Abstract
Purpose
To obtain whole lung morphometry measurements from 129Xe in a single breath‐hold with 3D multiple b‐value 129Xe diffusion‐weighted MRI (DW‐MRI) with an empirically optimized diffusion time and compressed sensing for scan acceleration.
Methods
Prospective three‐fold undersampled 3D multiple b‐value hyperpolarized 129Xe DW‐MRI datasets were acquired, and the diffusion time (Δ) was iterated so as to provide diffusive length scale (LmD) estimates from the stretched exponential model (SEM) that are comparable to those from 3He. The empirically optimized 129Xe diffusion time was then implemented with a four‐fold undersampling scheme and was prospectively benchmarked against 3He measurements in a cohort of five healthy volunteers, six ex‐smokers, and two chronic obstructive pulmonary disease patients using both SEM‐derived LmD and cylinder model (CM)‐derived mean chord length (Lm).
Results
Good agreement between the mean 129Xe and 3He LmD (mean difference, 2.2%) and Lm (mean difference, 1.1%) values was obtained in all subjects at an empirically optimized 129Xe Δ = 8.5 ms.
Conclusion
Compressed sensing has facilitated single‐breath 3D multiple b‐value 129Xe DW‐MRI acquisitions, and results at 129Xe Δ = 8.5 ms indicate that 129Xe provides a viable alternative to 3He for whole lung morphometry mapping with either the SEM or CM. Magn Reson Med 79:2986–2995, 2018. © 2017 The Authors Magnetic Resonance in Medicine published by Wiley Periodicals, Inc. on behalf of International Society for Magnetic Resonance in Medicine. This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.
Keywords: hyperpolarized 129Xe, lung morphometry, compressed sensing, stretched exponential model, hyperpolarized 3He
INTRODUCTION
The apparent diffusion coefficient (ADC) calculated from hyperpolarized 3He diffusion‐weighted MRI (DW‐MRI) has been shown to be sensitive to changes in lung microstructure 1, 2. The non‐Gaussian diffusion behavior of the gas in the lungs results in a non‐monoexponential signal attenuation with increasing b‐value 3. The signal decay is determined by experimental and physiological factors including gas diffusivity, diffusion gradient strengths and timings, and the complexity of alveolar microstructure, which together influence the measurement of ADC 4, 5. Theoretical diffusion models, such as the cylinder model (CM) 6, 7, stretched exponential model (SEM) 8, and q‐space analysis 9, have been proposed to model this non‐Gaussian diffusion behavior and derive estimates of alveolar length scales (i.e., morphometry) from multiple b‐value DW‐MRI acquisitions. Compressed sensing (CS) has enabled multiple b‐value 3He DW‐MRI for 3D whole lung morphometry mapping in a single breath‐hold 10 for quantitative regional assessment of lung microstructure.
With the limited availability of 3He gas 11, 129Xe provides a more cost‐effective alternative for pulmonary MRI, and with advancements in polarization levels 12, 13, recent studies have shown that comparable ventilation and microstructural information can be obtained using both nuclei 14, 15, 16, 17. DW‐MRI with 129Xe is, however, inherently more challenging due to the lower diffusivity and gyromagnetic ratio of 129Xe compared with 3He, resulting in longer diffusion gradient times, longer sequence echo time (TE) and repetition time (TR), and lower image SNR. Despite these challenges, theoretical models have been proposed for interpreting the 129Xe DW‐MRI signal from multiple b‐value acquisitions 18, and estimates of alveolar length scales have been derived from healthy subjects and chronic obstructive pulmonary disease (COPD) patients 19, 20, 21. However, the multiple b‐value interleaves in previous studies were acquired using noncontiguous, relatively thick 2D slices without whole lung coverage—and in some cases in separate breath‐holds—due to the associated long scan times. Furthermore, to our knowledge, no direct comparison of alveolar length scales derived from application of theoretical diffusion models of 3He and 129Xe in vivo have yet been presented.
In this study, compressed sensing acceleration methods developed for 3He 10 were adapted for 3D multiple b‐value 129Xe DW‐MRI in a single breath‐hold, and 3D morphometric maps of mean diffusive length scale (LmD) were generated using the SEM. Results were compared against equivalent 3D 3He LmD morphometric maps acquired with CS, and an optimal 129Xe diffusion time of Δ = 8.5 ms was derived empirically. Prospective acquisitions with the optimal 129Xe diffusion time were then benchmarked in healthy volunteers, ex‐smokers, and COPD patients with both SEM‐derived LmD and CM‐derived mean chord length (Lm) measurements.
THEORY
The Stretched Exponential Model
The non‐Gaussian signal decay from an imaging voxel can be modeled as the superposition of signals with different apparent diffusivities ( ):
| (1) |
where is the signal when , is the signal corresponding to a non‐zero b‐value, are all possible apparent diffusivities between 0 and (the free diffusion coefficient of 3He or 129Xe in air/N2), and is the probability distribution associated with the apparent diffusivities. The non‐Gaussian HP gas diffusion signal decay in the lungs can be well described by an SEM fit (Equation (2)) 22.
| (2) |
With 3He DW‐MRI, the SEM‐derived parameters of distributed diffusivity coefficient (DDC) and heterogeneity index (α) have been shown to be sensitive to changes in lung microstructure and are valid over a range of experimental conditions. DDC is dependent on diffusion time, while α has been demonstrated to be insensitive to lung inflation and experimental diffusion time 23. A numerical expression for can be estimated from the SEM‐derived parameters using the approach developed by Berberan‐Santos et al. 24:
| (3) |
where is 1/DDC, and is defined by
| (4) |
The parameters B and C are functions related to α, and parameters at specific α values can be found in Table 1 of Berberan‐Santos et al. 24. Interpolation can be used to derive the corresponding parameters B and C for other α values. The expression for can subsequently be related to a distribution of diffusion length scales associated with the different apparent diffusivities through the 1D diffusion equation LD = (2DΔ)½ (i.e., root mean squared displacements, where Δ is the diffusion time). The distributions should then represent the distribution of microscopic dimensions of the airways (i.e., the diffusion‐restricting boundaries) contained within a given voxel. These distributions can then be used to calculate the mean diffusion length scale (LmD) as a quantitative estimate of the mean acinar airway dimensions within a given voxel. The LmD metric should therefore be analogous to the calculation of mean linear intercept length (Lx) from histology.
