Abstract
Dynamic changes in DNA methylation and demethylation reprogram transcriptional outputs to instruct lineage specification during development. Here, we applied an integrative epigenomic approach to unveil DNA (hydroxy)methylation dynamics representing major endodermal lineage intermediates during pancreatic differentiation of human embryonic stem cells (hESCs). We found that 5-hydroxymethylcytosine (5hmC) marks genomic regions to be demethylated in the descendent lineage, thus reshaping the DNA methylation landscapes during pancreatic lineage progression. DNA hydroxymethylation is positively correlated with enhancer activities and chromatin accessibility, as well as the selective binding of lineage-specific pioneer transcription factors, during pancreatic differentiation. We further discovered enrichment of hydroxymethylated regions (termed ‘5hmC-rim’) at the boundaries of large hypomethylated functional genomic regions, including super-enhancer, DNA methylation canyon and broad-H3K4me3 peaks. We speculate that ‘5hmC-rim’ might safeguard low levels of cytosine methylation at these regions. Our comprehensive analysis highlights the importance of dynamic changes of epigenetic landscapes in driving pancreatic differentiation of hESC.
INTRODUCTION
DNA methylation is a stable and heritable epigenetic mark involved in the regulation of genome organization and gene transcription (1,2). Dynamic changes in DNA methylation are essential for reprogramming the transcriptional network during development (3–6). The covalent addition of a methyl group at the 5-carbon position of cytosine is primarily catalyzed by DNA methyltransferases (DNMTs) and often signals for transcriptional repression (7). The reversal of DNA methylation, or DNA demethylation, is achieved through a combination of both active and passive mechanisms (8). Active DNA demethylation is primarily mediated by the Ten-eleven translocation (TET) family of 2-oxoglutarate and iron-dependent dioxygenase that successively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), followed by thymine DNA glycosylase (TDG)-mediated base excision and repair to generate unmodified cytosine (9–12). TET-mediated 5mC oxidation is further implicated in passive DNA demethylation since DNMT1 activity might be reduced by up to 60-fold when the DNA substrate contains 5hmC, thereby leading to 5mC dilution during DNA replication and cell division (13–15). The DNA methylation homeostasis is exquisitely maintained through coordinated actions of DNMT and TET proteins, and is critical for shaping context-dependent epigenetic landscapes to orchestrate chromatin accessibility and gene expression (16,17). Genetic depletion of DMNTs or TETs has been shown to impair endoderm differentiation (10,18), indicating the importance of both DNA methylation and demethylation in regulating stem cell differentiation and lineage specification.
Directed differentiation of human embryonic stem cells (hESCs) provides a powerful in vitro model system for understanding how cells respond to extrinsic cues and intrinsic regulatory factors that instruct lineage specification and govern tissue or organ development (19). For example, stepwise differentiation of hESC to pancreatic endoderm (PE) can recapitulate essential steps of in vivo pancreatic development by exposing hESCs to different sets of extrinsic signaling molecules (Figure 1A). The pancreatic differentiation protocol involves the induction of hESC into definitive endoderm (DE), which gives rise to the primitive gut tube (GT). Following extrinsic cues, GT can be subsequently converted into posterior foregut (FG), followed by the generation of PE (20). This convenient approach enables us to obtain sufficient numbers of transitory lineage intermediates, which would otherwise be technically demanding to achieve in vivo with living embryos. This strategy allows us to capture the epigenetic states and gene expression profiles representative of each developmental stage, and to dissect how intrinsic regulatory factors, such as epigenomic modifiers and their catalytic products, remodel the transcriptional networks in response to extrinsic signals during transitions. Indeed, two recent elegant studies have shown that key histone modifications associated with polycomb proteins or enhancer activities are essential for hESC-to-PE differentiation (21,22). However, very little is known regarding the changes in the other arm of the epigenetic machinery, i.e. DNA methylation and demethylation, during the differentiation of hESC toward a terminal pancreatic fate in human. Furthermore, how DNA methylation and demethylation influence chromatin accessibility and transcription factor binding during this dynamic process remains largely undefined.
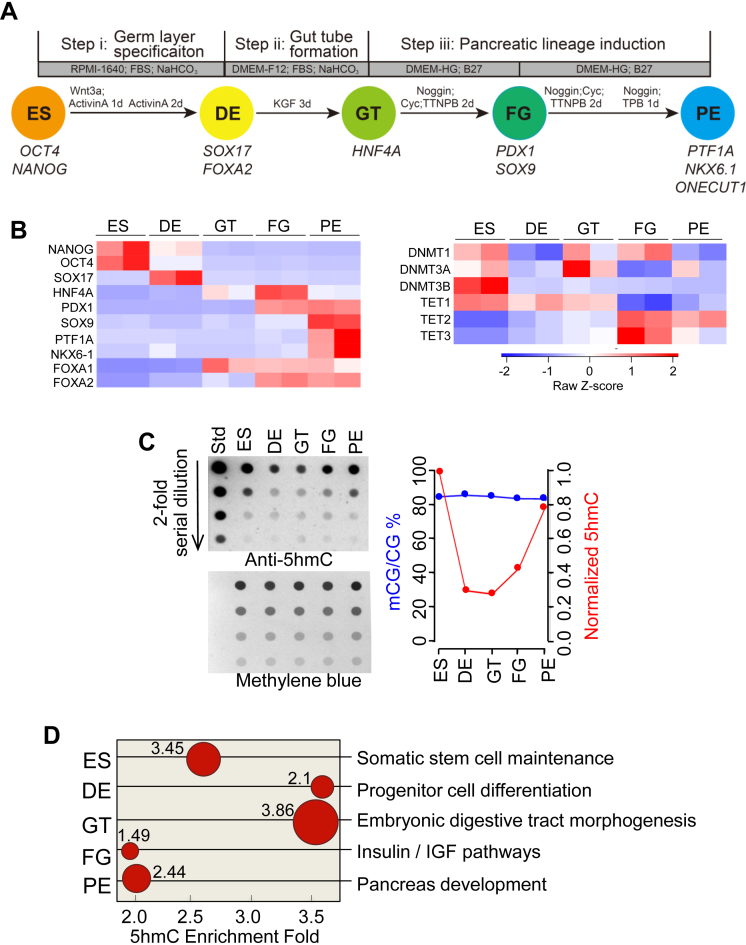
Figure 1.
Dynamic changes of gene expression, DNA methylation, hydroxymethylation, and chromatin accessibility during pancreatic differentiation of H1-hESCs. (A) Schematic illustrating directed differentiation of hESC toward a pancreatic fate. ES, embryonic stem cell; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. OCT4, octamer-binding transcription factor 4; SOX17, SRY (sex determining region Y)-box 17; HNF4A, hepatocyte nuclear factor 4; PDX1, pancreatic and duodenal homeobox 1; NKX6.1, NK6 homeobox 1. (B) Heatmaps depicting the expression of stage specific genes (left) and key genes involved in DNA methylation and demethylation (DNMTs and TETs; right) during the ES-to-PE progression. (C) Quantitation of dynamic changes in global 5mC and 5hmC levels during pancreatic differentiation of hESCs. (left) A dot-blot assay used to quantify 5hmC. The loading control was shown in the bottom panel by staining the blot with methylene blue to visualize the total amounts of input DNA. (right) Quantification of global 5mC (from WGBS) and 5hmC (from the dot-blot assay) levels at five defined stages of differentiation. (D) Gene ontology term enrichments for 5hmC peaks identified at each of the five defined differentiation stages. The representative functions for each stage were shown as red dots. The size of each circle (proportional to the number next to it) represents the –log10 (Binomial p values shown next to the circles); whereas the X-axis stands for the enrichment fold of 5hmC signals.
In this study, we performed whole-genome bisulfite sequencing (WGBS) (23), anti-CMS immunoprecipitation (CMS-IP)-based 5hmC profiling (24,25) and ATAC-seq (Assay for Transposase-Accessible Chromatin with high throughput sequencing) (26) to capture the dynamics of DNA methylome, DNA hydroxymethylome, and chromatin accessibility landscapes during stepwise pancreatic lineage specification of hESCs (Figure 1A). Through an integrated epigenomic and transcriptomic analysis, we uncovered previously underappreciated links between DNA methylation/demethylation (abbreviated thereafter as ‘(de)methylation’) and pancreatic lineage specification. Our results suggest 5hmC bookmarks DNA regions to be demethylated in the descendent lineage during the hESC-to-PE differentiation, which is positively correlated with enhancer activities and chromatin accessibility to facilitate lineage-specific and pioneer transcription factors (TFs) binding to induce gene expression. Furthermore, we discovered ‘5hmC-rim’ at the boundaries of large functional genomic regions, including super-enhancer, DNA methylation ‘canyon’ and broad H3K4me3 peaks, presumably to maintain these regions at low levels of cytosine methylation. Together our findings reveal the intricate interplays of methylome, hydroxymethylome, chromatin accessibility and transcriptional programming that ultimately dictate a pancreatic fate of differentiated hESCs.
MATERIALS AND METHODS
hESC culture and pancreatic differentiation
The hESC line H1 was obtained from WiCell Research Institute. H1 hESCs were maintained in mTeSR1™ (Stem Cell Technologies). To differentiate of H1 into pancreatic endoderm cells, we used a modified version of the previous published protocol (20–22). In brief, one day before differentiation, H1 cells were dissociated with Accutase™ (Innovative Cell Technologies) and seeded at a density of 150 000 cells/cm2 in mTeSR1 resulting in approximately 90% confluence after overnight culturing. Undifferentiated cells were washed in RPMI 1640 medium (Gibco) and then differentiated using a multi-step protocol with daily media feeding. Human activin A, mouse Wnt3a, human KGF (also known as FGF7), and human Noggin were purchased from R&D systems. Other media components included KAAD-Cyclopamine (Toronto Research Chemicals), and the retinoid analog TTNPB (Sigma Aldrich).
Step i: Germ layer specification (3 days). Cells were exposed to RPMI 1640 supplemented with GlutaMAX (Invitrogen), 0.2% FBS, 1.2 g/l NaHCO3, 100 ng/ml Activin A, 25 ng/ml Wnt3a for the first day. For the next 2 days, cells were cultured in RPMI 1640 with 0.5% FBS, 1.2 g/l NaHCO3, and 100 ng/ml Activin A.
Step ii: Gut tube formation (3 days). Cells were exposed to DMEM/F12 medium (Gibco) supplemented with GlutaMAX, 2% FBS, 2 g/l NaHCO3 and 50 ng/ml of KGF (also known as FGF7) for 3 days.
Step iii: Pancreatic lineage induction (total 5 days: 2 days for GT-to-FG transition and 3 days for FG-to-PE transition). Cells were cultured in DEME-HG medium (Gibco) supplemented with GlutaMAX, 1% B27 (Invitrogen), 100 ng/mL Noggin, 0.25 μM KAAD-Cyclopamine and 3 nM TTNPB for 4 days. For the last day, cells were exposed to DEME-HG medium (Gibco) supplemented with GlutaMAX, 1% B27 (Invitrogen), 100 ng/ml Noggin, and 500 nM TPB [(2S,5S)-(E,E)-8-(5-(4-(trifluoromethyl)phenyl)-2,4-pentadienoylamino)benzolactam] (Calbiochem®).
