Abstract
The CDK inhibitor p27Kip1 plays a central role in controlling cell proliferation and cell-cycle exit. p27Kip1 protein levels oscillate during cell-cycle progression and are regulated by mitogen or anti-proliferative signaling. The abundance of the protein is frequently determined by post-transcriptional mechanisms including ubiquitin-mediated proteolysis and translational control. Here, we report that the cold-inducible RNA-binding protein (CIRP) selectively binds to the 5′ untranslated region of the p27Kip1 mRNA. CIRP is induced, modified and relocalized in response to various stress stimuli and can regulate cell survival and cell proliferation particularly during stress. Binding of CIRP to the 5′UTR of the p27Kip1 mRNA significantly enhanced reporter translation. In cells exposed to mild hypothermia, the induction of CIRP correlated with increased translation of a p27Kip1 5′UTR reporter and with the accumulation of p27Kip1 protein. shRNA-mediated CIRP knockdown could prevent the induction of translation. We found that p27Kip1 is central for the decreased proliferation at lower temperature, since p27Kip1 KO mouse embryonic fibroblasts (MEFs) hardly increased their doubling time in hypothermic conditions, whereas wild-type MEFs significantly delayed proliferation in response to cold stress. This suggests that the CIRP-dependent p27Kip1 upregulation during mild hypothermia contributes to the cold shock-induced inhibition of cell proliferation.
INTRODUCTION
Various and in part conflicting endogenous and environmental signals and cues need to be integrated into the decision of cells to either proliferate or to withdraw from the cell cycle and enter into quiescence or terminally differentiate. The CDK inhibitor p27Kip1 plays a central role in these processes by controlling the CDK activation at the restriction point in G1 phase (1–3). Numerous signals impinge on p27 transcription, translation, stability or activity (1,4). Levels of p27 are critical to permit or restrict CDK activation and cell proliferation. Accordingly, p27 was found to be haplo-insufficient for tumor suppression (5). Mice deficient in p27 expression are characterized by multiorgan hyperplasia and increased body size and develop pituitary tumors spontaneously (6). Consistent with these observations, decreased p27 levels can correlate with a poor prognosis in various human cancers (1). Interestingly, a mutant p27 protein that fails to bind CDK/cyclin complexes possesses oncogenic properties (7).
p27 has an increasing number of CDK-independent functions. It regulates microtubule stability and it can prevent full activation of H-Ras and cell-cycle entry (6). The intrinsically unstructured protein influences cell migration and invasion by interacting with RhoA and stathmin (6). Recently, p27 was found to regulate transcription in a CDK-dependent and CDK-independent manner (6,8). Elevated levels of p27 can prevent CDK activation and cell-cycle progression (1,9). During G0 and G1 phase of the cell cycle, p27 binds to and regulates the activity of cyclin D/CDK4,6 and cyclin E/CDK2 complexes (1,10). Levels of p27 decline as cells progress over the restriction point. Cyclin/CDK complexes phosphorylate p27 on T187; the phosphorylated p27 is ubiquitinated by the SCF-Skp2 ubiquitin E3 ligase, triggering its proteasomal degradation (1). This degradation of p27 initiates a positive feedback loop that leads to robust CDK activation (3). Usually, p27 remains unstable throughout the remainder of the cell cycle, until CDK kinase activity declines in late mitosis, permitting the re-accumulation of p27. The feedback loop of CDK-induced p27 degradation consolidates the irreversibility of the transition from G1 toward S phase.
Multiple signals contribute to the control of p27 levels in G1 phase (1,4). In addition to transcriptional regulation, inactivation and degradation or cytoplasmic relocalization, translational control can regulate the p27 threshold prior to the restriction point passage. Interestingly, the abundance of p27 mRNA remains frequently constant throughout the cell cycle, whereas the rate of p27 translation is enhanced in quiescent cells (11–13) and can promote differentiation in various cell lines (14–16).
Both untranslated regions (UTRs) of the p27 transcript are targets of translational control. The 3′UTR contains binding sites for microRNAs (miRNAs) such as miR-221 and miR-222 (17), that lead to the destabilization of the transcript. Binding of miRNAs to the p27 transcript is modulated by RNA-binding proteins (RBPs) such as Dnd1, CPEB1 and PUM1, that prevent (18,19) or facilitate (20) the association of the miRNAs to the target regions in the p27 3′UTR.
The largest 5′UTR identified consists of 575 nt (21). Its sequence is highly conserved in vertebrates and the human and murine p27 5′UTRs share a sequence identity of 78%. The 5′UTR of the p27 mRNA is characterized by the presence of a conserved short upstream open reading frame (uORF), which partially overlaps with a cell-cycle regulatory element (CCRE). The CCRE is needed for increased translation of p27 during G1 phase (13). The major transcription start site is conserved in mice and humans and generates a 5′UTR of 472 nt in human cells (22). An internal ribosome entry site (IRES) was identified preceding the ATG start codon (16,21). However, due to cryptic promoter activities, the existence of the IRES had been an issue of debate (23–27). The p27 IRES was proposed to be responsible for impaired translation of p27 in patients with X-linked Dyskeratosis Congenita, caused by defective ribosomal RNA modification (28,29). A U-rich region (nucleotides −66 to −40) is located upstream of the initiation codon and has been demonstrated to be important for efficient translation in proliferating and non-proliferating cells (30) and represents a ribosome entry site (27). Several RBPs, including HuR (21), PTB (24), FBP1 (31) and CUGBP1 (32) bind to this region and modulate p27 translation.
RBPs play an important role in fine-tuning gene expression. A large and diverse family of RBPs is represented by the heterogeneous nuclear ribonucleoproteins (hnRNPs) (33), whose members are not only involved in processing pre-mRNAs into mature RNAs, but also act as trans-factors to influence mRNA export, localization, translation and stability (34).
We here identify cold-inducible RNA-binding protein (CIRP) (A18 hnRNP, Cirbp) as a novel p27 mRNA-binding protein. CIRP is composed of a conserved N-terminal RNA binding domain and several repeats of arginine-glycine-glycine within the C-terminal half. CIRP was the first cold shock protein identified in mammals (35,36). The protein becomes induced in response to mild hypothermia (e.g. lowering the temperature to 32°C), but not in severe hypothermia (e.g. 20°C and below). Other stress conditions including hypoxia and UV also induce CIRP protein expression (37–39). Stress such as UV irradiation (40), oxidative stress, osmotic pressure, heat shock or endoplasmic reticulum stress (41) can promote the translocation of CIRP from the nucleus to the cytoplasm or to cytoplasmic stress granules.
The functions of CIRP are cell type specific and have been linked to cellular and environmental states. In hypothermic conditions, CIRP regulates telomerase activity (42) and exerts anti-apoptotic effects in the brain (43–46); it mediates airway inflammatory response (47) and regulates spermatogenesis in the testis, where it is constitutively expressed (48–50). CIRP can regulate the ERK pathway and thereby acts as a pro-survival factor (51,52); moreover, it is involved in the control of the circadian rhythm (53–55). Interestingly, CIRP can also be released from cells and promote inflammatory responses in shock, sepsis, or upon brain damage, characterizing it as a damage-associated molecular pattern molecule (56–58). In unperturbed somatic cells, CIRP is predominantly nuclear. CIRP has been reported to bind to the 3′UTR of target transcripts, some of which are involved in the stress response, cell survival (40,59,60) or inflammation (47). Binding of CIRP to RPA2, TRX, ATR, HIF-1α and inflammatory cytokine transcripts stabilizes them and induces their translation, particularly under stress conditions (47,60).
CIRP mediates a cold-inducible growth arrest of diverse cell lines of rodent or human origin, including BALB/3T3 (35), NIH/3T3 or U2OS cells (48). However, in erythropoietin producing CHO cells, down regulation of CIRP failed to rescue hypothermic growth (61). Interestingly, CIRP knockout mice subsequently revealed that CIRP can even promote cell proliferation in mouse germ cells and in cultured mouse fibroblasts, where CIRP accelerates the G0 or G1- to S-phase transition (48).
Here, we report a novel pathway by which CIRP directly regulates p27. We found that CIRP binds to the 5′UTR of the p27 mRNA and enhances its translation. In mild hypothermia, increased CIRP leads to enhanced p27 mRNA translation and increased p27 protein expression in mouse embryonic fibroblasts (MEFs) and HEK293 cells. The increased p27 slows down cell proliferation, as knockout of p27 in MEFs prevents the cold-induced delay of cell proliferation. This novel and direct link between CIRP and p27 can contribute to the anti-proliferative functions of CIRP in hypothermic cells.
MATERIALS AND METHODS
Cell lines, cell culture and treatments
HEK293 and HEK293T cells, MCF7 cells and MEFs (obtained from Nisar P. Malek) were grown in Dulbecco’s Modified Eagle’s Medium (Sigma) supplemented with 10% Fetal Bovine Serum (PAA). Doxycycline (Sigma) was added to cells at a final concentration of 1 μg/ml. MG132 (Sigma) was added where indicated for 4–6 h at a final concentration of 10 μM.
DNA manipulations
The CIRP coding region of the EST clone MPMGp800M09529Q102 (RZPD, GmbH, Berlin) was isolated by polymerase chain reaction (PCR) and recombined into pDONOR-207 (Invitrogen). Sequence verified clones were used for LR recombination and generation of expression clones. CIRP-NES mutants were generated by fusing the minimal nuclear export sequence (NES) of the HIV-1 Rev protein (CTA CCA CCG CTT GAG AGA CTT ACT CTT) to the CIRP cDNA at the N-terminus or at the C-terminus by PCR.
Luciferase encoding reporter plasmids under control of the p27 UTRs or portions of these UTRs have been described (13). The pGL rev5′UTR construct contains the inverted (reverse – complement) p27 5′UTR (nt −1 to −575). The pGL N-myc construct was a kind gift from Anne Willis, Leicester.
