Abstract
Sae2 cooperates with the Mre11–Rad50-Xrs2 (MRX) complex to initiate resection of DNA double-strand breaks (DSBs) and to maintain the DSB ends in close proximity to allow their repair. How these diverse MRX-Sae2 functions contribute to DNA damage resistance is not known. Here, we describe mre11 alleles that suppress the hypersensitivity of sae2Δ cells to genotoxic agents. By assessing the impact of these mutations at the cellular and structural levels, we found that all the mre11 alleles that restore sae2Δ resistance to both camptothecin and phleomycin affect the Mre11 N-terminus and suppress the resection defect of sae2Δ cells by lowering MRX and Tel1 association to DSBs. As a consequence, the diminished Tel1 persistence potentiates Sgs1-Dna2 resection activity by decreasing Rad9 association to DSBs. By contrast, the mre11 mutations restoring sae2Δ resistance only to phleomycin are located in Mre11 C-terminus and bypass Sae2 function in end-tethering but not in DSB resection, possibly by destabilizing the Mre11–Rad50 open conformation. These findings unmask the existence of structurally distinct Mre11 domains that support resistance to genotoxic agents by mediating different processes.
INTRODUCTION
DNA double-strand breaks (DSBs) are repaired by two major pathways: non-homologous end-joining (NHEJ), which directly re-ligates the broken DNA ends, and homologous recombination (HR), which uses an intact homologous DNA sequence as a template to restore the genetic information at the break site (1,2). In order to repair a DSB by HR, the 5′ DNA strands on either side of the DSB have to be nucleolytically degraded through a process referred to as resection (3). The resulting 3′-ended single-stranded DNA (ssDNA) tails are first bound by the ssDNA binding complex Replication Protein A (RPA), which is then replaced by Rad51 to generate a right-handed helical filament that initiates HR (1,2).
The evolutionarily conserved MRX/MRN complex (Mre11–Rad50-Xrs2 in Saccharomyces cerevisiae, MRE11-RAD50-NBS1 in mammals) plays a central role in signaling, processing and repairing DNA DSBs (4). The N-terminal domain of the Mre11 subunit contains five conserved phosphoesterase motifs that are essential for both 3′-5′ double-stranded DNA (dsDNA) exonuclease and ssDNA endonuclease activities (5–11). Mre11 interacts with Rad50, which is an ATPase characterized by ATP-binding motifs that are located at both the N- and C-terminal regions of the protein and are separated by an intramolecular coiled-coil (12–15). The coiled-coil apex contains a motif that can dimerize thereby forming bridges that transiently tether broken DNA ends (16–20).
Structural studies of the catalytic domains of bacterial and archaeal Mre11–Rad50 complex have shown that, in the presence of ATP, the Rad50 dimer associates with the Mre11 nuclease site groove and blocks its access to DNA (21–23). By contrast, in the absence of ATP, Mre11 holds the Rad50 ATPase domains near the base of the coiled-coils and DNA becomes accessible to the Mre11 nuclease active site (22–24). These findings support a model in which ATP hydrolysis induces a switch between a closed state with occluded Mre11 nuclease domain, required for DNA end binding and tethering, to an open configuration with exposed Mre11 nuclease sites that can be engaged in DSB resection (25).
MRX function in DSB resection partially depends on Sae2 (CtIP in mammals and Ctp1 in Schizosaccharomyces pombe), which stimulates Mre11 endonuclease activity (26). Studies in budding yeast indicate that MRX and Sae2 initiate resection by catalyzing an endonucleolytic cleavage in the 5′-terminated strands at both DSB ends. Then, Mre11 exonuclease activity digests DNA in the 3′-5′ direction toward the DSB end, whereas processive nucleases, like Exo1 and Dna2, take over resection in the 5′-3′ direction from the gap (27–33). While Exo1 is able to release mononucleotide products from a dsDNA end (34), the resection activity of Dna2 requires the RecQ helicase Sgs1 (BLM in humans) that unwinds dsDNA in a 3′-5′ direction (28–30,32,33,35). In addition, the MRX complex promotes Exo1- and/or Sgs1-Dna2-mediated DSB resection by facilitating Exo1 and Sgs1 recruitment to the DSB ends independently of both Mre11 nuclease activity and Sae2 (36).
The endonucleolytic cleavage catalyzed by MRX-Sae2 is particularly important for initiating resection at DSBs, whose DNA ends are not accessible to Exo1 and Sgs1-Dna2 due to the presence of covalently end-bound proteins, such as the topoisomerase-like protein Spo11 that remains covalently linked to the 5′-terminated strands of meiotic DSBs (37).
The lack of Sae2 not only delays initiation of DSB resection, but it also causes enhanced Tel1 and Rad53 signaling activities (38,39). This persistent signaling contributes to the DNA damage hypersensitivity and the resection defects of Sae2-defective cells by promoting the binding to DSBs of Rad9, which in turn inhibits the resection activity of Sgs1-Dna2 (40–42). In fact, impairment of either Tel1 or Rad53 activity restores DNA damage resistance and resection in sae2Δ cells by decreasing the amount of DSB-bound Rad9, and therefore by relieving inhibition of Sgs1-Dna2 (42). Sae2 function in DNA damage resistance is also bypassed by lowering MRX association to DSBs (43,44). Whether this effect is due to a dampening of Tel1 signaling activity is unknown. In any case, sae2Δ cells display only mild resection defects (45), suggesting that resistance to DNA damaging agents involves other Sae2 functions besides its promotion of Mre11 nuclease to enhance resection. Consistent with this hypothesis, sae2Δ cells are defective in maintaining the DSB ends in close proximity (45,46).
To better understand how these diverse Sae2-MRX functions contribute to DNA damage resistance, we searched and characterized Saccharomyces cerevisiae mre11 mutants that suppress the sae2Δ hypersensitivity to phleomycin, which generates chemically complex DNA termini, and/or to camptothecin (CPT), which extends the half-life of topoisomerase 1 (Top1)-DNA cleavable complexes (Top1ccs) (47). We found that the mre11 mutations restoring sae2Δ resistance to both CPT and phleomycin also suppressed the resection defect of sae2Δ cells by lowering MRX and Tel1 association to DSBs. As a consequence, the decreased Tel1 persistence potentiated Sgs1-Dna2 by diminishing Rad9 association to DSBs. By contrast, the mre11 mutations restoring sae2Δ resistance only to phleomycin bypassed Sae2 function in end-tethering but not that in DSB resection, indicating that the hypersensitivity to phleomycin of sae2Δ cells does not reflect defective DNA-end processing per se. Interestingly, all the mre11 mutations suppressing sae2Δ resection defects caused amino acid changes in the Mre11 N-terminus, whereas the mutations specifically bypassing Sae2 function in end-tethering were located in the MRE11 region encoding the C-terminus, which is known to interact with Rad50 coiled-coil (21,22,24). We propose structural insights into the molecular effect of these mre11 mutations based on a computational model of Mre11–Rad50 and of both the dimerization and the Rad50 binding domains of Mre11, obtained by homology modeling and molecular dynamics.
MATERIALS AND METHODS
Yeast strains and growth conditions
Strain genotypes are listed in Supplementary Table S1. Strain JKM139, used to detect DSB resection, was kindly provided by J. Haber (Brandeis University, Waltham, USA). Strain tGI354, used to detect ectopic recombination, was kindly provided by G. Liberi (CNR, Pavia, Italy). Strain JYK40.6, used to detect end-tethering, was kindly provided by D.P. Toczyski (University of California, San Francisco, USA). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone) supplemented with 2% glucose (YEPD), 2% raffinose (YEPR) or 2% raffinose and 3% galactose (YEPRG). Gene disruptions were generated by one-step PCR disruption method. All the experiments have been performed at 27°C.
Search for mre11 mutants
Genomic DNA from a strain carrying the HPHMX gene located 250 bp downstream of the MRE11 stop codon was used as template to amplify by low-fidelity PCR a MRE11 region spanning from position –460 bp to +500 bp from the MRE11 coding region. Thirty independent PCR reaction mixtures were prepared, each containing 5U EuroTaq DNA polymerase (Euroclone), 10 ng genomic DNA, 500 ng each primer, 0.5 mM each dNTP (dATP, dTTP, dCTP), 0.1 mM dGTP, 0.5 mM MnCl2, 10 mM ßmercaptoethanol, 10 mM Tris–HCl (pH 9), 50 mM KCl and 1.5 mM MgCl2. The resulting PCR amplification products contained the MRE11 coding sequence and the HPHMX resistance gene were used to transform a sae2Δ mutant strain (YLL 1070.1). 3000 transformants were selected on YEPD medium added with Hygromycin B (0.3 mg/ml) and then assayed by drop tests for the ability to grow on YEPD plates containing phleomycin or camptothecin.
DSB resection and ectopic recombination
DSB end resection at the MAT locus in JKM139 derivative strains was analyzed on alkaline agarose gels, by using a single-stranded probe complementary to the unresected DSB strand, as previously described (48). Quantitative analysis of DSB resection was performed by calculating the ratio of band intensities for ssDNA and total amount of DSB products. DSB repair by ectopic recombination was detected by using the tGI354 strain as previously described (48). To determine the repair efficiency, the intensity of the uncut band at 2 h after HO induction (maximum efficiency of DSB formation), normalized with respect to a loading control, was subtracted to the normalized values of NCO and CO bands at the subsequent time points after galactose addition. The obtained values were divided by the normalized intensity of the uncut MATa band at time zero before HO induction (100%).
Plasmid religation assay
The centromeric plasmid pRS316 was digested with the BamH1 restriction enzyme before being transformed into the cells. Parallel transformation with undigested pRS316 DNA was used to determine the transformation efficiency. Efficiency of re-ligation was calculated by determining the number of colonies able to grow on medium selective for the plasmid marker and was normalized to the transformation efficiency for each sample. The re-ligation efficiency in mutant cells was compared to that of wild type cells that was set up to 100%.
ChIP and qPCR
ChIP analysis was performed as previously described (48). Quantification of immunoprecipitated DNA was achieved by quantitative real-time PCR (qPCR) on a Bio-Rad MiniOpticon apparatus. Triplicate samples in 20 μl reaction mixture containing 10 ng of template DNA, 300 nM for each primer, 2× SsoFast™ EvaGreen® supermix (Bio-Rad #1725201) (2× reaction buffer with dNTPs, Sso7d-fusion polymerase, MgCl2, EvaGreen dye, and stabilizers) were run in white 48-well PCR plates Multiplate™ (Bio-Rad #MLL4851). The qPCR program was as follows: step 1, 98°C for 2 min; step 2, 98°C for 5 s; step 3, 60°C for 10 s; step 4, return to step 2 and repeat 30 times. At the end of the cycling program, a melting program (from 65°C to 95°C with a 0.5°C increment every 5 s) was run to test the specificity of each qPCR. Data are expressed as fold enrichment at the HO-induced DSB over that at the non-cleaved ARO1 locus, after normalization of each ChIP signals to the corresponding input for each time point. Fold enrichment was then normalized to the efficiency of DSB induction.