ATAC-seq library preparation and data analysis
ATAC-seq library preparation was performed as described before (26). Briefly, nuclei were isolated in lysis buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) followed by centrifugation at 500 × g for 10 min at 4°C. Next, the transposition reaction was carried out by adding Nextera Tn5 Transposase from the Illumina Nextera DNA library preparation kit and incubated at 37°C for 30 min. Tagmentated DNAs were purified using the Qiagen MiniElute kit. Purified DNAs were amplified with the KAPA real-time library amplification kit followed by library purification using Agencourt Ampure XP beads. The quality of purified DNA libraries was checked by Agilent TapeStation and then subjected to highthroughput sequencing on an Illumina NextSeq instrument (150 cycle, pair-ended). ATAC-seq data analysis was performed as briefly described below. Bowtie2 with ‘-very-sensitive’ option was used to map the high-quality reads to hg19 version of human genome. The uniquely properly paired mapped reads were extracted for downstream analysis. MACS2 (27) with the ‘-nomodel’ and ‘-extsize 147’ was used to call ATAC peaks. Bedtools intersect was used to count the reads fall into peaks regions, and RPM (reads in peaks per million reads) was used to do the normalization.
WGBS library preparation and data analysis
Genomic DNA was isolated using a Qiagen DNeasy blood and tissue kit. Purified genomic DNA was sonicated into ∼300 bp using a Covaris focused ultrasonicator following the manufacturer's instructions. Sheared DNA was ligated with methylated adaptors using a TruSeq DNA library preparation kit (Illumina) followed by sodium bisulfite treatment (Zymo Research, EZ DNA methylation-lightning kit). Whole genome-wide bisulfite sequencing (WGBS) was performed by following a previous publication (23). Note that traditional bisulfite based DNA methylation profiling techniques will not be able to discriminate between 5mC and 5hmC (28). DNA libraries with methylated adapters were amplified using KAPA HiFi Uracil+ (Kapa Biosystems) polymerase with four PCR cycles. Amplified DNA fragments were purified by AmpuXP beads and sequenced using an Illumina NextSeq instrument (150 cycle, pair-ended). For data analysis, paired-end 75 bp reads were mapped against hg19 using bsmap (v2.89) (29) with paired mode and two allowable mismatches. In total, we identified 14.6 million CpG sites with coverage ≥10 reads and our downstream bioinformatic analyses were based on these CpG sites. MOABS (30) and BSeQC (31) were used to do the quality control and to calculate the methylation ratio for each CpG site. We defined DMRs as the regions between two samples that have an absolute difference of mean DNA methylation ratio of at least 20% and a false discovery rate (FDR) of <0.05. Annotation of DMRs were performed using HOMER software (32). BEDTools intersect was used to perform the overlap analysis between DMRs and histones peaks (33).
CMS-IP-seq library preparation and data analysis
CMS-IP-seq was performed as described previously (24,25). Bisulfite converted DNA libraries with methylated adapters were enriched using an in-house anti-CMS antibody bound to protein A/G dynabeads. The anti-CMS antibody has been successfully used to profile DNA hydroxymethylomes in embryonic or hematopoietic stem cells (24,25,34–36) Enriched fragments were cleaned up using the phenol/ chloroform/isoamyl-alcohol method and then amplified using KAPA HiFi Uracil+ (Kapa Biosystems) polymerase with 10 PCR cycles. Amplified libraries were purified by AmpuXP beads and then sequenced using an Illumina NextSeq instrument (150 cycle, pair-ended). Single-end reads were mapped to GRCh37/hg19 assembly using bsmap with the ‘-v 2 -n 1 -q 3 -r 0’ parameter. Duplicate reads were treated using macs2 filterdup with the ‘–keep-dup 2’ parameter. Bam2wig.py in RSeQC was used to transform the bam file to normalized bigWig files with the parameter ‘-t 2000000000’. Finally, the combined tracks for UCSC Genome Browser were generated. Macs2 was used to call 5hmC peaks for each stage with default setting. The BEDTools merge was used to merge 5hmC peaks from all the samples to create the consensus 5hmC peaks. We counted the reads numbers in the consensus 5hmC peaks in each sample using the filtered mapping bed files (output from macs2 filterdup function). To annotate the 5hmC peaks, we downloaded the elements’ regions (exon, intron, CpG island, 3′ UTR, 5′ UTR/promoters, Repeat and Intergenic) from the UCSC genome browser. We used BEDTools intersect to annotate 5hmC enriched peaks with overlapped elements regions. We defined the overlap by at least 1 bp overlap between 5hmC peaks and elements’ regions.
The raw reads count for each peak across all the stages were used as input for the DEGseq2 (R package) to call differential 5hmC peaks (FDR < 0.05) between different stages. PCA was performed by using DESeq2. GREAT analysis with single-nearest genes option was used to perform the functional annotation of 5hmC peaks.
Correlation analysis among 5hmC, ChIP-seq (histone marks) and ATAC-seq peaks
The ChIP-seq raw data for histone modifications (H3K4me1, H3K27Ac, H3K4me3 and H3K27me3) were downloaded from GSE54471 and E-MTAB-1086. The data analysis was similar to steps described above except that bowtie2 was used to map the ChIP-seq reads to GRCh37/hg19 assembly. Histones data across samples were normalized based on total mapped reads number. To test the correlations among 5hmC, histone marks and chromatin accessibility, we first used deepTools (37) to normalize all the 5hmC peaks to 1 kb and horizon heatmap plots were used to show the distribution of normalized 5hmC, histone marks and ATAC-seq signals within ±2 kb of 5hmC peaks. To correlate the 5hmC signals with histone modifications (H3K4me1, H3K27ac, H3K4me3 and H3K27me3), we first merged all the differential 5hmC peaks between adjacent differentiation stages to obtain the adjacent differential consensus peaks. Next, we counted the reads numbers in all samples for each 5hmC peak. At the same time, we counted the reads numbers of ChIP-seq in all samples for each 5hmC peak region. To compare the 5hmC and ChIP-seq signals across samples, we calculated the RPKM for 5hmC and histone marks. Based on 5hmC signals, we separated these regions to stage-specific high 5hmC regions (ES, DE, GT, FG and PE 5hmC high regions). The heatmaps for 5hmC and histone modifications were plotted for each stage high regions using the R package (gplots). To correlate the DMRs with histones markers (Supplementary Figure S3E), we performed the overlap analysis (at least 1 bp overlap) between DMRs and histone marker peaks at ES and GT stages, respectively. Bedtools shuffle was used to generate the random regions in the genome.
To quantify the correlation between 5hmC and histone marks, we calculated the Spearman's correlation coefficient between 5hmC and each of the four histone marks within 5hmC high regions in a time series manner using an in-house R script. To test if the enhancers (active/poised) correlated with 5hmC enrichment, we downloaded the enhancer regions from Wang et al (22). We subsequently used deepTools to calculate the average 5hmC signals within 1 kb (up- or down-) of enhancer midpoint (normalized bigwig minus input normalized bigWig) at each stage. R package gplot was used to plot heatmaps. In addition, an in-house script was used to calculate the ratio for genomic regions showing the same pattern of published H3K4me1/H3K27ac (22) signals with 5hmC signals. To compare the 5hmC level between active and poised enhancers, we used deepTools to plot the 5hmC and 5mC signals within 5 kb (5 kb up- or down-stream) of enhancer midpoint. To compare the GRO-Seq signals at enhancers (active/ poised) with and without 5hmC, we defined that if genomic region within 5 kb up- or down-stream of enhancers midpoint had at least 1 bp overlap with 5hmC peaks, these enhancers were counted as ‘with 5hmC’. Otherwise, they would be regarded as enhancers ‘without 5hmC’. The script bigWigAverageOverBed downloaded from UCSC Genome Browser was used to calculate the average GRO-Seq abundance at enhancers with and without 5hmC. To explore the potential involvement of 5hmC in regulating enhancer activity, we identified enhancers that underwent transition between active and poised states in adjacent stages. We used multiIntersectBed to identify the overlaps among the active and poised enhancers in the previous stage and in the current stage. For any enhancers defined at the previous stage as ‘active’ but became ‘poised’ at the current stage, we defined these regions as ‘active-to-poised’ transition enhancers if the overlapped regions were larger than 50 bp. The same criteria applied to the ‘poised-to-active’ transition enhancers. Next, we calculated the average 5hmC signals at these transition enhancers. The R package was used to plot the box plots.
Analysis on broad H3K4me3 peaks, super enhancers and DNA methylation canyon
MACS2 (with –broad parameter) was used to identify broad H3K4me3 peaks. The ROSE method (38) was used to identify super enhancers with H3K27ac signals as input. DNA methylation canyon was identified using an in-house script based on a Hidden Markov Model. We defined canyon as regions with their lengths larger than 3.5 kb and having an average methylation ratio less than 10%. DeepTools was used to plot 5hmC and 5mC signals within and up/dn 10 kb of broad H3K4me3 peaks, super-enhancers and DNA methylation canyons. DeepTools was used to calculate the 5hmC and 5mC signals distribution along broad H3K4me3 peaks, super enhancers and canyons. To establish the correlation between 5hmC and broad H3K4me3 peaks, we first clustered the broad H3K4me3 peaks into three groups based on their peak sizes: broad (≥3 kb), medium (1–3 kb), narrow (≤1 kb). We defined promoter regions as 2 kb upstream and 1 kb downstream from TSS. If the H3K4me3 broad peaks have at least 1 bp overlap with promoters, we defined this gene as broad H3K4me3 peaks associate gene. Boundaries were defined as the up/dn 100 bp of the exact boundary of broad H3K4me3 peaks, super enhancers and canyons. The 5hmC signals at the boundaries of broad H3K4me3 peaks were calculated for each group. The same analysis was applied to murine hematopoietic stem cells.
Statistical analysis
All the boxplot statistical significance was analyzed using the Wilcoxon signed-rank test. We used Kolmogorov-Smirnov test to calculate the significance for the curve analysis.