Generation of stable cell lines
HEK293 stable cells with tetracycline-inducible CIRP expression were generated using the Flp-In system. The CIRP sequence was cloned into a pTO destination vector (generated by Stephan Geley) and the plasmid was cotransfected with the pOG44 plasmid (Invitrogen) encoding the Flp recombinase into HEK293-Flp-In T-REx cells. Stable Flp-In expression cells were selected for antibiotic resistance.
For constitutive knockdown of CIRP, oligonucleotides (listed in Supplementary Table S1) were annealed and cloned into the BglII–HindIII site of the pENTR-THT III Gateway vector (62) and introduced into the lentiviral destination vector pHR-dest-SFFV-Puro by LR recombination. Lentiviral particles containing CIRP shRNA or control luciferase shRNA were generated in HEK293T packaging cells and used to infect target cells. Transduced cells were selected for puromycin resistance.
RNA–protein interactions
To identify proteins binding to a 32P-labeled full-length p27 5′UTR a northwestern screen was performed as described (21).
For protein–mRNA immunoprecipitation, cells were lysed in Polysome Lysis Buffer (PLB: 150 mM KCl, 5 mM MgCl2, 20 mM HEPES-KOH at pH 7.7, 1% NP-40, 1 mM Dithiothreitol (DTT), 100 units/ml RiboLock RNase Inhibitor, protease inhibitors). Protein G agarose beads (Merck) were pre-swollen in PLB supplemented with 5% bovine serum albumin and coated with antibody (FLAG M2 antibody, Sigma, CIRBP antibody, Proteintech or normal rabbit IgG, Santa Cruz Biotechnology) overnight at 4°C. Additionally, beads were saturated with 1 mg/ml yeast RNA before mixing with the cell lysate supernatants for IP. IP-tubes were incubated for 2–4 h at 4°C. After extensive washing with PLB, beads were resuspended in RLT buffer for RNA isolation (RNeasy Mini Kit, Qiagen). The RNA was reverse-transcribed using Maxima reverse transcriptase (Thermo Scientific) and random hexamer primers (Thermo Scientific). A total of 2 μl of the cDNA product were used as template for a p27 cDNA-specific PCR (using the same primers as for q-RT PCR). PCR products were analyzed on 1.5% agarose gels.
Reporter assays
HEK293 or HEK293T cells were transfected in 24-well plates using calcium–-phosphate precipitation with 45 ng of pGL3 reporter plasmid, 5 ng of Renilla DNA and 450 ng of control or CIRP or RBM3 encoding plasmid. Cells were harvested 24–30 h after transfection. Vectors used for bicistronic reporter assays have been previously described (21). A total of 250 ng of these plasmids were transfected along with 2.25 μg of control or CIRP expression plasmid into HEK293 cells grown in 6-well plates. Cells were harvested 24–36 h after transfection. Cell lysis and luciferase assays were performed using the Dual-Luciferase reagent (Promega) and a Lumat LB9501 luminometer (Berthold).
The pSP64 poly(A) vector (Promega) was used for cloning of monocistronic and bicistronic constructs containing Renilla luciferase (R luc) and/or firefly luciferase (F luc), with or without p27 5′UTR sequences upstream of the latter. These plasmids were used for in vitro transcription using the mMESSAGE mMACHINE SP6 kit (Ambion) to generate capped, poly(A)+ transcripts. The mRNA was incubated with TURBO DNase and purified using the RNA Cleanup procedure of the RNeasy Mini kit (Qiagen). The mRNA was transfected into HEK293 or HEK293T cells using the Lipofectamine 2000 reagent (Thermo Fisher Scientific). The 40 ng bicistronic RNA were transfected in 24-well plates; for monocistronic RNA reporter assays, 20 ng Renilla RNA and 40 ng firefly RNA were cotransfected in 24-well plates. Cells were harvested 8 h after transfection.
Quantitative RT-PCR
Total cellular RNA was prepared with the RNeasy Plus Mini Kit (Qiagen). First strand cDNA was synthesized from 1 μg total RNA using random hexamer primers and Maxima Reverse transcriptase (Thermo Scientific). qPCR was performed using the SensiFAST SYBR No-ROX Kit (Bioline) following the manufacturer’s protocol in a Rotor-Gene Q cycler (Qiagen) under the following conditions: 95°C 120 s; 40 cycles: 95°C 5 s, 60°C 10 s, 72°C 15 s (fluorescence acquisition). Relative RNA levels were calculated based on the comparative Delta Ct method (63), applying the formula 2Ct(control)-Ct(gene). Reaction efficiencies were estimated by serial dilution of cDNA: Ct values were plotted against the natural logarithm of the template concentration and the efficiencies were calculated from the slope of the regression line applying the equation E = 10[-1/slope]-1. PCR amplicon length was verified by agarose gel electrophoresis. TATA-Box binding protein (TBP) mRNA was used as endogenous control for normalization. Primer sequences for amplification of TBP, p27, F luc and (R luc are listed in Supplementary Table S2.
RNA interference
RNA interference was performed by transfecting pLKO.1-puro lentiviral vectors containing CIRP-specific shRNA (MISSION TRC shRNA TRCN0000017265, here abbreviated as shCIRP 1, and TRCN0000017267, abbreviated as shCIRP 2, obtained from Sigma) or scramble control shRNA (Addgene (Cambridge, MA, USA) plasmid #1864 (64)).
Protein analysis
Western blotting was performed according to standard procedures and described before (21). Antibodies detecting the following proteins or epitopes were used: β-tubulin (clone AA2, Sigma-Aldrich), p27 (clone 57, BD or HRP-conjugated p27 antibody, BD), CIRP (Proteintech), cyclin D3 (C-16, Santa Cruz), cyclin A (E67.1, Santa Cruz), FLAG (M2 mouse antibody or rabbit polyclonal antibody, Sigma-Aldrich). For quantitative analysis, Western blots were imaged and processed with ImageQuant LAS 4000 (GE Healthcare).
Fluorescence microscopy analysis
Cells were cultured on glass coverslips, transfected and fixed with 3% paraformaldehyde. Nuclei were stained with Hoechst 33342. Coverslips were mounted on glass slides with Mowiol (Sigma). Cells were examined by fluorescence microscopy (Axio Imager.M2, Zeiss); pictures were acquired using MetaVue software and processed with ImageJ.
Proliferation assay
Wild-type (wt) MEFs and p27 knockout (−/−) MEFs were seeded at a density of 0.5 × 105 cells/ml and grown overnight at 37°C. Then, cells were counted and shifted to a 32°C-incubator or kept at 37°C. Cells were trypsinized and counted at different time points. Each sample was analyzed in triplicate using a CASY Cell Counter (Roche). The doubling time was calculated with the program Roth V. 2006 <http://www.doubling-time.com/compute.php>.
Statistical analysis
The parametric Student's unpaired two-tailed t test was used for comparison between two sample groups. The data are presented as mean ± standard deviation (SD) or standard error of the mean (SEM).
RESULTS
CIRP binds selectively to the p27 mRNA
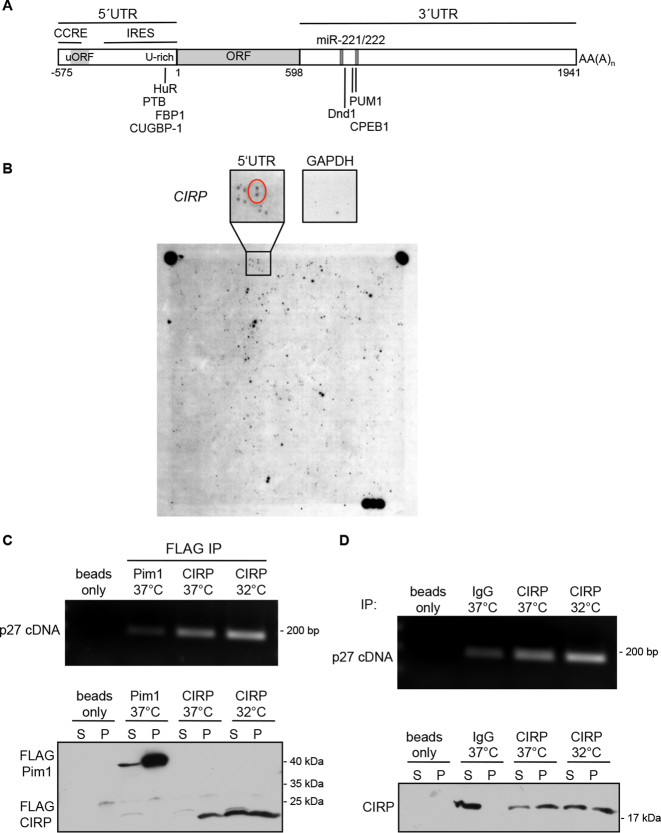
In order to identify novel p27 mRNA binding proteins, we hybridized a protein expression array with a radioactive labeled RNA probe of the full length p27 5′UTR (575 nt, Figure 1A). Bacteria expressing single human cDNAs were spotted onto this array in duplicates and in a specific pattern (21). The p27 5′UTR bound to extracts of cells expressing the cold inducible RNA binding protein CIRP, whereas a GAPDH control mRNA did not bind to these extracts (Figure 1B).
Figure 1.
CIRP binds to the p27 mRNA. (A) Schematic representation of the human p27 transcript. Regulatory elements and binding sites for proteins or miRNAs are indicated. (B) Identification of CIRP as a p27 5′UTR binding factor by northwestern blot of a protein array. Shown is the autoradiography of the nylon membrane, onto which a human protein expression library was spotted in duplicates and hybridized with a 32P-labeled p27 5′UTR. Each cDNA-expressing clone was spotted twice and in a specific pattern. The area of the filter containing the CIRP cDNA-expressing colonies is enlarged and the two spots detected by the p27 5′UTR probe are encircled. The membrane was stripped and hybridized with a control 32P-labeled GAPDH mRNA. (C) p27 mRNA is immunoprecipitated with FLAG-tagged CIRP. MCF-7 cells were transfected to express FLAG-CIRP and incubated at 32 or 37°C. CIRP was immunoprecipitated with mouse anti-FLAG antibodies. As control, FLAG-tagged Pim1 was precipitated. Upper panel: p27 mRNA co-immunoprecipitated with FLAG-IPs was recovered from the immunoprecipitates, subjected to reverse transcription and the p27 cDNA was amplified by PCR (33 cycles) and separated on an agarose gel. Lower panel: the immunoprecipitation was monitored in western blots of the supernatant (S) and the immunoprecipitated pellet (P) using rabbit anti-FLAG antibodies. (D) Endogenous p27 mRNA binds to endogenous CIRP. CIRP protein was immunoprecipitated from MCF-7 cells grown at either 37 or 32°C. Cell extracts were incubated with polyclonal CIRP antibodies or normal rabbit IgG as control. Upper panel: co-immunoprecipitated mRNA was recovered from the immunoprecipitates, subjected to reverse transcription and the p27 cDNA was amplified by PCR (33 cycles) and separated on an agarose gel. Lower panel: CIRP immunoprecipitation was monitored by western blots of the supernatant (S) and the immunoprecipitated pellet (P).
