Abstract
Activation of the RAS/ERK and its downstream signaling components is essential for growth factor–induced cell survival, proliferation, and differentiation. The Src homology-2 domain containing protein tyrosine phosphatase 2 (SHP2), encoded by protein tyrosine phosphatase, non-receptor type 11 (Ptpn11), is a positive mediator required for most, if not all, receptor tyrosine kinase–evoked RAS/ERK activation, but differentially regulates the PI3K/AKT signaling cascade in various cellular contexts. The precise mechanisms underlying the differential effects of SHP2 deficiency on the PI3K pathway remain unclear. We found that mice with myelomonocytic cell-specific [Tg(LysM-Cre);Ptpn11fl/fl mice] Ptpn11 deficiency exhibit mild osteopetrosis. SHP2-deficient bone marrow macrophages (BMMs) showed decreased proliferation in response to M-CSF and decreased osteoclast generation. M-CSF–evoked ERK1/2 activation was decreased, whereas AKT activation was enhanced in SHP2-deficient BMMs. ERK1/2, via its downstream target RSK2, mediates this negative feedback by negatively regulating phosphorylation of M-CSF receptor at Tyr721 and, consequently, its binding to p85 subunit of PI3K and PI3K activation. Pharmacologic inhibition of RSK or ERK phenotypically mimics the signaling defects observed in SHP2-deficient BMMs. Furthermore, this increase in PI3K/AKT activation enables BMM survival in the setting of SHP2 deficiency.—Wang, L., Iorio, C., Yan, K., Yang, H., Takeshita, S., Kang, S., Neel, B.G., Yang, W. An ERK/RSK-mediated negative feedback loop regulates M-CSF-evoked PI3K/AKT activation in macrophages.
Keywords: SHP2, MAPK, c-Fms, M-CSFR
Receptor tyrosine kinases (RTKs) regulate the growth, proliferation, differentiation, and survival of multiple cell types during development and in adulthood. Despite their pleiotropic actions, RTKs typically initiate a limited and common set of downstream signaling events. Most RTKs activate the small GTPase RAS, which, in turn, triggers a kinase cascade consisting of one or more RAF family members, MEK1/2, ERK1/2, and ERK-dependent kinases, such as ribosomal S6 kinase (RSK) family members (1,–4). Activated RAS also promotes the activation of class I PI3Ks (5), as do proteins that bind the regulatory subunits (e.g., p85α/β) of class I PI3Ks, which include some RTKs themselves and scaffolding adaptors, such as insulin receptor substrate-1 (IRS-1) (6) and growth factor receptor-bound protein 2–associated binding protein (GAB) (7,–9) family members. The lipid products of PI3K bind to pleckstrin homology domain-containing proteins and promote activation of the serine–threonine kinase AKT (and other pleckstrin homology domain-containing signaling components) (10). RTKs can also bind to and activate phospholipase C-γ, SRC family kinases (SFKs), and/or signal transducer and activator of transcription (STAT) family transcription factors (11).
A major question in RTK signal transduction is how activation of such a restricted number of downstream pathways can result in such diverse effects. Differential recruitment/activation of signal relay molecules and the cellular context in which RTK activation occurs may explain some of the distinct effects of RTKs. However, the mere number of RTKs and the paucity of downstream signaling cascades they evoke suggest that other mechanisms contribute to RTK specificity. Several studies show that the kinetics of downstream pathway activation/inactivation are critical determinants of the biologic response to RTK activation (12). Consequently, elucidating the mechanisms that control pathway dynamics, including negative feedback pathways, is critical for a detailed understanding of RTK action.
M-CSF is essential for monocyte/macrophage (Mϕ) and osteoclast proliferation, differentiation, viability, and motility (13,–15). Accordingly, op/op mice, which lack M-CSF, are deficient in most Mϕ populations and develop severe osteopetrosis (16). M-CSF signals via the class III RTK, the M-CSF receptor (M-CSFR; i.e., CSF1R, c-Fms), which, similar to other RTKs, binds multiple SH2 (and PTB) domain-containing signal-relay molecules that activate canonical downstream signaling cascades. M-CSF–evoked RAS/ERK activation reportedly is mediated via binding of SFKs, GRB2, and CBL1 to M-CSFR–Tyr559, –Tyr697/Tyr921, and –Tyr973, respectively (17,–20). M-CSF–induced PI3K/AKT activation is evoked by p85 binding to M-CSFR–Tyr721 (18, 21) and to GAB family adaptor proteins (22, 23). By contrast, M-CSFR–Tyr706 recruits and activates STAT1, whereas phosphorylation of Tyr559 promotes c-CBL activation, M-CSF receptor (M-CSFR) ubiquitination, and subsequent lysosomal trafficking and degradation of the receptor (24, 25). The prevailing model holds that the RAS/ERK pathway is important for M-CSF–evoked monocytic cell proliferation, whereas the PI3K/AKT pathway promotes cell survival (13), and STAT1 activation is important for Mϕ differentiation (26, 27). There is, however, some disagreement over details: for example, some reports implicate ERK activation in differentiation as well as proliferation (28), whereas others implicate PI3K/AKT activation in proliferation in addition to survival (29).
The SRC homology-2 domain-containing protein tyrosine phosphatase 2 (SHP2), encoded by the protein tyrosine phosphatase, non-receptor type 11 (PTPN11) gene, is required for optimal RAS/ERK pathway activation in most, if not all, RTK, cytokine and integrin signaling pathways (30, 31). SHP2 has variable effects on the PI3K/AKT pathway: in at least some cell types, SHP2 negatively regulates epidermal growth factor (EGF)–evoked PI3K/AKT activation by dephosphorylating the PI3K binding sites on GAB1 (32, 33). However, SHP2 is required for PI3K activation downstream of other RTKs (34,–36). Moreover, SHP2 has cell type– and receptor-specific effects on activation of STAT, RHO family GTPase, NF-κB, and nuclear factor of activated T cells (31, 37, 38). Reflecting its manifold effects on cell signaling, global Ptpn11 deletion in mice causes early embryonic lethality from trophoblast stem cell death (39), whereas postnatal SHP2 deletion has variably severe effects on the development and function of multiple cell and tissues (30, 31). Embryonic stem cell differentiation and chimera experiments implicate SHP2 in the earliest stages of embryonic and adult hematopoiesis, yet less is known about its role in specific hematopoietic cells (40, 41). In monocyte/ Mϕ lineage cells, for example, SHP2 is found in a signaling protein complex evoked by M-CSF and associates with GAB family adaptor proteins upon M-CSF stimulation (22). However, its precise role in M-CSF signaling, as well as its effects on monocyte/Mϕs and osteoclasts in vivo have remained unknown (15).
To address these issues, we generated mice with selective deletion of the Ptpn11 gene in monocyte/Mϕs and osteoclasts. As in other RTK signaling systems, we find that SHP2 is required for RAS/ERK pathway activation, although monocyte/Mϕ SHP2 deficiency has relatively minor effects, decreasing bone marrow Mϕ (BMM) proliferation and osteoclastogenesis. In the course of these studies, however, we uncovered a novel, ERK-mediated, negative-feedback pathway that controls phosphorylation of the M-CSFR on specific tyrosyl residues and thereby regulates the dynamics of M-CSF–evoked PI3K/AKT activation. Loss of this feedback pathway (because of defective RAS/ERK activation) might mitigate the effect of SHP2 deficiency on monocyte/Mϕ lineage cells and could provide potential escape mechanisms for cancer cells treated with SHP2 or MEK inhibitors.