Expression and purification of Mre11, Mre11-R522H, Rad50 and Xrs2
The pFB-Mre11–6xHis plasmid for Mre11 expression in insect cells was obtained from P. Cejka (University of Zurich, Switzerland). Bacmid and P3 baculovirus were prepared in Escherichia coli DH10Bac and Spodoptera frugiperda Sf9 cells, respectively, according to the manufacturer's protocols (Invitrogen). To express Mre11, 800 ml Trichoplusia ni High Five cells were grown to a density of 1 × 106 cells/ml and infected with 24 ml high-titer P3 baculovirus. After 44 h infection, cells were harvested, washed with PBS, snap frozen in liquid nitrogen, and stored at –80°C. All the subsequent purification steps were carried out at 4°C. The cell pellet (∼7 g from 800 ml of culture) was thawed and resuspended in 65 ml T buffer (25 mM Tris–HCl, pH 7.5, 10% glycerol, 0.5 mM EDTA) supplemented with fresh DTT (1 mM), Igepal (0.01%), KCl (300 mM) and protease inhibitors (5 μg/ml each of aprotinin, chymostatin, leupeptin and pepstatin A; 1 mM phenylmethylsulfonyl fluoride). After sonication for 1 min, the cell lysate was clarified by ultracentrifugation at 100 000×g for 45 min and mixed with 4 ml Ni-NTA agarose (Qiagen) together with 20 mM imidazole for 1 h. The resin was washed with 250 ml of T buffer containing 1 M KCl and 20 mM imidazole and bound proteins were eluted with 15 ml of 200 mM imidazole in T buffer containing 300 mM KCl. The eluate was diluted with an equal volume of T buffer and then applied onto a 4-ml Q Sepharose Fast Flow column (GE healthcare). The column was washed with 24 ml T buffer containing 150 mM KCl before being developed with an 80 ml gradient of KCl (150–500 mM) in T buffer. The fractions containing Mre11 were pooled, concentrated to 0.5 ml using an Amicon Ultra 30K filter (Millipore), and further fractionated in a 24-ml Superose 6 column (GE healthcare) in T buffer containing 300 mM KCl. The peak fractions were pooled, concentrated to ∼4 μg/μl and stored in small aliquots at -80°C. The typical yield of Mre11 was ∼2.5 mg from 800 ml of cell culture. The Mre11-R522H expression vector was constructed by site-directed mutagenesis of the pFB-Mre11–6xHis plasmid. The Mre11-R522H protein was purified using the procedure described above. Rad50 was overexpressed in yeast and purified as described previously (48). The pFB-Xrs2-Flag plasmid for Xrs2 expression in insect cells was obtained from P. Cejka, and the cell extract was prepared as described above for Mre11 purification. The clarified lysate was mixed with 6 ml anti-Flag affinity resin (Sigma) for 2 h incubation. After washing the resin with 250 ml of T buffer containing 500 mM KCl, bound proteins were eluted with 6 ml of T buffer containing 300 mM KCl and 250 ng/μl Flag peptide. The protein pool was diluted with 6 ml of T buffer and further fractionated in a 1-ml Mono S column (GE healthcare) with a 40 ml gradient of 100–400 mM KCl in T buffer. The Xrs2 peak fractions were pooled, concentrated to 0.5 ml using an Amicon Ultra 30K filter (Millipore), and further fractionated in a 24-ml Superose 6 column as described above for Mre11. The Xrs2 peak fractions were pooled, concentrated to ∼5 μg/μl and stored in small aliquots at –80°C. The typical yield of Xrs2 was ∼2.7 mg from 1 L of cell culture.
Affinity pull-down assay
To evaluate the assembly of the MRX complex, 0.9 μg His-tagged Mre11 or Mre11-R522H was mixed with Rad50 (1.5 μg) and Xrs2 (1 μg) in 30 μl T buffer containing 200 mM KCl, incubated on ice for 1 h, and imidazole was added to 20 mM. The reaction mixtures were incubated with Ni-NTA agarose for 1 h on ice with frequent tapping. The supernatant was removed and the affinity resin was washed three times with 50 μl of the same buffer. Then, bound proteins were eluted with 40 μl 2% SDS. The supernatant, wash, and eluate fractions were resolved by SDS-PAGE and proteins were revealed using the Oriole fluorescent gel stain (BioRad).
ATPase and nuclease assays
The ATPase assay was performed as described previously (48) with slight modifications. Briefly, Mre11 or Mre11-R522H, 100 nM each, was incubated with an equimolar concentration of Rad50 and Xrs2 in 12 μl of reaction buffer (25 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA) that also contained 75 mM KCl (final concentration), 100 bp dsDNA (200 nM), 300 μM of ATP, and 1.7 nM of [γ-32P]ATP at 30°C. At the indicated times, 2 μl of the reaction mixture was mixed with an equal volume of EDTA (0.5 M, pH 8.0) and placed on ice. Reaction samples were resolved by thin layer chromatography followed by phosphorimaging analysis as previously described (8). The nuclease activity of Mre11 or Mre11-R522H was tested on a DNA substrate with 5-nt 3′ overhangs and a nick in 12 μl reaction buffer (25 mM Tris–HCl, pH 7.5, 1 mM MnCl2, 1 mM DTT, 100 μg/ml BSA) containing 100 mM KCl (final concentration). After a 30 min incubation at 30°C, reaction mixtures were resolved in a denaturing polyacrylamide gel, followed by gel drying and phosphorimaging analysis.
Homology modeling
A BLASTX search of the Protein Data Bank (PDB) using Saccharomyces cerevisiae Mre11 (ScMre11) protein sequence as a query showed high sequence identity (52%) with Schizosaccharomyces pombe Mre11 (SpMre11) (PDB ID: 4FCX) (49) and also (50%) with Chaetomium thermophilum Mre11 (CtMre11) (PDB ID: 4YKE) (50). A homology based model of ScMre11 protein spanning endonuclease and capping domains (1–416 aa) was constructed based on the crystal structure coordinates of 4FCX. The pairwise alignment was performed using the homology module of Prime through the Maestro interface (Schrodinger, LLC, and New York, USA) (https://www.schrodinger.com/citations#Maestro). The pairwise alignment was improved manually by minor editing based on the secondary structure predictions as well, and the energy-driven modeling option was used in order to allow energy minimization using the force-field OPLS-2005. The initial structure for Rad50 binding domain (RBD) of ScMre11, spanning residues 443–526, was obtained by homology modeling based on the crystal coordinates of the RBD of CtMre11 in complex with Rad50 (PDB ID: 5DA9) (20). Since the sequence identity between the two proteins is limited in this domain, the pairwise alignment obtained using the Prime homology module was improved manually by minor editing based on the secondary structure predictions and imposing conservation of the relevant Rad50 interacting surface residues. The disordered connecting regions between the first model and the RBD model were designed ex novo by the energy-driven Prime homology module and then refined by Prime loops refinement module. The whole model of ScMre11, spanning residues 1–542, was then processed for molecular dynamics as a heterodimer with Rad50. A model for ScRad50 globular domain and initial coiled-coil regions, bound to ATP, was obtained by homology modeling as described above based on the crystal coordinates of CtRad50 (PDB ID: 5DA9), sharing 58% identity with ScRad50. An initial model of the tetrameric structure of ScMre11–Rad50 complex was obtained by superposition of Mre11 and Rad50 (ATP-bound) on the tetrameric complex structure from M. jannaschii (PDB ID: 5F3W) (51). This structure was then refined by molecular dynamics. A multiple alignment based on AL2CO structural conservation (52) of ScMre11 with Mre11 orthologs from Homo sapiens (PDB ID:3T1I), Schizosaccharomyces pombe (PDB ID:4FCX), Chaetomium thermophilum (PDB ID:4KYE and 5DA9 for RBD region), Methanococcus jannaschii (PDB ID:3AV0) and Pyrococcus furiosus (PDB ID:1II7) was built by UCSF Chimera software (http://www.cgl.ucsf.edu/chimera/).
Molecular dynamics
MD simulations were performed using gromacs 5.0.4 software package (www.gromacs.org) and the AMBER99 force field (53). During the simulations, the interactions between magnesium ions (Mg2+) and their coordinating residues were treated using non-bonded model with van der Waals and electrostatic terms present in the AMBER99 force field. Protein structures were soaked in a cubic box of SPC/E (Extended Single Point Charge) (54) water molecules and simulated using periodic boundary conditions. All protein atoms were at the distance of 1 nm from the box edges. The ionization state of residues was set to be consistent with physiological pH (7.0 pH ± 0.2). Na+ ions were added to all molecular systems as counterions to obtain electroneutral models. All systems were relaxed through a steepest descent minimization with a limit of 50,000 cycles. Temperature and pressure were equilibrated at 300 K and 1 atm with a NVT (isothermal-isochoric) simulation of 2 ns and a NPT (isothermal-isobaric) simulation of 100 ps, respectively. Productive MD simulations were carried out in the NPT ensemble at 300 K with a time step of 2 fs. Quality assessment of the MD simulations was performed by backbone root-mean-square deviation (RMSD) analysis (Supplementary Figure S1). Hydrogen bonds are computed and analyzed using the g_hbond tool of GROMACS. Hydrogen bonds are determined based on cutoffs for the angle Hydrogen-Donor-Acceptor (30°) and the distance donor-acceptor (3.5 Å). OH and NH groups are regarded as donors, O and N are acceptors. Salt bridges were analysed monitoring the distance between any of the oxygen atoms of acidic residues and the nitrogen atoms of basic residues along all the simulation. A salt bridge was considered formed if the distance between oxygen and nitrogen is within 4 Å.
