNA was prepared from the apexes and poly(A+)-rich RNA was enriched by chromatography on oligo-deoxythymidine cellulose, as described by Van Damme and Peumans (1993). A cDNA library was constructed with poly(A+)-rich RNA using the cDNA synthesis kit from Pharmacia. cDNA fragments were inserted into the EcoRI site of PUC18 (Pharmacia). The library was propagated in Escherichia coli XL1 Blue (Stratagene, La Jolla, CA).
Recombinant lectin clones were screened using a 32P-end-labeled degenerate oligonucleotide probe (17-mer, 5′ AARTARTCRAAYTCYTTRTG 3′ derived from the amino acid sequence HKEFDYF of the N terminus of CalsepRRP). In a later stage, cDNA clones encoding CalsepRRP were used as probes to screen for more cDNA clones. Hybridizations were done overnight as reported previously (Van Damme et al., 1996a). Colonies that produced positive signals were selected and rescreened at low density using the same conditions. Plasmids were isolated from purified single colonies on a miniprep scale using the alkaline lysis method described by Mierendorf and Pfeffer (1987) and sequenced by the dideoxy method (Sanger et al., 1977). DNA sequences were analyzed using programs from PC Gene (Intelligenetics, Mountain View, CA) and Genepro (Riverside Scientific, Seattle).
Northern-Blot Analysis
RNA electrophoresis was performed according to the method of Maniatis et al. (1982). Approximately 3 μg of poly(A+)-rich RNA was denatured in glyoxal and dimethylsulfoxide and separated in a 1.2% (w/v) agarose gel. Following electrophoresis, the RNA was transferred to Immobilon N membranes (Millipore, Bedford, MA) and the blot was hybridized using a random-primer-labeled cDNA insert or an oligonucleotide probe. Hybridization was performed as reported by Van Damme et al. (1992). An RNA ladder (0.16–1.77 kb) was used as a marker.
DNA Isolation and Southern-Blot Analysis
DNA was extracted from young leaves of C. sepium using the protocol described by Stewart and Via (1993). The DNA preparation was treated with RNase to remove any contaminating RNA. Approximately 50 μg of DNA was digested with restriction endonucleases and subjected to electrophoresis in a 0.8% (w/v) agarose gel. DNA was transferred to Immobilon N membranes (Millipore) and hybridized using the 32P-labeled cDNA insert encoding CalsepRRP. Hybridization was carried out at 60°C, as described previously (Van Damme et al., 1992).
PCR Amplification of Genomic DNA Fragments Encoding CalsepRRP
The reaction mixture for amplification of genomic DNA sequences contained 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.5 mm MgCl2, 100 mg/L gelatin, 0.4 mm of each dNTP, 2.5 units of Taq polymerase (Boehringer Mannheim, Basel), 50 ng to 500 μg of genomic DNA, and 20 μL of the appropriate primer mixture (20 μm) in a 100-μL reaction volume. The reaction was overlaid with 80 μL of mineral oil. After denaturation of the DNA for 5 min at 95°C, amplification was performed for 30 cycles through a regime of 1-min template denaturation at 92°C, followed by 1-min primer annealing at 60°C and 3-min primer extension at 72°C using an automatic thermal cycler (model 480, Perkin-Elmer/Applied Biosystems). The PCR primers were derived from both ends of the coding sequence of the cDNA clones encoding CalsepRRP.
Molecular Modeling
The amino acid sequence alignments were carried out on a Macintosh 5400/180 using the program SeqVu (J. Gardner, 1995, The Garvan Institute of Medical Research, Sydney). Multiple amino acid sequence alignments based on Clustal W (Thompson et al., 1994) were carried out using SeqPup (D.G. Gilbert, Biology Department, Indiana University, Bloomington) and modified manually according to the hydrophobic cluster analysis (HCA) data. The HCA (Gaboriaud et al., 1987; Lemesle-Varloot et al., 1990) was performed to delineate the structurally conserved regions along the amino acid sequences of CalsepRRP and the fungal RNase Rh from Rhizopus niveus (Horiuchi et al., 1988) used as a model. HCA plots were generated using the program HCA-Plot2 (Doriane, Paris).
Molecular modeling of C. sepium RNase was performed on a Silicon Graphics O2 R5000 workstation using the programs InsightII, Homology, and Discover (Molecular Simulations, San Diego). The atomic coordinates of RNase Rh (PDB code 1BOL) were taken from the Brookhaven Protein Data Bank (Kurihara et al., 1996) and used to build the three-dimensional model of CalsepRRP. Steric conflicts resulting from the replacement or the deletion of some residues in CalsepRRP were corrected during the model-building procedure using the rotamer library (Ponder and Richards, 1987) and the search algorithm implemented in the Homology program (Mas et al., 1992) to maintain proper side chain orientation. Energy minimization and relaxation of the loop regions was carried out by several cycles of steepest descent and conjugate gradient using the cvff forcefield of Discover. PROCHECK (Laskowski et al., 1993) was used to check the stereochemical quality of the three-dimensional model. In this respect, 76.6% of the residues of the modeled C. sepium RNase (78% for 1BOL) occurred in the most energetically favorable regions of the Ramachandran plot. The program TurboFrodo (Bio-Graphics, Marseille, France) was run on the O2 workstation to draw the Ramachandran plots and to perform the superimposition of the models. Cartoons were rendered using Molscript (Kraulis, 1991).
RESULTS
Resting Rhizomes of C. sepium Contain Several Abundant Proteins
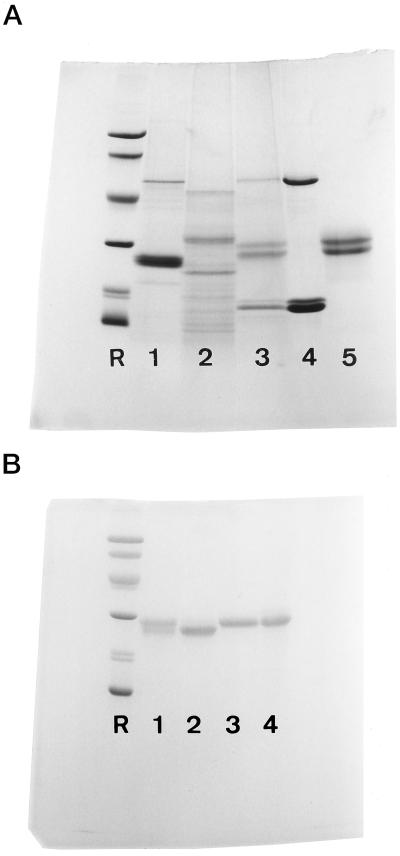
Crude extracts from resting C. sepium rhizomes contain four major polypeptides (Fig. 1A). Previous work revealed that the 15-kD polypeptide corresponds to the lectin subunit (Peumans et al., 1997), whereas the 45-kD polypeptide has tentatively been identified as a β-amylase based on its high sequence similarity to the β-amylase of sweet potato (W.J. Peumans, unpublished data). The two most abundant polypeptides of 26 and 28 kD, respectively, have not been identified yet. Since the abundance of both polypeptides and their molecular masses are reminiscent of those of the sporamins from sweet potato tubers, it seemed worthwhile to determine whether the underground storage organs of sweet potato and C. sepium accumulate the same or similar storage proteins. Therefore, the major rhizome proteins of C. sepium were isolated and characterized, and their corresponding cDNAs cloned. As is described below, these 26- to 28-kD proteins are not related to the sporamins but represent a novel class of plant storage proteins.
