Abstract
Post-transcriptional regulation is an important mechanism for modulating gene expression and is performed by numerous mRNA-binding proteins. To understand the mechanisms underlying post-transcriptional regulation, we investigated the phosphorylation status of 32 mRNA-binding proteins under glucose deprivation conditions in Saccharomyces cerevisiae. We identified 17 glucose-sensitive phosphoproteins and signal pathways implicated in their phosphorylation. Notably, phosphorylation of the eukaryotic translation initiation factor 4G (eIF4G) was regulated by both the Snf1/AMPK pathway and the target of rapamycin complex 1 (TORC1) pathway. The serine/threonine protein kinase Ksp1 has previously been suggested to be a downstream effector of TORC1, but its detailed function has rarely been discussed. We identified that Snf1/AMPK and TORC1 signalings converge on Ksp1, which phosphorylates eIF4G under glucose deprivation conditions. Ksp1-dependent phosphorylation of eIF4G regulates the degradation of specific mRNAs (e.g. glycolytic mRNAs and ribosomal protein mRNAs) under glucose deprivation conditions likely through the recruitment of Dhh1. Taken together, our results suggest that Ksp1 functions as a novel modulator of post-transcriptional regulation in yeast.
INTRODUCTION
Post-transcriptional regulation is a central component of gene expression that is typically regulated by mRNA-binding proteins. Three key aspects in the post-transcriptional regulation of mRNAs are control of mRNA localization, translation efficiency and mRNA degradation (1). (i) Subcellular localization of mRNAs enables cells to synthesize proteins at a discrete site, providing enhanced spatial and temporal regulation of gene expression (2). Accumulation of mRNAs in granules is another way for localization to modulate mRNA translation and degradation. Messenger ribonucleoprotein (mRNP) granules, which are composed of mRNAs and several mRNA-binding proteins, are dynamically assembled and disassembled in response to stresses, cell cycle and developmental programs. The best characterized mRNP granules are stress granules and mRNA processing bodies (P-bodies), which typically consist of translation initiation factors and mRNA decay factors, respectively, with translationally inactive mRNAs (3). (ii) Regulation of translation efficiency mainly targets the initiation step. Nitrogen starvation induces phosphorylation of eIF2α to inhibit formation of the 43S pre-initiation complex, resulting in global translational repression (4–6). Inactivation of mammalian target of rapamycin complex 1 (mTORC1) also induces global translational repression by dephosphorylation of 4E-BP1, which leads to destabilization of the eIF4F complex (7,8). Caf20 and Eap1 (yeast 4E-BPs) do not repress global translation, but instead regulate specific genes (9–11). In yeast, it has been reported that global translational repression under glucose starvation conditions is mediated by dissociation of eIF4G and eIF4A at the initiation step (12). (iii) The major pathway of mRNA degradation is 5′ to 3′ mRNA decay, which is promoted by exchanging the eIF4F complex for the Dcp1/Dcp2 mRNA decapping complex on the 5′ cap structure (13). Regulation of specific mRNA degradation in response to stress is related to gene utility under stress conditions. For instance, the degradation of the GAL gene mRNAs is promoted by replacing galactose medium with glucose medium. Moreover, the half-lives of ribosomal protein (RP) mRNAs are dramatically decreased with glucose starvation or rapamycin treatment, corresponding with global translational repression under these conditions (14).
The DEAD-box helicase Dhh1 is a conserved decapping activator that stimulates translational repression as well as mRNA decay. It is thought to play an important role in mRNP remodeling that mediates the transition from translation to mRNA decay (15,16). Although Dhh1 targets a wide range of mRNAs, it confers regulation of specific mRNA decay by cooperation with mRNA-binding proteins. Interaction between Dhh1 and Rbp1 promotes decay of porin mRNA, and recruitment of Dhh1 by Cth2 ARE-binding protein promotes decay of SDH4 mRNA in response to iron deficiency (17,18). Additionally, Dhh1 homolog Vad1 in Cryptococcus neoformans enhances decapping and degradation of ATG8 mRNA under normal growth conditions (19).
Since post-transcriptional regulation is modulated by various mRNA-binding proteins, modifications of mRNA-binding proteins play important roles in the regulation of mRNAs. As mentioned above, phosphorylation of eIF2α and 4E-BP1 is a well-known mechanism for regulating translation initiation. Recently, a high-throughput experiment revealed that signal transduction through Snf1 phosphorylates several mRNA decay factors that might lead to regulation of Snf1-dependent mRNAs (20). Moreover, cAMP-dependent protein kinase A (PKA)-dependent phosphorylation of Pat1 has been shown to regulate P-body formation (21). Therefore, deciphering modifications of mRNA-binding proteins is valuable for understanding mechanisms of post-transcriptional regulation.
eIF4G is a scaffold protein that assembles eIF4E and eIF4A to form the eIF4F complex, which plays a central role in translation initiation by recruiting 40S ribosomal subunit to mRNAs. Eukaryotic cells have two eIF4G isoforms, namely, eIF4G1 and eIF4G2. Several kinases are known to regulate phosphorylation of eIF4G in mammalian cells. PKCα directly phosphorylates Ser 1186 of eIF4G1 and regulates binding with Mnk1 (22–24). Serum-induced phosphorylation of Ser 1108, 1148 and 1192 is under control of the PI-3 kinase and FRAP/mTOR pathways, although they are not directly phosphorylated by mTOR. mTOR regulates the N-terminus of eIF4G1 to increase the accessibility of the C-terminus to unknown kinases (25). p21-activated kinase 2 (Pak2) is activated by stress conditions such as serum deprivation and hyperosmolarity, and directly phosphorylates Ser 896 of eIF4G1. This phosphorylation inhibits cap-dependent translation (26). Multipotential S6 kinase (MS6K) phosphorylates eIF4G1 in vitro (22). eIF4G2 is phosphorylated at Ser 1156 by calmodulin-dependent kinase I (27). Yeast eIF4G1 and eIF4G2 are encoded by TIF4631 and TIF4632, respectively. While diverse regulatory mechanisms for eIF4G have been discovered in mammalian cells, kinases for Tif4631 and Tif4632 have not yet been reported in yeast.
In this study, through analyzing the phosphorylation status of 32 mRNA-binding proteins under glucose deprivation conditions, we identified Ksp1 as a novel kinase of Tif4631 and Tif4632 in yeast. Phosphorylation of Tif4631 and Tif4632 by Ksp1 under glucose deprivation conditions was regulated by both the Snf1/AMPK and TORC1 pathways. Ksp1 also regulated the recruitment of Dhh1 to glycolytic mRNAs and RP mRNAs, leading to the degradation of specific mRNAs under glucose deprivation conditions. Taken together, we suggest that Ksp1 is a central kinase that modulates post-transcriptional regulation of specific mRNAs in yeast.
MATERIALS AND METHODS
Yeast strains, plasmids and culture conditions
Saccharomyces cerevisiae strains and plasmids used in this study are listed in Supplementary Table S1. Yeast cells were grown at 30°C in YPD (1% yeast extract, 2% peptone and 2% dextrose) or synthetic complete (SC) medium as indicated (28). For glucose deprivation, cells grown to mid-log phase (OD600 = 0.7∼1.0) were transferred to YP medium (1% yeast extract, 2% peptone) and cultured for 30 min. For rapamycin treatment, cells grown to mid-log phase were treated with 200 ng/ml of rapamycin and cultured for 30 min. For cycloheximide treatment, cells grown to mid-log phase were treated with a 100 μg/ml final concentration and cultured for 30 min.
Western blot analysis
Yeast cells grown to mid-log phase were harvested and disrupted with glass beads in lysis buffer (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 0.15% NP-40) containing 1 mM phenylmethylsulphonyl fluoride, protease inhibitors and phosphatase inhibitors. Lysates were clarified by centrifugation at 13 000 × g for 10 min at 4°C. After mixing with sodium dodecyl sulphate (SDS) sample buffer followed by boiling, same amounts of proteins were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE with Phos-tag was performed following the manufacturer’s instructions. The Phos-tag concentration used was 20 μM in all experiments. For alkaline phosphatase (AP) treatment, lysates were incubated with 20 units of AP (Roche, #11097075001) at 37°C for 30 min prior to boiling. Immunoblots were performed with HRP-conjugated rabbit anti-mouse IgG antibody (Sigma, A9044) for TAP tagged proteins, HRP-conjugated anti-c-Myc antibody (Santa Cruz, SC-40 HRP), anti-GFP antibody (Santa Cruz, SC-9996 HRP), anti-HA antibody (Santa Cruz, SC-7392 HRP) and anti-GST antibody (Santa Cruz, SC-138 HRP) for each tagged protein.
