Abstract
Tight cell-cycle regulation of the histone H4-K20 methyltransferase PR-Set7 is essential for the maintenance of genome integrity. In mammals, this mainly involves the interaction of PR-Set7 with the replication factor PCNA, which triggers the degradation of the enzyme by the CRL4CDT2 E3 ubiquitin ligase. PR-Set7 is also targeted by the SCFβ-TRCP ligase, but the role of this additional regulatory pathway remains unclear. Here, we show that Drosophila PR-Set7 undergoes a cell-cycle proteolytic regulation, independently of its interaction with PCNA. Instead, Slimb, the ortholog of β-TRCP, is specifically required for the degradation of the nuclear pool of PR-Set7 prior to S phase. Consequently, inactivation of Slimb leads to nuclear accumulation of PR-Set7, which triggers aberrant chromatin compaction and G1/S arrest. Strikingly, these phenotypes result from non-enzymatic PR-Set7 functions that prevent proper histone H4 acetylation independently of H4K20 methylation. Altogether, these results identify the Slimb-mediated PR-Set7 proteolysis as a new critical regulatory mechanism required for proper interphase chromatin organization at G1/S transition.
INTRODUCTION
An ordered progression through the cell cycle is essential to maintain genomic stability and prevents diseases such as cancer. This requires that the genome is faithfully replicated in a DNA synthesis (S) phase and each of the two resulting sets of sister chromatids are condensed and segregated properly to the two daughter cells during mitosis (M phase) (1). These cell-cycle events are tightly controlled and necessitate the concerted activity and timely regulation of a cohort of enzymes, including those that directly regulate the dynamic changes in chromatin structure critical for DNA replication, chromosome compaction and cell division (2). A well-known example is the balance exerted by the opposing action of histone H4 acetyltransferases (HAT) and deacetylases (HDAC) that modulates the levels of lysine acetylation on histone H4 and thus contributes to proper chromatin compaction during the cell cycle (3). Indeed, histone H4 acetylation is known to favor a more relaxed chromatin organization that is conducive to proper DNA replication initiation and S-phase progression (4). However, the mechanisms coordinating the activity of HAT and HDAC on histone H4 tail with the entry into S-phase still remain poorly understood.
The SET-domain methyltransferase PR-Set7 (also known as SET8, SETD8 or KMT5A) is another histone H4 modifying enzyme responsible for the monomethylation of histone H4 at lysine 20 (H4K20me1) and of several other non-histone substrates (5,6). In mammalian cells, loss and gain of function studies show that PR-Set7 is essential for the maintenance of genome stability, which involves the timely destruction of the enzyme during S-phase (7,8). This is mediated by ubiquitin-mediated proteolysis and requires the interaction of the enzyme with the DNA replication factor PCNA through a conserved PCNA-interacting (PIP) motif located upstream of the catalytic SET domain (9,10). PCNA serves as a cofactor to promote PR-Set7 interaction with the CRL4CDT2 E3 ubiquitin ligase, which earmarks PR-Set7 for ubiquitylation and degradation during S phase or upon DNA damage (10–14). PCNA-mediated degradation of mammalian PR-Set7 is essential for proper cell-cycle progression (14,15). Indeed, the mutation of the PIP-motif is sufficient to stabilize the enzyme and induces changes in chromatin compaction and DNA re-replication, which is partially due to the ability of PR-Set7 to stimulate the recruitment of pre-replication complex components on chromatin (13,16). In addition to the CRL4cdt2 pathway, the APCCdh1 and the F-box proteins Skp2 and β-TRCP of SCF ubiquitin E3 ligase complexes have also been reported to regulate PR-Set7 stability in human cells (15,17–19). However, because of the dominant effect of CRL4cdt2 pathway on PR-Set7 stability, it remains largely unclear whether these additional PR-Set7 degradation pathways play a critical role in PR-Set7 functions or whether they serve as fine-tuning system to regulate the abundance of the enzyme in different phases of the cell cycle.
Here, we have studied the functions of the Drosophila ortholog of PR-Set7 (20). As its mammalian counterpart, we show that Drosophila PR-Set7 is also subject to a proteolytic regulation during the cell cycle with the lowest levels from G1 to early S-phase. However, in contrast to mammals, a mutated PIP-motif neither stabilized PR-Set7 nor was critical for its functions in cell-cycle regulation during development. Thanks to the identification of a minimal functional sequence of PR-Set7 for proper cell proliferation, we confirmed that the catalytic activity of PR-Set7 is required for G2/M transition and revealed that targeting of the nuclear pool of this enzyme by Slimb, the ortholog of β-TRCP, is required for G1/S transition. Finally, we show that nuclear accumulation of PR-Set7 upon Slimb depletion led to abnormal chromatin compaction and DNA replication inhibition, thereby causing G1/S arrest. Strikingly, these phenotypes are driven by non-enzymatic PR-Set7 functions that negatively regulate the levels of histone H4 acetylation. Altogether, these results identify the Slimb-mediated degradation of PR-Set7 by itself as a new critical cell-cycle regulatory mechanism that ensures proper chromatin structure from G1 to S phase progression.
MATERIALS AND METHODS
Cell culture, establishment of stable cell lines, synchronization and RNA interference
Drosophila S2 (L2–4) or adherent S2R+ cells were grown in Drosophila Schneider's medium (InVitrogen) supplemented with 10% bovine serum and antibiotics (penicilline/ streptomycine). Cells were transfected according to manufacturer's instructions using Effectene transfection agent (Qiagen). Stable cell lines were selected with 200 μg/ml Hygromycin-B or 100 μg/ml of Blasticidine. RNA interference in cells was conducted as previously described. Briefly, double stranded RNA (dsRNA) was produced in vitro using T7 RNA production kit (Promega) from PCR products generated with T7-containing primers (Supplementary Table S1). S2 or S2R+ cells were plated in 6-well plate with 1 × 106 of cells in 1 ml of serum free medium per well with 10 μg of dsRNA incubated during 50 min and supplemented with 2 ml of complete medium. Cells were treated for 3–6 days before analysis. S2R+ Cell cycle arrest was induced by treating S2R+ cells at least 24 h with either 0.5 mM of mimosine (G1 arrest), 1 μM of hydroxyurea and 10 μM of aphidicolin (S phase arrest), 1.7 μM 20 hydroxyecdysone (G2 arrest), or 12 h of 30 μM of colcemid (M phase arrest) as described previously in (21).
Centrifugal elutriation of Drosophila S2 cells
1.5 × 109 S2 cells were grown in complete Schneider's Drosophila medium at a density of 3.5 × 106 cells/ml, collected, washed in ice-cold PBS, resuspended in 200 ml of PBS and injected in the sample injection loop of a centrifugal elutriator composed of a large chamber (40 ml) with a counterbalance at the opposite side in a JE-5.0 rotor in an Avanti J-26XP centrifuge (Beckman Coulter) (22). After 30 min centrifugation at 3500 rpm, necessary for the equilibration of cells in the chamber, fractions were collected by decreasing the rotor speed every 30 sec by 100 rpm until the rotor speed reached 2200 rpm. Cells present in each fraction were counted and separated into two pools for cell cycle analysis by FACS and to prepare total protein cell extracts for immunoblot.
Phosphoprotein purification
Qiagen Phosphoprotein purification columns (37 101) were used to isolate phosphoproteins according to manufacturer's protocol. Briefly, S2 cells extract was prepared using Phosphoprotein Lysis buffer, adjusted to at 0.1 mg/ml concentration and passed through the column. After collection of the flowthrough, columns were washed with Phosphoprotein Lysis buffer and elution buffer was applied several times to collect several eluted fractions before quantification and analysis by immunoblotting.
Total RNA preparation and Quantitative PCR
Total RNA was purified from 10 × 106 cells using lysis reagent (Qiazol, Qiagen) according to manufacturers instructions. Quantitative PCR (qPCR) was carried out in an ABI PRISM 7000 Sequence detection system in a 25 μl reaction with Power SYBR Green Master Mix (Applied Biosystems). Primers used to analyze PR-Set7 and Tubulin are liste in Supplementary Table S2.
Fluorescence-activated cell sorting (FACS) analysis
For cell-cycle analysis by flow cytometry, cells were washed in PBS, fixed with 1% formaldehyde in PBS + 0.1% Tween-20 for 10min. Cells were then permeabilized with PBS + 0.1% Tween20 for 15 min and resuspended in PBS containing RNAse A (Sigma) (200 μg/ml) for 30 min at 37°C. DNA was stained with Propidium Iodide (5 μg/ml, Sigma) in PBS overnight at 4°C. Samples were run on a FACS Calibur (Becton Dickinson) and data analysis was done using Flowjo software (Tree Star). Three independent experiments were analyzed for each cell line and condition.
BrdU incorporation for FACS analysis
Cells were incubated with 50 μM of BrdU for 2 h. Cells were then harvested in 200 μl ice-cold PBS after trypsinization. Fixed with 5 ml of ice-cold ethanol 70% added at low speed while vortexing. Cells were washed in (PBS + 0.5% Tween20), then incubated for 30 min with 1 ml of (2N HCl + 0.5% Triton) (protected from light). After cell wash in (PBS + 0.5% Tween20), cells were resuspended in 1 ml of BORAX (Na2B4O7 pH 8.5). Cells were washed and then incubated for 2 h in 200 μl of anti-BrdU (Becton Dickinson) diluted at 1/30 in (PBS + 0.5% Tween20 + 1% BSA). Cells were washed in (PBS + 0.5% Tween20 + 1% BSA) and then incubated for 2 h in 200 μl in anti-mouse FITC (Sigma) diluted at 1/300 in (PBS + 0.5% Tween20 + 1% BSA). Washed in (PBS + 0.5% Tween20 + 1% BSA) and then resuspended in 500 μl of PBS containing 200 μg/ml of RNAse (Sigma) (30 min incubation at 37°C). DNA was stained with 7-amino-actinomycin D (4 μg/ml) (Sigma) in PBS overnight at 4°C. Three independent experiments were analyzed for each cell line and condition.
DNA constructs
cDNA of PR-Set7 was amplified by PCR from BDGP clone (#LD12242) using Pfu ultra polymerase (Agilent) and first cloned into pTopo-ENTR vector (Invitrogen). Using Gateway technology we obtain FLAG-PR-Set7 (using pAWF vector, a vector used for ubiquitous cell expression of FLAG tagged proteins using Actin promoter) or GFP tagged PR-Set7 (using pPGW vector, a vector used for inducible expression of GFP tagged proteins in Drosophila using UAS/GAL4 driver system) using Gateway technology (Carnegie University). PR-Set7 cDNA was also cloned in frame with GFP into a Copper Sulfate inducible vector (pMT-GFP, metallothionein promoter) a generous gift of P. Heun. Mutant versions of PR-Set7 were obtained by site-directed mutagenesis using Pfu ultra polymerase. Primers used in this study are listed in Supplementary Table S3.
