Abstract
Background
Mutations in the CD46 gene account for an important proportion of patients with atypical hemolytic uremic syndrome (aHUS) who characteristically show multiple relapses, no response to plasma exchange and low recurrence risk in allograft. We screened for mutations in CD46 in patients with and without circulating anti-factor H (FH) antibodies–associated aHUS.
Methods
We estimated CD46 surface expression by flow cytometry and sequenced the CD46 gene in 23 and 56 patients with and without circulating anti-FH antibodies, respectively. Human Splicing Finder and PolyPhen2 were used for in silico prediction of pathogenicity.
Results
Two novel and three known (c.286 +2T > G, c.104G > A and c.565T > G) mutations in CD46 were found in nine (11.4%) patients; one patient had a variant of unknown significance and two patients presented during the first year of life. Novel intronic (c.1127 + 46C > G) and exonic (c.911C > T) mutations are proposed to activate cryptic splicing sites or alter protein conformation. Markedly reduced CD46 surface expression was found in homozygous states in five patients.
Conclusion
Patients with mutations in CD46 present at all ages, including the first year of life. Mutations in intron 2, (c.286 +2T > G) may be a potential hot spot in Indian children. Flow cytometry for CD46 expression is a satisfactory screening tool enabling early diagnosis.
Keywords: complement, hemolytic uremic syndrome, membrane cofactor protein
Introduction
Atypical hemolytic uremic syndrome (aHUS), characterized by microangiopathic hemolytic anemia and thrombocytopenia is an important cause of acute kidney injury (AKI) in children [1]. Mutations in genes encoding proteins of the complement and coagulation pathway, including factor H (FH), factor I (FI), factor B, membrane cofactor protein (MCP, CD46), C3, thrombomodulin and diacylglycerol kinase epsilon, predispose to the disease [1]. CD46, expressed on cell membranes, regulates the alternative complement pathway by serving as a cofactor for FI to mediate inactivation of C3b and C4b deposited on host cells [2]. The gene encoding this protein has 14 exons and is located in the regulator of complement activation cluster at 1q32 [2, 3]. Mutations in CD46 account for ∼5–20% of children with aHUS. These are typically older children and they have satisfactory outcomes with a low risk of recurrence in allografts [3–5].
We have previously reported that more than half of children with aHUS in India show autoantibodies to FH, resulting in inhibition of its regulatory action [6]. Most of these patients show a homozygous deletion in the gene encoding FH-related protein 1 (CFHR1) [6]. While genetic screening in a few patients did not show additional mutations in patients with antibody-associated aHUS [7–9], the precise frequency of CD46 mutations is unknown. We therefore prospectively screened a group of patients with aHUS, with or without anti-FH antibodies, to examine for mutations in the CD46 gene.
Materials and methods
From a cohort of patients with aHUS [6], we screened 23 and 56 consecutive patients with and without anti-FH antibody–associated HUS, respectively, for CD46 expression on neutrophils by flow cytometry and by sequencing of the CD46 gene. The diagnosis of HUS was based on the presence of AKI, microangiopathic hemolytic anemia and thrombocytopenia (platelets <150 000/mm3). Patients with prodromal dysentery, features of systemic pneumococcal infection, sepsis, systemic lupus or other collagen vascular diseases were excluded. Hematological remission was defined as a platelet count >150 000/mm3 and the absence of microangiopathic anemia. Disease relapse was considered when there was a new episode of illness after the patient was in remission for >2 weeks.
Flow cytometry was used to estimate CD46 expression on neutrophils and quantified as median fluorescence intensity (MFI). Briefly, 100 μL of ethylenediaminetetraacetic acid blood was incubated with 20 μL of fluorescein isothiocyanate–labeled anti-CD46 monoclonal antibody (Becton Dickinson Biosciences, San Jose, CA, USA) for 30 min at 4°C. Following erythrocyte lysis, cells were washed with phosphate buffered saline and pellets resuspended in 300 μL of staining buffer. A flow cytometer acquired, recorded and analysed 50 000 events using Diva software version 6.1.2 (BD Biosciences). Normal CD46 MFI expression was defined as 70–130% of the expression in 50 healthy controls.
