Abstract
Most human emerging infectious diseases originate from wildlife and bats are a major reservoir of viruses, a few of which have been highly pathogenic to humans. In some regions of Cameroon, bats are hunted and eaten as a delicacy. This close proximity between human and bats provides ample opportunity for zoonotic events. To elucidate the viral diversity of Cameroonian fruit bats, we collected and metagenomically screened eighty-seven fecal samples of Eidolon helvum and Epomophorus gambianus fruit bats. The results showed a plethora of known and novel viruses. Phylogenetic analyses of the eleven gene segments of the first complete bat rotavirus H genome, showed clearly separated clusters of human, porcine, and bat rotavirus H strains, not indicating any recent interspecies transmission events. Additionally, we identified and analyzed a bat bastrovirus genome (a novel group of recently described viruses, related to astroviruses and hepatitis E viruses), confirming their recombinant nature, and provide further evidence of additional recombination events among bat bastroviruses. Interestingly, picobirnavirus-like RNA-dependent RNA polymerase gene segments were identified using an alternative mitochondrial genetic code, and further principal component analyses suggested that they may have a similar lifestyle to mitoviruses, a group of virus-like elements known to infect the mitochondria of fungi. Although identified bat coronavirus, parvovirus, and cyclovirus strains belong to established genera, most of the identified partitiviruses and densoviruses constitute putative novel genera in their respective families. Finally, the results of the phage community analyses of these bats indicate a very diverse geographically distinct bat phage population, probably reflecting different diets and gut bacterial ecosystems.
Keywords: bat, virome, rotavirus H, bastrovirus, picobirnavirus, metagenomics
1. Introduction
Emerging infectious diseases have a serious impact on human health and our economy and unfortunately, their upward trend has not yet been halted (Mackey et al. 2014). About 60–80% of these emerging infections originate from wildlife including bats (Cleaveland et al. 2001; Taylor et al. 2001). Some bats (Order Chiroptera) have been implicated as a major reservoir of pathogenic zoonotic viruses such as rabies virus, Marburg virus, Severe Acute Respiratory Syndrome (SARS)- and Middle East Respiratory Syndrome (MERS)-related coronaviruses (CoVs), Nipah and Hendra viruses (Rupprecht et al. 1995; Chua et al. 2002; Lau et al. 2005; Leroy et al. 2005; Towner et al. 2007; Memish et al. 2013). Bats make up more than 20% of the ∼5,500 known terrestrial species of mammals (Nowak 1991; Vaughan and Ryan 2000) and have a combination of features that is believed to enhance their ability to facilitate virus evolution and transmission such as longevity, migratory activity, large and dense roosting communities, and close social interactions (Prendergast et al. 2002; Luis et al. 2013). Additionally, it has been speculated that some viruses which evolved with bats may use cellular receptors and biochemical pathways which are conserved in mammals that evolved later, thus enhancing their ability to transmit these viruses to other mammals including humans (Calisher et al. 2006). Apart from human behavioral changes (driven by increasing human populations) and spatial expansion of agriculture, direct contact with bats through hunting, selling, and/or eating might provide great opportunity for such zoonotic transmissions (Morse 2001), which is the case in Lysoka, Limbe and Moyuka in the Southwest Region of Cameroon.
Control and prevention of these emerging infections from bats and other wildlife entails a strategy of rapid pathogen identification, to determine their origin and control further spread to new hosts and new host species. Therefore, continuous surveillance to unravel the viral communities present in bats and other wildlife is of utmost importance to viral zoonosis prevention and control endeavors (Gerald et al. 2009). However, little or no data are available from Cameroon. In this study, we collected and analyzed fecal samples from two species of fruit bats (Eidolon helvum and Epomophorus gambianus), from the South West Region of Cameroon using high-throughput sequencing.
The gut virome typically contains both eukaryotic and prokaryotic viruses (phages), of which the latter usually represents the largest fraction of the gut virome in animals. However, the phage community in the bat gut has largely been ignored, or only the number of reads assigned to phages are briefly mentioned in some metagenomics data (Ge et al. 2012; Wu et al. 2012, 2016; Zheng et al. 2017). Here, we analyzed known and novel eukaryotic viruses, as well as the bat intestinal phage communities.
2. Results
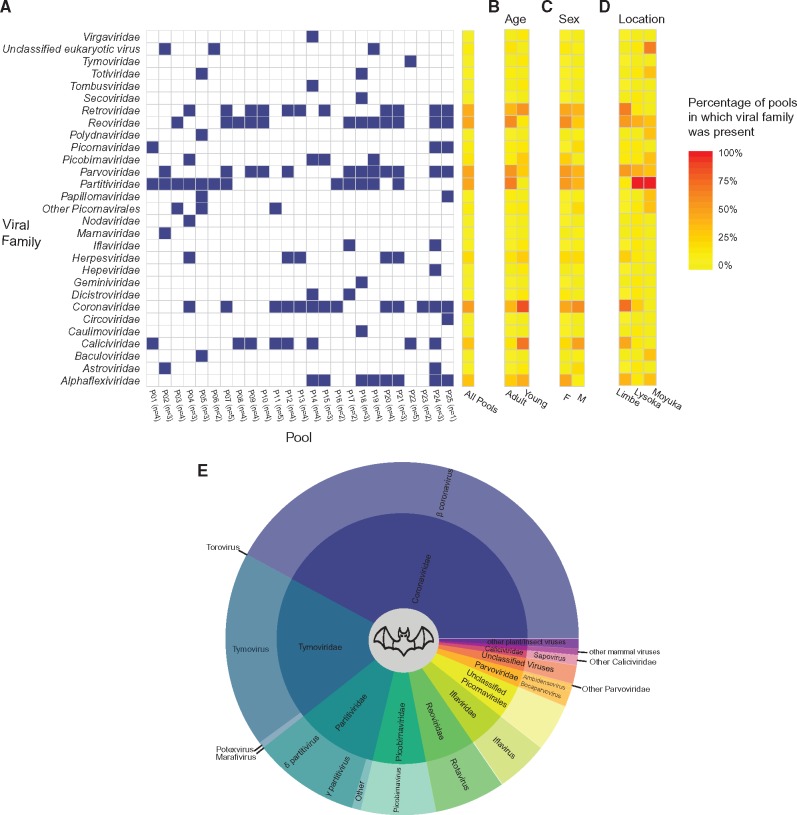
A total of eighty-seven fecal samples from two species of bat (E.helvum and E.gambianus) were collected from three localities of the South West Region of Cameroon: Limbe, Lysoka, and Muyuka (Fig. 1) (Yinda et al. 2016b). These samples were divided over twenty-five pools of one to five samples each, enriched for viral particles and then sequenced. Illumina sequencing yielded a total of 218 million reads and approximately half was trimmed out in the quality control and deduplication process. Out of the remaining 108 million reads, 14% could be assigned as viral while 86% was non-viral (bacterial, host, or dark matter). Most of the viral reads were assigned as phages (89%) and about 11% were eukaryotic viruses (as summarized in Table 1). Only a single pool was made from the samples from E.gambianus fruit bat, which generated mainly phage viral reads after sequencing. Therefore, all eukaryotic genomes described here were from E.helvum. Many of the eukaryotic viral reads belong to vertebrate infecting viral families/orders possessing known and potentially causative agents for gastroenteritis in humans. In particular, we identified members of the order Picornavirales and the families Caliciviridae, Reoviridae, Astroviridae, Picobirnaviridae, Circorviridae, and Parvoviridae. In addition, viruses with a high zoonotic potential belonging to the family Coronaviridae (Betacoronavirus), as well as viruses with a low potential of crossing the species barrier (Papillomaviridae) were detected (Fig. 2). Strikingly, most of the rotavirus (RV) (family Reoviridae) positive samples were detected in adult pools (92%, Fig. 2B). This is unlike in other animal (including humans) wherein RV mostly infect their young (Parashar et al. 2006). Moreover, we observed geographical differences in the percentage of pools in which eukaryotic viral families were present (Fig. 2D). These differences in the sampling sites (i.e. Limbe, Lysoka and Moyuka) maybe due to differences in the acute infections going on at the time of sampling or due to differences in the age of captured bats (all juvenile bats were from Limbe). A detailed analyses of the viruses belonging to the order Picornavirales and the families Papillomaviridae, Reoviridae (RVA) and Caliciviridae (Sapovirus) have been reported elsewhere (Yinda et al. 2016a, b, 2017a,b) Here, we analyzed the remaining viral sequences belonging to double-stranded RNA (families Reoviridae, Picobirnaviridae, and Partitiviridae), single-stranded RNA (families Astroviridae and Coronaviridae) and single-stranded DNA (families Parvoviridae and Circoviridae) viral groups. All the sizes of the contig (genomes) described in this study, together with their sequence coverage and closest GenBank hit can be found on Supplementary Table S1.
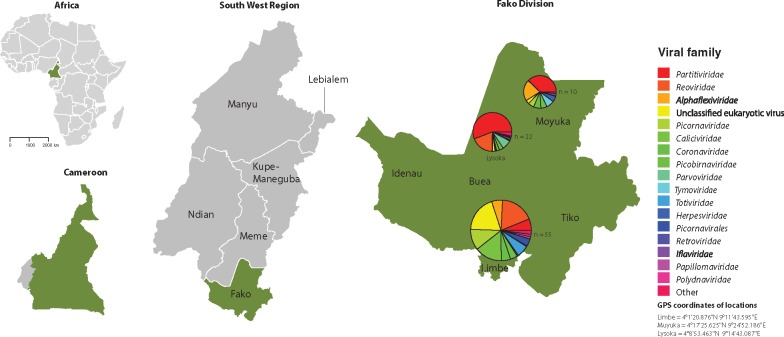
Figure 1.
Map of study site (Fako Division, South West Region, Cameroon). Pie shows the proportion of reads of eukaryotic viral families in different locations. The size of the pie is proportional to the amount of samples analyzed at each location. Maps were created in R (version 3.2.3) (R Core Team 2016), using the raster package (Wickham 2009) and the default plotting packages.
Table 1.