RESULTS
Dynamic changes of 5mC, 5hmC and chromatin accessibility during pancreatic differentiation of hESCs
We differentiated H1-hESC toward pancreatic progenitors by employing a previously established highly efficient differentiation protocol (20–22), in which cells synchronously progress stepwise through multiple lineage intermediates, including definitive endoderm (DE), primitive gut tube (GT) and posterior foregut (FG) (Figure 1A). This differentiation system consists of five stages including ES, DE, GT, FG, and pancreatic endoderm (PE), which cover three major endodermal developmental processes, germ layer specification, gut tube formation and pancreatic lineage induction. The differentiation efficiency was confirmed by immunostaining (Supplementary Figure S1A), flow cytometry analysis (Supplementary Figures S1B and C) and real-time quantitative PCR with state-specific markers (Supplementary Figure S1D), such as SOX17 and PDX1 (20–22). By monitoring the transcriptional profiles of each stage (ES, DE, GT, FG and PE) with RNA-seq, we confirmed the specific pancreatic lineage commitment, as most evidently reflected by the expression of stage-specific signature genes (21) (Figure 1B, Supplementary Figure S1E and Supplementary Table S1). To further confirm the desired differentiation status, we compared the RNA-seq data obtained from H1 hESC in the current study with previously published RNA-seq data based on another hESC line, CyT49 (21), which went through the similar pancreatic differentiation procedures. Our comparative transcriptomic analysis showed that H1 and CyT49 hESCs shared very similar gene expression profiles during pancreatic differentiation (Spearman's correlation coefficient ≥0.83; Supplementary Figure S1F), strongly attesting to the reliability and repeatability of our differentiation protocol, as well as the high quality of our acquired RNA-seq data.
Having confirmed the successful differentiation of hESC toward the pancreatic lineage, we next sought to profile the epigenetic modifications (5mC and 5hmC) on DNA and genome-wide chromatin accessibility. To do this, we collected cells at each differentiation stage to perform genome-wide profiling (Supplementary Table S2) of DNA methylome (WGBS), hydroxymethylome (with anti-CMS immunoprecipitation based sequencing; or CMS-IP-seq) and chromatin accessibility (with ATAC-seq). In WGBS analysis, we sequenced a total of 3.47 billion reads, covering a total of 14.6 billion of CpG dinucleotides (coverage ≥ 10) in the human genome with high correlation between two biological replicates (Supplementary Figure S1G, Supplementary Table S1). WGBS data of H1-ESC was obtained from the ENCODE database (39). In CMS-IP-seq, we identified 5hmC-enriched regions (HERGs) per differentiation stage (Supplementary Figure S1H, Supplementary Table S1) that covered 4.7–12.9% of the whole genome, with the average length of 5hmC peaks at ∼200 bp (Supplementary Figure S1I, Supplementary Table S1). The overall genomic distribution profiles of 5mC and 5hmC at annotated genomic regions at each stage (Supplementary Figure S1J) were similar to those observed in mouse embryonic stem cells, neurons and T lymphocytes as others and we have reported previously (23,24,34,40–43). In the coding regions, 5mC was depleted at transcriptional start sites (TSS) but enriched at genebody. By contrast, 5hmC was enriched at both TSS and genebody at all stages (Supplementary Figure S1K). In ATAC-seq, we collected a total of 164 million reads, which yielded on average 100 043 accessible regions during pancreatic differentiation from ES to PE (Supplementary Table S1). The majority of ATAC-seq peaks were enriched at promoters / 5′UTR (right, Supplementary Figure S1J). Together, the availability of these high-quality RNA-seq, WGBS, CMS-IP-seq and ATAC-seq datasets enabled us to identify transcriptomic and epigenomic changes associated with pancreatic lineage specification.
To obtain an overall view on the global changes in DNA hydroxymethylation, we quantified 5hmC levels at each differentiation stage by using a dot-blot assay (28,44). To our surprise, global 5hmC levels displayed a ‘U-shaped’ biphasic change during the ES-to-PE differentiation (Figure 1C). For the initial ES-to-DE transition, the global 5hmC level dropped by over 70% based on dot-blot assay results. This finding dovetails with the scenario seen in retinoic acid (RA)-induced differentiation of mouse ES cells (10), which might be due to reduced expression of TET1 and DNMTs (Figure 1B). In the subsequent lineage progression steps (GT-FG-PE), the global 5hmC levels underwent a gradual increase (Figure 1C), likely owing to the increased expression of TET2 and/ or TET3 to compensate TET1 downregulation (Figure 1B). The changes in average DNA methylation levels calculated from the WGBS data exhibited ∼2% fluctuation during this differentiation process (Figure 1C). The overall DNA methylation changes were relatively minor (<5%), suggesting that unlike global DNA demethylation during early embryonic development (45,46), DNA methylation alterations at selected loci might be essential for human ESC lineage commitment and differentiation. In parallel, we performed ATAC-seq to examine the chromatin accessibility at each of the five-defined differentiation stage. The alterations in chromatin accessibility showed a similar biphasic trend, albeit to a lesser extent, as the global changes in 5hmC during pancreatic differentiation (Supplementary Figure S1L).
Because most chromatin accessible regions are cis-acting transcriptional regulatory elements (e.g., proximal promoter and distal regulatory elements), we next focused on examining whether these regions, which were selected by high 5hmC (enrichment fold > 4) and enrichment of ATAC-seq signals (<1 kb to the center of 5hmC peaks) but low 5mC, are indeed functionally involved in defining and maintaining each differentiation stage. We performed Genomic Regions Enrichment of Annotations Tool (GREAT) (47) analysis on the selected genomic regions. We found that these chromatin accessible regions were significantly associated with signatures genes at each differentiation stage (Figure 1D). For example, the top 5000 HERGs at the PE stage were enriched at distal regulatory regions of genes associated with the development of the pancreas, exocrine system and endocrine pancreases (Supplementary Figure S1M). Collectively, these results demonstrate that 5mC and 5hmC undergo dynamic changes during endodermal lineage specification towards pancreas, with the most notable changes observed at the initial ES to DE transition, followed by gradual and reciprocal changes in 5mC and 5hmC during the subsequent lineage progression from the GT stage to the PE stage. These dynamic reconfigurations are closely associated with chromatin accessibility and occur most prominently at cis-acting transcriptional regulatory elements with annotated functions in regulating the differentiation of hESC toward a pancreatic fate.
Identification of differential 5mC- (DMRs) and 5hmC-enriched regions (DHMRs) between adjacent differentiation stages
DNA methylation homeostasis is important for temporal and spatial regulation of gene transcription during differentiation and development (48). To further explore the DNA methylation dynamics during pancreatic differentiation, we selected CpGs that displayed >20% changes of DNA methylation in three sequential developmental processes: (i) germ layer specification (ES versus DE), (ii) gut tube formation (DE versus GT) and (iii) pancreatic lineage induction (GT versus PE) (Figure 1A). We clustered differentially methylated CpGs (>20% changes in the ratio of mCG/CG) based on their methylation levels (from 0 to 100% with 10% as interval) and classified CpG sites into three categories (Figure 2A): low-methylation sites (LMS, 0–20% methylation), intermediate-methylation sites (IMS, 20–80% methylation) and high-methylation sites (HMS, 80–100% methylation). Among all the analyzed CpGs, we observed that IMS exhibited the most dynamic changes in DNA methylation whereas LMS and HMS remained relatively stable during pancreatic differentiation (Figure 2A). Commensurate with DNA methylation alterations, we detected a significant enrichment of IMS, but not in HMS, within 5hmC-enriched regions (HERGs) (Supplementary Figure S2A). Further analysis revealed that IMS located within HERGs were significantly enriched at distal regulatory regions (∼32 kb from TSS on average) bearing high chromatin accessibility (Figure 2B). These results clearly suggest that, rather than being randomly distributed across the genome, 5hmC is closely associated with dynamic DNA methylation alteration, especially at IMS of distal regulatory regions that have high chromatin accessibility.
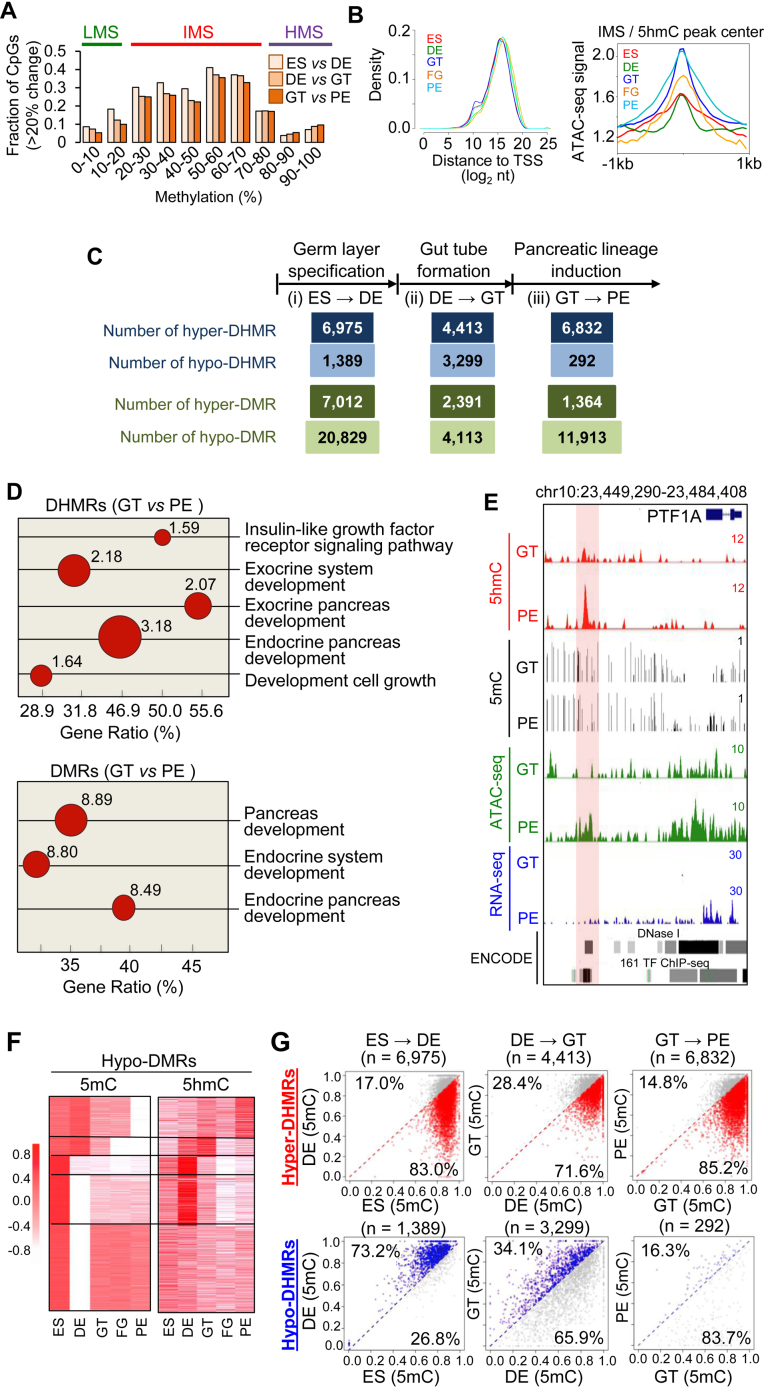
Figure 2.