Analysis of CD46 mutations
Genomic DNA was extracted from peripheral blood leukocytes of patients and 50 healthy controls, as previously described [10]. Genetic analysis was done by polymerase chain reaction amplification of coding exons and splice sites using 0.5 µM of each primer (Table 1), 50–100 ng DNA, 1.25 mM MgCl2, 0.25 mM of each dNTP (Invitrogen, Carlsbad, CA, USA) and 0.5 units of Taq polymerase (Invitrogen). The reactions were cycled on an ABI 9700 (thermocycler Applied Biosystems, Foster City, CA, USA) (Table 1). Amplified products were purified using Qiagen kits (Hilden, Germany) and sequenced on an ABI-3100 analyzer (Applied Biosystems). Nucleotide sequences were compared with the published cDNA sequences (GenBank ENSG00000117335).
Table 1.
Primer sequences used for CD46 screening
| Forward primer | Reverse primer | Annealing temperature (°C) | |
|---|---|---|---|
| Exon 1 | 5′- CTGTCCTGCAGCACTGGATG-3′ | 5′- CACGGCCTGCTGTGAGC-3′ | 62 |
| Exon 2 | 5′- AGGGCCTTTCTGTTTTTTCTG-3′ | 5′- GTAGTGGAATATGTACCCCAA-3′ | 54 |
| Exon 3 | 5′- ATTCCCACCCATTCAAAAGAG-3′ | 5′- GCCTATCTCCATAAAACATCC-3′ | 54 |
| Exon 4 | 5′- CCACCCCCTCAAACTACTGTAGTG-3′ | 5′- AGAAACCTCTTTGGGATCTTTGTTA-3′ | 62 |
| Exon 5 | 5′-CATTTCCTTTCCTCTTTTTC-3′ | 5′-ACACCTGCTTTGTTTATCTGTAGA-3′ | 52 |
| Exon 6 | 5′- GTCTCTGTTCACACTGGAAAT-3′ | 5′- TACATAACGTGCTAAGAACCC-3′ | 54 |
| Exon 7, 8 | 5′- CCAAGTGGTTGATCTTCTAAC-3′ | 5′- ATGGCTATACAAATGTCCTCC-3′ | 60 |
| Exon 9 | 5′- ATTGATAAGGCCCTGGTGAAT-3′ | 5′- CACACATACCCTAGAGCTTAA-3′ | 60 |
| Exon 10 | 5′- CCCTATGAGTTTAAAGGATTTTAAGCTT-3′ | 5′- CCTATGTTTGGGCACCTCATAA-3′ | 58 |
| Exon 11 | 5′- GGAGATCCATGTGTTCAACATCTT-3′ | 5′- TCGGTTTAACCAATTTACAAGCTG-3′ | 58 |
| Exon 12 | 5′- TTGACCACTGAAATGTAACCAACA-3′ | 5′- TGAAGCTGCACAAAAGCATGT-3′ | 60 |
| Exon 13 | 5′- ATCCCACTTGTTATGCTACTC-3′ | 5′- TGCCAATATCTCTTTGCTCAG-3′ | 60 |
| Exon 14 | 5′-TCATTTTCTGAATAGGCTTCTGGAAT-3′ | 5′-GCACTCATGAGAGTGAAACTA-3′ | 58 |
In silico prediction of pathogenicity of the variations was done using Human Splicing Finder (HSF) version 2.4.1 (http://www.umd.be/HSF/) to assess the effects of missense intronic and exonic variations on splicing [11]. PolyPhen-2 version 2.2.2 was also used to predict the impact of nonsynonymous exonic mutations on the structure and function of the protein [12]. Analysis was done using default threshold and cutoff values as mentioned in the software.
Factor H and anti-FH antibody assays
FH was measured by enzyme-linked immunosorbent assay (ELISA) [13] and C3 by nephelometry. Anti-FH antibody levels and rearrangements in the CFHR1–5 genomic region were analyzed by ELISA and multiplex ligation probe amplification (MLPA), respectively [6]. Anti-FH antibody levels >150 arbitrary units (AU)/mL were considered abnormal.