Table 1.
Summary of Subject Demographics and Pulmonary Function Test Data
| Subjects | Age | Sex | FEV1 (% pred) | FEV1/FVC (%) | TLC (% pred) | RV (% pred) | TLCO (% pred) | Smoking Pack Years |
|---|---|---|---|---|---|---|---|---|
| Healthy volunteers | ||||||||
| HV1 | 26 | Male | 102.9 | 81.2 | 105.6 | 107.0 | — | — |
| HV2 | 31 | Male | 102.0 | 82.8 | 100.3 | 85.0 | — | — |
| HV3 | 34 | Male | 77.0 | 88.0 | 91.7 | 107.2 | — | — |
| HV4 | 31 | Male | 105.0 | 87.0 | 91.6 | 70.7 | — | — |
| HV5 | 33 | Male | 85.1 | 76.0 | 84.0 | 74.1 | — | — |
| Ex‐smokers | ||||||||
| ES1 | 47 | Female | 86.7 | 68.8 | 108.4 | 105.3 | 93.0 | 30.0 |
| ES2 | 51 | Male | 95.2 | 69.2 | 106.7 | 100.9 | 97.2 | 30.0 |
| ES3 | 53 | Female | 90.1 | 59.7 | 130.0 | 139.2 | 99.2 | 4.1 |
| ES4 | 55 | Male | 107.7 | 67.5 | 132.0 | 127.0 | 86.5 | 10.0 |
| ES5 | 52 | Female | 90.9 | 71.0 | 101.9 | 106.6 | 89.3 | 25.0 |
| ES6 | 50 | Male | 111.6 | 96.1 | 109.0 | 89.4 | 98.4 | 22.5 |
| COPD patients | ||||||||
| COPD1 | 62 | Female | 39.6 | 36.5 | — | — | 37.4 | — |
| COPD2 | 64 | Female | 69.7 | 50.0 | — | — | 61.0 | — |
This method of calculating LmD differs from the method used to derive mean chord length (Lm) with the CM. In the CM, the underlying assumptions are that the acinar airways are considered cylindrical objects and thus the HP gas diffusion signal can be described by two anisotropic diffusion coefficients, longitudinal (DL) and transverse (DT). Phenomenological expressions were empirically optimized from Monte Carlo simulations to relate DL and DT to the cylindrical lung airway parameters, outer airway radii (R) and alveolar sleeve depth (h) 6, 25. Lm is subsequently derived from the alveoli surface area and volume based upon the geometrical parameters of R and h 7.
METHODS
All in vivo MRI experiments were performed under the approval of the UK National Research Ethics Committee and the local National Health Service research office. All CS simulations and lung morphometry calculations were implemented in‐house using MATLAB (MathWorks, Natick, Massachusetts, USA) software. The signal‐to‐noise ratio (SNR) for each dataset was computed in the magnitude images (b = 0) by dividing the mean signal of the entire segmented lung region by a region of background noise corrected for Rician distribution bias. It should be noted that SNR calculated from CS images present a biased measure of SNR, due to the denoising process associated with CS reconstruction.
3D Multiple b‐Value 129Xe DW‐MRI with CS
A fully sampled 3D 129Xe DW‐MRI dataset was acquired from a healthy male volunteer (HV1) on a 1.5 T (GE HDx) MR scanner using a flexible quadrature transmit/receive vest coil (Clinical MR Solutions, Brookfield, Wisconsin, USA) which was tuned to the Larmor frequency of 129Xe at 1.5 T (17.66 MHz). All lung imaging was performed at a lung volume of functional residual capacity plus 1L following inhalation of a dose of 800 mL enriched Xe [86% 129Xe, ∼30% polarization 12, 13] mixed with 200 mL of N2. Image acquisition parameters were: 3D spoiled gradient echo sequence; 2 × interleaves (b = 0, 12 s/cm2); elliptical‐centric phase encoding; in‐plane resolution = 64 × 52 (6.25 mm pixel dimension); 18 effective coronal slices (15 mm slice thickness); field of view = 40 × 32.5 × 27 cm3; TE/TR = 11.2/14.4 ms; diffusion time (Δ) = 5 ms (diffusion gradient strength = 22.7 mT/m, ramp time = 0.3 ms, plateau time = 3 ms, gap between lobes = 1.4 ms); flip angle = 2.2°; and bandwidth = ±6.97 KHz.
129Xe Δ = 5 ms was first chosen as it corresponds to the diffusion time originally proposed for 129Xe lung morphometry with the CM 18. This time was derived theoretically such that acinar airway geometrical parameters from the CM for 129Xe would be the same as those obtained with 3He 18, and these values have been subsequently used in 2D 129Xe DW‐MRI experimental studies 20, 21. Retrospective CS simulations of the fully sampled dataset with acceleration factors (AF) between 2 and 5 were implemented using the methodology described previously for 3He 10. The Wilcoxon signed‐rank test was employed to assess differences in fully sampled and retrospectively reconstructed ADC maps for each AF on a pixel‐by‐pixel basis.
The optimum k‐space sampling pattern for three‐fold undersampling was chosen based on the simulation results and was used for prospective acquisition of 3D 129Xe multiple b‐value DW‐MRI data from four healthy volunteers (HV1, HV2, HV3, HV4). Prospective data were acquired with an inhaled gas mixture of 750 mL 129Xe and 250 mL nitrogen, with imaging parameters as for the fully sampled acquisition other than the following: four interleaves (b = 0, 12, 20, 30 s/cm2); TE/TR = 11.7/15.0 ms; Δ = 5 ms (maximum diffusion gradient strength = 31.9 mT/m, ramp time = 0.3 ms, plateau time = 3.5 ms, gap = 0.9 ms); and flip angle = 2.7°. The AF of 3 reduces the scan time from 57 to 19 s. 129Xe LmD maps were calculated using the SEM, and results were compared with LmD maps derived from the same volunteers' lungs using 3He DW‐MRI as described by Chan et al. 10. 3He LmD at 3He Δ = 1.6 ms was chosen for comparison because healthy and COPD LmD values derived at this diffusion time have been demonstrated to match histologically derived healthy and COPD mean linear intercept values 26.