Metadata and number of raw reads, reads after trimming and viral reads per pool.
| Pool | No of samples | Sex | Age | Location | Total raw reads | Reads after trimming | Phage reads | Eukaryotic viral reads | Total viral reads | % viral reads | Others readsa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 4 | M | Adult | Lysoka | 7,681,616 | 1,267,653 | 831,900 | 6,038 | 837,938 | 66.10 | 429,715 |
| P2 | 3 | M | Adult | Lysoka | 4,006,702 | 2,040,250 | 349,971 | 64,544 | 414,515 | 20.32 | 1,625,735 |
| P3 | 4 | F | Adult | Lysoka | 7,882,440 | 1,548,502 | 624,087 | 76,032 | 700,119 | 45.21 | 848,383 |
| P4 | 3 | F | Adult | Lysoka | 9,608,680 | 4,786,763 | 639,351 | 41,507 | 680,858 | 14.22 | 4,105,905 |
| P5 | 3 | M | Adult | Moyuka | 7,898,316 | 2,115,775 | 694,518 | 936 | 695,454 | 32.87 | 1,420,321 |
| P6 | 2 | F | Adult | Moyuka | 7,277,474 | 936,328 | 139,335 | 376 | 139,711 | 14.92 | 796,617 |
| P7 | 5 | M | Adult | Limbe | 3,471,184 | 1,419,933 | 73,955 | 3,773 | 77,728 | 5.47 | 1,342,205 |
| P8 | 4 | M | Adult | Limbe | 8,153,542 | 3,999,576 | 488,794 | 52,173 | 540,967 | 13.53 | 3,458,609 |
| P9 | 4 | F | Adult | Limbe | 9,645,970 | 1,869,185 | 447,021 | 19,310 | 466,331 | 24.95 | 1,402,854 |
| P10 | 4 | F | Adult | Limbe | 9,337,264 | 4,321,908 | 1,334,750 | 45,970 | 1,380,720 | 31.95 | 2,941,188 |
| P11 | 5 | M | Young | Limbe | 8,693,798 | 3,746,818 | 1,377,106 | 59,786 | 1,436,892 | 38.35 | 2,309,926 |
| P12 | 4 | M | Young | Limbe | 3,775,248 | 1,788,839 | 7,574 | 711 | 8,285 | 0.46 | 1,780,554 |
| P13 | 4 | M | Young | Limbe | 12,589,288 | 5,804,209 | 2,833 | 2,005 | 4,838 | 0.08 | 5,799,371 |
| P14 | 4 | F | Young | Limbe | 11,047,740 | 5,081,387 | 765,542 | 29,594 | 795,136 | 15.65 | 4,286,251 |
| P15 | 3 | F | Young | Limbe | 17,437,336 | 7,587,472 | 7,534 | 106,990 | 114,524 | 1.51 | 7,472,948 |
| P16b | 2 | F | Adult | Lysoka | 8,040,290 | 3,252,902 | 466,407 | 105,950 | 572,357 | 17.60 | 2,680,545 |
| P17 | 3 | F | Adult | Lysoka | 11,702,558 | 8,950,127 | 1,649,086 | 153,810 | 1,802,896 | 20.14 | 7,147,231 |
| P18 | 3 | F | Adult | Lysoka | 8,844,716 | 5,697,807 | 13,708 | 1,217 | 14,925 | 0.26 | 5,682,882 |
| P19 | 4 | F | Adult | Muyoka | 6,786,714 | 3,994,014 | 545,295 | 1,603 | 546,898 | 13.69 | 3,447,116 |
| P20 | 4 | F | Adult | Limbe | 11,302,290 | 7,568,783 | 1,825 | 595 | 2,420 | 0.03 | 7,566,363 |
| P21 | 3 | F | Adult | Limbe | 11,222,122 | 6,642,675 | 3,148 | 2,923 | 6,071 | 0.09 | 6,636,604 |
| P22 | 5 | F | Young | Limbe | 11,107,052 | 8,112,574 | 667,730 | 334,067 | 1,001,797 | 12.35 | 7,110,777 |
| P23 | 2 | M | Adult | Limbe | 6,385,352 | 4,850,518 | 2,780 | 620,961 | 623,741 | 12.86 | 4,226,777 |
| P24 | 3 | M | Young | Limbe | 8,985,248 | 6,959,613 | 2,650,973 | 2,100 | 2,653,073 | 38.12 | 4,306,540 |
| P25 | 1 | F | Adult | Limbe | 5,535,248 | 4,343,684 | 5,846 | 15,795 | 21,641 | 0.50 | 4,322,043 |
These could be reads mapping to bacteria, host or dark matter.
Only pool with E. gambianus samples.
Figure 2.
Viral family content. The heat map shows the presence of eukaryotic viral families in feces from all 25 bat pools in relation to different parameters (A) Individual pools; (B) Age; (C) Sex; (D) Location. Blue square: presence of viral family in pool (>1% total reads of that pool); white square: absence of viral family in pool (<1% total reads of that pool). E. overview of the most abundant families and genera identified in bats in this study based on assigned reads. Low abundant mammalian viruses not in this figure are: Astroviridae, Circoviridae, Hepeviridae, Herpesviridae, Paramyxoviridae, and Papillomaviridae. Other low abundant plant/insect viruses not in the figure: Alphatetraviridae, Chrysoviridae, Dicistroviridae, Genomoviridae, Luteoviridae, Nodaviridae, Phycodnaviridae, and Picornavirales. The viruses of a family that could not be assigned to any known genus are referred to as others. Families represented by less than 100 reads were excluded.
3. Double-stranded RNA viral sequences
3.1 Identification of the first near complete bat rotavirus H genome
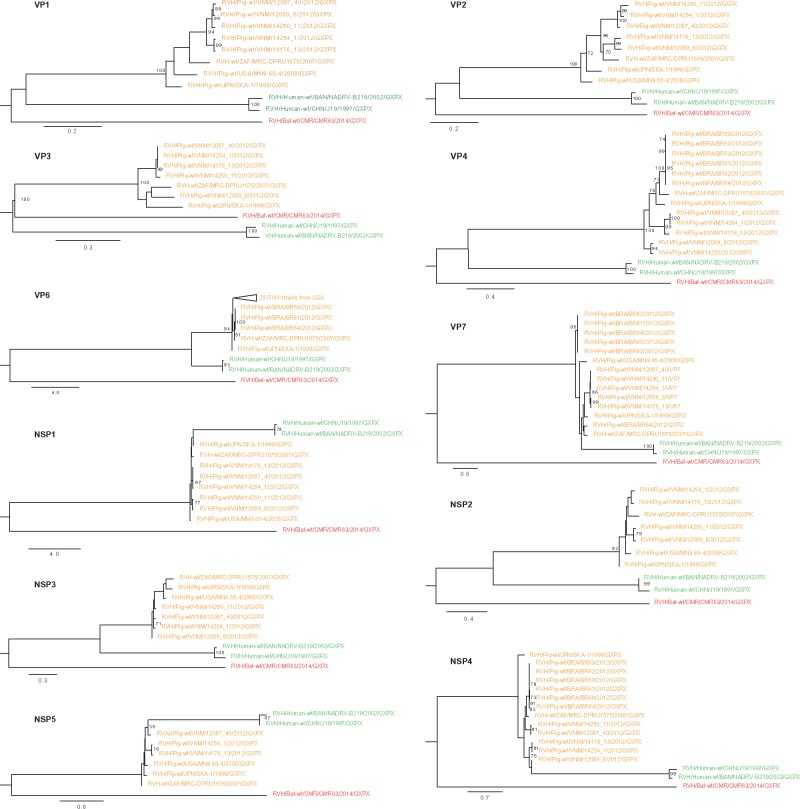
RVs are major enteric pathogen causing severe dehydrating diarrhea mostly in the young of humans and animals worldwide (Bridger et al. 1998). RVs belong to the family Reoviridae and the genus RV consists of nine species, A to I (Attoui et al. 2012; Mihalov-Kovács et al. 2015). RVAs are the most common of all the species having a wide host range including humans. Unlike RVA, RVH, initially referred to as Novel adult diarrhoea RV was mainly identified in humans (China and Bangladesh) and pigs from Brazil, USA, and South Africa (Alam et al. 2007; Jiang et al. 2008; Nagashima et al. 2008; Marthaler et al. 2014; Molinari et al. 2015). We identified RVH reads in two different bat pools, including one near complete genome. Based on genetic relatedness, all the eleven bat RVH segments recovered were distantly related to those of human (19–74% aa identity) and porcine (29–80% aa identity) RVH strains. The eleven bat RVH gene segments were more or less equally distantly related to both human and porcine RVH strains. Similarly, based on the phylogenetic analysis, bat, porcine, and human RVH strains formed three distinct sub-clusters distantly related to each other (Fig. 3). A recent paper described partial sequences of VP1, VP3 and VP4 RVH sequences (268, 340, and 202 nt, respectively) from South Korean bat fecal samples (Kim et al. 2016). These Korean bat RVH strains were only distantly related to human, porcine and the Cameroonian bat RVH strains (nt identity of 68–71%) described here, suggesting a broad genetic diversity of bat RVH strains (Supplementary Fig. S1). RVH was first identified as a cause of outbreaks of gastroenteritis in humans in China in 1997 and in a sporadic case of adult diarrhea in Bangladesh (Alam et al. 2007; Jiang et al. 2008). It has also been increasingly identified in pigs but this might simply be as a results of improved surveillance rather than recent spread. The current identification of RVH in bats further expands its host range and opens new perspetives on the evolutionary origin and history of this pathogen. It has been clearly associated with gastroenetritis in humans and less clearly in pigs as most known infections occur in co-infections with other RV species such as RVA, RVB, and RVC (Marthaler et al. 2014). Whether RVH causes diseases in bats is currently unknown.
Figure 3.
Phylogenetic trees constructed from nucleotide sequences of strain CMR62 and all other (near) complete RVH genome segments in GenBank. Tree was constructed using the GTR + G+I nucleotide substitution model using RAxML, with the autoMRE flag, which enables a posteriori bootstopping analysis. Trees were midpoint rooted for purposes of clarity. Only bootstrap values >70% are shown except at branches and clusters including the novel bat virus. Bars indicate nucleotide substitutions per site. Red, novel bat RVH; orange, porcine RVH; green, human RVH.