Fly stocks and handling, transgenic lines
Flies were raised on standard corn meal/yeast extract medium. pr-set7 null mutant allele (pr-set720) was a generous gift of Ruth Steward (20,23,24). Transgenic lines expressing GFP tagged versions of PR-Set7 were obtained from conventional P element transgenesis (Best Genes Inc.) and characterized. Transgenic lines for specific RNAi were obtained from VDRC (Vienna) (25). The construct identification numbers are: 34590 (PR-Set7), 107825 (Slimb). These different homozygous lines were crossed with the following GAL4 driver lines: ptc-GAL4 (# 65661), Actin-GAL4 (#4414), eyeGAL4 (#5535). Ubx-FLP; FRT82B-GFP/TM6 and FRT82Bslimb9H4–17/TM6tb were crossed to generate mitotic clones specifically in wing discs of third instar larvae. All crosses were raised at 25°C.
Small-scale biochemical fractionation
Small-scale biochemical fractionation was performed as described previously (26). Briefly, 20–30 millions of cells were collected, washed in ice-cold PBS and resuspended in buffer A (10 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol and protease inhibitor cocktail (Roche)). Triton X-100 was added (0.1% final concentration), the cells were incubated on ice for 5 min, and nuclei (fraction P1) were collected by centrifugation (5 min, 1300g). The supernatant S1 (soluble cytsolosic fraction) was collected and cleared by centrifugation (5 min, 20 000g). The fraction P1 was washed in buffer A and lysed for 30 min in buffer B (3 mM EDTA, 0.2 mM EGTA), and insoluble chromatin (P3) and soluble fraction (S2, nucleosoluble proteins) were separated by centrifugation (5 min, 1700g). The P3 fraction was washed once in buffer B and resuspended in sodium dodecyl sulfate (SDS)–Laemmli buffer (62.5 mM Tris pH 6.8 + 25% glycerol + 2% SDS + 0.01% Bromophenol blue + 710 mM β-mercaptoethanol) and boiled for 10 min. Three independent experiments were analyzed for each cell line and condition.
In vivo ubiquitylation assay
S2 cells stably expressing FLAG-tagged PR-Set7 were grown in complete medium at 2–5 × 106/ml concentration upon the addition of MG132 (final concentration: 25 μM). Cells were harvested 5 h later in lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA, 10 mM Iodoacetamide, 25 μM MG132, 0.1% Triton, 5% glycerol) supplemented with 0.1 mM PMSF and protease cocktail inhibitor (Roche) to make total cell extract. 1% SDS was then added to the cell lysate before a 10 min boiling at 95°C. Triton was added to the denaturing lysate to final concentration of 1%. Denaturing cell lysate was incubated for 4 h with the anti-FLAG M2 conjugated magnetic beads (Sigma) or with only protein G conjugated magnetic beads (Mock) at 4°C. Beads were washed three times for 10 min in Lysis Buffer and were resuspended in (SDS)–Laemmli buffer before immunoblot analysis. Two independent experiments were analyzed for each cell line and condition.
Co-immunoprecipitation
Total cell lysates from S2 cells expressing FLAG-tagged PR-Set7 were made by resuspension of cell pellet in lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 1% triton, 5% glycerol) supplemented with 0.1 mM PMSF and protease inhibitor cocktail (Roche). After 20 min at 4°C, lysates were cleared by centrifugation at maximum speed during 5 min. Lysates were incubated with anti-Flag M2 conjugated magnetic beads (Sigma) or protein G conjugated magnetic beads (mock) overnight at 4°C on a rotating wheel. After several washes, magnetic beads were resuspended in 30 μl of TBS buffer (10 mM Tris, 150 mM NaCl) containing 5 μg of FLAG peptide and incubated at 4°C during 2 h to allow elution of proteins from magnetic beads. Proteins were then denaturated by the addition of (SDS)–Laemmli buffer followed by 10 min boiling before immunoblot analysis.
Immunoblot
Immunoblotting experiments were carried out by separating proteins with SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotting onto nitrocellulose membranes in blotting buffer (20 mM Tris, 192 mM glycine, 20% ethanol). Membranes were incubated during 1 h at room temperature in Blocking Buffer (TBS, Tween 0.2%, 5% non-fat dry milk) followed by overnight incubation with primary antibodies diluted in blocking buffer. Membranes were washed three times in washing buffer (TBS-Tween 0.2%) and then incubated for 2 h with the appropriate secondary antibodies coupled with horseradish peroxidase (HRP) diluted in blocking buffer before a final cycle of washes in washing buffer. Immunoblot signals were visualized by chemiluminescence using the Immobilon Western Chemiluminescence HRP substrate (Millipore). The following primary antibodies used were: mouse anti ubiquitin antibody (1/500, Santa Cruz), Mouse anti Flag M2 (1/1000, Sigma), Guinea Pig anti Slimb (1/1000, a gift from G. Rogers), Rabbit anti PR-Set7 (1/1000, Novus Biological), mouse anti Tubulin (1/10 000, Sigma), rabbit anti GFP (1/1000, Sigma), rabbit anti H3 (1/10000, Abcam), rabbit anti H4 (1/10000, Cell signaling), rabbit anti H3S10p (1/1000, Diagenode), rabbit anti H4K20me1 (1/1000, Cell Signaling). The following secondary antibodies were used: anti mouse-HRP (1/10 000, Cell Signaling), anti rabbit-HRP (1/10000, Cell Signaling) and anti Guinea Pig-HRP (1/10 000, Abcam).
Immunostaining and microscopy
The anterior parts of 5–10 third instar larvae, which contain the majority of larval imaginal discs were dissected, pooled in Eppendorf tubes and fixed in 4% formalhedyde in PBT (PBS, 0.1% Tween20) for 20 min at room temperature (RT). After several washes in PBT, larvae were blocked in PBS-Tr (PBS, 0.3% Triton) with 10% Normal Donkey Serum (NDS) for 2 h at RT. They were incubated overnight at 4°C with rabbit anti PR-Set7 antibody (1/200), with rabbit anti-H3S10p antibody (Millipore, 1/1000) or with rabbit anti-H4K16ac (Active motif, 1/500). Larval tissues were washed in PBS-Tr, blocked 1 h in PBS-Tr-10% NDS for 1 h at RT and with an anti-rabbit Cy3 (Jackson Laboratories) diluted 1/500 in PBS-Tr-10% NDS for 1 h at RT. After several washes in PBS-Tr DNA was counterstained with DAPI and separated imaginal discs were mounted on slides in Prolong Antifade Medium (Molecular Probes). For immunostaining on Drosophila cells, ∼1 × 106 of cells were deposited on poly-lysine slides and incubated during 2 h at RT. Fixation was performed with 4% formaldehyde in PBT during 15 min. Cells were washed and blocked in PBT–10% NDS during 1 h. Anti-Rabbit PR-Set7 antibody was deposited on slides at 1/200 dilution in PBT–10% NDS. Slides were incubated in humid chamber overnight at 4°C. After several washes in PBS-Tr slides were incubated during 45 min with Anti Rabbit Alexa 488 diluted to 1/1000 at RT. After several washes, DNA was counterstained with DAPI and slides were mounted in Prolong Antifade Medium. Images were collected with Leica SP5 microscope equipped with a Plan/Apo 63 × 1.4 NA oil immersion objective. Each image (512 × 512 pixels) was saved in TIFF format upon sequential acquisition of 2–3 fluorochromes and analysed using ImageJ software. Images presented in the manuscript correspond to representative examples generated using Photoshop software. For analysis of chromosome structure shown in Figures 6 and 7, a minimum of 200 nuclei was analyzed per condition. Three independent experiments were performed for each cell line and each condition in order to calculate a mean value. Student t-test was applied to measure the significance of the results.
Figure 6.
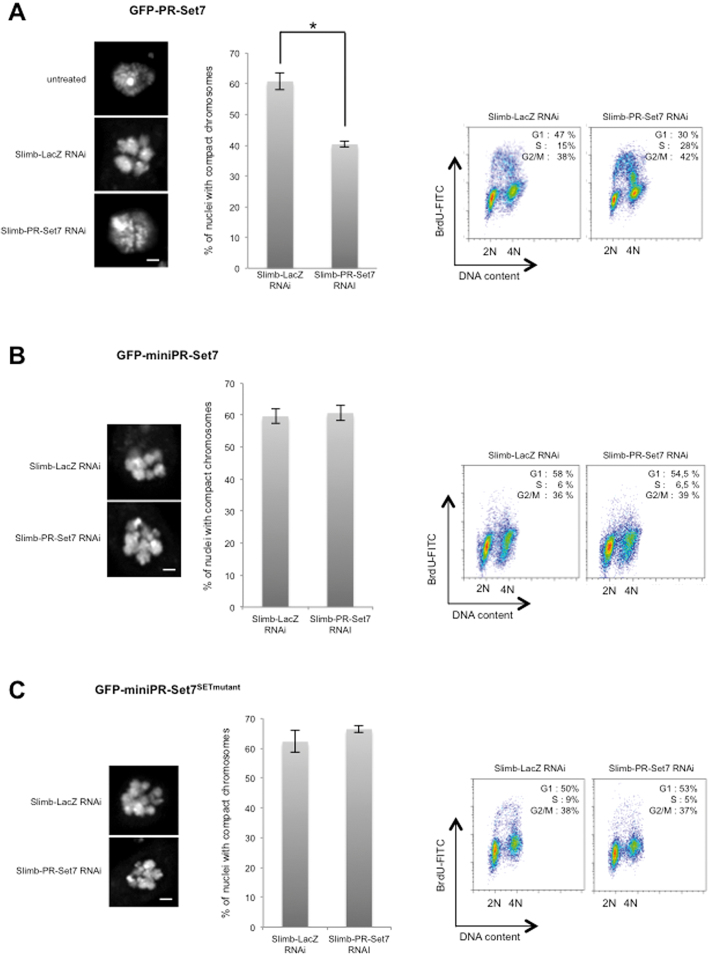
Degradation of PR-Set7 at G1/S transition induced by the SCFSlimb complex contributes to S phase entry and chromatin relaxation. (A–C) Left panels: Representative images of nuclei stained with DAPI in different S2 cells expressing different GFP-tagged PR-Set7 versions (as indicated on top) treated with different RNAi combinations as indicated. Scale bars represent 1 μm. Middle panels: Charts representing the percentage of nuclei with compact chromosomes in different S2 cells lines treated with Slimb/LacZ or Slimb/PR-Set7 RNAi as indicated. The number of nuclei analyzed per RNAi condition was approximately 200. Bars represent standard deviation of the mean calculated from three independent experiments. Star in (A) corresponds to P-value < 0.05 (Student's t-test). Right panels: Representative example of FACS analysis showing cell cycle profiles upon BrdU incorporation in the different S2 cells treated with different combinations upon different RNAi conditions as indicated, n = 3. Percentages of cells in the different phases of the cell cycle are indicated for each FACS profile.