Empirical Determination of Optimal 129Xe Diffusion Time
With the aim of obtaining the best agreement between 129Xe and 3He lung morphometry results [rather than simply using the 129Xe Δ = 5 ms proposed by Sukstanskii and Yablonskiy 18], HV1 was imaged at additional diffusion times (Δ = 5, 7, 8, and 10 ms). 129Xe Δ = 10 ms was chosen as it corresponds to the same 1D characteristic free diffusion length ( ∼530 µm) as experienced in the benchmark 3He experiment (assuming , and ΔHe = 1.6 ms). Each additional scan was acquired with the same gas mixture and b‐values as the previous prospective CS acquisitions at 129Xe Δ = 5 ms, and LmD maps were calculated from each dataset.
Benchmarking of Empirically Optimized 129Xe Diffusion Time
The empirically optimized diffusion time (129Xe Δ = 8.5 ms [see Results]) was then benchmarked against 3He equivalent measurements for lung morphometry mapping over different ranges of acinar length scales that are experienced with smoking‐related emphysema. Five healthy volunteers (age, 31.0 ± 3.1 years), six ex‐smokers (age, 51.3 ± 2.7 years), and two COPD patients (age, 63.0 ± 1.4 years, GOLD II‐IV) were recruited for this preliminary study. Subject demographics and pulmonary function test (PFT) data for each subject are summarized in Table 1.
Each subject was imaged with 3D multiple b‐value 129Xe DW‐MRI, using 750 mL of inhaled 129Xe and the following imaging parameters: TE/TR = 14.0/17.3 ms; maximum DW gradient strength = 32.6 mT/m; Δ = 8.5 ms; ramp time = 0.3 ms; plateau time = 2.3 ms; gap = 5.6 ms; and flip angle = 3.1°. Using 129Xe Δ = 8.5 ms, the duration of three‐fold undersampled CS scans was increased by 3 s due to the increased diffusion time. Therefore, four‐fold undersampling (AF = 4) was now implemented in the subsequent prospective CS acquisitions to further reduce the breath‐hold to 16 s, similar to the 15 s acquisition for 3He 10, and to demonstrate the clinical viability of this sequence. 3D 3He DW‐MRI was acquired in same‐day scan sessions for all subjects (except for HV1–HV3, for whom 3He data were acquired approximately 1 year previously), with experimental parameters described previously 10. 129Xe and 3He LmD maps were derived and compared in each subject.
Finally, the applicability of 129Xe Δ = 8.5 ms to CM derivations of lung morphometry parameters was assessed. The 129Xe‐based CM phenomenological expressions are optimized for 129Xe Δ = 5 ms; however, if the same theoretical free diffusion length is probed with both nuclei (i.e., ΔHe = 1.6 ms and ΔXe = 10 ms), the original 3He‐based phenomenological expressions should in theory be applicable for derivation of 129Xe lung morphometry parameters 18. Initial CM analysis of 129Xe DW‐MRI data in healthy subjects at 129Xe Δ = 8.5 ms and 129Xe Δ = 10 ms, suggested that, as with the SEM, more consistent 129Xe lung morphometry results were obtained with 129Xe Δ = 8.5 ms (see Discussion). The 3D multiple b‐value 129Xe DW‐MRI data at 129Xe Δ = 8.5 ms was therefore analyzed using the 3He‐based CM phenomenological expressions 7, and the 129Xe mean chord length (Lm) was hence derived and compared with 3He‐derived Lm for each subject in the preliminary study.
RESULTS
3D Multiple b‐Value 129Xe DW‐MRI with CS
The SNR of the fully sampled 129Xe DW‐MRI dataset was 25. Optimal k‐space undersampling patterns for different AFs were determined through CS simulations. Retrospectively reconstructed datasets from each optimal undersampling pattern showed a small increase in mean absolute error (MAE) of normalized signal intensity value for the b = 0 data (from 2.27% at AF = 2 to 4.25% at AF = 5), indicating a good preservation of image details with increased AF (Fig. 1). Whole lung mean ADC histograms and single slice ADC maps generated from the reconstructed CS datasets also demonstrated a good preservation of quantitative information and low MAEADC (Fig. 2). Wilcoxon signed‐rank tests for each AF found no significant differences (P > 0.05) between CS‐reconstructed and fully sampled ADC maps on a pixel‐by‐pixel basis, confirming preservation of quantitative information and indicating that CS is suitable for 3D 129Xe multiple b‐value DW‐MRI.
Figure 1.
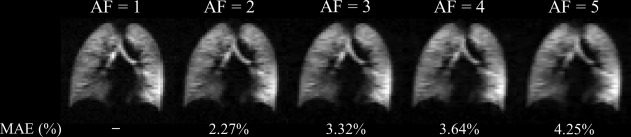
CS simulation results for 3D 129Xe DW‐MRI. Reconstructed magnitude image (b = 0) for each AF, with corresponding MAE values (AF = 1; fully sampled dataset [SNR = 25]).
Figure 2.
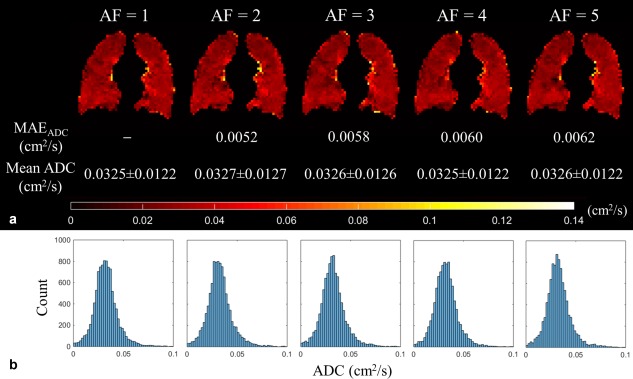
ADC results for 3D 129Xe DW‐MRI CS simulations. (a) Single‐slice ADC maps with the MAEADC, and mean global ADC values for each AF. (b) Corresponding whole lung ADC histograms for each AF.