3.2 Identification of novel picobirna-like RNA-dependent RNA polymerase-encoding genome segments using the invertebrate mitochondrial genetic code
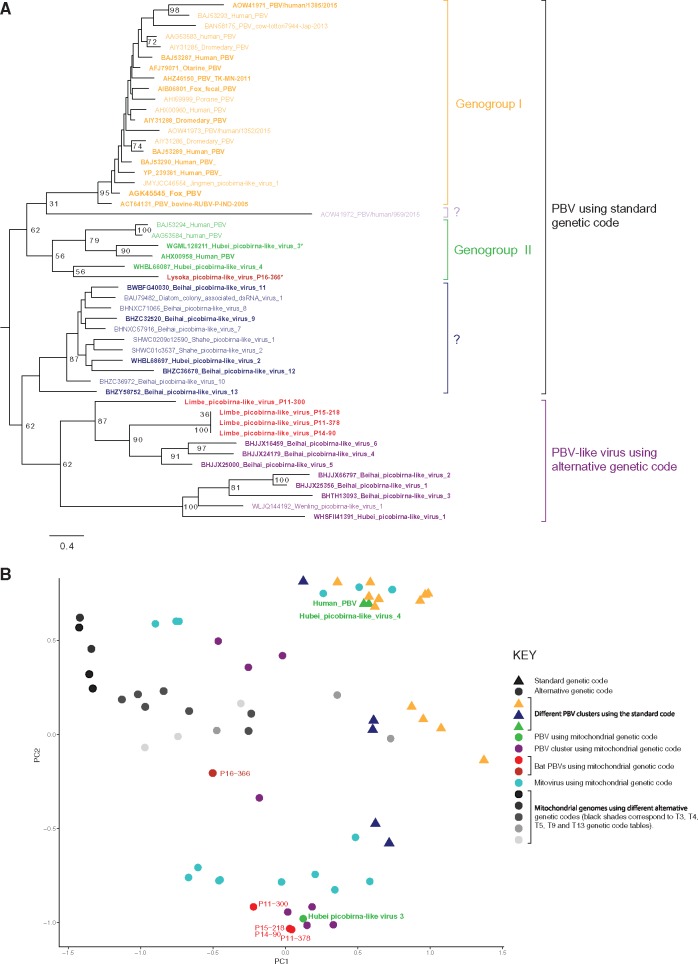
Picobirnavirus (PBV) is the only genus in the family Picobirnaviridae, and their members consist of non-enveloped, icosahedral virions containing a bisegmented double-stranded RNA genome of about 4 kb. The larger segment (≈2.2–2.7 kb) encodes a capsid precursor, and the smaller segment (≈1.2–1.9 kb) encodes the RNA-dependent RNA polymerase (RdRp) (Rosen et al. 2000). PBVs have been identified in feces of several mammals, birds, and invertebrates, and in the respiratory tract of pigs and humans (Pereira et al. 1988; Smits et al. 2011, 2012; Mondal and Majee 2014;). PBVs are currently classified into two genogroups (Smits et al. 2011; Malik et al. 2014) and recently, a large number of novel PBV-like sequences from insects have been described (Shi et al. 2016). Here, we identified five contigs with a PBV-like RdRp sequence. Surprisingly, these RdRP open reading frames (ORFs) could not be identified using the standard genetic translation code, and only when alternative genetic codes were used, a single large ORF could be identified. All these alternative genetic codes are those of invertebrate mitochondria and have in common a reassignment of the TGA stop to another amino acid (e.g. a tryptophan). An RdRp-based phylogenetic tree indicated that all PBV-like sequences in GenBank globally fall into two main clusters. One cluster mainly contains PBVs that use standard genetic code for protein translation, while the other clade contains PBV-like sequences that make exclusive use of the alternative genetic code for translation (Fig. 4A). Exceptions to this rule are the Chinese strain (Hubei picobirna-like virus 3) and the Cameroonian strain (Lysoka picobirna-like virus P16-366), both of which use an alternative genetic code, despite clustering within PBVs that use standard genetic code. In the clade of PBV-like viruses that use the alternative genetic code, three of the novel bat strains, P11-378, P14-90, and P15-218 share >98% aa identity and clustered with Beihai picobirna-like Virus 4, 5, and 6 identified in hermit crabs in China (Fig. 4A). The distant phylogenetic clustering of these strains with crustacean viruses, suggest either that these viruses are true crustacean viruses (instead of bat infecting viruses), or (more likely) that there is a lack of available sequence data of this virus from other potential host species, which does not allow us to make a reasonable assumption about its true host species at this moment. Only for the Cameroonian strain P16-366, a capsid sequence could be identified in our metagenomics data. In an attempt to identify the ‘missing’ highly divergent capsid sequences we used a custom build database containing all known PBV capsid genes and used HMMER to search the sequencing pools in which we found the PBV RdRps. However, no capsid sequences could be identified. As an internal control, we used the same procedure to successfully identify the previously identified capsid sequence in the pool containing picobirna-like virus P16-366. Furthermore, for the PBV-like RdRp sequences derived from crustaceans and also using the alternative codons, no capsids had been described either (Shi et al. 2016). This could also be because of the inability of the used methods (DIAMOND and HMMER motif finder search; Finn et al. 2011; Buchfink et al. 2015) to detect highly divergent capsid sequences.
Figure 4.
(A) Phylogenetic tree of the RdRp amino acid sequence of PBVs. Tree was constructed using the LG + G amino acid model using RAxML, with the autoMRE flag, which enables a posteriori bootstopping analysis. Tree was midpoint rooted for purposes of clarity. Only bootstrap values >70% are shown except at branches and clusters including the novel bat viruses. Bars indicate amino acid substitutions per site. Red strains: sequences from this study; orange strains: Genogroup I PBVs; green strains: genogroup II PBVs; light purple and blue: unclassified PBVs that use standard genetic code; purple: PBV-like viruses that use alternative genetic code; strains with asterisk: PBVs that use alternative genetic code in Genogroup II. (B) PCA based on the genetic code usage of viral and mitochondrial genomic sequences. Graphs represent separation of groups using the most influential factors and points represent values for individual sequences. P11-300, P11-378, P14-90, P15-218, and P16-366 are the novel Cameroonian PBVs.
The identification of these PBV-like RdRp sequences without an apparent capsid is reminiscent to that of mitoviruses (family Narnaviridae), which are known to infect the mitochondria of fungal species (Polashock and Hillman 1994; Hong et al. 1999). Their genome also consists of a double-stranded RNA element that encodes only an RdRp but does not form encapsulated viral particles. This virus genome is transmitted horizontally through mating (anastomosis) or vertically from mother to daughter cells (Shackelton and Holmes 2008). Apart from mitoviruses, only damselfish virus-like agents (a DNA virus) are known to infect mitochondria, although their mode of replication and transmissions is yet to be delineated (Schmale et al. 2009). To further investigate if indeed these PBV-like strains could be infecting the mitochondrion, we performed a principle component analysis (PCA) of the codon usage bias of different known mitochondrial genome sequences, mitoviruses, classical PBVs and PBV-like viruses using an alternative genetic code (Shi et al. 2016). Based on this analysis, the four PBV-like sequences identified in this study (P11-300, P11-378, P14-90, and P15-218) clustered closely together with other viruses using the alternative genetic code, such as mitoviruses, and other in crustacean/insect PBV-like viruses (Fig. 4B). The Cameroonian strain P16-366, clustered more closely with mitochondrial genomes (Fig. 4B). Most of the PBVs using a standard genetic code, clustered away from sequences using the alternative genetic codes, although there were some exceptions.
Based on the following combined observations for the Cameroonian PBV-like sequences P11-300, P11-378, P14-90, and P15-218: (1) a mitochondria like genetic code needed to translate the RdRp, (2) a potential absence of PBV-like capsid (after HMMER motif finder search; Eddy 2011), and (3) PCA analyses clustering them closely with mitoviruses, we speculate that these four PBV-like RdRp sequences might have a similar life style as mitoviruses, without an extracellular virion phase. However, it is not fully clear how to explain the observation that strain P16-366 uses an alternative genetic code, clusters with other sequences using the alternative code in the PCA analyses, and yet clusters with classical PBVs in the phylogenetic tree in genogroup II, and has a capsid sequence in the high-throughput sequencing data. Further analyses will have to shed light on this hypothesis.
3.3 Identification of novel divergent partiti-like viruses
Partitiviruses are viruses of the family Partitiviridae possessing a 30–35-nm diameter spherical particle which contains a bisegmented double-stranded RNA genome (Nibert et al. 2014). The larger genome segment (dsRNA1) codes for the RdRp while the smaller segment (dsRNA2) encodes one (or two) coat protein(s) (Buck and Ghabrial 1991). Also, these segments are encapsidated in separate virus particles, which must therefore, simultaneously infect the host for successful propagation (Buck and Kempson-Jones 1973; Szegő et al. 2010). Partitiviruses are known to infect plants, fungi and an apicomplexan protist of the genus Cryptosporidium and are classified into five genera based on phylogenetic analysis: Alphapartitivirus, Betapartitivirus, Gammapartitivirus, Deltapartitivirus, and Cryspovirus (Nibert et al. 2014). Here we identified thirty-nine partiti-like RdRp sequences. Surprisingly, only eight of the strains fall within these established genera: five of which clusters with fig cryptic virus (genus Deltapartitivirus) while the other three are closely related to viruses of the genus Gammapartitivirus. Twenty-eight of the sequences form several new and divergent clusters with a varying degree of relatedness to recently described unclassified invertebrate partiti-like viruses from China (Shi et al. 2016) (Supplementary Fig. S2). Overall, these data suggest a far greater diversity of viruses in this family, than was previously recognized, warranting the creation of multiple novel genera inside the family Partitiviridae. Here, capsid sequences were recovered only for fig crytic-like partitiviruses of the Deltapartitivirus genus and not for other sequences. This is most likely due to high genetic divergence, not allowing their identification using DIAMOND search. Given that partitiviruses are known to infect plants, fungi, and protist, it is most likely that these viruses are derived from the plants and fruits on which the bats feed.