Figure 7.
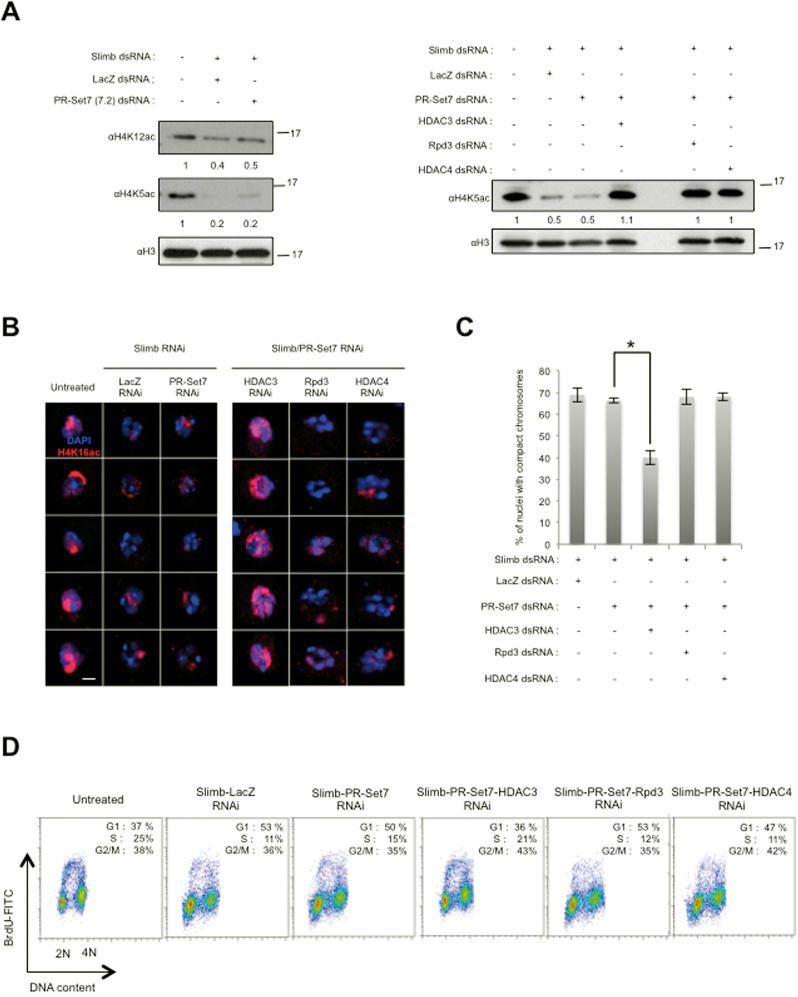
the SCFSlimb mediated degradation of PR-Set7 is necessary to counteract non-enzymatic functions of PR-Set7 in order to allow proper H4 acetylation. (A) Immunoblot analysis of whole cell extracts made from S2 cells expressing GFP-miniPR-Set7SETmutant upon treatment with different combinations of dsRNA as indicated on the top. Antibodies are indicated on the left. Relative amount of H4K5ac and H4K12ac normalized to H3 are indicated above each panel. (B) Representative images of nuclei stained with αH4K16ac (Red) DAPI (blue) in GFP-miniPR-Set7SETmutant S2 cells in different RNAi conditions as indicated on the top of each lane. Scale bars represent 1 μm. (C) Charts representing the percentage of nuclei with compact chromosomes in GFP-miniPR-Set7SETmutant S2 cells treated different combination of dsRNA as indicated above. The number of nuclei analyzed per RNAi condition was approximately 150. Bars represent standard deviation of the mean calculated from three independent experiments. Star corresponds to P-value < 0.05 (Student's t-test). (D) Representative example of FACS analysis showing cell cycle profiles upon BrdU incorporation in the different S2 cells treated with different combinations upon different RNAi conditions as indicated, n = 3.
RESULTS
Drosophila PR-Set7 is targeted for degradation by the ubiquitin-proteasome pathway from G1 to early S phase
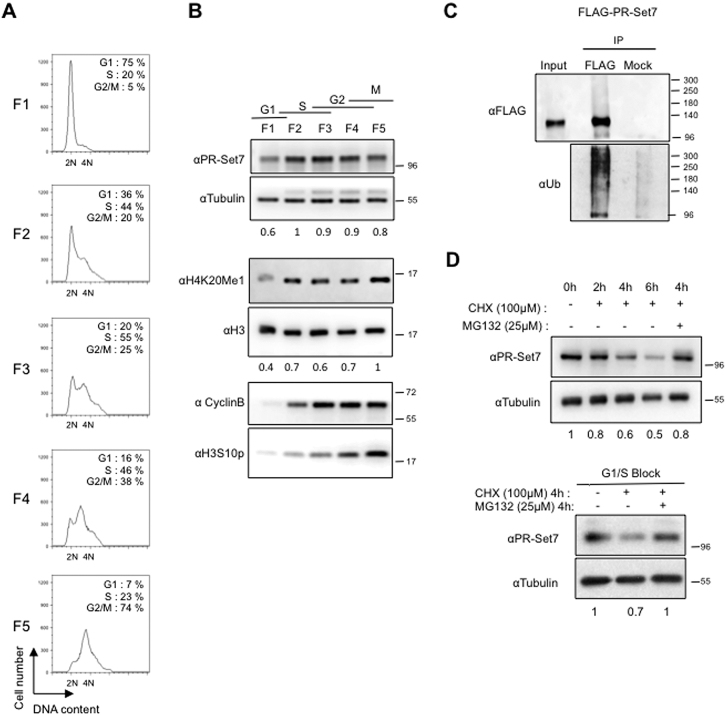
We first investigated whether the cell-cycle oscillations of PR-Set7 and H4K20 monomethylation mark (H4K20me1) described in mammalian cells are evolutionary conserved in Drosophila. To this end, PR-Set7 protein and histone H4K20me1 levels were analyzed through out the cell cycle in Drosophila S2 cells. Logarithmically growing S2 cells were separated according to cell size by centrifugal elutriation, allowing the isolation of cells in different cell-cycle phases with minimal physiological perturbation (22). The measure of DNA content by flow cytometry confirmed the differential cell-cycle distribution in each elutriated fraction (Figure 1A, left panels). Immunoblotting analyses of whole-cell extracts from each elutriated fraction showed that the levels of PR-set7 and H4K20me1 fluctuate during the cell cycle in Drosophila cells (Figure 1B). Previous studies in mammalian cells showed that the lowest levels of PR-Set7 and H4K20me1 were observed during S phase (7,8). In contrast, our data indicate that this down-regulation occurs earlier during the cell cycle in Drosophila. PR-Set7 and H4K20me1 lowest levels being observed in the elutriated fraction enriched in G1 and early S phase cells (Figure 1B). Relative quantification of PR-Set7 normalized to tubulin protein levels revealed that the amount of the enzyme was reduced by 40% in this fraction (Figure 1B). Of note, similar results were obtained with chemically synchronized cells that did not result from pr-set7 gene expression changes (Supplementary Figure S1A). Altogether, these results suggest that the cell-cycle oscillation of Drosophila PR-Set7 protein is likely caused by ubiquitin/proteasome-dependent degradation. To verify this, S2 cells expressing a FLAG-tagged version of PR-Set7 (FLAG-PR-Set7) were lysed under denaturing conditions and anti-FLAG immuno-complexes were probed with anti-ubiquitin antibody. As shown in Figure 1C, polyubiquitylated forms of FLAG-PR-Set7 were easily detected in these experimental conditions. To determine whether this PR-Set7 polyubiquitylation is associated with a proteasome-mediated degradation, we measured the half-life of endogenous PR-Set7 following inhibition of new protein synthesis by the protein-synthesis inhibitor cycloheximide (CHX) in asynchronous S2 cells and in adherent S2R+ cells enriched in G1 and early S-phase upon mimosine treatment (21). Synchronization of mimosine-treated S2R+ cells was verified by FACS analysis (Supplementary Figure S1B). In both CHX-treated asynchronous and mimosine-synchronized cells, the decrease in PR-Set7 protein levels was prevented by addition of the proteasome inhibitor MG132 (Figure 1D). Altogether these results show that the cell-cycle proteolytic regulation of PR-Set7 is conserved in Drosophila except that the lowest levels of the enzyme are observed from G1 to early S phase.
Figure 1.
Cell-cycle regulation of Drosophila PR-Set7 depends on the proteasome. (A) Representative examples of cell cycle profile obtained by FACS showing DNA content of cells in different fractions (F1 to F5) collected by elutriation from an asynchronous population of Drosophila S2 cells. Percentages of cells in the different phases of the cell cycle are indicated for each FACS profile. n = 2. (B) Immunoblot analysis of whole cell extracts made from fractions (F1 to F5) shown in (A). Cell cycle phases and antibodies used are indicated on the top and the left, respectively. Relative amounts of PR-Set7 and H4K20me1 normalized respectively to Tubulin and H3 are indicated. α-Cyclin B and α-H3S10P antibodies were used to discriminate fractions enriched in cells in G2 phase or in Mitosis. (C) Immunoblot analysis of immunoprecipitates (IP) done from lysates of S2 cells expressing ubiquitously (actin promoter) FLAG-PR-Set7 using αFLAG coupled or protein G coupled (Mock) dynabeads revealed with α-FLAG (top) and α-Ubiquitin (bottom) specific antibodies. Input lane corresponds to 10% of lysates used per IP. Note that 10% of the eluted material from the IP was loaded on gel to detect FLAG protein. (D) PR-Set7 stability analyzed in an asynchronous population of S2 cells (upper panel) or in adherent S2R+ cells that can be treated with mimosine to induce a cell-cycle arrest at G1/S transition (lower panel). Cells were treated with cycloheximide (CHX) alone or in combination with the proteasome inhibitor MG132 for the indicated times, before harvesting and immunoblot analysis of total cell extracts. Antibodies used are indicated on the left. Relative amounts of PR-Set7 normalized to Tubulin are indicated.