MATERIALS AND METHODS
Reagents
M-CSF and granulocyte M-CSF (GM-CSF) were purchased from PeproTech (Rocky Hill, NJ, USA) and erythropoietin (EPO) was purchased from Janssen-Ortho (Markham, ON, Canada). UO126 and LY294002 were from Calbiochem (San Diego, CA, USA), and rapamycin was from Sigma-Aldrich (St. Louis, MO, USA). The RSK inhibitor fluoromethylketone (Fmk) was synthesized as described previously (42). Rabbit pAbs against murine M-CSFR (c-Fms) were a gift from Dr. S. A. Courtneidge (Knight Cancer Institute, Oregon Health & Science University, Portland, OR, USA). Rabbit pAbs against the C terminus of M-CSFR, SHP2, and the p85 subunit of PI3K were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), rabbit pAbs against PKA substrates (R/KXXS/T), p-ERK1/2 (Try202Thr208), p-AKT (Ser473), p-p90RSK(Ser380), p-p70S6 kinase, p90RSK2, and p–M-CSFR (Tyr721) were from Cell Signaling Technology Inc. (Danvers, MA, USA), anti-phosphotyrosine mAbs (clone 4G10) were from Thermo Fisher Scientific (Waltham, MA, USA), and anti–FLAG-epitope antibodies were purchased from Sigma-Aldrich. Phycoerythrin-conjugated anti-F4/80, CD11b, and CD115 antibodies were purchased from eBioscience (San Diego, CA, USA) and Thermo Fisher Scientific, respectively. Recombinant RSK2 is from SignalChem (Richmond, BC, Canada). Horseradish peroxidase–labeled anti-mouse and rabbit secondary antibodies and protein-A and -G beads were purchased from GE Healthcare Life Sciences (Little Chalfont, United Kingdom). For Li-Cor–based Western blotting, goat anti-rabbit IR-Dye 800 and 680, goat anti-mouse IR-Dye 800 and 680 were used. All antibodies were diluted in 1 × TBS/5% nonfat milk for immunoblotting experiments. pMX(puro)-based constructs harboring EPO receptor (EPOR)/M-CSFR–wild-type (WT; FLAG-epitope-tagged) were previously described (19). EPOR/M-CSFR chimeras containing Ser711 to Ala and Ser728 to Ala mutations were constructed with the QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and the following primers: (forward) 5′-ATATGTGCGCAGGGACGCTGGCTTCTCCAGTCAGGGTG-3′ and (reverse) 5′-CACCCTGACTGGAGAAGCCAGCGTCCCTGCGCAC-3′; (forward) 5′-CGTGGAGATGAGGCCTGTCGCGACTTCTTCAAGTGACTCC-3′ and (reverse) 5′-GGAGTCACTTGAAGAAGTCGCGACAGGCCTCATCTCCACG-3′. Point mutations were confirmed by DNA sequencing. Recombinant M-CSFR and RSK2 proteins were purchased from SignalChem.
Generation of Ptpn11 conditional-deletion allele
The Ptpn11-floxed allele (43), Tg(Rosa26-ZsGreen1) (44) and Tg(LysM-Cre) (45) were described previously. All animal work was reviewed and approved by the Rhode Island Hospital Institutional Animal Care and Use Committee (A3922-01) and was performed in accordance with Public Health Service policy on the humane care and use of laboratory animals. PCR conditions for genotyping are available upon request. All experiments herein used mutant mice and littermate controls on a C57BL/6 × 129/Svj, mixed genetic background. To assess the role of SHP2 in myelomonocytic cells, including Mϕs, mice bearing a Ptpn11-floxed allele were bred with Tg(LysM-Cre) mice to generate Ptpn11fl/+;Tg(LysM-Cre) (hereafter SHP2mΦCTR) and Ptpn11fl/fl;Tg(LysM-Cre) (hereafter, SHP2mϕKO) mice, respectively (Fig. 1A). To trace the fate of myelomonocytic cells in the SHP2mϕCTR and SHP2mϕKO mice in vivo and in vitro, Tg(Rosa26-ZsGreen1) reporter was crossed to SHP2mϕCTR and SHP2 mϕKO mice in some studies.
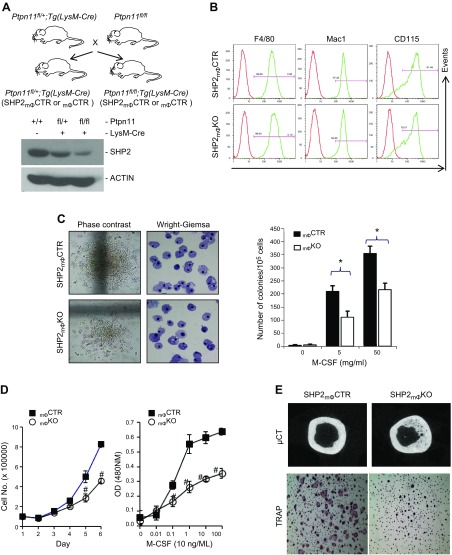
Figure 1.
SHP2 deficiency compromises myelomonocytic cell proliferation but is dispensable for differentiation. A) Diagram showing breeding scheme for generating Ptpn11+/+, Ptpn11fl/+; Tg(LysM-Cre) (SHP2mΦCTR) and Ptpn11fl/fl; Tg(LysM-Cre) (SHP2mϕKO) mice. Panels show SHP2 (top) and actin (bottom) immunoblots of BMM lysates (20 µg). Actin serves as a loading control. B) Flow cytometric analysis of cell surface-marker expression in SHP2mϕCTR and SHP2mϕKO BMMs. C) Mϕ colony assays (CFU-M) in SHP2mϕCTR and SHP2mϕKO mice. Phase contrast views of colony morphology and Wright-Giemsa stain of cytospin preparation from colonies (left). M-CSF dose–response curve (right). Note that SHP2 deficiency results in fewer and smaller CFU-Ms with preserved Mϕ differentiation, as well as diminished sensitivity to M-CSF (n = 3). *P < 0.05 (Student’s t test). D) SHP2 deficiency decreases BMM proliferation and M-CSF sensitivity. Shown are proliferation (left) and dose–response (WST1 assay; right) curves for SHP2mϕCTR and SHP2mϕKO BMMs. #P < 0.05 (Student’s t test). E) SHP2 deficiency causes mild osteopetrosis. Representative micro-computed tomography images (top) show increased thickness of the cortical bone of an 8-mo-old SHP2mϕKO mouse, compared with an SHP2mϕCTR mouse. In vitro osteoclastogenesis assay showing that SHP2mϕKO BMMs are defective in generating TRAP+ osteoclasts in response to M-CSF (10 ng/ml) and RANKL (100 ng/ml). Shown is 1 of 2 experiments with similar results.
Cell culture
BMMs from 6- to 9 wk-old, WT, SHP2 mϕCTR and SHP2 mϕKO mice were derived and cultured for 7–9 d in DMEM containing 10% fetal bovine serum (FBS) and 1/100 volume of CMG14-12 culture supernatant, as described previously (19). Platinum-E packaging cells were cultured in the presence of puromycin and blasticidin, as described previously (46). RAW264.7 Mϕs were cultured in DMEM supplemented with 10% FBS. All cell cultures contained 1% penicillin/streptomycin. To generate BMMs and RAW264.7 cells expressing EPOR/M-CSFR chimeras, platinum-E cells were transiently transfected with pMX(puro)-based constructs with Effectene (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Viruses were collected 48 h posttransfection and were used to infect bone marrow (BM) cells seeded the previous day in the presence of M-CSF (10 ng/ml) and polybrene (4 µg/ml). Infected cells were subsequently cultured in M-CSF and 2 µg/ml puromycin to select and expand transductants (19, 47).
Proliferation, differentiation, and cell cycle assays
BMMs were washed once in 1× PBS, starved in DMEM with 0.2% FBS for 12 h, and harvested by treatment with trypsin (0.25%)–EDTA for 5 min. For growth curves, 2 × 105 BMMs were seeded in triplicate in 6-cm dishes, and cell number was counted with a hemocytometer on the successive 5 d. For dose–response curves, cells were seeded in triplicate in flat-bottom, 96-well tissue culture plates (2 × 104 cells/well) at 37°C. Each well contained 100 µl DMEM supplemented with 10% FCS and various doses of M-CSF. After 2 d of culture, 10 µl (1/10 vol) of WST-1 reagent (F. Hoffmann-La Roche, Basel, Switzerland) was added, and the absorbance of the formazan product was measured at 440 nm. For colony-forming unit–Mϕ (CFU-M) assays, BM cells were placed in semisolid, methylcellulose medium containing M-CSF (10 ng/ml); cultures were incubated for 7 d at 37°C, and colonies containing ≥50 cells were enumerated using a dissection microscope. After counting, cells were recovered by cytocentrifugation and stained with Wright-Giemsa.