In vitro kinase assay
BY4741 cells and cells integrated with KSP1-HA or ksp1-K47D-HA were grown in YPD medium at 30°C to mid-log phase and then were subjected to glucose deprivation for 30 min. Cells were harvested and lysed with glass beads in lysis buffer containing protease inhibitors and phosphatase inhibitors. After centrifugation at 13 000 × g for 10 min, cell lysates were incubated with anti-HA antibody (Santa Cruz, SC-7392) and protein A-Sepharose beads for each 2 h at 4°C. Tif4631 and Tif4632 were cloned into a pGEX4T-1 plasmid and were purified from Escherichia coli by glutathione S-transferase (GST) affinity purification. Kinase reactions were performed in kinase buffer (25 mM HEPES, pH 7.5, 10 mM MgCl2 and 50 μM adenosine triphosphate (ATP)). Immunoprecipitated Ksp1 and purified GST-Tif4631 or Tif4632 were incubated with or without 4 μCi [γ-32P]-ATP for 15 min at 30°C. The reactions were terminated by addition of SDS-PAGE sample buffer followed by heating at 95°C for 10 min. Eluted protein samples were subjected to conventional SDS-PAGE or Phos-tag SDS-PAGE. Autoradiography signals were detected by BAS-2500 (Fuji Film), and western blotting was performed with appropriate antibodies.
Measurement of mRNA half-life
For thiolutin treatment, cells were grown to mid-log phase in YPD medium. A total of 3 μg/ml of thiolutin was used to treat cell cultures with or without glucose deprivation. For doxycycline treatment, cells harboring HB0529 or HB0530 plasmid were grown in SC-Leu media for overnight and diluted with YPD medium to an OD600 = 0.2. Upon growing to mid-log phase, 15 μg/ml of doxycycline was used to treat cell cultures with or without glucose deprivation. Samples were taken at 0, 5, 10, 15 and 20 min, and RNA was extracted by using an RNeasy mini kit (Qiagen). cDNA was synthesized using random primers, and quantitative real-time polymerase chain reaction (qRT-PCR) was performed on Quantstudio 3 (Applied Biosystems). mRNA levels were normalized to SCR1, and mRNA half-life was estimated by fitting a one-phase exponential decay curve to data by non-linear regression.
RNA immunoprecipitation
Cell extracts were prepared as described above with lysis buffer containing 100 units/ml RNasin (Promega) and then diluted to a concentration of approximately 5 mg/ml. Input samples were collected at this point. Diluted extracts were incubated with anti-c-Myc antibody (Santa Cruz, SC-40) for 2 h at 4°C, followed by addition of protein A-Sepharose beads (GE healthcare, #17513801). After incubation for 2 h, beads were washed five times with lysis buffer. RNA was extracted from immunoprecipitates and 3% of input samples using TRIzol (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized using oligo(dT) primers, and qRT-PCR was performed on Quantstudio 3. mRNA levels of immunoprecipitation samples were normalized to input samples.
RESULTS
Identification of 17 mRNA-binding proteins phosphorylated in a glucose-dependent manner
Recently, Parker and colleagues identified 120 mRNA-binding proteins in yeast and reported widespread changes in their subcellular localization during stress (29). Notably, they observed that 32 mRNA-binding proteins were translocalized from the cytoplasm to P-bodies or stress granules under glucose deprivation conditions. Given that a conserved aspect of the eukaryotic stress response is aggregation of non-translating mRNPs into P-bodies and stress granules (30), it is plausible that these 32 mRNA-binding proteins showing translocation to P-bodies or stress granules during stress may be enriched with proteins involved in the regulation of mRNAs. To determine how mRNA-binding proteins regulate mRNAs, we set out to analyze the phosphorylation status of these 32 proteins under glucose deprivation conditions, because phosphorylation is one of the major post-translational modifications regulating protein functions.
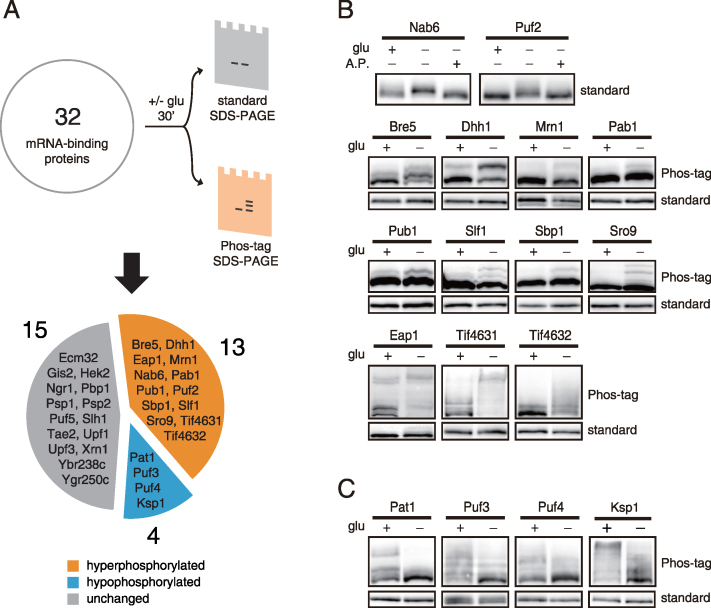
We investigated the phosphorylation status of 32 mRNA-binding proteins under both normal and glucose deprivation conditions (Figure 1A). Phosphorylation was analyzed by a mobility shift assay using standard SDS-PAGE and Phos-tag SDS-PAGE, which enhances the mobility retardation of phosphorylated proteins (31). To our surprise, 17 of the 32 mRNA-binding proteins showed changes in phosphorylation status under glucose deprivation conditions; 13 of them were hyperphosphorylated and four of them were hypophosphorylated (Figure 1B and C; Supplementary Figure S1). We confirmed that the mobility shift of these 17 proteins was caused by phosphorylation through AP treatment (Figure 1B and Supplementary Figure S2). Interestingly, for some proteins such as Pab1, Pub1, Slf1, Sbp1 and Sro9, phosphorylation could be detected in only a small fraction of the proteins even with Phos-tag SDS-PAGE. It is presumable that a minor fraction of these proteins may be subject to phosphorylation under certain conditions and thus contribute to specific regulation of target mRNAs. Overall, the identification of numerous mRNA-binding proteins phosphorylated in a glucose-dependent manner suggests the possibility that phosphorylation may play a role in modulating their function in the post-transcriptional regulation of mRNAs.
Figure 1.
Identification of 17 glucose-sensitive phosphoproteins from 32 mRNA-binding proteins. (A) Scheme of phosphoprotein screening. C-terminally TAP-tagged strains for 32 mRNA-binding proteins were subjected to glucose deprivation for 30 min and analyzed by standard SDS-PAGE and Phos-tag SDS-PAGE. Proteins with changed or unchanged phosphorylation status are indicated in the pie chart. (B) Thirteen proteins that were hyperphosphorylated under glucose deprivation conditions. AP treatments were for 30 min at 37°C. (C) Four proteins that were hypophosphorylated under glucose deprivation conditions. The results from Phos-tag SDS-PAGE and standard SDS-PAGE are indicated beside the blots.