Identification and features of differential 5mC- (DMRs) and 5hmC-enriched regions (DHMRs) between adjacent pancreatic differentiation stages. (A) Fractions of differentially-methylated CpG sites (cut-off set as over 20% changes in DNA methylation during lineage transition) plotted against their methylation status at 10% intervals. We clustered these CpGs based on their methylation levels to yield three categories: low- (<20%), intermediate- (20–80%) and high- (>80%) methylation CpG sites (LMS, IMS and HMS, respectively). (B) The distribution profile and ATAC-seq signals of IMS located within 5hmC-enriched regions (HERGs) at each stage. (Left) The density of IMS within HERGs plotted on the basis of their distance to TSS (as log2 nucleotides) at each differentiation stage. (Right) Normalized ATAC-seq signals relative to the center (1 kb up- or down-stream) of IMS within HERGs. (C) Numbers of differentially methylated regions (DMRs) or hydroxymethylated regions (DHMRs) that show upregulation (defined as hyper-DMR or hyper-DHMR) or downregulation (termed as hypo-DMR or hypo-DHMR) of 5mC/5hmC signals when cells differentiate into the descendent lineage intermediate (e.g. ES-to-DE (i, germ layer specification), DE-to-GT (ii, gut tube formation), or GT-to-PE (iii, pancreatic lineage induction)). Threshold was set by two parameters: FDR (false discovery rate) ≤0.05 and log2 (fold change) ≥1. (D) Enriched Gene Ontology terms for DHMRs (top) and DMRs (bottom) identified during the GT-to-PE transition. DHMRs and DMRs were notably enriched at distal-regulatory regions of genes that are associated with pancreas development and digestive tract morphogenesis. The size of circles corresponds to the value of –log10 (Binomial p values shown next to the dots). (E) Genome browser view of the PTF1A locus that showed increase in DNA hydroxymethylation (5hmC, red), chromatin accessibility (ATAC-seq trace; green) and gene expression (blue), but a decrease in DNA methylation (5mC; black), during the transition from GT to PE. (F) Clustering of 5mC and 5hmC signals within Hypo-DMRs at the five defined stages during pancreatic differentiation of hESCs. Red color means high 5mC or 5hmC; White color represents low 5mC or 5hmC. (G) Dot plots showing dynamic alternations in DNA methylation within hyper- or hypo-DHMRs between two adjacent differentiation stages. X-axis showed 5mC levels at the indicated differentiation stage; while Y-axis showed 5mC levels at the descendent differentiation stage. (Top) Increased DNA hydroxymethylation (Hyper-DHMRs) signified an overall reduction in DNA methylation during the ES-to-DE, DE-to-GT or GT-to-PE lineage progression. The red dots represent CpGs with reduced DNA methylation in hyper-DHMRs when cells progress toward the descendent differentiation stage. The grey dots represent CpGs with increased DNA methylation. (Bottom) Decrease in DNA hydroxymethylation (Hypo-DHMRs) may not necessarily align with increased DNA methylation during pancreatic lineage progression. The blue dots represent CpGs with increased DNA methylation within Hypo-DHMRs between two adjacent lineage intermediates; while the grey dots represent CpGs with reduced DNA methylation. The number represents the percentage of CpG sites in the corresponding categories.
To further characterize the dynamics of DNA (hydroxy)methylation during the ES-to-PE differentiation, we set out to identify differentially methylated (DMRs; false discovery rate (FDR) ≤ 0.05; minimum change in the fraction of methylated CpG sites (ΔmCG) = 20%) and hydroxymethylated regions (DHMRs; FDR ≤ 0.05; fold change ≥ 2-fold) within sequential developmental processes (Figure 2C). We divided both DMRs and DHMRs into two categories: hypo-DMR/DHMR (regions showing reduced DNA methylation/ hydroxymethylation during the lineage progression) and hyper-DMR/DHMR (increased DNA methylation / hydoxymethylation). The identified DMRs displayed an average median |ΔmCG| of 46.1% (Supplementary Figure S2B); whereas DHMRs exhibited an averaged enrichment of 4.73 (calculated as log2 fold change; ES: 4.95; DE: 4.87; GT: 4.78; FG: 4.29; PE: 4.77) during pancreatic lineage specification (Supplementary Figure S2C). We compared the numbers of hypo- or hyper-DMRs / DHMRs during three endodermal developmental processes (Figure 2C), and found that a large fraction of genomic regions exhibited a significant increase in 5hmC (numbers of hyper-DHMRs > numbers of hypo-DHMRs; with the ratio over 1.3–23.4-fold; Figure 2C) and a decrease in 5mC (as reflected in the ratio of hypo-DMRs over hyper-DMRs by over 2.3–4.5-fold; Figure 2C) in all three transition steps. These findings seem to be inconsistent with the observation of a global decrease in 5hmC during the ES-to-DE differentiation. To investigate the discrepancy, we plotted all identified 5hmC peaks at ES and DE stages based on their FDR and fold-change during the ES-to-DE transition (Supplementary Figure S2D). Although most peaks showed reduction in 5hmC levels during ES-to-DE differentiation, when we filtered with our criteria for DHMR (FDR ≤ 0.05; fold change ≥ 2-fold), more peaks fell into the category of hyper-DHMR compared to hypo-DHMR (2.5% versus 0.5%). Furthermore, we also confirmed the hyper- and hypo-DHMR status at selected loci by using the oxBS method (49,50) at ES and DE stages, which reflected the DNA hydroxymethylation status at single-base resolution (Supplementary Figure S2E). These observations well explained the discrepancy seen between the changes of global 5hmC and DHMRs identified from CMS-IP-Seq data.
To explore whether DMRs and DHMRs are associated with specific differentiation stages, we performed GREAT analysis on DMRs and DHMRs identified during pancreatic lineage induction (step iii), and found that these regions were significantly (binomial p value < 0.01) correlated with genes implicated in pancreatic and digestive tract development (Figure 2D). Principal component analysis (PCA) analysis also revealed sharp separation in DHMRs and DMRs when cells progressed from the current stage to the descendent lineage (Supplementary Figure S2F). Similarly, PCA analysis on RNA-seq and ATAC-seq results also showed clear separation among the three developmental steps (Supplementary Figure S2F). A notable example is illustrated in Figure 2E with a focus on the epigenetic changes at the distal regulatory region of a gene encoding pancreas transcription factor 1 (PTF1A), which is known to be essential for pancreatic organogenesis (51,52). During pancreatic lineage induction, we detected a significant increase in 5hmC, accompanied by reduced 5mC distribution and increased chromatin accessibility (Figure 2E) at this genomic locus, which was correlated with significant upregulation of PTF1A expression by ∼103-fold. Altogether, our findings demonstrated dynamic alternations in the epigenetic landscapes, including DNA methylation, hydroxymethylation and chromatin accessibility, that are closely associated with hESC differentiation toward pancreatic endoderm.
To gain further insights into the dynamics and relations between DNA methylation and hydroxymethylation during pancreatic differentiation, we systematically compared 5mC and 5hmC levels within hyper- and hypo-DMRs / DHMRs. First, we observed an overall negative correlation between 5mC and 5hmC in hypo-DMRs: the decrease of 5mC was largely accompanied by 5hmC enrichment during three developmental steps (Figure 2F), but there is no overt correlation between 5mC and 5hmC signals in hyper-DMRs during lineage specification (Supplementary Figure S2G). Second, we assessed the DNA methylation levels in hyper-DHMRs or hypo-DHMRs (Figure 2G). Hyper-DHMRs exhibited an overall reduction of DNA methylation during all three pancreatic development processes (Figure 2G, upper panels): 83.0%, 71.6% and 85.2% of hyper-DHMRs (with increased DNA hydroxymethylation) showed reduced DNA methylation during the transition from ES to DE, DE to GT and GT to PE, respectively. An explicit example illustrating such reciprocal changes (acquisition of 5hmC with concomitant loss of 5mC) was observed during GT to PE progression (left, Supplementary Figure S2H). The opposite scenario was not consistently seen in all transition stages because we failed to detect such reciprocal changes within hypo-DHMRs during DE-to-GT or GT-to-PE progression (Figure 2G, lower panels). For a significant fraction of hypo-DHMRs, the decrease of 5hmC was accompanied by reduced DNA methylation (as exemplified in Supplementary Figure S2H, right panel). Consistently, we found that 5hmC-enriched regions (HERGs) displayed overall lower DNA methylation level (ks.test; P < 2.2e−16) compared with 5hmC-depleted regions (Supplementary Figure S2I) at all five differentiation stages. Collectively, these findings suggest that 5hmC marks genomic regions that undergo DNA demethylation (as reflected by a decrease in 5mC) in the descendent differentiation stage. By contrast, the decreases of 5hmC may not be necessarily correlated with increased DNA methylation during the ES-to-PE progression.
DNA hydroxymethylation is enriched at enhancers at each differentiation stage
Since IMS with high 5hmC signal (Supplementary Figure S2A) are enriched at distal regulatory regions (e.g., enhancers) with high chromatin accessibility (Figure 2B), we hypothesized that DNA hydroxymethylation is closely involved in regulating enhancer activities. To test this, we examined the histone modification status within HERGs because enhancer activity can be inferred based on histone mark deposition (22). We centered on all 5hmC-enriched regions identified from each stage, and then analyzed chromatin accessibility (ATAC-seq) and enrichment of histone modifications (mono-methylation of histone H3 lysine 4 (H3K4me1), tri-methylation of histone H3 lysine 4 (H3K4me3), acetylation of histone H3 lysine 27 (H3K27ac), tri-methylation of histone H3 lysine 27 (H3K27me3)) identified from previous studies at those regions (21,22). We observed a significant overlap of HERGs with H3K4me1 and chromatin accessible regions at each stage, but to a lesser extent with H3K27ac (Figure 3A, Supplementary Figure S3A). The other two histone marks, H3K4me3 and H3K27me3, showed much less enrichment with 5hmC peaks at each differentiation stage (Figure 3A, Supplementary Figure S3A). To further study the correlation between 5hmC and other epigenetic marks during hESC differentiation, we clustered 5hmC profiles to identify stage-specific HERGs among all identified DHMRs, and correlated them with the enrichment patterns of histone marks, including H3K4me1, H3K27ac, H3K4me3 and H3K27me3, as well as 5mC (Figure 3B). As most intuitively reflected in the heat maps (Figure 3B) but more accurately quantified with Spearman's correlation coefficients (Figure 3C, Supplementary Figures S3A and B), 5hmC was positively correlated with H3K4me1 and H3K27ac (average Spearman correlations: 0.51 for H3K4me1 and 0.39 for H3K27ac), but showed a weaker correlation with H3K4me3 and H3K27me3 at each stage (Figure 3C, Supplementary Figure S3B). H3K4me1 and H3K27ac are known to be enriched at enhancers with high chromatin accessibility to control the lineage specification during pancreatic differentiation (22). When perusing the distal regulatory region of PDX1 (a key transcription factor for pancreas development), we observed a gradual increase in 5hmC, with simultaneous enrichment of H3K4me1 and H3K7ac during lineage progression from DE to PE (Figure 3D).