In order to investigate if CIRP associates with the p27 mRNA in vivo, FLAG-tagged CIRP was immunoprecipitated from MCF-7 cells. As a control, FLAG-tagged Pim1 kinase was overexpressed in MCF-7 cells and immunoprecipitated. p27 mRNA which associated with the immune precipitates was amplified by RT-PCR. The p27 mRNA could be detected in CIRP immune complexes (Figure 1C and Supplementary Figure S1A), supporting the hypothesis that CIRP might selectively bind to the p27 mRNA. Interestingly, incubation at low temperature (24 h at 32°C) increased FLAG-CIRP protein levels (Figure 1C, lower panel). This resulted in an increase of co-immunoprecipitated p27 mRNA (Figure 1C, upper panel and Supplementary Figure S1A).
To explore if endogenous CIRP binds to endogenous p27 mRNA, we immunoprecipitated CIRP protein from MCF-7 cells. In these endogenous IPs, p27 mRNA co-precipitated with CIRP (Figure 1D and Supplementary Figure S1B). CIRP protein levels were increased by hypothermia (32°C, 24 h). Despite similar amounts of precipitated CIRP protein (due to limiting amounts of antibodies), we noticed an increase of co-precipitated p27 mRNA at low temperature, which might reflect improved binding of the p27 transcript to CIRP. Since the assay is not quantitative, further studies need to confirm the enhanced CIRP/p27 mRNA binding at low temperature and determine the potential molecular mechanisms.
CIRP increases p27 5′UTR-mediated translation
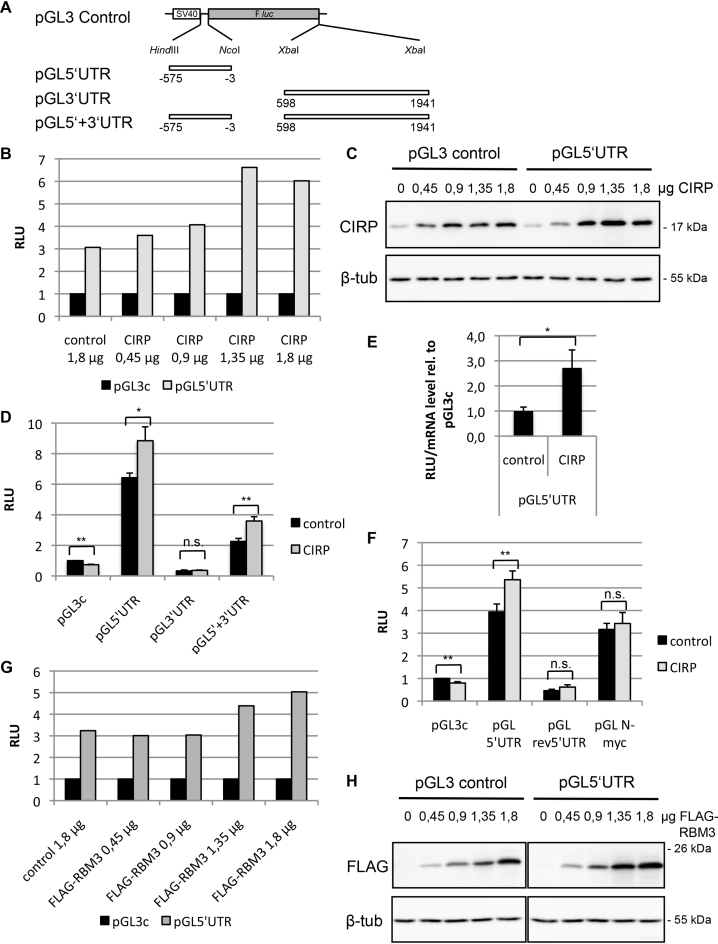
CIRP has been proposed to function as an RNA chaperone and to facilitate translation, especially at low temperatures (37). We therefore wished to determine if CIRP binding to the p27 mRNA might alter its translation. We used pGL3-derived luciferase reporter plasmids, where the luciferase coding region was inserted either downstream of the p27 5′UTR or upstream of the p27 3′UTR or in combination with both UTR sequences (Figure 2A). These plasmids were cotransfected with a CIRP expression vector or the empty control vector. Increasing amounts of transfected CIRP vector resulted in increased CIRP protein expression (Figure 2C). CIRP expression led to a stimulation of reporter translation in the presence of the p27 5′UTR and this stimulation was dose-dependent (Figure 2B). No increase in reporter translation was observed for the pGL3-control plasmid, whereas CIRP enhanced the expression of the pGL5′UTR reporter (by a factor of 1.9, when adjusted to the inhibitory effect of CIRP on pGL3-control) (Figure 2D). A similar increase of reporter translation occurred in the presence of both p27 UTRs, whereas the p27 3′UTR did not significantly alter reporter translation (Figure 2D). These data support the hypothesis that CIRP stimulates p27 5′UTR-mediated translation. To exclude potential transcriptional effects, we determined the luciferase mRNA levels by quantitative RT-PCR. Cells were transfected with pGL3c and pGL5′UTR in presence or absence of CIRP and relative luciferase activity was adjusted for reporter mRNA levels. The normalized reporter activity of pGL5′UTR was significantly induced by CIRP (Figure 2E). This experiment demonstrates that CIRP stimulates pGL5′UTR reporter expression post-transcriptionally and does not alter mRNA stability.
Figure 2.
CIRP enhances p27 5′UTR-mediated translation. (A) Reporter plasmids used for luciferase assays. The p27 5′UTR and 3′UTR were inserted upstream or downstream of the F luc coding region (gray box) as indicated in the pGL3-control plasmid. The open box represents the SV40 promoter. (B and C) HEK293 cells were transfected with reporter plasmid pGL3-control or pGL5′UTR, different amounts of CIRP expression plasmid or empty control plasmid, and Renilla luciferase as a normalization control. Each sample was trypsinized and divided into two portions for subsequent luciferase activity measurement (B) or western blot analysis (C). Relative luciferase activity (RLU) was calculated as ratio of firefly and Renilla light units; the ratio for pGL3c was adjusted to 1 for comparison. β-tubulin was monitored as loading control in the western blot. (D) HEK293T cells were transfected with reporter plasmid, CIRP or empty control plasmid and Renilla luciferase expression plasmid. The ratio of firefly and Renilla luciferase activities was calculated (RLU). The ratio for pGL3c/control was adjusted to 1. Data are shown as mean ± SD, n = 3 independent experiments. Unpaired two-tailed t-test was used to compare the average RLU of the different reporter constructs in absence or presence of overexpressed CIRP: *P < 0.05, **P < 0.01. (E) HEK293 cells were transfected with reporter plasmid, CIRP or empty control plasmid and Renilla luciferase. Luciferase activities and luciferase RNA levels were determined. The relative luciferase activity (RLU, ratio of firefly and Renilla luciferase activities) was adjusted to the relative mRNA level (firefly RNA levels were determined using Renilla RNA as normalization control). The normalized RLU of pGL5′UTR in absence or presence of overexpressed CIRP was compared to pGL3c and pGL5′UTR/control was set to 1. Shown is the mean ± SD, n = 3 independent experiments. *P < 0.05. (F) HEK293 cells were transfected with reporter plasmids containing different UTRs upstream of the F luc coding region, CIRP or empty control plasmid and Renilla luciferase. pGLrev5′UTR contains the reverse-complement p27 5′UTR upstream of F luc, and pGL N-myc contains the 317 nt N-myc IRES sequence upstream of Fluc. RLU was calculated as described above (B). The ratio for pGL3c/control was set to 1. Data are shown as mean ± SD, n = 3 independent experiments. *P < 0.05, **P < 0.01. (G and H) HEK293 cells were transfected with pGL3-control or pGL5′UTR, increasing amounts of FLAG-RBM3 or empty control plasmid and Renilla luciferase as a normalization control. Samples were prepared and analysed as described above (B, C).
To investigate the specificity of CIRP-induced reporter regulation, we employed the reverse—complement p27 5′UTR sequence (rev5′UTR) or the N-myc 5′UTR in reporter assays. The rev5′UTR sequence with the same length as p27 5′UTR (575 nt) contains three ATGs, one of which is located at position [−357 −355] and has an in-frame stop codon at position [−114 −112]. Both, the wt and the rev 5′UTR are predicted to form secondary structures with similarly low folding free energy (structure prediction using the program mfold, data not shown). Whereas CIRP stimulated the p27 5′UTR-mediated reporter translation, overexpressed CIRP did not significantly increase reporter expression in the presence of the inverted p27 5′UTR (Figure 2F). The N-myc 5′UTR of 317 nt contains another IRES element (65). Consistent with earlier observations using a dicistronic vector (65), the N-myc 5′UTR increased expression of the reporter. Co-expression of CIRP did not significantly alter reporter expression (Figure 2F).
The cold shock-, RNA binding-protein RBM3 (RNA-binding motif protein 3) is a paralog of CIRP with high sequence homology especially within the RNA-binding domain (74% amino acid identity). Both proteins are induced upon cold shock (66) or low oxygen (39). To investigate if RBM3 can also regulate p27 translation, we transfected increasing amounts of RBM3 encoding expression plasmids. Increased transfected vector led to increased RBM3 protein expression (Figure 2H). Similar to CIRP, RBM3 could also enhance reporter translation in the presence of the p27 5′UTR (Figure 2G).
