Abstract
To investigate the mechanism by which fusicoccin (FC) induces the activation of the plasma membrane (PM) H+-ATPase, we used phenylarsine oxide (PAO), a known inhibitor of protein tyrosine-phosphatases. PAO was supplied in vivo in the absence or presence of FC to radish (Raphanus sativus L.) seedlings and cultured Arabidopsis cells prior to PM extraction. Treatment with PAO alone caused a slight decrease of PM H+-ATPase activity and, in radish, a decrease of PM-associated 14-3-3 proteins. When supplied prior to FC, PAO drastically inhibited FC-induced activation of PM H+-ATPase, FC binding to the PM, and the FC-induced increase of the amount of 14-3-3 associated with the PM. On the contrary, PAO was completely ineffective on all of the above-mentioned parameters when supplied after FC. The H+-ATPase isolated from PAO-treated Arabidopsis cells maintained the ability to respond to FC if supplied with exogenous, nonphosphorylated 14-3-3 proteins. Altogether, these results are consistent with a model in which the dephosphorylated state of tyrosine residues of a protein(s), such as 14-3-3 protein, is required to permit FC-induced association between the 14-3-3 protein and the PM H+-ATPase.
The H+-ATPase is the most important ion pump in the plant plasma membrane (PM), playing a crucial role in several aspects of plant physiology; it is regulated in vivo by several endogenous and environmental factors (Marré et al., 1993; Palmgren, 1998). One of the best characterized modulators to date is the phytotoxin fusicoccin (FC), which rapidly and strongly stimulates electrogenic proton extrusion in a variety of higher plants (Marré, 1979; Marré et al., 1992; Palmgren, 1998). The FC-induced activation of H+-ATPase causes a shift in the pH optimum of the enzyme toward more alkaline values and a decrease in the apparent Km for the substrate Mg-ATP (Rasi-Caldogno and Pugliarello, 1985; Rasi-Caldogno et al., 1986, 1993; De Michelis et al., 1991; Johansson et al., 1993; Olivari et al., 1993; Lanfermeijer and Prins, 1994).
Different experimental approaches have allowed the identification of a C-terminal autoinhibitory domain on the H+-ATPase of higher plants. This domain is involved in FC-induced activation of the H+-ATPase, which depends on a conformational modification of the enzyme, leading to the displacement of the C terminus (Palmgren et al., 1990, 1991; Johansson et al., 1993; Rasi-Caldogno et al., 1993; Regenberg et al., 1995).
It has recently been shown that the activation of the PM H+-ATPase by FC is caused by the FC-induced association between the C-terminal domain of the enzyme and member(s) of the 14-3-3 protein family (Oecking et al., 1997; Baunsgaard et al., 1998; Fullone et al., 1998; Piotrowsky et al., 1998; Oecking and Hagemann, 1999). On the other hand, the C-terminal domain of the PM H+-ATPase does not contain any sequence similar to the known 14-3-3 binding motifs RSX1,2pSXP (Muslin et al., 1996; Yaffe et al., 1997) in which the phosphorylation of the Ser residue is crucial for the binding of 14-3-3 to target proteins. Recent data suggest that in spinach, the phosphorylation of Thr-948 localized at the C terminus could modulate the FC-induced association between 14-3-3 proteins and the C terminus domain of PM H+-ATPase (Olsson et al., 1998). However, 14-3-3 proteins also bind to the H+-ATPase independently of FC (Fullone et al., 1998), and this FC-independent binding is phosphorylation dependent and also occurs in a truncated H+-ATPase lacking the C-terminal domain (Jahn et al., 1998).
To understand the physiological factors modulating the interaction between the PM H+-ATPase and 14-3-3 proteins, we used an inhibitor of Tyr-specific protein phosphatase (PTP) to investigate the possible involvement of phosphorylation-dephosphorylation of Tyr residues. We show that in vivo treatment of radish (Raphanus sativus L.) seedlings and cultured Arabidopsis cells with phenylarsine oxide (PAO), a known inhibitor of PTPs (Garcia-Morales et al., 1990; Heimovaara-Dijkstra et al., 1996; Knetsch et al., 1996), inhibits the formation of the FC-H+-ATPase-14-3-3 complex and thus the FC-induced activation of the PM H+-ATPase.
MATERIALS AND METHODS
Plant Material and in Vivo Treatments
The method for germination of radish (Raphanus sativus L. cv Tondo Rosso Quarantino, Ingegnoli, Milan) seeds was previously published (De Michelis et al., 1996). In vivo treatments with FC (5 μm, final concentration) and/or PAO were performed after 21 h of germination for the times specified in the figure legends. After the treatments the seedlings were frozen at −80°C. PAO was dissolved in dimethyl sulfoxide (DMSO) (10 mm stock solution), and supplied at the final concentrations indicated in the figure legends.
Cell-suspension cultures of Arabidopsis ecotype Landsberg were grown as described in Curti et al. (1993). In vivo treatments with FC and/or PAO were performed by adding the two effectors to the culture medium at the final concentrations and for the times specified in the figure legends. At the end of the treatments the cells were collected by centrifuging the samples twice at 1,000g for 5 min.
Isolation of PM
A PM-enriched fraction from germinating radish seedlings was obtained by an aqueous two-phase partitioning system, as previously described (Rasi-Caldogno et al., 1995).