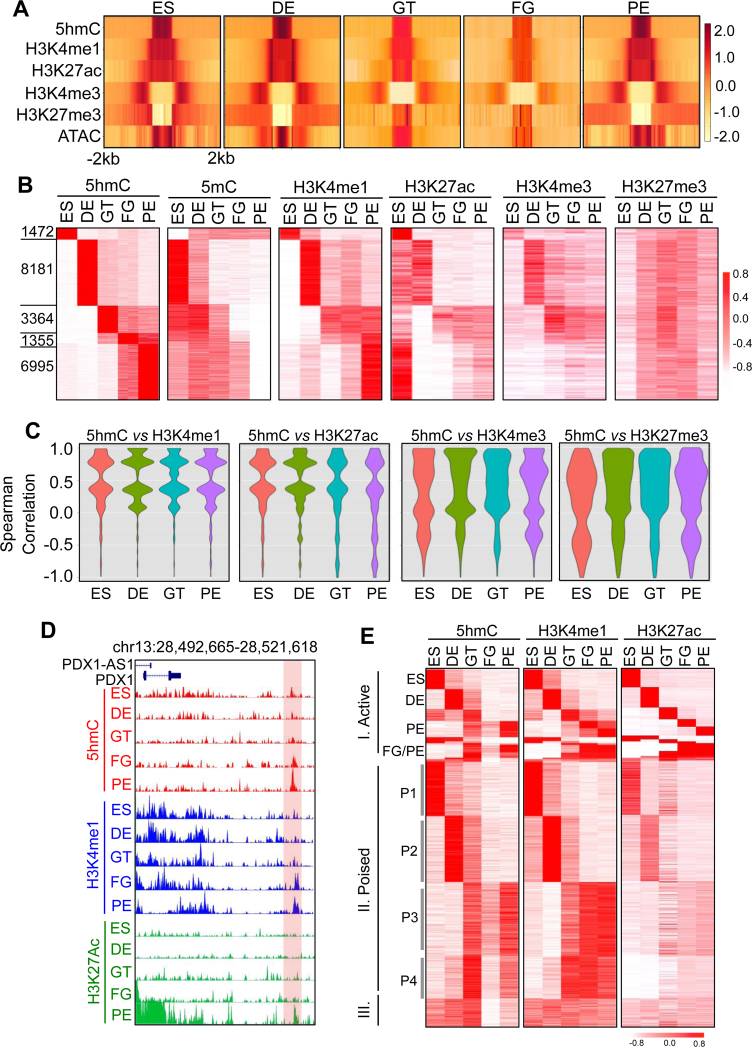
Figure 3.
5hmC is enriched with selected histone marks at enhancers. (A) Heatmaps depicting enrichment patterns of histone marks (H3K4me1, H3K27ac, H3K4me3 and H3K27me3) relative to the center of 5hmC peaks (±2 kb). Chromatin accessibility measured by ATAC-seq was also included at the bottom row. The yellow-to-red color scale shown on the right indicates the enrichment from low to high. (B) Clustering of 5hmC, 5mC, ChIP-Seq (H3K4me1, H3K27ac, H3K4me3 and H3K27me3) signals based on stage-specific 5hmC enriched regions. The numbers on the left represent the amounts of stage-specific 5hmC peaks. Each row represents individual 5hmC peak, and each column represents the corresponding differentiation stage. The color scale from white to red represents the enrichment from low to high. (C) Violin plots for the distribution of time-series Spearman correlation coefficients between the two indicated comparison groups (stage-specific 5hmC peaks versus one of the following four histone marks: H3K4me1, H3K27ac, H3K4me3, or H3K27me3). 5hmC showed a strong correlation with H3K4me1 or H3K27ac. (D) Genome-browser view of distal-regulatory regions close to PDX1. A genomic region with gradual increase in 5hmC, H3K4me1 and H3K27ac signals during pancreatic differentiation was highlighted in light red. (E) 5hmC, H3K4me1 and H3K27ac distribution patterns relative to enhancer types (I, active; II. poised, or III. others/both). Enhancers were clustered based on H3K4me1 and H3K27ac enrichment from a previous publication (22).
Enhancers identified in pancreas differentiation from hESCs have been classified into three groups based on the enrichment status of H3K4me1 and/or H3K27ac: (i) active in a stage-restricted manner, (ii) poised but remaining inactive and (iii) constitutively active (22). We further plotted the 5hmC enrichment signatures based on H3K4me1 / H3K27ac status in these three groups of enhancers. We observed highly consistent enrichment patterns between 5hmC and H3K4me1 (and to a lesser extent with H3K27ac) in all three types of histone-marked enhancers across five differentiation stages (Figure 3E, Supplementary Figures S3C and D). Furthermore, consistent with our analysis in Figure 2F, genomic regions marked as stage-specific HERGs showed reduction in DNA methylation at the same differentiation stage (Figure 2F; 5hmC panel versus 5mC panel). To further examine the correlation between dynamic changes of DNA methylation and histone marks, we selected genomic regions that displayed decreased (hypo-) or increased (hyper-) DNA methylation during the transition from ES to GT and then counted corresponding histone marks (H3K4me1 and H3K27ac, respectively). We found a strong negative correlation between DNA methylation and H3K4me1/ H3K27ac enrichment (Supplementary Figure S3E and F), suggesting a strong correlation between DNA demethylation and enhancer activity establishment during the differentiation process. Together, these findings indicate that 5hmC marks genomic regions for DNA demethylation at enhancers and is associated with enhancer establishment during pancreatic lineage commitment.
DNA hydroxymethylation is positively correlated with enhancer activities during hESC differentiation
To further dissect the role of 5hmC in modulating enhancer activities during pancreatic differentiation of hESC, we examined 5hmC distribution patterns in both poised and active enhancers (22). 5hmC was enriched in both poised and active enhancers, with enrichment at active enhancers more pronounced across the five differentiation stages (Figure 4A, top). Similar enrichment at enhancers was also observed in chromatin accessible regions as using ATAC-seq (Supplementary Figure S4A). On the other hand, we observed an overall hypomethylation in both poised and active enhancers, with active enhancers showing lower methylation levels than poised enhancers (Figure 4A, bottom). This phenomenon was further confirmed by comparing our 5hmC analysis with published GRO-seq data (22). GRO-seq is a global nuclear run-on sequencing technique to analyze nascent RNA transcription that reflects enhancer activities. We found that 5hmC-marked enhancers displayed higher transcriptional activities (reflected by higher GRO-seq signals) than 5hmC-depleted enhancers in all differentiation stages (Figure 4B). To further reveal the dynamics of 5hmC changes at poised and active enhancers during differentiation, we selected regulatory genomic regions that experienced changes in enhancer activities (active to poised status; or vice versa) between adjacent differentiation stages and examined their 5hmC enrichment status (Figure 4C, Supplementary Figure S4B). Our results suggested a strong correlation between dynamic enhancer activity and changes in 5hmC enrichment. 5hmC enrichment was reduced when active enhancers became poised ones in the descendent differentiation stage; and vice versa. In parallel, we compared the 5hmC enrichment within each developmental process (ES versus DE, DE versus GT or GT versus PE) with H3K4me1 enrichment and locations of engaged Pol II along actively transcribed genes as revealed by GRO-seq (22). Again, a similar positive correlation was observed between 5hmC enrichment and increase of H3K4me1 or GRO-seq signals at each developmental step (Figure 4D). Enhancers are usually regarded as highly accessible regions to facilitate transcription factors (TFs) binding. To further examine the correlation between 5hmC and chromatin accessibility during the differentiation from hESC to pancreatic progenitors, we compared the dynamic changes of 5hmC with chromatin accessibility measured by ATAC-seq between ES and PE. Again, we found a significant positive correlation between 5hmC and ATAC-seq signals: genomic regions with increased hydroxymethylation showed higher chromatin accessibility, as indicated by an increase in ATAC-seq signals, and vice versa (Figure 4E). Finally, to examine whether alterations in 5hmC and chromatin accessibility were associated with enhancer activities, we analyzed the GRO-seq signals and overlapped 5hmC/ATAC-seq peaks within enhancers marked by H4K3me1. Our analysis showed a clear positive correlation between 5hmC enrichment and nascent transcription activity (GRO-seq) or chromatin accessibility at enhancers (Figure 4F, Supplementary Figure S4C). In aggregate, results from our unbiased and systematic bioinformatic analysis indicate that DNA hydroxymethylation is closely associated with chromatin accessibility and enhancer activities when hESCs undergo directed differentiation toward pancreatic endoderm.
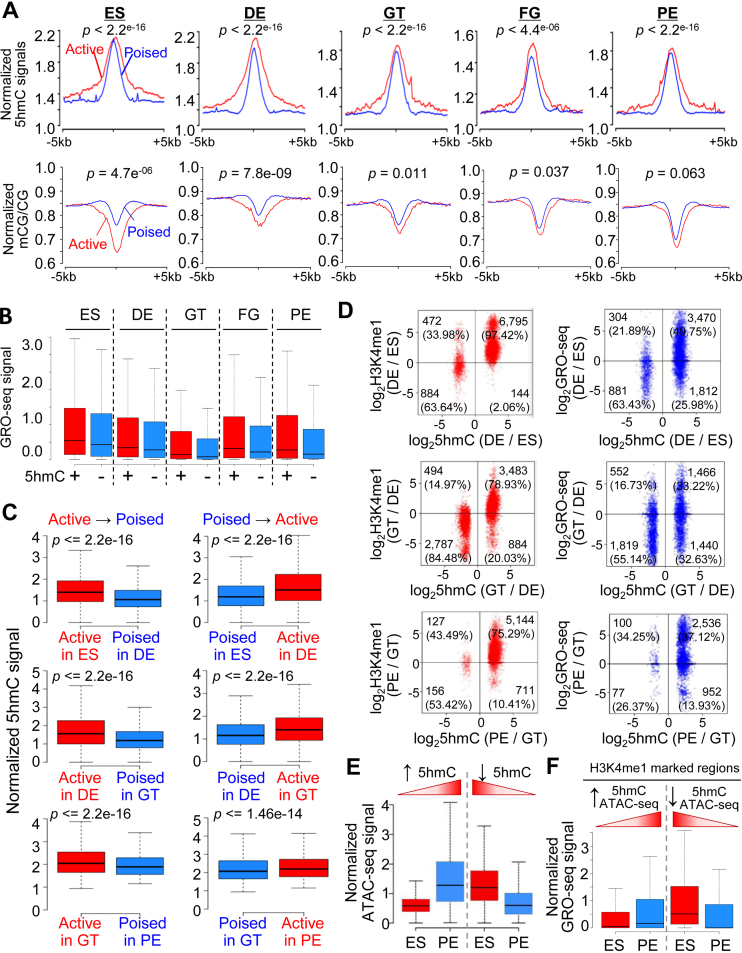
Figure 4.