Prospective 3D 129Xe multiple b‐value DW‐MRI was acquired in four healthy volunteers with AF = 3 and 129Xe Δ = 5 ms, and resulting ADC and LmD maps were compared with previously calculated lung microstructural maps acquired using 3D 3He multiple b‐value DW‐MRI. Mean SNR for the four prospective 129Xe datasets was 40. The prospective CS whole lung mean 129Xe ADC value for volunteer HV1 (0.0329 cm2/s) was very similar (+1.2% difference) to the fully sampled mean ADC value (0.0325 cm2/s) that was obtained for CS simulations. Example 129Xe and 3He LmD maps from the comparative slices in HV1 are shown in Figure 3 and a summary of mean ADC and LmD values for each volunteer is provided in Table 2. At 129Xe Δ = 5 ms, mean 129Xe LmD values for all subjects were ∼50 µm smaller than the corresponding mean 3He values.
Figure 3.
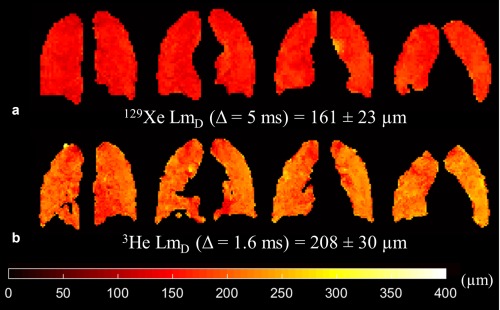
Prospective CS results for a healthy volunteer (HV1) (SNR = 30). (a) Example 129Xe LmD maps derived from 3D multiple b‐value 129Xe DW‐MRI. (b) Example 3He LmD maps in comparative slices demonstrate the mismatch in LmD values between the two nuclei.
Table 2.
Summary of Whole Lung Mean ADC and LmD Values for Four Healthy Volunteers Derived from Prospective 3D Multiple b‐Value 129Xe and 3He DW‐MRI With CS
| Subjects | 129Xe ADC (cm2/s) (Δ = 5 ms) | 129Xe LmD (µm) (Δ = 5 ms) | 3He ADC (cm2/s) (Δ = 1.6 ms) | 3He LmD (µm) (Δ = 1.6 ms) |
|---|---|---|---|---|
| HV1 | 0.033 ± 0.012 | 161 ± 23 | 0.182 ± 0.085 | 208 ± 30 |
| HV2 | 0.039 ± 0.012 | 176 ± 20 | 0.196 ± 0.077 | 223 ± 24 |
| HV3 | 0.030 ± 0.011 | 157 ± 19 | 0.166 ± 0.068 | 205 ± 23 |
| HV4 | 0.030 ± 0.011 | 156 ± 18 | 0.169 ± 0.065 | 210 ± 20 |
Empirical Determination of Optimal 129Xe Diffusion Time
A strong positive linear correlation (r = 0.998, P < 0.001) was established between 129Xe LmD and diffusion times, and at Δ = 8.5 ms the 129Xe LmD value best matched the volunteer's 3He LmD value (Fig. 4a). In contrast to LmD, mean 129Xe ADC decreased with increasing diffusion time; a 12.5% decrease in mean 129Xe ADC was observed from Δ = 5 ms to 10 ms. The relationship between 129Xe ADC and diffusion time was nonlinear, however, and best fitted a logarithmic function (R2 = 0.961) (Fig. 4b).
Figure 4.
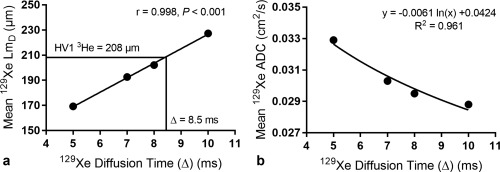
Global mean 129Xe LmD and ADC results at different 129Xe diffusion times for one healthy volunteer. (a) A strong linear dependence in 129Xe diffusion time and mean 129Xe LmD value was observed. At 129Xe Δ=8.5 ms, the 129Xe LmD matches the volunteer's corresponding 3He LmD value. (b) Mean 129Xe ADC decreases with increasing diffusion time in a nonlinear logarithmic relationship.
Benchmarking of Empirically Optimized 129Xe Diffusion Time
The mean 3He and 129Xe SNR of the b = 0 image for all preliminary study subjects was 32 and 65, respectively. A summary of 129Xe LmD and corresponding 3He LmD values are shown in Table 3. An improved matching of mean 129Xe and 3He LmD was obtained with the empirically optimized diffusion time, and this is visible in example LmD maps from three representative subjects (Fig. 5). A difference in LmD of less than 7% was observed in all subjects, with a mean difference (129Xe − 3He) in all subjects of −2.2%. Figure 6a shows a very strong correlation (r = 0.987, P < 0.001) between individual lung 3He and 129Xe mean LmD values in all subjects. LmD values fall around the line of equality, and this good agreement was confirmed by Bland‐Altman analysis (Fig. 6b) of individual lung LmD values, where a mean bias of −2.1% (−4.8 µm) for 129Xe mean LmD with a 95% confidence interval of −6.7% to 2.5% (−14.8 to 5.2 µm) was observed.
Table 3.
Summary of 129Xe Whole Lung SEM‐Derived LmD and CM‐Derived Lm Values for Healthy Volunteers, Ex‐smokers, and COPD Patients Acquired With AF = 4 and 129Xe Δ = 8.5 ms and Their Corresponding 3He Mean Lung Morphometry Values (AF = 3, 3He Δ = 1.6 ms)
| Subjects | Stretched Exponential Model | Cylinder Model (3He‐based) | ||||
|---|---|---|---|---|---|---|
| 129Xe LmD (µm) | 3He LmD (µm) | LmD Difference (%) | 129Xe Lm (µm) | 3He Lm (µm) | Lm Difference (%) | |
| Healthy volunteers | ||||||
| HV1 | 205 | 208 | −1.4 | 183 | 183 | 0.0 |
| HV2 | 218 | 224 | −2.7 | 222 | 210 | +5.6 |
| HV3 | 206 | 205 | +0.5 | 196 | 171 | +12.5 |
| HV4 | 200 | 210 | −4.8 | 173 | 178 | −3.1 |
| HV5 | 192 | 205 | −6.3 | 164 | 170 | −3.6 |
| Mean HV | 204 | 210 | −2.9 | 188 | 182 | +2.3 |
| Ex‐smokers | ||||||
| ES1 | 232 | 234 | −0.9 | 259 | 222 | +14.3 |
| ES2 | 230 | 234 | −1.7 | 254 | 240 | +5.3 |
| ES3 | 234 | 236 | −0.8 | 266 | 250 | +6.0 |
| ES4 | 245 | 246 | −0.4 | 326 | 335 | −2.7 |
| ES5 | 221 | 231 | −4.3 | 222 | 226 | −2.1 |
| ES6 | 217 | 215 | +0.9 | 217 | 201 | +7.2 |
| Mean ES | 230 | 233 | −1.2 | 257 | 246 | +4.7 |
| COPD patients | ||||||
| COPD1 | 317 | 323 | −1.9 | 639 | 671 | −5.0 |
| COPD2 | 251 | 263 | −4.6 | 318 | 381 | −19.8 |
| Mean COPD | 284 | 293 | −3.2 | 478 | 526 | −12.4 |
| Overall mean | — | — | −2.2 | — | — | +1.1 |
Figure 5.