4. Single-stranded RNA viruses
4.1 Identification of bat astrovirus and a novel bat bastro-like virus
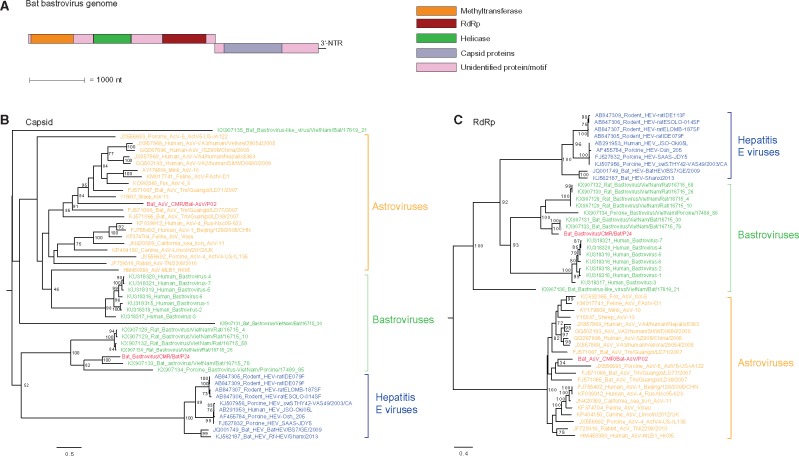
Recently, viral genomes with characteristics of both astro- and hepatitis E-like genomes (called bastrovirus) were identified in human stool samples from the Netherlands. Analysis of these genomes revealed they are made up of two ORFs: ORF1 contain non-structural domains that share identity with domains from members of the family Hepeviridae while ORF2 contain structural protein that shares the highest sequence identity with members of the Astroviridae (Oude Munnink et al. 2016). Further, unpublished sequences of bastrovirus and bastro-like viruses have been identified from rats, pigs, and bats from Vietnam. Here we describe a novel bastro-like virus in bats from Cameroon and analysis of the genome showed the same genome organization as those found in humans, rats, and other bats (Fig. 5A). A phylogeny based on the capsid (Fig. 5B) showed a cluster of human bastrovirus closest to astrovirus and distantly related to HEV strains. Most of the animal bastroviruses form a monophyletic clade except for the two highly divergent strains Bat_Bastrovirus-like_virus/VietNam/Bat/17819_21 and Bat_Bastrovirus/VietNam/Bat/16715_30. Inside this bastrovirus cluster, strains cluster according to their bat, rat or porcine host species, including the Cameroonian (Bat_Bastrovirus/CMR/Bat/P24) and Vietnamese (Bastrovirus/VietNam/Bat/16715_78) bat bastrovirus strains (77% amino acid similarity). The RdRp phylogenetic tree shows that bastroviruses and hepatitis E viruses form a unique clade, distinct from astroviruses (Fig. 5C). All bastroviruses form a monophyletic clade, and again they cluster per host species, including the Cameroonian and Vietnamese bat strains (KX907133 and KX907131, 77% amino acid identity). The only exception was Bat_Bastrovirus-like_virus/VietNam/Bat/17819_21, forming a distinct outgroup.
Figure 5.
Genome organization of bat bastrovirus (A). Phylogenetic tree of the capsid (B) and RdRp (C) amino acid sequence of bastrovirus and members of the Astroviridae and Hepeviridae families. Trees were constructed using the LG + G amino acid model using RAxML, with the autoMRE flag, which enables a posteriori bootstopping analysis. Trees were midpoint rooted for purposes of clarity. Only bootstrap values greater than 70% are shown except at branches and clusters including the novel bat virus. Bars indicate amino acid substitutions per site. Red taxa, sequences obtained in this study; orange clade, astroviruses; blue clade, hetatitis E viruses, green clades, bastroviruses.
Therefore, the capsid and the RdRp regions of bastrovirus are more phylogenetically related to those of astroviruses and hepatitis E viruses, respectively, confirming the previous findings, and the hypothesis of an (ancient?) recombination event (Oude Munnink et al. 2016). Furthermore, the observation that the Cameroonian strain CMR/Bat/P24 clustered with two bat strains from Vietnam in the RdRp tree and only with strain, Bat_Bastrovirus/VietNam/Bat/16715_30 in the capsid tree, suggests the occurrence of additional recombination events within the bastroviruses.
4.2 Identification of novel Betacoronavirus
CoVs are enveloped single-stranded, positive sense RNA viruses that belong to the Order Nidovirales, Family Coronaviridae and Subfamily Coronavirinae. This subfamily is made up of four genera: Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus (Adams and Carstens 2012). Gamma-CoV and Delta-CoV mainly contain bird viruses as well as a few mammalian viruses, whereas Alpha- and Beta-CoV infect mainly mammalian species (Drexler et al. 2014). Since bats were implicated as reservoir hosts for SARS-related CoVs, subsequent studies in bats have identified a great diversity of Alpha-CoV and Beta-CoV (Lau et al. 2005; Li et al. 2005). Furthermore, it has also been established that bats are potential reservoir host for the human CoV 229E and MERS-CoV, although it is now more plausible that the intermediate host of human MERS-CoV is dromedary camel (Annan et al. 2013; Chan et al. 2015; Corman et al. 2015; Sabir et al. 2015). The genus Beta-CoV is further classified into lineages A–D, and has been identified in a wide variety of hosts including humans, numerous domestic and peridomestic animals, and multiple bat species (41, 42). Infection of E. helvum with lineage D Beta-CoV has been reported at high frequency in Kenya and recently in Nigeria (Tong et al. 2009; Tao et al. 2012; Leopardi et al. 2016). Here we identified five near complete CoV genomes from E.helvum fruit bats in Cameroon. All the strains identified here belong to the lineage Beta-D. Phylogenetic analysis of at least 700 nt of the representative sequences of the subfamily Coronovirinae showed that in the Beta-D lineage, all strains (including four novel strains from this study) are generally clustered by host species (Supplementary Fig. S3). One exception was the Cameroonian strain CMR66, which clustered with Rousettus spp, D Beta-CoV strains, indicating that despite the existence of bat host-CoV co-evolution, there have been occasional interspecies events either within the Chiropteran order or to other orders (Tao et al. 2017).
5. Single-strand DNA viruses
5.1 Identification of novel viruses of the family Parvoviridae
Parvoviridae is a family of small non-enveloped viruses encoded by a single-stranded DNA genome of ∼4–6 kb with two ORFs: ORF1 encodes the non-structural protein(s) which has replicase activity, while ORF2 encodes the structural or capsid protein(s) (King et al. 2012).The family is further divided into the subfamilies Parvovirinae and Densovirinae. Members of the subfamily Parvovirinae infect vertebrates, while members of the subfamily Densovirinae infect invertebrates (Liu et al. 2011; Cotmore et al. 2014). The subfamily Parvovirinae is comprised of ten genera: Dependoparvovirus, Copiparvovirus, Bocaparvovirus, Amndoparvovirus, Aveparvovirus, Protoparvovirus, Tetraparvovirus, Erythroparvovirus, Marinoparvovirus, and Chapparvovirus (Cotmore et al. 2014; Phan et al. 2015; Yang et al. 2016). The subfamily Densovirinae is comprised of five genera: Ambidensovirus, Brevidensovirus, Hepandensovirus, Iteradensovirus, and Penstyldensovirus (Cotmore et al. 2014). We identified two parvovirus and five densovirus genomes in this study. The parvoviruses identified are BtBoV/CMR/2014 and BtPV/CMR/2014, which clustered together with members of the genera Bocaparvovirus and Chapparvovirus, respectively (Supplementary Fig. S4). Strain BtBoV/CMR/2014 was rather divergent from all other bocaviruses with <45% aa similarity with the closest bocavirus BBoV-SX/CHN/2010 (HQ223038) and BBoV-H18/CHN/2008 (HQ291308). On the other hand, BtPV/CMR/2014 had an aa identity of 49–65% with members of the genus Chapparvovirus and clustered closest to Ghanaian E.helvum parvovirus (BtPV2/GHN/2009, JX885610) with an aa identity of 65%. Both novel parvoviruses share <85% amino acid identity in the NS1 protein and therefore likely constitute new species within their respective genera (Cotmore et al. 2014).
We also identified five sequences of the subfamily Densovirinae named as Cameroonian bat densovirus 1-5 (CMRBtDV1-5). We had near complete genomes of CMRBtDV1-CMRBtDV4 (genome lengths of 3,823, 4,327, 4,327, and 5,017 nt respectively) but only 1,761 nt of CMRBtDV5. CMRBtDV1-CMRBtDV3 had a genome organization of two ORFs, ambisense to each other (Supplementary Fig. S5A) while that of CMRBtDV4 surprisingly showed four ORFs (two non-structural and two structural) instead of the typical two ORFs for ambidensoviruses (Supplementary Fig. S5B). This type of genome origination was also noticed in the closely related Dysaphis plantaginea densovirus isolate, DplDNV (FJ040397; Ryabov et al. 2009) and therefore this is unlikely to be a sequencing artifact. Among all the novel densoviruses, the closest to a known densoviruses was CMRBtDV4, which was nested within the genus Ambidensovirus with an aa identity of 52% with DpLDNV (Ryabov et al. 2009). The other four identified densoviruses formed two distinct distantly related clades with less than 20% aa identity to all other DVs (Supplementary Fig. S6). CMRBtDV2 and CMRBtDV3 are almost 99% identical and closest to CMRBtDV1 (44% aa identity), but with just 17% and 18% aa identity to CMRBtDV4 and CMRBtDV5, respectively. These two clades (CMRBtDV1, CMRBtDV2 and CMRBtDV3, and CMRBtDV5) constitute putative novel genera in the subfamily Densovirinae since both share <30% identity with all other DVs.
Parvoviruses have been identified in fecal and blood samples of bats around the world (Canuti et al. 2011; Ge et al. 2012; Kemenesi et al. 2015; Lau et al. 2016); however, none of these have been associated with any specific diseases (also the situation here). It is possible that these parvoviruses strains infect these bats without causing any diseases, as it is the case for many bat viruses. Unlike parvoviruses, the presence of densoviruses in bats is very unlikely to be associated with infection given that they have not been shown to infect vertebrates (Liu et al. 2011; Cotmore et al. 2014).
5.2 Identification of a novel single-stranded circular DNA virus
Circular Rep-encoding ssDNA (CRESS-DNA) viruses are a diverse group of viruses known to infect a wide range of hosts including crustaceans, fungi, a variety of plants (family Germiniviridae and Nanoviridae) and vertebrate species (family Circoviridae) (TFF et al. 2013; Yu et al. 2010). Recent studies have identified CRESS-DNA sequences in other environments including aquatic settings, insects and stool of animals (Dayaram et al. 2014; Zawar-Reza et al. 2014; Ng et al. 2015). Generally, the genomes of most CRESS-DNA viruses are 2–3 kb in length. Genomes of CRESS-DNA generally encode both a replication initiator protein (Rep) and a capsid protein (Cap), and contain a DNA stem loop structure required for the initiation of DNA replication (Stenger et al. 1991). Here we were able to obtain complete genomes of two CRESS-DNA sequences in different pools tentatively named Bat CyV-LimbeP14/CMR/2014 and Bat CyV-LysokaP4/CMR/2014 with genome length of 1,784 and 1,791 nt, respectively. Both genomes contain two ORFs encoding a Rep and capsid protein with small intergenic region between the 3′ ends of major ORFs. They also contain the circovirus nonanucleotide motif TAGTATTAC at the apex of a potential stem–loop structure (Supplementary Fig. S7A). They share 99% identity on the Rep gene and both have the highest amino acid and nucleotide identity (75 and 70%, respectively) to a human cyclovirus strain from Pakistan (GQ404845_CyV-PK5034/PAK/2007). The phylogenetic analysis of the Rep sequences (cyclovirus, Supplementary Fig. S7B) showed that both identified genomic sequences clustered with the above mentioned human strain. Also, cycloviruses have been found in bats from China and the USA (Li et al. 2010; Ge et al. 2011). Given that these were insect eating bats, the authors postulated that cycloviruses found in bat feces might infect insects consumed by bats rather than the bats themselves. This same assumption was used to explain the presence of cyclovirus found in humans stool (Rosario et al. 2012). Following this argument, their presence in fruit-eating bats highlight the possibility of consumption of insect-contaminated fruits. However, the real hosts of these viruses are yet to be determined.