DNA hydroxymethylation is positively correlated with enhancer activities and chromatin accessibility. (A) 5hmC (top) and 5mC (bottom) enrichment profiles at poised (blue) and active (red) enhancers during ES-to-PE differentiation. The normalized 5hmC or 5mC levels were plotted within 5 kb up- or downstream of the midpoint of pre-identified poised or active enhancers at each differentiation stage. (B) Normalized GRO-seq signals at enhancers with (red) or without (blue) 5hmC peaks at each differentiation stages. (C) Normalized 5hmC enrichment at enhancers undergoing active-to-poised status transition (left), or poised-to-active status transition (right), during the pancreatic lineage progression (ES-to-DE, DE-to-GT, or GT-to-PE). (D) Dot plots depicting the ratios (DE over ES, GT over DE or PE over GT) of H3K4me1 ChIP-seq reads densities (left) or GRO-seq signals (right) against the ratio of 5hmC signals during pancreatic lineage progression. Changes in 5hmC were positively correlated with alterations in H3K4me1 or GRO-seq signals during differentiation. The X-axis represents the log2(fold change of 5hmC), and the Y-axis represents the log2(fold change of H3K4me1 or GRO-seq signals) between the two indicated adjacent differentiation stages. The numbers of peaks were indicated in each quadrant, with the percentages calculated by using the amounts of DHMRs identified from two adjacent stages as the denominator for each developmental step. (E) Normalized ATAC-seq signals in genomic regions that showed increased (left) or reduced (right) DNA hydroxymethylation during the transition from ES to PE. ATAC-seq signals were positively correlated with 5hmC intensities. (F) Normalized GRO-seq signals at H3K4me1-deposited enhancers that showed increase or decrease in both 5hmC and ATAC-seq signals (identified from panel E). GRO-Seq signals were positively correlated with 5hmC/ATAC-seq densities.
DNA hydroxymethylation is selectively correlated with the binding of key TFs at chromatin accessible regions during pancreatic lineage specification
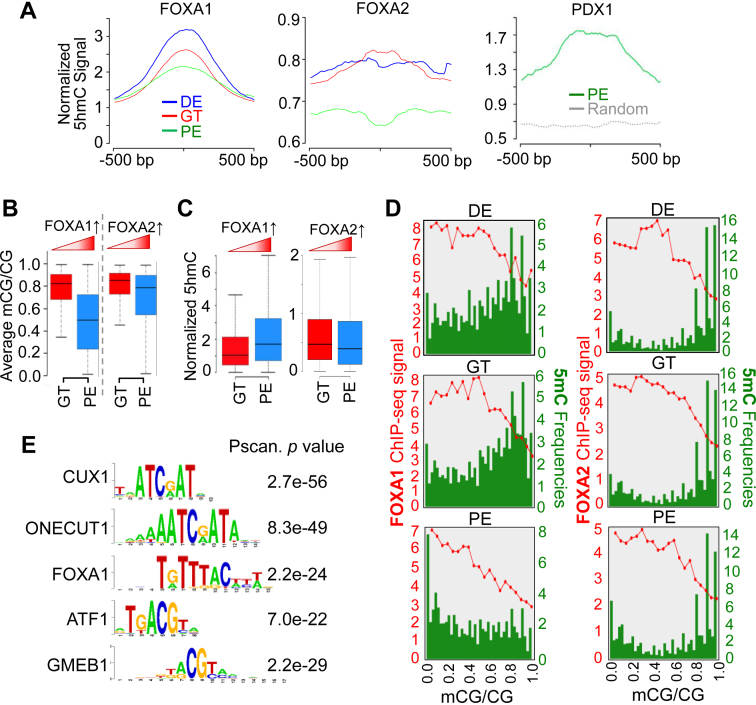
Given that key developmental TF usually binds to chromatin accessible regions with active/ poise enhancers, we further scrutinized 5hmC enrichment at two pioneer TFs, FOXA1 and FOXA2, both of which belong to the same winged helix transcription factor family (53) that is critical for endoderm differentiation (22). Although 5hmC was enriched within both FOXA1 and FOXA2 binding regions (Figure 5A) with high chromatin accessibility (Supplementary Figure S5A), its enrichment at FOXA1 binding regions was significantly more pronounced and consistent across DE, GT and PE stages (Figure 5A). We have demonstrated that 5hmC marks DNA demethylated regions in the genome during pancreatic lineage specification (Figure 2). Additionally, previous studies have shown that alterations in DNA (de)methylation affect TF binding (54). Therefore, we reasoned that varying DNA methylation statuses at FOXA1 and FOXA2 loci might have differential impact on FOXA1 and FOXA2 binding to their target genes during lineage progression. To test this, we compared the dynamic changes of 5hmC and 5mC within the corresponding regions at the GT and PE stages. After integrating existing FOXA1 and FOXA2 ChIP-seq data (22) with our own DNA methylome and hydroxymethylome data, we found distinct 5mC dynamics at enhancers that showed increased FOXA1 or FOXA2 binding (Figure 5). More specifically, enhancers with increased FOXA1 binding, but not those with enhanced FOXA2 binding, exhibited significant reduction in DNA methylation (Figure 5B) with concomitant increase in DNA hydroxymethylation during the GT to PE progression (Figure 5C). Next, we further analyzed the correlation of DNA methylation with FOXA1/FOXA2 binding by dividing their binding regions based on DNA methylation levels at GT, DE and PE stages (Figure 5D). Our analysis revealed that (1) FOXA1 binding sites displayed relatively low levels of DNA methylation compared to FOXA2 binding sites; (2) FOXA1 binding sites showed a reduction in DNA methylation, while DNA methylation patterns in FOXA2 binding regions largely remained unaltered during DE to PE transition (Figure 5D).
Figure 5.
Dynamic DNA methylation alterations are selectively associated with the binding of key TFs during ES-to-PE differentiation. (A) Normalized 5hmC signals plotted according to the clustered peaks (500 bp up- or downstream) of FOXA1 (left), FOXA2 (middle) and PDX1 (right) binding sites at DE (blue), GT (red) and PE (green) stages. (B and C) Average 5mC signals (B) and normalized 5hmC signals (C) at enhancers that exhibited increased FOXA1 (left) or FOXA2 (right) binding during GT-to-PE differentiation. Enhanced FOXA1 binding to the genome was correlated with reduced DNA methylation and increased 5hmC. Such correlation was not observed between FOXA2 binding and 5mC/ 5hmC. (D) Quantification of DNA methylation levels (green) within FOXA1 or FOXA2 binding sites (revealed by ChIP-seq intensities; red) at the indicated pancreatic differentiation stages. Green bars represent the frequencies of CpGs sites in FOXA1 or FOXA2 binding sites (at 5% methylation ratio intervals). Red curves indicate FOAX1 or FOXA2 ChIP-seq signals within the corresponding methylation intervals. (E) Identification of key TF binding motifs within 5hmC enriched regions at the PE stage by the Pscan software (55).
To further explore whether 5hmC enrichment associates the binding of pancreatic lineage-specific TFs during pancreatic differentiation, we analyze 5hmC enrichment at PDX1 enriched regions at the PE stage (22) and observed a significant enrichment of 5hmC signals at PDX1 binding sites (Figure 5A). Furthermore, we performed Pscan-ChIP (55) motif analysis on HERGs at the PE stage. In addition to the FOXA1 binding motif, Pscan analysis revealed the overrepresentation of binding motifs for CUX1 and ONECUT1, ATF1 and GMEB1 within HERGs (Figure 5E). CUX1 and ONECUT1 are also reported as key TFs for normal pancreas function (56,57). Interestingly, the consensus DNA binding motifs for ATF1 and GMEB1 contain CpG sites at the center of their binding sites, raising the possibility that the DNA methylation status might influence the binding of ATF1 or GMEB1 toward their target sequences. Our findings imply that 5hmC located at enhancers is associated with altered DNA methylation status within TF binding sites to facilitate the binding of potential DNA methylation-sensitive TFs, thereby regulating the transcription of pancreatic lineage specific genes, a speculation that warrants further investigation.
5hmC ‘rims’ demarcate large functional genomic domains to maintain a low methylation status
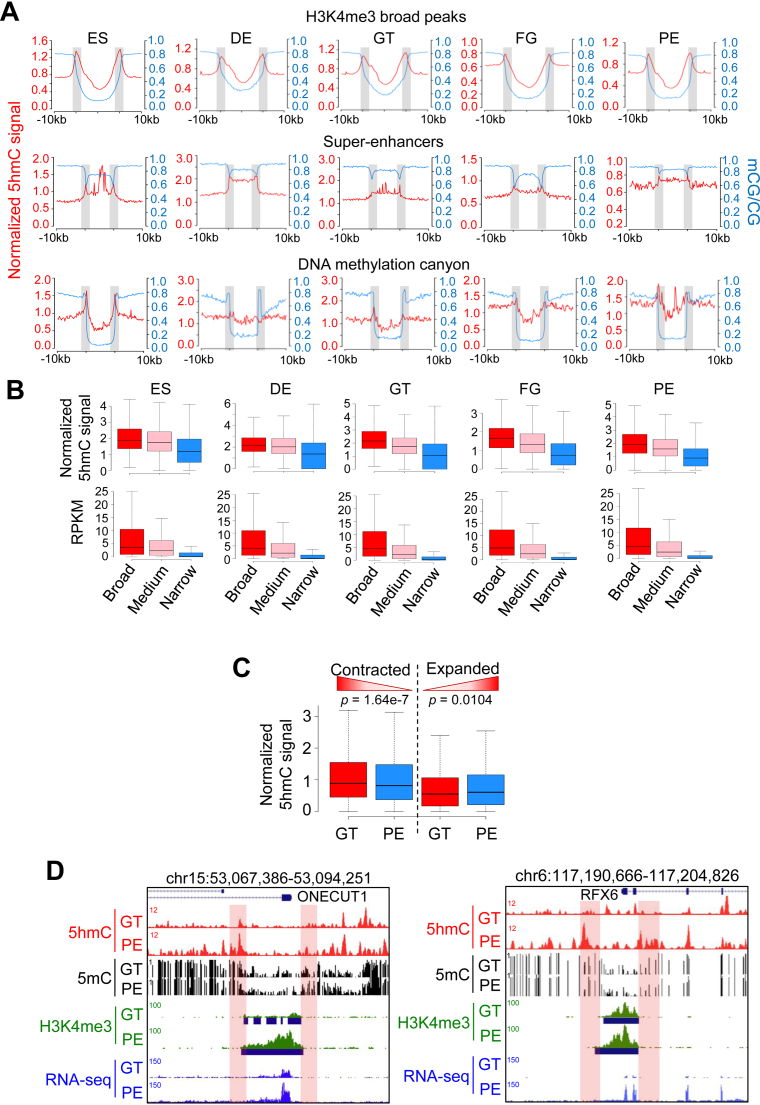
The enrichment of 5hmC at the edge of large genomic regions with low levels of DNA methylation (designated as ‘canyons’; methylation ratio ≤ 20%, length ≥ 3.5k) has been reported in hematopoietic stem cells (HSC) (58). 5hmC enrichment is closely associated with the dynamic changes of the canyon size (58). To obtain more insights into 5hmC and 5mC enrichment at large functional genomic domains during hESC differentiation toward pancreatic endoderm, we examined 5hmC distributions at typical large functional genomic domains, including super-enhancer (Supplementary Figure S6A) (59), broad H3K4me3 (60,61) and DNA methylation canyons. Interestingly, 5hmC was prominently enriched at the boundaries (designated as 5hmC ‘rims’ in contrast to DNA methylation ‘canyons’) of all the analyzed large functional genomic domains, while 5mC levels were relatively low within these regions (Figure 6A). This feature was most striking in broad H3K4me3 peaks, which are regarded as one of the key epigenetic signatures for tumor suppression in normal cells and are anti-correlated with 5mC during maternal-to-zygotic transition (60,61). Because the length of H3K4me3-marked genomic regions was closely associated with transcriptional elongation and enhancer activity, we examined 5hmC enrichment based on the length of broad H3K4me3 peaks (Supplementary Figure S6B). We found that the 5hmC enrichment at the boundaries was positively correlated with the length of the broad H3K4me3 peaks at all five stages (Figure 6B, upper panel). Consistent with two early studies (60,61), the length of broad H3K4me3 peaks was positively correlated with the expression of their corresponding genes (Figure 6B, lower panel). We also observed a striking enrichment of H3K4me1 at the edge of broad H3K4me3 peaks (Supplementary Figure S6C), a finding that is concordant with the positive correlation between 5hmC and H3K4me1 revealed by our bioinformatics analyses (Figure 3A).