Figure 1.
A, SDS-PAGE of crude extracts from underground storage organs of different Convolvulaceae species. Samples were loaded as follows: 1, crude extract (100 μL) from sweet potato tubers; 2, crude extract (100 μL) from C. arvensis rhizomes; 3, crude extract (100 μL) from C. sepium rhizomes; 4, purified β-amylase and lectin (25 μg each) from C. sepium rhizomes; 5, purified CalsepRRP (25 μg). All samples were reduced with 2-mercaptoethanol. Molecular mass reference proteins (lane R) were lysozyme (14 kD), soybean trypsin inhibitor (20 kD), carbonic anhydrase (30 kD), ovalbumin (43 kD), bovine serum albumin (67 kD), and phosphorylase b (94 kD). B, SDS-PAGE of purified CalsepRRP. Lanes 1 and 2 were loaded with 25 μg of total CalsepRRP and unglycosylated CalsepRRP, respectively. Glycosylated samples (25 μg each) loaded in lanes 3 and 4 correspond to the fractions desorbed from the ConA column with 0.2 m α-methylpyranoside and Tris-HCl (pH 10), respectively. All samples were reduced with 2-mercaptoethanol. Molecular mass reference proteins are the same as in A.
Isolation and Characterization of CalsepRRP
CalsepRRP was purified using a combination of conventional protein purification techniques. Unreduced and reduced (with 2-mercaptoethanol) samples of the final preparation yielded two polypeptides of 26 and 28 kD, respectively, upon SDS-PAGE (Fig. 1A). The native protein eluted as a single symmetrical peak with an apparent molecular mass of about 30 kD upon gel filtration chromatography on a Superose 12 column (results not shown), indicating that it consists of a single polypeptide of 26 or 28 kD. N-terminal amino acid sequencing yielded exactly the same sequence (GHKEF DYFTL ALTWS GTELL) for both the 26- and 28-kD polypeptide, suggesting that CalsepRRP consists of two closely related isoforms. To determine whether the difference in molecular mass between the two polypeptides is due to differences in glycosylation, blotted CalsepRRP was stained for carbohydrate using periodic acid Schiff's reagent. The 28-kD polypeptide clearly contained carbohydrate, whereas the 26-kD polypeptide was apparently devoid of covalently bound sugars (results not shown). It is concluded, therefore, that the CalsepRRP polypeptides are only partly glycosylated.
The glycosylated and unglycosylated isoforms were separated by affinity chromatography on immobilized ConA (Fig. 2). About half of the total CalsepRRP was retained on the column. Of the bound proteins, only a small portion was desorbed with 0.2 m methylmannopyranoside. The remainder of the bound CalsepRRP eluted upon washing the column with Tris buffer at pH 10, under which conditions ConA is reversibly inactivated and therefore releases all bound glycoproteins. SDS-PAGE confirmed that the unbound CalsepRRP fraction consists mainly of unglycosylated 26-kD subunits, whereas the fractions desorbed with both methylmannopyranoside and Tris, pH 10, consist exclusively of glycosylated 28-kD subunits (Fig. 1B). Mass spectrometry of the unglycosylated and glycosylated CalsepRRP polypeptides yielded a value of 23,380 D and approximately 25,000 D, respectively.
Determination of the carbohydrate content of both affinity-purified CalsepRRP fractions yielded a value of 4.1% and 4.3% (by mass), respectively. Assuming a molecular mass of 170 D per monosaccharide, the number of sugar residues amounts to about seven per polypeptide. Taking into consideration that typical N-linked plant glycans consist of six to eight monosaccharide residues, we assume that the 28-kD CalsepRRP contains a single oligosaccharide side chain. The occurrence of a single N-glycan is also in good agreement with the difference in molecular mass between the unglycosylated and glycosylated forms (as determined by mass spectrometry).
Molecular Cloning of CalsepRRP
A search of the database indicated that the N-terminal sequence of CalsepRRP exhibits sequence similarity to the N terminus of plant RNases. To check whether the sequence similarity between the rhizome protein and plant RNases extends beyond the N terminus, cDNAs encoding CalsepRRP were isolated and sequenced. Screening of a cDNA library constructed with poly(A+)-rich RNA from rhizome apexes using a synthetic oligonucleotide derived from the amino acid sequence of the CalsepRRP yielded multiple positive clones of approximately 1 kb. Sequence analysis of CalsepRRP1 revealed that this clone contains an open reading frame of 777 bp encoding a 259-amino acid precursor with one putative initiation codon at position 7 of the deduced amino acid sequence (Fig. 3A). Translation starting with this Met residue results in a protein of 253 amino acids with a calculated molecular mass of 27,766 D. A comparison of the deduced amino acid sequence of CalsepRRP1 and the N-terminal sequence of the protein revealed a perfect match between residues G29–L48 except for the Cys at position 47. Sequence analysis of multiple CalsepRRP cDNAs revealed minor differences in the deduced amino acid sequences. However, the overall sequence similarity between the different cDNA clones ranges from 95% to 99% at the deduced amino acid level.
Figure 3.
A, Deduced amino acid sequence of the RNase-related potein from C. sepium. Since the Met at position 7 is probably the first amino acid, the residues preceding this Met are shown in lowercase. The sequence corresponding to the N-terminal sequence of the protein is underlined. Putative N-glycosylation sites are indicated in bold. The arrowheads indicate the positions of the intron sequences. B, Sequence comparison of RNase Rh from R. niveus and CalsepRRP. Deletions (gaps) have been introduced to maximize the homology, and identical residues are boxed.