Results
Of 79 patients screened, 53 were boys; the median age at evaluation was 5.9 [interquartile range (IQR) 1.5–9.6] years. All had microangiopathic hemolysis and AKI (dialysis in 80%); Stage 2 hypertension (43.0%) and significant proteinuria (59.4%) were common. The median blood level of C3 was 73.5 (IQR 57.5–93.3) mg/dL and the anti-FH antibody level in 23 patients with antibody-associated aHUS was 5573 (IQR 1078–10 734) AU/mL. Patients were treated with plasma exchange (46.8%), plasma infusions (13.9%) or both (8.9%); those with anti-FH antibodies also received therapy with prednisolone (n = 23) and cyclophosphamide (n = 10), intravenous rituximab (n = 4) and mycophenolate mofetil (n = 9). Relapses were seen in 15 (18.9%) patients and a family history of a similar illness was present in 9.
Patients with CD46 mutations
Genetic analysis revealed sequence alterations in the coding or intronic region of the CD46 gene in 10 (12.7%) patients (Table 2). These patients were between 6 months and 15 years of age, with median age of 4 (IQR 1.6–7.5) years; two were <12 months. At a median follow-up of 27 (IQR 11–42) months, hematological remission and renal recovery were achieved in all, with estimated glomerular filtration rate of 46–100 mL/1.73 m2/min. Five patients had 1–11 relapses 6 months to 5 years from the initial illness. Familial occurrence was present in five children from three families. Complement C3 levels were low (<90 mg/dL) in three patients; one also had borderline low levels of FH at 124 mg/L (normal 150–320 mg/L). Nine patients were negative for anti-FH antibodies. Only one patient showed anti-FH antibodies (6277 AU/mL) with homozygous deletion of CFHR1/3 (Patient 1; Table 2). The phenotype of this patient was not particularly different from those with isolated anti-FH antibodies.
Table 2.
Clinical and biochemical features in patients with aHUS associated with CD46 mutation
| Patient | Age, sex | Features | Hypertension; proteinuria | Hemoglobin (g/dL; platelet/mm3) | Peak creatinine (mg/dL) | Serum C3 (mg/dL)a | FH (mg/dL)b; antibody (AU/mL) | Relapses | eGFR (mL/min/1.73 m2) at follow-up |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 9 years, M | Fever, oliguria | Stage 2; 1+ | 6.1; 670 00 | 5.30 | 131 | 200; 6277 | 1 (after 1 year) | 70 after 1.5 years |
| 2 | 5 years, M | Dengue, melena, high transaminases | Stage 2; 4+ | 6.0; 80 000 | 5.93 | 99 | 124; 62 | None | 100 after 3 months; normal blood pressure |
| 3 | 8 years, F | Pallor, oliguria | NA | NA | NA | NA | 171; <37 | None | 95 after 3 years; normal blood pressure |
| 4 | 6 months, M | Fever, diarrhea, high transaminases | None | 5.7; 59 000 | 3.20 | 96 | 151; <37 | None | 46 after 1 month; normal blood pressure |
| 5 | 6 years, M | Fever | Stage 2; 2+ | 9.9; 300 000 | 4.60 | 180 | 152; <37 | NA | NA |
| 6 | 1.7 years, M | NA | NA | NA | NA | 97 | 162; 85 | 11 (interval 0.5–2 years) | 90 after 14 years |
| 7 | 1.5 years, M | Oliguria, high transaminases | Stage 1; 4+ | 5.5; 21 000 | 0.95 | 57 | 199; 43 | 1 (after 5 years) | 75 after 5 years |
| 8 | 10 months, F | Fever, oliguria, hematuria | Stage 1; 3+ | 4.9; 34 000 | 1.42 | 73 | 200; 57 | NA | NA |
| 9 | 3 years, M | Diarrhea, pallor, oliguria | None, 1+ | 7.3; 161 000 | 4.9 | 45 | 170; 77.4 | 1 (after 6 months) | 95 after 14 months |
| 10 | 15 years, M | Fever, cough, coryza | Stage 1; 2+ | 8.6, 80 000 | 8.3 | 136 | 180; 54.8 | 1 (after 3 years) | 90 after 3 .5 years; normal blood pressure |
Therapy included PD or hemodialysis; Patient 1 also received plasma exchanges and received immunosuppressive agents; Patients 2, 7, and 8 received plasma infusions.
Patients 2 and 3 (previously reported in Bhatia et al. [14]) and 7 and 8 are siblings.