Figure 6.
‘5hmC-rim’ identified at the boundaries of large functional genomic regions with low methylation. (A) Normalized 5hmC (red) and 5mC (blue) intensities within 10 kb up- and down-stream of H3K4me3 broad peaks (top), super-enhancers (middle), or DNA methylation ‘canyon’ (bottom) at five differentiation stages. Enrichment of 5hmC (with ‘5hmC-rim’ marked by gray bars) was most notable at the boundaries of these large genomic regions with low methylation levels. (B) Normalized 5hmC signals at the boundaries (top) and the corresponding genes expression levels (RPKM calculated from RNA-seq data; bottom) of broad H3K4me3 peaks with TSS. TSS-containing broad H3K4me3 peaks were categorized into 3 categories: broad (≥3 kb), medium (1–3 kb) and narrow (≤1 kb). (C) Normalized 5hmC signals at the boundaries of contracted (decease in peak size) or expanded (increase in peak size) H3K4me3 broad peaks during the GT-to-PE transition. (D) Examples of promoters (at the ONECUT1 and RFX6 loci) that underwent dynamic changes in DNA hydroxymethylation at the boundaries of H3K4me3 broad peaks (red), H3K4me3 broad peak size (green), as well as gene expression (blue), during the GT-to-PE lineage progression. Increase in 5hmC was correlated with reduced DNA methylation, as well as increased sizes of H3K4me3 ChIP-seq and RNA-seq reads.
Broad H3K4me3 peaks located within TSS are closely associated with gene expression (60,61). We therefore divided broad H3K4me3 peaks into two categories: outside TSS and within TSS. Approximately 60–70% of broad H3K4me3 peaks (ES, 62.14%; DE, 58.97%; GT, 62.79%; FG, 67.45% and PE, 67.58%) fell within TSS across the five stages (Supplementary Figures S6D and E). To further investigate the relationship between 5hmC at the edge of peaks and the length of broad H3K4me3 peaks during pancreatic differentiation, we compared 5hmC enrichment and H3K4me3 broad peak size between two representative stages (GT vs PE) that showed significant difference in DNA hydroxymethylation (Figure 2D). We first selected the broad H3K4me3 peaks with at least 50% overlap and the change of lengths ≥20% between the GT and PE stages, and then correlated them with 5hmC alterations at the boundaries of these selected regions. Our analysis suggested a positive correlation between changes in 5hmC and the size of broad H3K4me3 peaks: contraction of the broad H3K4me peak size was correlated with a reduction in 5hmC; whereas increased 5hmC level was associated with expanded broad H3K4me3 peak sizes (Figure 6C). Notably, we observed a significant increase of 5hmC level (74.36%) at the boundaries of broad H3K4me3 peaks, accompanied by the expansion of peak sizes (3.6-fold) at the promoter of ONECUT1 (a key TF involved in regulating pancreas development) (57) and upregulation of gene expression during GT-to-PE differentiation (Figure 6D). A similar scenario was also visualized at the promoter of RFX6 (Figure 6D), the expression of which is highly restricted to adult human islet cells and is essential for islet formation and insulin production (62). Taken together, the enrichment of 5hmC at the boundaries of broad H3K4me3 peaks that are located within promoters is positively associated with the histone mark peak size and the corresponding gene expression.
To further test whether the enrichment of 5hmC at large functional genomic regions is a general feature in mammals other than human, we examined the enrichment of 5hmC at DNA methylation canyon and broad H3K4me3 peaks in murine hematopoietic stem cells (HSCs). Consistent with scenarios seen in human cells, we observed 5hmC ‘rims’ situating at the edge of large functional genomic regions (Supplementary Figures S6F-G), thus reinforcing the conclusion that 5hmC ‘rims’ mark the low-methylated large genomic regions and might safeguard these regions to prevent DNA methylation.
DISCUSSION
In the current study, we performed epigenome and transcriptome profiling to map the dynamic changes of DNA methylation, hydroxymethylation, chromatin accessibility and gene transcription during the differentiation of hESC toward pancreatic endoderm. Our study based on a stepwise in vitro pancreas differentiation system, along with integrative (epi)genome profiling results published by Sander's group (21,22), provides a comprehensive reference epigenomic map for studying the role of DNA methylation and demethylation, as well as chromatin accessibility and histone modifications, during hESC differentiation and lineage specification.
Our study has unveiled a previously unappreciated biphasic change in the global levels of 5hmC during hESC differentiation toward the pancreatic lineage. PCA analysis showed distinct 5hmC and 5mC signatures at each differentiation stage, suggesting dynamic but unique DNA methylation homeostasis during this differentiation process. During the initial germ layer specification step (ES-to-DE), the global level of 5hmC is remarkably reduced with a concomitant increase in the global DNA methylation, a trend that is consistent with previous studies and is probably needed to silence the pluripotency factors (63,64). However, an opposing scenario is visualized during the later differentiation step (pancreatic lineage induction: GT→FG→PE): 5hmC undergoes gradual increase together with enhanced chromatin accessibility; but 5mC gradually declines at each transition step. It is well-known that DNA methylation acts as an epigenetic barrier between cellular lineages during cellular reprogramming (3,65,66). The global increase of 5hmC might facilitate cells overcoming the DNA methylation barrier and increase the chromatin accessibility for TFs binding (16) to aid pancreatic lineage progression. Indeed, our detailed analysis on DMRs and DHMRs between adjacent differentiation stages has revealed significantly more hypo-DMRs (reduced DNA methylation when progressing toward the next differentiation stage) and hyper-DHMRs (increased hydroxymethylation during transition) compared with the numbers of hyper-DMRs or hypo-DHMRs. Furthermore, a close scrutiny of the orders and correlations of 5hmC with 5mC during lineage progression leads to the general conclusion that genomic regions exhibiting 5hmC enrichment are positively correlated with regions showing 5mC decrease, and that DNA hydroxymethylation probably signals for the removal of the methyl group at cytosine (as indirectly reflected by a decrease in 5mC signals at the same sites) during ES-to-PE differentiation. However, reduced DNA hydroxymethylation is not necessarily associated with increased DNA methylation. This also partially explains why deletion of Tet proteins in certain cell types (e.g. B cells or HSCs) only exerts subtle effects on the upregulation of DNA methylation (35,67).
Our integrative epigenetic study has revealed varying degrees of changes in DNA methylation at different regions of the human genome during the ES-to-PE differentiation. DNA methylation at both high- and low-methylated CpG sites (HMS or LMS) remains largely stable, while genomic regions (such as enhancers) with intermediate methylation levels (IMS) exhibit the most dynamic changes in DNA methylation during pancreatic lineage progression. During our analysis, we observed that IMS, but not HMS, preferred to enrich at HERGs, suggesting an important role of 5hmC in selectively bookmarking genomic regions during the pancreas differentiation procedure. Furthermore, IMS enriched HERGs are found to be prominently accumulated at distal regulatory regions (e.g. enhancers) with high chromatin accessibility, like enhancers. Our analysis further revealed that 5hmC enrichment is positively correlated with enhancer activities and active transcription, indicating an important function of 5hmC in regulating enhancer activity and nascent transcription. But it remains to be further clarified if changes in the DNA hydroxymethylation landscapes serve as the cause or as the consequence of altered enhancer activities. This can be ideally tested by using recently developed epigenome editing tools that combine the use of catalytically-dead Cas9 with catalytic domains derived from TET to enable targeted DNA methylation editing (68–70). This approach has been used to demonstrate that targeting DNA demethylation at the distal enhancer of MyoD can affect its expression and thereby facilitate myogenic reprogramming (68). Similar experiments can be performed at key enhancers identified in the current study to examine how epigenetic regulation of enhancer activity dictate the fate of hESCs during pancreatic differentiation.
Our analysis unveils a strong positive correlation of DNA hydroxymethylation with chromatin accessibility alteration during ES-to-PE differentiation, implicating that 5hmC might be involved in modulating the function of key DNA regulatory elements during differentiation. We observed enrichment of essential pancreas specific TFs (e.g. PDX1, FOXA1) at 5hmC enriched regions. Motif analysis in 5hmC enriched regions has led to the discovery of more TFs, such as CUX1 and ONECUT1, ATF1, and GMEB1, some of which are known to be critical for pancreatic development and function and now seem to be subjected to epigenetic regulation. Most interestingly, we observed differential enrichment of 5hmC at pioneer TFs, FOXA1 and FOXA2, which are essential for endodermal lineage specification. FOXA1 and FOXA2 belong to the forkhead transcription factors that play indispensable roles during pancreas development by regulating PDX1 gene expression (71). FOXA1 binding sites, but not FOXA2 binding sites, show a significant enrichment of 5hmC signals at later differentiation stages (DE, GT and PE). This seems to be associated with the differences in the DNA methylation status at the corresponding binding sites. FOXA1 binding sites displayed relatively low DNA methylation but showed dynamic DNA demethylation changes during differentiation. By contrast, DNA methylation within FOXA2 binding regions remains relatively high and stable; and therefore, changes in DNA methylation exert less effect on FOXA2 binding. Recent studies have pointed to a possible link between FOXA1 and DNA demethylation in the transcriptional pioneering process (72), an independent piece of evidence suggesting that 5hmC deposition and the subsequent DNA demethylation is essential for FOXA1 binding. It has been suggested that not all the hypomethylated enhancers are always active in adult tissues, likely owing to inherited epigenetic memory (17). Our results suggest that dynamic changes in 5hmC and DNA demethylation at enhancers are closely associated with enhancer activities and might be essential for the binding of DNA methylation-sensitive TFs during transcriptional regulation.