To test the effect of Ptpn11 deletion on differentiation, BMMs were cultured for 5–7 d, harvested with nonenzymatic cell dissociation solution (Sigma-Aldrich), and resuspended in ice-cold PBS plus 2% FBS. For staining with biotinylated F4/80 and FITC-conjugated CD11b (Mac-1) antibodies, cells were initially incubated in Fcγ-block antibodies (1:100 dilution) on ice for 30 min, followed by incubation with primary antibodies (1:200 dilution) on ice for 30 min. Cells were then washed twice in staining solution and incubated with streptavidin–phycoerythrin (1:500 dilution) for a further 30 min on ice. For cell-cycle assays, BMMs were cultured in DMEM supplemented with 10% FBS and 10 ng/ml M-CSF, and bromodeoxyuridine (BrdU; 10 µM) was added 1 h before removal of the cells with nonenzymatic cell-dissociation solution. BMMs were then fixed, permeabilized, and stained with anti-BrdU antibodies and 7-aminoactinomycin D (1 µl/106 cells), and analyzed with a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Biochemical analyses
BMMs were starved for 16 h in DMEM supplemented with 0.2% FBS, and then stimulated with the indicated growth factors. After stimulation, cells were lysed in NP-40 buffer [0.5% NP40, 150 mM NaCl, 1 mM EDTA, 50 mM Tris (pH 7.4)], supplemented with a protease inhibitor cocktail (1 mM PMSF, 1 mM NaF, 1 mM sodium orthovanadate, 10 μg/ml aprotinin, 0.5 μg/ml antipain, and 0.5 μg/ml pepstatin). Immunoprecipitations were performed on cleared lysates, as described previously (39). For immunoblotting, total cell lysates (TCLs; 30–50 µg) were resolved by SDS-PAGE, transferred to PVDF membranes, and incubated with primary antibodies at 4°C for 2 h to overnight (according to the manufacturer’s instructions), followed by horseradish peroxidase–conjugated secondary antibodies or goat anti-rabbit/mouse IR-Dye 800 or -600 antibodies for 1 h. Detection was by enhanced chemiluminescence (Amersham Biosciences, Chalfont, United Kingdom) or Li-Cor scanning.
For immune complex PI3K assays, M-CSFR immunoprecipitates from starved and M-CSF–stimulated BMMs were washed and subjected to a lipid kinase assay, as described (48). Reactions were terminated by adding 200 μl of 1 normal (N) hydrochloric acid, extracted with 200 μl methanol/chloroform (1:1), and 50 μl of the organic phase were resolved by thin layer chromatography in methanol/chloroform/H2O/NH4OH (20:23:4:1) on LK6D plates (Whatman, Maidstone, United Kingdom). Plates were exposed to X-ray film, and phospholipid species were identified by co-chromatographed standards.
For protein kinase assays, recombinant M-CSFR cytoplasmic domain (500 µg) was incubated with recombinant human RSK2 (0.1 µg) for 30 min in the presence of γ-32P-ATP (3000 Ci/mM) in kinase buffer (25 mM 4-morpholinepropanesulfonic acid [pH 7.2], 12.5 mM β-glycerophosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA and 25 mM DTT). Samples were then boiled, subjected to SDS-PAGE, transferred onto a PVDF membrane, and autoradiographed. Bands corresponding to the M-CSFR were excised, hydrolyzed in 5.7 N hydrochloric acid for 1 h at 110°C, dried in a Speedvac, and resuspended in 5 μl pH 1.9 buffer containing a mixture of nonradioactive phosphoserine, phosphothreonine, and phosphotyrosine. Phospho–amino acids were separated by 2-dimensional electrophoresis/chromatography, according to a published protocol (49). For phosphorylation of peptide arrays, 23 peptides spanning all serine residues in the M-CSFR cytoplasmic domain were synthesized by using standard 9-fluorenylmethoxycarbonyl chemistry on a modified cellulose membrane with an Intavis MultiPep Spot peptide arrayer (Cologne, Germany), as described in Hilpert et al. (50) and Leung et al. (51). Peptide-based kinase assays were also performed as described in Warner et al. (52).
RESULTS
Generation of mice with conditional Ptpn11 deletion in myelomonocytic cells
Global SHP2 deficiency causes embryonic lethality in mice (39, 53, 54). To assess the role of SHP2 in Mϕs, we crossed our Ptpn11-floxed allele (43) to Tg(LysM-Cre) mice (45). As expected, SHP2 levels were reduced by ∼50 and >90% in BMMs from SHP2mϕCTR and SHP2mϕKO mice, respectively, compared with WT control levels (Fig. 1A). No truncation variants were detected by immunoblotting with antibodies directed against either the N or C terminus of SHP2 (43).
SHP2 is required for myelomonocytic cell proliferation but not for differentiation
SHP2mϕKO mice were born at the expected Mendelian ratio and appeared healthy. Blood counts and differentials, measured at 6–8 wk of age, were comparable for SHP2mϕCTR and SHP2mϕKO mice. Preliminary experiments showed that there was no difference in phenotype between WT, SHP2mϕCTR (hemizygous) and Ptpn11fl/fl or Tg(LysM-Cre) parental mice; consequently, we used SHP2mϕCTR and SHP2mϕKO mice for most experiments. BMMs (Fig. 1B), as well as cells from BM, spleen, and peritoneal cavity of SHP2mϕKO mice (Supplemental Fig. S1), showed normal expression of myelomonocytic cell-surface markers, indicating that Mϕ differentiation was unaffected by SHP2 deficiency. Likewise, cells from M-CSF–dependent colonies showed normal monocytic morphology (Fig. 1C). By contrast, the number and size of CFU-M were reduced in SHP2mϕKO mice, suggesting that SHP2 is required for optimal M-CSF–driven proliferation (Fig. 1C). Indeed, proliferation in response to a saturating dose of M-CSF was impaired in SHP2mϕKO BMMs. M-CSF responsiveness was also diminished, as indicated by a rightward shift in the dose–response curve (Fig. 1D). Consistent with these data and the monocyte/Mϕ origin of osteoclasts, SHP2mϕKO mice showed defective osteoclastogenesis in response to M-CSF and receptor activator for NF-κB ligand (RANKL) in vitro and developed mild osteopetrosis at >7 mo old (Fig. 1E). We concluded that SHP2 was required for optimal proliferation of Mϕ progenitors but is dispensable for their differentiation to mature Mϕ populations. SHP2 also contributes to the generation of optimal levels of osteoclasts.
SHP2 deficiency impairs RAS/ERK activation but enhances the PI3K/AKT pathway
To explore the molecular basis for defective proliferation of SHP2-deficient BMMs, we assayed known M-CSF–evoked signaling pathways. BMMs from SHP2mϕCTR and SHP2mϕKO mice were starved for 16 h, and then stimulated with M-CSF (30 ng/ml). As expected, SHP2mϕCTR BMMs showed robust and sustained activation of RAS (assayed by GST-Raf-RBD binding) and ERK (adjudged by immunoblotting with phospho-specific antibodies). By contrast (but similar to the effects of SHP2 deficiency in many other RTKs and cytokine signaling pathways), RAS and ERK activation were transient in SHP2mϕKO BMMs (Fig. 2A). In control BMMs, AKT activation (assessed by Ser473 phosphorylation) peaked at 5 min and then declined to basal levels by 30 min. In SHP2mϕKO BMMs, however, AKT activation was enhanced and sustained. These differences in ERK and AKT activation could not be explained by altered M-CSFR (c-Fms) levels; surface M-CSFR (CD115) expression (Fig. 1B) was unaffected by SHP2 deficiency, and M-CSFR protein, assessed by immunoblotting, declined at a comparable rate after M-CSF stimulation of SHP2mϕCTR and SHP2mϕKO BMMs. SHP2 deficiency also had no effect on M-CSF–evoked STAT3 or STAT1 activation (Supplemental Fig. S2A).
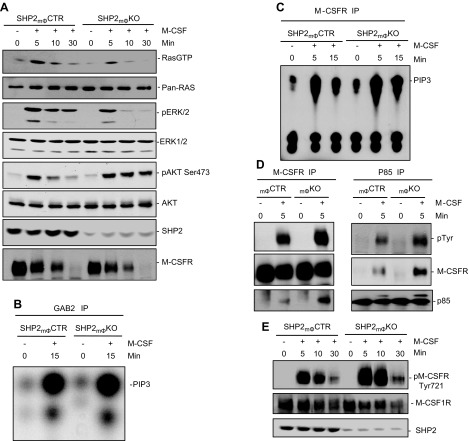
Figure 2.
SHP2 promotes M-CSF–evoked RAS/ERK pathway, but negatively regulates M-CSFR–associated PI3K and AKT activation. A) BMMs were starved for 14 h, then either stimulated with M-CSF (30 ng/ml) for the indicated times or left untreated. TCLs from SHP2mϕCTR and SHP2mϕKO BMMs were analyzed by GST-Raf-RBD binding assay to assess RAS loading (800 µg) or by immunoblotting (30 µg) with the indicated antibodies. B, C) GAB2- and M-CSFR-associated PI3K assays from SHP2mϕCTR and SHP2mΦKO BMMs. Note that SHP2 deficiency leads to increased M-CSFR–associated (C), but not GAB2-associated (B) PI3K activity. D) SHP2 deficiency increases M-CSFR association with PI3K. Lysates (800 µg) from starved or M-CSF-stimulated BMMs were immunoprecipitated (IP) with anti–M-CSFR antibodies, followed by immunoblotting for the p85 subunit of PI3K (D, left), or immunoprecipitated with anti-p85 antibodies, followed by immunoblotting with M-CSFR or anti-pTyr antibodies (D, right). Note the increased M-CSFR/PI3K association and slight increase in M-CSFR tyrosyl phosphorylation in SHP2mϕKO BMMs. E) Increased phosphorylation of M-CSFR on Tyr721 in SHP2mϕKO BMMs. TCLs (40 µg) from the indicated BMM samples were immunoblotted with p-Tyr721-specific antibodies or M-CSFR antibodies as a control for loading. Data shown are representative of 3 independent experiments.