According to the rules for protein processing of Von Heijne (1986), a signal peptide can be cleaved between amino acids 28 and 29 of the CalsepRRP precursor, resulting in a polypeptide of 24,892 D (225 amino acids) with an pI of 4.42. Since the mature (unglycosylated) polypeptide has a molecular mass of only 23,380 D, we assume that the CalsepRRP precursor is also processed at the C terminus by the removal of a propeptide of 13 residues. Due to the C-terminal processing, mature CalsepRRP contains only one putative glycosylation site at position 131 (Asn-Ile-Ser). The fact that only half of the total CalsepRRP is glycosylated indicates that even the accessible site is only partly glycosylated.
Northern-Blot Analysis
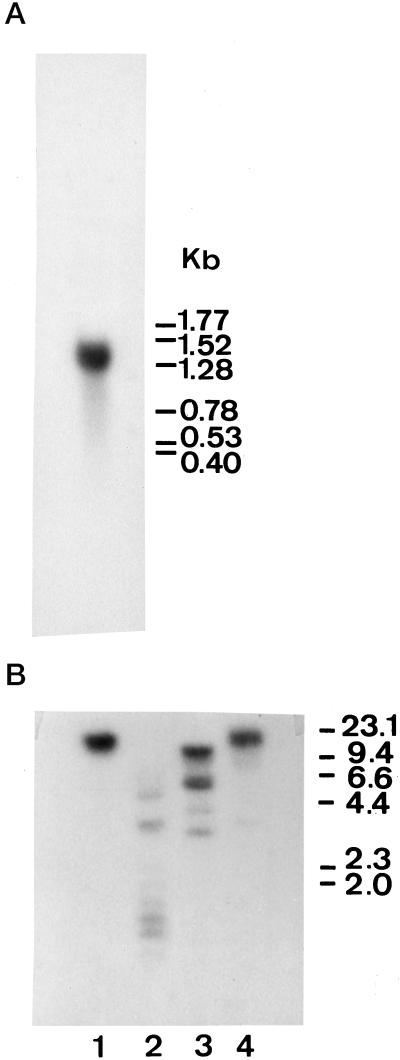
Northern-blot analysis was performed to determine the total length of the mRNA encoding CalsepRRP. Hybridization of the blot using the synthetic oligonucleotide as a probe yielded one band of approximately 1.3 kb (Fig. 4A). This result was identical when hybridization was performed using the random-primer-labeled cDNA clone and was consistent with the length of the cDNA clones analyzed.
Figure 4.
A, Northern blot of poly(A+)-rich RNA isolated from C. sepium rhizomes. The blot was hybridized using the labeled cDNA insert CalsepRRP1. Numbers on the right show RNA size (kb). B, Southern-blot analysis of genomic DNA isolated from young leaves of C. sepium. DNA was digested with BamHI (lane 1), EcoRI (lane 2), HindIII (lane 3), and PstI (lane 4), and hybridized using the labeled cDNA insert CalsepRRP1. The DNA sizes are indicated on the right.
Southern-Blot Analysis
Genomic DNA isolated from young leaves of C. sepium was digested with the restriction enzymes BamHI, EcoRI, HindIII, and PstI, and analyzed by gel electrophoresis. As shown in Figure 4B, hybridization of the blot with the labeled cDNA CalsepRRP1 revealed only one or a few bands. Since of all the restriction enzymes used, only HindIII is known to have one cleavage site in the coding sequence of the mature protein, the results of the Southern-blot analysis show that the RNase-related protein is encoded by one or a few related genes.
Analysis of Genomic Fragments Encoding CalsepRRP
PCR amplification of genomic DNA fragments encoding CalsepRRP yielded PCR products of approximately 2,200 bp. Sequence analysis revealed a sequence virtually identical to the sequence of the cDNA interspersed with seven intron sequences of 356, 99, 257, 285, 102, 123, and 117 nucleotides, respectively (Fig. 3A).
Sequence Similarity with Other Proteins
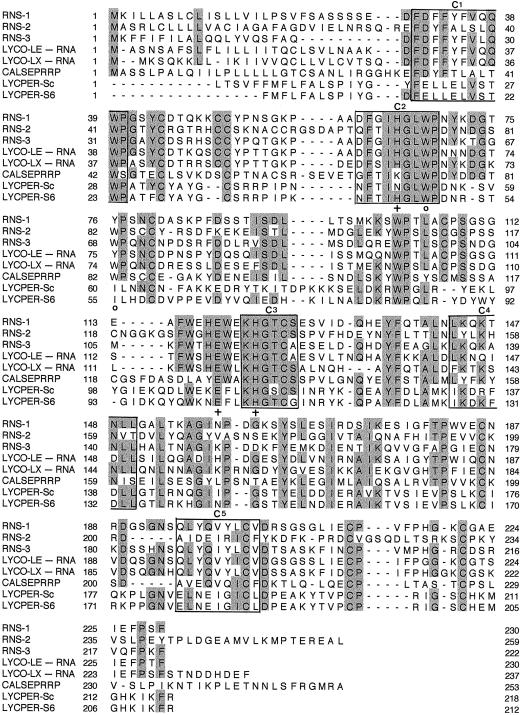
A search in the database revealed striking sequence similarity between the CalsepRRP sequence and RNases. The highest degree of sequence similarity was found with RNases RNS2 from Arabidopsis (GenBank accession no. M98336, 55.7% sequence similarity), RNase T2 from Cicer arietinum (GenBank accession no. AJ012689, 41.4% sequence similarity), and RNASE LE and LX from tomato (Lycopersicon esculentum) (GenBank accession nos. X79337 and X79338, 37.0% and 30.4% sequence similarity, respectively). Sequence comparison of different sequences encoding S-RNases and S-RNase-like proteins from several plant species and CalsepRRP shows that some regions in the sequence are highly conserved (Fig. 5).
Figure 5.
Comparison of the amino acid sequences of RNases and RNase-related proteins from Arabidopsis (RNS1, RNS2, and RNS3), L. esculentum (Lyco-LE-RNA and Lyco-LX-RNA), L. peruvianum (LYCPER-Sc and LYCPER-S6), and C. sepium (CALSEPRRP). Gaps (deletions) were introduced to maximize the homologies, and identical residues are boxed in gray. The three charged residues involved in the active site of RNases are indicated by (+), and the two aromatic residues, which presumably maintain the conformational stability of the site, are indicated by (°). The sequences of the five conserved regions (C1–C5) identified for plant RNase are boxed.