RESULTS
Identification of mre11 alleles that suppress the DNA damage hypersensitivity of sae2Δ cells
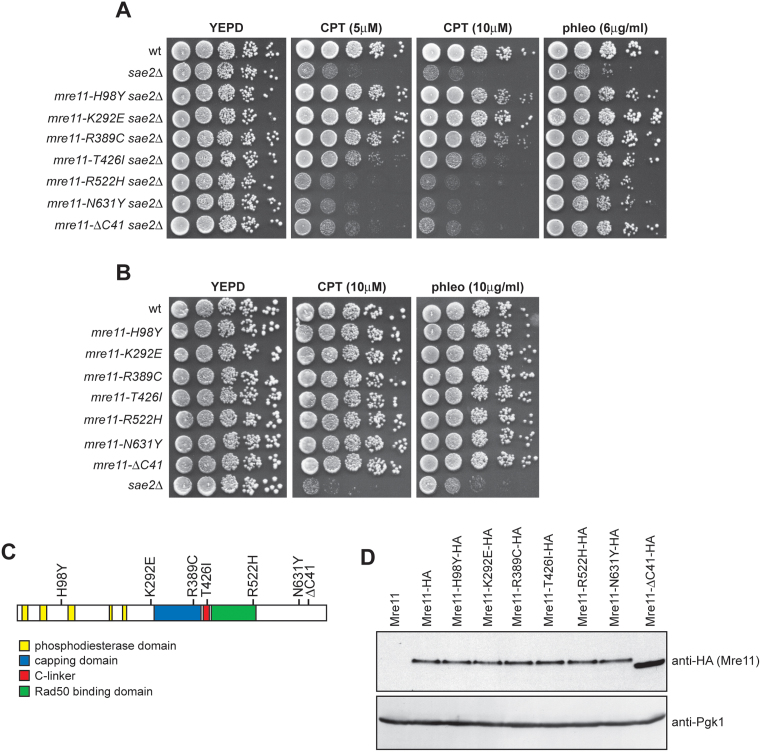
MRX and Sae2 are involved in resection of DSB ends and in maintaining the DSB ends tethered to each other (4). In order to understand how these diverse Sae2-MRX functions contribute to DNA damage resistance, we used low-fidelity PCR to random mutagenize the MRE11 gene, and then searched for mre11 alleles that suppressed the hypersensitivity of sae2Δ cells to different DNA damaging agents such as CPT and/or phleomycin. Linear MRE11 PCR products were transformed into sae2Δ cells in order to replace the corresponding MRE11 wild type sequence with the mutagenized DNA fragments (see Materials and Methods for details). Transformant clones showing higher viability than the untransformed strain in the presence of CPT and/or phleomycin were chosen for further characterization. Subsequent sequencing and genetic analysis identified 7 single mre11 mutations that suppressed the CPT and/or phleomycin sensitivity of sae2Δ cells (Figure 1A). Among them, the mre11-H98Y, mre11-K292E, mre11-R389C and mre11-T426I alleles suppressed the hypersensitivity of sae2Δ cells to both CPT and phleomycin, whereas the mre11-R522H, mre11-N631Y and mre11-ΔC41 alleles suppressed only the hypersensitivity of sae2Δ cells to phleomycin (Figure 1A). No mre11 alleles able to suppress only the sensitivity of sae2Δ cells to CPT have been identified. The inability of the Mre11-R522H, Mre11-N631Y and Mre11-ΔC41 variants to restore CPT resistance of sae2Δ cells is not due to hypersensitivity of the corresponding mutant cells to DNA damaging agents, as mre11-R522H, mre11-N631Y and mre11-ΔC41 cells did not show reduced survival to any tested drug in the presence of fully functional Sae2 (Figure 1B).
Figure 1.
mre11 mutations suppress the sensitivity of sae2Δ cells to CPT and/or phleomycin. (A, B) Exponentially growing cultures were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without CPT or phleomycin at the indicated concentrations. (C) Position of the amino acid changes in the Mre11 sequence. (D) Protein extracts prepared from exponentially growing cells were analyzed by western blot using anti-HA antibody. The filter was also hybridized with anti-Pgk1 antibodies as loading control.
Structural studies have shown that Mre11 can be divided into an N- and a C-terminal domain linked by an extended connecting loop (21,22,24) (see Supplementary Figure S2 for a structural alignment of Mre11 orthologs showing the positions of the conserved domains). Interestingly, the mutations restoring both CPT and phleomycin resistance cause amino acid changes in the Mre11 N-terminus, whereas the mutations restoring only phleomycin resistance are located in the Mre11 C-terminus coding region (Figure 1A and C), suggesting that these two Mre11 domains reflect distinct MRX functions. Consistent with this hypothesis, independent searches for mre11 mutations that bypass Sae2 function in CPT resistance by two other laboratories identified changes in H37, Q70, T74, L77, L89, E101 and P110 residues, all located in Mre11 N-terminus, as suppressors of the CPT sensitivity of sae2Δ cells (43,44). Curiously, our mre11 mutations did not come up in these screens, while we did not find the H37, Q70 and P110 substitutions that were identified independently in both the above screens (43,44).
With the exception of the mre11-ΔC41 allele, which results in the formation of a stop codon at position 651 leading to a truncated protein lacking 41 amino acids at the C-terminus, all the other Mre11 mutant variants carried single amino acid substitutions due to missense mutations. HA epitope tagging of the mutant proteins followed by western blot analysis revealed that the amount of Mre11-ΔC41 variant was higher than that of wild type Mre11, while the amount of all the other Mre11 mutant proteins were unaltered (Figure 1D).
Suppression of the CPT hypersensitivity of sae2Δ cells correlates with restored DSB resection
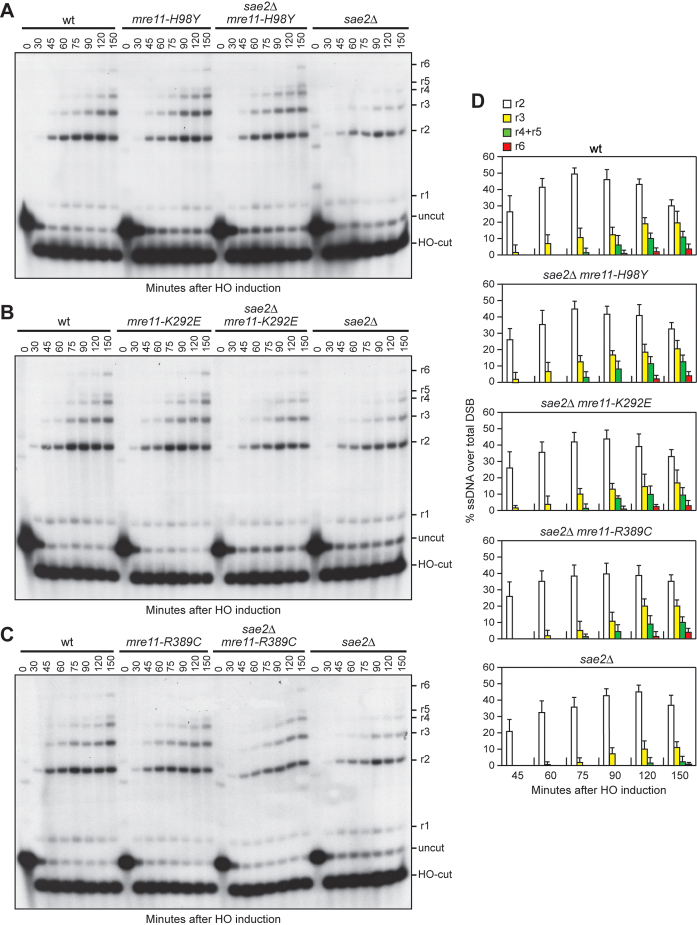
To investigate whether the above mutations also suppressed the DSB resection defect of sae2Δ cells, we introduced the mutations in a strain (JKM139), where expression of the HO endonuclease from a galactose-inducible promoter leads to the generation of a single DSB at the MAT locus (55). Due to the lack of the homologous donor sequences HML and HMR, this DSB cannot be repaired by HR. The extent of suppression in the derivative strains was similar to that observed in W303 background (Supplementary Figure S3). Because ssDNA cannot be cleaved by restriction enzymes, we directly monitored ssDNA formation at the irreparable HO-cut by following the loss of SspI restriction fragments. Southern blot analysis was performed under alkaline conditions, using a single-stranded probe that anneals to the 3′ end at one side of the break. As the mre11-T426I mutation suppressed the DNA damage hypersensitivity of sae2Δ only slightly and the mre11-ΔC41 allele increased the corresponding protein level, only the mre11-H98Y, mre11-K292E, mre11-R389C, mre11-R522H and mre11-N631Y mutations were examined.
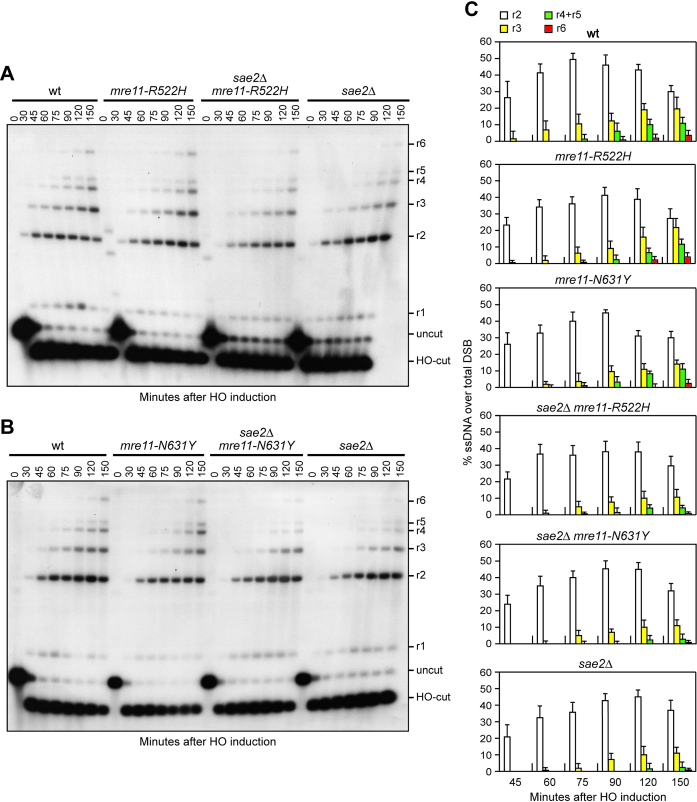
Resection of the HO-induced DSB occurred more efficiently in sae2Δ mre11-H98Y, sae2Δ mre11-K292E and sae2Δ mre11-R389C cells compared to sae2Δ cells (Figure 2A–D), indicating that Mre11-H98Y, Mre11-K292E and Mre11-R389C suppress the resection defect caused by the lack of Sae2. By contrast, the presence of the mre11-R522H and mre11-N631Y alleles did not suppress the resection defect of sae2Δ cells (Figure 3A–C). Rather, mre11-R522H and mre11-N631Y cells showed a very slight resection defect in the presence of wild type Sae2 (Figure 3A–C). As the Mre11-H98Y, Mre11-K292E and Mre11-R389C mutant proteins also restored CPT resistance in sae2Δ cells, whereas Mre11-R522H and Mre11-N631Y did not, suppression of sae2Δ CPT sensitivity appears to correlate with restored DSB resection.
Figure 2.
Mre11-H98Y, Mre11-K292E and Mre11-R389C suppress the resection defect of sae2Δ cells. (A–C) DSB resection. YEPR exponentially growing cell cultures were transferred to YEPRG at time zero. SspI-digested genomic DNA separated on alkaline agarose gel was hybridized with a single-stranded MAT probe that anneals with the unresected strand. 5′-3′ resection progressively eliminates SspI sites, producing larger SspI fragments (r1 through r6) detected by the probe. (D) Densitometric analysis. The experiment as in (A–C) was independently repeated and the mean values are represented with error bars denoting S.D. (n = 3).
Figure 3.
Mre11-R522H and Mre11-N631Y do not suppress the resection defect of sae2Δ cells. (A, B) DSB resection. YEPR exponentially growing cell cultures were transferred to YEPRG at time zero. SspI-digested genomic DNA separated on alkaline agarose gel was treated as in Figure 2. (C) Densitometric analysis. The experiment as in (A, B) was independently repeated and the mean values are represented with error bars denoting S.D. (n = 3).