SHP2 negatively regulates EGF-induced AKT activation by dephosphorylating the p85 binding site on the adaptor protein GAB1 (32, 33, 55, 56). Surprisingly, PI3K activity associated with GAB2, the major GAB family member in BMMs, was unaffected in SHP2-deficient BMMs (Fig. 2B). PI3K also binds directly to the M-CSFR (18, 21), and indeed, M-CSFR–associated PI3K activity was enhanced and sustained in SHP2mϕKO BMMs (Fig. 2C). That increase was accompanied by a slight increase in overall tyrosyl phosphorylation of M-CFSR (Fig. 2D, left) and a more substantial increase in phosphorylation of the binding site (Tyr721) for p85 on the receptor (18, 21) (Fig. 2E). M-CSFR/p85 association was also enhanced (Fig. 2D, right). Taken together, these findings strongly suggest that increased AKT activation in SHP2-deficient BMMs is due to increased M-CSFR phosphorylation on its binding site for p85.
SHP2 regulates M-CSFR tyrosyl phosphorylation indirectly via its effect on ERK activation
The simplest explanation for the above results would be that SHP2 dephosphorylates Tyr721 (and perhaps other sites) on the M-CSFR. However, SHP2 has not been reported to associate with M-CSFR (13, 22, 23), and we did not find evidence for SHP2-catalyzed dephosphorylation of the receptor (Supplemental Fig. S6). We wondered whether increased M-CSF–evoked PI3K/AKT activity in SHP2-deficient BMMs might be the indirect consequence of their defective ERK activation. Indeed, whereas pretreatment of WT BMMs with the PI3K/mammalian target of rapamycin (mTOR) inhibitor LY294002 blocked M-CSF–evoked AKT activation, the MEK inhibitor UO126 enhanced AKT activation (Fig. 3A and Supplemental Fig. S2C). That effect was receptor specific because MEK inhibition did not affect AKT activation in response to GM-CSF (Fig. 3B). Similar to the effects of SHP2 deficiency, UO126 pretreatment resulted in increased M-CSF–evoked PI3K activation, increased phosphorylation of Tyr721, and increased association of p85 with the M-CSFR (Fig. 3C, D).
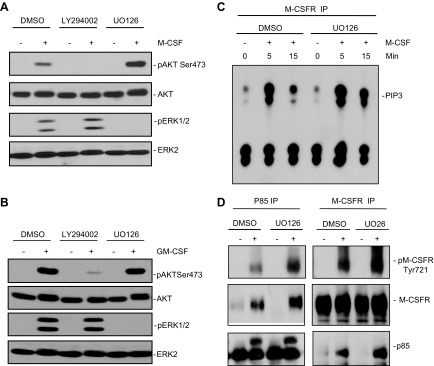
Figure 3.
Blocking ERK activation increases M-CSF–evoked M-CSFR–Tyr721 phosphorylation and PI3K activation. A, B) WT BMMs were starved; pretreated with the MEK inhibitor UO126 (10 µM), the PI3K/mTOR inhibitor LY294002 (10 µM), or vehicle (DMSO) for 1 h; and then were either left unstimulated (−) or stimulated (+) for 10 min with 30 ng/ml M-CSF (A) or 50 ng/ml GM-CSF (B). AKT and ERK activation were assessed in TCLs (30 µg) by immunoblotting with the indicated p-specific antibodies. Blocking ERK activation by MEK inhibitor treatment increases M-CSF-, but not GM-CSF–evoked AKT activation, whereas inhibiting M-CSF or GM-CSF–evoked PI3K activation did not affect ERK activation. C, D) MEK inhibitor treatment increases M-CSFR–associated PI3K activity and M-CSFR Tyr721 phosphorylation and association with the p85 subunit of PI3K. WT BMMs were starved, pretreated for 1 h with vehicle (DMSO), UO126 (10 µM), or LY294002 (10 µM); and then stimulated with M-CSF (30 ng/ml) for the indicated times. M-CSFR immunoprecipitates (from 1 mg TCL) were subjected to PI3K assays (C). M-CSFR and p85 immunoprecipitates were also immunoblotted with the indicated antibodies (D). Data shown are representative of the 3 independent experiments.
Previous studies of other cell types and RTK signaling pathways have shown that ERK or the ERK-activated kinase RSK (1) can phosphorylate and inhibit TSC2, thereby promoting mTOR activity. mTOR, in turn, activates p70S6 kinase (p70S6K), which can inhibit PI3K/AKT activation via a well-known negative feedback loop (57, 58). However, treatment of BMMs with rapamycin, which inhibits TORC1 (and p70S6K) activation, did not enhance AKT phosphorylation (Supplemental Fig. S2B), indicating that abrogation of the p70S6K feedback pathway cannot account for the effects of MEK inhibition on M-CSF–evoked AKT activation in BMMs. Thus, RAS/ERK pathway activation in BMM tempers PI3K activation via a novel, negative-feedback pathway that regulates phosphorylation of Tyr721 of M-CSFR.
RSK2 modulates M-CSFR phosphorylation and AKT activation
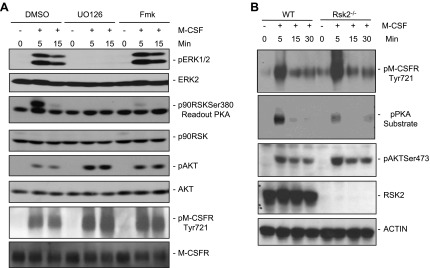
We considered the possibility that ERK might phosphorylate M-CSFR and thereby affect Tyr721 phosphorylation and PI3K association. There are, however, no obvious ERK phosphorylation sites (YXXXS/TP) (59,–61) in the M-CSFR cytoplasmic domain, and we did not observe an interaction between ERK and M-CSFR in cotransfection/coimmunoprecipitation assays. We next asked whether ERK-dependent regulation of M-CSFR signaling might be mediated via an ERK-dependent kinase. Indeed, treatment of WT BMMs with the RSK-specific inhibitor Fmk (42) resulted in enhanced M-CSF–evoked AKT activation and M-CSFR Tyr721 phosphorylation to extents similar to that evoked by UO126 treatment (Fig. 4A). Consistent with those observations, M-CSF–evoked M-CSFR Tyr721 phosphorylation and subsequent AKT activation were enhanced, whereas Ser phosphorylation within R/KXXS/T motifs of M-CSFR, assessed by PKA substrate phosphorylation, was impaired, in Rsk2−/− BMMs, compared with WT controls (Fig. 4B).
Figure 4.
RSK2 regulates M-CSFR Tyr721 phosphorylation and AKT activation. A) WT BMMs were starved; pretreated with UO126 (10 µM), the RSK inhibitor Fmk (6 µM), or vehicle (DMSO) for 1 h; and then either stimulated with M-CSF (30 ng/ml) or left unstimulated. TCLs (30 µg) were immunoblotted with the indicated antibodies. B) BMMs from Rsk2−/− mice or littermate controls (WT) were starved, and then stimulated with M-CSF (30 ng/ml) for the indicated times (+) or left unstimulated (−). TCLs (30 µg) were subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. Data shown are a representative of the 3 independent experiments.
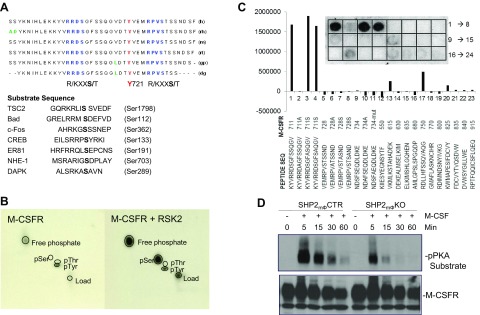
Inspection of the M-CSFR cytoplasmic domain revealed 2 highly conserved serine residues that fell within the consensus motifs for RSK phosphorylation; interestingly, those motifs, YVRRDS711G and MRPVS728T, also flank Tyr721 (Fig. 5A). Despite substantial effort, we were unable to map RSK-dependent phosphorylation sites on the M-CSFR in vivo, most likely because the tryptic peptides containing the putative sites did not fly well in mass spectrometry experiments. However, recombinant human RSK2 phosphorylated the M-CSFR cytoplasmic domain in vitro (Fig. 5B). Therefore, as an alternative approach to identifying RSK phosphorylation sites, we generated a peptide array spanning the entire M-CSFR cytoplasmic domain and assessed the ability of recombinant RSK2 to phosphorylate those peptides. Remarkably, only peptides containing Ser711 were phosphorylated by RSK2, whereas Ser711 > Ala-substituted peptides were refractory to phosphorylation (Fig. 5C). Consistent with those observations, immunoblotting with phospho-PKA substrate antibodies revealed a reduction in M-CSF–evoked M-CSFR phosphorylation in SHP2-deficient BMMs, compared with SHP2- sufficient controls (Fig. 5D).