CalsepRRP Has No RNase Activity
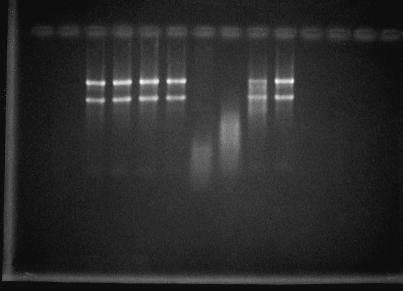
The obvious sequence similarity of CalsepRRP to previously cloned plant RNases raised the question of whether the most abundant protein of the C. sepium rhizome is a catalytically active RNase. Therefore, total CalsepRRP was subjected to a series of activity tests. Preliminary RNase activity assays based on the release of perchloric-acid-soluble oligonucleotides from yeast RNA indicated that CalsepRRP exhibits no detectable RNase activity. Since 1,000-fold-diluted crude extracts from tobacco styles still yielded a high activity in the same assay, it could be concluded that CalsepRRP has no measurable exonuclease activity. To test the possible endonuclease activity of CalsepRRP, a highly sensitive electrophoretic method was developed based on the visualization of fragments generated from ribosomal RNA. As shown in Figure 6, total CalsepRRP even at concentrations exceeding 5 μm was not able to degrade total ribosomal RNA from elderberry, whereas a 100-fold-diluted extract from tobacco styles caused an almost complete degradation of the same RNA. To test the possible effect of the pH, the RNase test was also performed at different pH values. However, no activity could be observed in a pH range between 2.0 and 10.0 (results not shown). The same tests have also been performed with the purified unglycosylated proteins and the two glycosylated fractions obtained by affinity chromatography. Since no RNase activity could be detected, these results clearly indicate that CalsepRRP is devoid of both endo- and exonuclease activity, and therefore cannot be considered a RNase.
Figure 6.
RNase-like activity of CalsepRRP. Lanes 1 through 4 show RNA treated with CalsepRRP at 5 μm, 500 nm, 50 nm, and 5 nm, respectively. Lanes 5 through 8 show RNA treated with tobacco RNase at different concentrations. Lanes 5 through 8 show a 10-, 100-, 1,000-, and 10,000-fold dilution of the tobacco extract, respectively.
CalsepRRP Has No Ribosome-Inactivating Activity
The absence of a detectable RNase activity does not preclude that CalsepRRP can affect RNA by another enzymatic activity. Therefore, the possible ribosome-inactivating activity of CalsepRRP was tested using rabbit reticulocyte ribosomes as a substrate. No activity comparable to that of the genuine ribosome-inactivating proteins could be detected. Even at high concentrations (5 μm), CalsepRRP did not generate the so-called Endo fragments (Endo et al., 1987), whereas a clear positive reaction was obtained with the type 1 RIP from Iris bulb at a concentration as low as 0.2 nm (results not shown). These results demonstrate that CalsepRRP has no N-glycosidase activity comparable to that of genuine ribosome-inactivating proteins.
CalsepRRP Homologs Do Not Occur in the Underground Storage Organs of the Convolvulaceae Species Sweet Potato and Convolvulus arvensis (Bindweed)
The discovery of a highly abundant, catalytically inactive RNase homolog in rhizomes of C. sepium raised the question of whether similar proteins also occur in the underground storage organs of other Convolvulaceae species. To address this question, the overall protein composition of crude extracts from sweet potato tubers and C. arvensis rhizomes was compared with that of C. sepium. As shown in Figure 1A, neither the sweet potato nor the C. arvensis extract contained detectable amounts of polypeptides with a molecular mass similar to that of CalsepRRP. The predominant 25-kD polypeptide in the sweet potato extract corresponded to sporamin, whereas the second most intensively stained polypeptide was β-amylase. N-terminal sequencing of the major 23-, 32-, and 47-kD polypeptides present in the crude extract from the C. arvensis rhizomes yielded no sequence similarity to CalsepRRP. Attempts to isolate CalsepRRP homologs from sweet potato tubers and C. arvensis rhizomes (using the same procedures for the purification of CalsepRRP) were also unsuccessful. Similarly, screening of a cDNA library constructed with mRNA from developing C. arvensis rhizomes using the cDNA clone encoding CalsepRRP as a probe yielded no positive clones, whereas cDNAs encoding the C. arvensis lectin and β-amylase could easily be identified using the respective heterologous probes from C. sepium (E.J.M. Van Damme, unpublished data).
Developmental Regulation of CalsepRRP
The high abundance of CalsepRRP in resting rhizomes of C. sepium is reminiscent of that of typical storage proteins from vegetative storage tissues. To determine whether CalsepRRP behaves as a storage protein with respect to its developmental regulation, the accumulation and disappearance of the protein was followed during rhizome formation and degradation, respectively. SDS-PAGE analysis of crude extracts from rhizome samples collected during rhizome formation (from the beginning of September until early December) clearly indicated that the relative abundance of the CalsepRRP polypeptides increases during early fall to reach a maximum around mid-October. Thereafter, no changes occur in the relative concentrations of the major rhizome proteins. Analysis of extracts from different parts of a long resting rhizome further demonstrated that CalsepRRP is present all over the rhizome and its relative concentration is comparable in all parts. A similar SDS-PAGE analysis of rhizome samples collected from plants grown in a greenhouse during early spring showed that the relative abundance of CalsepRRP rapidly decreases as soon as the plant resumes growth and consumes the storage compounds of the rhizome. Furthermore, CalsepRRP was only detected in rhizomes and was absent from C. sepium leaves and flowers. These observations strongly suggest that CalsepRRP is a typical VSP.
Molecular Modeling
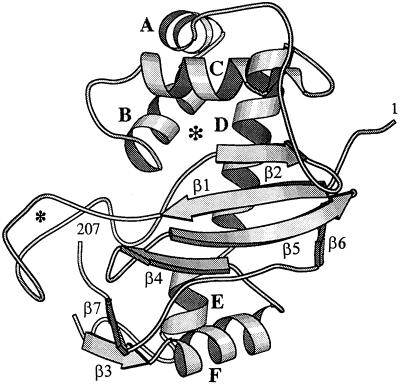
CalsepRRP has been modeled using the coordinates of the RNase Rh from R. niveus, the structure of which has been resolved by x-ray diffraction analysis (Kurihara et al., 1996). Although CalsepRRP differs from RNase Rh by an N-terminal deletion of 19 residues and an insertion of 16 residues (Fig. 3B), the HCA plots of both proteins are fairly similar (results not shown). As a result, the secondary structure features can accurately be delineated along the amino acid sequence of CalsepRRP. The three-dimensional model of CalsepRRP constructed from the x-ray coordinates of RNase Rh exhibits an overall fold very similar to that of the fungal enzyme. CalsepRRP contains seven strands of β-sheet (β1–β7) connected by turns and loops to six α-helices (A–F) (Fig. 7). The main differences between CalsepRRP and RNase Rh are an extra loop of 16 residues at the N terminus and a deletion of 10 residues in the middle of the CalsepRRP polypeptide. This deletion brings α-helices C and D very close to each other.