AU/mL, arbitrary units/mL; eGFR, estimated glomerular filtration rate; FH, complement factor H; HD, hemodialysis; F, female; M, male; NA, not available.
Serum C3 normal range 90–130 mg/dL. b FH normal range 150–320 mg/dL.
Sequence changes found included a previously reported splice-site mutation c.286 + 2T > G (intron 2) in five patients (three families) and missense mutations c.104G > A (exon 2) and c.565T > G (exon 5) in one patient each. Novel intronic (c.475 + 33A > G and c.1127 + 46C > G) and exonic (c.911C > T) variations were found in four patients (Table 3, Figure 1). Sibling pairs 2–3 and 7–8 with familial disease showed a homozygous c.286 + 2T > G splice-site mutation; in both families, the parents were consanguineous. One patient (Patient 9) had a homozygous c.104G > A missense mutation resulting in substitution of tyrosine for cysteine at codon 35; his affected sibling had died before genetic evaluation. A previously reported c.565T > G mutation was present in a heterozygous state in Patient 10 that resulted in substitution of aspartic acid for tyrosine at codon 189 in exon 5 (Table 3).
Table 3.
CD46 cell surface expression and genetic analysis in patients with aHUS
| Patients | MFI, % | cDNA position of mutation; amino acid change | In silico prediction/functional studies | Classification | Zygosity |
|---|---|---|---|---|---|
| 1 | 59.4 | Intron 12; c.1127 + 46C>Ga | Activation of intronic cryptic acceptor site; potential alteration of splicing | Novel | Heterozygous |
| 2 | 10.4 | Intron 2; c.286 + 2T>Gc | Defective splicing causing protein truncation | Previously reported | Homozygous |
| 3 | 10.4 | Intron 2; c.286 + 2T>Gc | Defective splicing causing protein truncation | Previously reported | Homozygous |
| 4 | 69.4 | Exon 8; c.911C>Ta,b, Missense (p.S304F) | Alteration of exonic splicing enhancer site; possibly damaging | Novel | Heterozygous |
| 5 | – | Intron 4; c.475 + 33A>Ga | Activation of intronic cryptic donor site; potential alteration of splicing | Variant of unknown significance, MAF <0.01% (ExAC) | Heterozygous |
| 6 | 9.5 | Intron 2; c.286 + 2T>Gc | Defective splicing causing protein truncation | Previously reported | Heterozygous |
| 7 | 12.8 | Intron 2; c.286 + 2T>Gc | Defective splicing causing protein truncation | Previously reported | Homozygous |
| 8 | 16.0 | Intron 2; c.286 + 2T>Gc | Defective splicing causing protein truncation | Previously reported | Homozygous |
| 9 | 8.5 | Exon 2; c.104G>Ad, Missense (p.C35Y) | Formation of aberrant precursor protein; alteration of exonic splicing enhancer site | Previously reported | Homozygous |
| 10 | 58.2 | Exon 5; c.565T>Gb,c, Missense (p.Y189D) | Formation of aberrant precursor protein, probably damaging | Previously reported, MAF 0.02% (1000 genome) | Heterozygous |
Disease occurred sporadically in all, except Patients 9 and sibling pairs Patients 2–3 [14] and 7–8 whose parents were heterozygous for the same mutation.
ExAC, Exome Aggregation Consortium; MAF, minor allele frequency by public databases; MFI, median fluorescence intensity.
In silico prediction by Human Splicing Finder (HSF) version 2.4.1.
In silico prediction by PolyPhen-2.
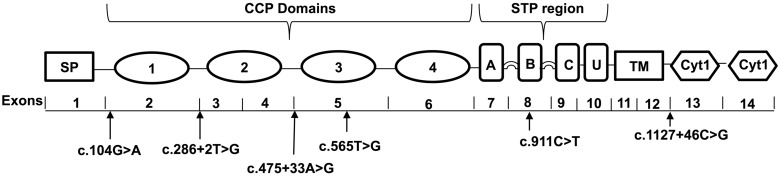
Fig. 1.
Schematic representation of CD46 showing variations identified in the present study. CD46 has 14 exons and encodes a protein with a signal peptide (SP) region of 34 amino acids, 4 complement control proteins (CCP 1–4) repeats that perform regulatory functions, alternatively spliced region for o-glycosylation (A, B and C), a segment of undefined function (U), a transmembrane domain (TM) and two alternatively spliced cytoplasmic tails (CYT-1, CYT-2).