Figure 5.
RSK2 phosphorylates M-CSFR in vitro. A) Sequence of section of M-CSFR cytoplasmic domain, showing position of 2 potential RSK sites (consensus motif R/KXXS/T) flanking the p85 binding site, Tyr721. Note that these sites are conserved in mouse (m), human (h), monkey (rh), rat (rt), guinea pig (gp), and dog (dg). B) M-CSFR phosphorylation by RSK2 in vitro. Recombinant M-CSFR cytoplasmic domain was incubated with recombinant RSK2 in the presence of γ32P-ATP, as described in Materials and Methods. The reaction products were resolved by SDS-PAGE, and the phosphorylated M-CSFR band was subjected to phospho–amino acid analysis. C) Peptide library-based kinase assays. Peptides corresponding to the 23 serine residues of M-CSFR cytoplasmic domain were subjected to spot-blot kinase assays with recombinant RSK2. Phosphorylated peptides were visualized and quantified with a phosphorimager. Note that only peptides containing Ser711 were phosphorylated to a significant extent. D) M-CSFR is inducibly phosphorylated on site or sites that conform to RSK consensus motif. Lysates (1 mg) from SHP2mϕCTR or SHP2mϕKO BMMs that was starved and stimulated with M-CSF (30 ng/ml) for the indicated times (+) or left unstimulated (−) were immunoprecipitated with anti–M-CSFR antibodies and then immunoblotted with anti–p-PKA substrate antibodies (which recognize the sequence R/KXXpS/pT).
Phosphorylation of Ser711 does not influence the phosphorylation of Tyr721 in M-CSFR and its associated AKT activation
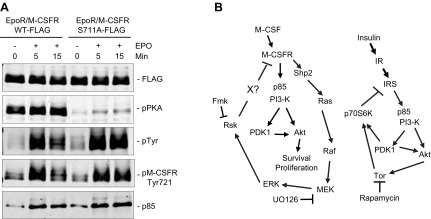
These experiments suggested that RSK2 phosphorylates Ser711 on the M-CSFR. To test that hypothesis and to determine the effect of preventing Ser711 phosphorylation on M-CSF–evoked AKT activation, we used retroviral gene transduction to express chimeric receptors comprising the EPOR extracellular domain, linked to the transmembrane and cytoplasmic domains of WT M-CSFR or M-CSFR carrying a Ser711 > Ala mutation (Supplemental Fig. S3); both chimeras also contained a FLAG-tag at their amino termini. Use of those chimeras allowed us to probe the effects of M-CSFR mutations in cells expressing endogenous M-CSFR. Initial experiments were carried out with the murine Mϕ cell line RAW264.7. After starvation, transduced cells were stimulated with EPO (to selectively activate the chimeric receptor), and the chimeric receptors were immunoprecipitated using anti-FLAG antibodies and subjected to immunoblotting. Mutation of the Ser711 > Ala showed significantly reduced M-CSFR immunoreactivity with phospho-PKA substrate antibodies (Fig. 6A), providing strong evidence that Ser711 is phosphorylated in vivo. Notably, Ser711 was phosphorylated even in the absence of EPO stimulation, arguing against MCSF- and RSK-dependent phosphorylation of that site. Nevertheless, M-CSFR Tyr721 phosphorylation and M-CSFR/p85 association were increased/sustained upon M-CSF (EPO) stimulation in cells expressing the Ser711A mutant chimeras (Fig. 6A) Importantly, the WT and S711A chimeras were expressed at comparable levels, as revealed by anti-FLAG immunoblotting, and M-CSF–evoked AKT activation was sustained in cells expressing the S711A mutant, consistent with its increased Tyr721 phosphorylation (Supplemental Fig. S4A). These data indicate that S711 phosphorylation constrains Tyr 721 phosphorylation, but phosphorylation of Ser711 does not appear to be M-CSF/RSK dependent. Hence, although RSK2 can phosphorylate Ser711 in vitro and Ser711 phosphorylation constrains the extent and duration of Tyr 721 in cells, RSK does not phosphorylate Ser711 in vivo. How RSK controls Tyr721 phosphorylation remains unclear.
Figure 6.
Loss of RSK2-mediated feedback pathway helps promote proliferation and survival of SHP2-deficient BMMs. A) Mutation of Ser711 on M-CSFR leads to increased phosphorylation on M-CSFR Tyr721 and enhanced M-CSF–evoked AKT activation. RAW264.7 cells expressing the indicated EPOR/M-CSFR chimera were starved (−) and then stimulated with 5 U/ml EPO for the indicated times (+). Cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with the indicated antibodies. Note that the Ser711-Ala mutant shows decreased reactivity with anti-pPKA antibodies and increased reactivity with anti-pTyr and -pTyr721 antibodies. Data shown are representative of the 3 independent experiments. B) Model for RSK-mediated, negative-feedback pathway in comparison with p70S6 kinase-mediated feedback signaling loop. See text for details.
Loss of the ERK/RSK-mediated negative feedback loop promotes survival of SHP2-deficient BMMs
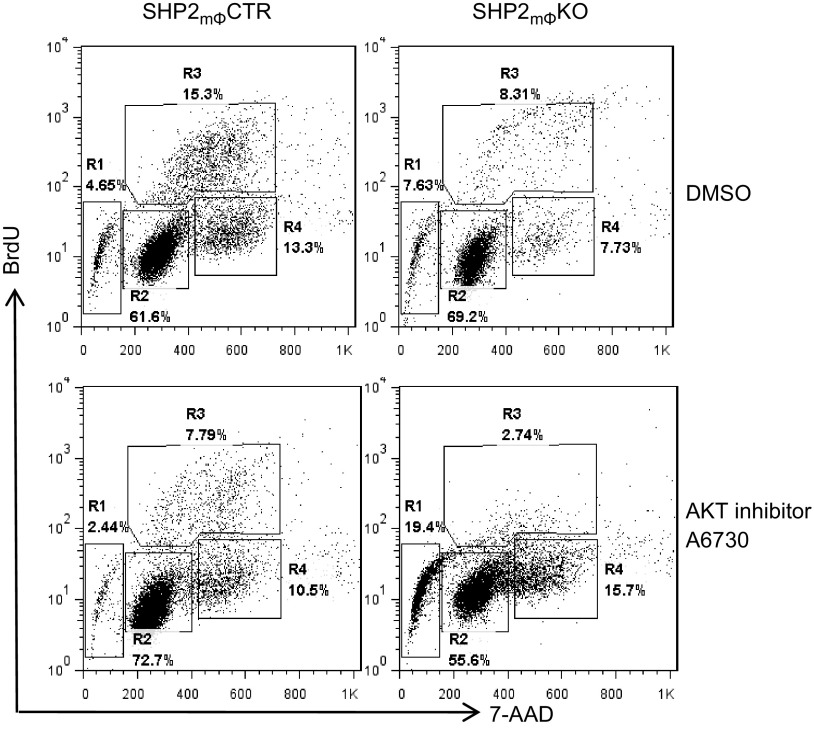
ERK and RSK each phosphorylate several proteins important for cellular proliferation and survival (1, 12, 62), and SHP2mϕKO BMMs have substantially diminished RAS/ERK/RSK activation. Mϕ progenitors in SHP2mϕKO mice do show diminished proliferation, yet those defects and the effects of monocyte/Mϕ SHP2 deficiency on the whole organism are fairly mild. We wondered whether the increase in PI3K/AKT pathway activation in SHP2mϕKO BMMs (because of the loss of RSK-mediated, negative-feedback signaling) might mitigate the effects of SHP2 deficiency on ERK and RSK activation. To test that hypothesis, SHP2mϕCTR and SHP2mϕKO BMMs were treated with the AKT inhibitor A6730 (10 µM) in the presence of murine M-CSF (10 ng/ml) for 48 h, and cell-cycle distribution was assessed. AKT inhibition decreased the percentage of SHP2mϕCTR BMMs in the S phase by ∼50%, but did not increase the number of sub-G1 (apoptotic) cells (Fig. 7, left). By contrast, under normal growth conditions, SHP2mϕKO BMMs showed ∼2-fold fewer S and G2/M cells, and a slightly increased number of sub-G1 cells, consistent with their decreased proliferation rate (Fig. 7, top). Addition of the AKT inhibitor (10 µM) caused a further ∼3-fold decrease in S and ∼2-fold increase in G2/M phase cells and evoked substantial apoptosis, with the sub-G1 population rising to ∼19% in SHP2mΦKO [compared with ∼2% in SHP2mΦCTR) BMMs (Fig. 7, right)]. These data suggest that increased activation of the PI3K/AKT pathway in SHP2mϕKO BMMs (because of impaired RSK-mediated, negative feedback) helps to protect them from the decrease in proliferation and survival that would ordinarily accompany defective RAS/ERK pathway activation. Our data also suggest that enhanced AKT activation promotes cell proliferation, possibly by accelerating the G2/M transition. This observation comports with a previous report showing that AKT deficiency in MEFs attenuates the transition from G2/M to G1 (63). Consistent with that notion, combined inhibition of both the RAS/ERK and PI3K/AKT pathways in WT BMMs (by treatment with UO126 and LY294002) caused a dramatic increase in cell death (Supplemental Fig. S5), comparable to the effects of AKT inhibition in SHP2-deficient BMMs (Fig. 7). Our data are consistent with a previous report showing synergism between the ERK1/2 and PI3K pathways in the regulation of Mϕ proliferation (64).