The PKA, Snf1/AMPK and TORC1 pathways are involved in phosphorylation of the 17 mRNA-binding proteins
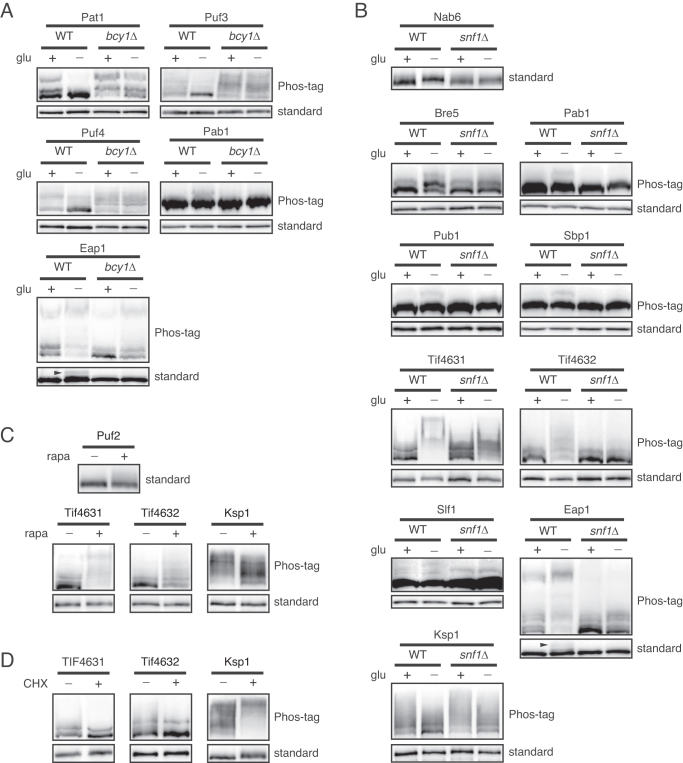
To explore which signaling pathway is involved in phosphorylation of these 17 mRNA-binding proteins, we investigated the implication of three major signaling pathways that are regulated by glucose starvation, i.e. the PKA, Snf1/AMPK and TORC1 pathways (32,33). First, we tested the implication of the PKA pathway by using bcy1Δ mutants, in which PKA is constitutively activated regardless of glucose availability. Among the 17 proteins, three hypophosphorylated proteins (Pat1, Puf3 and Puf4) and two hyperphosphorylated proteins (Pab1 and Eap1) under glucose deprivation conditions recovered their normal phosphorylation status in bcy1Δ mutants (Figure 2A), suggesting that these proteins are regulated by PKA. Consistent with this result, it has been shown that Pat1 is a substrate of PKA, and its dephosphorylation with glucose deprivation is mediated by inhibition of PKA (21). Given that Puf3 and Puf4 have PKA consensus sites at Ser 56 and Thr 205, respectively, it is likely that PKA directly phosphorylates Puf3 and Puf4.
Figure 2.
The PKA pathway, Snf1/AMPK pathway and TORC1 pathway regulate phosphorylation of mRNA-binding proteins under glucose deprivation conditions. (A and B) BCY1 or SNF1 was deleted from TAP-tagged strains for 17 glucose-sensitive phosphoproteins, and their phosphorylation status was compared with that of WT cells under glucose deprivation conditions. The results from Phos-tag SDS-PAGE and standard SDS-PAGE are indicated beside the blots. (A) PKA-dependent phosphoproteins found in this study. Dephosphorylation of Eap1 in bcy1Δ strain was detected only in standard SDS-PAGE. Arrowhead, phosphorylated Eap1. (B) Snf1/AMPK-dependent phosphoproteins found in this study. (C) TORC1-dependent phosphoproteins found in this study. Cells were treated with 200 ng/ml of rapamycin for 30 min and subjected to western blotting. (D) Cycloheximide-sensitive phosphoproteins found in this study. Cells were treated with 100 μg/ml cycloheximide for 30 min and subjected to western blotting.
Activation of the Snf1 pathway is another major change in signal transduction under glucose deprivation conditions (34). Snf1 is the only member of the adenosine monophosphate-activated protein kinase (AMPK) family in yeast, which has the serine/threonine protein kinase activity. We investigated whether phosphorylation of the 17 mRNA-binding proteins is dependent on Snf1 signaling. We observed that hyperphosphorylation of nine proteins (Bre5, Eap1, Nab6, Pab1, Pub1, Sbp1, Slf1, Tif4631 and Tif4632) and hypophosphorylation of one protein (Ksp1) induced by glucose deprivation was abolished or alleviated in snf1Δ mutants (Figure 2B), suggesting that phosphorylation of these proteins under glucose deprivation conditions is regulated by Snf1 signaling. This result is consistent with a previous report that Snf1 regulates mRNA decay by phosphorylating numerous mRNA decay factors (20). Furthermore, among the nine proteins hyperphosphorylated under glucose deprivation conditions in a Snf1-depedent manner, five proteins (Eap1, Nab6, Pub1, Tif4631 and Tif4632) overlapped with previous results (20).
Though nutrient control of TORC1 has been widely studied under nitrogen starvation conditions, TORC1 is effectively inactivated also by glucose starvation (33). We checked the phosphorylation status of the 17 mRNA-binding proteins under treatment of rapamycin, a specific inhibitor of TORC1, to investigate whether the changes in phosphorylation are mediated by inactivation of TORC1. Among the 17 proteins, three proteins (Puf2, Tif4631 and Tif4632) were hyperphosphorylated and one protein (Ksp1) was hypophosphorylated under rapamycin treatment (Figure 2C). To confirm that phosphorylation of these proteins is dependent on TORC1 activity, we examined whether phosphorylation of these proteins is influenced by cycloheximide, which is known to hyperactivate TORC1 signaling (35). Phosphorylation of Tif4631 and Tif4632 was slightly decreased whereas phosphorylation of Ksp1 was increased under cycloheximide treatment (Figure 2D), demonstrating that phosphorylation of these proteins is regulated by TORC1.
Ksp1 directly phosphorylates eIF4G
TIF4631 and TIF4632 encode yeast eIF4G1 and eIF4G2, which are isoforms of eIF4G. We observed that both Tif4631 and Tif4632 are phosphorylated under glucose deprivation (Figure 1B). Phosphorylation of Tif4631 and Tif4632 was independent of PKA activity and dependent on Snf1 signaling (Figure 2B and Supplementary Figure S3A). Given this, we examined whether Snf1 directly phosphorylates eIF4G. However, phosphorylation of Tif4631 and Tif4632 by Snf1 was not observed in an in vitro kinase assay (data not shown), indicating that Snf1 is not the direct kinase for eIF4G. Consistent with this observation, neither Tif4631 nor Tif4632 has the consensus Snf1 recognition sequence, ΦXRXXSXXXΦ (Φ; hydrophobic residues). Inactivation of TORC1 also induced phosphorylation of Tif4631 and Tif4632 (Figure 2C), suggesting the presence of a kinase for eIF4G that is inhibited by TORC1. Taken together, these results support the idea that eIF4G is phosphorylated by a kinase that is regulated by both Snf1/AMPK and TORC1 pathways.
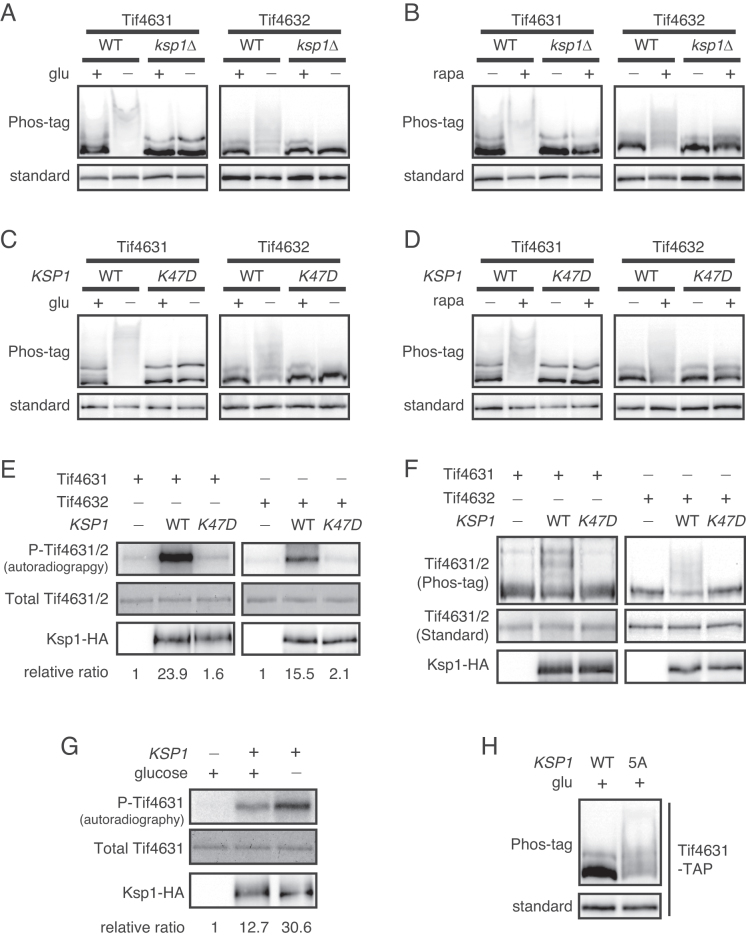
To identify a kinase for eIF4G, we next focused on Ksp1, a serine/threonine protein kinase in the casein kinase II subfamily. Ksp1 was the only kinase included in the 17 mRNA-binding proteins showing changes in phosphorylation status under glucose deprivation conditions (Figure 1C). Ksp1 was hypophosphorylated under glucose deprivation conditions in a PKA-independent and Snf1-dependent manner (Figure 2B and Supplementary Figure S3B). Ksp1 was also phosphorylated by TORC1 signaling (Figure 2C and D), as previously reported (36–38). Although some phenotypes of ksp1Δ mutants, such as defects in filamentous growth and increased autophagy, have been reported, the specific roles and substrates of Ksp1 have not yet been identified (39,40). To check whether Ksp1 is involved in phosphorylation of eIF4G, we examined phosphorylation of Tif4631 and Tif4632 in ksp1Δ mutants. Under both glucose deprivation and rapamycin treatment conditions, deletion of KSP1 dramatically decreased hyperphosphorylation of Tif4631 and Tif4632 (Figure 3A and B). To determine whether ksp1Δ mutation has a broad effect on phosphorylation of mRNA-binding proteins, we analyzed phosphorylation of the remaining 14 mRNA-binding proteins phosphorylated in a glucose-dependent manner. Among these 14 proteins, only Bre5 showed a slight decrease in phosphorylation in ksp1Δ mutants (Supplementary Figure S4), suggesting that Ksp1 has a specific role in eIF4G phosphorylation.