Figure 7.
Schematic drawing of the three-dimensional model of CalsepRRP built from the x-ray coordinates of R. niveus RNase Rh (1BOL). The strands of β-sheet are indicated by gray arrows. N- (1) and C-terminal (207) residues are labeled. The small asterisk indicates the extra loop of 16 residues at the N terminus of the CalsepRRP polypeptide chain. The seven strands of β-sheet (β1–β7) and six α-helices (A–F) are indicated on the model. The asterisk shown in bold indicates the location of the active site. The drawing was rendered using Molscript (Kraulis, 1991).
In addition, a C-terminal deletion of three residues in the CalsepRRP polypeptide allows strands β5 and β6 to collapse. CalsepRRP contains 12 Cys residues (versus 10 in RNase Rh). Four of these residues are close enough to create two disulfide bonds (Cys-58-Cys-108 and Cys-170-Cys-198), which are homologous to the disulfide bridges between Cys-63-Cys-112 and Cys-182-Cys-213, respectively, of RNase Rh (Kurihara et al., 1996). The three other disulfide bonds of RNase Rh (between Cys-3-Cys-20, Cys-10-Cys-53, and Cys-19-Cys-120, respectively) have no counterparts in CalsepRRP. Mature CalsepRRP contains a single putative N-glycosylation site (Asn-131-Ile-132-Ser-133). According to the model, this site is located at the beginning of the exposed α-helix E, and is therefore accessible for glycosylation (which is confirmed by the fact that about 50% of the CalsepRRP polypeptides are glycosylated). A very similar three-dimensional model was obtained for the self-incompatibility RNase (S3-RNase) from wild tomato (Lycopersicon peruvianum), which contains three disulfide bridges and a single N-glycosylation site occupied by (Man)3 oligomannosidic-type glycans (Parry et al., 1998). In addition, a similar three-dimensional fold was also suggested for S-RNases from the Rosaceae species Japanese pear (Pyrus pyrifolia) and apple (Malus × domestica) (Ishimizu et al., 1998).
RNase Rh belongs to the 2′,3′-cyclizing RNases, which break phosphodiester bonds of RNA to release 2′,3′-cyclic nucleotides that are subsequently hydrolyzed to give 3′-nucleotides. These RNases are subdivided in three distinct groups differing from each other in molecular mass and amino acid sequence (Kurihara et al., 1996). CalsepRRP is most closely related to the group of fungal RNases of high molecular mass (24 kD) that comprises RNase T2 (Aspergillus oryzae), RNase M (Aspergillus saitori), and RNase Rh (R. niveus), as well as the S- and S-like RNases from higher plants (Sanda et al., 1985a, 1985b). According to chemical modification and site-directed mutagenesis experiments (Sanda et al., 1985a, 1985b; Ohgi et al., 1993), His-46, His-109, and Glu-105 form the active site of RNase Rh (Fig. 8A).
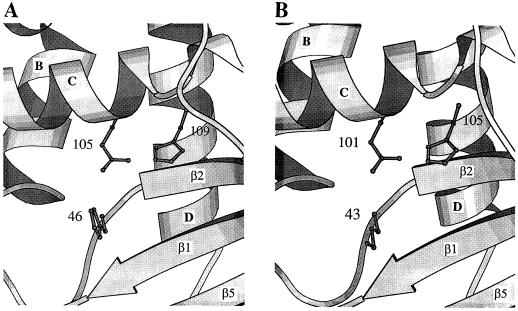
Figure 8.
Comparison of the active site of R. niveus RNase Rh (A) and CalsepRRP (B). The figures show the side chains of the His-46, Glu-105, and His-109 of R. niveus RNase Rh, and the Lys-43, Glu-101, and His-105 of CalsepRRP protruding toward the center of the flattened cleft forming the active site. The models are similarly oriented as in Figure 7. The drawing was rendered using Molscript (Kraulis, 1991).
Structural and molecular modeling also suggested that the active site is surrounded by a hydrophobic pocket consisting of the two aromatic residues, Trp-49 and Tyr-57, which possibly participate in the RNase activity (Kurihara et al., 1996). Tyr-57 is believed to preserve the active site conformation through a stacking interaction with the imidazole ring of His-109 and hydrogen bonding to the side chain of Glu-105. In CalsepRRP, the basic residues Lys-43 and His-105 and the carboxylic residue Glu-101 correspond to His-46, His-109, and Glu-105 of RNase Rh, respectively (Fig. 8B). Due to the substitution of His by Lys at position 43, a shift of more than 3.0 Å occurs in the location of the positive charge associated with these charged residues. This charge dislocation apparently accounts for the lack of RNase activity of CalsepRRP, even though Trp-46 (which replaces Trp-49 of RNase Rh) is similarly hydrogen bonded to Glu-101 (homologous to Glu-105 of RNase Rh) and displays a stacking interaction with His-105 (homologous to His-109 of RNase Rh).
DISCUSSION
This paper describes the isolation, characterization, molecular cloning, and molecular modeling of the most abundant protein from resting rhizomes of C. sepium. Native CalsepRRP is a monomeric protein occurring as a natural mixture of glycosylated and unglycosylated forms. According to the deduced amino acid sequence, CalsepRRP is synthesized as a preproprotein. The presence of a characteristic signal peptide and the partial glycosylation indicate that CalsepRRP is synthesized on the endoplasmic reticulum and follows the secretory pathway. Mass spectroscopy further suggests that the CalsepRRP precursor is cleaved post-translationally by the removal of a C-terminal peptide. Cleavage of this sequence rich in hydrophobic and acidic amino acids suggests a vacuolar localization of CalsepRRP and is in agreement with preliminary results of immunolocalization studies.