Arabidopsis cells harvested from 6-d-old subcultures were homogenized in ice-cold extraction medium (2 mL/1 g of fresh weight); a microsomal fraction was obtained essentially as previously described (Rasi-Caldogno et al., 1995). A highly purified PM fraction was obtained by a two-step aqueous two-phase partitioning system containing 6.2% (w/w) Dextran T500 (Pharmacia Biotech, Piscataway, NJ), 6.2% (w/w) PEG 3350 (Sigma-Aldrich, St. Louis), 11% (w/w) Suc, 5 mm potassium phosphate buffer (pH 7.8), and 1 mm or 5 mm KCl in the first- and second-phase systems, respectively. The final upper phase was diluted 5-fold with 1 mm Bis-Tris propane-4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (BTP-HEPES), pH 7.0, 0.1 mm EGTA, 3 mm dithiothreitol (DTT), 10% (v/v) glycerol, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 0.1 mg mL−1 polyoxyethylene 20 cetyl ether (Brij 58), and centrifuged at 48,000g for 35 min. The obtained pellets were resuspended in 10% (v/v) glycerol, 0.5 mm DTT, and 1 mm 3-(N-morpholino)-propanesulfonic acid (MOPS)-KOH (pH 7), and frozen at −80°C. Membrane proteins were assayed according to the method of Markwell et al. (1978).
PM H+-ATPase Activity
The PM H+-ATPase activity of PM isolated from cultured Arabidopsis cells was assayed in 0.2 mm EGTA, 50 mm KNO3, 3 mm MgSO4, 5 mm (NH4)2SO4, 0.1 mm ammonium molybdate, 1 μg mL−1 oligomycin, 0.1 mg mL−1 Brij 58, 5 μm carbonyl cyanide p-(trifluoromethoxy)phenyl-hydrazone, 40 mm BTP-2-(N-morpholino)-ethanesulfonic acid (BTP-MES) (pH 6.0–6.7), or BTP-HEPES (pH 6.8–7.5), 2 units mL−1 pyruvate kinase, 1 mm phosphoenolpyruvate (PEP), and 1 mm ATP (unless otherwise specified).
The PM H+-ATPase activity of PM isolated from germinating radish seedlings was assayed in the same assay medium without pyruvate kinase or PEP but with 3 mm ATP and 5 mm MgSO4.
PMs (5–20 μg of protein) were incubated in 250 μL of assay medium and the reaction was carried out for 60 min at 30°C. Released Pi was determined as described in De Michelis and Spanswick (1986). The PM H+-ATPase activity was evaluated as the difference between total activity and that measured in the presence of 100 μm vanadate.
FC Radioimmunoassay
Antiserum against bovine serum albumin (BSA)-conjugated dideacetyl-FC was kindly supplied by P. Aducci and M. Marra (Dipartimento di Biologia, Università di Roma Tor Vergata, Rome). [3H]FC (0.7 kBq pmol−1) was a generous gift of Prof. G. Randazzo (Università di Napoli, Italy). The FC radioimmunoassay was performed as previously described (De Michelis et al., 1996).
FC Binding
FC binding was assayed essentially as described in Rasi-Caldogno et al. (1993), except that membranes (100 μg of protein) were incubated in 1 mm MOPS-KOH, pH 7, 10% (v/v) glycerol, 0.5 mm DTT, 5 mm MgSO4, and 0.1 mg mL−1 Brij 58 (200 μL of final volume) in the presence of 5 nm [3H]dihydrofusicoccin ([3H]FC). Unspecific binding, evaluated in the presence of 10 μm unlabeled FC, was subtracted from all of the binding values to obtain specific binding.
SDS-PAGE
Samples were treated as reported in Rasi-Caldogno et al. (1993). SDS-PAGE was performed essentially according to the method of Laemmli (1970). About 10 to 20 μg of protein were loaded onto a 4% to 20% gradient polyacrylamide Tris-Gly Ready Mini Gel (catalog no. 161-0903, Bio-Rad Laboratories, Hercules, CA), and subjected to electrophoresis under standard conditions.
Western-Blot Analysis
After SDS-PAGE, the polypeptides were electrophoretically transferred to a 0.2-μm nitrocellulose membrane (reference no. 401 391, Schleicher & Schull, Keene, NH). The blot was incubated for 2 h with anti-N terminus H+-ATPase polyclonal antibody diluted 1:1,000 (Olivari et al., 1998) or with anti-14-3-3 polyclonal antibody diluted 1:1,000 (Marra et al., 1994), kindly supplied by P. Aducci and M. Marra (Dipartimento di Biologia, Università di Roma Tor Vergata, Rome). Immunodecoration was performed with anti-rabbit IgG conjugated with alkaline-phosphatase (Sigma-Aldrich) for H+-ATPase and with anti-rabbit IgG conjugated with horseradish peroxidase (catalog no. 170-6463, Bio-Rad) for 14-3-3 proteins. Detection of the PM H+-ATPase was performed by using 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium (BCIP-NBT) alkaline phosphatase substrate (B-5655, Sigma-Aldrich). Detection of the 14-3-3 proteins was performed with an enhanced chemiluminescence system (RPN 2209, Amersham-Pharmacia Biotech, Uppsala).
Statistics
Data reported in the figures are the results from one experiment with three replicates, representative of at least two experiments, each performed on an independent PM preparation; se of the assays did not exceed 3% of the measured values.
RESULTS
Effect of PAO on PM H+-ATPase Activity
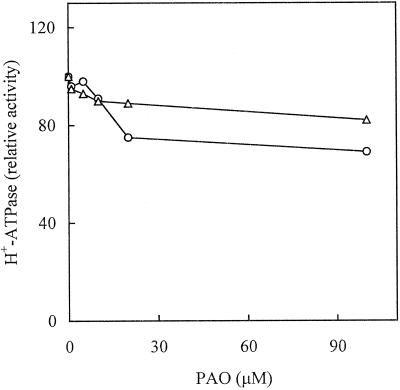
In a first set of experiments, we tested the effect of PAO supplied in vivo to radish seedlings and cultured Arabidopsis cells on the PM H+-ATPase activity. Figure 1 shows the H+-ATPase activity (measured at pH 6.4) in PM isolated from both plant materials after 3 h of incubation in the presence of different concentrations of PAO. In both plant materials, treatment with PAO caused a partial decrease of PM H+-ATPase activity. In radish, the highest concentration of PAO tested (100 μm) decreased the enzyme activity by about 30%, while in Arabidopsis the same concentration of PAO decreased the PM H+-ATPase activity by less than 20%. PAO supplied in vitro to PM isolated from both plant materials was completely ineffective (data not shown).