Figure 3.
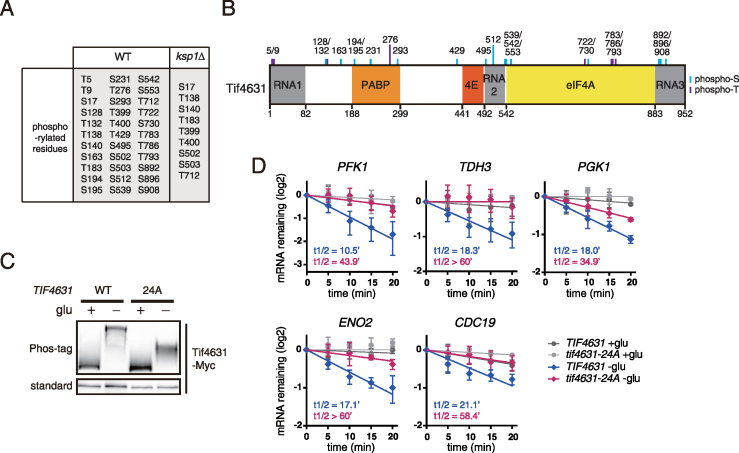
Ksp1 is a novel kinase of eIF4G. (A and B) Phosphorylation of eIF4G in ksp1Δ cells. KSP1 was deleted from TIF4631-TAP and TIF4632-TAP strains. (C and D) Phosphorylation of eIF4G in cells expressing the kinase-dead allele ksp1-K47D. KSP1 (WT) or ksp1-K47D alleles were integrated into ksp1Δ cells expressing Tif4631-TAP or Tif4632-TAP. (E and F) In vitro kinase assay of Ksp1 with Tif4631 and Tif4632 as substrates. (E) In vitro kinase assay using radioisotope labeling. Upper panel, incorporation of 32P into substrates; middle panel, the amount of substrates visualized by Coomassie blue staining; lower panel, the amount of Ksp1 kinase. The relative ratio of phosphorylated Tif4631/Tif4632 to total Tif4631/Tif4632 is shown below each lane. (F) In vitro kinase assay using Phos-tag SDS-PAGE. Upper panel, western blotting of substrates using Phos-tag SDS-PAGE; middle panel, western blotting of substrates using standard SDS-PAGE; bottom panel, the amount of Ksp1 kinase. (G) In vitro kinase assay of Ksp1 purified under normal growth conditions and glucose deprivation conditions. Tif4631 was used to detect the activity of Ksp1 under each condition. The relative ratio of phosphorylated Tif4631 to total Tif4631 is shown below each lane. (H) Phosphorylation of Tif4631 in ksp1–5A mutant. KSP1 (WT) and ksp1–5A alleles were integrated into ksp1Δ cells expressing Tif4631-TAP, followed by western blot analysis.
To examine whether phosphorylation of Tif4631 and Tif4632 is dependent on the kinase activity of Ksp1, we expressed wild-type (WT) KSP1 and kinase-dead allele of KSP1, ksp1-K47D (41), in ksp1Δ cells. Expression of WT KSP1 successfully restored phosphorylation of Tif4631 and Tif4632, whereas expression of ksp1-K47D did not (Figure 3C and D). This result indicates that the kinase activity of Ksp1 is essential for phosphorylation of Tif4631 and Tif4632 under glucose deprivation and rapamycin treatment conditions.
Next, we performed in vitro kinase assays to evaluate the possibility that Ksp1 acts as a direct kinase of Tif4631 and Tif4632. We immunoprecipitated Ksp1-HA and Ksp1-K47D-HA from glucose-starved yeast cells and performed in vitro kinase reactions with E. coli purified GST-Tif4631 and GST-Tif4632. Phosphorylation of Tif4631 and Tif4632 was detected in two ways: by radioisotope labeling (Figure 3E) and by Phos-tag SDS-PAGE (Figure 3F). Both methods showed that Tif4631 and Tif4632 were phosphorylated when they were incubated with Ksp1, but not with Ksp1-K47D, indicating that Ksp1 directly phosphorylates Tif4631 and Tif4632.
Ksp1 is activated by dephosphorylation
The above results that glucose deprivation induces Ksp1-dependent phosphorylation of eIF4G prompted us to ask whether the kinase activity of Ksp1 is increased by glucose deprivation. To answer this question, we performed in vitro kinase assays with Ksp1 immunoprecipitates obtained from non-starved or glucose-starved yeast cells. Tif4631 was markedly more phosphorylated by Ksp1 from glucose-starved cells than Ksp1 from non-starved cells (Figure 3G), suggesting that dephosphorylated form of Ksp1, which is induced by glucose deprivation, is more active.
According to previous studies (36–38), Thr 526, Ser 529, 624, 814 and 827 of Ksp1 are putative target residues for TORC1. To examine whether dephosphorylation of Ksp1 induced by inactivation of TORC1 increases the kinase activity of Ksp1, we constructed a non-TORC1-phosphorylatable mutant of Ksp1 (Ksp1–5A; Ksp1[T526A,S529A,S624A,S814A,S827A]). Expression of Ksp1–5A increased phosphorylation of Tif4631 under normal growth conditions (Figure 3H), indicating that dephosphorylation of Ksp1 stimulates phosphorylation of eIF4G. Taken together, these results suggest that glucose deprivation activates Ksp1 at least in part by suppressing TORC1.
Ksp1 promotes the degradation of specific mRNAs by recruiting Dhh1 to the target mRNAs
The preceding results suggest the possibility that Ksp1 may be involved in the post-transcriptional regulation of mRNAs via eIF4G under glucose deprivation conditions. To further investigate the role of Ksp1 under glucose deprivation conditions, we first monitored translation efficiency by using polysome profiling to determine whether Ksp1 modulates translation repression under glucose deprivation conditions. When WT cells were subjected to glucose deprivation, the ratio of polysome to 80S monosome (P/M) was decreased from 4.71 ± 0.25 to 0.24 ± 0.03 (Supplementary Figure S5A and B), indicating that translation is globally repressed under glucose deprivation conditions. Deletion of KSP1 only slightly, though significantly, increased the P/M ratio to 0.59 ± 0.02 under glucose deprivation conditions, suggesting that Ksp1 does not play an essential role in global translation repression. We also examined the eIF4G–eIF4A interaction that is known to modulate translation under glucose deprivation conditions, but did not observe any significant effect of Ksp1 on the eIF4G–eIF4A interaction (Supplementary Figure S5C and D). These results suggest that Ksp1-mediated phosphorylation of eIF4G does not regulate global translation.