PCNA-interacting (PIP) motif of Drosophila PR-Set7 is not required for its degradation and is dispensable for development
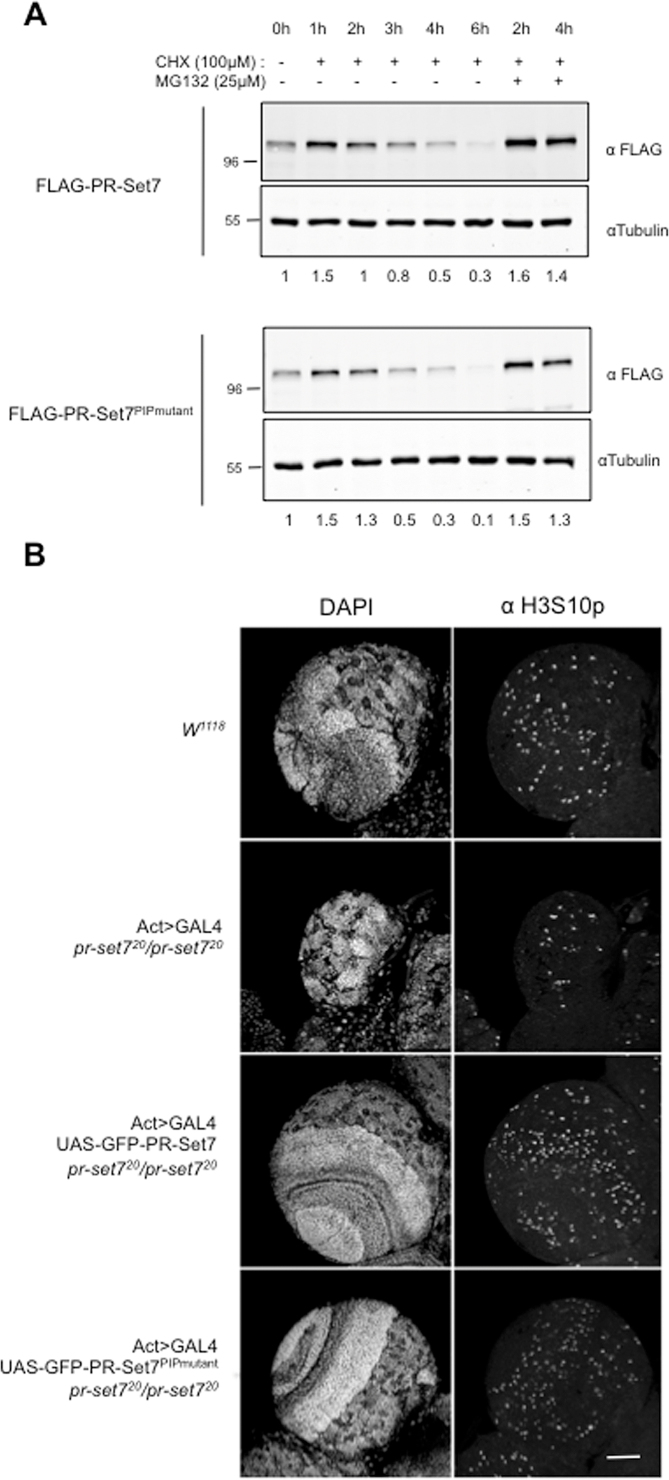
In mammalian cells, the proteasome-mediated degradation of PR-Set7 mainly depends on its interaction with PCNA via an evolutionary conserved PCNA-interacting (PIP) motif located upstream of the catalytic SET domain (20,27). Since the interaction between PR-Set7 and PCNA is conserved in Drosophila (28), we asked whether the PIP-motif is required for the proteolytic regulation of Drosophila PR-Set7. To address this question, we repeated cycloheximide chase experiments with S2 cell lines stably expressing either FLAG-tagged wild type PR-Set7 (FLAG-PR-Set7) or a FLAG-tagged PR-Set7 PIP-motif mutant (FLAG-PR-Set7PIPmutant) deficient for the interaction with PCNA (7,11,29). The results are shown in Figure 2A. As expected, FLAG-PR-Set7 exhibited a half-life and MG132-sensitivity similar to those of the endogenous protein (compare Figures 1D and 2A). Most surprising, however, was that FLAG-PR-Set7PIPmutant displayed similar behavior (Figure 2A, lower panels), suggesting that the PIP-motif PCNA-dependent pathway is not involved in the cell-cycle oscillation of Drosophila PR-Set7 protein.
Figure 2.
PCNA Interacting (PIP) motif of PR-Set7 is not necessary for PR-Set7 degradation and is dispensable for Drosophila development. (A) Immunoblot analysis of whole cell extracts prepared from S2 cells expressing ubiquitously (actin promoter) a wild type (FLAG-PR-Set7, upper panel) or a PIP mutant version of PR-Set7 (FLAG-PR-Set7PIPmutant, lower panel) treated with cycloheximide (CHX) alone or in combination with the proteasome inhibitor MG132 as indicated. Antibodies and relative amounts of each FLAG tagged PR-Set7 protein normalized to Tubulin are indicated on the left and above each panel, respectively. (B) Optic lobes of third instar larvae of different genotypes, as indicated on the left, were stained with DAPI (left panel) and the mitotic marker anti-H3S10p (right panel) and analyzed by immunofluorescence. W1118 corresponds to a wild type control line. Act>GAL4 indicates the expression of GAL4 protein by the actin promoter. pr-set720 corresponds to a loss of function allele of pr-set7. UAS-GFP-PR-Set7 and UAS-GFP-PR-Set7PIPmutant corresponds to transgenic constructs under the control of an UAS sequence. Scale bar represents 50μm.
To determine whether the PCNA interaction, via the PIP-motif, is nevertheless required for PR-Set7 functions during development, we generated transgenic animals expressing either GFP-tagged PR-Set7 or GFP-tagged PR-Set7PIPmutant under the inducible UAS/GAL4 system (30). We then determined whether the expression of these GFP-tagged proteins could rescue cell growth in developing tissues of flies homozygous null mutant for pr-set7 (allele pr-set720) (24). Staining with anti-histone H3 Serine 10 phosphorylation (H3S10p) and DAPI showed that the brain optic lobes, which correspond to regions of high cell-division rates, were almost missing in homozygous pr-set7 null larvae as compared to w1118 control line (Figure 2B, compare upper panels). In contrast, expression of GFP-PR-Set7 as well as GFP-PR-Set7PIPmutant was able to rescue cell proliferation and proper organization in larval brains (Figure 2B, middle and lower panels). In line with this, GFP-PR-Set7 or GFP-PR-Set7PIPmutant were both effective to restore proper H4K20me1 levels and to prevent the lethality of homozygous prset7 null larvae (Supplementary Figure S2). Altogether, these results demonstrate that the PR-Set7 PIP-motif is neither essential for the proteolytic regulation of the enzyme, nor for its functions during Drosophila development.
Identification of the minimal Drosophila PR-Set7 sequence required for proper cell-cycle progression
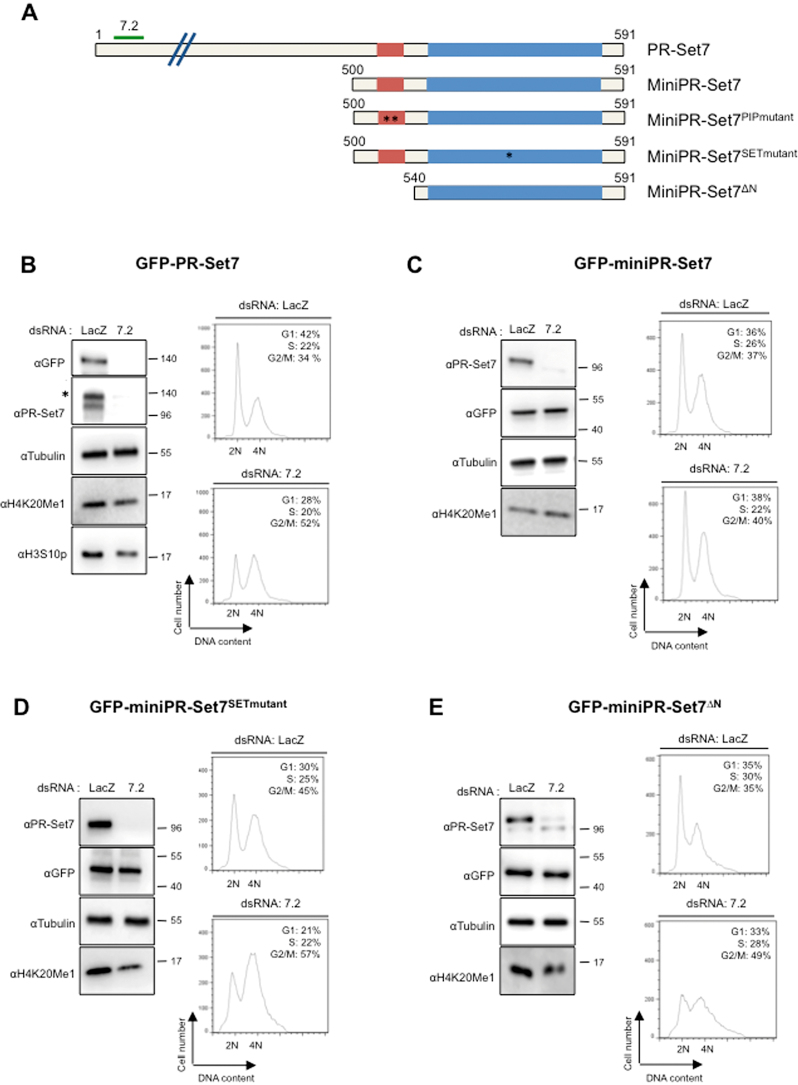
To identify the regions of Drosophila PR-Set7 necessary for proper cell proliferation, we examined the ability of different GFP-tagged PR-Set7 mutants to bypass the cell-cycle defects induced by the loss of endogenous PR-Set7 in Drosophila S2 cells. Figure 3A shows the Drosophila wild type version of GFP-tagged PR-Set7 (GFP-PR-Set7) and of the four different GFP-PR-Set7 mutants tested. In contrast to GFP-PR-Set7, all these mutants missed the sequence targeted by the PR-Set7 dsRNA (named 7.2) and were therefore naturally resistant to the 7.2 dsRNA-mediated PR-Set7 silencing. The wild type and the GFP-PR-Set7 mutants were stably expressed in S2 cells that were then treated with either 7.2 dsRNA or an irrelevant LacZ dsRNA as a control. After 5 days, the efficacy of RNAi treatment was measured by immunoblotting and the progression through the cell cycle was examined by flow cytometry, as shown in Supplementary Figure S3 and in Figure 3B to E. In agreement with previous results (23,29), Drosophila PR-Set7 depletion in parental S2 cells triggered a decrease in H4K20me1 followed by an accumulation of cells with 4N or near 4N DNA content (Supplementary Figure S3A). These cells were likely arrested in late S and G2 phases as indicated by the low levels of the mitotic marker H3S10p in 7.2 treated cells compared to control cells (Supplementary Figure S3A). Whereas a similar cell-cycle phenotype was observed in GFP-PR-Set7 expressing cells treated with 7.2 dsRNA (Figure 3B), expression of the GFP-miniPR-Set7 mutant, which is deleted of the first 500 amino acids of PRSet7, was able to maintain proper H4K20me1 levels and proper cell-cycle progression (Figure 3C). These results indicate that the PR-Set7 C-terminal region of 91 amino acids, which harbors the SET domain and an upstream sequence of 40 amino acids (residues 500–540), is necessary and sufficient for the cell-cycle functions of PR-Set7.