Figure 7.
SHP2 deficiency sensitizes BMM to apoptosis evoked by AKT inhibitor A6730. Randomly growing BMMs from SHP2mΦCTR and SHP2mϕKO mice were treated with DMSO or the AKT inhibitor A6730 (10 µM). BrdU (10 µM) was added to the culture medium for 1 h, and cells were stained with anti-BrdU antibodies and 7-aminoactinomycin D (7-AAD) and subjected to flow cytometric cell-cycle analysis. Regions 1, 2, 3, and 4 (R1, 2 3, and 4) represent sub-G1, G0/G1, S phase, and G2/M cell populations, respectively. Note that, compared with SHP2mϕCTRs, SHP2mϕKO BMMs are hypersensitive to AKT inhibition. Data shown are representative of the 3 independent experiments.
DISCUSSION
SHP2 is a key mediator of signaling by most, if not all, RTKs and cytokine receptors (30, 31), yet its role in specific hematopoietic lineages remains largely unexplored. By studying the effects of selective deletion of a floxed Ptpn11 allele in the presence of a LysM-Cre transgene, we found that SHP2, although dispensable for myelomonocytic differentiation, is required for optimal proliferation of monocyte/Mϕ progenitors. Because these progenitors also give rise to osteoclasts, SHP2 is required for efficient osteoclast production in vitro, and SHP2mϕKO mice develop mild osteopetrosis as they age. In attempting to elucidate the molecular basis for those mild defects, we identified a novel, negative feedback pathway in which the ERK-dependent kinase RSK2 regulates the phosphorylation of the binding site for p85 on the M-CSFR, Tyr721, and possibly other tyrosyl residues (Fig. 6B). That feedback pathway, in turn, controls the extent and duration of M-CSFR–evoked PI3K and AKT activation, and its absence (and the resultant increase in AKT activation) likely contributes to the relatively mild effects of SHP2 deficiency in the monocyte/Mϕ lineage.
SHP2mΦKO mice exhibit significant defects in myelomonocytic cell proliferation in vitro. BM from these mice yields fewer CFU-M than does WT BM, indicating fewer monocytic progenitors. Moreover, those progenitors have diminished M-CSF responsiveness, as indicated by their rightward-shifted, dose–response curve. SHP2mΦKO BMMs show similar proliferative defects, attributable to their diminished M-CSF sensitivity. SHP2-deficient mice also develop mild, age-related osteopetrosis and show defective osteoclast generation in vitro. Mice lacking the SHP2-binding protein GAB2 also have osteopetrosis, although that phenotype was attributed to defective RANKL, not M-CSF, signaling (65). However, overall, despite those in vitro defects, SHP2mϕKO mice were remarkably healthy, with peripheral blood counts within reference range and no apparent increased tendency to infection, presumably reflecting the ability of healthy homeostatic mechanisms to compensate for the defective M-CSF responsiveness.
Similar to other RTK signaling pathways and other cell types, SHP2 deficiency in BMMs results in decreased RAS/ERK activation in response to M-CSF stimulation. Previous studies have implicated the RAS/ERK and PI3K/AKT pathways in M-CSFR–driven proliferation and/or survival (13). Given that AKT activation is not diminished in SHP2mϕKO BMMs (but is, instead, enhanced; see below), diminished RAS/ERK activation probably explains, at least in large part, their defective proliferation. Previous work implicated STAT1 and STAT3 in BM cell differentiation toward the myelomonocytic lineage (66, 67). SHP2 does not affect M-CSF–evoked STAT1 and STAT3 phosphorylation, and consistent with that finding, Mϕ differentiation is apparently unaffected in SHP2mϕKO mice.
SHP2-deficient BMMs show increased AKT activity, which, by analogy to studies of other signaling pathways (32, 33, 56), we initially expected to be due to increased GAB binding to p85. However, GAB2-associated PI3K activity (and p85 association) is comparable in SHP2mϕCTR and SHP2mϕKO BMMs. Instead, we found that M-CSFR–associated PI3K activation was enhanced in SHP2mϕKO BMMs because of increased M-CSFR Tyr721 phosphorylation and p85 recruitment.
Several lines of evidence suggest that an indirect, RSK-mediated, negative-feedback pathway (Fig. 6B) explains the increased Tyr721 phosphorylation and enhanced PI3K/AKT activation in the absence of SHP2. First, the effects of MEK (U0126) or RSK (Fmk) inhibitors on overall M-CSFR tyrosyl phosphorylation, Tyr721 phosphorylation, p85 association with M-CSFR, and AKT activation are comparable to those of SHP2 deficiency. RSK2 deficiency also enhanced M-CSF–evoked AKT activation, arguing against off-target effects of the pharmacologic inhibitors and suggesting that, of the 4 known RSK family members (1), RSK2 specifically mediates this negative feedback pathway (Fig. 4B). Second, GM-CSF–evoked AKT activation was unaffected by SHP2 deficiency, which suggests that receptor-proximal events are involved in the negative regulatory mechanism. Although RSK can phosphorylate M-CSFR on Ser711 in vitro, and Ser711 phosphorylation controls the extent and duration of Tyr721 phosphorylation, immunoblotting experiments indicate that Ser711 is constitutively phosphorylated. Furthermore, UO126 treatment does not affect Ser711 phosphorylation (Supplemental Fig. S4B). Therefore, the RSK-mediated negative-feedback pathway must be mediated by another, yet unidentified, protein, and Ser711 must be phosphorylated by another kinase. PKA is a good candidate for the latter because BMMs treated with PKA activator 8-bromo cAMP show reduced tyrosyl phosphorylation of M-CSFR (68).
Our findings have important implications for understanding Mϕ and osteoclast regulation by M-CSFR, for RTK signaling in general and for the potential use of MEK inhibitors for cancer therapeutics. The concomitant loss of the RSK2-mediated feedback pathway probably helps explain the relatively mild effects of impaired RAS/ERK activation in the monocyte/Mϕ lineage. Consistent with that notion, SHP2mϕKO BMMs are strongly sensitized to PI3K or AKT inhibitor treatment (Fig. 7). It is increasingly clear that the duration and magnitude of downstream pathway activation are key determinants of signal specificity by RTKs and other types of receptors. Feedback pathways probably have an important role in regulating those pathway properties. In that regard, the RSK2-mediated pathway that we identified is reminiscent of the now well-characterized p70S6K-catalyzed, negative-feedback regulation of insulin/insulin-like growth factor receptor signaling (4, 57). Finally, the RAS/ERK pathway is up-regulated in many tumors, which has prompted the development of MEK inhibitors as potential antineoplastic agents (69, 70). If the RSK2-mediated, negative-feedback pathway (or an analogous MEK-dependent pathway) were operative in cancer cells, they might “escape” from MEK-inhibitor treatment because of increased AKT activation. A similar escape mechanism via inactivation of the p70S6K-mediated negative-feedback pathway has been documented in some cancer cells treated with Tor inhibitors, such as rapamycin (71). Alternatively, inactivation of RSK2-mediated, negative feedback (by mutations or epigenetic mechanisms) might allow cancer cells to benefit from an increased activation of the PI3K/AKT pathway. Notably, signals emanating from M-CSFR pTyr721 mediate Mϕ enhancement of tumor cell invasion (72). Importantly, analysis of the signaling pathways downstream of the M-CSFR has demonstrated an important role for both ERK1/2 and PI3K signaling, not only in maintaining Mϕs but also in preserving their noninflammatory, protumoral state (73). Further studies will be required to elucidate the detailed mechanism by which RSK2-mediated negative feedback controls M-CSFR signaling and to assess the generality of its role in RTK regulation and cancer.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Changqi Sun (Rhode Island Hospital) for technical assistance in constructing pMX(puro) expression constructs. This work was supported in part by U.S. National Institutes of Health (NIH) National Cancer Institute Grants R01CA114945 and R37 CA49152 (to B.G.N.), NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants R21AR57156, RO1AR066746, and NIH National Center for Research Resources Grant P20RR025179 (to W.Y.). This study was also aided by a grant from the Arthritis National Research Foundation (to W.Y.). B.G.N. was a Canada Research Chair, Tier 1, and this research was also funded, in part, by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. The authors declare no conflicts of interest.