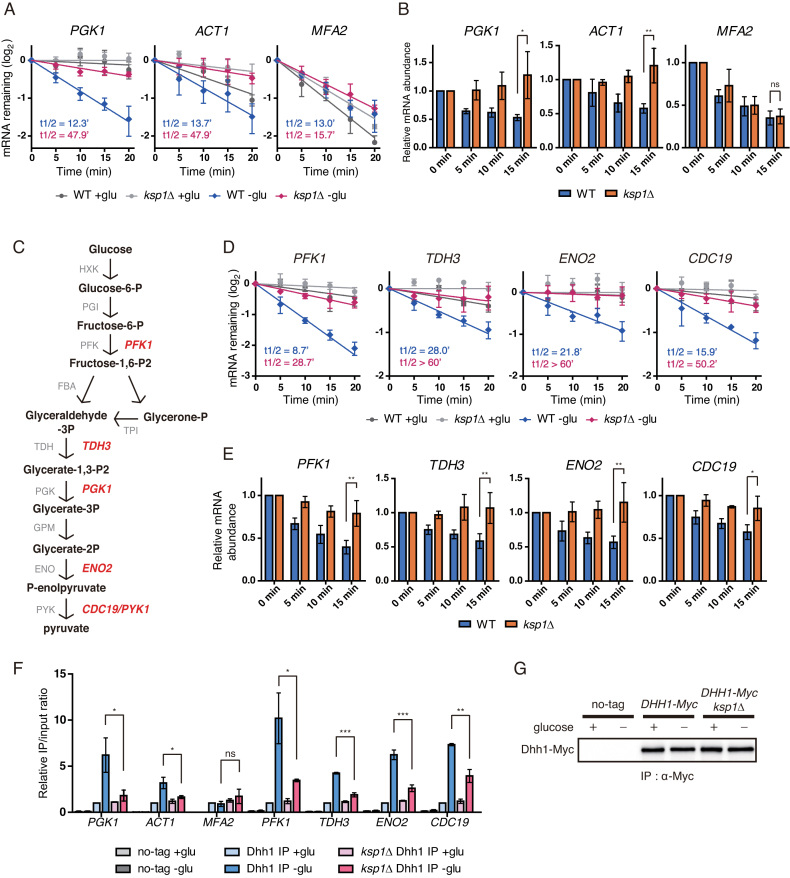
The regulation of mRNA stability is one of the major mechanisms to control gene expression at a post-transcriptional level. Next, we tested whether Ksp1 is involved in the degradation of mRNAs that are specifically regulated under glucose deprivation conditions. It has been reported that PGK1 and ACT1 mRNAs, which are stable under normal growth conditions, are specifically degraded under glucose starvation (42). We used thiolutin to block transcription and measured the half-lives of PGK1 and ACT1 mRNAs in WT and ksp1Δ cells. As reported previously, the half-lives of PGK1 and ACT1 mRNAs were decreased with glucose deprivation (Figure 4A). Strikingly, deletion of KSP1 significantly increased the half-lives of PGK1 and ACT1 mRNAs under glucose deprivation conditions. Consistent with a previous report (42), MFA2 mRNA, an mRNA decay reporter in yeast (43,44), was unstable regardless of growth conditions. Furthermore, the stability of MFA2 mRNA varied only slightly between WT and ksp1Δ cells (Figure 4A). To confirm the physiological relevance of Ksp1-dependent mRNA degradation, we measured the mRNA abundance under glucose deprivation conditions over time without transcriptional perturbation. Consistent with the above results, the mRNA levels of PGK1 and ACT1 were decreased over time after glucose deprivation in WT cells but not in ksp1Δ cells (Figure 4B). In contrast, MFA2 showed a decrease in mRNA level upon glucose deprivation in both WT and ksp1Δ cells. Taken together, these results suggest that Ksp1 promotes selective degradation of specific mRNAs under glucose deprivation conditions.
Figure 4.
Specific mRNAs are targeted by KSP1-dependent mRNA decay. (A) Decay curves of PGK1, ACT1 and MFA2 after thiolutin treatment in WT and ksp1Δ cells under glucose deprivation conditions. X-axis, time after thiolutin treatment. (B) Time-course measurement of mRNA levels of PGK1, ACT1 and MFA2 in WT and ksp1Δ cells after glucose deprivation. X-axis, time after glucose deprivation. (C) The glycolysis pathway and its enzymes in yeast. Glycolytic genes tested in this study are represented in red. (D) Decay curves of glycolytic mRNAs after thiolutin treatment in WT and ksp1Δ cells. X-axis, time after thiolutin treatment. (E) Time-course measurement of glycolytic mRNA levels in WT and ksp1Δ cells after glucose deprivation. X-axis, time after glucose deprivation. (F) RNA immunoprecipitation using an anti-c-Myc antibody to immunoprecipitate Dhh1-Myc and its associated transcripts. Dhh1-Myc was immunoprecipitated from each strain, and its associated mRNAs were analyzed by qRT-PCR. (G) Western blotting of immunoprecipitated Dhh1 from each strain. For (A) and (D), data were fitted to one-phase exponential decay curve to determine mRNA half-lives. For (B) and (E), cDNA was synthesized using random primers, and mRNA levels were normalized to SCR1. Relative mRNA levels were calculated using the 2−ΔΔCt method. Error bars indicate SD of experiments performed in triplicate. P-values were determined by Student’s t-test (***P < 0.005; **P < 0.01; *P < 0.05; ns, P > 0.05).
We next asked what kinds of mRNAs are targeted by Ksp1-dependent degradation under glucose deprivation conditions. PGK1 is a 3-phosphoglycerate kinase that participates in the glycolysis pathway. Given this fact and that the half-life of PGK1 mRNA was decreased under glucose deprivation conditions, we supposed that mRNAs of glycolytic genes might be destabilized with glucose starvation. To test this hypothesis, we selected four more genes—PFK1, TDH3, ENO2 and CDC19—involved in different steps of glycolysis (Figure 4C) and measured their mRNA half-lives in WT and ksp1Δ cells. As expected, all of four glycolytic mRNAs were stable under normal conditions and glucose deprivation induced their degradation (Figure 4D). Like PGK1, the mRNA stability of these genes was not decreased in ksp1Δ cells in the face of glucose deprivation, suggesting that Ksp1 positively regulates the degradation of glycolytic mRNAs under glucose deprivation conditions. Time-course measurements of mRNA levels after glucose deprivation also indicated that Ksp1 is required for the observed decreases in the glycolytic mRNA levels under glucose deprivation conditions (Figure 4E).
Specific mRNA degradation under certain conditions has been reported to be promoted by binding of mRNA decay activators to the target mRNAs (17–19). To further understand the mechanism by which Ksp1 promotes specific degradation of glycolytic mRNAs under glucose deprivation conditions, we examined interaction between the mRNA decay activator Dhh1 and glycolytic mRNAs. The DEAD-box helicase Dhh1 is a central modulator of mRNP remodeling that mediates the transition from translation to mRNA decay (15,45). Recruitment of Dhh1 to mRNA represses translation and promotes mRNA degradation. We analyzed binding of Dhh1 to mRNAs under glucose deprivation conditions using RNA-immunoprecipitation (RIP) assays. Binding of Dhh1 to glycolytic mRNAs was considerably increased with glucose deprivation (Figure 4F). In contrast, binding of Dhh1 to MFA2 mRNA, whose degradation is independent of Ksp1, was not increased at all. Remarkably, deletion of KSP1 significantly decreased the interactions between Dhh1 and glycolytic mRNAs under glucose deprivation conditions. The protein level of immunoprecipitated Dhh1 was not affected by deletion of KSP1 (Figure 4G). These results support the hypothesis that Ksp1 promotes the degradation of glycolytic mRNAs under glucose deprivation conditions by inducing Dhh1 recruitment to the glycolytic mRNAs.
Ksp1-dependent phosphorylation of eIF4G promotes the degradation of glycolytic mRNAs under glucose deprivation conditions
Given the above results that Ksp1 phosphorylates eIF4G under glucose deprivation conditions, we investigated whether Ksp1-dependent mRNA degradation is mediated by phosphorylation of eIF4G. To identify residues that are phosphorylated by Ksp1, we analyzed phosphorylation of Tif4631 under glucose deprivation conditions by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and compared the phosphorylated residues between WT and ksp1Δ cells. Tif4631 was highly phosphorylated under glucose deprivation conditions, as expected from Phos-tag SDS-PAGE results. In WT cells, we identified 33 phosphorylated residues of Tif4631 under glucose deprivation conditions, and nine of them were also detected in ksp1Δ cells (Figure 5A).
Figure 5.