CIRP regulates p27 expression in vivo
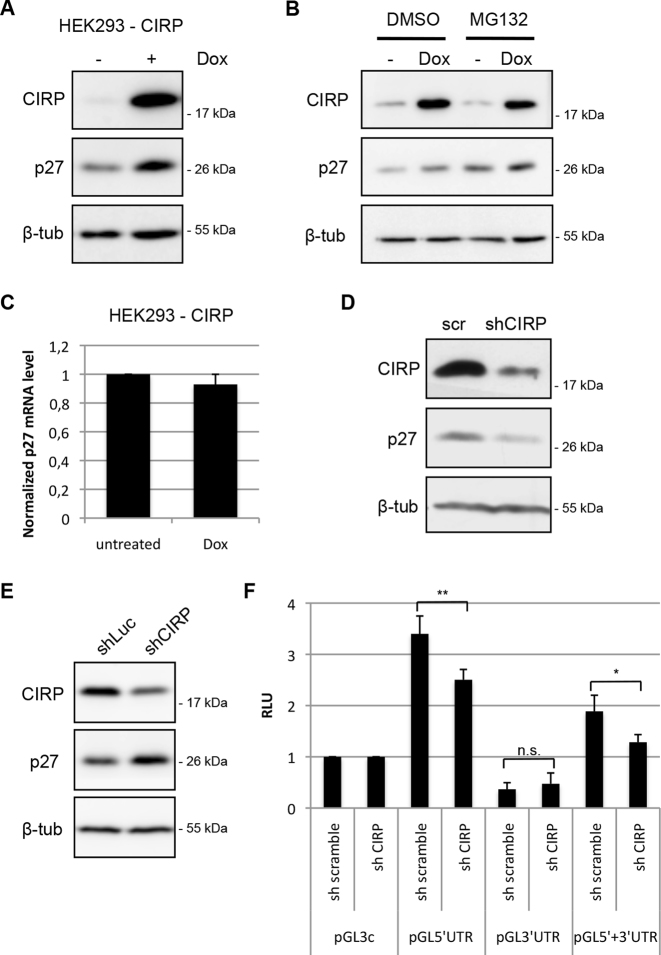
CIRP is induced by a number of stress pathways including cold, hypoxia, UVB, UVC, cisplatin or the neurotoxin domoic acid (35,38,39,67,68). Depending on the cell type and stress stimulus, activation of CIRP can promote or attenuate cell proliferation (35,48,52,60,61,69–71). The different response of cell lines to CIRP does not always correlate with the levels of CIRP, p27 or other cell-cycle regulatory proteins (48). Since CIRP can bind the p27 mRNA and enhance reporter translation, we wished to determine if induced CIRP expression increases p27 expression. To exclude the potential influence of other stress-induced factors on p27, we generated a HEK293 cell line where CIRP expression could be induced by doxycycline. Consistent with the reporter translation in transient transfection assays, the induction of CIRP by doxycycline caused an increase in p27 protein expression (Figure 3A). Increased p27 protein levels might originate from increased mRNA transcription, translation or increased p27 stability. The stability of p27 is enhanced in G1 arrested cells (11,72). However, induction of CIRP led only to a minor increase (1–3%) in cells in G1 phase following up to six days of CIRP induction (Supplementary Figure S2). This indicated that the increase in p27 protein is not caused by cells accumulating in the G1 phase of the cell cycle. To investigate a potential contribution of proteasomal degradation of p27, we repeated the experiment in presence of the proteasome inhibitor MG132. If CIRP would impair p27 proteasomal degradation, the protein should accumulate to similar levels in presence of MG132. However, MG132 increased the level of p27 in both, the absence or presence of doxycycline and p27 levels were still increased following induction of CIRP (Figure 3B). This excludes inhibition of p27 proteasomal degradation as a potential cause of the CIRP-mediated upregulation of p27. Alternatively, CIRP might induce transcription of p27 or stabilize the p27 mRNA. We therefore determined the abundance of the p27 mRNA by quantitative RT-PCR. Doxycycline-mediated CIRP expression did not increase p27 mRNA level, but rather slightly reduced it (Figure 3C). These data support the model that CIRP overexpression can induce p27 expression in HEK293 cells by enhancing its translation.
Figure 3.
Expression of CIRP can induce p27. (A) A HEK293 cell line where CIRP expression can be induced by doxycycline was generated. Doxycycline (Dox) was added to proliferating HEK293-CIRP cells to the final concentration of 1 μg/ml for 48 h. Induced CIRP, p27 and tubulin were monitored using immunoblot analysis of protein extracts. (B) HEK293-CIRP inducible cells were treated with doxycycline to induce CIRP as described in (A) and with 10 μM MG132 or DMSO as solvent control for 5 h. Expression of CIRP, p27 and β-tubulin was analyzed by immunoblotting. (C) Total RNA was isolated from HEK293-CIRP inducible cells grown for 48 h in the absence or presence of doxycycline and p27 mRNA was quantified by qPCR. Shown is the mean of three experiments with error bars representing the SD. (D) Immunoblot analysis of MCF7 cells transfected with lentiviral plasmids expressing scramble shRNA or CIRP targeting shRNA; transfected cells were selected with puromycin for 5 days before harvesting. (E) HEK293T cell lines stably expressing CIRP shRNA or F luc shRNA were generated by lentiviral transduction and selection of infected cells. Immunoblot analysis of these cell lines is shown. (F) HEK293T cells were transfected with firefly reporter plasmids containing p27 UTRs, scramble shRNA or CIRP shRNA encoding plasmid and Renilla luciferase and were harvested after 44–48 h. Relative luciferase activity (RLU) was determined as ratio of firefly and Renilla luciferase activities; pGL3c activity was adjusted to 1 for comparison. Data are shown as mean ± SD, n = 4 independent experiments. Unpaired two-tailed t-test was used to compare the average RLU of the different reporter constructs in the presence of scramble or CIRP shRNA: *P < 0.05, **P < 0.01.
To evaluate the role of endogenous CIRP on p27 expression in the absence of stress, we performed a number of knockdown experiments and obtained mixed results. In most experiments, transient knockdown of CIRP by transfection of lentiviral derived plasmids expressing CIRP-shRNA led to a reduction of p27 expression in MCF-7 cells (Figure 3D) or HEK293 cells (Supplementary Figure S3A), consistent with the hypothesis that CIRP promotes p27 translation. In other experiments however we observed no regulation or even an increase in p27 upon transient depletion of CIRP by shRNA, siRNA or doxycycline-inducible shRNA expression in an HRT-18 stable cell line (Supplementary Figure S3B and C).
It had been reported previously that CIRP−/− MEF cell lines proliferate faster when CIRP is reintroduced, and it was proposed that decreased p27 causes this acceleration (48). We did not see a consistent variation of p27 expression levels in CIRP−/− MEFs compared to wt MEFs (data not shown). Using lentiviral transduction we generated and selected CIRP-specific shRNA- or control luciferase shRNA- expressing HEK293T cells. In these cells, reduced CIRP protein correlated with increased p27 protein (Figure 3E).
These apparently inconsistent data might be explained by the complex role of CIRP by binding to many different mRNAs or by its differential localization in the nucleus, cytoplasm and stress granules (41,73). CIRP may also regulate proteins, including Mirk/Dyrk1b (48) and Erk1/2 (70), that alter p27 expression by multiple pathways. To investigate if translation is affected by CIRP depletion, we analyzed luciferase activity in cells coexpressing reporter constructs containing p27 UTRs and CIRP shRNA or control shRNA, thereby restricting the analysis on translation and excluding post-translational regulation of p27. shRNA-mediated CIRP knockdown reproducibly reduced the activity of pGL5′UTR and pGL5′+3′UTR, supporting the hypothesis that CIRP downregulation decreases p27 translation even in the absence of stress (Figure 3F).