Figure 1.
Effect of increasing concentrations of PAO on the PM H+-ATPase activity. Radish seedlings (○) and cultured Arabidopsis cells (▵) were treated with PAO for 180 min prior to PM isolation. PM H+-ATPase activity, assayed at pH 6.4, is expressed as a percentage of that measured in the absence of PAO (0.30 μmol min−1 mg−1 protein for radish seedlings; 1.40 μmol min−1 mg−1 protein for Arabidopsis).
Effect of PAO on FC-Induced Activation of the PM H+-ATPase
In all plant materials tested, FC fed in vivo strongly stimulated the PM H+-ATPase. This activation causes a shift in the pH optimum of the enzyme toward more alkaline values and a decrease in the apparent Km for the substrate Mg-ATP (Rasi-Caldogno and Pugliarello, 1985; Rasi-Caldogno et al., 1986; Schulz et al., 1990; De Michelis et al., 1991; Johansson et al., 1993; Olivari et al., 1993). Thus, FC-induced stimulation of the PM H+-ATPase is much stronger when the enzyme activity is assayed at pH values typical of the cytoplasm of a plant cell (pH 7.5) than at the relatively acidic pH optimum for enzyme activity (pH 6.4–6.6). Therefore, the ratio between the activity measured at pH 6.4 and that measured at pH 7.5 (pH ratio) is lower in PM isolated from plant material treated in vivo with FC compared with that measured in untreated tissue, and the pH ratio is an useful parameter with which to monitor the activation state of the PM H+-ATPase (Olivari et al., 1998).
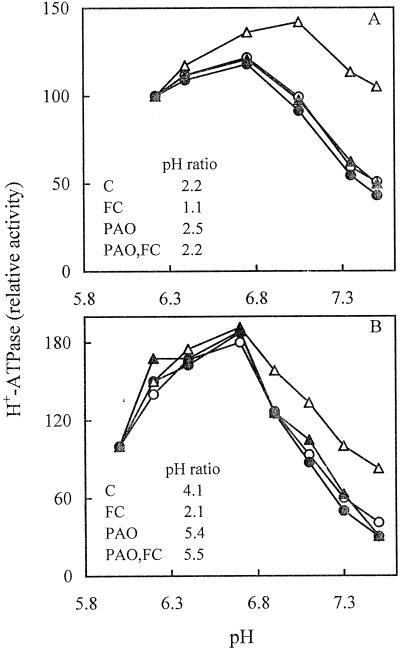
We determined whether PAO affected the FC-induced activation of the PM H+-ATPase. Radish seedlings and cultured Arabidopsis cells were untreated or treated in vivo with PAO alone, 5 μm FC alone, or pretreated with PAO followed by FC. The pH dependence of the enzyme activity in isolated PMs was then analyzed. Figure 2 shows that in both plant materials pretreatment with 100 μm PAO completely prevented the FC-induced increase of PM H+-ATPase activity at pHs above the optimum, so that the pH curve of PMs from PAO-pretreated cells became similar to that of control PMs. This is highlighted by the pH ratio values (inserts to Fig. 2), which become similar to those of control PM in radish seedlings (Fig. 2A), and even higher in cultured Arabidopsis cells (Fig. 2B). Figure 2 also shows that, even in the absence of a treatment with FC, the inhibition of the PM H+-ATPase activity caused by pretreatment with 100 μm PAO slightly increased with the increase of pH, so that the pH ratio for the H+-ATPase activity of PMs from PAO-pretreated cells becomes higher than that of control PMs. These results suggest that treatment with PAO, aside from inhibiting FC-induced activation of the PM H+-ATPase, also decreases its basal activation state.
Figure 2.
Effect of PAO on the pH dependence of the PM H+-ATPase activity. PM was isolated from radish seedlings (A) and cultured Arabidopsis cells (B) untreated (○, C), pretreated with 100 μm PAO for 180 min (●, PAO), pretreated with 5 μm FC for 120 min (▵, FC), or pretreated with 100 μm PAO for 60 min followed by a treatment with 5 μm FC for 120 min (▴, PAO,FC). PM H+-ATPase activity is expressed as a percentage of that measured at pH 6.2 for radish seedlings (C = 0.31; PAO = 0.21; FC = 0.28; PAO,FC = 0.21 μmol min−1 mg−1 protein) and at pH 6.0 for Arabidopsis (C = 0.56; PAO = 0.46; FC = 0.46; PAO,FC = 0.27 μmol min−1 mg−1 protein).
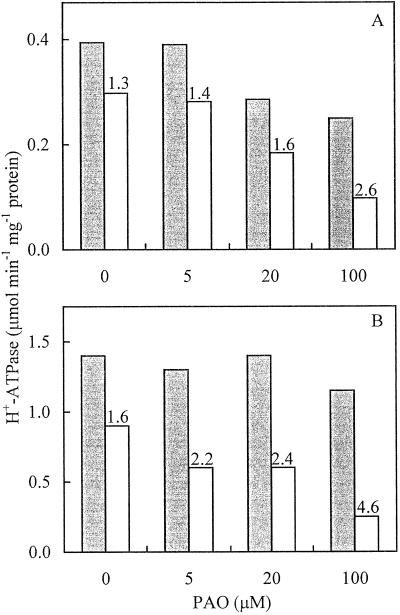
Figure 3 shows the dose-response plot of the PM H+-ATPase activity assayed at pH 6.4 and 7.5 (and the correspondent pH ratios). Radish seedlings (Fig. 3A) and cultured Arabidopsis cells (Fig. 3B) were pretreated in vivo with different concentrations of PAO followed by treatment with 5 μm FC. In both plant materials, the decrease of the PM H+-ATPase activity was more dramatic when assayed at pH 7.5 at all concentrations of PAO tested, so that the pH ratio progressively increased with PAO concentration. The inhibition of FC-induced activation of the PM H+-ATPase was half maximal at about 30 μm PAO, a concentration only slightly higher than those used to inhibit Tyr phosphorylation in animal cells or plant protoplasts (Garcia-Morales et al., 1990; Heimovaara-Dijkstra et al., 1996; Knetsch et al., 1996).