Ksp1-dependent phosphorylation of eIF4G regulates mRNA decay of glycolytic genes. (A) The phosphorylated residues of Tif4631 under glucose deprivation conditions in WT and ksp1Δ cells. Immunoprecipitated Tif4631-TAP was analyzed by LC-MS/MS after tryptic digestion. (B) The potent residues that are phosphorylated by Ksp1. Twenty-four residues that were detected only in WT cells, not in ksp1Δ cells, are depicted above domain information of Tif4631. (C) Phosphorylation of Tif4631–24A under glucose deprivation conditions. C-terminally c-Myc tagged TIF4631 (WT) or tif4631–24A alleles were integrated into tif4631Δ cells. Cells were subjected to glucose deprivation for 30 min and analyzed by standard SDS-PAGE and Phos-tag SDS-PAGE. (D) Decay curves of glycolytic mRNAs after thiolutin treatment in TIF4631 and tif4631–24A cells. Data were fitted to one-phase exponential decay curve to determine mRNA half-lives. X-axis, time after thiolutin treatment. Error bars indicate SD of experiments performed in triplicate.
To evaluate the effect of Ksp1-dependent phosphorylation of Tif4631 on the degradation of glycolytic mRNAs, we introduced alanine substitution mutations within Tif4631 at 24 residues whose phosphorylation was not detected in ksp1Δ cells (Figure 5B). Cells with Tif4631–24A mutant exhibited little, if any, difference in growth (data not shown), suggesting that the biological function of Tif4631 is not disturbed by alanine substitution at 24 residues. As expected, Tif4631–24A mutant was strongly downshifted under glucose deprivation conditions, compared to WT Tif4631 (Figure 5C). Then we examined the degradation of glycolytic mRNAs in cells with WT Tif4631 or Tif4631–24A mutant. Cells expressing Tif4631–24A exhibited significantly retarded degradation of glycolytic mRNAs under glucose deprivation conditions, compared to cells with WT Tif4631 (Figure 5D). These results indicate that Ksp1-mediated phosphorylation of eIF4G induces the degradation of specific mRNAs under glucose deprivation conditions.
Ksp1 regulates the degradation and translation efficiency of RP mRNAs under glucose deprivation conditions
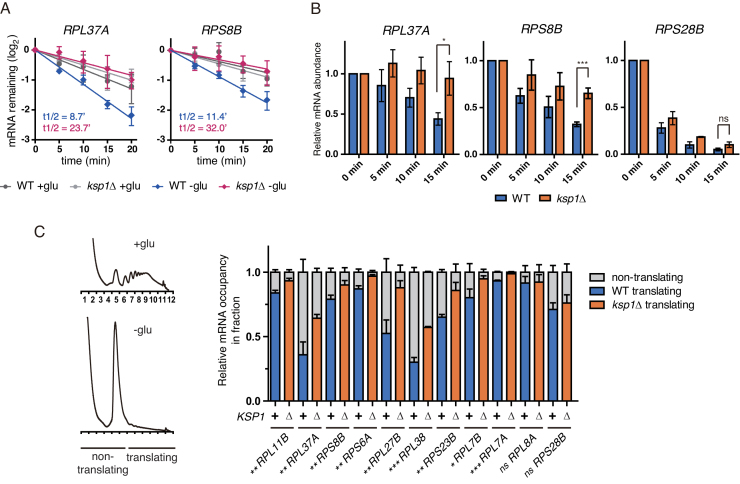
Above, we showed that phosphorylation of eIF4G was induced by both glucose deprivation and rapamycin treatment in a Ksp1-dependent manner. A recent study regarding mRNA half-life profiling discovered that acute glucose starvation, as well as rapamycin treatment, enhances the degradation of RP mRNAs (14). Given these, we investigated whether Ksp1 also targets RP mRNAs to promote their degradation. Among 137 RP genes in yeast, we randomly chose 11 to compare mRNA decay rates in WT and ksp1Δ cells in the face of glucose deprivation. All of the tested RP genes, with the exception of RPS28B, exhibited increased mRNA half-lives in both ksp1Δ and tif4631–24A cells under glucose deprivation conditions (Supplementary Figure S6A). Given that the degradation of RPS28B mRNA is independent of Dhh1 (46), these results are consistent with our hypothesis that Ksp1 stimulates the degradation of specific mRNAs under glucose deprivation conditions by recruiting Dhh1. However, it has been reported that inhibition of transcription by chemicals such as thiolutin and 1,10-phenanthroline may lead to the destabilization of RP mRNAs (14). To exclude the possibility of interfering effects of these chemicals, we examined the mRNA decay rate of RPL37A and RPS8B by using a Tet-off system that can block transcription without cell perturbation. Similar to the above results, glucose deprivation led to rapid degradation of RPL37A and RPS8B mRNAs in WT cells while their degradation was significantly retarded in ksp1Δ cells under glucose deprivation conditions (Figure 6A). Time-course measurements of mRNA levels after glucose deprivation also showed that the mRNA levels of RP genes, with the exception of RPS28B, decreased more slowly in ksp1Δ cells than in WT cells (Figure 6B and Supplementary Figure S6B). These results suggest that Ksp1 promotes glucose deprivation-induced degradation of RP mRNAs.
Figure 6.
Ksp1 regulates the degradation and translation of RP mRNAs under glucose deprivation conditions. (A) Decay curves of RPL37A and RPS8B after doxycycline treatment in WT and ksp1Δ cells. Data were fitted to one-phase exponential decay curve to determine mRNA half-lives. X-axis, time after doxycycline treatment. (B) Time-course measurement of RP mRNA levels in WT and ksp1Δ cells after glucose deprivation. cDNA was synthesized using random primers, and mRNA levels were normalized to SCR1. Relative mRNA levels were calculated using the 2−ΔΔCt method. X-axis, time after glucose deprivation. (C) The translation efficiency of RP mRNAs in WT and ksp1Δ cells under glucose deprivation conditions. RNA was extracted from translating and non-translating fractions, and cDNA was synthesized using oligo(dT) primers. The translation efficiency of each mRNA was calculated by the relative amount of the mRNA in the translating fraction and represented as blue (WT) or orange (ksp1Δ) in graph. Error bars indicate SD of experiments performed in triplicate. P-values were determined by Student’s t-test (***P < 0.005; **P < 0.01; *P < 0.05; ns, P > 0.05).
It has been reported that Dhh1 promotes not only mRNA decay but also translational repression (15,53). Furthermore, our ribosome profiling results showed that translation is slightly increased in ksp1Δ cells with glucose deprivation (Supplementary Figure S5A). Given these results, we hypothesized that Ksp1 may regulate the translation efficiency of its target mRNAs under glucose deprivation conditions. To evaluate this hypothesis, we purified mRNAs from non-translating fractions (fractions 1–6; NT) and translating fractions (fractions 7–12; T), and compared the T/(T+NT) ratio of each mRNA (Figure 6C). The T/(T+NT) ratios of all tested RP mRNAs, with the exception of RPL8A and RPS28B, were significantly increased in ksp1Δ cells under glucose deprivation conditions (Figure 6C). These results suggest that Ksp1 also regulates the translation efficiency of RP mRNAs under glucose deprivation conditions. Intriguingly, however, the T/(T+NT) ratios of glycolytic mRNAs were not increased in ksp1Δ cells (Supplementary Figure S7). It is likely that the regulatory mechanisms of translation are diverse among different genes (see ‘Discussion’ section).
DISCUSSION
In this study, we analyzed the phosphorylation status of mRNA-binding proteins to investigate how they are regulated under stress conditions. A total of 17 proteins among 32 tested mRNA-binding proteins showed changes in phosphorylation status with glucose deprivation, suggesting the existence of dynamic post-transcriptional regulation of mRNAs under this condition. We focused on Tif4631 and Tif4632 (yeast eIF4G1 and eIF4G2), whose phosphorylation is induced by not only glucose deprivation but also rapamycin treatment. We found that both activation of the Snf1/AMPK pathway and inhibition of the TORC1 pathway induce phosphorylation of eIF4G under glucose deprivation conditions. Furthermore, we identified Ksp1 as a novel kinase of eIF4G. Ksp1 was activated under glucose deprivation conditions, and non-TORC1-phosphorylatable Ksp1 increased phosphorylation of eIF4G. Taken together, these results support a model for regulation of eIF4G wherein the Snf1/AMPK pathway and the TORC1 pathway co-regulate Ksp1 under glucose deprivation conditions, leading to dephosphorylation and activation of Ksp1, which then phosphorylates eIF4G (Supplementary Figure S8). Ksp1-dependent phosphorylation of eIF4G induces the degradation of specific mRNAs including glycolytic mRNAs. In addition, Ksp1 is also required for the degradation and translational repression of RP mRNAs under glucose deprivation conditions.