Figure 3.
Identification of a minimal version of PR-Set7 sufficient for cell cycle control. (A) Schematic representation of PR-Set7 and different mutant versions of PR-Set7 used in this study. Red and blue rectangles represent the PIP and the catalytic SET domains, respectively. Green line above PR-Set7 named 7.2 indicates the targeted region by double stranded RNA (dsRNA) used to deplete PR-Set7. MiniPR-Set7 encodes the last 91 amino acids of PR-Set7. MiniPR-Set7PIPmutant harbors a double mutation (F523A, F524A) in the PIP domain as indicated by stars. MiniPR-Set7SETmutant harbors a single mutation (R634P) in the catalytic SET domain as indicated by a star. MiniPR-Set7ΔN encodes only the catalytic SET domain of PR-Set7. Each of these versions of PR-Set7 was GFP tagged, stably transfected in S2 cells and expressed under the control of metallothionein promoter inducible with CuSO4. (B–E) Left panels: Immunoblot analysis of whole cell extracts derived from S2 cell lines expressing each GFP tagged version of PR-Set7 (as indicated on the top of each panel) treated during 5 days either with LacZ (control) or PR-Set7 (7.2) dsRNA as indicated. Antibodies used are indicated on the left for each panel. Star in PR-Set7 immunoblot in (B) indicates the GFP tagged version of PR-Set7. Right panels: Representative examples of cell cycle profiles obtained by FACS showing DNA content of S2 cell lines expressing the different GFP tagged versions of PR-Set7 as indicated, 5 days after the addition of LacZ or 7.2 dsRNA as indicated on the top, n = 3. Percentages of cells in the different phases of the cell cycle are indicated for each panel.
In contrast to fly homozygous null mutant for pr-set7 (20,24), a mutation of lysine 20 of histone H4 into alanine is viable into the adulthood (31), which questions whether the SET domain is essential for PR-Set7 cell-cycle functions. We addressed this critical issue by depleting the endogenous PR-Set7 in S2 cells expressing the GFP-miniPR-Set7SETmutant mutant, in which an arginine to a proline substitution at position 634 is sufficient to abolish the activity of the SET domain (20,27). As shown in Figure 3D (upper panel), GFP-miniPR-Set7SETmutant expression naturally led to higher percentage of control cells with 4N DNA content, likely caused by a slight dominant negative effect of this mutant. Nevertheless, a strong G2/M arrest was still observed in 7.2 dsRNA–treated GFP-miniPR-Set7SETmutant cells (Figure 3D, lower panel), showing indeed that Drosophila PR-Set7 methylase activity is necessary for G2 to M phase progression. In addition to the SET domain, we also found that the N-terminal region of GFP-mini-PR-Set7 (residues 500–540 of PR-Set7) is required for G2/M progression (Figure 3E). Of note, the PIP-motif is included in this 40 amino acid region (Figure 4A). However, consistent with the results in Figure 2, the mutation of this motif did not abrogate the ability of GFP-miniPR-Set7 to rescue the PR-Set7 loss of function phenotype in S2 cells (Supplementary Figure S3B). Thus, the minimal sequence of Drosophila PR-Set7 necessary for proper cell cycle progression involves the SET domain and a novel upstream element located between the residues 500–540.
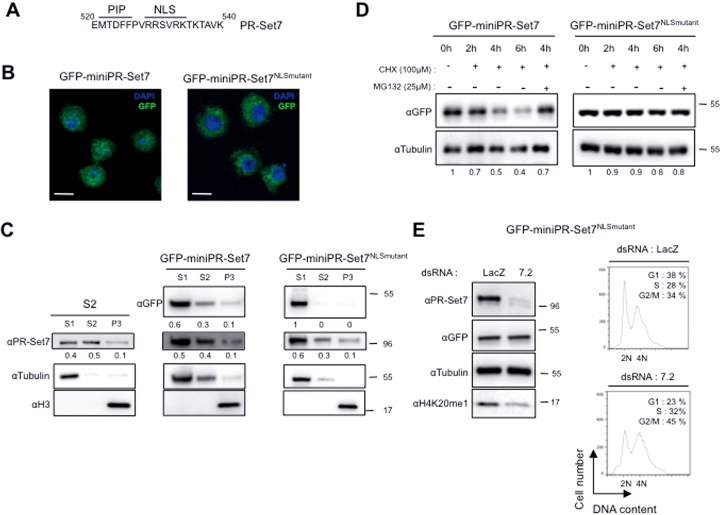
Figure 4.
Nuclear fraction of PR-Set7 is important for cell cycle control and is targeted by proteasome. (A) Sequence of PR-Set7 (aa 520–540) showing the PIP domain and a putative NLS sequence as indicated. (B) S2 cells expressing GFP-mini-PR-Set7 (left) or GFP-mini-PR-Set7NLSmutant (right) were stained with DAPI (shown in blue) and analyzed by immunofluorescence. Green signals correspond to GFP tagged version of miniPR-Set7 as indicated on the top. Bars represent 5 μm. (C) Immunoblot analysis of sub-cellular fractions (S1: cytosoluble, S2: nuclear soluble, P3: pellet containing chromatin) resulting from biochemical fractionation of parental S2 cells (left panel) or S2 cells expressing GFP-miniPRSet7 (middle panel) or GFP-miniPR-Set7NLSmutant (right panel). Antibodies used are indicated on the left. Percentages of PR-Set7 and GFP tagged proteins present in the different fractions are indicated above each panel. αTubulin and αH3 antibodies were used to validate efficacy of fractionation. (D) Immunoblot analysis with the indicated antibodies of whole cell extracts prepared from S2 cells expressing GFP-mini-PR-Set7 (upper panel) or GFP-miniPR-Set7NLSmutant (lower panel) treated with cycloheximide (CHX) alone or in combination with the proteasome inhibitor MG132 for the indicated times. Relative amounts of the different GFP-tagged proteins normalized to Tubulin are indicated. (E) Left: Immunoblot analysis with the indicated antibodies of whole cell extracts derived from GFP-miniPR-Set7NLSmutant expressing S2 cells treated with LacZ (control) or PR-Set7 (7.2) specific dsRNA as indicated. Right panel: Representative examples of cell cycle profiles obtained by FACS showing DNA content of S2 cells expressing GFP-miniPR-Set7NLSmutant following 5 days of RNA interference against LacZ (control) or PR-Set7 (7.2), n = 3.
Nuclear import of Drosophila PR-Set7 is important for its cell-cycle function and its proteasome-mediated degradation
Close examination of the PR-Set7 region between the residues 500–540 revealed that this region, in addition to the PIP-motif, contains a short conserved sequence (RRSVRK) that resembles a Nuclear Localization Signal (NLS) (Figure 4A). To determine whether this sequence functions indeed as a NLS, we first examined by fluorescence the cellular localization of GFP-miniPR-Set7 and GFP-miniPR-set7NLSmutant, in which the RRSVRK sequence was mutated into AASVAA. Whereas GFP-miniPR-Set7 displayed both cytoplasmic and nuclear localization, GFP-miniPR-set7NLSmutant was exclusively found in the cytoplasm (Figure 4B). To confirm these results and determine quantitatively which amount of PR-Set7 is effectively localized into the nucleus, cell lysates from S2 cells expressing or not GFP-miniPR-Set7 or GFP-miniPR-Set7NLSmutant were fractionated into three samples: (i) fraction (S1) with soluble cytosolic proteins, (ii) fraction (S2) with soluble nuclear components and (iii) fraction (P3) containing insoluble chromatin and nuclear matrix components (26). Proper fractionation was confirmed by immunoblotting with antibodies against histone H3 and tubulin. As shown in Figure 4C (left and middle panels) and consistent with the fluorescence staining in Figure 4B, both endogenous PR-Set7 and GFP-miniPR-Set7 were found equally distributed between S1 and S2 fractions with only a small amount of the enzyme in the P3 fraction. In sharp contrast, GFP-miniPR-Set7NLSmutant was only found into the cytoplasmic S1 fraction (Figure 4C, right panels). Altogether these results demonstrate that the RRSVRK sequence constitutes an intrinsic NLS that is essential for the nuclear import of PR-Set7.
To determine the role of this NLS-mediated nuclear import in PR-Set7 functions, we first examined the kinetics of degradation of GFP-miniPR-Set7 and GFP-miniPR-Set7NLSmutant upon CHX treatment. The results are shown in Figure 4D. Whereas GFP-miniPR-Set7 was degraded as the endogenous PR-Set7 (compare Figures 1D and 4D), miniPR-Set7NLSmutant was very stable even after 6 h of cycloheximide (CHX) treatment. These results suggest that the proteolytic degradation of PR-Set7 is restrained to the nuclear pool of the enzyme. We next examined the cell-cycle progression of GFP-miniPR-Set7NLSmutant cells upon loss of endogenous PR-Set7 by flow cytometry. Conspicuously, depletion of endogenous PR-Set7 in 7.2 treated GFP-miniPR-Set7NLSmutant cells led to an accumulation of cells with 4N along with a decrease in the level of H4K20me1 compared to LacZ (control) RNAi treated cells (Figure 4D). Altogether, these results show that the import of PR-Set7 into the nucleus is necessary for its cell-cycle function as well as its proteasome-mediated degradation.