Figure 3.
Inhibiting effect of PAO on FC-induced activation of the PM H+-ATPase. PM H+-ATPase activity was measured at pH 6.4 (gray bars) or pH 7.5 (white bars) in PM isolated from radish seedlings (A) and cultured Arabidopsis cells (B) treated with increasing concentrations of PAO for 60 min, followed by a treatment with 5 μm FC for 120 min. Numbers on top of the white bars represent the corresponding pH ratios.
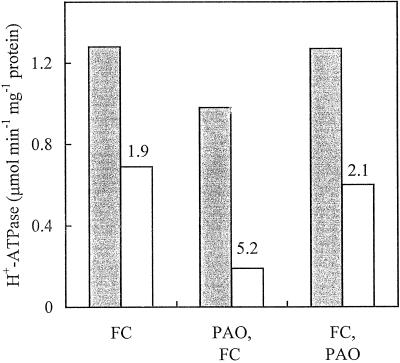
We then evaluated whether FC was able to prevent the effect of PAO. Figure 4 shows the effect of PAO supplied to cultured Arabidopsis cells prior to or after FC on the PM H+-ATPase activity assayed at pH 6.4 and 7.5 and on the corresponding pH ratios. The results indicate that, when supplied after FC, PAO was unable to reverse the FC-induced activation of the PM H+-ATPase.
Figure 4.
PAO is unable to revert FC-induced activation of the PM H+-ATPase. PM H+-ATPase activity was measured at pH 6.4 (gray bars) or pH 7.5 (white bars) in PM isolated from cultured Arabidopsis cells pretreated with 5 μm FC for 180 min (FC), pretreated with 100 μm PAO for 60 min followed by a treatment for 120 min with FC (PAO,FC), or pretreated with 5 μm FC for 60 min followed by a treatment for 120 min with 100 μm PAO (FC,PAO). Numbers on top of the white bars represent the corresponding pH ratios.
Effect of PAO on FC Binding to the PM
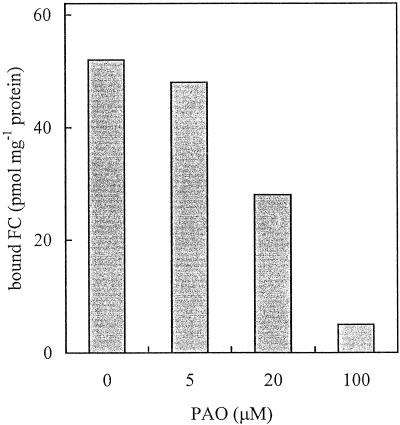
To better investigate which step of the mechanism leading to FC-induced activation of the PM H+-ATPase was affected by PAO, we tested the effect of the inhibitor on FC binding to the PM upon in vivo and in vitro treatment with FC. Figure 5 shows the effect of pretreatment with increasing concentrations of PAO on FC binding to the PM upon treatment of radish seedlings with 5 μm FC. Data obtained indicate that pretreatment with PAO inhibited FC binding to the PM with the same concentration dependence as monitored for FC-induced activation of the PM H+-ATPase (compare with Fig. 3A). Moreover, in PM isolated from cultured Arabidopsis cells treated with PAO, the binding of FC supplied in vitro to PM was almost completely inhibited compared with that measured on control PM: 0.21 pmol FC mg−1 protein versus 1.46 pmol FC mg−1 protein, respectively.
Figure 5.
Effect of PAO on FC binding to the PM. PM was isolated from radish seedlings treated with increasing concentrations of PAO followed by a treatment with 5 μm FC. The amount of FC bound to PM upon in vivo treatment was evaluated by FC radioimmunoassay.
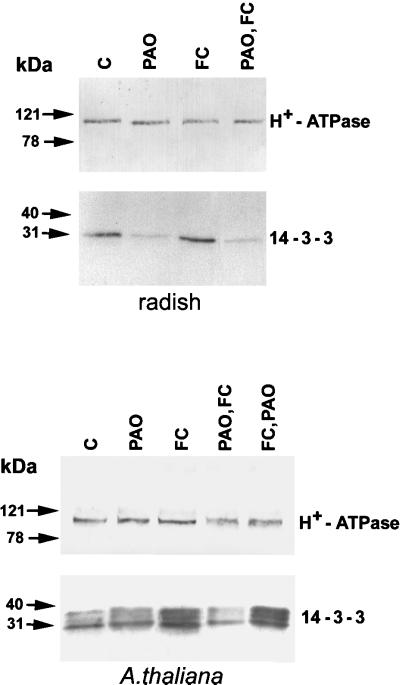
It has been shown that FC binding to the PM involves an interaction between the C terminus of the H+-ATPase and members of the 14-3-3 protein family (Oecking et al., 1997; Baunsgaard et al., 1998; Fullone et al., 1998; Piotrowsky et al., 1998; Oecking and Hagemann, 1999) and that FC strongly enhances the amount of PM-associated 14-3-3 proteins (De Michelis et al., 1996; Jahn et al., 1997; Oecking et al., 1997, Olivari et al., 1998). Thus, we checked the effect of PAO on the amount of 14-3-3 proteins associated with the PM. Figure 6 shows western analysis of PM isolated from radish seedlings or cultured Arabidopsis cells untreated or treated with PAO alone, FC alone, or PAO supplied prior to or after the treatment with FC. The blots were immunodecorated with polyclonal antibodies raised against the N-terminal domain of the PM H+-ATPase (Olivari et al., 1998) or against 14-3-3 proteins (Marra et al., 1994). In the first case, the antibody identified a single band of 100 kD, the intensity of which was similar in the different lanes, indicating that PAO had no major effect on the PM H+-ATPase amount. In radish, the intensity of the band at 30 kD, identified by the anti-14-3-3 antibody, was reduced by treatment with PAO supplied in vivo alone, indicating that PAO decreases the association of 14-3-3 proteins with the PM. In Arabidopsis, the band at 30 kD was barely affected by the same treatment. As reported for different plant materials (Korthout and de Boer, 1994; Oecking et al., 1994; Jahn et al., 1997; Baunsgaard et al., 1998; Fullone et al., 1998; Olivari et al., 1998), FC strongly increased the amount of 14-3-3 associated with the PM both of radish and moreso of Arabidopsis. Pretreatment with PAO completely prevented the FC-induced increase of PM-associated 14-3-3 proteins in both plant materials. On the contrary, when supplied after FC, PAO was unable to reverse the FC-induced increase of 14-3-3 proteins associated with the PM.