6. Bacteriophages
There were 498 contigs annotated as phages by VirSorter. Further comparison to GenBank showed that they belonged to the Myoviridae (74 contigs), Podoviridae (22 contigs), and Siphoviridae (20 contigs) families, whereas the rest were unassigned (382 contigs). To assess differences in the bacteriophage communities of different bats, we compared the bacteriophage richness between female and male bats (Supplementary Fig. S8A, P = 0.657); adult and young bats (Supplementary Fig. S8B, P = 0.109) and between the three locations sampled (Supplementary Fig. S8C). Interestingly, Lysoka presented a significantly higher richness than Limbe, but not Moyuka, which is likely due to the low sample size from Moyuka bats. To further look into the location differences, we then clustered the three different locations on a PCoA using a binary jaccard matrix distance (Supplementary Fig. S8D). We observed a distinct clustering of the three locations (R2 = 0.3553, P = 0.001). Even though we cannot determine the putative drivers of these differences, we can speculate that these locations might have different fruits accessible, leading to a distinct gut flora in the bats. Furthermore, the diversity of these phages also reflects the bacterial flora harbored inside the bats, suggesting that bats from these locations might have quite diverse microbiota. However, given the disparity in the sampling number at different sites, this should be interpreted with caution. Using searches with CRISPR sequences against the phage community revealed that the most likely hosts of these phages are bacteria of the genera Enterobacter, Enterococcus, Escherichia, Klebsiella, Veillonella, and Salmonella (Table 2).
Table 2.
Results of CRISPR sequences blast against the phage community revealing the presence of bacteria of the genera Enterobacter, Enterococcus, Escherichia, Klebsiella, Veillonella, and Salmonella.
| Pool | Contig sizea | Spacer CRISPR sequence | E-valueb | Accession no.c | Bacteria species | Family |
|---|---|---|---|---|---|---|
| P01 | 37308 | CCAGCGGGAATGTGCCGGGTTCAACTGGACGC | 7.05e-09 | NZ_CP007731 | Klebsiella pneumoniae subsp. pneumoniae KPNIH27 | Enterobacteriaceae |
| P01 | 18455 | CTGTTGGCAAGCCAGGATCTGAACAATACCGT | 4.21e-11 | NZ_CP018957 | Escherichia coli strain Ecol_316 | Enterobacteriaceae |
| P01 | 22199 | CCTGATTGATGGCTTCTTTGATGTCAAACCGA | 4.21e-11 | NZ_CP018957 | Escherichia coli strain Ecol_316 | Enterobacteriaceae |
| P02 | 41734 | GGTTCAAATCCTCTCGTGCCGACCAAAAACAC | 4.21e-11 | NC_009800 | Escherichia coli HS | Enterobacteriaceae |
| P02 | 38384 | GGTCCGACTGCGAGCGGTCAGAAATCAATTTTAGGG | 3.24e-13 | NC_013520 | Veillonella parvula DSM 2008 | Veillonellaceae |
| P02 | 41734 | AGCCAGTCCGCATCTCGCCAAAAACCGTTAAC | 4.21e-11 | NC_013850 | Klebsiella michiganensis strain KCTC 1686 | Enterobacteriaceae |
| P02 | 41734 | CCAGCGGGAATGTGCCGGGTTCAACTGGACGC | 4.21e-11 | NZ_CP007731 | Klebsiella pneumoniae subsp. pneumoniae KPNIH27 | Enterobacteriaceae |
| P02 | 41734 | ATCGTGGCGATCAGGTGTGACACCTCGGAAGA | 5.45e-10 | NZ_CP017184 | Enterobacter cloacae complex ‘Hoffmann cluster IV’ strain DSM 16690 | Enterobacteriaceae |
| P07 | 12819 | GCAAAATACCAGGTCGCCCCAATCTGTCGCGA | 1.96e-09 | NC_016612 | Enterococcus faecalis str. Symbioflor 1 | Enterobacteriaceae |
| P07 | 29346 | ATGAACTAAATGTTTCAGATTCTCAAATAA | 6.55e-09 | NC_019770 | Salmonella enterica subsp. enterica serovar Montevideo | Enterococcaceae |
| P16 | 22046 | AGCCAGTCCGCATCTCGCCAAAAACCGTTAAC | 4.21e-11 | NC_013850 | Klebsiella variicola At-22 | Enterobacteriaceae |
| P16 | 22046 | CCAGCGGGAATGTGCCGGGTTCAACTGGACGC | 4.21e-11 | NZ_CP007731 | Klebsiella pneumoniae subsp. pneumoniae KPNIH27 | Enterobacteriaceae |
| P16 | 22046 | TAGCCAACCTGTAAGTAATCGTGATGAGTTGC | 4.21e-11 | NZ_CP019403 | Salmonella enterica subsp. enterica serovar Apapa str. SA20060561 | Enterobacteriaceae |
| P17 | 8125 | GAGGGGAATGGACAAGGAACGGAGAACACTTCAGACCA | 2.78e-14 | NC_013520 | Veillonella parvula DSM 2008 | Veillonellaceae |
| P18 | 19208 | AAGAGGAAAAGGAATTTCGCCACTGTATATGG | 4.21e-11 | NZ_CP007222 | Salmonella enterica subsp. enterica serovar Montevideo | Enterobacteriaceae |
Contigs identified as phage by Virsorter.
E-value the CRISPR spacer and phage.
Accession number of bacteria species corresponding to the CRISPR spacer.
7. Discussion/Conclusion
In the recent past, there has been an increased interest in bat viruses from all over the world because of the implications of bats as a reservoir for deadly viruses like rabies virus, Marburg virus, SARS- and MERS-related CoVs, and Nipah and Hendra virus. Many virome studies have identified numerous viruses in different viral families around the world (Donaldson et al. 2010; Li et al. 2010; Ge et al. 2012; He et al. 2013; Ng et al. 2013; Chen et al. 2014; O’Shea et al. 2014; Wu et al. 2016). Most of these studies have been conducted in China and the USA and most of the viruses identified from Africa have been limited to individual virus screening or isolation (Razafindratsimandresy et al. 2009; Drexler et al. 2012, 2013; Weiss et al. 2012; Baker et al. 2013; Quan et al. 2013; Corman et al. 2015; Tao et al. 2017; Waruhiu et al. 2017). Of all these, only two studies report screening of bat viruses in Cameroon with the identification of hepaciviruses, pegiviruses and alphaherpesviruses (Razafindratsimandresy et al. 2009; Quan et al. 2013). Different from these previous reports, this study characterize the entire fecal virome of one of the most eaten bat species (E.helvum) in Cameroon (Mickleburgh et al. 2009) using high-throughput sequencing.
Most of the viral reads recovered were assigned to bacteriophages and this is consistent with previous reports as reviewed in (Clokie et al. 2011). The other viral reads were eukaryotic and belonged to viruses of the order Picornavirales and the families Astroviridae, Caliciviridae, Circorviridae, Coronaviridae, Papillomaviridae, Parvoviridae Picobirnaviridae, and Reoviridae. Apart from CoVs, one strain of RVA (Yinda et al. 2016b), bat parvovirus and bat cyclovirus identified here were rather divergent from known viruses, suggesting that there is still a large number of unidentified viruses present in bats. Only one pool (P16, Table 1) was constituted of E.gambianus samples. Though this pool had a large amount of viral reads, most were phages and almost no complete mammalian virus was present in the pool. Therefore, all the novel eukaryotic viruses described here were identified in E. helvum and since only one pool was made from E. gambianus fruit bat, no reasonable comparison could be made between viruses identified in the different bat species. The presence of a vast spectrum of mammalian viruses in E. helvum reveals that this bat species may indeed also act as reservoirs for diverse mammalian viruses in Cameroon.
The identification of Rotavirus H (RVH) in bats from Cameroon and Korea (Kim et al. 2016), further broadens the breath of viruses found in bats so far. Given that this is the first near complete RVH genome identification in bats, more in vivo and extensive molecular epidemiologic studies are required to completely understand their genetic diversity and geographical spread. Furthermore, the phylogeny of RVH shows clear distinct clades of human, porcine and bat indicating that no interspecies transmission of this virus occurred in the recent past. In the same light, the presence of bastrovirus previously described only in humans (Oude Munnink et al. 2016) further indicates the increasing breath of viruses in bats. On the other hand, the bastrovirus capsid and the RdRp trees show different topologies and are phylogenetically related to astroviruses and hepatitis E viruses, respectively. This is in line with previous findings that hypothesize a recombination event between members of the family Astroviridae and Hepeviridae in the distant past (Oude Munnink et al. 2016). Furthermore, we observed phylogenetic incongruence within the bat bastroviruses, suggesting the occurrence of additional recombination events. These recombinations indicate that multiple bastroviruses may co-circulate and also co-infect the same host. A better understanding of the evolutionary history, host range and zoonotic potential of bastroviruses will be revealed without a doubt with ongoing and future virome studies in humans and animals. Whether or not these strains of RVH and bastrovirus can cause diseases in bats still need to be investigated.