Drosophila PR-Set7 proteolytic degradation is controlled by the SCFSlimb complex
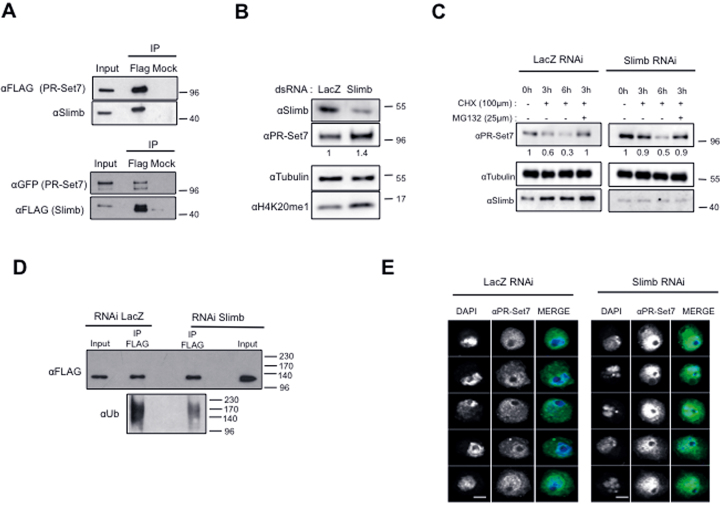
To gain insights into the mechanisms that regulate the stability of PR-Set7, we first sought to identify which complexes containing E3 ubiquitin ligase(s) promotes the effective targeting of PR-Set7 to the proteasome. In addition to the PCNA-mediated E3 ligase complex CRL4Cdt2, recent studies showed that mammalian PR-Set7 is also a substrate of APCCDH1 and SCFβ-TRCP E3 ubiquitin complexes (15,18). Compared to Cyclin B, which is a known target of the APCCDH1 (32), we did not observe any significant change in PR-Set7 protein levels in S2 cells depleted for Fizzy-related (Fzr), the Drosophila ortholog of CDH1 (Supplementary Figure S4A). In contrast, we identified a functional link between PR-Set7 and Slimb, the fly homolog of β-TRCP (33). Indeed, we observed that rough phenotype mediated by PR-Set7 RNAi in Drosophila eye was rescued by the co-depletion of Slimb (Supplementary Figure S4B) and by immunoprecipitation assays in S2 cells that ectopically expressed FLAG-PR-Set7 protein can associate with endogenous Slimb, (Figure 5A, upper panels). Likewise, FLAG-tagged Slimb efficiently immunoprecipitated GFP-tagged wild-type and mini-PR-Set7 but not with the cytoplasmic mini-PR-Set7NLSmutant (Figure 5A, lower panels and Supplementary Figure S4C). Finally, immunostaining with anti-PRSet7 antibody revealed a strong accumulation of PR-Set7 into the nuclei of S2 cells (Figure 5E) and of wing disc cells upon Slimb depletion (Supplementary Figures S4D and S4E), thereby suggesting that Slimb is a critical determinant of PR-Set7 stability in Drosophila nuclei. To verify this hypothesis, we first examined by immunoblotting the levels of endogenous PR-Set7 and of GFP-miniPRset7 in S2 cells treated with control (LacZ) or Slimb dsRNA. We found that the basal level and half-life of both proteins were significantly increased in Slimb depleted cells compared to control cells (Figure 5B, C and Supplementary Figure S5A, B). Accordingly, polyubiquitylated forms of FLAG-PR-Set7 were found reduced upon Slimb depletion (Figure 5D).
Figure 5.
the SCFSlimb complex targets the nuclear pool of PR-Set7. (A) Immunoblot analysis for the indicated antibodies of immunoprecipitates (IP) done from lysates of S2 cells expressing FLAG-PR-Set7 (upper panel) or GFP-PR-Set7 and FLAG-Slimb (lower panel) using FLAG specific or irrelevant (Mock) antibodies. Input lane corresponds to 10% of cell lysates used per IP. (B) Immunoblot analysis for the indicated antibodies of whole cell extracts derived from S2 cells treated with LacZ as a negative control or Slimb specific dsRNA as indicated. Antibodies used are indicated on the left. (C) Immunoblot analysis of whole cell extracts derived from S2 cells incubated with LacZ (control, left) or Slimb dsRNA (right) during 5 days and treated with cycloheximide (CHX) alone or in combination with MG132 as indicated. Antibodies used are indicated in the left. Relative amounts of PR-Set7 normalized to tubulin in each RNAi condition are indicated. (D) Immunoblot analysis of immunoprecipitates (IP) done from lysates of S2 cells expressing ubiquitously (actin promoter) FLAG-PR-Set7 using αFLAG coupled or protein G coupled (Mock) dynabeads revealed with α-FLAG (top) and α-Ubiquitin (bottom) specific antibodies upon LacZ (control, left) or Slimb RNAi treatment during 5 days. Input lane corresponds to 10% of lysates used per IP. Note that 10% of the IP material was loaded on gel to detect FLAG protein. (E) Representative examples of S2 cells incubated with LacZ (left panels) or Slimb dsRNA during 4 days stained with PR-Set7 antibody (green) and DAPI (gray) and analyzed by immunofluorescence. Scale bars represent 3μm.
Since multiple evidences have demonstrated that protein ubiquitylation and degradation by the SCFSlimb complex occur on phosphorylated substrates, we assessed whether Drosophila PR-Set7 is phosphorylated as its mammalian counterpart (18). Total cell lysates from miniPR-Set7-GFP expressing cells were applied on Qiagen Phosphoprotein column that specifically retain phosphorylated proteins. Specific analysis by immunoblot of the column bound fractions as compared to unbound fraction (Flowthrough, FT) revealed that endogenous PR-Set7 and mini-PR-Set7 proteins are indeed phosphorylated (Supplementary Figure S5C). As it has been previously reported that CKIα and GSK3β are often involved in Slimb-mediated degradation (18,34–36), we next analyzed whether these kinases contribute to PR-Set7 degradation. A significant increase in the steady-state levels of GFP-PR-Set7 was observed in CKIα-depleted S2 cells (Supplementary Figure S5D), leading to an accumulation of the enzyme into nuclei (Supplementary Figure S5E). Altogether, these results show that SCFSlimb complex, likely via CKIα-mediated phosphorylation, negatively regulates PR-Set7 nuclear functions in Drosophila cells.
Slimb-mediated PR-Set7 degradation ensures G1/S transition and proper interphase chromatin compaction by preventing non-enzymatic PR-Set7 nuclear functions
Inhibition of Slimb functions triggers a G1/S arrest associated with a strong compaction of interphase chromosomes, which has been related to an improper accumulation of the condensin II subunit Cap-H2 on chromatin (36,37). Since PR-Set7 has been also implicated in chromatin structure maintenance (38,39) and, as shown above, it is a Slimb substrate degraded in G1 and early S-phase, we examined whether an abnormal stabilization of PR-Set7 could also contribute to the defects in chromatin structure and cell-cycle progression in Slimb-depleted cells. To this end, we performed RNAi treatment during seven days by combining Slimb dsRNA with an irrelevant LacZ or 7.2 (PR-Set7) dsRNA in S2 cells expressing GFP-miniPR-Set7 that is naturally resistant to PR-Set7 RNAi treatment (see Figure 3C). The cell line expressing wild-type GFP-PR-Set7 was used as a control. To circumvent the G2/M arrest induced by PR-Set7 depletion, which might indirectly interfere with the appearance of Slimb-associated cell cycle phenotypes, cells lines were first treated with Slimb dsRNA during 4 days to induce a robust G1/S arrest (Supplementary Figure S6A). Slimb-depleted cell were then treated with lacZ (control) or PR-Set7 dsRNA for 3 days. As expected, combination of Slimb with lacZ dsRNA led to Slimb depletion followed by an increase in endogenous PR-Set7 as well as in GFP-PR-Set7 proteins (Supplementary Figures S6B-C, compare lanes 1 and 2), whereas the combination of Slimb with 7.2 dsRNA was sufficient to prevent the accumulation of endogenous PR-Set7 and wild-type GFP-PR-Set7 but not of GFP-miniPR-Set7 (Supplementary Figures S6B and S6C). Chromosome morphology was then examined by DAPI staining and cell cycle progression was monitored by FACS analysis after BrdU incorporation. As shown in Figure 6A and B, the appearance of dense and compact chromosomes in 60% of treated cells and a G1/S arrest were observed in GFP-PRSet7 and GFP-miniPRSet7 cells treated with Slimb/LacZ RNAi. Note of, an aberrant chromosome compaction phenotype was also observed in CKIα-depleted S2 cells that displayed a PR-Set7 nuclear accumulation (Supplementary Figure S5E). Upon Slimb/PR-Set7 RNAi treatment, both G1/S arrest and chromosome compaction were significantly reduced in GFP-PRSet7 cells but not in cells expressing the RNAi-resistant GFP-miniPRSet7 (Figure 6A and B). These results show that Slimb-mediated PR-Set7 degradation contributes to interphase chromatin relaxation and G1/S transition in proliferating Drosophila cells.
To further gain insights into the mechanisms by which PR-Set7 can negatively regulate G1/S transition and favor chromosome compaction, we asked whether the PR-Set7 methylase activity was required by performing the same RNAi experiments in cells expressing the catalytic inactive GFP-miniPR-Set7SETmutant (Supplementary Figure S6D). Strikingly, the presence of GFP-miniPR-Set7SETmutant in Slimb/PR-Set7 RNAi treated cells was sufficient to maintain G1/S arrest and abnormal chromosomal compaction (Figure 6C), suggesting that both phenotypes occur independently of PR-Set7 catalytic activity. Histone H4 acetylation is known to favor chromatin relaxation and DNA replication (4,40,41) and Drosophila PR-Set7 has been associated with histone deacetylase (HDAC) activity in chromatin assembly assays (42). We therefore hypothesized that the chromatin compaction and G1/S arrest phenotypes upon PR-Set7 stabilization could be related to changes in histone H4 acetylation induced by improper HDAC activity. Consistent with this possibility, immunoblotting and immunofluorescence experiments showed a decrease in H4K5ac, H4K12ac and H4K16ac levels upon GFP-miniPR-Set7SETmutant stabilization in Slimb-depleted cells (Figure 7A and B, left panels). To determine whether this decrease in H4 acetylation might contribute to the appearance of chromatin compaction and G1/S arrest induced by PR-Set7 stabilization, GFP-miniPR-Set7SETmutant cells depleted for Slimb and endogenous PR-Set7 were treated with dsRNA directed against either histone deacetylases HDAC3, Rpd3/HDAC1 or HDAC4 mRNAs and the levels of H4K5 and H4K16 acetylation, the chromatin status and the cell-cycle distribution of these cells were then examined as described above. As shown in Figure 7A (right panels) and Figure 7B (right panels), although HDAC3, Rpd3 or HDAC4 RNAi treatment was sufficient to increase H4K5ac levels in Slimb/PR-Set7 RNAi treated GFP-miniPR-Set7SETmutant cells, only HDAC3 depletion restored proper H4K16 acetylation in these cells. In line with these results, we found that HDAC3 depletion, but not of Rpd3 or HDAC4, reduced significantly the percentage of cells with aberrant chromosomal compaction and restored proper G1/S progression (Figure 7C and D). Altogether, these results reveal that SCFSlimb-mediated PR-Set7 proteolytic degradation is required to inhibit non-enzymatic PR-Set7 functions, likely to avoid inappropriate HDAC3 activity, and thus ensures proper acetylation at different lysines on histone H4 tail and a chromatin relaxation threshold that is necessary for G1/S progression.