Figure 6.
Effect of PAO on the amount of PM-associated 14-3-3 proteins. PM was isolated from radish seedlings or cultured Arabidopsis cells untreated (C), pretreated with 100 μm PAO (PAO), pretreated with 5 μm FC (FC), pretreated with 100 μm PAO followed by a treatment with 5 μm FC (PAO,FC), or with 5 μm FC followed by a treatment with 100 μm PAO (FC,PAO). After SDS-PAGE and western blotting, immunodecoration was performed with an anti-N-terminal H+-ATPase polyclonal antibody and with an anti-14-3-3 polyclonal antibody.
Effect of PAO on the Activation of the PM H+-ATPase of Arabidopsis by GF 14-6 14-3-3 and FC
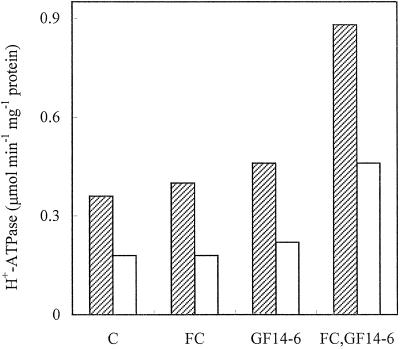
All data presented so far indicate that PAO abolishes the FC-induced activation of the PM H+-ATPase by hampering the association between 14-3-3 proteins and the enzyme. The simplest hypothesis to explain these data is that PAO affects the phosphorylation state of relevant Tyr residues located on the amino acid sequences of the PM H+-ATPase or of 14-3-3 proteins. The availability of recombinant 14-3-3 (Fullone et al., 1998) allowed us to determine whether the PM H+-ATPase from Arabidopsis cells pretreated with PAO maintained the capability to respond to FC when supplied with 14-3-3. Figure 7 shows the PM H+-ATPase activity of control PM and PM from PAO-treated cells measured in the presence or absence of 5 μm FC alone or supplied with 20 μg mL−1 GF 14-6 14-3-3. The data indicate that, as for several plant materials (Blum et al., 1988; Olivari et al., 1993), in vitro addition of FC essentially did not stimulate the H+-ATPase activity, while the simultaneous supply of GF 14-6 14-3-3 and FC to control PM caused a dramatic activation: by about 100%, of the H+ATPase, which is similar to that reported in yeast expressing Arabidopsis PM H+-ATPase AHA2 (Baunsgaard et al., 1998). In PM from PAO-treated cells, the simultaneous supply of GF 14-6 14-3-3 and FC caused the activation of the H+ATPase to a similar extent monitored for control PM, suggesting that PAO treatment does not affect the H+ATPase responsiveness to FC. The addition of GF 14-6 14-3-3 alone slightly increased the activity of the enzyme of PM from both control and PAO-treated cells.
Figure 7.
Effect of recombinant GF 14-6 and FC on the PM H+-ATPase activity of PM isolated from cultured Arabidopsis cells untreated (hatched bars) or treated with 100 μm PAO (white bars). PMs (1.5 μg of proteins) were incubated with or without 5 μm FC and with or without 2 μg of GF 14-6 in 50 μL of assay medium (pH 7.3) without ATP, PK, and PEP at 30°C for 30 min. The volume was adjusted to 100 μL with assay medium added with pyruvate kinase, PEP, and ATP (0.3 mm final concentration); the reaction was carried out for 60 min at 30°C.
DISCUSSION
In this paper we have shown that PAO, a known inhibitor of PTPs (Garcia-Morales et al., 1990; Heimovaara-Dijkstra et al., 1996; Knetsch et al., 1996), supplied in vivo to radish seedlings or cultured Arabidopsis cells, completely prevents the FC-induced activation of PM H+-ATPase. In fact, the pH dependence of H+-ATPase activity of PM from cells treated first with PAO then with FC becomes similar to that of control PM (Fig. 2). This reveals an increasing effect at pH values above the optimum of the H+-ATPase activity, where the FC-induced activation is much stronger and more relevant is the autoinhibitory effect of C-terminal domain (Rasi-Caldogno and Pugliarello, 1985; Rasi-Caldogno et al., 1986; Schulz et al., 1990; De Michelis et al., 1991, 1993; Johansson et al., 1993; Olivari et al., 1993, 1998). The effect of PAO is due to the inhibition of FC binding to PM (Fig. 5); in fact, both FC binding and inhibition of the FC-induced activation of H+-ATPase show the same dose-response relationship, being completely abolished by 100 μm PAO (Figs. 3 and 5). Treatment with 100 μm PAO also suppressed the FC-induced increase of PM-associated 14-3-3 proteins (Fig. 6). Our results indicate that PAO prevents the FC-induced association between 14-3-3 proteins and the C terminus of the H+ATPase.