For several viruses, there seems to be a taxon (species or genus)-specific host restriction. For bat CoVs, a single epidemiological unit of continental populations of E.helvum seems to exist (Gloza-Rausch et al. 2008; Peel et al. 2013; Razanajatovo et al. 2015). Also, the parvovirus strain BtPV/CAM/2004 is genetically related to a Ghanaian E.helvum strain and a similar observations was made for RVA, where a RVA strain from Cameroonian E.helvum bat was related to a known Kenyan strain (Yinda et al. 2016b). However, the presence of the novel E.helvum CoV strain CMR66 in an exclusively Rousettus spp. clade (Supplementary Fig. S4) shows an example of interspecies transmission between bat species. Such occasional interspecies events either within the Chiropteran order or to other orders, account for emerging viral zoonotic outbreaks like SARS and MERS (Tao et al. 2017). This is particularly easy in regions where there is close proximity between human and bats as is the case in the Southwest Region of Cameroon (Mickleburgh et al. 2009).
In addition to these well studied virus families, most of the novel densoviruses and partitiviruses are phylogenetically divergent from existing genera of their respective sub-family (Densovirinae) or family (Partitiviridae). This indicates the need for the creation of more genera within this (sub) family.
Additionally, we described for the first time in bat, picobirna-like sequences that use an alternative invertebrate mitochondrial genetic code. This observation together with our PCA analyses and the lack of an identifiable capsid sequence suggests that these PBV-like sequences might behave like mitoviruses, infecting mitochondria and not having an extracellular virion phase. Though experimental characterization of these viruses is needed to test this hypothesis, recent identification of bacterial ribosomal binding site in PBV genomes may suggest that prokaryotes are the potential hosts of PBVs (Krishnamurthy and Wang 2018). Therefore, the question on what the true host of PBVs is still to be fully answered.
Eidolon helvum mainly feeds on fruit juice. In addition, they are reported to chew up soft wood and bark, apparently to obtain moisture (Happold 1987). The region were these samples were collected are regions characterized by the presence of plantations and dense tropical forest which represent their prime habitat (Ossa et al. 2012; Benneh and DeLancey 2017). Therefore, these identified plant/insect viruses might just reflect the consumption of virus infected fruits or leaves containing insects, larvae, or eggs infected with these viruses. However, the virome of these plants and insects are yet to be investigated, hence no final conclusion can be made on the true host origin of these potential plant/fungi/insect associated viruses.
Nearly all animal virome studies ignore the bacteriophage content of stools. Here, we investigated the bat phageome, which revealed a great geographical diversity in the bat phage populations. Furthermore, the corresponding potential host bacterial species of these phages are similar to those that also live in the human gut. This raises the question if bacteriophages can also be transmitted from animals to humans. Whether this occurs and whether or not this might influence the human gut microbiota remains the subject for further studies.
Noticeably, all bats from which samples were collected appeared healthy and showed no overt signs of disease, further suggesting that bats can either co-adapt or tolerate diverse viruses through their unique metabolic and immune systems (Shi 2010).
8. Methods
8.1 Sample collection and preparation
Fecal samples were collected from Cameroonian fruit-eating bats, E.helvum and E.gambianus from Limbe (4°1′20.876″N 9°11′43.595″E), Lysoka (4°8′53.463″N 9°14′43.087″E), and Moyuka (4°17′25.625″N 9°24′52.186″E) in Southwest Region of Cameroon between December 2013 and May 2014. Bats were captured around fruit trees at night using mist nets as previously described in Yinda et al. (2016a,b). Morphological features (weight, forearm length, sex, reproductive state, and age [adult or juvenile]) of captured bats were assessed and used to determine the species as described in (Yinda et al. 2017b). None of the captured animals showed physical signs of disease. Samples were temporally kept at −20°C at Biotechnology Unit, University of Buea, Cameroon and later transferred to the Laboratory of Clinical and Epidemiological Virology, Leuven, Belgium and stored at −80°C. A total of eighty-seven fecal samples were collected (eighty-five from E.helvum and two from E.gambianus) from which twenty-five pools (of one to five samples per pool) were made based on age, sex, and location. To enrich for viral particles these pools were treated using the NetoVIR protocol (Conceicao-Neto et al. 2015) and sequenced on the Illumina HiSeq 2500 platform for 300 cycles (2 × 150 bp paired ends).
8.2 Genomic and phylogenetic analysis
NGS reads were analyzed as described in (Yinda et al. 2016a,b) Briefly, after raw reads were trimmed and de novo assembled using trimmomatic and SPAdes, respectively (Bankevich et al. 2012; Bolger et al. 2014), the assembled contigs were annotated by DIAMOND with the sensitive option using the GenBank’s non-redundant (nr) database (Buchfink et al. 2015). From the contigs, ORFs were identified and further analyzed for conserved motifs identification in the amino acid sequences using NCBI's conserved domain database (CDD) (Marchler-Bauer et al. 2015) and/or Pfam (Finn et al. 2014). To rule out the possibility of false positive, the presence of some of the viruses were confirmed on original samples by PCR (bastrovirus, CoV (66/2014/CMR, N704-P13), densovirus (CAMBtDV2, CAMBtDV4) and RVH; list of primers in Supplementary Table S2). Additionally, we checked for common contigs across our NGS runs which may indicate contamination from other samples or commercial kits and these were excluded from the analysis. Amino acid alignments were used for all trees except CoV and RVH alignments and for each of these nt alignments, a test for substitution saturation was performed using Dambe (Xia 2017). Alignments of viral sequences were made with Muscle implemented in MEGA7 (Molecular Evolutionary Genetics Analysis version 7) (Kumar et al. 2016) or MAFFT (Multiple Alignment using Fast Fourier Transform) (for Picobirnaviridae and Partiviridae, because of the high genetic diversity in these families) (Katoh et al. 2002). After trimming with trimAL (Capella-Gutiérrez et al. 2009), substitution models were determined using ModelGenerator (Keane et al. 2006) and phylogenetic trees constructed using RAxML (Stamatakis 2014), with the autoMRE flag, which enables a posteriori bootstopping analysis. All trees were visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and midpoint rooted for purposes of clarity. Only bootstrap values >70% are shown except at branches and clusters including the described bat virus. The choice of the sequence type (nucleotide or amino acid) for alignment and phylogenetic analysis was based on the genetic relatedness between the novel strains and reference strains, and the classification criteria of the viral family/genera to which the virus strain belongs.
8.3 Codon usage bias and PCA
To find support for the hypothesis that picobirna-like viruses possessing a genome that uses alternative codons, would infect mitochondria, we used a codon usage bias analysis. Briefly, we obtained PBVs, mitoviruses and mitochondria sequences that exhibit different genetic codes (3, 4, 5, 9, 10, 13, 14, 21, and 24 [https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi]) from GenBank. From these sequences, we obtained the codon usage bias using the codon usage tool (Stothard 2000). We excluded codons that vary between genetic codes and genetic codes for which there is only one sequence (10, 14, 21, and 24). A list of the accession numbers of the sequences used for PCA is in Supplementary Table S3. PCA was performed and the figure was created in R (R Core Team 2016) using the built-in stats and ggplot2 (Wickham 2009) packages.
8.4 Phageome analysis
For identification of bacteriophages, scaffolds >1 kb were classified using VirSorter (decontamination mode; Roux et al. 2015). Only scaffolds assigned to Categories 1 and 2 were considered bacteriophage contigs and were filtered for redundancy at 95% nt identity over 70% of the length using Cluster Genomes (Dios et al. 2014). Then, trimmed reads from each bat sample were mapped using Bowtie 2 (Langmead and Salzberg 2012) to the bacteriophage contigs and the generated BAM files were filtered to remove reads that aligned at <95% identity using BamM (http://ecogenomics.github.io/BamM/). Abundance tables were obtained and normalized for total number of reads. For the richness comparison, Mann-Whitney tests were used and for the clustering, an Adonis test was performed. All downstream analysis were done in R Core Team (2016) using the vegan package (Oksanen et al. 2017). Furthermore, to identify the potential corresponding bacterial species of the bacteriophage contigs identified by VirSorter, a database of this contigs was made to which a nucleotide BLASTn search (100% identity without gaps) was performed using a fasta file of CRISPR sequences (Grissa et al. 2007) as query.
8.5 Ethics approval and consent to participate
Ethical authorization for the protocol and the use of animal samples was obtained from the Cameroon National Ethics Committee, Yaoundé. All animal experiments were performed in accordance with the Ministry’s National Ethics Committee guidelines.
Data availability
All sequences were deposited in GenBank (accession numbers are in Supplementary Table S1). Raw reads were submitted to the NCBI’s Short Read Archive (SRA) under the project ID PRJNA344863.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
Supplementary Material
Acknowledgements
This work was supported by KU Leuven Grant EJX-C9928-StG/15/020BF awarded to Jelle Matthijnssens. C.K.Y. was supported by the Interfaculty Council for Development Cooperation (IRO) from the KU Leuven. N.C.N. and L.B. were supported by a PhD grant of Flanders Innovation & Entrepreneurship (VLAIO Vlaanderen).