Mre11-H98Y, Mre11-K292E and Mre11-R389C require Sgs1-Dna2 to suppress the DNA damage hypersensitivity of sae2Δ cells
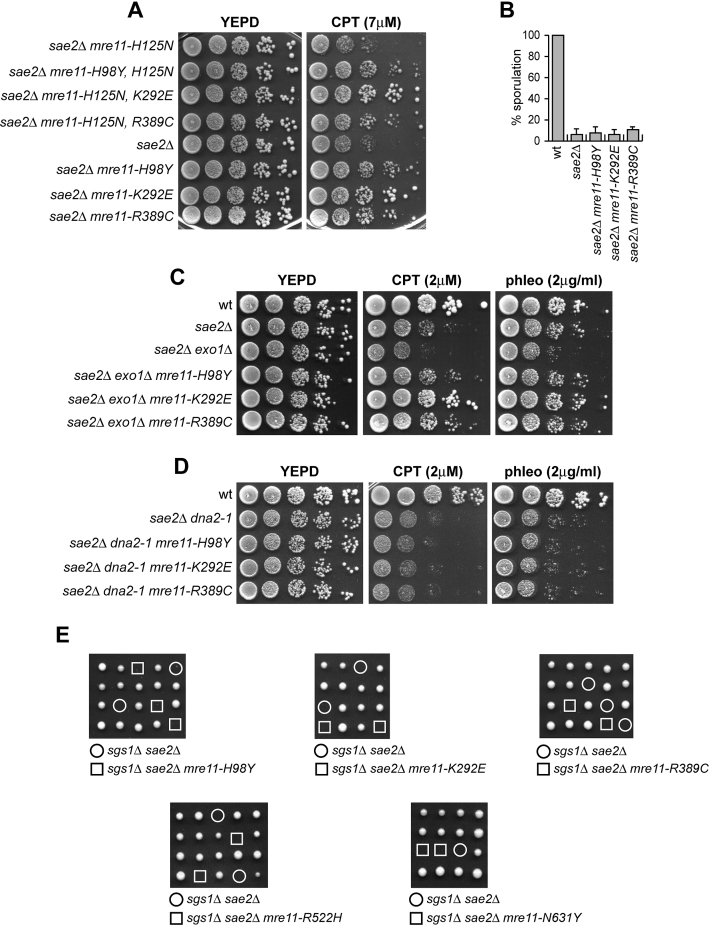
Sae2 activates the Mre11 endonuclease to incise the 5′ strand, followed by resection from the nick by Exo1 and Sgs1-Dna2 in a 5′-3′ direction (26–28). In addition, MRX promotes the binding of Exo1 and Sgs1-Dna2 at the DSB ends independently of Mre11 nuclease activity and Sae2 (36). Thus, Mre11-H98Y, Mre11-K292E and Mre11-R389C could restore DSB resection in sae2Δ cells either because they possess a Sae2-independent endonuclease activity or because they potentiate the Exo1 and/or Sgs1-Dna2 downstream nucleases. To distinguish between these possibilities, first we asked whether the suppressor effect of the Mre11 variants was eliminated by abolishing Mre11 nuclease activity through a point mutation changing His125 to Asn in one of the Mre11 phosphoesterase motifs (10). As shown in Figure 4A, the Mre11-H98Y, Mre11-K292E and Mre11-R389C variants also carrying the H125N amino acid substitution still suppressed the hypersensitivity to genotoxic agents of sae2Δ cells, indicating that suppression does not require Mre11 nuclease activity.
Figure 4.
Suppression of sae2Δ DNA damage sensitivity by Mre11-H98Y, Mre11-K292E and Mre11-R389C depends on Sgs1-Dna2. (A, C, D) Exponentially growing cultures were serially diluted (1:10) and each dilution was spotted out onto YEPD plates with or without CPT or phleomycin at the indicated concentrations. (B) Sporulation efficiency of diploid strains homozygous for the indicated mutations. Data are expressed as percentage of sporulation relative to wild type that was set up at 100%. Plotted values are the mean values with error bars denoting S.D. (n = 3). (E) Meiotic tetrads were dissected on YEPD plates that were incubated at 25°C, followed by spore genotyping.
The endonucleolytic cleavage catalyzed by MRX-Sae2 is absolutely required to initiate resection at meiotic DSBs, whose 5′-terminated strands are not accessible to exonucleases because they are covalently bound by the topoisomerase-like protein Spo11 (9,31,37). Consistent with the finding that Mre11-H98Y, Mre11-K292E and Mre11-R389C do not bypass the requirement for Sae2 to activate Mre11 nuclease, the mre11-H98Y, mre11-K292E and mre11-R389C alleles were unable to suppress the sporulation defect of sae2Δ/sae2Δ diploid cells (Figure 4B), suggesting that they fail to remove Spo11 from meiotic DSBs.
We then asked whether the mre11-H98Y, mre11-K292E and mre11-R389C suppression effects might depend on Dna2 and/or Exo1. Although the lack of Exo1 exacerbated the sensitivity to DNA damaging agents of sae2Δ cells, sae2Δ exo1Δ mre11-H98Y, sae2Δ exo1Δ mre11-K292E and sae2Δ exo1Δ mre11-R389C triple mutant cells were more resistant to genotoxic agents than sae2Δ exo1Δ double mutant cells (Figure 4C), indicating that the suppressor effect of the analyzed mre11 alleles is Exo1-independent. By contrast, Mre11-H98Y, Mre11-K292E and Mre11-R389C failed to suppress the sensitivity to DNA damaging agents of sae2Δ cells carrying the hypomorphic dna2–1 allele (Figure 4D), suggesting that their suppressor effect requires Dna2 activity.
The lack of Sgs1 impairs viability of sae2Δ cells even in the absence of genotoxic agents. This synthetic lethality is likely due to defects in DSB resection, as it is suppressed by either EXO1 overexpression or KU deletion (56). Consistent with a requirement of Sgs1-Dna2 for the bypass of Sae2 function, tetrad dissection of diploids heterozygous for sae2Δ, sgs1Δ and the mre11 mutations under analysis did not allow to find viable sae2Δ sgs1Δ mre11-H98Y, sae2Δ sgs1Δ mre11-K292E or sae2Δ sgs1Δ mre11-R389C spores (Figure 4E), indicating that Mre11-H98Y, Mre11-K292E and Mre11-R389C do not restore viability of sae2Δ sgs1Δ cells. Altogether, these findings indicate that Mre11-H98Y, Mre11-K292E and Mre11-R389C require Sgs1-Dna2 to restore DNA damage resistance in sae2Δ cells.
Mre11-H98Y, Mre11-K292E and Mre11-R389C relieve Rad9-mediated inhibition of Sgs1-Dna2 activity
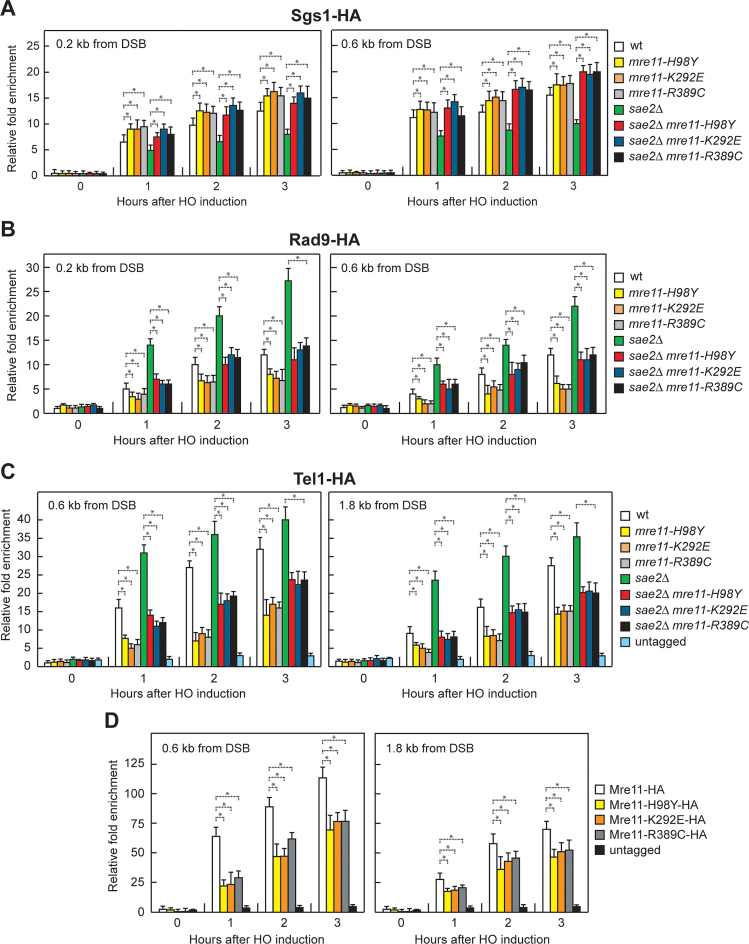
The lack of Sae2 increases Rad9 association to DSBs, which in turn inhibits the resection activity of Sgs1-Dna2 by restricting the access of Sgs1 to the DSB ends (40,41). The DNA damage sensitivity and the resection defect of Sae2-deficient cells are suppressed by both the lack of Rad9 and the expression of a hypermorphic Sgs1 variant that escapes Rad9-mediated inhibition (40,41). We then asked whether Mre11-H98Y, Mre11-K292E and Mre11-R389C potentiate Sgs1-Dna2 by lowering Rad9 association to DSBs. As expected (40), the decrease in Sgs1 association to DSBs in sae2Δ cells was concomitant with an increased Rad9 persistence at DSBs (Figure 5A and B). The mre11-H98Y, mre11-K292E and mre11-R389C mutations increased Sgs1 association (Figure 5A) and decreased Rad9 association at DSBs (Figure 5B) both in the presence and in the absence of Sae2. Together with the observation that suppression of sae2Δ hypersensitivity to genotoxic agents requires Sgs1-Dna2, these findings strongly suggest that Mre11-H98Y, Mre11-K292E and Mre11-R389C restore both DNA damage resistance and DSB resection in sae2Δ cells by decreasing Rad9 association close to the DSB, and therefore by relieving the inhibition of Sgs1-Dna2 resection activity.
Figure 5.
Effects of H98Y, K292E and R389C substitutions on Sgs1, Rad9, Tel1 and Mre11 association to DSBs. (A–D) ChIP analysis. Exponentially growing YEPR cell cultures were transferred to YEPRG at time zero. Relative fold enrichment of the indicated fusion proteins at the indicated distances from the HO cleavage site was determined after ChIP with anti-HA antibody and subsequent qPCR analysis. Plotted values are the mean values with error bars denoting S.D. (n = 3). *P< 0.05 (Student's t-test).