The intronic c.475 + 33A > G and c.901 + 36A > G variations were predicted to activate an intronic cryptic donor splice site, with variation in consensus value (ΔCV) of −55.1% and +17.1%, respectively. These are reported as very rare variants with a minor allele frequency of <0.01% in public databases (EXaC and 1000 Genome). The patient with c.901 + 36A > G variation showed almost normal CD46 surface expression (93.2%), therefore we do not consider this variation to be clinically significant. Flow cytometry could not be performed in the other patient with a c.475 + 33A > G variation, therefore this variant is of unknown significance.
The heterozygous intronic c.1127 + 46C > G and exonic c.911C > T sequence change was not reported in public databases and in 50 healthy Indian controls, indicating an association with the disease condition. The intronic c.1127 + 46C > G change was predicted to activate an intronic cryptic acceptor splice site (ΔCV +52.9% and +1492.6% by HSF and MaxEnt, respectively). The exonic mutation c.911C > T, resulting in a missense substitution of phenylalanine for serine, was predicted to be ‘possibly damaging’, with a score of 0.87 on PolyPhen-2, in addition to disrupting an exonic splicing enhancer site. CD46 expression was decreased (∼50%) in both these patients. CD46 single nucleotide polymorphisms rs2724374 and rs11118580 were present with equal frequency in patients and 50 healthy controls.
Surface expression of CD46 in patients with homozygous mutations was ∼<10% of normal MFI (severely deficient) (Table 3). While heterozygous mutation is expected to result in ∼50% expression of the protein, Patient 6 (Table 3) and his asymptomatic 5-year-old brother, both with a heterozygous c.286 + 2T > G mutation, had markedly reduced expression (9.5% and 16%, respectively).
Discussion
We describe the clinical features and CD46 mutations in Indian children with aHUS. These patients presented at a relatively young age and had a relapsing course; all attained hematological remission and none progressed to end-stage renal failure. The frequency of CD46 mutations was 16.1% (9/56) and 4.3% (1/23) in anti-FH antibody–negative and–positive patients, respectively. This finding is similar to the previously reported frequency of 5–20% in antibody-negative patients [4, 5] and a rare report in an antibody-positive patient [17]. We found three previously reported pathogenic mutations and two novel mutations in nine patients; one other patient showed a variant of unknown significance.
Most CD46 mutations reported in the literature are missense and only ∼11% cause splicing defects [2]. Consistent with this finding, all exonic mutations found in the current study were missense mutations. Decreased surface expression due to the missense exonic c.911C > T mutation may lead to an abnormal conformation of the mutated protein that abolishes its proper expression on the cell membrane. Previous studies report that the exonic c.104G > A and c. 565T > G mutations lead to an aberrant precursor CD46 protein [3, 5, 16]. The exonic c.911C > T mutation is also predicted to alter splicing regulatory signals, leading to the formation of a defective protein. Splicing regulatory elements including exonic splicing enhancers and silencers are required for accurate pre-mRNA splicing [18]. While exonic mutations cause defects in encoded proteins, ∼25% might also alter splicing regulatory signals [19], similar to that predicted for the exonic c.911C > T. Three of the six mutations identified in the current study were predicted to alter pre-mRNA splicing, consistent with the role of aberrant mRNA processing in causing diseases [20]. Intronic mutations, such as c.1127 + 46C > G in this study, that introduce new or cryptic splice sites may result in partial inclusion of intronic or partial or complete exclusion of exonic sequences [21].
The reported intronic splice site mutation c.286 + 2T > G was seen in five of the present patients. Functional studies have shown this change results in abnormal splicing of exon 2 and deletion of 48 amino acids from the short consensus repeats 1–2 of CD46 protein [15]. This might impair CD46 protein production and/or trafficking to the cell surface, decreasing its expression [22], as noted in our patients and their asymptomatic siblings. Given its presence in multiple patients in this small cohort, this change might represent a mutational hot spot.