CIRP enhances translation of a bicistronic reporter but fails to regulate translation of transfected mRNA
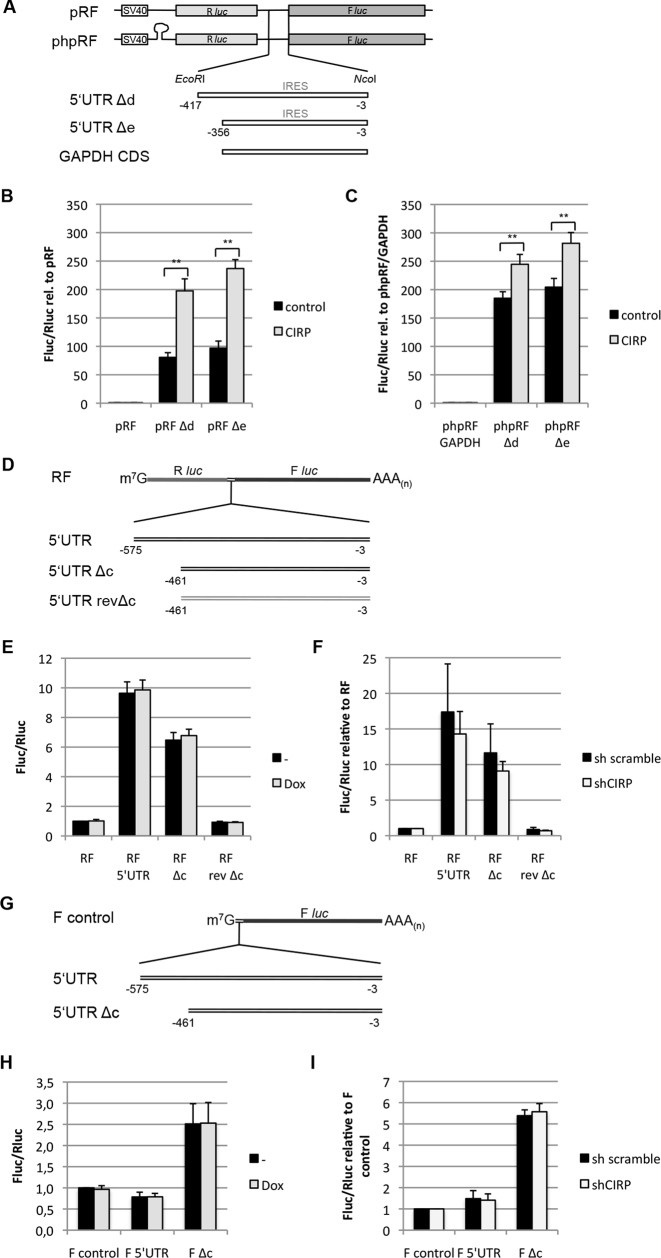
To further characterize the CIRP-dependent enhanced translation of p27, we used bicistronic reporter plasmids. These plasmids encode the Renilla and the F luc on a single mRNA transcript. The p27 5′UTR contains an IRES that can induce translation of a downstream open reading frame (ORF) of a bicistronic transcript (16,21,25). Two IRES-containing fragments of the p27 5′UTR (Δd, −417 to −3 and Δe, −356 to −3; Figure 4A) were cloned between these cistrons. The full length 5′UTR of p27 was not analyzed because it contains a cryptic promoter activity that can be detected in northern blots (21). Using this bicistronic expression system, we found that CIRP could strongly enhance translation of the downstream ORF in the presence of p27 5′UTR sequences in the intercistronic region (Figure 4B). In addition, we used a second plasmid that contains a stable palindrome sequence inserted upstream of the Renilla luciferase ORF (phpRF), generating a stable hairpin that inhibits cap-dependent translation. This inhibited Renilla luciferase translation, but presence of the intracistronic p27 5′UTR rendered the downstream F luc translation resistant to this hairpin (21). Under these conditions, CIRP could stimulate translation of the downstream cistron; however the increase was weaker than in the absence of the hairpin (Figure 4C). These data confirm that CIRP can act as a positive regulator of p27 translation and indicate that it might promote p27 cap-independent translation initiation.
Figure 4.
CIRP can enhance translation of a downstream cistron of a bicistronic mRNA. (A) Bicistronic reporter plasmids used for luciferase assays. The p27 5′UTR Δd and Δe fragments or a fragment of the GAPDH coding sequence were inserted in the intercistronic region of pRF and phpRF plasmids. These plasmids contain the Renilla (Rluc, light gray box) and the firefly (Fluc, dark gray box) luciferase coding regions under a single SV40 promoter (open box). (B and C) HEK293 cells were transfected with pRF (B) or phpRF (C) and CIRP or empty control plasmid. The ratio of firefly and Renilla luciferase activities was calculated and is shown relative to pRF or phpRF-GAPDH activity. Data are shown as mean ± SD, n = 3 independent experiments. *P < 0.05, **P < 0.01. (D) Schematic representation of in vitro transcribed, capped and polyadenylated bicistronic reporter mRNAs that were used for luciferase assays. The full length p27 5′UTR, the Δc fragment or a fragment with reverse-complementary Δc sequence (rev Δc) were inserted in the intercistronic region of the RF RNA construct. (E) HEK293-CIRP inducible cells were grown in the absence or presence of doxycycline for 24 h before transfection with bicistronic reporter RNAs. Cells were harvested 8 h after transfection and firefly and Renilla luciferase activities were determined. The ratio of F luc and R luc activities was calculated and expressed relative to RF-untreated samples. Data represent the mean ± SEM, n = 6 independent experiments. (F) HEK293 cells were transfected with plasmids encoding scramble shRNA or CIRP-specific shRNA 40 h before transfection with bicistronic reporter RNAs; puromycin was added to a 1 μg/ml concentration 24 h after shRNA transfection to inhibit the growth of non-trasfected cells. The ratio of F luc and R luc activities was determined and expressed relative to RF. Data are shown as mean ± SD, n = 4 independent experiments. (G) Schematic representation of monocistronic reporter mRNAs. The full length p27 5′UTR and the Δc fragment were inserted upstream of the F luc coding sequence. The mRNA was in vitro transcribed, capped and polyadenylated. (H) HEK293-CIRP inducible cells were grown in the absence or presence of doxycycline for 24 h before transfection with firefly reporter RNA constructs and Renilla luciferase mRNA. The ratio of F luc and Renilla luciferase activities was calculated and expressed relative to firefly (F) control untreated samples. The data represent the mean ± SD of n = 4 independent experiments. (I) HEK293 cells were transfected with plasmids encoding scramble or CIRP-specific shRNA 40 h before transfection with monocistronic reporter RNAs; puromycin was added to the culture medium as described for (F). Cells were harvested 8 h after RNA transfection. The ratio of F luc and Renilla luciferase activities was determined and expressed relative to firefly (F) control. Data are shown as mean ± SD, n = 4 independent experiments.
The bicistronic reporter constructs used in the experiments described above have been scrutinized because they may harbor intrinsic promoter activities (23,26). In fact, deleting the SV40 promoter from the pRF backbone resulted in a much stronger reduction of the Renilla reporter activity than the firefly reporter activity, supporting the hypothesis that there might be a cryptic promoter activity in the Renilla ORF or in the p27 5′UTR (MK, data not shown). To exclude the problem of potential weak cryptic promoters, we performed RNA transfection experiments using in vitro synthesized capped RNAs (Figure 4D and G). One potential disadvantage of this technique is that mRNA-binding proteins can not sequentially associate co-transcriptionally with the mRNA and that the isolated mRNA is transfected in the cytoplasm, where some nuclear RBPs may not be able to bind. Bicistronic luciferase reporter mRNAs containing p27 5′UTR fragments were transfected into HEK293-CIRP cells. This cell line was generated to express a doxycycline-inducible CIRP. Bicistronic translation was quantified based on Renilla and firefly luciferase activities. Similar to previous observations in breast cancer cell lines (25,27), the p27 5′UTR strongly stimulated translation of the downstream cistron. The full length 5′UTR led to a 8- to 12-fold increase in the firefly/Renilla luciferase activity ratio, and the truncated 461 nt IRES fragment Δc led to a 5- to 8-fold increase in the firefly/Renilla ratio (Figure 4E). This increase in translation supports the finding of an IRES in the p27 5′UTR. The reverse complemented Δc fragment (rev Δc) was inserted as a control; its presence in the intercistronic region did not stimulate translation but rather slightly reduced firefly translation (Figure 4E). Surprisingly, induction of CIRP by doxycycline failed to further stimulate F luc translation (Figure 4E). Incubation of the cells at 32°C to induce endogenous CIRP expression did also not significantly alter p27 5′UTR-mediated reporter translation (Supplementary Figure S4A). In a second experiment, we tested if knockdown of CIRP using shRNA might impair bicistronic RNA translation. We employed HEK293 cells that had been transfected with CIRP shRNA or scramble shRNA 42 h before mRNA transfection. Transient knockdown of CIRP could only slightly reduce bicistronic translation driven from the p27 5′UTR or the Δc fragment, but a similar reduction (about 15%) was observed for the rev Δc fragment (Figure 4F). Two different CIRP-targeting shRNAs had similar effects on reporter translation (Supplementary Figure S4B) and both reduced CIRP protein levels (Supplementary Figure S4D).
Lack of strong regulation of transfected mRNA by CIRP might be specific for bicistronic plasmids or be a general property of transfected mRNA molecules, especially if they contain strong secondary structures as predicted for the p27 5′UTR. To test this hypothesis, we generated monocistronic RNAs containing the p27 5′UTR or Δc fragment upstream of the F luc coding sequence (Figure 4G). The monocistronic RNA reporter assays in HEK293-CIRP inducible cells revealed a slightly inhibitory effect of the full length p27 5′UTR on reporter translation. A shorter 5′UTR fragment Δc, lacking a functional uORF, could stimulate translation 2.5-fold (Figure 4H). Still, CIRP overexpression by doxycycline addition did not alter F luc activity, neither in presence of the p27 5′UTR nor in presence of the Δc fragment, compared to the untreated controls (Figure 4H). To investigate whether the absence of regulation by CIRP might be caused by already abundant endogenous CIRP, these experiments were repeated in HEK293 cells previously transfected with CIRP shRNA or scramble shRNA. Translation of monocistronic reporter RNAs containing p27 5′UTRs was not affected by CIRP knockdown (Figure 4I and Supplementary Figure S4C), indicating that CIRP cannot stimulate translation of transfected mRNAs containing p27 5′UTR sequences. These experiments can be explained by the hypothesis that nuclear or co-transcriptional mRNA–protein complex assembly may be essential for CIRP-mediated translational regulation of p27 5′UTR- containing mRNAs. It may however also be that transfection of the isolated highly structured p27 5′UTR mRNA prevents protein binding and fails to unfold in the cytoplasm.