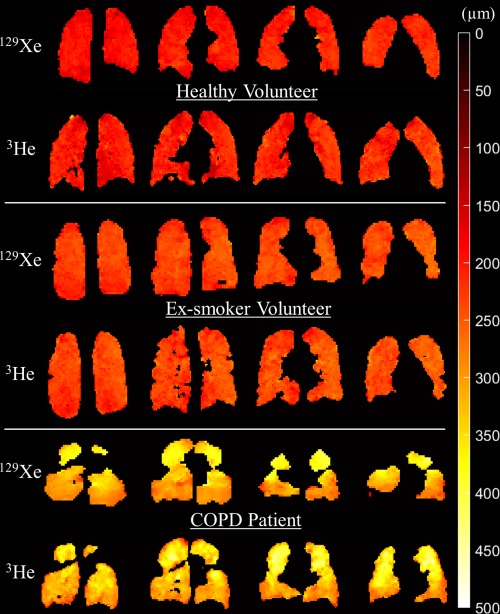
Comparison of 129Xe and 3He example slice LmD maps for a representative healthy, ex‐smoker, and COPD subject. 129Xe LmD maps derived using 3D multiple b‐value 129Xe DW‐MRI at an empirically optimized diffusion time Δ = 8.5 ms demonstrate good agreement with 3He LmD maps. 3D 129Xe DW‐MRI mean SNR was 37, 44, 80 for the representative healthy volunteer, ex‐smoker, and COPD patient, respectively.
Figure 6.
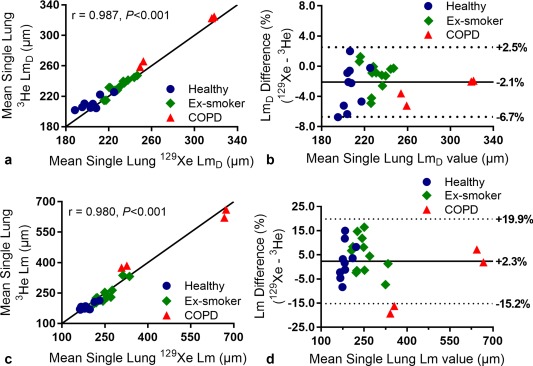
(a) Comparison of 129Xe and 3He mean single (left and right) lung LmD values for all subjects. The solid line represents the line of equality. (b) Bland‐Altman analysis of mean single lung LmD values. The percentage difference (129Xe – 3He) between the two nuclei is plotted against the mean single lung LmD value of the two nuclei for all subjects. The solid line represents the mean percentage difference, and the two dotted lines indicate the 95% (±1.96 SD) difference range. (c) Comparison of 129Xe and 3He mean single lung Lm values derived from the cylinder model for all subjects. Both 129Xe and 3He data were analyzed with the 3He‐based cylinder model. The solid line represents the line of equality. (d) Corresponding Bland‐Altman analysis of mean single lung Lm values.
The mean difference in 129Xe and 3He CM Lm values was +1.1% (Table 3), demonstrating a similar level of agreement in CM‐derived Lm at 129Xe Δ = 8.5 ms as SEM‐derived LmD. 3He and 129Xe CM single lung Lm values were also strongly correlated (r = 0.980, P < 0.001) (Fig. 6c), and Bland‐Altman analysis of mean single lung Lm values indicates a mean bias of +2.3% in 129Xe Lm values with a 95% confidence interval of −15.2% to 19.9% (Fig. 6d).
DISCUSSION
3D Multiple b‐Value 129Xe DW‐MRI with CS
CS has enabled the acquisition of 3D multiple b‐value 129Xe DW‐MRI in a single breath‐hold for the generation of whole lung maps of alveolar diffusion length scale with a voxel size of 6.25 × 6.25 × 15 mm3. Retrospectively undersampled 129Xe datasets demonstrated good preservation of image details and microstructural information with increased undersampling. MAE and MAEADC values from 129Xe CS simulations were similar to those reported with 3He 10. The presence of image blurring in the fully sampled 129Xe images is likely the result of elliptical‐centric phase encode ordering used with 129Xe in contrast to sequential encoding used previously with 3He. Elliptical‐centric phase encoding maximizes SNR at the consequence of increased image blurring with a RF depolarization k‐space filter that originates from the center of k‐space 27. The full width at half maximum values of retrospectively undersampled 129Xe ADC histograms decreased with AF; this trend matches the results of 3He CS simulations 10 and demonstrates decreased spatial heterogeneity associated with the de‐noising reconstruction process of CS. However, this loss of spatial heterogeneity did not result in a statistically significant difference between fully sampled ADC and undersampled CS ADC maps.
Prospective three‐fold undersampled 3D multiple b‐value 129Xe DW‐MRI was acquired in four healthy volunteers at Δ = 5 ms. The difference of + 1.2% between CS (0.0329 cm2/s) and fully sampled mean 129Xe ADC (0.0325 cm2/s) for one volunteer (HV1) was similar to the small differences we reported previously between fully sampled and CS undersampled 2D and 3D 3He ADC values 10, 28. The whole lung mean 129Xe ADC value for all four healthy volunteers (∼0.033 cm2/s) was also consistent with previously reported healthy subject ADC values, with b = 12 s/cm2 at 1.5 T 29. The observed mean LmD mismatch of approximately 50 µm between 3He and 129Xe suggests that the 129Xe diffusion time of Δ = 5 ms, previously proposed for in vivo lung morphometry with the CM 18, is not applicable for 129Xe lung diffusion length scale measurements derived from the SEM.