Mre11-H98Y, Mre11-K292E and Mre11-R389C bind poorly to DSBs and decrease Tel1 association to DSBs
The lack of Sae2 enhances Tel1 signaling activity (38,39), which is responsible for the increased Rad9 association at DSBs in sae2Δ cells (42). Thus, we asked whether the low Rad9 association to DNA ends in mre11-H98Y, mre11-K292E and mre11-R389C cells is due to decreased Tel1 binding to the same DNA ends. Indeed, the mre11-H98Y, mre11-K292E and mre11-R389C mutations decreased the amount of Tel1 bound at the HO-induced DSB both in the presence and in the absence of Sae2 (Figure 5C), suggesting that Mre11-H98Y, Mre11-K292E and Mre11-R389C reduce Rad9 association/persistence to DSBs by impairing Tel1 recruitment to the same DNA ends.
MRX is required for loading Tel1 to DSBs through a direct interaction between Tel1 and Xrs2 (57–59). Thus, the mre11-H98Y, mre11-K292E and/or mre11-R389C mutations might reduce Tel1 association/persistence to DSBs either because they impair Mre11-Xrs2 interaction or because the corresponding mutant MRX complexes are poorly recruited to DSBs. Similar amount of Xrs2-Myc could be detected in immunoprecipitates of HA-tagged Mre11, Mre11-H98Y, Mre11-K292E and Mre11-R389C (Supplementary Figure S4), strongly suggesting that the reduced Tel1 association at DSBs is not due to defective Mre11-Xrs2 interaction. Rather, the amount of Mre11-H98Y, Mre11-K292E and Mre11-R389C associated to the HO-induced DSB turned out to be lower than that of wild type Mre11 (Figure 5D). Thus, these Mre11 variants appear to reduce Tel1 association at DSBs because they are poorly recruited to DNA ends. As a consequence, the reduced Tel1 binding leads to decreased Rad9 recruitment at the DSB ends, and therefore to the relieve of Rad9-mediated inhibition of Sgs1-Dna2 activity, which can compensate for the lack of Sae2 in DNA damage resistance and resection.
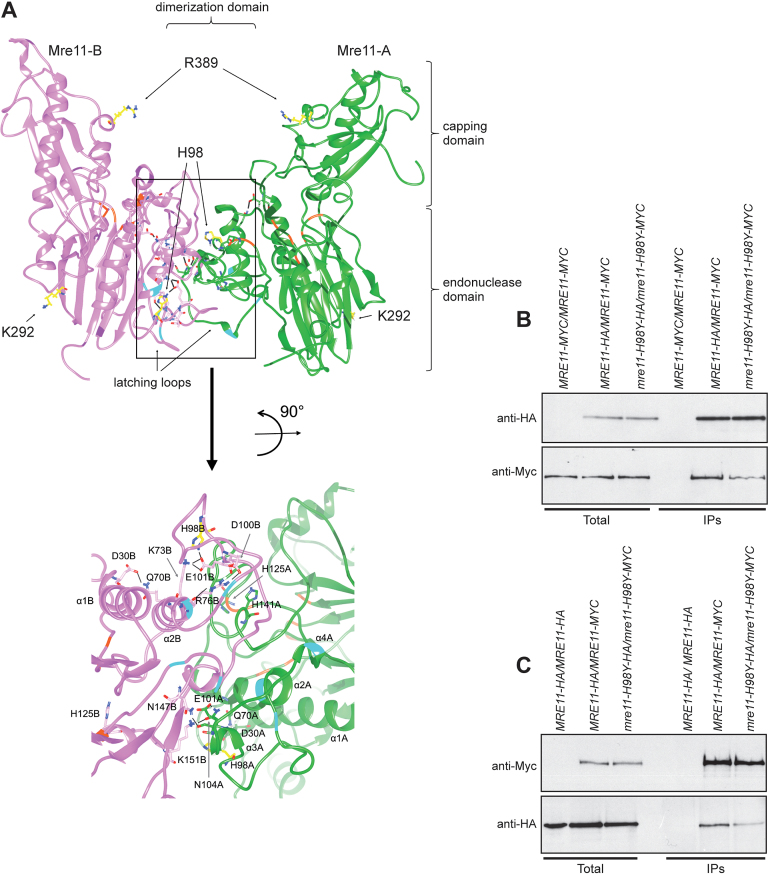
Mre11-H98Y affects Mre11 dimer formation
To map the location of the H98Y, K292E and R389C amino acid substitutions within the Mre11 structure, we used the tertiary structure of the S. pombe Mre11 (SpMre11) counterpart (49) as a template to generate a preliminary molecular model of S. cerevisiae Mre11 (ScMre11), which was refined further by a molecular dynamic simulation (500ns) (see Materials and Methods for details). Ensuing analyses indicated that the altered residues are located in three different regions that are not in mutual close proximity: H98 is located at the interface of the two ScMre11 subunits, K292 is situated on the other side of the protein surface respect to H98, and R389 is in the capping domain (Figure 6A). Subsequently, a model for the ScMre11–Rad50 heterotetrameric structure was refined by molecular dynamics (Supplementary File 1), and then the ScMre11 homodimer structure obtained by this simulation was analysed further by molecular dynamics (Supplementary File 2). These analyses allow to construct a model for the Mre11-Mre11 interface, comprising the latching loops (aa 80–118) together with α1 and α2 helices and the mostly disordered regions spanning residues 130–151 of each monomer, stabilizing the Mre11 dimeric structure and constituting an intermolecular dimerization domain (Figure 6A). This domain was largely uncharacterized due to the lack of structural information on the disordered regions, which are hardly resolved by crystallography. We mapped all the interactions of H98 and K292 residues with the rest of the dimeric ScMre11 protein, monitoring the persistence of hydrogen bonds and salt bridges (Supplementary Figure S5) (Mre11 monomers are indicated as A and B). The two ScMre11 monomers did not show a symmetrical behavior. In particular, both H98 and K292 establish a persistent intra-chain hydrogen bond during the entire simulation, with H98 contacting Y99 and E101 in Mre11-A and Mre11-B, respectively (Supplementary Figure S5A), whereas K292 interacting with D284 in both Mre11 monomers (Supplementary Figure S5B). Moreover, H98 is involved in a salt bridge with E101-B (Supplementary Figure S5C) and it also establishes an inter-chain hydrogen bond with L134-A (Supplementary Figure S5A). Altogether, these findings suggest that a complex network of dynamic interactions exists between residues within the dimerization domain and that mutations impairing some of these interactions would improve the mobility of the disordered regions, likely affecting the stability of the dimerization interface.
Figure 6.
Mre11-H98Y alters Mre11 dimer formation. (A) Structural prediction of S. cerevisiae Mre11 dimer, obtained by homology modelling and refined by molecular dynamics. The two Mre11 monomers are shown in green (Mre11-A) and pink (Mre11-B). The position of endonuclease, capping and dimerization domains are shown. The lateral chain is shown for the residues (in yellow) whose mutations were described in the text. A close-up view of the dimerization domain is shown at the bottom. On the ribbon, positions involved in direct interaction with Xrs2 are in cyan, while positions constituting the active site are in orange. Black bars represent the hydrogen bonds/salt bridges existing in this particular conformation. (B, C) Mre11 dimer formation. Protein extracts prepared from exponentially growing diploid cells with the indicated genotypes were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (Total) or after immunoprecipitation (IPs) with anti-HA antibody (B) or anti-Myc antibody (C).
While the role of K292 is difficult to envisage, these observations predicted that H98 is directly implicated in the stabilization of Mre11 dimerization domain (Figure 6A), and its substitution with Y might lower the affinity between the two Mre11 subunits by disrupting hydrogen bonds at the dimerization interface. We directly tested this prediction by immunoprecipitating Mre11 with anti-HA or anti-Myc antibody from protein extracts of MRE11-HA/MRE11-MYC and mre11-H98Y-HA/mre11-H98Y-MYC diploid cells. The amount of Mre11-H98Y-Myc detected by anti-MYC antibodies in immunoprecipitates of Mre11-H98Y-HA was reduced compared to that of wild type Mre11-Myc detected in immunoprecipitates of Mre11-HA (Figure 6B). Furthermore, the amount of Mre11-H98Y-HA detected by anti-HA antibodies in immunoprecipitates of Mre11-H98Y-Myc was reduced compared to that of wild type Mre11-HA detected in immunoprecipitates of Mre11-Myc (Figure 6C). As Mre11 dimer formation is required for MRX association to DSBs (15), this defect in the stabilization of the Mre11 dimer interface could explain the decreased Mre11-H98Y association to DSBs.
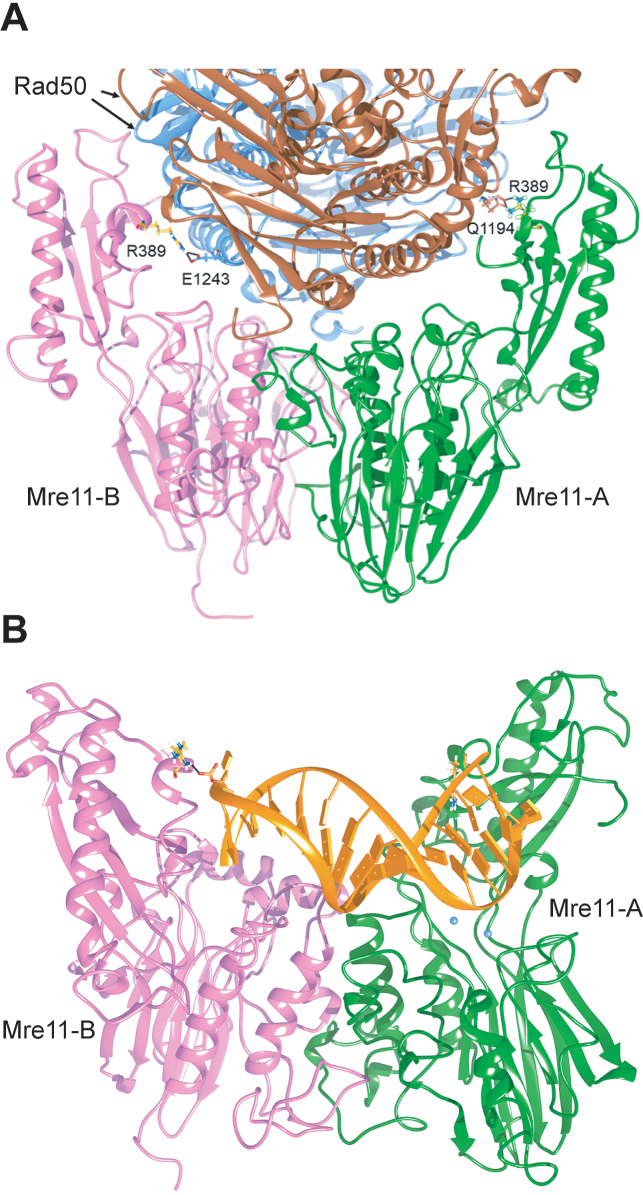
R389 mediates the interactions of Mre11 capping domain with both Rad50 and DNA
Residue R389 of Mre11 is localized in the capping domain, facing Rad50 in the ScMre11–Rad50 heterotetramer (Figure 7A). Again, the interaction is not symmetric, involving different residues (E1243 or Q1194) on the different Rad50 subunits. The dynamics of ScMre11 homodimer indicate that R389-A and R389-B do not show any functionally relevant interaction with the rest of the dimeric system (Supplementary Figure S5D and E), although R389-B is involved in a persistent salt bridge (Supplementary Figure S5C). On the other hand, due to the localization of this residue in the capping domain, which has been reported to be involved in the interaction with branched DNA in Pyrococcus furiosus Mre11 (PfMre11) crystal structure (15), R389 could also be involved in DNA binding. In fact, R389 is one of a pair of arginine residues (the other being R390) exposed in the correct position to interact with DNA, at least as inferred by superposition with DNA branched bound-PfMre11 structure (PDB: 3DSD) (15). To test this hypothesis, we performed a MD simulation of 300 ns by including a dsDNA in the model, in order to reconstruct a structure that could reproduce the conformation reached after ATP hydrolysis by Rad50 but before resection of DNA ends. DNA has been added on the ScMre11 complex using the structure of Methanococcus jannaschii Mre11 (MjMre11) dimer bound to a dsDNA end (PDB: 4TUG) (60) as a template. During the simulation, dsDNA binds to residues (such as K62 and N24) that correspond to positions already reported to interact with dsDNA in PfMre11 (15). Interestingly, R389, which was not previously identified as a possible DNA binding site due to the difficulty in aligning the capping domains of PfMre11 and eukaryotic Mre11 (15), forms hydrogen bonds with the phosphate groups of DNA in both Mre11 subunits, with R389 of subunit B contacting the phosphodiesteric bridge of the 3′ terminus (Figure 7B). The R389C substitution, which removes the arginine positive charge, would impair the DNA‐binding interface on the capping domain of Mre11 due to the loss of the attractive force between the arginine and the phosphate group.