The overall data of mRNA stability suggest that Ksp1 is a central kinase in regulation of specific mRNA degradation in the face of glucose deprivation. All tested mRNAs whose degradation is enhanced under glucose deprivation conditions were targeted by Ksp1, except the mRNA whose degradation is independent of Dhh1. Thus, it would be interesting to investigate whether Ksp1 generally targets mRNAs whose degradation is enhanced by glucose deprivation in a Dhh1-dependent manner.
Ksp1 was first identified from a screen to find suppressors of a prp20 mutant in 1996 (41), and has been suggested as a kinase required for filamentous growth (39,47). However, given that the function of Ksp1 in filamentous growth is likely mediated by its translocation to the nucleus, it would be distinguished from the function of Ksp1 in post-transcriptional regulation in the cytoplasm. Indeed, Ksp1 did not translocate to the nucleus under our experimental conditions (29), unlike in filamentous growth-inducible conditions. Another function of Ksp1 has been proposed by Klionsky and colleagues to regulate autophagy via activation of the TORC1 pathway (40). Ksp1 has several PKA consensus phosphorylation sites, among which Ser 624, 827 and 884 are dephosphorylated under rapamycin treatment via inactivation of PKA (36–38). Klionsky and Umekawa reported that phosphorylation at Ser 624 and 827 of Ksp1 by PKA is responsible for activation of TORC1 (40). However, these residues were not responsible for phosphorylation of Tif4631 (Supplementary Figure S9), suggesting that the function of Ksp1 in autophagy is also unrelated to its function in post-transcriptional regulation. Taken together, our results suggest that the novel role of Ksp1 in post-transcriptional regulation is distinguishable from known functions of Ksp1 in filamentous growth and autophagy.
The mammalian homolog of Ksp1 has not yet been reported. Among known kinases of mammalian eIF4G, only Pak2 is activated under stress conditions (26). However, we do not regard Pak2 as the mammalian homolog of Ksp1 because (i) homology between the two proteins is low; (ii) although Pak2 is regulated by the TSC1/2 complex, a negative regulator of mTOR, its regulation is independent of mTOR (48); and (iii) Pak2-mediated phosphorylation of eIF4G decreases the global translation rate (26), whereas Ksp1-mediated phosphorylation does not. It has been reported that mTOR regulates the N-terminus region of eIF4G, leading to phosphorylation at Ser 1108, 1148 and 1192 of eIF4G (25). The mechanism underlying how mTOR regulates the N-terminus of eIF4G has not yet been elucidated. It would be interesting to investigate whether mTOR utilizes a Ksp1-like kinase in regulating eIF4G, as observed in yeast.
Although we provide evidence in this study that phosphorylation of eIF4G by Ksp1 promotes the recruitment of Dhh1 to the target mRNAs, the precise mechanism underlying how phosphorylation of eIF4G regulates Dhh1 is not yet known. Given that eIF4G is a scaffold protein that modulates translation with its interactors, it is likely that the recruitment of Dhh1 is regulated by one of the eIF4G interactors. Ded1 is an RNA helicase that has direct interactions with eIF4G to form Ded1–eIF4F–RNA complex at the early translation initiation step (49). Interestingly, GST pull-down assays showed that the interaction affinity of Ded1 with Tif4631 was decreased under glucose deprivation conditions but was restored by deletion of KSP1 (Supplementary Figure S10A). Co-immunoprecipitation experiments showed similar results to those obtained by GST pull-down assays (Supplementary Figure S10B). These observations suggest that Ded1 has a lower affinity for phosphorylated Tif4631 under glucose deprivation conditions. The physiological role of Ded1 is controversial. It stimulates translation by rewinding 5′-UTR structure to increase the access of 48S pre-initiation complex, and thereby translation is impaired in Ded1-defective mutant (50,51). On the other hand, overexpression of Ded1 inhibits translation (52). Moreover, Ded1 has been shown to stimulate the formation of stress granules and P-bodies. Taken together, it seems that Ded1 modulates the mRNA status between translational activation, translational repression and accumulation. One plausible model for explaining how Ded1 works in translation is that Ded1 promotes transition from a P-body mRNP into a Ded1–eIF4F–mRNA complex (53). We found that glucose deprivation induces RNA-mediated interaction between Tif4631 and Dhh1, which is abolished in ksp1Δ cells (Supplementary Figure S10C and D). Given that Tif4631–Ded1 interaction is in inverse relation to Tif4631–Dhh1 interaction in our results, it is presumable that the decrease in Tif4631–Ded1 interaction may lead to the recruitment of Dhh1 to the Tif4631-associated mRNAs under glucose deprivation conditions.
A recent study that categorized 2767 yeast mRNAs using the RIP-seq enrichment profiles for the closed loop complex components (eIF4G1, eIF4G2 and Pab1) and 4E-BPs highlights diverse mechanisms for the translation initiation (54). Although both glycolytic mRNAs and RP mRNAs have high ribosome occupancies and thus are heavily translated, their translation initiation is differently regulated. Many RP mRNAs are included in a group (Group III) that is enriched specifically with all of the closed loop complex components, indicating that this group represents mRNAs that rely on the closed loop complex for protein synthesis. In contrast, almost all of glycolytic mRNAs are under-represented with the closed loop complex components except Pab1, suggesting that Pab1 plays a key role in the translation of glycolytic mRNAs in a closed loop-independent manner. Given the different effects of Ksp1 on the translation efficiency of glycolytic mRNAs and RP mRNAs (Figure 6C and Supplementary Figure S7), it is likely that Ksp1-dependent recruitment of Dhh1 cannot inhibit translation of mRNAs that interact poorly with the closed loop complex. Consistent with this notion, the translation efficiency of RPL8A mRNA, which is exceptionally included in the same group (Group I) with glycolytic mRNAs (54), was not affected by Ksp1 (Figure 6C). RPS28B mRNA, whose translation efficiency was also independent of Ksp1 (Figure 6C), is classified in a different subgroup from the majority of RP mRNAs. MFA2 mRNA is not included in the group of glycolytic mRNAs nor in the group of RP mRNAs.