The superfamily of plant RNases is subdivided into S- and S-like RNases (Green, 1994; Richman et al., 1997). S-RNases, which were originally described as S-proteins associated with gametophytic self-incompatibility in Solanaceae species, are an extended group of basic RNases occurring in high concentrations in the transmitting tract of the styles of self-incompatible Rosaceae, Solanaceae, and Scrophulariaceae species. It is generally accepted that S-RNases play an important role in self-incompatibility (Green, 1994; Royo et al., 1994). S-Like (or non-S-) RNases are widespread among both monocot and dicots, where they occur in various tissues. In contrast to the basic S-RNases, all S-like RNases except the seed RNases from the Cucurbitaceae species Momordica charantia and Luffa cylindrica have an acidic pI. Based on its overall amino acid sequence, CalsepRRP is more closely related to the S-like RNases than to the S-RNases, which is in accordance with the location of the C. sepium protein in the rhizomes. In contrast to the S-like RNases, CalsepRRP shows a very complex gene structure. Analysis of the genomic sequence revealed the presence of seven intron sequences in CalsepRRP, whereas RNases from Rosaceae and Solanaceae species contain only one single intron, the position of which coincides with intron 3 in the CalsepRRP sequence (Broothaerts et al., 1995; Matton et al., 1995).
Molecular modeling confirmed that CalsepRRP has the same overall three-dimensional fold as the fungal and plant RNases. However, in spite of the obvious structural similarity and high sequence similarity with catalytically active fungal and plant RNases, CalsepRRP is completely devoid of RNase activity. To explain this apparent lack of enzymatic activity, the sequence of CalsepRRP was compared with that of the catalytically active S- and S-like RNases. All plant RNases contain five highly conserved regions designated C1 to C5 (Ioerger et al., 1991; Green, 1994), two of which resemble the sequences of the active site of fungal RNases. In addition, two pairs of Cys residues that form disulfide bonds are conserved among all known S- and S-like RNases. At present, only a limited number of experiments have been performed to establish which amino acids are involved in the catalytic activity of plant RNases. Based on these results and sequence comparisons with the RNases from A. oryzae and R. niveus, it is generally accepted that the two His residues located in the conserved sequences C2 and C3 are required for catalytic activity (Kawata et al., 1990; Ishimizu et al., 1995, 1996; Kurihara et al., 1996; Parry et al., 1997). A close examination of the sequence of CalsepRRP indicated that it contains all five conserved regions (C1–C5) as well as the conserved Cys residues. However, although the His residue in the region C3 is present, CalsepRRP lacks the His residue in C2. The substitution of this His by Lys can explain why CalsepRRP does not possess RNase activity.
CalsepRRP is not the first example of an “inactive” plant RNase. It has previously been demonstrated that the substitution of His 33 in C2 by an Asn at the active site of an S-RNase from a self-compatible accession of L. peruvianum results in the loss of catalytic activity (which itself abolishes the self-incompatibility of the plant) (Royo et al., 1994). However, to the best of our knowledge, CalsepRRP is the first “inactive” S-like RNase to be identified and purified. Moreover, the availability of large quantities of CalsepRRP enabled thorough studies of the possible residual enzymatic activity. It should be emphasized that a (very low) residual RNase activity can only be excluded by using a combination of a highly sensitive enzymatic assay and a high protein concentration in the test. From this point of view, our results provide convincing evidence that the substitution of a His by a Lys residue completely abolishes the catalytic activity of plant RNases. It should be mentioned here that a maize cDNA clone encoding a putative S-like RNase that lacks the active-site His residues has been identified (GenBank accession no. U66241). However, since the putative protein has not been isolated, it remains to be demonstrated that it corresponds to a catalytically inactive S-like RNase.
Several lines of evidence indicate that CalsepRRP is a VSP. First, CalsepRRP is the most abundant protein in the C. sepium rhizomes, representing over 50% of the total protein content. Moreover, apart from the cytoplasmic β-amylase and lectin, CalsepRRP is the only abundant rhizome protein. Second, CalsepRRP accumulates exclusively in the rhizomes. Third, the accumulation and disappearance of the protein during rhizome formation and degradation, respectively, fits exactly that of a typical storage protein. Fourth, CalsepRRP probably has no other function than a storage role because its enzymatic activity is completely lost. To the best of our knowledge, the C. sepium rhizome is the only documented plant tissue that accumulates large quantities of a catalytically inactive homolog of plant RNases. Even in the underground storage organs of two close relatives of C. sepium (sweet potato and C. arvensis), no CalsepRRP homolog was found.
The identification of a catalytically inactive RNase homolog as a major VSP is important in view of the possible origin of storage proteins. In the past, several putative storage proteins that possess an enzymatic or other biological activity have been identified. Many examples have been described of protease inhibitors, lectins, ribosome-inactivating proteins, and enzymes that are major proteins in either seeds or vegetative storage tissues, and meet all of the requirements to be considered as genuine storage proteins (except that they do have a well-defined biological activity). There are, however, also examples of storage proteins that are clearly related to genuine enzymes, protease inhibitors, or lectins, but either have no or a strongly reduced activity. Patatin, for example, is a poorly active lipid acyl hydrolase (Andrews et al., 1988). Similarly, the acid phosphatase activity of the soybean VSP is 2 to 4 orders of magnitude lower than that of a tomato homolog (Staswick, 1994).
Sweet potato sporamin definitely exhibits trypsin inhibition activity, but has a low specific activity compared with genuine trypsin inhibitors. The same holds true for the garlic bulb (storage protein) lectins, which are about 2 orders of magnitude less active than their relatives from the leaves (Smeets et al., 1997). A few cases have also been reported of storage proteins that are closely related to lectins but devoid of carbohydrate-binding activity. For example, the major bark storage protein of Cladrastis lutea (yellow wood) shares high sequence identity with bark lectin, but lacks functional carbohydrate-binding sites because of a three amino acid residue insertion in the normal carbohydrate-binding site (Van Damme et al., 1995a). Similarly, one of the major bark storage proteins of elderberry is a close relative of the bark lectins, which lost its carbohydrate-binding activity as a result of the substitution of a few amino acids in the normal binding sites (Van Damme et al., 1997b).