Treatment with PAO also caused a slight general decrease of PM H+-ATPase activity, which was particularly evident in the absence of FC (Fig. 1). This effect, which requires PAO concentrations higher than those effective on FC-induced activation of the enzyme, may reflect some nonspecific action of PAO on metabolism (Gibson et al., 1989), which would require further investigation. It is noteworthy, however, that in the absence of FC, PAO also causes a decrease of the activation state of the PM H+-ATPase (compare the pH ratios of PM from PAO-treated cells with that of control PM in Fig. 2) and, at least in radish, this correlates with a reduced amount of 14-3-3 proteins associated with the PM (Fig. 6). This result suggests that the association of 14-3-3 proteins may be important in regulating the PM H+-ATPase activity independently of FC.
The inhibitory effect of PAO was completely prevented when it was supplied after FC (Fig. 4), suggesting that once the FC-induced activation of H+-ATPase is established, the modulatory site(s) inhibited by PAO is no more accessible or becomes less relevant.
One simple interpretation of these results is that PAO affects the phosphorylation state of relevant Tyr residues located on the sequences of the PM H+-ATPase, of the 14-3-3 proteins, or both. The availability of a recombinant, nonphosphorylated GF 14-6 14-3-3 isoform allowed us to show that in PM from Arabidopsis cells pretreated with PAO, the H+-ATPase maintains the capability to respond to FC when supplied in vitro with 14-3-3 (Fig. 7). This result suggests that the phosphorylation state of Tyr residues located on the PM H+-ATPase may not be involved in the association of 14-3-3 with the enzyme, and that the dephosphorylation of 14-3-3-located specific Tyr residues would be required to permit the FC-induced activation of the PM H+-ATPase. Searching Tyr-kinase phosphorylation site motifs on the 14-3-3 protein sequence in several isoforms of different plant materials using the consensus pattern [RK]-X(2,3)-[DE]-X(2,3)-Y (Hunter, 1982; Patschinsky et al., 1982; Cooper et al., 1984), we found a highly conserved putative Tyr kinase phosphorylation site KMKGDYYRY, which was also completely conserved in the 14-3-3 GF14-6 isoform from maize (Fullone et al., 1998). It is tempting to speculate that de-phosphorylation of this site may be required to allow the FC-induced association between 14-3-3 proteins and the PM H+-ATPase C terminus. However, we cannot rule out the possibility that PAO modifies a third partner protein, which, directly or indirectly, modifies 14-3-3 proteins. Work is in progress to discriminate between these possibilities.
Phosphorylation of Tyr residues is an ubiquitous, highly conserved modulatory mechanism involved in mitogen-activated protein kinase (MAPK) cascade among eukaryotes (Anderson et al., 1990; Posada et al., 1991). In animal and yeast systems, PTPs play important roles in a number of signal tranduction pathways involving the modulation of MAPKs (Neel and Tonks, 1997). In higher plants, recent results have shown that Tyr phosphorylation is critical for the activation of MAPKs involved in a variety of signal transduction pathways (Suzuki and Shinshi, 1995; Hirt, 1997; Zhang and Klessing, 1997). In barley, PAO prevents the activation of a MAPK in response to abscisic acid (Knetsch et al., 1996), indicating that a PTP is involved in the MAPK pathway in plants. Recently, an Arabidopsis cDNA clone has been isolated that encodes a PTP (AtPTP1) that may function in stress responses (Xu et al., 1998). Molecular characterization of PTPs in Arabidopsis, pea, and soybean has been recently published (Fordham-Skelton et al., 1999), suggesting that their role may not be as MAPK phosphatases and indicating that the function of these PTPs in higher plants is still unclear. The PAO-inhibited PTPs involved in FC-induced activation of PM H+-ATPase could be the final step of a signal transduction pathway that modulates the association between 14-3-3 proteins and the H+-ATPase, by affecting the phosphorylation state of Tyr residues of the enzyme or, more likely, of 14-3-3 proteins.
Modulation of PM H+-ATPase by modification of its phosphorylation state has been reported to occur both in vivo and in vitro; however, to date, only Ser and Thr residues have been involved (Sussman, 1994; Xing et al., 1996; Lino et al., 1998; Olsson et al., 1998). In particular, different phosphorylation sites identified in the H+-ATPase sequence have been shown to be involved in the interaction with the 14-3-3 proteins. The activation of the PM H+-ATPase by FC is determined by the FC-induced association between the C-terminal domain of the enzyme and member(s) of the 14-3-3 protein family (Oecking et al., 1997; Baunsgaard et al., 1998; Fullone et al., 1998; Piotrowsky et al., 1998; Oecking and Hagemann, 1999). The C-terminal domain of the PM H+-ATPase does not contain any sequence similar to the known 14-3-3-binding motifs RSX1,2pSXP (Muslin et al., 1996; Yaffe et al., 1997), in which the phosphorylation of the Ser residue is crucial for the binding of 14-3-3 to target proteins. Recent data suggest that in spinach the phosphorylation of Thr-948 localized at the C terminus could modulate the FC-induced association between 14-3-3 proteins and the C-terminal domain of PM H+-ATPase (Olsson et al., 1998). However, 14-3-3 proteins also bind to the H+-ATPase independently of FC (Fullone et al., 1998), and this FC-independent binding is phosphorylation dependent and occurs in a truncated H+-ATPase lacking the C-terminal domain (Jahn et al., 1998). Recently, Marra et al. (2000) identified a sequence of the PM H+-ATPase highly conserved in different isoforms of different plants and localized in the cytosolic stretch connecting transmembrane segments 8 and 9, which mimics the known 14-3-3 binding motif (Muslin et al., 1996; Yaffe et al., 1997). A phosphopeptide corresponding to such a sequence bound a 14-3-3 protein and inhibited FC binding and FC-induced activation of the PM H+-ATPase; all of these effects were dependent on the phosphorylation of a Ser residue.