Calorie restriction extends lifespan in a wide spectrum of organisms including yeast, worms and mice (55). In yeast, calorie restriction causes metabolic shift from glycolysis toward respiration, leading to extension of lifespan (56). Mutants of glycolytic pathway exhibit prolonged lifespan in yeast, and impaired glycolytic pathway induced by 2-deoxy-D-glucose also extends lifespan in Caenorhabditis elegans (57), indicating the causal effect of glycolysis on lifespan. Intriguingly, although ksp1Δ cells show increased autophagy and decreased TORC1 activity, which are known to promote lifespan extension, both chronological and replicative lifespan of ksp1Δ cells are significantly reduced (58–60). In this study, we observed that the degradation of glycolytic mRNAs is impaired in ksp1Δ cells under glucose deprivation conditions. Presumably, the maintenance of elevated glycolytic mRNA levels may be related to lifespan reduction in ksp1Δ cells. It remains to be elucidated whether and how the function of Ksp1 in eIF4G phosphorylation and mRNA decay contribute to the regulation of lifespan in yeast.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ji-Sook Han for generously providing the plasmids for the Tet-off system, V. Narry Kim and Jong-Eun Park for technical assistance in polysome profiling and Gyubum Lim for technical assistance in western blotting. We also thank members of the Huh laboratory for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [2015R1A2A1A01007871]. Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Buchan J.R. mRNP granules: assembly, function, and connections with disease. RNA Biol. 2014; 11:1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kloc M., Zearfoss N.R., Etkin L.D.. Mechanisms of subcellular mRNA localization. Cell. 2002; 108:533–544. [DOI] [PubMed] [Google Scholar]
- 3. Decker C.J., Parker R.. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012; 4:a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinnebusch A.G., Natarajan K.. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 2002; 1:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cherkasova V.A., Hinnebusch A.G.. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003; 17:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaman S., Lippman S.I., Zhao X., Broach J.R.. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008; 42:27–81. [DOI] [PubMed] [Google Scholar]
- 7. Gingras A.C., Gygi S.P., Raught B., Polakiewicz R.D., Abraham R.T., Hoekstra M.F., Aebersold R., Sonenberg N.. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999; 13:1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hay N., Sonenberg N.. Upstream and downstream of mTOR. Genes Dev. 2004; 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- 9. Altmann M., Schmitz N., Berset C., Trachsel H.. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997; 16:1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cosentino G.P., Schmelzle T., Haghighat A., Helliwell S.B., Hall M.N., Sonenberg N.. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000; 20:4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cridge A.G., Castelli L.M., Smirnova J.B., Selley J.N., Rowe W., Hubbard S.J., McCarthy J.E.G., Ashe M.P., Grant C.M., Pavitt G.D.. Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins. Nucleic Acids Res. 2010; 38:8039–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castelli L.M., Lui J., Campbell S.G., Rowe W., Zeef L.A.H., Holmes L.E.A., Hoyle N.P., Bone J., Selley J.N., Sims P.F.G. et al. Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated. Mol. Biol. Cell. 2011; 22:3379–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwartz D.C., Parker R.. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2000; 20:7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munchel S.E., Shultzaberger R.K., Takizawa N., Weis K.. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell. 2011; 22:2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coller J., Parker R.. General translational repression by activators of mRNA decapping. Cell. 2005; 122:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R.. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001; 7:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang L.C., Lee F.J.S.. The RNA helicase Dhh1p cooperates with Rbp1p to promote porin mRNA decay via its non-conserved C-terminal domain. Nucleic Acids Res. 2012; 40:1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pedro-Segura E., Vergara S.V., Rodriguez-Navarro S., Parker R., Thiele D.J., Puig S.. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 2008; 283:28527–28535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu G., McQuiston T., Bernard A., Park Y.-D., Qiu J., Vural A., Zhang N., Waterman S.R., Blewett N.H., Myers T.G. et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 2015; 17:930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Braun K.A., Vaga S., Dombek K.M., Fang F., Palmisano S., Aebersold R., Young E.T.. Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci. Signal. 2014; 7:ra64. [DOI] [PubMed] [Google Scholar]
- 21. Ramachandran V., Shah K.H., Herman P.K.. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell. 2011; 43:973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tuazon P.T., Merrick W.C., Traugh J.A.. Comparative analysis of phosphorylation of translational initiation and elongation factors by seven protein kinases. J. Biol. Chem. 1989; 264:2773–2777. [PubMed] [Google Scholar]
- 23. Morley S.J., Traugh J.A.. Differential stimulation of phosphorylation of initiation factors eIF-4F, eIF-4B, eIF-3, and ribosomal protein S6 by insulin and phorbol esters. J. Biol. Chem. 1990; 265:10611–10616. [PubMed] [Google Scholar]
- 24. Dobrikov M., Dobrikova E., Shveygert M., Gromeier M.. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase C{alpha} regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 2011; 31:2947–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raught B., Gingras A.C., Gygi S.P., Imataka H., Morino S., Gradi A., Aebersold R., Sonenberg N.. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000; 19:434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ling J., Morley S.J., Traugh J.A.. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J. 2005; 24:4094–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qin H., Raught B., Sonenberg N., Goldstein E.G., Edelman A.M.. Phosphorylation screening identifies translational initiation factor 4GII as an intracellular target of Ca(2+)/calmodulin-dependent protein kinase I. J. Biol. Chem. 2003; 278:48570–48579. [DOI] [PubMed] [Google Scholar]
- 28. Sherman F. Getting started with yeast. Meth. Enzymol. 2002; 350:3–41. [DOI] [PubMed] [Google Scholar]
- 29. Mitchell S.F., Jain S., She M., Parker R.. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013; 20:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buchan J.R., Parker R.. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009; 36:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T.. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics. 2006; 5:749–757. [DOI] [PubMed] [Google Scholar]
- 32. Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012; 192:73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hughes Hallett J.E., Luo X., Capaldi A.P.. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics. 2014; 198:773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanz P. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 2003; 31:178–181. [DOI] [PubMed] [Google Scholar]
- 35. Binda M., Peli-Gulli M.P., Bonfils G., Panchaud N., Urban J., Sturgill T.W., Loewith R., De Virgilio C.. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009; 35:563–573. [DOI] [PubMed] [Google Scholar]
- 36. Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R.. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009; 23:1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oliveira A.P., Ludwig C., Zampieri M., Weisser H., Aebersold R., Sauer U.. Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci. Signal. 2015; 8:rs4. [DOI] [PubMed] [Google Scholar]
- 38. Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., Hall M.N.. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 2010; 21:3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bharucha N., Ma J., Dobry C.J., Lawson S.K., Yang Z., Kumar A.. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol. Biol. Cell. 2008; 19:2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umekawa M., Klionsky D.J.. Ksp1 kinase regulates autophagy via the target of rapamycin complex 1 (TORC1) pathway. J. Biol. Chem. 2012; 287:16300–16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fleischmann M., Stagljar I., Aebi M.. Allele-specific suppression of a Saccharomyces cerevisiae prp20 mutation by overexpression of a nuclear serine/threonine protein kinase. Mol. Gen. Genet. 1996; 250:614–625. [DOI] [PubMed] [Google Scholar]
- 42. Jiao X., Xiang S., Oh C., Martin C.E., Tong L., Kiledjian M.. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010; 467:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Decker C.J., Parker R.. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993; 7:1632–1643. [DOI] [PubMed] [Google Scholar]
- 44. Muhlrad D., Parker R.. Premature translational termination triggers mRNA decapping. Nature. 1994; 370:578–581. [DOI] [PubMed] [Google Scholar]
- 45. Carroll J.S., Munchel S.E., Weis K.. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J. Cell Biol. 2011; 194:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haimovich G., Medina D.A., Causse S.Z., Garber M., Millán-Zambrano G., Barkai O., Chávez S., Pérez-Ortín J.E., Darzacq X., Choder M.. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013; 153:1000–1011. [DOI] [PubMed] [Google Scholar]
- 47. Laxman S., Tu B.P.. Multiple TORC1-associated proteins regulate nitrogen starvation-dependent cellular differentiation in Saccharomyces cerevisiae. PLoS One. 2011; 6:e26081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alves M.M., Fuhler G.M., Queiroz K.C.S., Scholma J., Goorden S., Anink J., Spek C.A., Hoogeveen-Westerveld M., Bruno M.J., Nellist M. et al. PAK2 is an effector of TSC1/2 signaling independent of mTOR and a potential therapeutic target for Tuberous Sclerosis complex. Sci. Rep. 2015; 5:14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hilliker A., Gao Z., Jankowsky E., Parker R.. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell. 2011; 43:962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chuang R.Y., Weaver P.L., Liu Z., Chang T.H.. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997; 275:1468–1471. [DOI] [PubMed] [Google Scholar]
- 51. Sen N.D., Zhou F., Ingolia N.T., Hinnebusch A.G.. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015; 25:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beckham C., Hilliker A., Cziko A.-M., Noueiry A., Ramaswami M., Parker R.. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell. 2008; 19:984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hilliker A., Gao Z., Jankowsky E., Parker R.. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell. 2011; 43:962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costello J., Castelli L.M., Rowe W., Kershaw C.J., Talavera D., Mohammad-Qureshi S.S., Sims P.F.G., Grant C.M., Pavitt G.D., Hubbard S.J. et al. Global mRNA selection mechanisms for translation initiation. Genome Biol. 2015; 16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heilbronn L.K., Ravussin E.. Calorie restriction and aging: review of the literature and implications for studies in humans. Am. J. Clin. Nutr. 2003; 78:361–369. [DOI] [PubMed] [Google Scholar]
- 56. Lin S.J., Kaeberlein M., Andalis A.A., Sturtz L.A., Defossez P. A., Culotta V.C., Fink G.R., Guarente L.. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002; 418:344–348. [DOI] [PubMed] [Google Scholar]
- 57. Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M.. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007; 6:280–293. [DOI] [PubMed] [Google Scholar]
- 58. Powers R.W., Kaeberlein M., Caldwell S.D., Kennedy B.K., Fields S.. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006; 20:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Garay E., Campos S.E., González de la Cruz J., Gaspar A.P., Jinich A., Deluna A.. High-resolution profiling of stationary-phase survival reveals yeast longevity factors and their genetic interactions. PLoS Genet. 2014; 10:e1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao W., Zheng H. Z., Zhou T., Hong X.S., Cui H.J., Jiang Z.W., Chen H.J., Zhou Z.J., Liu X.G.. CTT1 overexpression increases the replicative lifespan of MMS-sensitive Saccharomyces cerevisiae deficient in KSP1. Mech. Ageing Dev. 2017; 164:27–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.