Figure 7.
R389 at the Mre11–Rad50 and Mre11-DNA interfaces. (A) Residue R389 of Mre11 is localized at the interface between Mre11 and Rad50 in the ScMre11–Rad50 heterotetramer. The position of R389 (in yellow) is shown on each Mre11 monomer constituting a tetramer with Rad50 subunits (in light blue and brown). The interaction is not symmetric, involving different residues (E1243 or Q1194) on the different Rad50 subunits. (B) Interaction of R389 (in yellow) of dimeric ScMre11 (in pink and green) with dsDNA (in orange). Black line indicates the salt bridge between R389 and the phosphodiesterase bridge at the 3′ terminus of the dsDNA end. Mg2+ ions in the active site in one monomer are indicated in light blue.
Mre11-R522H bypasses Sae2 function in end-tethering and HR
The mre11-R522H and mre11-N631Y mutations restore phleomycin resistance of sae2Δ cells but neither CPT resistance nor DSB resection (Figures 1A and 3A–C), suggesting that different Sae2 functions are involved in survival to CPT and phleomycin treatment. Consistent with this finding, Mre11-R522H and Mre11-N631Y did not cause a decrease of Rad9 (Supplementary Figure S6A) and Tel1 association to DSBs (Supplementary Figure S6B). Furthermore, they failed to restore viability of sae2Δ sgs1Δ cells (Figure 4E), possibly because they did not restore Sae2 function in DSB resection.
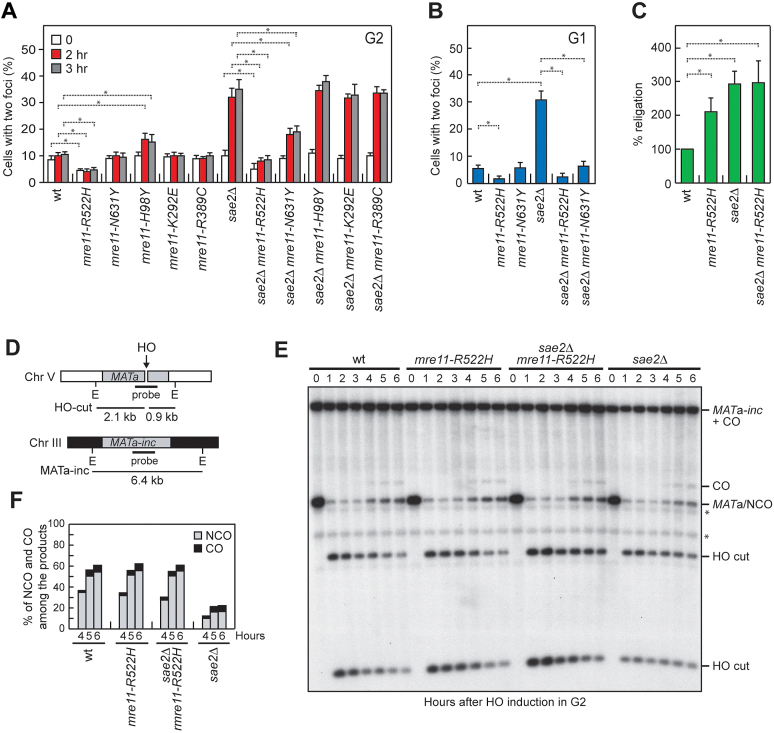
In addition to promoting DNA-end resection, MRX and Sae2 are also required to maintain the DSB ends adjacent to each other to facilitate DSB repair by both HR and NHEJ (45,46,61,62). Thus, we investigated whether Mre11-R522H and Mre11-N631Y can suppress the DSB end-tethering defect of sae2Δ cells. We used a yeast strain where the DNA proximal to an irreparable HO-inducible DSB can be visualized by binding of a LacI-GFP fusion protein to multiple repeats of the LacI repressor binding site that is integrated at a distance of 50 kb on both sides of the HO cut site (61). HO expression was induced by galactose addition to cell cultures that were arrested in G2 with nocodazole and kept blocked in G2 by nocodazole treatment in order to ensure that all cells would arrest in metaphase. Most wild type cells showed a single LacI-GFP focus after HO induction, indicating their ability to maintain the broken DNA ends together (Figure 8A). Consistent with previous results (45,46), sae2Δ cells showed an increase of cells with two LacI-GFP spots after HO induction compared to wild type cells (Figure 8A). Strikingly, the amounts of cells showing two LacI-GFP spots after HO induction was decreased in both sae2Δ mre11-R522H and sae2Δ mre11-N631Y compared to sae2Δ cells, with mre11-R522H showing the strongest effect (Figure 8A). By contrast, the mre11-H98Y, mre11-K292E and mre11-R389C alleles did not suppress the end-tethering defect of sae2Δ cells (Figure 8A), possibly because of a reduced MRX association to DSBs.
Figure 8.
Mre11-R522H suppresses the end-tethering and the HR defects of sae2Δ cells. (A, B) DSB end-tethering. Exponentially growing YEPR cell cultures were arrested in G2 with nocodazole (A) or in G1 with α-factor (B) at time zero and transferred to YEPRG in the presence of nocodazole or α-factor, respectively. 200 cells for each strain were analyzed to determine the percentage of cells showing two LacI-GFP foci. Plotted values are the mean values with error bars denoting S.D. (n = 3). *P< 0.05 (Student's t-test). (C) Plasmid re-ligation assay. Cells were transformed with BamH1-linearized or uncut pRS316 plasmid DNA. Data are expressed as percentage of re-ligation relative to wild type that was set up at 100% after normalization to the corresponding transformation efficiency with the uncut plasmid. Plotted values are the mean values with error bars denoting S.D. (n = 3). *P< 0.05 (Student's t-test). (D) System to detect ectopic recombination. HO generates a DSB at a MATa DNA sequence inserted on chromosome V, while the homologous MATa-inc region on chromosome III cannot be cut by HO and is used as a donor for HR-mediated repair. E, EcoRI. (E) YEPR cell cultures arrested in G2 with nocodazole were transferred to YEPRG at time zero in the presence of nocodazole. Southern blot analysis of EcoRI-digested genomic DNA with the MATa probe depicted in D. * indicates cross hybridization signals. (F) Densitometric analysis of CO versus NCO repair bands at the indicated times after HO induction.
To confirm suppression of the end-tethering defect of sae2Δ cells by Mre11-R522H and Mre11-N631Y, we evaluated the frequency of two LacI-GFP foci after HO expression in α-factor-arrested cells that were kept arrested in G1 by α-factor in the presence of galactose. About 30% of G1-arrested sae2Δ cells showed two LacI-GFP foci 1 hour after HO induction and this frequency dramatically decreased in sae2Δ mre11-R522H and sae2Δ mre11-N631Y double mutant cells (Figure 8B).
Interestingly, the percentage of mre11-R522H cells showing two LacI-GFP spots was decreased compared to wild type cells even in the presence of functional Sae2 (Figure 8A and B), indicating that the MR522HRX complex possesses increased efficiency of DNA tethering. Consistent with a higher degree of tethering activity, mre11-R522H mutant cells were more efficient than wild type cells in re-ligating a plasmid that was linearized before being transformed into the cells (Figure 8C).
The maintenance of the DSB ends tethered to each other is important to repair a DSB by both NHEJ and HR (44–46,48). As both sae2Δ and mre11-R522H sae2Δ cells exhibit increased NHEJ frequency possibly due to reduced DSB resection (Figure 8C), it is unlikely that the restored end-tethering in mre11-R522H sae2Δ and mre11-N631Y sae2Δ cells may lead to phleomycin resistance by increasing NHEJ efficiency. Thus, we tested the effect of these mutations on the ability of sae2Δ cells to repair a DSB by HR.
In the canonical HR pathway, the 3′-ended ssDNA tail invades an intact duplex homologous, creating a loop structure (D-loop). The displaced ssDNA can then anneal with the complementary sequence on the other side of the DSB to generate a double Holliday junction, whose random cleavage yield to noncrossover (NCO) and crossover (CO) products (1,2). Alternatively, if the ssDNA strand is displaced by the D-loop, its annealing with the 3′ ssDNA end at the other end of the DSB leads to the generation of NCO products in a process called synthesis-dependent strand-annealing (SDSA) (1,2). As the end-tethering activity of MRX was shown to be particularly important for SDSA-mediated DSB repair (48), we monitored CO and NCO formation in sae2Δ cells. We used a strain that carries a MATa gene on chromosome V that can be cleaved by a galactose-inducible HO endonuclease and repaired by using an uncleavable MATa (MATa-inc) gene on chromosome III as template (Figure 8D) (63). This repair event was shown to generate NCO and CO outcomes, with COs being ∼5% among the overall repair events (63). Galactose was added to induce HO production and then it was maintained in the medium to cleave the HO sites that were eventually reconstituted by NHEJ. Wild type and sae2Δ cells appeared to have similar percentage of COs (Figure 8E and F). By contrast, the 3 kb MATa band resulting from NCO recombination events re-accumulated less efficiently in sae2Δ cells compared to wild type cells, while the mre11-R522H mutation restored wild type levels of NCO products in sae2Δ cells (Figure 8E and F). As most NCO products are generated by the SDSA mechanism and the end-tethering activity is important to support DSB repair by SDSA (48), this finding suggests that the restored end-tethering conferred by Mre11-R522H can suppress the sae2Δ phleomycin sensitivity by increasing the efficiency of SDSA-mediated DSB repair.