References
- Adams M. J., Carstens E. B. (2012) ‘Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses (2012)’, Archives of Virology, 157: 1411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. M. et al. (2007) ‘Genetic Analysis of an ADRV-N-like Novel Rotavirus Strain B219 Detected in a Sporadic Case of Adult Diarrhea in Bangladesh’, Archives of Virology, 152: 199–208. [DOI] [PubMed] [Google Scholar]
- Annan A. et al. (2013) ‘Human betacoronavirus 2c EMC/2012-Related Viruses in Bats, Ghana and Europe’, Emerging Infectious Diseases, 19: 456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attoui H. et al. (2012) ‘Family: Reoviridae’, in King A.M.Q., Adams M. J., Carstens E. B., Lefkowitz E. J. (eds.) Virus Taxonomy: Ninth Report of the ICTV, pp. 541–563. Elsevier Amsterdam: Academic Press. [Google Scholar]
- Baker K. S. et al. (2013) ‘Novel, Potentially Zoonotic Paramyxoviruses from the African Straw-Colored Fruit Bat Eidolon Helvum’, Journal of Virology, 87: 1348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A. et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’, Journal of Computational Biology: A Journal of Computational Molecular Cell Biology, 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benneh G., DeLancey M. W. (2017) ‘Cameroon: Plant and Animal Life’ In: Encyclopædia Britannica. Encyclopædia Britannica, Inc; https://www.britannica.com/place/Cameroon/Plant-and-animal-life [Google Scholar]
- Bolger A. M. et al. (2014) ‘Trimmomatic: A Flexible Trimmer for Illumina Sequence Data’, Bioinformatics (Oxford, England), 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger J. C. et al. (1998) ‘Determinants of Rotavirus Host Range Restriction–a Heterologous Bovine NSP1 Gene Does Not Affect Replication Kinetics in the Pig’, Virology, 245: 47–52. [DOI] [PubMed] [Google Scholar]
- Buchfink B. et al. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60.Nature Research. [DOI] [PubMed] [Google Scholar]
- Buck K. W., Ghabrial S. A. (1991) ‘Family Partitiviridae’, in Francki R. I., Fauquet C. M., Knudson D. L., Brown F. (eds.) Classification and Nomenclature of Viruses: FifthReport of the International Committee on Taxonomy of Viruses, pp. 208–11. Springer, Verlag: New York. [Google Scholar]
- Buck K. W., Kempson-Jones G. F. (1973) ‘Biophysical Properties of Penicillium Stoloniferum Virus S’, The Journal of General Virology, 18: 223–35. [DOI] [PubMed] [Google Scholar]
- Calisher C. H. et al. (2006) ‘Bats: Important Reservoir Hosts of Emerging Viruses’, Clinical Microbiology Reviews, 19: 531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuti M. et al. (2011) ‘Two Novel Parvoviruses in Frugivorous New and Old World Bats’, (J.-P. Vartanian, Ed.)’, PLoS One, 6: e29140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S. et al. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F. W. et al. (2015) ‘Middle East Respiratory Syndrome Coronavirus: Another Zoonotic Betacoronavirus Causing SARS-like Disease’, Clinical Microbiology Reviews, 28: 465–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. (2014) ‘DBatVir: The Database of Bat-Associated Viruses’, Database : The Journal of Biological Databases and Curation, 2014: bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K. B. et al. (2002) ‘Isolation of Nipah Virus from Malaysian Island Flying-Foxes’, Microbes and Infection, 4: 145–51. [DOI] [PubMed] [Google Scholar]
- Cleaveland S. et al. (2001) ‘Diseases of Humans and Their Domestic Mammals: Pathogen Characteristics, Host Range and the Risk of Emergence’, Philosophical Transactions of the Royal Society B: Biological Sciences, 356: 991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie M. R. J. et al. (2011) ‘Phages in Nature’, Bacteriophage, 1: 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao-Neto N. et al. (2015) ‘Modular Approach to Customise Sample Preparation Procedures for Viral Metagenomics: A Reproducible Protocol for Virome Analysis’, Scientific Reports, 5: 16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M. et al. (2015) ‘Evidence for an Ancestral Association of Human Coronavirus 229E with Bats’, (S. Schultz-Cherry, Ed.)’, Journal of Virology, 89: 11858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S. F. et al. (2014) ‘The Family Parvoviridae ’, Archives of Virology, 159: 1239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A. et al. (2014) ‘Novel Circular DNA Viruses Identified in Procordulia Grayi and Xanthocnemis Zealandica Larvae Using Metagenomic Approaches’, Infection, Genetics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 22: 134–41. [DOI] [PubMed] [Google Scholar]
- Dios F. et al. (2014) ‘DNA Clustering and Genome Complexity’, Computational Biology and Chemistry, 53: 71–8. [DOI] [PubMed] [Google Scholar]
- Drexler J. F. et al. (2012) ‘Bats Host Major Mammalian Paramyxoviruses’, Nature Communications, 3: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J. F. et al. (2013) ‘Bats Carry Pathogenic Hepadnaviruses Antigenically Related to Hepatitis B Virus and Capable of Infecting Human Hepatocytes’, Proceedings of the National Academy of Sciences of the United States of America, 110: 16151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J. F. et al. (2014) ‘Ecology, Evolution and Classification of Bat Coronaviruses in the Aftermath of SARS’, Antiviral Research, 101: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2011) ‘Accelerated Profile HMM Searches’, PLOS Computational Biology, 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E. F. et al. (2010) ‘Metagenomic Analysis of the Viromes of Three North American Bat Species: Viral Diversity among Different Bat Species That Share a Common Habitat’, Journal of Virology, 84: 13004–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. (2011) ‘HMMER Web Server: Interactive Sequence Similarity Searching’, Nucleic Acids Research, 39: W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. (2014) ‘Pfam: The Protein Families Database’, Nucleic Acids Research, 42: D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. et al. (2011) ‘Genetic Diversity of Novel Circular ssDNA Viruses in Bats in China’, The Journal of General Virology , 92: 2646–53. [DOI] [PubMed] [Google Scholar]
- Ge X. et al. (2012) ‘Metagenomic Analysis of Viruses from Bat Fecal Samples Reveals Many Novel Viruses in Insectivorous Bats in China’, Journal of Virology, 86: 4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald K. T. et al. (Eds). (2009) ‘National Research Council (US) Committee on Achieving Sustainable Global Capacity for Surveillance and Response to Emerging Diseases of Zoonotic Origin’ Sustaining Global Surveillance and Response to Emerging Zoonotic Diseases. Washington, DC: National Academies Press (US; ). [PubMed] [Google Scholar]
- Gloza-Rausch F. et al. (2008) ‘Detection and Prevalence Patterns of Group I Coronaviruses in Bats, Northern Germany’, Emerging Infectious Diseases, 14: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I. et al. (2007) ‘The CRISPRdb Database and Tools to Display CRISPRs and to Generate Dictionaries of Spacers and Repeats’, BMC Bioinformatics, 8: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happold D. C. (1987) The Mammals of Nigeria. New York: Oxford University Press. [Google Scholar]
- He B. et al. (2013) ‘Virome Profiling of Bats from Myanmar by Metagenomic Analysis of Tissue Samples Reveals More Novel Mammalian Viruses’, (H. Tse, Ed.)’, PLoS One, 8: e61950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y. et al. (1999) ‘Multiple Mitochondrial Viruses in an Isolate of the Dutch Elm Disease FungusOphiostoma Novo-Ulmi’, Virology, 258: 118–27. [DOI] [PubMed] [Google Scholar]
- Jiang S. et al. (2008) ‘Molecular Characterization of a Novel Adult Diarrhoea Rotavirus Strain J19 Isolated in China and Its Significance for the Evolution and Origin of Group B Rotaviruses’, Journal of General Virology, 89: 2622–9. [DOI] [PubMed] [Google Scholar]
- Katoh K. (2002) ‘MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform’, Nucleic Acids Research, 30: 3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M. et al. (2006) ‘Assessment of Methods for Amino Acid Matrix Selection and Their Use on Empirical Data Shows That Ad Hoc Assumptions for Choice of Matrix Are Not Justified’, BMC Evolutionary Biology, 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenesi G. et al. (2015) ‘Genetic Diversity and Recombination Within bufaviruses: Detection of a Novel Strain in Hungarian Bats’, Infection, Genetics and Evolution, 33: 288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K. et al. (2016) ‘Detection of Severe Acute Respiratory Syndrome-like, Middle East Respiratory Syndrome-like Bat Coronaviruses and Group H Rotavirus in Faeces of Korean Bats’, Transboundary and Emerging Diseases, 63: 365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. et al. (2012) Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press. [Google Scholar]
- Krishnamurthy S. R., Wang D. (2018) ‘Extensive Conservation of Prokaryotic Ribosomal Binding Sites in Known and Novel Picobirnaviruses’, Virology, 516: 108–14. [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. (2016) ‘MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. P. et al. (2005) ‘Severe Acute Respiratory Syndrome Coronavirus-like Virus in Chinese Horseshoe Bats’, Proceedings of the National Academy of Sciences of the United States of America, 102: 14040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. P. et al. (2016) ‘Identification and Interspecies Transmission of a Novel Bocaparvovirus among Different Bat Species in China’, The Journal of General Virology, 97: 3345–58. [DOI] [PubMed] [Google Scholar]
- Leopardi S. et al. (2016) ‘The Close Genetic Relationship of Lineage D Betacoronavirus from Nigerian and Kenyan Straw-Colored Fruit Bats (Eidolon Helvum) Is Consistent with the Existence of a Single Epidemiological Unit across Sub-Saharan Africa’, Virus Genes, 52: 573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E. M. et al. (2005) ‘Fruit Bats as Reservoirs of Ebola Virus’, Nature, 438: 575–6. [DOI] [PubMed] [Google Scholar]
- Li L. et al. (2010) ‘Bat Guano Virome: Predominance of Dietary Viruses from Insects and Plants plus Novel Mammalian Viruses ’, Journal of Virology, 84: 6955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. (2005) ‘Bats Are Natural Reservoirs of SARS-like Coronaviruses’, Science, 310: 676. [DOI] [PubMed] [Google Scholar]
- Liu H. et al. (2011) ‘Widespread Endogenization of Densoviruses and Parvoviruses in Animal and Human Genomes ’, Journal of Virology, 85: 9863–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A. D. et al. (2013) ‘A Comparison of Bats and Rodents as Reservoirs of Zoonotic Viruses: Are Bats Special?’, Proceedings: Biological Sciences, 280: 20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey T. K. et al. (2014) ‘Emerging and Reemerging Neglected Tropical Diseases: A Review of Key Characteristics, Risk Factors, and the Policy and Innovation Environment’, Clinical Microbiology Reviews, 27: 949–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y. S. et al. (2014) ‘Epidemiology, Phylogeny, and Evolution of Emerging Enteric Picobirnaviruses of Animal Origin and Their Relationship to Human Strains’, Biomed Res Int, 2014: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. (2015) ‘CDD: NCBI’s Conserved Domain Database’, Nucleic Acids Research, 43: D222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D. et al. (2014) ‘Widespread Rotavirus H in Commercially Raised Pigs, United States’, Emerging Infectious Diseases, 20: 1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z. A. et al. (2013) ‘Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia’, Emerging Infectious Diseases, 19: 1819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickleburgh S. et al. (2009) ‘Bats as Bushmeat: A Global Review’, Oryx, 43: 217–34. [Google Scholar]
- Mihalov-Kovács E. et al. (2015) ‘Candidate New Rotavirus Species in Sheltered Dogs, Hungary’, Emerging Infectious Diseases, 21: 660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari B. L. et al. (2015) ‘Genetic Variability of VP6, VP7, VP4, and NSP4 Genes of Porcine Rotavirus Group H Detected in Brazil’, Virus Research, 197: 48–53. [DOI] [PubMed] [Google Scholar]
- Mondal A., Majee S. (2014) ‘Novel Bisegmented Virus (Picobirnavirus) of Animals, Birds and Humans’, Asian Pacific Journal of Tropical Disease, 4: 154–8. [Google Scholar]
- Morse S. S. (2001) ‘Factors in the Emergence of Infectious Diseases BT - Plagues and Politics: Infectious Disease and International Policy’, in Price-Smith A. T. (ed.), pp. 8–26. Palgrave Macmillan UK: London.