Glossary
- BM
bone marrow
- BMM
bone marrow macrophage
- BrdU
bromodeoxyuridine
- CFU-M
colony-forming unit–macrophage
- EGF
epidermal growth factor
- EPO
erythropoietin
- EPOR
erythropoietin receptor
- FBS
fetal bovine serum
- Fmk
fluoromethylketone
- GAB
growth factor receptor-bound protein 2–associated binding protein
- GM-CSF
granulocyte M-CSF
- KO
knockout
- M-CSFR
M-CSF receptor
- Mϕ
macrophage
- mTOR
mammalian target of rapamycin
- PTPN11
protein tyrosine phosphatase, non-receptor type 11
- RSK
ribosomal S6 kinase
- RTK
receptor tyrosine kinase
- SFK
SRC family kinase
- SHP2
Src homology-2 domain containing protein tyrosine phosphatase 2
- STAT
signal transducer and activator of transcription
- TCL
total cell lysate
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
B. G. Neel and W. Yang conceived and designed the experiments; L. Wang, C. Iorio, K. Yan, H. Yang, and W. Yang performed the experiments; S. Takeshita and S. Kang provided key reagents for this study; B. G. Neel and W. Yang analyzed the data and wrote the paper; and all authors read and approve the final version of this manuscript.
REFERENCES
- 1.Anjum R., Blenis J. (2008) The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 2.McKay M. M., Morrison D. K. (2007) Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113–3121 [DOI] [PubMed] [Google Scholar]
- 3.Raman M., Chen W., Cobb M. H. (2007) Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 4.Sebolt-Leopold J. S., Herrera R. (2004) Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4, 937–947 [DOI] [PubMed] [Google Scholar]
- 5.Ramjaun A. R., Downward J. (2007) Ras and phosphoinositide 3-kinase: partners in development and tumorigenesis. Cell Cycle 6, 2902–2905 [DOI] [PubMed] [Google Scholar]
- 6.Engelman J. A., Luo J., Cantley L. C. (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Rohrschneider L. R. (2002) The gift of Gab. FEBS Lett. 515, 1–7 [DOI] [PubMed] [Google Scholar]
- 8.Gu H., Neel B. G. (2003) The “Gab” in signal transduction. Trends Cell Biol. 13, 122–130 [DOI] [PubMed] [Google Scholar]
- 9.Simister P. C., Feller S. M. (2012) Order and disorder in large multi-site docking proteins of the Gab family—implications for signalling complex formation and inhibitor design strategies. Mol. Biosyst. 8, 33–46 [DOI] [PubMed] [Google Scholar]
- 10.Manning B. D., Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemmon M. A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy L. O., Blenis J. (2006) MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 31, 268–275 [DOI] [PubMed] [Google Scholar]
- 13.Pixley F. J., Stanley E. R. (2004) CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14, 628–638 [DOI] [PubMed] [Google Scholar]
- 14.Teitelbaum S. L., Ross F. P. (2003) Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 15.Stanley E. R., Chitu V. (2014) CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 6, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida H., Hayashi S., Kunisada T., Ogawa M., Nishikawa S., Okamura H., Sudo T., Shultz L. D., Nishikawa S. (1990) The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345, 442–444 [DOI] [PubMed] [Google Scholar]
- 17.Alonso G., Koegl M., Mazurenko N., Courtneidge S. A. (1995) Sequence requirements for binding of Src family tyrosine kinases to activated growth factor receptors. J. Biol. Chem. 270, 9840–9848 [DOI] [PubMed] [Google Scholar]
- 18.Van der Geer P., Hunter T. (1993) Mutation of Tyr697, a GRB2-binding site, and Tyr721, a PI 3-kinase binding site, abrogates signal transduction by the murine CSF-1 receptor expressed in Rat-2 fibroblasts. EMBO J. 12, 5161–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeshita S., Faccio R., Chappel J., Zheng L., Feng X., Weber J. D., Teitelbaum S. L., Ross F. P. (2007) c-Fms tyrosine 559 is a major mediator of M-CSF-induced proliferation of primary macrophages. J. Biol. Chem. 282, 18980–18990 [DOI] [PubMed] [Google Scholar]
- 20.Rohde C. M., Schrum J., Lee A. W. (2004) A juxtamembrane tyrosine in the colony stimulating factor-1 receptor regulates ligand-induced Src association, receptor kinase function, and down-regulation. J. Biol. Chem. 279, 43448–43461 [DOI] [PubMed] [Google Scholar]
- 21.Reedijk M., Liu X., van der Geer P., Letwin K., Waterfield M. D., Hunter T., Pawson T. (1992) Tyr721 regulates specific binding of the CSF-1 receptor kinase insert to PI 3′-kinase SH2 domains: a model for SH2-mediated receptor-target interactions. EMBO J. 11, 1365–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Jenkins B., Shin J. L., Rohrschneider L. R. (2001) Scaffolding protein Gab2 mediates differentiation signaling downstream of Fms receptor tyrosine kinase. Mol. Cell. Biol. 21, 3047–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee A. W., States D. J. (2000) Both src-dependent and -independent mechanisms mediate phosphatidylinositol 3-kinase regulation of colony-stimulating factor 1-activated mitogen-activated protein kinases in myeloid progenitors. Mol. Cell. Biol. 20, 6779–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee P. S., Wang Y., Dominguez M. G., Yeung Y. G., Murphy M. A., Bowtell D. D., Stanley E. R. (1999) The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 18, 3616–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y., Song D., Cai Y., Yu W., Yeung Y. G., Stanley E. R. (2011) A CSF-1 receptor phosphotyrosine 559 signaling pathway regulates receptor ubiquitination and tyrosine phosphorylation. J. Biol. Chem. 286, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak U., Nice E., Hamilton J. A., Paradiso L. (1996) Requirement for Y706 of the murine (or Y708 of the human) CSF-1 receptor for STAT1 activation in response to CSF-1. Oncogene 13, 2607–2613 [PubMed] [Google Scholar]
- 27.Novak U., Harpur A. G., Paradiso L., Kanagasundaram V., Jaworowski A., Wilks A. F., Hamilton J. A. (1995) Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood 86, 2948–2956 [PubMed] [Google Scholar]
- 28.Suzu S., Hiyoshi M., Yoshidomi Y., Harada H., Takeya M., Kimura F., Motoyoshi K., Okada S. (2007) M-CSF-mediated macrophage differentiation but not proliferation is correlated with increased and prolonged ERK activation. J. Cell. Physiol. 212, 519–525 [DOI] [PubMed] [Google Scholar]
- 29.Lee A. W., States D. J. (2006) Colony-stimulating factor-1 requires PI3-kinase-mediated metabolism for proliferation and survival in myeloid cells. Cell Death Differ. 13, 1900–1914 [DOI] [PubMed] [Google Scholar]
- 30.Grossmann K. S., Rosário M., Birchmeier C., Birchmeier W. (2010) The tyrosine phosphatase Shp2 in development and cancer. Adv. Cancer Res. 106, 53–89 [DOI] [PubMed] [Google Scholar]
- 31. Neel, B. G., Chan, G., Dhanji, S. (2009) SH2 domain-containing protein-tyrosine phosphatases. In Handbook of Cell Signaling, 2nd ed. (Bradshaw, R. and Dennis, E., eds.), pp. 771–809, Academic Press, San Diego, CA. [Google Scholar]
- 32.Zhang S. Q., Tsiaras W. G., Araki T., Wen G., Minichiello L., Klein R., Neel B. G. (2002) Receptor-specific regulation of phosphatidylinositol 3′-kinase activation by the protein tyrosine phosphatase Shp2. Mol. Cell. Biol. 22, 4062–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattoon D. R., Lamothe B., Lax I., Schlessinger J. (2004) The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol. 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaeper U., Vogel R., Chmielowiec J., Huelsken J., Rosario M., Birchmeier W. (2007) Distinct requirements for Gab1 in Met and EGF receptor signaling in vivo. Proc. Natl. Acad. Sci. USA 104, 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agazie Y. M., Hayman M. J. (2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol. Cell. Biol. 23, 7875–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C. J., O’Rourke D. M., Feng G. S., Johnson G. R., Wang Q., Greene M. I. (2001) The tyrosine phosphatase SHP-2 is required for mediating phosphatidylinositol 3-kinase/Akt activation by growth factors. Oncogene 20, 6018–6025 [DOI] [PubMed] [Google Scholar]