The obvious evolutionary relationships between biologically inactive/poorly active storage proteins and “normally active” enzymes/bioactive proteins strongly suggest that (some) storage proteins may be derived from genes that originally encoded proteins with a well-defined enzymatic or other biological activity. According to Staswick (1994), duplicate copies of genes encoding enzymes or other biologically active proteins could be free to acquire a promoter that directs abundant expression according to the storage needs of a specific organ. Afterward, these genes may have lost some or all of the biological activity associated with their previous function. Van Damme et al. (1998) proposed a similar mechanism to explain the evolution of the Allium lectins from a common ancestor. The main evolutionary line of these proteins leads to a group of highly active leaf-specific lectins that are markedly conserved among all Allium species and are believed to play a role in the plant's defense against sucking insects. All side branches that diverged from the main evolutionary line consist of lectins with low or no carbohydrate-binding activity that are expressed at high levels in bulbs and behave as typical VSPs. It is believed, therefore, that conservation and/or enhancement of carbohydrate-binding activity was the most important selection criterion in the evolution of the presumed defense-related leaf lectins. The evolution of the diverging groups clearly followed different criteria. As a result, many Allium species now contain storage proteins with a residual lectin activity.
NOTE ADDED IN PROOF
Recently a gene encoding an RNase S-like homolog from barley (Genbank accession no. AF182129) was identified. These results will be published (Gausing K [2000] A barley gene (rsh1) encoding an RNase S-like homologue specifically expressed in young light-grown leaves. Planta [in press]).
Footnotes
This work was supported in part by grants from the Katholieke Universiteit Leuven (no. OT/98/17), Centre National de la Recherche Scientifique and the Conseil Régional de Midi-Pyrénées, and the Fund for Scientific Research-Flanders (grant no. G.0223.97). W.J.P. is Research Director and E.J.M.V.D. is a postdoctoral fellow of this fund. Q.H. acknowledges the receipt of a doctoral scholarship from the Research Council of the Katholieke Universiteit Leuven.
LITERATURE CITED
- Andrews DL, Beames B, Summers MD, Park WD. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J. 1988;252:199–206. doi: 10.1042/bj2520199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts W, Janssens GA, Proost P, Broekaert WF. cDNA cloning and molecular analysis of two self-incompatibility alleles from apple. Plant Mol Biol. 1995;27:499–511. doi: 10.1007/BF00019317. [DOI] [PubMed] [Google Scholar]
- Brown PH, Ho T-HD. Barley aleurone layers secrete a nuclease in response to gibberellic acid: purification and partial characterization of the associated ribonuclease, deoxyribonuclease, and 3′ nucleotidase activities. Plant Physiol. 1986;82:8021–8026. doi: 10.1104/pp.82.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman GD, Chen TH, Ernst SG, Fuchigami L. Photoperiod control of poplar bark storage protein accumulation. Plant Physiol. 1991;96:686–692. doi: 10.1104/pp.96.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wald DB, Mason HS, Mullet JH. The soybean vegetative storage proteins VSPα and VSPβ are acid phosphatases active on polyphosphates. J Biol Chem. 1992;267:15958–15964. [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugar and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes: the site and the characteristics of the modification in 28S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyze amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Green PJ. The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:421–445. [Google Scholar]
- Hankins CN, Kindinger JI, Shannon LM. The lectins of Sophora japonica: II. Purification, properties, and N-terminal amino acid sequences of five lectins from bark. Plant Physiol. 1988;86:67–70. doi: 10.1104/pp.86.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, Yanai K, Takagi M, Yano K, Wakabayashi E, Sansa A, Mines S, Ohgi K, Irie M. Primary structure of a base non-specific ribonuclease from Rhizopus niveus. J Biochem. 1988;103:408–418. doi: 10.1093/oxfordjournals.jbchem.a122284. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-H. Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod. 1991;4:81–87. [Google Scholar]
- Ishimizu T, Endo T, Yamaguchi-Kabata Y, Nakamura KT, Sakiyama F, Norioka S. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Lett. 1998;440:337–342. doi: 10.1016/s0014-5793(98)01470-7. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Miyagi M, Norioka S, Liu Y-H, Clarke AE, Sakiyama F. Identification of histidine 31 and cysteine 95 in the active site of self-incompatibility associated S6-RNase in Nicotiana alata. J Biochem. 1995;118:1007–1013. doi: 10.1093/jb/118.5.1007. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Norioka S, Kanai M, Clarke AE, Sakiyama F. Location of cysteine and cystine residues in S-ribonucleases associated with gametophytic self-incompatibility. Eur J Biochem. 1996;242:627–635. doi: 10.1111/j.1432-1033.1996.0627r.x. [DOI] [PubMed] [Google Scholar]
- Kawata Y, Sakiyama F, Hayashi F, Kyogoku Y. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem. 1990;187:255–262. doi: 10.1111/j.1432-1033.1990.tb15303.x. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Kurihara H, Nonaka T, Mitsui Y, Ohgi K, Irie M, Nakamura KT. The crystal structure of ribonuclease Rh from Rhizopus niveus at 2.0 Å resolution. J Mol Biol. 1996;255:310–320. doi: 10.1006/jmbi.1996.0025. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JN. PROCHECK: a program to check the stereochemistry of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lemesle-Varloot L, Henrissat B, Gaboriaud C, Bissery V, Morgat A, Mornon JP. Hydrophobic cluster analysis: procedure to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990;72:555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- Maeshima M, Sasaki T, Asahi Y. Characterization of the major proteins in sweet potato tuber roots. Phytochemistry. 1985;24:1899–1902. [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Mas MT, Smith KC, Yarmush DL, Aisaka K, Fine RM. Modeling the anti-CEA antibody combining site by homology and conformational search. Proteins Struct Funct Genet. 1992;14:483–498. doi: 10.1002/prot.340140409. [DOI] [PubMed] [Google Scholar]
- Matton DP, Mau S-L, Okamoto S, Clarke AE, Newbigin E. The S-locus of Nicotiana alata: genomic organization and sequence analysis of two S-RNase alleles. Plant Mol Biol. 1995;28:847–858. doi: 10.1007/BF00042070. [DOI] [PubMed] [Google Scholar]
- Mierendorf RC, Pfeffer D. Direct sequencing of denatured plasmid DNA. Methods Enzymol. 1987;152:556–562. doi: 10.1016/0076-6879(87)52061-4. [DOI] [PubMed] [Google Scholar]