Empirical Determination of Optimal 129Xe Diffusion Time
Mean 129Xe ADC values (at b = 12 s/cm2) decreased nonlinearly with increasing diffusion time; a trend observed previously in 3He ADC measurements 4, 30. The logarithmic relationship observed between 129Xe ADC and diffusion time also matches the trend observed for 3He ADC 30. The SEM‐derived LmD values exhibited a strong positive linear dependence with Δ over the range of 5–10 ms. The dependence of LmD on Δ reflects the changes in the theoretical characteristic free diffusion lengths probed for each experiment. At Δ = 10 ms, corresponding to the characteristic free diffusion length of 129Xe ( = 530 µm) which is identical to the free diffusion length of 3He in air for the diffusion times used by Chan et al. 10, a mismatch of LmD values was still observed in the data from three healthy volunteers (Fig. 7).
Figure 7.
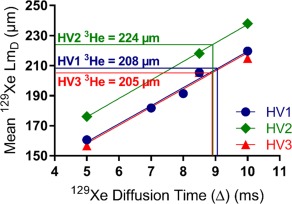
Mean 129Xe LmD results at different 129Xe diffusion times for three healthy volunteers. A strong linear dependence in 129Xe diffusion time and mean 129Xe LmD value was obtained for HV1 (r = 0.98, P = 0.015). When the Δ = 8.5 ms results for HV1 was considered, the diffusion time Δ = 9.1 ms corresponded to the subject's 3He LmD value. A similar diffusion time trend was observed in the other two healthy volunteers (HV2 and HV3).
This mismatch suggests that even at the same characteristic free diffusion length there may be inherent differences in the specific diffusion dephasing regime of the respective gas in the lung alveoli which makes this assumption of Gaussian relation between diffusion length and diffusion time inexact. The differences in diffusion dephasing regime stems from intrinsic properties (i.e., gyromagnetic ratio and diffusivity) of each gas, and thus leads to different mechanisms that contribute to non‐Gaussian diffusion signal behaviors that are not accounted for in the calculation of characteristic free diffusion length. For example, differences in the diffusional dephasing regime due to microscopic background susceptibility gradients may exist between 129Xe and 3He at the same field strength due to the smaller gyromagnetic ratio of 129Xe. These effects on diffusive length scales are similar to the effect of different B0 field strengths on 3He ADC values 5.
Benchmarking of Empirically Optimized 129Xe Diffusion Time
The decision to further accelerate with four‐fold undersampling was motivated by the need to reduce the breath‐hold duration incurred with 129Xe diffusion times > 5 ms. To verify that good agreement in LmD values was obtained with three‐ and four‐fold undersampling, all five healthy volunteers were imaged with an additional AF = 3 129Xe CS acquisition at 129Xe Δ = 8.5 ms. A slice‐by‐slice comparison of mean LmD values for the five healthy volunteers was performed, and Bland‐Altman analysis confirmed a mean bias of +1.5% (+2.9 µm) for AF = 4. The 95% confidence interval of −6.9% to + 10.0% (−13.4 to 19.3 µm) was within typical standard deviation values of lung LmD values in healthy volunteers. This slight increase in mean slice LmD values obtained with AF = 4 is likely the result of CS reconstruction error associated with increased undersampling. In addition, the broad 95% confidence interval range could also be explained by inexact coregistration of image slices due to slight changes in subject position between the AF = 3 and AF = 4 scan sessions. However, the small increase in LmD justifies that implementation of AF = 4 in prospective acquisitions with 129Xe Δ = 8.5 ms. The reduction of scan time to within 16 s is more tolerable for a wider range of subjects, therefore AF = 4 will be used in all subsequent 3D multiple b‐value 129Xe DW‐MRI acquisitions.
Using the empirically optimized diffusion time, 129Xe‐derived LmD values demonstrated improved matching with 3He LmD at 129Xe Δ = 8.5 ms than at 129Xe Δ = 5 ms. The mean difference between whole lung 129Xe and 3He LmD values across all subjects was −2.2%, and the mean bias in individual lung 129Xe LmD values was −2.1%. 129Xe Δ = 8.5 ms was derived from preliminary data, and this small bias may suggest that a different optimal diffusion time (slightly longer than Δ = 8.5 ms) could be used to bring the bias toward 0%. Considering Δ = 8.5 ms LmD for HV1, a 129Xe Δ = 9.1 ms was found to match the volunteer's 3He LmD value (Fig. 7). Additionally, when the previous 129Xe Δ = 5 and 8.5 ms results for HV2 and HV3 are considered in conjunction with an additional acquisition at 129Xe Δ = 10 ms, a similar optimal diffusion time of around 9 ms was obtained as well (Fig. 7). Nevertheless, the observed bias of −2.1% is equivalent to the same‐day reproducibility error (2.1%) 31 of Lm values calculated from multiple b‐value 3He DW‐MRI using the CM. This indicates that any mismatch between 3He and 129Xe LmD values at the 129Xe Δ = 8.5 ms is of the order of same‐day reproducibility error, and we conclude that comparable lung morphometry maps can be obtained with 129Xe.
One limitation of this study is that the 129Xe diffusion time was optimized based upon the LmD results from healthy volunteers only. In subjects with emphysematous changes to alveolar length scales, a different relationship between 129Xe LmD and diffusion time may exist. However, the strong agreement between 129Xe and 3He LmD results from the subsequent prospective acquisitions in healthy volunteers, ex‐smokers, and COPD patients suggests that 129Xe Δ = 8.5–9 ms is valid across a range of alveolar sizes subject to age and smoking‐related emphysema.