R522 contributes to the stabilization of the Mre11–Rad50 binding domain
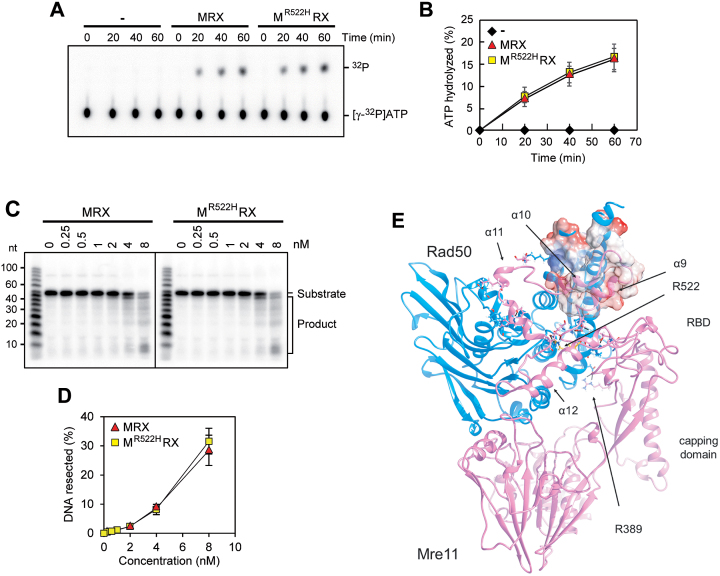
Because Mre11-R522H showed the strongest effect in the bypass of the end-tethering Sae2 function and possessed an increased end-tethering activity by itself, we analyzed the effect of this mutation at biochemical and structural levels. The R522 residue is located in the Rad50 binding domain (RBD) that, together with the capping domain, mediates the interaction between Mre11 and ATP-bound Rad50 (21). ATP hydrolysis by Rad50 induces the change from a closed MRX conformation, required for end-tethering, to an open configuration that promotes Mre11 nuclease activity (25). After ATP hydrolysis, the Mre11 capping domain dissociates from Rad50, leaving the RBD as the only Mre11–Rad50 interface (21). We then expressed the Mre11-R522H protein in insect cells and purified it to near homogeneity (Supplementary Figure S7A) to test the purified mutant protein for interaction with Rad50 and Xrs2. As shown in Supplementary Figure S7B, Mre11-R522H retained the ability to associate with Rad50 and Xrs2 and, importantly, the MR522HRX complex showed normal ATPase activity (Figure 9A and B) and nuclease activity (Figure 9C and D). These results are consistent with the very minor resection defect of mre11-R522H cells (Figure 3A and C).
Figure 9.
Functional and structural characterization of Mre11-R522H. (A, B) ATP hydrolysis by wild type MRX or MR522HRX mutant (100 nM of each) in the presence of 100-bp dsDNA (200 nM) (A). The results from three independent experiments were quantified (B). The mean values are represented with error bars denoting S.D. (n = 3). (C, D) The nuclease activity of wild type MRX or MR522HRX complex was tested on the 5′-labeled 95 bp dsDNA (8 nM) with 5 nt 3′ overhangs and a nick 55 nt away from the labeled end. The results were quantified, based on the product at the gel bottom, with error bars denoting S.D. (n = 3). (E) A model for Mre11 comprising Rad50 binding domain (RBD) was optimized by molecular dynamics. The position of residue R522 (in yellow) and of residues involved in Mre11–Rad50 interaction are shown. The Coulomb potential surface (red, negative charge; white, hydrophobic surface; blue, positive charge) is represented for the coiled-coiled region of Rad50 and Mre11 α9 and α10, which mainly share hydrophobic interactions.
As ScMre11 RBD structure was not available, we obtained a model for a heterodimer ScMre11–Rad50 comprising Mre11 RBD by molecular dynamics (Figure 9E) (see Materials and Methods for details). The energetically favored structure for ScMre11 RBD was actually not identical to the Mre11 structure resolved for the thermophilic filamentous fungus Chaetomium thermophilum (PDB ID: 5DA9). ScMre11 RBD binds not only to the Rad50 coiled-coiled region but also to the globular domain (Figure 9E). Two helices, α9 and α10, take contact with Rad50 coiled-coil region, mainly via hydrophobic interactions, while a third helix, α11, interacts with the globular region of Rad50. A fourth helix, α12, is involved in stabilization of the structure, mainly by residue R522 indeed, which strongly binds to both E443 (at the beginning of α9 helix) and E494 (at the end of α11 helix). The R522H amino acid substitution would allow maintenance of only a few of these interactions, conferring more mobility to the whole RBD. Consistent with the finding that Mre11-R522H is still capable to associate with Rad50 (Supplementary Figure S7B), the increased RBD mobility is not sufficient to impair the association between Mre11 and ATP-bound Rad50, which in fact depends on both the RBD and the capping domains. However, it could lead to a slight destabilization of the Mre11–Rad50 interaction only after ATP hydrolysis, when the RDB remains the sole Mre11–Rad50 interface. As a consequence, a defect in stabilizing the interaction between Mre11 and ADP-bound Rad50 might lead to increased association at DSBs of MRX in its ATP-bound conformation, and therefore to a more efficient end tethering.
DISCUSSION
Here we identified and characterized mre11 mutations that suppress the hypersensitivity to phleomycin and/or CPT of sae2Δ cells. The mre11-H98Y, mre11-K292E and mre11-R389C mutations, which restore resistance to both CPT and phleomycin, also increase the resection efficiency of sae2Δ cells. By contrast, the mre11-R522H and mre11-N631Y mutations, which restore resistance only to phleomycin, bypass Sae2 function in end-tethering but not in end-resection. These findings indicate that an end-tethering defect by itself is not sufficient to account for the CPT hypersensitivity of sae2Δ cells, whereas Sae2 function in end-resection can be dispensable to repair phleomycin-induced DNA lesions.
CPT cytotoxicity is due to the reversibly trapping of Top1 on nicked DNA intermediates (47). The endonucleolytic activity of MRX has been implicated in the nucleolytic removal of Top1-DNA adducts (64–67). Our findings that Mre11-H98Y, Mre11-K292E and Mre11-R389C do not bypass the requirement for Sae2 in either Spo11 removal or activation of Mre11 endonuclease strongly suggest that the suppression of sae2Δ CPT sensitivity is not due to a more efficient endonucleolytic removal of Top1ccs. Rather, Mre11-H98Y, Mre11-K292E and Mre11-R389C were found to increase the resection efficiency of sae2Δ cells by potentiating the Sgs1-Dna2 resection machinery. Since DNA replication and/or transcription can convert the Top1ccs that are reversibly trapped by CPT into DSBs and/or unusual replication intermediates (47,68), both of which require homologous recombination to be repaired/resolved, a more efficient DNA-end resection by the above mutant variants could restore CPT resistance in sae2Δ cells by facilitating the repair of these secondary DNA lesions.
By contrast, Sae2-MRX function in end-tethering appears to be more important to repair phleomycin-induced rather than CPT-induced DNA lesions. As phleomycin leads directly to DNA cleavage and DSB repair by HR has been shown to require limited amount of ssDNA at DSB ends (69,70), the amount of ssDNA generated in sae2Δ cells may be sufficient to repair by HR phleomycin-induced DSBs, whose DNA ends are readily accessible to the resection machinery. By contrast, a more efficient and fast resection could be required to repair by HR CPT-induced lesions, which are accessible to the resection nucleases only once DNA replication and/or transcription have processed the Top1-DNA cleavage complexes.
The primary cause of the resection defect of sae2Δ cells is a reduction of the Sgs1-Dna2 activity that is due to enhanced Rad9 binding to DSBs. Mre11-H98Y, Mre11-K292E and Mre11-R389C were found to restore DNA damage resistance and resection in sae2Δ cells by relieving Rad9-mediated inhibition of the Sgs1-Dna2 resection machinery. In fact, these mutant variants poorly bind to DNA ends and this defect causes decreased Tel1 association to DSBs. Consequently, diminished Tel1 binding leads to a decreased amount of Rad9 bound at DSBs and therefore to the relief of Sgs1-Dna2 inhibition.
Structural studies on Mre11–Rad50 complex from archaea and bacteria have shown that Mre11 has an extended structure that can be divided into two functional domains connected by a poorly structured linker (C-linker): a nuclease module at the N-terminus, which comprises the phosphodiesterase and the capping domains, and a Rad50-binding domain at the C-terminus that binds the Rad50 coiled-coils (21,22,24). Notably, the mre11 mutations restoring both CPT resistance and efficient DSB resection in sae2Δ cells are all located upstream of the C-linker domain. This observation is further strengthened by two other works that identified changes in amino acids (H37, Q70, T74, L77, L89, E101, P110) all located in Mre11 N-terminus as suppressors of the CPT sensitivity of sae2Δ cells (43,44). The common feature of all these mre11 mutations is a reduction of Mre11 association to DSBs (Figure 5D) (43,44). Structural studies by homology modeling and molecular dynamics of ScMre11–Rad50 indicate that the H98Y and R389C amino acid substitutions reduce Mre11 association to DSBs by different means. In particular, the H98Y substitution alters the formation of hydrogen bonds in the Mre11 dimer interface and reduces in vivo Mre11 dimer formation, which is known to facilitate MRX-DNA association (15). Interestingly, our inter-chain hydrogen bond analysis of the ScMre11 dimer interface indicates that other residues play similar roles, such as Q70 and E101, whose substitutions have been shown to bypass Sae2 function in CPT resistance possibly by lowering MRX association to DSBs (43,44). By contrast, the R389 residue is involved in mediating the interactions of the ScMre11 capping domain with both Rad50 and branched DNA. Substitution of R389 with C could alter the Mre11 DNA‐binding interface by losing the attractive force between the arginine and the phosphate group of DNA. In addition, it could overcome the need of a salt bridge rupture during the ATP-driven rotation of the Rad50 subunits, thus facilitating the conformational transition of Mre11 and Rad50 to an open configuration that possesses a lower DNA binding affinity (25). Consistent with this hypothesis, substitution of residues mediating the interaction between the Mre11 capping domain and Rad50 have been shown to facilitate the disengagement of Mre11 capping domain and disclosure of the Mre11 endonucleolytic site (21).
By contrast, the mre11-R522H and mre11-N631Y mutations, which are located at the C-terminus, restore resistance only to phleomycin and bypass Sae2 function in end-tethering but not in end-resection, suggesting that an end-tethering defect by itself is sufficient to account for the hypersensitivity of sae2Δ cells to phleomycin. Despite causing reduced end-tethering, the lack of Sae2 leads to increased NHEJ frequency (71) that can be explained by the reduced 5′ resection, as the extent of nucleolytic degradation can be an impediment for NHEJ. Therefore, it is unlikely that the increased end-tethering activity conferred by the mre11-R522H and mre11-N631Y mutations might suppress the phleomycin sensitivity of sae2Δ cells by increasing the NHEJ efficiency. Rather, we found that sae2Δ cells are specifically defective in DSB repair by SDSA and that the mre11-R522H mutation suppresses this defect. Given that the end-tethering function of MRX is particularly important for DSB repair by SDSA, possibly because it facilitates the annealing of the displaced strand to the other DSB end (48), the increased phleomycin resistance conferred by the mre11-R522H and mre11-N631Y mutations is likely due to a more efficient DSB repair by SDSA.