DISCUSSION
In this study, we provide evidence that Drosophila PR-Set7 undergoes a proteasome-mediated degradation during the cell cycle that is highly active from G1 to early S phase (Figure 1). Contrary to its human counterpart, this proteolytic regulation does not involve the conserved PCNA-interacting (PIP) motif, which appears dispensable for PR-Set7 functions during Drosophila development (Figure 2). Expression of different mutant RNAi-resistant mutants reveals that PR-Set7 controls cell growth by exerting positive and negative roles at distinct phases of the cell cycle. While the nuclear localization of a catalytically active PR-Set7 is important for G2/M progression (Figures 3 and 4), we show that the specific degradation of the nuclear pool of PR-Set7 by the E3 ligase complex SCFSlimb contributes to DNA replication and chromatin relaxation by ensuring proper histone H4 acetylation during G1 phase (Figures 5–7). Finally, we show that the G1/S arrest and the aberrant chromatin structure induced by PR-Set7 stabilization does not involve its catalytic activity (Figure 7), thereby revealing a non-enzymatic role of PR-Set7 in the regulation of chromatin structure and cell cycle progression.
Previous studies in both mammalian and Drosophila models showed that loss of PR-Set7 perturbs S-phase progression leading to a G2/M arrest with chromosomal anomalies and DNA damage (8,10,12,13). It has been assumed that the appearance of these cell-cycle defects in mammalian cells was related to the absence of PR-Set7-induced histone H4K20 methylation and the accumulation of unmodified H4K20. However, the use of histone replacement system to interrogate directly the function of specific lysine residues in Drosophila revealed that the mutation of histone H4 lysine 20 to alanine (H4K20A) has no impact on cell-cycle progression and on completion of development (31). Although exhibiting some DNA damage in wing discs, H4K20A mutant flies (29) are mainly viable in contrast to pr-set7 null flies that die at the pupal stage (20,24), thereby questioning whether the methylase activity of Drosophila PR-Set7 is required for cell-cycle progression. Our results show that a minimal functional version of Drosophila PR-Set7 (miniPR-Set7) harboring a mutation in the SET domain was unable to rescue the G2/M arrest induced by silencing of endogenous PR-Set7 (Figure 3). Thus, PR-Set7 catalytic activity is necessary for proper entry into mitosis likely through the methylation of H4K20 and non-histones substrates that remain to be identified. However, our results also indicate that the SET domain of PR-Set7 is not sufficient for G2/M transition. An additional short upstream sequence, corresponding to a nuclear localization signal (NLS) is also required. Consistent with this, mutation of this sequence prevents the localization of miniPR-Set7 into the nucleus and abolishes its ability to rescue loss of PR-Set7 functions (Figure 4). Clearly, the subcellular localization of Drosophila PR-Set7 appears as an important regulatory mechanism since half of the enzyme is cytoplasmic and that proteolytic degradation targets preferentially the nuclear fraction of PR-Set7. Interestingly, the NLS sequence is conserved between Drosophila and mammals (our unpublished data), suggesting that the control of PR-Set7 cellular localization could also constitute an important mode of regulation of the enzyme in mammals. In line with this, the distribution of human PR-Set7 seems to vary greatly between human cell types. While the protein is exclusively found in the nucleus of U2OS cell lines (13,43), the distribution of the enzyme appears both cytoplasmic and nuclear in Hela cells (17,19). Whether deregulation of the mechanisms controlling PR-Set7 subcellular distribution could contribute to cancer progression is therefore an interesting issue to explore, since up-regulation of PR-Set7 is considered as a poor prognosis factor in neuroblastoma and breast tumors (44–46).
In mammals, proper cell cycle progression also involves a tight regulation of PR-Set7 enzymatic activity through the timely destruction of the enzyme at specific phases of the cell cycle. This is mainly orchestrated by the PCNA-interacting motif of PR-Set7 that drives its ubiquitylation by CRL4cdt2 and its subsequent proteolytic degradation during S-phase. Indeed, expression of a PIP-mutated PR-Set7 protein resistant to the PCNA-mediated CRL4Cdt2-dependent destruction in human U2OS cells causes a premature chromatin compaction followed by DNA re-replication during S-phase (10,11,13). This DNA re-replication, which can be abolished by a second mutation in the catalytic SET domain, reflects the ability of PR-Set7-mediated H4K20me1 and its subsequent conversion to higher H4K20me states by Suv4–20h enzymes to stimulate the recruitment on chromatin of pre-replication complexes that define functional replication origins along the genome (16,47). In this context, the finding that proteolytic degradation of Drosophila PR-Set7 during the cell cycle is conserved but occurs independently of the PCNA interacting PIP motif was unexpected (Figure 2). Consistent with a specific requirement of the PIP-motif in the regulation of mammalian PR-Set7 replication functions, it has been recently revealed that Drosophila PR-Set7 is dispensable for the licensing and the activation of replication origins (29). This could also explain why Drosophila suv4–20 null mutant is viable compared to the loss of their mammalian counterparts (48–50). It is widely believed that proper setting of replication origins on chromatin requires different combinations of genetic and epigenetic features, which might differ between organisms according to the evolution of genome complexity. Likely, the contribution of the PIP motif and PCNA interaction in the cell-cycle proteolytic regulation of PR-Set7 has emerged with a role of H4K20 methylation and Suv4–20h in the control of replication origins in mammalian genomes.
In contrast to the PIP-mediated PR-Set7 degradation, our results show that the proteolytic regulation of PR-Set7 controlled by the casein kinase (CKI)-induced SCFβ-TRCP pathway is conserved in Drosophila. This is demonstrated by the specific interaction of nuclear PR-Set7 with Slimb, the Drosophila ortholog of β-TRCP, and by the stabilization of PR-Set7 upon Slimb or CKI silencing. However, in contrast to its mammalian counterpart, we were not able to determine a canonical SLIMB degron motif (D/ES/TpGxxS/Tp) suggesting that, like described for other Slimb substrates (34,35), the degradation of Drosophila PR-Set7 might occur through multiple CKI phosphorylation sites that remain to be identified. Clearly, Slimb-mediated degradation constitutes a main regulatory mechanism of PR-Set7 stability in vivo as demonstrated by the nuclear accumulation of the enzyme in both Drosophila S2 cells and in larval tissues upon the loss of Slimb (Figures 5 and 6). In mammals, it has been proposed that the degradation of PR-Set7 mediated by β-TRCP, in addition to reduce the levels of PR-Set7 in response to DNA damage, could be somehow required for cell cycle progression (18). However due to the prominent role of the PIP-mediated proteolytic degradation of PR-Set7 in mammalian cells, the exact function of the CKI/SCFβ-TRCP mediated degradation of PR-Set7 remains unclear. Here, thanks to the fact that the PIP-motif is dispensable for Drosophila PR-Set7 degradation, we were able to reveal that SCFβ-TRCP mediated degradation of PR-Set7 is conserved and plays a critical role and in the regulation of chromatin structure and cell proliferation in Drosophila. We first observed that stabilization of PR-Set7 following the silencing of Slimb led to impairment of cell cycle progression from G1 to S phase, suggesting that degradation of PR-Set7 during G1 phase and early S phase is crucial for the onset of DNA replication. The reason of this G1 arrest upon PR-Set7 stabilization is probably related to abnormal interphase chromatin organization, where chromosomes tend to individualize and form highly compact structures. The SCFSlimb complex was already known to participate in the proper organization of interphase chromosomes through the specific targeting of Cap-H2, an essential protein of the condensin II complex (37,51,52). Indeed, massive chromatin compaction and improper individualization of chromosomes in interphase induced by the silencing of Slimb can be completely abolished by the co-depletion of Cap-H2. Here we show that besides, its crucial function to fine tune the chromatin level of CapH2 throughout the cell cycle, the SCFSlimb complex may contribute to interphase chromatin relaxation prior to S phase entry by targeting PR-Set7 to the proteasome. This is demonstrated by the fact that the percentage of nuclei with highly compact chromosomes observed upon Slimb silencing significantly decreases upon the co-depletion of PR-Set7 (Figure 6). So far, the regulation of chromatin structure mediated by PR-Set7 has been systematically attributed to its catalytic activity on H4K20. Therefore, it has been notably shown that H4K20me1 could promote the recruitment of Cap-D3 and Cap-G2, two other members of condensin II complex (53). Although we cannot rule out this possibility, our results clearly demonstrate that neither abnormal chromatin compaction, nor G1/S arrest observed upon the silencing of Slimb, was due to unscheduled catalytic activity of PR-Set7 at G1/S transition. Indeed, the presence of a catalytic dead version of miniPR-Set7 is sufficient to prevent chromatin relaxation and S phase entry in Slimb/PR-Set7 RNAi treated cells (Figure 6). It rather seems that the presence of PR-Set7 protein itself at G1/S transition could inhibit proper H4 acetylation since PR-Set7 stabilization strongly decreases H4K5, H4K12 and H4K16 acetylation (Figure 7). How the accumulation of PR-Set7 in the nucleus at G1/S transition could perturb H4 acetylation? Since it is known that proper H4 acetylation is regulated by the balance between the actions of histone acetyltransferases (HAT) and deacetylases (HDAC), two non-mutually exclusive possibilities could be proposed. First, the binding of PR-Set7 on the N-terminal tail of H4 might inhibit the activity of HATs. In mammals, it is actually the case for HBO1 an H4-specific histone acetylase that is required for DNA replication by promoting the licensing of the MCM complex at origins (40,41). The expression of a small portion of PR-Set7 called histone binding domain (HBD), is sufficient to prevent HBO1-dependent H4 acetylation and to inhibit DNA replication (19). The second possibility is that PR-Set7 could cooperate with HDAC enzymes and guide their binding on chromatin. We rather favor this second scenario because our results show that the chromatin compaction and G1/S arrest induced by the stabilization of catalytic-dead PR-Set7 mutant can be specifically reverted by the depletion of HDAC3 and the restoration of both H4K5 and H4K16 acetylation (Figure 7). These results are consistent with several previous studies showing that (i) HDAC3 contributes to proper balance of H4K16 acetylation and cell proliferation (54) and (ii) the presence of PR-Set7 in Drosophila embryos extracts may facilitate histone H4 deacetylation during in vitro chromatin assembly assays via HDAC activity (42). Thus, besides its methyltransferase activity, which is crucial for G2/M transition, PR-Set7 has also non-enzymatic functions that need to be properly controlled by the CKIα and SCFSlimb complex. Since the SCFβ-TRCP-mediated degradation of PR-Set7 also occurs in mammalian cells (18), it is likely that this proteolytic degradation is also necessary to inhibit non-enzymatic functions of mammalian PR-Set7 in order to promote the onset of DNA replication. Unraveling these non-enzymatic functions of PR-Set7 in the control of the human cell cycle is an important issue for future studies, particularly in the context of cancer where PR-Set7 up-regulation appears as a poor prognosis factor (44–46).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Microscopy (MRI) and Drosophila facilities (BioCampus Montpellier, CNRS, INSERM, Université Montpellier, France), R. Steward and C. Rogers for the generous gift of the null allelle of pr-set7 and of Slimb antibody, C. Bonne-Andrea and E. Taillebourg for their helpful technical advices for the detection of ubiquitylated proteins, E. Fabbrizio and F. Izard for critical readings of the manuscript.