The empirically optimized 129Xe Δ = 8.5 ms used in our study is significantly longer than the diffusion time used in other 129Xe lung morphometry studies. In Sukstanskii and Yablonskiy 18, 129Xe Δ = 5 ms was chosen and CM phenomenological expressions for acinar airway geometrical parameters were also recalibrated for 129Xe such that lung morphometry results matched those of 3He. However, it was noted that if the same theoretical free diffusion length is probed with both nuclei, the 3He‐based phenomenological expressions can be applied to derive 129Xe lung morphometry parameters 18. In a small subset of the preliminary study cohort (HV1–HV4), the assumption that, like the SEM, the CM will yield more comparable lung morphometry results at 129Xe Δ = 8.5 ms than with 129Xe Δ = 10 ms was explored. 129Xe Δ = 8.5 and 10 ms data were analyzed with 3He‐based CM parameters, and derived Lm was compared with 3He‐derived Lm values. A mean difference of 4.3% was obtained between 129Xe Δ = 8.5 ms Lm and 3He Lm, whereas at 129Xe Δ = 10 ms the difference was larger (11.5%). These results, albeit in a small subset of subjects, support the implementation of the 3He‐based CM with 129Xe DW‐MRI at 129Xe Δ = 8.5 ms.
The mean 3He Lm values for healthy volunteers (∼180 µm), ex‐smokers (∼250 µm), and COPD patients (∼500 µm) were consistent with previously reported 3He Lm values 7, 32, 33. The mean 129Xe Lm for ex‐smokers (with 129Xe Δ = 8.5 ms) are also in agreement with previous 129Xe Lm values reported at 3 T obtained with 129Xe Δ = 5 ms 20, 21. The 129Xe Lm for the GOLD II COPD subject (318 µm) is also comparable to the 129Xe Lm (∼350 µm) reported in COPD patients (GOLD I‐III) 20, 21. When 129Xe Lm from the 129Xe Δ = 8.5 ms data was evaluated with 3He‐based CM, an overall mean difference of +1.1% and +2.3% was obtained for whole lung and individual lung 129Xe and 3He Lm values, respectively. This small bias is of a similar magnitude as that observed with SEM‐derived LmD and therefore suggests that 129Xe lung morphometry results obtained with 129Xe Δ = 8.5 ms are comparable to 3He results analyzed with both the cylinder and stretched exponential models.
Conclusions
With limited availability of 3He, there is a strong motivation to evaluate functional and structural information that can be derived from the readily available and cheaper 129Xe gas isotope. Compressed sensing has facilitated acquisition of single‐breath 3D multiple b‐value 129Xe DW‐MRI for whole lung morphometry mapping. SEM‐derived LmD demonstrated a linear dependence with diffusion time, and the best agreement between 129Xe and 3He LmD results was obtained with an empirically optimized 129Xe Δ = 8.5 ms. Prospective CS acquisitions were used to validate 129Xe Δ = 8.5 ms in healthy volunteers, ex‐smokers, and COPD patients, and a strong agreement (mean LmD bias of −2.2%) in 129Xe and 3He LmD values was obtained. A similar level of agreement (mean Lm bias of +1.1%) was obtained with CM‐derived Lm, indicating that 129Xe DW‐MRI acquired with 129Xe Δ = 8.5 ms is a viable alternative to 3He for 3D whole lung morphometry assessment with both cylinder and stretched exponential models.
ACKNOWLEDGMENTS
We thank Juan Parra‐Robles for useful discussions of preliminary results.
The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research or the Department of Health.
REFERENCES
- 1. Saam BT, Yablonskiy DA, Kodibagkar VD, Leawoods JC, Gierada DS, Cooper JD, Lefrak SS, Conradi MS. MR imaging of diffusion of 3He gas in healthy and diseased lungs. Magn Reson Med 2000;44:174–179. [DOI] [PubMed] [Google Scholar]
- 2. Salerno M, Altes TA, Brookeman JR, de Lange EE, Mugler 3rd JP. Dynamic spiral MRI of pulmonary gas flow using hyperpolarized (3)He: preliminary studies in healthy and diseased lungs, Magn Reson Med 2001;46:667–677. [DOI] [PubMed] [Google Scholar]
- 3. Ajraoui S, Parra‐Robles J, Deppe M, Teh K, Parnell SR, Owers‐Bradley J, Wild JM. Experimental Investigation of Non‐Gaussian Diffusion in Hyperpolarized 3He MRI of Lungs. In Proceedings of the 17th Annual Meeting of ISMRM, Honolulu, Hawaii, USA, 2009. p. 2178.
- 4. Fichele S, Paley MN, Woodhouse N, Griffiths PD, van Beek EJ, Wild JM. Investigating 3He diffusion NMR in the lungs using finite difference simulations and in vivo PGSE experiments. J Magn Reson 2004;167:1–11. [DOI] [PubMed] [Google Scholar]
- 5. Parra‐Robles J, Ajraoui S, Marshall H, Deppe MH, Xu X, Wild JM. The influence of field strength on the apparent diffusion coefficient of 3He gas in human lungs, Magn Reson Med 2012;67:322–325. [DOI] [PubMed] [Google Scholar]
- 6. Yablonskiy DA, Sukstanskii AL, Leawoods JC, Gierada DS, Bretthorst GL, Lefrak SS, Cooper JD, Conradi MS. Quantitative in vivo assessment of lung microstructure at the alveolar level with hyperpolarized 3He diffusion MRI, Proc Natl Acad Sci U S A 2002;99:3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yablonskiy DA, Sukstanskii AL, Woods JC, Gierada DS, Quirk JD, Hogg JC, Cooper JD, Conradi MS. Quantification of lung microstructure with hyperpolarized 3He diffusion MRI. J Appl Physiol (1985) 2009;107:1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parra‐Robles J, Marshall H, Hartley R, Brightling CE, Wild J. Quantification of Lung Microstructure in Asthma Using a 3He Fractional Diffusion Approach. In Proceedings of the 22nd Annual Meeting of ISMRM, Milan, Italy, 2014. p. 3529.
- 9. Shanbhag DD, Altes TA, Miller GW, Mata JF, Knight‐Scott J. q‐Space analysis of lung morphometry in vivo with hyperpolarized 3He spectroscopy, J Magn Reson Imaging 2006;24:84–94. [DOI] [PubMed] [Google Scholar]
- 10. Chan HF, Stewart NJ, Parra‐Robles J, Collier GJ, Wild JM. Whole lung morphometry with 3D multiple b‐value hyperpolarized gas MRI and compressed sensing, Magn Reson Med 2017;77:1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shea DA, Morgan D. The helium‐3 shortage: supply, demand, and options for Congress. Washington, DC: Congressional Research Service; 2010. [Google Scholar]
- 12. Norquay G, Parnell SR, Xu X, Parra‐Robles J, Wild JM. Optimized production of hyperpolarized 129Xe at 2 bars for in vivo lung magnetic resonance imaging, J Appl Phys 2013;113:044908. [Google Scholar]
- 13. Norquay G, Collier GJ, Rao M, Maunder A, Rodgers OI, Stewart NJ, Wild JM. Large‐Scale Production of Highly‐Polarized 129Xe. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, Hawaii, USA, 2017. p. 2140.