The Mre11-R522 residue is located in the RBD domain that, together with the capping domain, mediates the interaction between Mre11 and ATP-bound Rad50 (21). The conformational change induced by ATP hydrolysis leads to dissociation of the Mre11 capping domain from Rad50, which remains associated to Mre11 mainly through the RBD (21). We found that the R522H substitution affects neither MRX complex formation nor Rad50 ATPase activity, while our structural analysis reveals that it might confer more mobility to the whole RBD. Based on this finding and on the observation that Mre11-R522H possesses an increased tethering activity by itself, we speculate that the substitution of R522 with H might lead to a slight destabilization of the Mre11–Rad50 interaction that could be unmasked only after ATP hydrolysis, when the RBD becomes essential to maintain Mre11 fastened to Rad50 in the ADP-bound state. As a consequence, a defect in accommodating the ATP-hydrolysis-dependent conformational transitions of the complex could lead to increased association at DSBs of MRX in its ATP-bound conformation and therefore to a more efficient end tethering.
In summary, we provide evidence that two structurally distinct N-terminal and C-terminal domains of Mre11 have different roles in supporting the functions of the MRX complex in DNA damage resistance. Given the strong evolutionarily conservation of the MRX complex, these structure-function relationships might also apply to other systems including human cells and their understanding could improve our knowledge of the consequences of MRX dysfunctions in human diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to G. Lucchini and M. Clerici for critical reading of the manuscript and P. Cejka, J. Haber, G. Liberi and D.P. Toczyski for strains and plasmids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG 19783]; Progetti di Ricerca di Interesse Nazionale (PRIN) [2015 to M.P.L.]; US National Institutes of Health [RO1 ES015632 to P.S.]; Fondazione Italiana Ricerca sul Cancro (FIRC) (to C.C.). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro.
Conflict of interest statement. None declared.
REFERENCES
- 1. Daley J.M., Gaines W.A., Kwon Y., Sung P.. Regulation of DNA pairing in homologous recombination. Cold Spring Harb. Perspect. Biol. 2014; 6:a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Villa M., Cassani C., Gobbini E., Bonetti D., Longhese M.P.. Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell. Mol. Life Sci. 2016; 73:3655–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gobbini E., Cassani C., Villa M., Bonetti D., Longhese M.P.. Functions and regulation of the MRX complex at DNA double-strand breaks. Microb. Cell. 2016; 3:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bressan D.A., Olivares H.A., Nelms B.E., Petrini J.H.. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998; 150:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., Ohta K.. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998; 17:6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paull T.T., Gellert M.. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998; 1:969–979. [DOI] [PubMed] [Google Scholar]
- 8. Trujillo K.M., Yuan S.S., Lee E.Y., Sung P.. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998; 273:21447–21450. [DOI] [PubMed] [Google Scholar]
- 9. Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., Ogawa T.. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998; 95:705–716. [DOI] [PubMed] [Google Scholar]
- 10. Moreau S., Ferguson J.R., Symington L.S.. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trujillo K.M., Sung P.. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 2001; 276:35458–35464. [DOI] [PubMed] [Google Scholar]
- 12. de Jager M., van Noort J., van Gent D.C., Dekker C., Kanaar R., Wyman C.. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001; 8:1129–1135. [DOI] [PubMed] [Google Scholar]
- 13. Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A.. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001; 105:473–485. [DOI] [PubMed] [Google Scholar]
- 14. Moncalian G., Lengsfeld B., Bhaskara V., Hopfner K.P., Karcher A., Alden E., Tainer J.A., Paull T.T.. The Rad50 signature motif: essential to ATP binding and biological function. J. Mol. Biol. 2004; 335:937–951. [DOI] [PubMed] [Google Scholar]
- 15. Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G. et al. . Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008; 135:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hopfner K.P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A., Karcher A., Henderson B., Bodmer J.L., McMurray C.T. et al. . The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002; 418:562–566. [DOI] [PubMed] [Google Scholar]
- 17. Lobachev K., Vitriol E., Stemple J., Resnick M.A., Bloom K.. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004; 14:2107–2112. [DOI] [PubMed] [Google Scholar]
- 18. Wiltzius J.J., Hohl M., Fleming J.C., Petrini J.H.. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 2005; 12:403–407. [DOI] [PubMed] [Google Scholar]
- 19. Hohl M., Kwon Y., Galván S.M., Xue X., Tous C., Aguilera A., Sung P., Petrini J.H.. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 2011; 18:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seifert F.U., Lammens K., Stoehr G., Kessler B., Hopfner K.P.. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016; 35:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim H.S., Kim J.S., Park Y.B., Gwon G.H., Cho Y.. Crystal structure of the Mre11–Rad50-ATPγS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011; 25:1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams G.J., Williams R.S., Williams J.S., Moncalian G., Arvai A.S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P. et al. . ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011; 18:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Möckel C., Lammens K., Schele A., Hopfner K.P.. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012; 40:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lammens K., Bemeleit D.J., Möckel C., Clausing E., Schele A., Hartung S., Schiller C.B., Lucas M., Angermüller C., Söding J. et al. . The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011; 145:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deshpande R.A., Williams G.J., Limbo O., Williams R.S., Kuhnlein J., Lee J.H., Classen S., Guenther G., Russell P., Tainer J.A. et al. . ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014; 33:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50-Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 27. Mimitou E.P., Symington L.S.. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008; 455:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11–Rad50-Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L. et al. . Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010; 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia V., Phelps S.E., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C.. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011; 25:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F. et al. . DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014; 53:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tran P.T., Erdeniz N., Dudley S, Liskay R.M.. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair. 2002; 1:895–912. [DOI] [PubMed] [Google Scholar]
- 35. Kao H.I., Campbell J.L., Bambara R.A.. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004; 279:50840–50849. [DOI] [PubMed] [Google Scholar]
- 36. Shim E.Y., Chung W.H., Nicolette M.L., Zhang Y., Davis M., Zhu Z., Paull T.T., Ira G., Lee S.E.. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010; 29:3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keeney S., Kleckner N.. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Usui T., Ogawa H., Petrini J.H.. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001; 7:1255–1266. [DOI] [PubMed] [Google Scholar]
- 39. Clerici M., Mantiero D., Lucchini G., Longhese M.P.. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006; 7:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bonetti D., Villa M., Gobbini E., Cassani C., Tedeschi G., Longhese M.P.. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 2015; 16:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrari M., Dibitetto D., De Gregorio G., Eapen V.V., Rawal C.C., Lazzaro F., Tsabar M., Marini F., Haber J.E., Pellicioli A.. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 2015; 11:e1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gobbini E., Villa M., Gnugnoli M., Menin L., Clerici M., Longhese M.P.. Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 2015; 11:e1005685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen H., Donnianni R.A., Handa N., Deng S.K., Oh J., Timashev L.A., Kowalczykowski S.C., Symington L.S.. Sae2 promotes DNA damage resistance by removing the Mre11–Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1880–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Puddu F., Oelschlaegel T., Guerini I., Geisler N.J., Niu H., Herzog M., Salguero I., Ochoa-Montaño B., Viré E., Sung P. et al. . Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 2015; 34:1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clerici M., Mantiero D., Lucchini G., Longhese M.P.. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005; 280:38631–38638. [DOI] [PubMed] [Google Scholar]
- 46. Lee K., Zhang Y., Lee S.E.. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008; 454:543–546. [DOI] [PubMed] [Google Scholar]
- 47. Pommier Y., Sun Y., Huang S.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell. Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cassani C., Gobbini E., Wang W., Niu H., Clerici M., Sung P., Longhese M.P.. Tel1 and Rif2 regulate MRX functions in end-tethering and repair of DNA double-strand breaks. PLoS Biol. 2016; 14:e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schiller C.B., Lammens K., Guerini I., Coordes B., Feldmann H., Schlauderer F., Möckel C., Schele A., Strässer K., Jackson S.P. et al. . Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012; 19:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seifert F.U., Lammens K, Hopfner KP.. Structure of the catalytic domain of Mre11 from Chaetomium thermophilum. Acta Crystallogr. Struct. Biol. Commun. 2015; 71:752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Y., Sung S., Kim Y., Li F., Gwon G., Jo A., Kim A.K., Kim T., Song O.K., Lee S.E. et al. . ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016; 35:743–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pei J., Grishin N.V.. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001; 17:700–712. [DOI] [PubMed] [Google Scholar]
- 53. Wang J., Cieplak P., Kollman P.A.. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules. J. Comp. Chem. 2000; 21:1049–1074. [Google Scholar]
- 54. Berendsen H.J.C., Grogera J.R., Straatsma T.P.. The missing term in effective pair potentials. J. Phys. Chem. 1987; 91:6269–6271. [Google Scholar]
- 55. Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E.. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998; 94:399–409. [DOI] [PubMed] [Google Scholar]
- 56. Mimitou E.P., Symington L.S.. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010; 29:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakada D., Matsumoto K., Sugimoto K.. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003; 17:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Falck J., Coates J., Jackson S.P.. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434:605–611. [DOI] [PubMed] [Google Scholar]
- 59. You Z., Chahwan C., Bailis J., Hunter T., Russell P.. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005; 25:5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sung S., Li F., Park Y.B., Kim J.S., Kim A.K., Song O.K., Kim J., Che J., Lee S.E., Cho Y.. DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA. EMBO J. 2014; 33:2422–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaye J.A., Melo J.A., Cheung S.K., Vaze M.B., Haber J.E., Toczyski D.P.. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 2004; 14:2096–2106. [DOI] [PubMed] [Google Scholar]
- 62. Nakai W., Westmoreland J., Yeh E., Bloom K., Resnick M.A.. Chromosome integrity at a double-strand break requires exonuclease 1 and MRX. DNA Repair. 2011; 10:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Saponaro M., Callahan D., Zheng X., Krejci L., Haber J.E., Klein H.L., Liberi G.. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010; 6:e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu C., Pouliot J.J., Nash H.A.. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:14970–14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hartsuiker E., Neale M.J., Carr A.M.. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 2009; 33:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sacho E.J., Maizels N.. DNA repair factor MRE11/RAD50 cleaves 3’-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J. Biol. Chem. 2011; 286:44945–44951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee K.C., Padget K., Curtis H., Cowell I.G., Moiani D., Sondka Z., Morris N.J., Jackson G.H., Cockell S.J., Tainer J.A. et al. . MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol. Open. 2012; 1:863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M.. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012; 19:417–423. [DOI] [PubMed] [Google Scholar]
- 69. Ira G., Haber J.E.. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002; 22:6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jinks-Robertson S., Michelitch M., Ramcharan S.. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993; 13:3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee K., Lee S.E.. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007; 176:2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.