One of the most striking findings made from our integrative analysis is the discovery of ‘5hmC-rim’, a unique epigenetic feature that is characterized by striking enrichment of 5hmC at the boundaries of large functional genomic regions, including super-enhancer (59), DNA methylation ‘canyon’ (58) and broad H3K4m3-enriched regions (60,61) that exhibit low DNA methylation. 5hmC-rim is also found in mouse HSCs (58), indicating that this could be a general epigenetic signature in various tissues and cell types. We speculate that heavy deposition of 5hmC mediated by TET proteins at these boundaries might guard the maintenance of low methylation at these genomic domains, but the exact function and the underlying molecular mechanism remain to be further delineated. Accumulating evidence has suggested that TET-mediated 5hmC generation is involved in regulating chromatin accessibility during vertebrate development (16). Furthermore, Flavahan et al demonstrated that increased DNA methylation could prevent CTCF binding and interfere with the interaction of topological domains in gliomas (73). Reduction of 5hmC, presumably due to TET loss of function, is a general feature in both solid tumors and hematological malignancies (44,74). The 5hmC-rim might be closely involved in modulating chromatin accessibility and topological domains in the genome during hESC differentiation. The presence of 5hmC-rim at broad H3K4me3 peaks raises the possibility that TET-mediated hydroxymethylation at the boundary might prevent the shortening of H3K4me3, which is often observed in tumor (60). To extrapolate these findings to pathological conditions, disruption of DNA methylation and demethylation balance, owing to mutations or altered expression of DNMTs or TET proteins, will very likely cause aberrant transcription by affecting enhancer activity, transcription factor binding and possibly the re-organization of large functional genomic regions.
To conclude, the present study has provided an atlas of DNA methylomes, DNA hydroxymethylomes, transcriptomes and genome-wide chromatin accessible maps that represent five typical stages of pancreatic differentiation from hESC to pancreatic endoderm (PE). Knowledge gained from our integrative studies not only provides useful information to guide the efficient production of functional pancreatic progenitors for stem cell-based therapies for diabetes, but also facilitates the understanding of pathogenic mechanisms underlying pancreatic diseases.
DATA AVAILABILITY
The RNA-seq, WGBS, CMS-IP-seq and ATAC-Seq datasets have been deposited into GEO under accession number GSE97992.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for Dr Jianjun Shen and the MD Anderson next-generation sequencing core at Smithville (CPRIT RP120348 and RP170002), and the Genomics & Bioinformatics core in Faculty of Health Sciences at the University of Macau for highthroughput sequencing service.
Author contributions: Y.H. and Y.Z. conceived the project. Y.H., R.X. and D.S. directed and oversaw the project. X.W. performed in vitro hESC pancreatic differentiation and ATAC-seq library preparation. M.J. and W.H. performed dotblot analysis. M.J., Y.Z., W.C. and L.G. prepared RNA-seq, CMS-IP and WGBS-seq sequencing libraries. J.L. and D.S. performed integrative data analysis. W.M. and W.C. provided intellectual inputs. Y.H. and Y.Z. wrote the manuscript with all the other authors participating in discussion, data interpretation and manuscript editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Prevention and Research Institute of Texas [RR140053 to Y.H., to RP170660 to Y.Z.]; Innovation Award from American Heart Association [16IRG27250155 to Y.H.]; John S. Dunn Foundation Collaborative Research Award (to Y.H.); National Science Foundation of China (31701276 to R.X.); University of Macau Start-up Research Grant [SRG2014-00030-FHS to R.X.]; National Institute of Health [R01GM112003 to Y.Z.]; Welch Foundation [BE-1913 to Y.Z.]; American Cancer Society [RSG-16-215-01-TBE to Y.Z.]; Texas A&M University start-up funds (to Y.H. and D.S.). Funding for open access charge: Texas A&M University Open Access to Knowledge Fund (OAKFund), supported by the University Libraries and the Office of the Vice President for Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 2. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002; 16:6–21. [DOI] [PubMed] [Google Scholar]
- 3. De Carvalho D.D., You J.S., Jones P.A.. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010; 20:609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 5. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al. . Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O’Malley R., Castanon R., Klugman S. et al. . Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011; 471:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bestor T.H., Ingram V.M.. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:5559–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li E., Zhang Y.. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014; 6:a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. . Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G. et al. . Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011; 8:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. et al. . Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hashimoto H., Liu Y., Upadhyay A.K., Chang Y., Howerton S.B., Vertino P.M., Zhang X., Cheng X.. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012; 40:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inoue A., Zhang Y.. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011; 334:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji D., Lin K., Song J., Wang Y.. Effects of Tet-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol. Biosyst. 2014; 10:1749–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bogdanovic O., Smits A.H., de la Calle Mustienes E., Tena J.J., Ford E., Williams R., Senanayake U., Schultz M.D., Hontelez S., van Kruijsbergen I. et al. . Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 2016; 48:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B.. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013; 45:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dai H.Q., Wang B.A., Yang L., Chen J.J., Zhu G.C., Sun M.L., Ge H., Wang R., Chapman D.L., Tang F. et al. . TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016; 538:528–532. [DOI] [PubMed] [Google Scholar]
- 19. Kaestner K.H. An epigenomic road map for endoderm development. Cell Stem Cell. 2015; 16:343–344. [DOI] [PubMed] [Google Scholar]
- 20. Rezania A., Bruin J.E., Xu J., Narayan K., Fox J.K., O’Neil J.J., Kieffer T.J.. Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells. 2013; 31:2432–2442. [DOI] [PubMed] [Google Scholar]
- 21. Xie R., Everett L.J., Lim H.W., Patel N.A., Schug J., Kroon E., Kelly O.G., Wang A., D’Amour K.A., Robins A.J. et al. . Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013; 12:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang A., Yue F., Li Y., Xie R., Harper T., Patel N.A., Muth K., Palmer J., Qiu Y., Wang J. et al. . Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015; 16:386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. . Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P. et al. . Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011; 473:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Y., Pastor W.A., Zepeda-Martinez J.A., Rao A.. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat. Protoc. 2012; 7:1897–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., Rao A.. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010; 5:e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xi Y., Li W.. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009; 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun D., Xi Y., Rodriguez B., Park H.J., Tong P., Meong M., Goodell M.A., Li W.. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 2014; 15:R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin X., Sun D., Rodriguez B., Zhao Q., Sun H., Zhang Y., Li W.. BSeQC: quality control of bisulfite sequencing experiments. Bioinformatics. 2013; 29:3227–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsagaratou A., Aijo T., Lio C.W., Yue X., Huang Y., Jacobsen S.E., Lahdesmaki H., Rao A.. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E3306–E3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lio C.W., Zhang J., Gonzalez-Avalos E., Hogan P.G., Chang X., Rao A.. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. Elife. 2016; 5:e18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X., Su J., Jeong M., Ko M., Huang Y., Park H.J., Guzman A., Lei Y., Huang Y.H., Rao A. et al. . DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 2016; 48:1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W.. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011; 473:398–402. [DOI] [PubMed] [Google Scholar]
- 41. Wu H., D’Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y.. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011; 25:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K.. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011; 473:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X. et al. . Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011; 29:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R. et al. . Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010; 468:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kohli R.M., Zhang Y.. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013; 502:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A.. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012; 484:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu H., Zhang Y.. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014; 156:45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., Balasubramanian S.. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012; 336:934–937. [DOI] [PubMed] [Google Scholar]
- 50. Booth M.J., Ost T.W., Beraldi D., Bell N.M., Branco M.R., Reik W., Balasubramanian S.. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013; 8:1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K.. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998; 12:3752–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V.. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002; 32:128–134. [DOI] [PubMed] [Google Scholar]
- 53. Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H.. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005; 435:944–947. [DOI] [PubMed] [Google Scholar]
- 54. Zhu H., Wang G., Qian J.. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016; 17:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zambelli F., Pesole G., Pavesi G.. PscanChIP: Finding over-represented transcription factor-binding site motifs and their correlations in sequences from ChIP-Seq experiments. Nucleic Acids Res. 2013; 41:W535–W543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krug S., Kuhnemuth B., Griesmann H., Neesse A., Muhlberg L., Boch M., Kortenhaus J., Fendrich V., Wiese D., Sipos B. et al. . CUX1: a modulator of tumour aggressiveness in pancreatic neuroendocrine neoplasms. Endocr. Relat. Cancer. 2014; 21:879–890. [DOI] [PubMed] [Google Scholar]
- 57. Lynn F.C., Smith S.B., Wilson M.E., Yang K.Y., Nekrep N., German M.S.. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10500–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jeong M., Sun D., Luo M., Huang Y., Challen G.A., Rodriguez B., Zhang X., Chavez L., Wang H., Hannah R. et al. . Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 2014; 46:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen K., Chen Z., Wu D., Zhang L., Lin X., Su J., Rodriguez B., Xi Y., Xia Z., Chen X. et al. . Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015; 47:1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H. et al. . Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016; 537:558–562. [DOI] [PubMed] [Google Scholar]
- 62. Smith S.B., Qu H.Q., Taleb N., Kishimoto N.Y., Scheel D.W., Lu Y., Patch A.M., Grabs R., Wang J., Lynn F.C. et al. . Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010; 463:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen T., Ueda Y., Dodge J.E., Wang Z., Li E.. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003; 23:5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jackson M., Krassowska A., Gilbert N., Chevassut T., Forrester L., Ansell J., Ramsahoye B.. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 2004; 24:8862–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Constantinides P.G., Jones P.A., Gevers W.. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977; 267:364–366. [DOI] [PubMed] [Google Scholar]
- 66. Kallin E.M., Rodriguez-Ubreva J., Christensen J., Cimmino L., Aifantis I., Helin K., Ballestar E., Graf T.. Tet2 facilitates the derepression of myeloid target genes during CEBPalpha-induced transdifferentiation of pre-B cells. Mol. Cell. 2012; 48:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. An J., Gonzalez-Avalos E., Chawla A., Jeong M., Lopez-Moyado I.F., Li W., Goodell M.A., Chavez L., Ko M., Rao A.. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 2015; 6:10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu X., Lu H., Chen T., Nallaparaju K.C., Yan X., Tanaka S., Ichiyama K., Zhang X., Zhang L., Wen X. et al. . Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 2016; 14:1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maeder M.L., Angstman J.F., Richardson M.E., Linder S.J., Cascio V.M., Tsai S.Q., Ho Q.H., Sander J.D., Reyon D., Bernstein B.E. et al. . Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013; 31:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee M., Li J., Liang Y., Ma G., Zhang J., He L., Liu Y., Li Q., Li M., Sun D. et al. . Engineered split-TET2 enzyme for inducible epigenetic remodeling. J. Am. Chem. Soc. 2017; 139:4659–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gao N., LeLay J., Vatamaniuk M.Z., Rieck S., Friedman J.R., Kaestner K.H.. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008; 22:3435–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y., Zhang D., Li Q., Liang J., Sun L., Yi X., Chen Z., Yan R., Xie G., Li W. et al. . Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat. Genet. 2016; 48:1003–1013. [DOI] [PubMed] [Google Scholar]
- 73. Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suva M.L., Bernstein B.E.. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016; 529:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang H., Liu Y., Bai F., Zhang J.Y., Ma S.H., Liu J., Xu Z.D., Zhu H.G., Ling Z.Q., Ye D. et al. . Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013; 32:663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq, WGBS, CMS-IP-seq and ATAC-Seq datasets have been deposited into GEO under accession number GSE97992.