Cytoplasmic CIRP mutants cannot stimulate p27 5′UTR-mediated translation
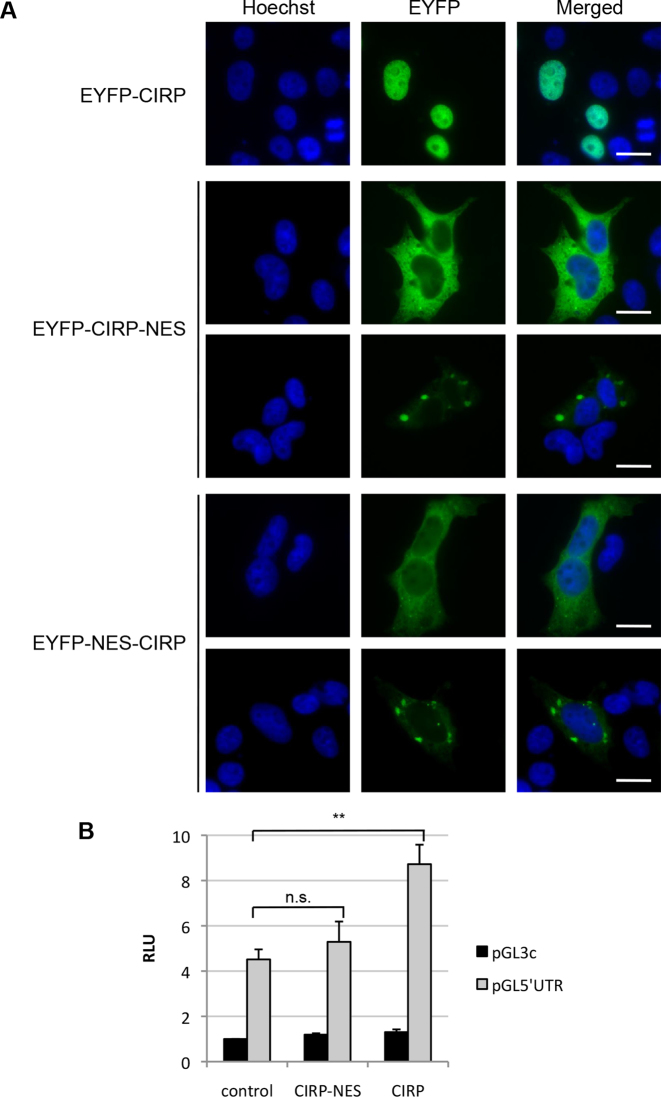
CIRP is mainly localized in the nucleus of most cell types, except frog oocytes (74) and mouse round spermatids (50) where it was found cytoplasmic. The protein can shuttle between nucleus and cytoplasm (41,75). RNA transfection experiments (see Figure 4) suggested the possibility that the site of loading of CIRP on the p27 mRNA might be important to permit CIRP to regulate p27 translation. To investigate if cytoplasmic CIRP can alter p27 translation, we generated a cytoplasmic mutant of CIRP by adding a nine amino acids NES to the N-terminus or C-terminus of CIRP cDNA. When expressed in HEK293T cells, the EYFP-CIRP fusion protein was mostly nuclear, whereas the EYFP-NES-CIRP protein was predominantly found in the cytoplasm (Figure 5A). Of note, in over 50% of the cells the fusion protein accumulated in cytoplasmic foci, likely representing stress granules (identified by positive TIA-1 co-staining; data not shown). If CIRP needs to interact with p27 5′UTR in the nucleus to stimulate p27 translation, then a predominantly cytoplasmic mutant might be incapable to regulate a reporter gene containing p27 5′UTR. Luciferase reporter assays revealed that the CIRP-NES mutant was unable to alter reporter translation, whereas the wt CIRP significantly enhanced pGL5′UTR reporter activity when compared to the control (Figure 5B). These data support the model that CIRP must interact with the p27 mRNA already in the nucleus in order to regulate p27 translation.
Figure 5.
Cytoplasmic CIRP mutants fail to regulate reporter translation via p27 5′UTR. (A) Microscopy analysis of HEK293T cells overexpressing EYFP-tagged wt and NES-mutants of CIRP. The NES was inserted either at the C-terminus (CIRP-NES) or at the N-terminus (NES-CIRP) of CIRP. Cells were fixed and CIRP localization was analyzed by fluorescence microscopy. Nuclei were stained with Hoechst (blue). Scale bar: 20 μm. (B) HEK293 cells were transfected with pGL3c or pGL5′UTR, a control plasmid encoding EYFP or wt CIRP or CIRP-NES mutant, and Renilla luciferase for normalization. Relative luciferase activity (RLU) was calculated as ratio of firefly and Renilla luciferase activities; pGL3c/control activity was adjusted to 1. Data are shown as mean ± SD, n = 3 independent experiments. Unpaired two-tailed t-test was used to compare the average RLU of pGL5′UTR in the absence or presence of overexpressed CIRP-NES mutant or wt CIRP: *P < 0.05, **P < 0.01.
CIRP stimulates p27 translation in mild hypothermia
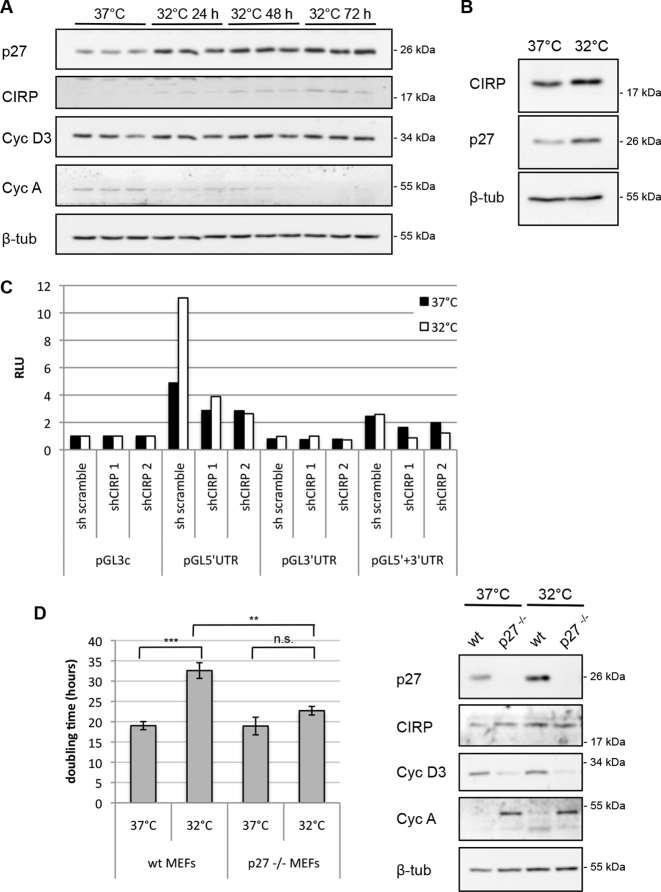
Mild hypothermic conditions of 32 to 33°C lead to a reduction of metabolic activity and to a decrease of cell proliferation and a rapid induction of CIRP (37). Cells accumulate in response to mild cold shock mainly in the G1 phase of the cell cycle (37). If CIRP stimulates p27 translation, it should induce p27 expression and p27 could mediate the cell-cycle delay. Interestingly, we observed that p27 protein levels were increased when CIRP was induced in MEFs following mild hypothermia of 24, 48 or 72 h. Whereas cyclin D3 level remained largely unchanged, cyclin A level slowly declined, indicating reduced S-phase cells (Figure 6A). Similarly to MEFs, HEK293 cells induced CIRP and p27 upon cold shock (Figure 6B). In order to investigate whether p27 translation was induced during mild hypothermia, we employed reporter assays and CIRP knockdown under mild hypothermic conditions. Luciferase reporter translation by the p27 5′UTR was stronger enhanced at 32°C than at 37°C. Importantly, this stimulation was diminished by knockdown of CIRP using two separate shRNAs (Figure 6C). In contrast, reporter translation by the p27 3′UTR remained unchanged in mild cold shock and in presence or absence of CIRP (Figure 6C). These data support the hypothesis that CIRP causes increased p27 translation in mild hypothermia. This increase is mediated by the p27 5′UTR and not by its 3′UTR. Similarly, pGL5′UTR reporter translation was enhanced in Hela cells grown at 32°C (Supplementary Figure S5A). We also analyzed p27 synthesis at 37 or 32°C in pulse labeling experiments with 35S-methionine and 35S-cysteine. Cells were incubated with control shRNA or CIRP shRNA. Immunoprecipitation and autoradiography of newly synthesized 35S-labeled p27 protein revealed increased p27 synthesis at 32°C (Supplementary Figure S5B and C). Importantly, the induction of p27 at 32°C was abrogated by CIRP shRNA, demonstrating that CIRP is a key protein inducing p27 synthesis in mild hypothermic conditions (Supplementary Figure S5B).
Figure 6.
CIRP induces p27 translation in mild hypothermia. (A) MEFs were grown at 37°C (control, normothermia) or incubated at 32°C (mild hypothermia) for the times indicated. The expression of p27, CIRP, cyclin D3, cyclin A and β-tubulin was determined by western blot. (B) HEK293 cells were moved to 32°C and incubated for 40 h. Protein extracts from these cells or cells grown at 37°C were subjected to immunoblot analysis for CIRP, p27 and β-tubulin. (C) HEK293 cells were transfected with CIRP shRNA or scramble shRNA expression plasmids, luciferase reporter plasmids (depicted in Figure 2A) and Renilla luciferase for normalization control. 25 h after transfection, one aliquot of cells was incubated at 32°C for additional 25 h. Relative luciferase activity (RLU) was determined as ratio of firefly and Renilla luciferase activities and normalized for the activity of pGL3-control. One representative experiment out of four is shown. (D) Average doubling time of wt MEFs and p27−/− MEFs grown at 37 or 32°C. Wt MEFs and p27−/− MEFs were seeded and their proliferation was followed by counting the cells at timepoints starting 5–10 h after shifting to the lower temperature incubator and for 72 h. Cell numbers obtained during the exponential growth phase were used to calculate the doubling time. Data are shown as mean ± SEM, n = 5 independent experiments. Unpaired two-tailed t-test was used to compare the average doubling time of MEFs at 37°C and at 32°C and of p27−/− MEFs at 37°C and at 32°C; *P < 0.05, **P < 0.01, ***P < 0.001. The difference between the average doubling time of MEFs at 32°C and p27−/− MEFs at 32°C is also statistically significant, based on the same test. A representative immunoblot analysis of these cells is shown (right panels). Cells were collected after 8 h at 32°C and the expression of p27, CIRP, cyclin D3, cyclin A and β-tubulin was determined.
To determine the physiological role of the induction of p27 upon cold shock, we compared the proliferation of p27-knockout (p27−/−) MEFs with wt MEFs at 37°C and in mild cold shock. Whereas wt MEFs significantly reduced their proliferation rate in mild cold shock (1.7 fold), p27−/− MEFs showed only a minor reduction of proliferation (1.2-fold). Especially under mild hypothermic conditions, p27−/− MEFs proliferated significantly faster than wt MEFs (Figure 6D). This indicates that p27 is, at least in part, responsible for the cold-induced reduction of cell proliferation. Interestingly, binding of the p27 transcript to CIRP seemed to be enhanced in mild hypothermia (Figure 1C), which might contribute to the regulation of p27 translation by CIRP.