- 37.Chan G., Kalaitzidis D., Neel B. G. (2008) The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 38.Fornaro M., Burch P. M., Yang W., Zhang L., Hamilton C. E., Kim J. H., Neel B. G., Bennett A. M. (2006) SHP-2 activates signaling of the nuclear factor of activated T cells to promote skeletal muscle growth. J. Cell Biol. 175, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W., Klaman L. D., Chen B., Araki T., Harada H., Thomas S. M., George E. L., Neel B. G. (2006) An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev. Cell 10, 317–327 [DOI] [PubMed] [Google Scholar]
- 40.Qu C. K., Feng G. S. (1998) Shp-2 has a positive regulatory role in ES cell differentiation and proliferation. Oncogene 17, 433–439 [DOI] [PubMed] [Google Scholar]
- 41.Qu C. K., Nguyen S., Chen J., Feng G. S. (2001) Requirement of Shp-2 tyrosine phosphatase in lymphoid and hematopoietic cell development. Blood 97, 911–914 [DOI] [PubMed] [Google Scholar]
- 42.Cohen M. S., Zhang C., Shokat K. M., Taunton J. (2005) Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science 308, 1318–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W., Wang J., Moore D. C., Liang H., Dooner M., Wu Q., Terek R., Chen Q., Ehrlich M. G., Quesenberry P. J., Neel B. G. (2013) Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling. Nature 499, 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 46.Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 47.Feng X., Takeshita S., Namba N., Wei S., Teitelbaum S. L., Ross F. P. (2002) Tyrosines 559 and 807 in the cytoplasmic tail of the macrophage colony-stimulating factor receptor play distinct roles in osteoclast differentiation and function. Endocrinology 143, 4868–4874 [DOI] [PubMed] [Google Scholar]
- 48.Gu H., Saito K., Klaman L. D., Shen J., Fleming T., Wang Y., Pratt J. C., Lin G., Lim B., Kinet J. P., Neel B. G. (2001) Essential role for Gab2 in the allergic response. Nature 412, 186–190 [DOI] [PubMed] [Google Scholar]
- 49.Sefton B. M. (2001) Phosphoamino acid analysis. Curr. Protoc. Protein. Sci. Chapter 13, Unit13 13. [DOI] [PubMed] [Google Scholar]
- 50.Hilpert K., Winkler D. F., Hancock R. E. (2007) Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 [DOI] [PubMed] [Google Scholar]
- 51.Leung G. C., Murphy J. M., Briant D., Sicheri F. (2009) Characterization of kinase target phosphorylation consensus motifs using peptide SPOT arrays. Methods Mol. Biol. 570, 187–195 [DOI] [PubMed] [Google Scholar]
- 52.Warner N., Wybenga-Groot L. E., Pawson T. (2008) Analysis of EphA4 receptor tyrosine kinase substrate specificity using peptide-based arrays. FEBS J. 275, 2561–2573 [DOI] [PubMed] [Google Scholar]
- 53.Saxton T. M., Henkemeyer M., Gasca S., Shen R., Rossi D. J., Shalaby F., Feng G. S., Pawson T. (1997) Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 16, 2352–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arrandale J. M., Gore-Willse A., Rocks S., Ren J. M., Zhu J., Davis A., Livingston J. N., Rabin D. U. (1996) Insulin signaling in mice expressing reduced levels of Syp. J. Biol. Chem. 271, 21353–21358 [DOI] [PubMed] [Google Scholar]
- 55.Edouard T., Combier J. P., Nédélec A., Bel-Vialar S., Métrich M., Conte-Auriol F., Lyonnet S., Parfait B., Tauber M., Salles J. P., Lezoualc’h F., Yart A., Raynal P. (2010) Functional effects of PTPN11 (SHP2) mutations causing LEOPARD syndrome on epidermal growth factor-induced phosphoinositide 3-kinase/AKT/glycogen synthase kinase 3beta signaling. Mol. Cell. Biol. 30, 2498–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montagner A., Yart A., Dance M., Perret B., Salles J. P., Raynal P. (2005) A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J. Biol. Chem. 280, 5350–5360 [DOI] [PubMed] [Google Scholar]
- 57.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrington L. S., Findlay G. M., Lamb R. F. (2005) Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 59.Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 60.Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 61.Tanoue T., Nishida E. (2003) Molecular recognitions in the MAP kinase cascades. Cell. Signal. 15, 455–462 [DOI] [PubMed] [Google Scholar]
- 62.Lawrence M. C., Jivan A., Shao C., Duan L., Goad D., Zaganjor E., Osborne J., McGlynn K., Stippec S., Earnest S., Chen W., Cobb M. H. (2008) The roles of MAPKs in disease. Cell Res. 18, 436–442 [DOI] [PubMed] [Google Scholar]
- 63.Kandel E. S., Skeen J., Majewski N., Di Cristofano A., Pandolfi P. P., Feliciano C. S., Gartel A., Hay N. (2002) Activation of Akt/protein kinase B overcomes a G(2)/m cell cycle checkpoint induced by DNA damage. Mol. Cell. Biol. 22, 7831–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu W., Chen J., Xiong Y., Pixley F. J., Yeung Y. G., Stanley E. R. (2012) Macrophage proliferation is regulated through CSF-1 receptor tyrosines 544, 559, and 807. J. Biol. Chem. 287, 13694–13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wada T., Nakashima T., Oliveira-dos-Santos A. J., Gasser J., Hara H., Schett G., Penninger J. M. (2005) The molecular scaffold Gab2 is a crucial component of RANK signaling and osteoclastogenesis. Nat. Med. 11, 394–399 [DOI] [PubMed] [Google Scholar]
- 66.Novak U., Nicholson S., Bourette R. P., Rohrschneider L. R., Alexander W., Paradiso L. (1998) CSF-1 and interferon-γ act synergistically to promote differentiation of FDC-P1 cells into macrophages. Growth Factors 15, 159–171 [DOI] [PubMed] [Google Scholar]
- 67.Hamilton J. A. (1997) CSF-1 signal transduction. J. Leukoc. Biol. 62, 145–155 [DOI] [PubMed] [Google Scholar]
- 68.Wilson N. J., Cross M., Nguyen T., Hamilton J. A. (2005) cAMP inhibits CSF-1-stimulated tyrosine phosphorylation but augments CSF-1R-mediated macrophage differentiation and ERK activation. FEBS J. 272, 4141–4152 [DOI] [PubMed] [Google Scholar]
- 69.Sebolt-Leopold J. S. (2004) MEK inhibitors: a therapeutic approach to targeting the Ras-MAP kinase pathway in tumors. Curr. Pharm. Des. 10, 1907–1914 [DOI] [PubMed] [Google Scholar]
- 70.Hersey P., Bastholt L., Chiarion-Sileni V., Cinat G., Dummer R., Eggermont A. M., Espinosa E., Hauschild A., Quirt I., Robert C., Schadendorf D. (2009) Small molecules and targeted therapies in distant metastatic disease. Ann. Oncol. 20(Suppl 6), vi35–vi40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dowling R. J., Topisirovic I., Fonseca B. D., Sonenberg N. (2010) Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta 1804, 433–439 [DOI] [PubMed] [Google Scholar]
- 72.Sampaio N. G., Yu W., Cox D., Wyckoff J., Condeelis J., Stanley E. R., Pixley F. J. (2011) Phosphorylation of CSF-1R Y721 mediates its association with PI3K to regulate macrophage motility and enhancement of tumor cell invasion. J. Cell Sci. 124, 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caescu C. I., Guo X., Tesfa L., Bhagat T. D., Verma A., Zheng D., Stanley E. R. (2015) Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood 125, e1–e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.