CD46 mutations are usually inherited as heterozygous changes [1]. However, homozygous mutations were reported in 6 of 214 (2.8%) patients with aHUS in the French cohort, often in patients with onset in childhood [5/89 (5.6%)] rather than in adults [1/125 (0.8%)] [23]. Only 2 of 273 (0.7%) aHUS patients in the Italian cohort showed homozygous CD46 mutations [5]. In the present report, homozygous mutations were seen in 5 of 79 (6.3%) children with aHUS. Homozygous mutations result in extremely low surface expression of CD46 on leukocytes (<10% MFI) [1–3, 23], as confirmed in this study.
One patient with a novel heterozygous exonic variation (c.911C > T) and another with a homozygous splice site mutation (c.286 + 2T > G) presented at the ages of 6 and 10 months, respectively. The onset of aHUS during the first year of life in two of the present patients is noteworthy, since an early presentation is exceptional in patients with CD46 deficiency [4]. No patient in the French cohort [24] and only 1 of 18 patients in the Italian registry [5] presented before 1 year of age. Our findings suggest that, like patients with CFH, DGKE or CFI mutations, homozygous and heterozygous CD46 mutations may present during infancy. The presence of a diarrheal prodrome in two patients highlights the need to recognize infectious triggers. Infections with Shigella flexneri [25] and shiga toxin–producing Escherichiacoli O157:H7 [15, 26] have been associated with CD46 deficiency–associated aHUS. Renal recovery without progression to end-stage renal disease in our patients confirms the satisfactory renal prognosis of patients with CD46 mutations, especially in those with disease onset in childhood compared with adults [24]. We also observed a high rate of relapses (15 relapses in 7 patients), in agreement with the risk of relapses in patients with underlying CD46 defects [5].
The present report has certain limitations. First, the study was limited to a small number of subjects, comprising ∼10% patients in the database, limiting its generalizability to the larger cohort. Second, we did not screen for concomitant mutations in other genes known to be associated with aHUS. A mutation in CD46 in combination with changes in the CFH, CFI or C3 gene is reported in 2.4% of a cohort of 795 patients from the International Registry of aHUS and registries from France, Spain and the UK, without differences in features in those with isolated CD46 and combined mutations [27]. Finally, while in silico prediction of pathogenicity was done, we did not sequence cDNA from the mRNA or perform functional studies to confirm that the novel variations were likely to affect splicing and cause disease.
Nevertheless, our findings suggest that homozygous and heterozygous mutations in the CD46 gene are an important cause of aHUS in Indian children. Children might present at all ages, including infancy, with HUS being triggered by an infectious illness. Patients with homozygous mutations show significantly reduced surface expression of CD46 on neutrophils. Flow cytometry for CD46 expression is a satisfactory screening tool that enabled detection in all patients with CD46 mutations in this series where this test could be performed. Being a more rapid method than in-depth genetic analysis, flow cytometry may influence early treatment decisions, keeping in mind that a negative result does not exclude the presence of CD46 mutations. Identification of patients with CD46 deficiency has implications for management, since they show multiple relapses and are unlikely to respond to plasma exchanges.
Acknowledgements
Collaboration between the All India Institute of Medical Sciences, New Delhi and Laboratoire d'Immunologie, Hôpital Européen Georges Pompidou, Paris, France, was possible through the Indo-French Centre for the Promotion of Advanced Research (CEFIPRA) Project 4703.
Funding
The study was supported by the Department of Biotechnology, Government of India, 102/IFD/SAN/PR2624/2010–11 Collaboration between the All India Institute of Medical Sciences, New Delhi and Laboratoire d'Immunologie, Hôpital Européen Georges Pompidou, Paris, France, was possible through the Indo-French Centre for the Promotion of Advanced Research (CEFIPRA) Project 4703.
Conflict of interest statement
None declared.