DISCUSSION
We report here that the cold-inducible RBP CIRP binds to the 5′UTR of the p27 mRNA. Using reporter plasmids, we demonstrate that binding of CIRP to the 5′UTR enhances reporter translation. Interestingly, binding of CIRP to the p27 mRNA appears to be enhanced in mild hypothermic conditions. Under these conditions, we also observe a CIRP-dependent increase in reporter translation and an accumulation of p27 protein. We conclude therefore that CIRP-mediated enhanced translation of the p27 mRNA can contribute to the accumulation of p27 in mild hypothermia. This may be of significant physiological relevance, since p27 is able to delay cell proliferation of murine embryonic fibroblasts in mild hypothermic conditions, whereas MEFs lacking p27 largely abolish the cold-induced delay of cell proliferation.
The determination of a transcript-specific function of RBPs can be challenging since these proteins frequently interact with a large number of RNAs and evoke complex physiological responses. While reporter assays demonstrated that CIRP enhances p27 5′UTR-mediated translation, the analysis of the physiological consequences of CIRP knockdown on p27 and cell-cycle progression is more complex. Knockdown of CIRP always resulted in decreased reporter translation, but resulted not always in decreased p27 protein levels. There are a number of potential causes that might explain these observations. First, CIRP is subject to post-translational modifications including arginine methylation and phosphorylation (41,59) and is recruited to stress granules in response to stress or upon overexpression (41,76). CIRP might regulate target transcripts by recruiting or releasing them into or from stress granules. In fact, we have observed that the p27 mRNA and CIRP can colocalize in stress granules following arsenic stress, indicating that CIRP might also recruit the p27 mRNA to stress granules (our unpublished data). Second, CIRP can regulate proteins involved in pathways modulating p27 stability, thereby indirectly regulating p27. It was described that direct binding of Mirk/Dyrk1b kinase by CIRP stimulates proliferation of immature germ cells. This interaction directly regulates p27, as it prevents p27 phosphorylation by Mirk/Dyrk1b kinase and eventually reduces p27 stability and expression (48). In addition, CIRP was shown to activate the Erk pathway (51,52,70), which controls cell proliferation by modulating key cell-cycle regulators including p27. Furthermore, CIRP might act through the regulation of additional factors that alter p27 transcription, mRNA stability, translation, or its activity or stability. Of note, it was reported earlier that CIRP can bind to a large number of transcripts (49,54,55), including mRNAs that encode other cell-cycle regulatory proteins (54). Regulation of cyclin E1 (76) and cyclin D1 (48,70) by CIRP was reported. These proteins can bind to p27 and alter its stability. Hence, reporter assays represent an option to separate translational control of p27 by CIRP from potential alternative direct or indirect pathways controlling p27. Finally, additional p27 mRNA-binding proteins might compete with CIRP for p27 mRNA binding, especially in the absence of stress. Of note, hypothermia seems to induce the CIRP/p27 mRNA binding.
The cell type and context-specific outcome of CIRP depletion or knockout on cell proliferation had been observed and discussed earlier, when it was found that CIRP can either prevent or promote cell proliferation (48). One critical factor that could contribute to these opposing consequences on cell proliferation following CIRP deletion could be p27, where CIRP impinges on several pathways of p27 regulation.
Whereas CIRP has been described to alter protein expression by binding to the 3′UTR of mRNAs, where it can stabilize the transcripts (40,59,60), only one study reported binding of CIRP to both 5′ and 3′UTRs of a transcript, Prm2 (49). The p27 mRNA is the first transcript where CIRP binding to the 5′UTR seems sufficient to alter translation. Binding may involve multiple binding sites like for the cyclin E mRNA, where CIRP binding was found to take place in the 3′UTR and in the coding region (76). U-rich regions were proposed to preferentially bind CIRP (35,49), in addition to a 51-nucleotides signature motif in 3′ UTRs (77) or an 8 nt-motif in CIRP-binding clusters that are located in exonic regions, especially coding sequence and 3′UTR (55). Other studies failed to identify CIRP-specific sequence motifs and proposed that structural motifs or other positional determinants confer binding specificity (54). We also tried to map a CIRP binding site(s) using an array of 5′ and 3′ truncations of the p27 5′UTR (13), but found that deletions from both ends of the p27 5′UTR reduced CIRP-dependent regulation of the reporter. The only fragment of the 5′UTR that could be deleted included 100 nt of the very 5′ end containing the CCRE and the uORF (data not shown). This indicates that the integrity of the structure of the 5′UTR, the presence of multiple binding sites, or binding of CIRP-interacting proteins to the p27 mRNA might determine CIRP binding to the p27 transcript.
We have shown here that CIRP induces p27 translation especially in mild hypothermic conditions. Under these conditions, we did not observe stress granules formation (data not shown). Initial experiments indicate that CIRP recruitment into stress granules leads to the colocalization of the p27 mRNA and reduced p27 translation (our unpublished data). It will be interesting to determine the molecular basis for this regulation in the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephan Geley for providing plasmids (pENTR THT shLuc, pTO destination vectors) and cell lines and for his help and critical discussion. We also thank Anne Willis (Leicester) for the pGL N-myc construct, Jun Fujita (Kyoto) for kindly providing the CIRP−/− MEF cell lines and Nisar P. Malek (Tübingen) for the p27−/− MEFs.
Author Contributions: M.R. designed the study, performed all experiments with the exception of Figure 1B and Supplementary Figure S5B and C, analyzed the data and wrote the manuscript. M.K. performed the experiments shown in Figure 1B and Supplementary Figure S5B and C, generated plasmids and provided intellectual input. L.H. conceived the study and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Cancer Research Tyrol (Österreichische Krebshilfe Tirol); Austrian Science Fund FWF (Doctoral college W1101 and project P24031).
Conflict of interest statement. None declared.
REFERENCES
- 1. Chu I.M., Hengst L., Slingerland J.M.. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008; 8:253–267. [DOI] [PubMed] [Google Scholar]
- 2. Hitomi M., Yang K., Guo Y., Fretthold J., Harwalkar J., Stacey D.W.. p27Kip1 and cyclin dependent kinase 2 regulate passage through the restriction point. Cell Cycle. 2006; 5:2281–2289. [DOI] [PubMed] [Google Scholar]
- 3. Jakel H., Peschel I., Kunze C., Weinl C., Hengst L.. Regulation of p27 (Kip1) by mitogen-induced tyrosine phosphorylation. Cell Cycle. 2012; 11:1910–1917. [DOI] [PubMed] [Google Scholar]
- 4. Hnit S.S., Xie C., Yao M., Holst J., Bensoussan A., De Souza P., Li Z., Dong Q.. p27(Kip1) signaling: transcriptional and post-translational regulation. Int. J. Biochem. Cell Biol. 2015; 68:9–14. [DOI] [PubMed] [Google Scholar]
- 5. Fero M.L., Randel E., Gurley K.E., Roberts J.M., Kemp C.J.. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature. 1998; 396:177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma S.S., Pledger W.J.. The non-canonical functions of p27(Kip1) in normal and tumor biology. Cell Cycle. 2016; 15:1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Besson A., Hwang H.C., Cicero S., Donovan S.L., Gurian-West M., Johnson D., Clurman B.E., Dyer M.A., Roberts J.M.. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 2007; 21:1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orlando S., Gallastegui E., Besson A., Abril G., Aligue R., Pujol M.J., Bachs O.. p27Kip1 and p21Cip1 collaborate in the regulation of transcription by recruiting cyclin-Cdk complexes on the promoters of target genes. Nucleic Acids Res. 2015; 43:6860–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philipp-Staheli J., Payne S.R., Kemp C.J.. p27(Kip1): regulation and function of a haploinsufficient tumor suppressor and its misregulation in cancer. Exp. Cell Res. 2001; 264:148–168. [DOI] [PubMed] [Google Scholar]
- 10. Larrea M.D., Liang J., Da Silva T., Hong F., Shao S.H., Han K., Dumont D., Slingerland J.M.. Phosphorylation of p27Kip1 regulates assembly and activation of cyclin D1-Cdk4. Mol. Cell. Biol. 2008; 28:6462–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hengst L., Reed S.I.. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996; 271:1861–1864. [DOI] [PubMed] [Google Scholar]
- 12. Agrawal D., Hauser P., McPherson F., Dong F., Garcia A., Pledger W.J.. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol. Cell. Biol. 1996; 16:4327–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gopfert U., Kullmann M., Hengst L.. Cell cycle-dependent translation of p27 involves a responsive element in its 5′-UTR that overlaps with a uORF. Hum. Mol. Genet. 2003; 12:1767–1779. [DOI] [PubMed] [Google Scholar]
- 14. Millard S.S., Yan J.S., Nguyen H., Pagano M., Kiyokawa H., Koff A.. Enhanced ribosomal association of p27(Kip1) mRNA is a mechanism contributing to accumulation during growth arrest. J. Biol. Chem. 1997; 272:7093–7098. [DOI] [PubMed] [Google Scholar]
- 15. Larocque D., Galarneau A., Liu H.N., Scott M., Almazan G., Richard S.. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005; 8:27–33. [DOI] [PubMed] [Google Scholar]
- 16. Miskimins W.K., Wang G., Hawkinson M., Miskimins R.. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 2001; 21:4960–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafre S.A. et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007; 26:3699–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A. et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007; 131:1273–1286. [DOI] [PubMed] [Google Scholar]
- 19. Galardi S., Petretich M., Pinna G., D’Amico S., Loreni F., Michienzi A., Groisman I., Ciafre S.A.. CPEB1 restrains proliferation of Glioblastoma cells through the regulation of p27(Kip1) mRNA translation. Sci. Rep. 2016; 6:25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R.. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010; 12:1014–1020. [DOI] [PubMed] [Google Scholar]