The physiological roles of such a complex modulation of the PM H+-ATPase involving phosphorylation mechanisms requires the dissection of the single steps of the putative cascade and the identification and characterization of the enzyme activities implicated.
ACKNOWLEDGMENTS
The authors are grateful to Patrizia Aducci (Dipartimento di Biologia, Università di Roma Tor Vergata, Rome) and Mauro Marra (Università degli Studi del Sannio, Benevento, Italy) for the generous gift of antisera anti-FC and anti-14-3-3 and of recombinant GF 14-6.
Footnotes
This work was supported by Ministero per le Risorse Agricole, Alimentari e Forestali in the frame of the “Piano Nazionale per le Biotecnologie Vegetali.”
LITERATURE CITED
- Anderson NG, Maller JL, Tonks NK, Sturgill TW. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HAAJ, de Boer AH, Palmgren MG. The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- Blum W, Key G, Weiler EW. ATPase activity in plasmalemma-rich vesicles isolated by aqueous two-phase partitioning from Vicia faba mesophyll and epidermis: characterization and influence of abscissic acid and fusicoccin. Physiol Plant. 1988;72:279–287. [Google Scholar]
- Cooper JA, Esch FS, Taylor SS, Hunter T. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinase in vivo and in vitro. J Biol Chem. 1984;259:7835–7841. [PubMed] [Google Scholar]
- Curti G, Massardi F, Lado P. Synergistic activation of plasma membrane H+-ATPase in Arabidopsis thaliana cells by turgor decrease and by fusicoccin. Physiol Plant. 1993;87:592–600. [Google Scholar]
- De Michelis MI, Rasi-Caldogno F, Pugliarello MC, Olivari C. Fusicoccin binding to its plasma membrane receptor and the activation of the plasma membrane H+-ATPase: II. Stimulation of the H+-ATPase in a plasma membrane fraction purified by phase partitioning. Bot Acta. 1991;104:265–271. [Google Scholar]
- De Michelis MI, Rasi-Caldogno F, Pugliarello MC, Olivari C. Fusicoccin binding to its plasma membrane receptor and the activation of the plasma membrane H+-ATPase: III. Is there a direct interaction between the fusicoccin receptor and the plasma membrane H+-ATPase? Plant Physiol. 1996;110:957–964. doi: 10.1104/pp.110.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michelis MI, Spanswick RM. H+-pumping driven by the vanadate-sensitive ATPase in membrane vesicles from corn roots. Plant Physiol. 1986;81:542–547. doi: 10.1104/pp.81.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Skipsey M, Eveans MI, Edwards R, Gatehouse JA. Higher plant tyrosine-specific phosphatases (PTPs) contain novel amino-terminal domains: expression during embryogenesis. Plant Mol Biol. 1999;39:593–605. doi: 10.1023/a:1006170902271. [DOI] [PubMed] [Google Scholar]
- Fullone MR, Visconti S, Marra M, Fogliano V, Aducci P. Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J Biol Chem. 1998;273:7698–7702. doi: 10.1074/jbc.273.13.7698. [DOI] [PubMed] [Google Scholar]
- Garcia-Morales P, Minami Y, Luong E, Klausner RD, Samelson LE. Tyrosine phosphorylation in T cells is regulated by phosphatase activity: studies with phenylarsine oxide. Proc Natl Acad Sci USA. 1990;87:9255–9259. doi: 10.1073/pnas.87.23.9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AE, Noel RJ, Herlihy JT, Ward WF. Phenylarsine oxide inhibition of endocytosis: effect on asialofetuin internalization. Am J Physiol. 1989;257:C182–C184. doi: 10.1152/ajpcell.1989.257.2.C182. [DOI] [PubMed] [Google Scholar]
- Heimovaara-Dijkstra S, Nieland TJF, van der Meulen RM, Wang M. Abscissic acid-induced gene-expression requires the activity of protein(s) sensitive to the protein-tyrosine phosphatase inhibitor phenylarsine oxide. Plant Growth Regul. 1996;18:115–123. [Google Scholar]
- Hirt H. Multiple roles of MAP kinases in plant signal transduction. Trends Biochem Sci. 1997;2:11–15. [Google Scholar]
- Hunter T. Synthetic peptide substrates for a tyrosine protein kinase. J Biol Chem. 1982;257:4843–4848. [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Drumm K, Palmgren MG. Multiple interaction of 14-3-3 proteins with the plant plasma membrane H+-ATPase. In: Tester M, Morris C, Davies J, editors. Proceedings of the 11th International Workshop on Plant Membrane Biology. UK: Cambridge; 1998. p. 24. [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Bruntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson F, Sommarin M, Larsson C. Fusicoccin activates the plasma membrane H+-ATPase by a mechanism involving the C-terminal inhibitory domain. Plant Cell. 1993;5:321–327. doi: 10.1105/tpc.5.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch MLW, Wang M, Snaar-Jagalska BE, Heimovaara-Dijkstra S. Abscissic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell. 1996;8:1061–1067. doi: 10.1105/tpc.8.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthout HAAJ, de Boer AH. A fusicoccin binding protein belongs to the family of 14-3-3 brain protein homologs. Plant Cell. 1994;6:1681–1692. doi: 10.1105/tpc.6.11.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriofage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Prins HBA. Modulation of H+-ATPase activity by fusicoccin in plasma membrane vesicles from oat (Avena sativa L.) roots. Plant Physiol. 1994;104:1277–1285. doi: 10.1104/pp.104.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino B, Baizal-Aguirre VM, Gonzàles de la Vara LE. The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta. 1998;204:352–359. doi: 10.1007/s004250050266. [DOI] [PubMed] [Google Scholar]