Author contributions: A.Z., C.F., Y.P., C.G. and E.J. performed experiments and analyzed data. C.G. conceived the project and supervised the study. E.J. and C.S. provided conceptual advice on study and interpretation of the data. E.J. and C.G. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
INCA/Plan-Cancer [C13106FS]; Reégion Languedoc-Roussillon (LR); Labex Epigenmed; Institutional Support was provided by the Institut National de la Santé et de la Recherche Médicale (INSERM); Centre National de la Recherche Scientifique (CNRS); Y.P. was supported by a PhD Fellowship from «La ligue nationale contre le cancer». Funding for open access charge: IRCM [INSERM U1194].
Conflict of interest statement. None declared.
REFERENCES
- 1. Abbas T., Keaton M.A., Dutta A.. Genomic Instability in Cancer. Cold Spring Harbor Perspect. Biol. 2013; 5:a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raynaud C., Mallory A.C., Latrasse D., Jégu T., Bruggeman Q., Delarue M., Bergounioux C., Benhamed M.. Chromatin meets the cell cycle. J. Exp. Bot. 2014; 65:2677–2689. [DOI] [PubMed] [Google Scholar]
- 3. Hartl T., Boswell C., Orr-Weaver T.L., Bosco G.. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma. 2007; 116:197–214. [DOI] [PubMed] [Google Scholar]
- 4. Ruan K., Yamamoto T.G., Asakawa H., Chikashige Y., Kimura H., Masukata H., Haraguchi T., Hiraoka Y.. Histone H4 acetylation required for chromatin decompaction during DNA replication. Sci. Rep. 2015; 5:12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck D.B., Oda H., Shen S.S., Reinberg D.. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012; 26:325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi X., Kachirskaia I., Yamaguchi H., West L.E., Wen H., Wang E.W., Dutta S., Appella E., Gozani O.. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell. 2007; 27:636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tardat M., Murr R., Herceg Z., Sardet C., Julien E.. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 2007; 179:1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jørgensen S., Elvers I., Trelle M.B., Menzel T., Eskildsen M., Jensen O.N., Helleday T., Helin K., Sørensen C.S.. The histonemethyltransferase SET8 is required for S-phase progression. J. Cell Biol. 2007; 179:1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A.. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell. 2010; 40:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Centore R.C., Havens C.G., Manning A.L., Li J.-M., Flynn R.L., Tse A., Jin J., Dyson N.J., Walter J.C., Zou L.. CRL4Cdt2-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell. 2010; 40:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jørgensen S., Eskildsen M., Fugger K., Hansen L., Larsen M.S.Y., Kousholt A.N., Syljuåsen R.G., Trelle M.B., Jensen O.N., Helin K. et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 2011; 192:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dulev S., Tkach J., Lin S., Batada N.N.. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep. 2014; 15:1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E.. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010; 12:1086–1093. [DOI] [PubMed] [Google Scholar]
- 14. Oda H., Hübner M.R., Beck D.B., Vermeulen M., Hurwitz J., Spector D.L., Reinberg D.. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell. 2010; 40:364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu S., Wang W., Kong X., Congdon L.M., Yokomori K., Kirschner M.W., Rice J.C.. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010; 24:2531–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck D.B., Burton A., Oda H., Ziegler-Birling C., Torres-Padilla M.-E., Reinberg D.. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012; 26:2580–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Houston S.I., McManus K.J., Adams M.M., Sims J.K., Carpenter P.B., Hendzel M.J., Rice J.C.. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 2008; 283:19478–19488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z., Dai X., Zhong J., Inuzuka H., Wan L., Li X., Wang L., Ye X., Sun L., Gao D. et al. SCF(β-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015; 6:10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yin Y., Yu V.C., Zhu G., Chang D.C.. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle. 2008; 7:1423–1432. [DOI] [PubMed] [Google Scholar]
- 20. Nishioka K., Rice J.C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002; 9:1201–1213. [DOI] [PubMed] [Google Scholar]
- 21. Brownlee C.W., Klebba J.E., Buster D.W., Rogers G.C.. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 2011; 195:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banfalvi G., Trencsenyi G., Ujvarosi K., Nagy G., Ombodi T., Bedei M., Somogyi C., Basnakian A.G.. Supranucleosomal organization of chromatin fibers in nuclei of Drosophila S2 cells. DNA Cell Biol. 2007; 26:55–62. [DOI] [PubMed] [Google Scholar]
- 23. Sakaguchi A., Joyce E., Aoki T., Schedl P., Steward R.. The histone H4 lysine 20 monomethyl mark, set by PR-Set7 and stabilized by L(3)mbt, is necessary for proper interphase chromatin organization. PLoS ONE. 2012; 7:e45321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakaguchi A., Steward R.. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J. Cell Biol. 2007; 176:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007; 448:151–156. [DOI] [PubMed] [Google Scholar]
- 26. Wysocka J., Reilly P.T., Herr W.. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 2001; 21:3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang J., Feng Q., Ketel C.S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y.. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002; 12:1086–1099. [DOI] [PubMed] [Google Scholar]
- 28. Sahashi R., Crevel G., Pasko J., Suyari O., Nagai R., Saura M.M., Yamaguchi M., Cotterill S.. DNA polymerase α interacts with PrSet7 and mediates H4K20 monomethylation in Drosophila. J. Cell. Sci. 2014; 127:3066–3078. [DOI] [PubMed] [Google Scholar]
- 29. Li Y., Armstrong R.L., Duronio R.J., MacAlpine D.M.. Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Res. 2016; 44:7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duffy J.B. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002; 34:1–15. [DOI] [PubMed] [Google Scholar]
- 31. McKay D.J., Klusza S., Penke T.J.R., Meers M.P., Curry K.P., McDaniel S.L., Malek P.Y., Cooper S.W., Tatomer D.C., Lieb J.D. et al. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell. 2015; 32:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grosskortenhaus R., Sprenger F.. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell. 2002; 2:29–40. [DOI] [PubMed] [Google Scholar]
- 33. Wojcik E.J., Glover D.M., Hays T.S.. The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr. Biol. 2000; 10:1131–1134. [DOI] [PubMed] [Google Scholar]
- 34. Smelkinson M.G., Kalderon D.. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr. Biol. 2006; 16:110–116. [DOI] [PubMed] [Google Scholar]
- 35. Smelkinson M.G., Zhou Q., Kalderon D.. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev. Cell. 2007; 13:481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen H.Q., Nye J., Buster D.W., Klebba J.E., Rogers G.C., Bosco G.. Drosophila casein kinase I alpha regulates homolog pairing and genome organization by modulating condensin II subunit Cap-H2 levels. PLoS Genet. 2015; 11:e1005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buster D.W., Daniel S.G., Nguyen H.Q., Windler S.L., Skwarek L.C., Peterson M., Roberts M., Meserve J.H., Hartl T., Klebba J.E. et al. SCF Slimb ubiquitin ligase suppresses condensin II–mediated nuclear reorganization by degrading Cap-H2. J. Cell Biol. 2013; 201:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brustel J., Tardat M., Kirsh O., Grimaud C., Julien E.. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 2011; 21:452–460. [DOI] [PubMed] [Google Scholar]
- 39. Beck D.B., Oda H., Shen S.S., Reinberg D.. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012; 26:325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miotto B., Struhl K.. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008; 22:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miotto B., Struhl K.. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010; 37:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scharf A.N.D., Meier K., Seitz V., Kremmer E., Brehm A., Imhof A.. Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Mol. Cell. Biol. 2009; 29:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beck D.B., Burton A., Oda H., Ziegler-Birling C., Torres-Padilla M.-E., Reinberg D.. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012; 26:2580–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Veschi V., Liu Z., Voss T.C., Ozbun L., Gryder B., Yan C., Hu Y., Ma A., Jin J., Mazur S.J. et al. Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell. 2017; 31:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang F., Sun L., Li Q., Han X., Lei L., Zhang H., Shang Y.. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012; 31:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dhami G.K., Liu H., Galka M., Voss C., Wei R., Muranko K., Kaneko T., Cregan S.P., Li L., Li S.S.-C.. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol. Cell. 2013; 50:565–576. [DOI] [PubMed] [Google Scholar]
- 47. Brustel J., Kirstein N., Izard F., Grimaud C., Prorok P., Cayrou C., Schotta G., Abdelsamie A.F., Déjardin J., Méchali M. et al. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017; 36:2726–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T.. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004; 18:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jorgensen S., Schotta G., Sorensen C.S.. Histone H4 Lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013; 41:2797–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phalke S., Nickel O., Walluscheck D., Hortig F., Onorati M.C., Reuter G.. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009; 41:696–702. [DOI] [PubMed] [Google Scholar]
- 51. Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012; 26:1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rana V., Bosco G.. Condensin regulation of genome architecture: condensing regulation of genome architecture. J. Cell. Physiol. 2017; 232:1617–1625. [DOI] [PubMed] [Google Scholar]
- 53. Liu W., Tanasa B., Tyurina O.V., Zhou T.Y., Gassmann R., Liu W.T., Ohgi K.A., Benner C., Garcia-Bassets I., Aggarwal A.K. et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010; 466:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lv W.-W., Wei H.-M., Wang D.-L., Ni J.-Q., Sun F.-L.. Depletion of histone deacetylase 3 antagonizes PI3K-mediated overgrowth of Drosophila organs through the acetylation of histone H4 at lysine 16. J. Cell Sci. 2012; 125:5369–5378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.