- 21. Kullmann M., Gopfert U., Siewe B., Hengst L.. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002; 16:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coleman J., Hawkinson M., Miskimins R., Miskimins W.K.. The major transcription initiation site of the p27Kip1 gene is conserved in human and mouse and produces a long 5′-UTR. BMC Mol. Biol. 2001; 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Z., Dong Z., Han B., Yang Y., Liu Y., Zhang J.T.. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005; 33:3763–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cho S., Kim J.H., Back S.H., Jang S.K.. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005; 25:1283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang H., Coleman J., Miskimins R., Srinivasan R., Miskimins W.K.. Cap-independent translation through the p27 5′-UTR. Nucleic Acids Res. 2007; 35:4767–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cuesta R., Martinez-Sanchez A., Gebauer F.. miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol. Cell. Biol. 2009; 29:2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coleman J., Miskimins W.K.. Structure and activity of the internal ribosome entry site within the human p27 Kip1 5′-untranslated region. RNA Biol. 2009; 6:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoon A., Peng G., Brandenburger Y., Zollo O., Xu W., Rego E., Ruggero D.. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006; 312:902–906. [DOI] [PubMed] [Google Scholar]
- 29. Bellodi C., Krasnykh O., Haynes N., Theodoropoulou M., Peng G., Montanaro L., Ruggero D.. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010; 70:6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Millard S.S., Vidal A., Markus M., Koff A.. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 2000; 20:5947–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng Y., Miskimins W.K.. Far upstream element binding protein 1 activates translation of p27Kip1 mRNA through its internal ribosomal entry site. Int. J. Biochem. Cell Biol. 2011; 43:1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng Y., Miskimins W.K.. CUG-binding protein represses translation of p27Kip1 mRNA through its internal ribosomal entry site. RNA Biol. 2011; 8:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G.. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993; 62:289–321. [DOI] [PubMed] [Google Scholar]
- 34. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 35. Nishiyama H., Itoh K., Kaneko Y., Kishishita M., Yoshida O., Fujita J.. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 1997; 137:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishiyama H., Higashitsuji H., Yokoi H., Itoh K., Danno S., Matsuda T., Fujita J.. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997; 204:115–120. [DOI] [PubMed] [Google Scholar]
- 37. Fujita J. Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1999; 1:243–255. [PubMed] [Google Scholar]
- 38. Sheikh M.S., Carrier F., Papathanasiou M.A., Hollander M.C., Zhan Q., Yu K., Fornace A.J. Jr. Identification of several human homologs of hamster DNA damage-inducible transcripts. Cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J. Biol. Chem. 1997; 272:26720–26726. [DOI] [PubMed] [Google Scholar]
- 39. Wellmann S., Buhrer C., Moderegger E., Zelmer A., Kirschner R., Koehne P., Fujita J., Seeger K.. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J. Cell Sci. 2004; 117:1785–1794. [DOI] [PubMed] [Google Scholar]
- 40. Yang C., Carrier F.. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 2001; 276:47277–47284. [DOI] [PubMed] [Google Scholar]
- 41. De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., Gueydan C.. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 2007; 313:4130–4144. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y., Wu Y., Mao P., Li F., Han X., Zhang Y., Jiang S., Chen Y., Huang J., Liu D. et al. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2015; 44:761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang G., Zhang J.N., Guo J.K., Cai Y., Sun H.S., Dong K., Wu C.G.. Neuroprotective effects of cold-inducible RNA-binding protein during mild hypothermia on traumatic brain injury. Neural Regen. Res. 2016; 11:771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang H.T., Xue J.H., Zhang Z.W., Kong H.B., Liu A.J., Li S.C., Xu D.G.. Cold-inducible RNA-binding protein inhibits neuron apoptosis through the suppression of mitochondrial apoptosis. Brain Res. 2015; 1622:474–483. [DOI] [PubMed] [Google Scholar]
- 45. Saito K., Fukuda N., Matsumoto T., Iribe Y., Tsunemi A., Kazama T., Yoshida-Noro C., Hayashi N.. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010; 1358:20–29. [DOI] [PubMed] [Google Scholar]
- 46. Liu A., Zhang Z., Li A., Xue J.. Effects of hypothermia and cerebral ischemia on cold-inducible RNA-binding protein mRNA expression in rat brain. Brain Res. 2010; 1347:104–110. [DOI] [PubMed] [Google Scholar]
- 47. Juan Y., Haiqiao W., Xie W., Huaping H., Zhong H., Xiangdong Z., Kolosov V.P., Perelman J.M.. Cold-inducible RNA-binding protein mediates airway inflammation and mucus hypersecretion through a post-transcriptional regulatory mechanism under cold stress. Int. J. Biochem. Cell Biol. 2016; 78:335–348. [DOI] [PubMed] [Google Scholar]
- 48. Masuda T., Itoh K., Higashitsuji H., Higashitsuji H., Nakazawa N., Sakurai T., Liu Y., Tokuchi H., Fujita T., Zhao Y. et al. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:10885–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xia Z., Zheng X., Zheng H., Liu X., Yang Z., Wang X.. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 2012; 586:3299–3308. [DOI] [PubMed] [Google Scholar]
- 50. Xia Z.P., Zheng X.M., Zheng H., Liu X.J., Liu G.Y., Wang X.H.. Downregulation of cold-inducible RNA-binding protein activates mitogen-activated protein kinases and impairs spermatogenic function in mouse testes. Asian J. Androl. 2012; 14:884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sakurai T., Itoh K., Higashitsuji H., Nonoguchi K., Liu Y., Watanabe H., Nakano T., Fukumoto M., Chiba T., Fujita J.. Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase. Biochim. Biophys. Acta. 2006; 1763:290–295. [DOI] [PubMed] [Google Scholar]
- 52. Artero-Castro A., Callejas F.B., Castellvi J., Kondoh H., Carnero A., Fernandez-Marcos P.J., Serrano M., Ramon y Cajal S., Lleonart M.E.. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol. Cell. Biol. 2009; 29:1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nishiyama H., Xue J.H., Sato T., Fukuyama H., Mizuno N., Houtani T., Sugimoto T., Fujita J.. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem. Biophys. Res. Commun. 1998; 245:534–538. [DOI] [PubMed] [Google Scholar]
- 54. Morf J., Rey G., Schneider K., Stratmann M., Fujita J., Naef F., Schibler U.. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012; 338:379–383. [DOI] [PubMed] [Google Scholar]
- 55. Liu Y., Hu W., Murakawa Y., Yin J., Wang G., Landthaler M., Yan J.. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 2013; 3:2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qiang X., Yang W.L., Wu R., Zhou M., Jacob A., Dong W., Kuncewitch M., Ji Y., Yang H., Wang H. et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat. Med. 2013; 19:1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rajayer S.R., Jacob A., Yang W.L., Zhou M., Chaung W., Wang P.. Cold-inducible RNA-binding protein is an important mediator of alcohol-induced brain inflammation. PLoS One. 2013; 8:e79430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou M., Yang W.L., Ji Y., Qiang X., Wang P.. Cold-inducible RNA-binding protein mediates neuroinflammation in cerebral ischemia. Biochim. Biophys. Acta. 2014; 1840:2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang R., Weber D.J., Carrier F.. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 2006; 34:1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang E.T., Parekh P.R., Yang Q., Nguyen D.M., Carrier F.. Heterogenous ribonucleoprotein A18 (hnRNP A18) promotes tumor growth by increasing protein translation of selected transcripts in cancer cells. Oncotarget. 2016; 7:10578–10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hong J.K., Kim Y.G., Yoon S.K., Lee G.M.. Down-regulation of cold-inducible RNA-binding protein does not improve hypothermic growth of Chinese hamster ovary cells producing erythropoietin. Metab. Eng. 2007; 9:208–216. [DOI] [PubMed] [Google Scholar]
- 62. Sigl R., Ploner C., Shivalingaiah G., Kofler R., Geley S.. Development of a multipurpose GATEWAY-based lentiviral tetracycline-regulated conditional RNAi system (GLTR). PLoS One. 2014; 9:e97764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 64. Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 65. Jopling C.L., Willis A.E.. N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene. 2001; 20:2664–2670. [DOI] [PubMed] [Google Scholar]
- 66. Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J.H., Itoh K., Matsuda T., Fujita J.. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 1997; 236:804–807. [DOI] [PubMed] [Google Scholar]
- 67. Brochu C., Cabrita M.A., Melanson B.D., Hamill J.D., Lau R., Pratt M.A., McKay B.C.. NF-kappaB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1beta. PLoS One. 2013; 8:e57426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ryan J.C., Morey J.S., Ramsdell J.S., Van Dolah F.M.. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience. 2005; 136:1121–1132. [DOI] [PubMed] [Google Scholar]
- 69. Zeng Y., Kulkarni P., Inoue T., Getzenberg R.H.. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J. Cell. Biochem. 2009; 107:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jian F., Chen Y., Ning G., Fu W., Tang H., Chen X., Zhao Y., Zheng L., Pan S., Wang W. et al. Cold inducible RNA binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via Erk signaling pathway. Oncotarget. 2016; 7:9175–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Q., Wang Y.Z., Zhang W., Chen X., Wang J., Chen J., Luo W.. Involvement of cold inducible RNA-binding protein in severe hypoxia-induced growth arrest of neural stem cells in vitro. Mol. Neurobiol. 2017; 54:2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pagano M., Tam S.W., Theodoras A.M., Beer-Romero P., Del Sal G., Chau V., Yew P.R., Draetta G.F., Rolfe M.. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995; 269:682–685. [DOI] [PubMed] [Google Scholar]
- 73. Zhu X., Buhrer C., Wellmann S.. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. 2016; 73:3839–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matsumoto K., Aoki K., Dohmae N., Takio K., Tsujimoto M.. CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes. Nucleic Acids Res. 2000; 28:4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Aoki K., Ishii Y., Matsumoto K., Tsujimoto M.. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 2002; 30:5182–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo X., Wu Y., Hartley R.S.. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol. Carcinog. 2010; 49:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang R., Zhan M., Nalabothula N.R., Yang Q., Indig F.E., Carrier F.. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J. Biol. Chem. 2010; 285:8887–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.