- Markwell MAK, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Marra M, Fullone MR, Fogliano V, Pen J, Mattei M, Masi S, Aducci P. The 30-kilodalton protein present in purified fusicoccin receptor preparations is a 14-3-3 like protein. Plant Physiol. 1994;106:1497–1501. doi: 10.1104/pp.106.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M, Olivari C, Visconti S, Albumi C, Aducci P, De Michelis MI. A phosphopeptide corresponding to the cytosolic stretch connecting transmembrane segments 8 and 9 of the plasma membrane H+-ATPase binds 14-3-3 proteins and inhibits fusicoccin induced activation of the H+-ATPase. Plant Biol. 2000;2:1–6. [Google Scholar]
- Marré E. Fusicoccin: a tool in plant physiology. Annu Rev Plant Physiol. 1979;30:273–288. [Google Scholar]
- Marré E, Bellando M, Beffagna N, Marré MT, Romani G, Vergani P. Synergisms, additive and non additive factors regulating proton extrusion and intracellular pH. Curr Top Plant Biochem Physiol. 1992;11:213–230. [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- Oecking C, Eckerskorn C, Weiler EW. The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 1994;352:163–66. doi: 10.1016/0014-5793(94)00949-x. [DOI] [PubMed] [Google Scholar]
- Oecking C, Hagemann K. Association of 14-3-3 proteins with the C-terminal autoinhibitory domain of the plant plasma membrane H+-ATPase generates a fusicoccin-binding complex. Planta. 1999;207:480–482. [Google Scholar]
- Oecking C, Piotrowski M, Hagermeier J, Hagemann K. Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J. 1997;12:441–453. [Google Scholar]
- Olivari C, Meanti C, De Michelis MI, Rasi-Caldogno F. Fusicoccin binding to its plasma membrane receptor and the activation of the plasma membrane H+-ATPase: IV. Fusicoccin induces the association between the plasma membrane H+-ATPase and the fusicoccin receptor. Plant Physiol. 1998;116:529–537. doi: 10.1104/pp.116.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari C, Pugliarello MC, Rasi-Caldogno F, De Michelis MI. Characteristics and regulatory properties of the H+-ATPase in a plasma membrane fraction purified from Arabidopsis thaliana. Bot Acta. 1993;106:13–19. [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998;118:551–555. doi: 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. Proton gradients and plant growth: role of the plasma membrane H+-ATPase. Adv Bot Res. 1998;28:1–70. [Google Scholar]
- Palmgren MG, Larsson C, Sommarin M. Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J Biol Chem. 1990;265:13423–13426. [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem. 1991;266:20470–20475. [PubMed] [Google Scholar]
- Patschinsky T, Hunter T, Esch FS, Cooper JA, Sefton BM. Analysis of the sequence of amino acids surrounding site of tyrosine phosphorylation. Proc Natl Acad Sci USA. 1982;79:973–977. doi: 10.1073/pnas.79.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowsky M, Morsomme P, Boutry M, Oecking C. Complementation of the Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene generates a highly abundant fusicoccin binding site. J Biol Chem. 1998;273:30018–30023. doi: 10.1074/jbc.273.45.30018. [DOI] [PubMed] [Google Scholar]
- Posada J, Sanghera J, Pelech S, Aebersold R, Cooper J. Tyrosine phosphorylation and activation of homologous protein kinases during oocyte maturation and mitogenic activation of fibroblasts. Mol Cell Biol. 1991;11:2517–2528. doi: 10.1128/mcb.11.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasi-Caldogno F, Carnelli A, De Michelis MI. Identification of the plasma membrane Ca2+-ATPase and its autoinhibitory domain. Plant Physiol. 1995;108:105–113. doi: 10.1104/pp.108.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasi-Caldogno F, De Michelis MI, Pugliarello MC, Marrè E. H+-pumping driven by the plasma membrane ATPase in membrane vesicles from radish: stimulation by fusicoccin. Plant Physiol. 1986;82:121–125. doi: 10.1104/pp.82.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasi-Caldogno F, Pugliarello MC. Fusicoccin stimulates the H+-ATPase of plasmalemma in isolated membrane vesicles from radish. Biochem Biophys Res Commun. 1985;133:280–285. doi: 10.1016/0006-291x(85)91872-8. [DOI] [PubMed] [Google Scholar]
- Rasi-Caldogno F, Pugliarello MC, Olivari C, De Michelis MI. Controlled proteolysis mimics the effect of fusicoccin on the plasma membrane H+-ATPase. Plant Physiol. 1993;103:391–398. doi: 10.1104/pp.103.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Villalba JM, Lanfermeijer FC, Palmgren MG. C-terminal deletion analysis of plant plasma membrane H+-ATPase: yeast as a model for solute transport across the plasma membrane. Plant Cell. 1995;7:1655–1666. doi: 10.1105/tpc.7.10.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, Oelgemoller E, Weiler EW. Fusicoccin action in cell-suspension cultures of Corydalis sempervirens Pers. Planta. 1990;183:83–91. doi: 10.1007/BF00197571. [DOI] [PubMed] [Google Scholar]
- Sussman MR. Molecular analysis of proteins in the plant plasma membrane. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:211–234. [Google Scholar]
- Suzuki K, Shinshi H. Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell. 1995;7:639–647. doi: 10.1105/tpc.7.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing T, Higgins VJ, Blumwald E. Regulation of plant defense response to fungal pathogens: two types of protein kinase in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell. 1996;8:555–564. doi: 10.1105/tpc.8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Fu HH, Gupta R, Luan S. Molecular characterization of a tyrosine- specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SK, Cantley LC. The structural basis for 14-3-3:phoshopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Klessing DF. Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]