- Mignery GA, Pikaard CS, Hannapel DJ, Park WD. Isolation and sequence analysis of cDNAs for the major potato tuber protein, patatin. Nucleic Acids Res. 1984;12:7987–8000. doi: 10.1093/nar/12.21.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgi K, Horiuchi H, Watanabe H, Iwama M, Takagi M, Irie M. Role of Asp51 and Glu105 in the enzymatic activity of a ribonuclease from Rhizopus. J Biochem. 1993;113:219–224. doi: 10.1093/oxfordjournals.jbchem.a124029. [DOI] [PubMed] [Google Scholar]
- Parry S, Newbigin E, Craik D, Nakamura KT, Bacic A, Oxley D. Structural analysis and molecular model of a self-incompatibility RNase from wild tomato. Plant Physiol. 1998;116:463–469. doi: 10.1104/pp.116.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry S, Newbigin E, Currie G, Bacic A, Oxley D. Identification of active-site histidine residues of a self-incompatibility ribonuclease from a wild tomato. Plant Physiol. 1997;115:1421–1429. doi: 10.1104/pp.115.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans WJ, Nsimba-Lubaki M, Peeters B, Broekaert WF. Isolation and partial characterization of a lectin from Aegopodium podagraria rhizomes. Planta. 1985;164:75–82. doi: 10.1007/BF00391028. [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJM. Lectins as plant defense proteins. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans WJ, Winter HC, Bemer V, Van Leuven F, Goldstein IJ, Truffa-Bachi P, Van Damme EJM. Isolation of a novel plant lectin with an unusual specificity from Calystegia sepium. Glycoconj J. 1997;14:259–265. doi: 10.1023/a:1018502107707. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. Tertiary templates for proteins: use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Richman AD, Broothaerts W, Kohn JR. Self-incompatibility RNases from three plant families: homology or convergence? Am J Bot. 1997;84:912–917. [PubMed] [Google Scholar]
- Royo J, Kunz C, Kowyama Y, Anderson M, Clarke AE, Newbigin E. Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-incompatibility in Lycopersicon peruvianum. Proc Natl Acad Sci USA. 1994;91:6511–6514. doi: 10.1073/pnas.91.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda A, Takizawa Y, Irie M. Carboxymethylation of a ribonuclease from Rhizopus sp. Chem Pharm Bull. 1985a;33:4515–4521. [Google Scholar]
- Sanda A, Takizawa Y, Iwama M, Irie M. Modification of a ribonuclease from Rhizopus sp. with 1-cyclohexyl-3-(2-morpholinyl-(4)-ethyl) carbodiimide p-toluenesulfonate. J Biochem. 1985b;98:125–132. doi: 10.1093/oxfordjournals.jbchem.a135250. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets K, Van Damme EJM, Verhaert P, Barre A, Rougé P, Van Leuven F, Peumans WJ. Isolation, characterization and molecular cloning of the mannose-binding lectins from leaves and roots of garlic (Allium sativum L.) Plant Mol Biol. 1997;33:223–234. doi: 10.1023/a:1005717020021. [DOI] [PubMed] [Google Scholar]
- Staswick PE. Developmental regulation and the influence of plant sinks on vegetative storage protein gene expression in soybean leaves. Plant Physiol. 1989a;89:309–315. doi: 10.1104/pp.89.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Preferential loss of an abundant storage protein from soybean pods during seed development. Plant Physiol. 1989b;90:1252–1255. doi: 10.1104/pp.90.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Storage proteins of vegetative plant tissues. Annu Rev Plant Physiol Mol Biol. 1994;45:303–322. [Google Scholar]
- Stewart CN, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques. 1993;14:748–750. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Allen AK, Peumans WJ. Related mannose-specific lectins from different species of the family Amaryllidaceae. Physiol Plant. 1988;73:52–57. [Google Scholar]
- Van Damme EJM, Barre A, Bemer V, Rougé P, Van Leuven F, Peumans WJ. A lectin and a lectin-related protein are the two most prominent proteins in the bark of yellow wood (Cladrastis lutea) Plant Mol Biol. 1995a;29:579–598. doi: 10.1007/BF00020986. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Peumans WJ. Molecular cloning of the bark and seed lectins from the Japanese pagoda tree (Sophora japonica) Plant Mol Biol. 1997a;33:523–536. doi: 10.1023/a:1005781103418. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Van Leuven F, Peumans WJ. Isolation and molecular cloning of a novel type 2 ribosome-inactivating protein with an inactive B chain from elderberry (Sambucus nigra) bark. J Biol Chem. 1997b;272:8353–8360. doi: 10.1074/jbc.272.13.8353. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Smeets K, Torrekens S, Van Leuven F, Rougé P, Peumans WJ. The bark lectin of Robinia pseudoacacia contains a complex mixture of isolectins: characterization of the proteins and the cDNA clones. Plant Physiol. 1995b;107:833–843. doi: 10.1104/pp.107.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Verhaert P, Rougé P, Peumans WJ. Molecular cloning of the mitogenic mannose/maltose-specific rhizome lectin from Calystegia sepium. FEBS Lett. 1996a;397:352–356. doi: 10.1016/s0014-5793(96)01211-2. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Briké F, Winter HC, Van Leuven F, Goldstein IJ, Peumans WJ. Molecular cloning of two different mannose-binding lectins from tulip bulbs. Eur J Biochem. 1996b;236:419–427. doi: 10.1111/j.1432-1033.1996.00419.x. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ. Cell-free synthesis of lectins. In: Gabius H-J, Gabius S, editors. Lectins and Glycobiology. Berlin: Springer Verlag; 1993. pp. 458–468. [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P. Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit Rev Plant Sci. 1998;17:575–692. [Google Scholar]
- Van Damme EJM, Smeets K, Torrekens S, Van Leuven F, Goldstein IJ, Peumans WJ. The closely related homomeric and heterodimeric mannose-binding lectins from garlic are encoded by one-domain and two-domain lectin genes, respectively. Eur J Biochem. 1992;206:413–420. doi: 10.1111/j.1432-1033.1992.tb16941.x. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Smeets K, Torrekens S, Van Leuven F, Peumans WJ. The mannose-specific lectins from ramsons (Allium ursinum L.) are encoded by three sets of genes. Eur J Biochem. 1993;217:123–129. doi: 10.1111/j.1432-1033.1993.tb18226.x. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Van Leuven F, Peumans WJ. Isolation, characterization and molecular cloning of the bark lectins from Maackia amurensis. Glycoconj J. 1997c;14:449–456. doi: 10.1023/a:1018595300863. [DOI] [PubMed] [Google Scholar]
- Von Heijne G. A method for predicting signal cleavage sites. Nucleic Acids Res. 1986;11:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KW, Chen JC, Lin MI, Chen YM, Lin CY. Functional activity of sporamin from sweet potato (Ipomoea batatas Lam.): a tuber storage protein with trypsin inhibitory activity. Plant Mol Biol. 1997;33:565–570. doi: 10.1023/a:1005764702510. [DOI] [PubMed] [Google Scholar]