- Nagashima S. et al. (2008) ‘Whole Genomic Characterization of a Human Rotavirus Strain B219 Belonging to a Novel Group of the Genus Rotavirus’, Journal of Medical Virology, 80: 2023–33. [DOI] [PubMed] [Google Scholar]
- Ng T. F. F. et al. (2013) ‘Distinct Lineage of Vesiculovirus from Big Brown Bats, United States’, Emerging Infectious Diseases, 19: 1978–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. F. F. et al. (2015) ‘A Diverse Group of Small Circular ssDNA Viral Genomes in Human and Non-Human Primate Stools’, Virus Evolution, 1: vev017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M. L. et al. (2014) ‘Taxonomic Reorganization of Family Partitiviridae and Other Recent Progress in Partitivirus Research’, Virus Research, 188: 128–41. [DOI] [PubMed] [Google Scholar]
- Nowak R. (1991) ‘Order Chiroptera’, in Walker’s Mammals of the World , Vol. 1, 5th edn., pp. 190–4. Johns Hopkins University Press: Baltimore. [Google Scholar]
- O’Shea T. J. et al. (2014) ‘Bat Flight and Zoonotic Viruses’, Emerging Infectious Disease Journal, 20: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J. et al. (2017) ‘vegan: Community Ecology Package’. https://CRAN.R-project.org/package=vegan
- Ossa G. et al. (2012) ‘The Movement Ecology of the Straw-Colored Fruit Bat, Eidolon Helvum, in Sub-Saharan Africa Assessed by Stable Isotope Ratios’, (T. Deschner, Ed.)’, PLoS One, 7: e45729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B. B. et al. (2016) ‘A Novel Astrovirus-like RNA Virus Detected in Human Stool’, Virus Evolution, 2: vew005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar U. D. et al. (2006) ‘Rotavirus and Severe Childhood Diarrhea’, Emerging Infectious Diseases, 12: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel A. J. et al. (2013) ‘Continent-Wide Panmixia of an African Fruit Bat Facilitates Transmission of Potentially Zoonotic Viruses’, Nature Communications, 4: 2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira H. G. et al. (1988) ‘Novel Viruses in Human Faeces’, Lancet (London, England), 2: 103–4. [DOI] [PubMed] [Google Scholar]
- Phan T. G. et al. (2015) ‘Sesavirus: Prototype of a New Parvovirus Genus in Feces of a Sea Lion’, Virus Genes, 50: 134–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polashock J. J., Hillman B. I. (1994) ‘A Small Mitochondrial Double-Stranded (Ds) RNA Element Associated with a Hypovirulent Strain of the Chestnut Blight Fungus and Ancestrally Related to Yeast Cytoplasmic T and W dsRNAs’, Proceedings of the National Academy of Sciences of the United States of America, 91: 8680–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast B. J. et al. (2002) ‘Periodic Arousal from Hibernation Is Necessary for Initiation of Immune Responses in Ground Squirrels’, American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 282: R1054–62. [DOI] [PubMed] [Google Scholar]
- Quan P.-L. et al. (2013) ‘Bats Are a Major Natural Reservoir for Hepaciviruses and Pegiviruses’, Proceedings of the National Academy of Sciences of the United States of America, 110: 8194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2016) ‘R: A Language and Environment for Statistical Computing’. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Razafindratsimandresy R. et al. (2009) ‘Partial Molecular Characterization of Alphaherpesviruses Isolated from Tropical Bats’, Journal of General Virology, 90: 44–7. [DOI] [PubMed] [Google Scholar]
- Razanajatovo N. H. et al. (2015) ‘Detection of New Genetic Variants of Betacoronaviruses in Endemic Frugivorous Bats of Madagascar’, Virology Journal, 12: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K. et al. (2012) ‘Diverse Circular ssDNA Viruses Discovered in Dragonflies (Odonata: Epiprocta)’, Journal of General Virology, 93: 2668–81. [DOI] [PubMed] [Google Scholar]
- Rosen B. I. et al. (2000) ‘Cloning of Human Picobirnavirus Genomic Segments and Development of an RT-PCR Detection Assay’, Virology, 277: 316–29. [DOI] [PubMed] [Google Scholar]
- Roux S. et al. (2015) ‘VirSorter: mining viral signal from microbial genomic data’, PeerJ, 3: e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht C. E. et al. (1995) ‘The Ascension of Wildlife Rabies: A Cause for Public Health Concern or Intervention?’, Emerging Infectious Diseases, 1: 107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov E. V. et al. (2009) ‘Densovirus Induces Winged Morphs in Asexual Clones of the Rosy Apple Aphid, Dysaphis Plantaginea’, Proceedings of the National Academy of Sciences of the United States of America, 106: 8465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J. S. M. et al. (2015) ‘Co-Circulation of Three Camel Coronavirus Species and Recombination of MERS-CoVs in Saudi Arabia’, Science, 351: 81 LP–4. [DOI] [PubMed] [Google Scholar]
- Schmale M. C. et al. (2009) ‘25. A Parasite of Mitochondria? a Virus-like Agent in Neurogenic Tumors of a Tropical Marine Fish’, Mitochondrion, 9: 67. [Google Scholar]
- Shackelton L. A., Holmes E. C. (2008) ‘The Role of Alternative Genetic Codes in Viral Evolution and Emergence’, Journal of Theoretical Biology, 254: 128–34. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2016) ‘Redefining the Invertebrate RNA Virosphere’, Nature, 540: 539–43. [DOI] [PubMed] [Google Scholar]
- Shi Z. (2010) ‘Bat and Virus’, Protein and Cell, 1: 109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S. L. et al. (2011) ‘Genogroup I and II Picobirnaviruses in Respiratory Tracts of Pigs’, Emerging Infectious Diseases, 17: 2328–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S. L. et al. (2012) ‘Picobirnaviruses in the Human Respiratory Tract’, Emerging Infectious Diseases, 18: 1539–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics (Oxford, England), 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D. C. et al. (1991) ‘Replicational Release of Geminivirus Genomes from Tandemly Repeated Copies: Evidence for Rolling-Circle Replication of a Plant Viral DNA’, Proceedings of the National Academy of Sciences of the United States of America, 88: 8029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. (2000) ‘The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences’, Biotechniques, 28: 1102–4. [DOI] [PubMed] [Google Scholar]
- Szegő A. et al. (2010) ‘The Genome of Beet Cryptic Virus 1 Shows High Homology to Certain Cryptoviruses Present in Phylogenetically Distant Hosts’, Virus Genes, 40: 267–76. [DOI] [PubMed] [Google Scholar]
- Tao Y. et al. (2012) ‘Genomic Characterization of Seven Distinct Bat Coronaviruses in Kenya’, Virus Research, 167: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y. et al. (2017) ‘Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History’, Journal of Virology, 91: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L. H. et al. (2001) ‘Risk Factors for Human Disease Emergence’, Philosophical Transactions of the Royal Society B: Biological Sciences, 356: 983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TFF N. et al. (2013) ‘Metagenomic Identification of a Nodavirus and a Circular ssDNA Virus in Semi-Purified Viral Nucleic Acids from the Hepatopancreas of Healthy Farfantepenaeus Duorarum Shrimp ’, Diseases of Aquatic Organisms, 105: 237–42. [DOI] [PubMed] [Google Scholar]
- Tong S. et al. (2009) ‘Detection of Novel SARS-like and Other Coronaviruses in Bats from Kenya’, Emerging Infectious Diseases, 15: 482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J. S. et al. (2007) ‘Marburg Virus Infection Detected in a Common African Bat’, PLoS One, 2: e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan T., Ryan J. C. N. (2000) Mammalogy, 4th edn Toronto: Brooks Cole. [Google Scholar]
- Waruhiu C. et al. (2017) ‘Molecular Detection of Viruses in Kenyan Bats and Discovery of Novel Astroviruses, Caliciviruses and Rotaviruses’, Virologica Sinica, 32: 101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. et al. (2012) ‘Henipavirus-Related Sequences in Fruit Bat Bushmeat, Republic of Congo’, Emerging Infectious Diseases, 18: 1536–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wu Z. et al. (2012) ‘Virome Analysis for Identification of Novel Mammalian Viruses in Bat Species from Chinese Provinces’, Journal of Virology, 86: 10999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. (2016) ‘Deciphering the Bat Virome Catalog to Better Understand the Ecological Diversity of Bat Viruses and the Bat Origin of Emerging Infectious Diseases’, ISME Journal, 10: 609–20. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. (2017) ‘DAMBE6: New Tools for Microbial Genomics, Phylogenetics, and Molecular Evolution’, The Journal of Heredity, 108: 431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. et al. (2016) ‘A Novel Rodent Chapparvovirus in Feces of Wild Rats’, Virology Journal, 13: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C. K. et al. (2016a) ‘A Single Bat Species in Cameroon Harbors Multiple Highly Divergent Papillomaviruses in Stool Identified by Metagenomics Analysis’, Virology Reports, 6: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C. K. et al. (2016b) ‘Novel Highly Divergent Reassortant Bat Rotaviruses in Cameroon, without Evidence of Zoonosis’, Scientific Reports, 6: 34209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C. K. et al. (2017a) ‘Highly Diverse Population of Picornaviridae and Other Members of the Picornavirales, in Cameroonian Fruit Bats’, BMC Genomics, 18: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C. K. et al. (2017b) ‘Novel Highly Divergent Sapoviruses Detected by Metagenomics Analysis in Straw-Colored Fruit Bats in Cameroon’, Emerging Microbes and Infections, 6: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. et al. (2010) ‘A Geminivirus-Related DNA Mycovirus That Confers Hypovirulence to a Plant Pathogenic Fungus’, Proceedings of the National Academy of Sciences of the United States of America, 107: 8387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawar-Reza P. et al. (2014) ‘Diverse Small Circular Single-Stranded DNA Viruses Identified in a Freshwater Pond on the McMurdo Ice Shelf (Antarctica)’, Infection, Genetics and Evolution, 26: 132–8. [DOI] [PubMed] [Google Scholar]
- Zheng X. et al. (2017) ‘Viral Metagenomics of Six Bat Species in Close Contact with Humans in Southern China’, Archives of Virology, 163: 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences were deposited in GenBank (accession numbers are in Supplementary Table S1). Raw reads were submitted to the NCBI’s Short Read Archive (SRA) under the project ID PRJNA344863.