- 14. Kirby M, Ouriadov A, Svenningsen S, Owrangi A, Wheatley A, Etemad‐Rezai R, Santyr GE, McCormack DG, Parraga G. Hyperpolarized 3He and 129Xe magnetic resonance imaging apparent diffusion coefficients: physiological relevance in older never‐ and ex‐smokers. Physiol Rep 2014;2:e12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirby M, Svenningsen S, Kanhere N, Owrangi A, Wheatley A, Coxson HO, Santyr GE, Paterson NA, McCormack DG, Parraga G. Pulmonary ventilation visualized using hyperpolarized helium‐3 and xenon‐129 magnetic resonance imaging: differences in COPD and relationship to emphysema. J Appl Physiol (1985) 2013;114:707–715. [DOI] [PubMed] [Google Scholar]
- 16. Kirby M, Svenningsen S, Owrangi A, et al. Hyperpolarized 3He and 129Xe MR imaging in healthy volunteers and patients with chronic obstructive pulmonary disease. Radiology 2012;265:600–610. [DOI] [PubMed] [Google Scholar]
- 17. Svenningsen S, Kirby M, Starr D, Leary D, Wheatley A, Maksym GN, McCormack DG, Parraga G. Hyperpolarized (3) He and (129) Xe MRI: differences in asthma before bronchodilation. J Magn Reson Imaging 2013;38:1521–1530. [DOI] [PubMed] [Google Scholar]
- 18. Sukstanskii AL, Yablonskiy DA. Lung morphometry with hyperpolarized 129Xe: theoretical background. Magn Reson Med 2012;67:856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruppert K, Quirk JD, Mugler JP III, Altes TA, Wang C, Miller GW, Ruset IC, Mata JF, Hersman FW, Yablonskiy DA. Lung Morphometry Using Hyperpolarized Xenon‐129: Preliminary Experience. In Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, Victoria, Australia, 2012, p. 3152.
- 20. Ouriadov A, Farag A, Kirby M, McCormack DG, Parraga G, Santyr GE. Lung morphometry using hyperpolarized (129) Xe apparent diffusion coefficient anisotropy in chronic obstructive pulmonary disease, Magn Reson Med 2013;70:1699–1706. [DOI] [PubMed] [Google Scholar]
- 21. Ouriadov A, Farag A, Kirby M, McCormack DG, Parraga G, Santyr GE. Pulmonary hyperpolarized Xe morphometry for mapping xenon gas concentrations and alveolar oxygen partial pressure: proof‐of‐concept demonstration in healthy and COPD subjects. Magn Reson Med 2015;74:1726–1732. [DOI] [PubMed] [Google Scholar]
- 22. Parra‐Robles J, Ajraoui S, Wild JM. Modelling Non‐Gaussian 3He Diffusion Signal Behaviour Using a Fractional Dynamics Approach. In Proceedings of the 18th Annual Meeting of ISMRM, Stockholm, Sweden, 2010. p. 2538.
- 23. Parra‐Robles J, Marshall H, Wild JM. Characterization of 3He Diffusion in Lungs Using a Stretched Exponential Model. In Proceedings of the 21st Annual Meeting of ISMRM, Salt Lake City, Utah, USA, 2013. p. 820.
- 24. Berberan‐Santos MN, Bodunov EN, Valeur B. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential). Chem Phys 2005;315:171–182. [Google Scholar]
- 25. Sukstanskii AL, Yablonskiy DA. In vivo lung morphometry with hyperpolarized 3He diffusion MRI: theoretical background. J Magn Reson 2008;190:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woods JC, Choong CK, Yablonskiy DA, Bentley J, Wong J, Pierce JA, Cooper JD, Macklem PT, Conradi MS, Hogg JC. Hyperpolarized 3He diffusion MRI and histology in pulmonary emphysema. Magn Reson Med 2006;56:1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wild JM, Paley MN, Viallon M, Schreiber WG, van Beek EJ, Griffiths PD. k‐space filtering in 2D gradient‐echo breath‐hold hyperpolarized 3He MRI: spatial resolution and signal‐to‐noise ratio considerations, Magn Reson Med 2002;47:687–695. [DOI] [PubMed] [Google Scholar]
- 28. Ajraoui S, Lee KJ, Deppe MH, Parnell SR, Parra‐Robles J, Wild JM. Compressed sensing in hyperpolarized 3He lung MRI. Magn Reson Med 2010;63:1059–1069. [DOI] [PubMed] [Google Scholar]
- 29. Kaushik SS, Cleveland ZI, Cofer GP, et al. Diffusion‐weighted hyperpolarized 129Xe MRI in healthy volunteers and subjects with chronic obstructive pulmonary disease. Magn Reson Med 2011;65:1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gierada DS, Woods JC, Bierhals AJ, et al. Effects of diffusion time on short‐range hyperpolarized (3)He diffusivity measurements in emphysema. J Magn Reson Imaging 2009;30:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quirk JD, Chang YV, Yablonskiy DA. In vivo lung morphometry with hyperpolarized (3) He diffusion MRI: reproducibility and the role of diffusion‐sensitizing gradient direction. Magn Reson Med 2015;73:1252–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quirk JD, Lutey BA, Gierada DS, Woods JC, Senior RM, Lefrak SS, Sukstanskii AL, Conradi MS, Yablonskiy DA. In vivo detection of acinar microstructural changes in early emphysema with (3)He lung morphometry. Radiology 2011;260:866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quirk JD, Sukstanskii AL, Woods JC, Lutey BA, Conradi MS, Gierada DS, Yusen RD, Castro M, Yablonskiy DA. Experimental evidence of age‐related adaptive changes in human acinar airways. J Appl Physiol (1985) 2016;120:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]