References
- 1. Kavanagh D, Goodship TH, Richards A.. Atypical hemolytic uremic syndrome. Semin Nephrol 2013; 33: 508–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liszewski MK, Atkinson JP.. Complement regulator CD46: genetic variants and disease associations. Hum Genomics 2015; 9: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richards A, Kathryn Liszewski M, Kavanagh D. et al. Implications of the initial mutations in membrane cofactor protein (MCP; CD46) leading to atypical hemolytic uremic syndrome. Mol Immunol 2007; 44: 111–122 [DOI] [PubMed] [Google Scholar]
- 4. Loirat C, Fremeaux-Bacchi V.. Atypical hemolytic uremic syndrome. Orphanet J Rare Dis 2011; 6: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noris M, Caprioli J, Bresin E. et al. Relative role of genetic complement abnormalities in sporadic and familial aHUS and their impact on clinical phenotype. Clin J Am Soc Nephrol 2010; 5: 1844–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinha A, Gulati A, Saini S. et al. Prompt plasma exchanges and immunosuppressive treatment improves the outcomes of anti-factor H autoantibody-associated hemolytic uremic syndrome in children. Kidney Int 2014; 85: 1151–1160. [DOI] [PubMed] [Google Scholar]
- 7. Dragon-Durey MA, Sethi SK, Bagga A. et al. Clinical features of anti-factor H autoantibody-associated hemolytic uremic syndrome. J Am Soc Nephrol 2010; 21: 2180–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abarrategui-Garrido C, Martinez-Barricarte R, Lopez-Trascasa M. et al. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood 2009; 114: 4261–4271 [DOI] [PubMed] [Google Scholar]
- 9. Hofer J, Janecke AR, Zimmerhackl LB. et al. Complement factor H-related protein 1 deficiency and factor H antibodies in pediatric patients with atypical hemolytic uremic syndrome. Clin J Am Soc Nephrol 2013; 8: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller SA, Dykes DD, Polesky HF.. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desmet FO, Hamroun D, Lalande M. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramensky V, Bork P, Sunyaev S.. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dragon-Durey MA, Loirat C, Cloarec S. et al. Anti-factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 2005; 16: 555–563 [DOI] [PubMed] [Google Scholar]
- 14. Bhatia D, Khandelwal P, Sinha A. et al. Incomplete penetrance of CD46 mutation causing familial atypical hemolytic uremic syndrome. Pediatr Nephrol 2015; 30: 2215–2220 [DOI] [PubMed] [Google Scholar]
- 15. Fremeaux-Bacchi V, Moulton EA, Kavanagh D. et al. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol 2006; 17: 2017–2025 [DOI] [PubMed] [Google Scholar]
- 16. Caprioli J, Noris M, Brioschi S. et al. Genetics of HUS: the impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood 2006; 108: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore I, Strain L, Pappworth I. et al. Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 2010; 115: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pagani F, Buratti E, Stuani C. et al. A new type of mutation causes a splicing defect in ATM. Nat Genet 2002; 30: 426–429 [DOI] [PubMed] [Google Scholar]
- 19. Sterne-Weiler T, Sanford JR.. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol 2014; 15: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammond SM, Wood MJ.. Genetic therapies for RNA mis-splicing diseases. Trends Genet 2011; 27: 196–205 [DOI] [PubMed] [Google Scholar]
- 21. Wang Z, Burge CB.. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 2008; 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fremeaux-Bacchi V, Arzouk N, Ferlicot S. et al. Recurrence of HUS due to CD46/MCP mutation after renal transplantation: a role for endothelial microchimerism. Am J Transplant 2007; 7: 2047–2051 [DOI] [PubMed] [Google Scholar]
- 23. Fremeaux-Bacchi V, Fakhouri F, Garnier A. et al. Genetics and outcome of atypical hemolytic uremic syndrome: a nationwide French series comparing children and adults. Clin J Am Soc Nephrol 2013; 8: 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Couzi L, Contin-Bordes C, Marliot F. et al. Inherited deficiency of membrane cofactor protein expression and varying manifestations of recurrent atypical hemolytic uremic syndrome in a sibling pair. Am J Kidney Dis 2008; 52: e5–e9 [DOI] [PubMed] [Google Scholar]
- 25. Brocklebank V, Wong EK, Fielding R. et al. Atypical haemolytic uremic syndrome associated with a CD46 mutation triggered by Shigella flexneri. Clin Kidney J 2014; 7: 286–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alberti M, Valoti E, Piras R. et al. Two patients with history of STEC-HUS, posttransplant recurrence and complement gene mutations. Am J Transplant 2013; 13: 2201–2206 [DOI] [PubMed] [Google Scholar]
- 27. Bresin E, Rurali E, Caprioli J. et al. Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol 2013; 24: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]