Abstract
During its life cycle, Trypanosoma brucei shuttles between a mammalian host and the tsetse fly vector. In the mammalian host, immune evasion of T. brucei bloodstream form (BSF) cells relies on antigenic variation, which includes monoallelic expression and periodic switching of variant surface glycoprotein (VSG) genes. The active VSG is transcribed from only 1 of the 15 subtelomeric expression sites (ESs). During differentiation from BSF to the insect-resident procyclic form (PCF), the active ES is transcriptionally silenced. We used mass spectrometry-based interactomics to determine the composition of telomere protein complexes in T. brucei BSF and PCF stages to learn more about the structure and functions of telomeres in trypanosomes. Our data suggest a different telomere complex composition in the two forms of the parasite. One of the novel telomere-associated proteins, TelAP1, forms a complex with telomeric proteins TbTRF, TbRAP1 and TbTIF2 and influences ES silencing kinetics during developmental differentiation.
INTRODUCTION
The telomeric ends of eukaryotic chromosomes are protected by nucleoprotein complexes (1). The telomeric protein complex in mammals, called shelterin, consists of six core subunits: TRF1, TRF2 and POT1, which bind directly to the telomeric TTAGGG repeats, and three additional proteins TIN2, TPP1 and RAP1, which are associated by protein–protein interactions. This complex and its accessory factors are central players in the maintenance of genome integrity by shielding the chromosome ends from unwanted DNA repair activities (2). Telomeres are actively elongated in cancer and germ cells by the enzyme telomerase, a process involving the shelterin complex (3) and the direct telomere-binding protein HOT1 (4). In yeast, telomeric protein complexes are different. While Saccharomyces cerevisiae telomeres are bound by scRAP1, which interacts with RIF1 and RIF2, Schizosaccharomyces pombe features a telomeric complex with at least six subunits (5). In trypanosomes, the causative agent of sleeping sickness in humans and nagana in animals, thus far three telomeric proteins have been characterized: TbTRF, TbRAP1 and TbTIF2 (6–8).
In both yeasts and human, it has been observed that telomeres can be tethered to the nuclear periphery (9,10) and exert a gene regulatory effect by forming a heterochromatic structure that reversibly suppresses the transcription of their nearby subtelomeric proximal genes. This telomere position effect (TPE) or telomeric silencing relies on epigenetic regulation by histone modifications (11,12). In S. cerevisiae, histone deacetylases (Sir) interacting with scRAP1 spread hypoacetylation of histones H3 and H4 along the chromosome to establish silent chromatin (13). Similarly, TPE in mammals is known to involve SIRT6, a histone deacetylase of the Sir2 family (14).
In microbial pathogens, virulence gene expression can be regulated by the telomere structure as those genes are often found adjacent to them (15). In trypanosomes, the developmentally regulated subtelomeric variant surface glycoprotein (VSG) genes are the main virulence determinants in the mammalian-infectious bloodstream form (BSF). The cell surface of the BSF parasite is densely covered with a single species of VSG. To escape the mammalian host immune response, trypanosomes depend on antigenic variation of its VSG coat. Antigenic variation in trypanosomes is characterized by monoallelic transcription and by switching of the VSG gene (16). Key structures of antigenic variation are 15 specialized subtelomeric transcription units, the expression sites (ESs). Each ES contains an RNA polymerase (pol) I-driven ES promoter, usually located 40–60 kb upstream of the single VSG gene at the telomere (17). At any given time only one ES is transcriptionally active in the BSF of the parasite, while the remaining 14 ESs are silenced (17). The monoallelic VSG expression is mediated by an extranucleolar pol I-containing expression site body, ensuring that only one isoform of VSG is displayed on the cell surface (18). The expressed VSG type can be changed by either a transcriptional or a recombination switch (19).
There is an intricate connection between the subtelomeric VSG genes and the telomeric proteins in trypanosomes. The telomere-binding proteins TbTRF and TbTIF2 suppress VSG switching (8,20,21). TbRAP1 promotes ES silencing by controlling the telomeric chromatin structure (22). Depletion of TbRAP1 leads to derepression of ES-linked VSG genes in BSF and PCF cells. In contrast to the yeast TPE mechanism, the TbRAP1-mediated VSG silencing in trypanosomes seems to act independently of Sir proteins, as depletion of the trypanosome Sir2 homolog does not prevent VSG silencing (23). The studies done on TbTRF, TbTIF2 and TbRAP1 provide information on how trypanosome telomeres maintain the inactive state of ESs and suppress recombination. If and how telomere-binding proteins contribute to ES silencing initiation has not yet been studied.
ES silencing initiation plays an important role during developmental transition of BSF parasites. Trypanosoma brucei is transmitted by the tsetse fly vector. In the insect vector, BSF trypanosomes differentiate to procyclic form (PCF) trypanosomes and replace their VSG coat with procyclin (24). Thus, during developmental transition the active ES is repressed to stop VSG transcription (25). During this process chromatin restructuring takes place (26). The active ES promoter undergoes rapid repositioning to the nuclear envelope where it is silenced, presumably by chromatin condensation (27,28). Less is known about how the developmental silencing process is initiated, timed and regulated on a DNA level. It has been demonstrated that ES transcriptional activity and differentiation are mechanistically linked (29). Transcriptional ES attenuation can initiate the differentiation process whereby ES transcription stops before the chromatin condensates (30). Bromodomain proteins, which bind acetylated lysine residues of histones and control gene expression by interacting with the transcriptional machinery, were shown to counteract the differentiation process of BSF to PCF parasites (31).
However, control of transcription and chromatin organization must be temporally fine-tuned during life cycle differentiation. Each process must take place with specific kinetics to ensure a coordinated ES silencing, and thus likely involves further regulatory factors.
Here, we show that the novel telomere-binding protein TelAP1 is part of the TbTRF–TbRAP1–TbTIF2 complex in BSF cells and forms a separate complex in PCF cells. This provides the first evidence for developmental differences in the telomere complex in trypanosomes. Further analysis showed that TelAP1 influences the kinetics of ES silencing during early events of the developmental transition from BSF to PCF.
MATERIALS AND METHODS
Trypanosome cell lines and cultivation
Monomorphic T. brucei BSFs (strain Lister 427, antigenic type MITat 1.2 clone 221a) were cultured in HMI-9 medium with 10% heat-inactivated fetal calf serum (FCS) (Sigma) at 37°C and 5% CO2 (32). Cells of single marker (SM) (33) or 2T1 (34) background co-expressing the T7 RNA polymerase and tetracycline (Tet) repressor were used to generate the BSF cell lines for this study.
PCFs (strain 427) were cultured in modified SDM-79 with 10% heat-inactivated FCS (Sigma) at 27°C (35). Here, 29–13 or wild-type (WT) procyclic cells were used to generate transgenic procyclic cell lines. The 29–13 procyclic cells co-express T7 RNA polymerase and the Tet repressor (33).
Cell densities of BSF and PCF cultures were determined using a Coulter Counter Z2 particle counter (Beckman Coulter). Transfections and drug selections were carried out as described previously (36).
Transgenic trypanosome cell lines
SM TelAP1 RNAi, 29-13 TelAP1 RNAi
For RNAi-mediated depletion of TelAP1 in BSF and PCF cells the Gateway recombination system was used as described in (37). A TelAP1 fragment (position 187–627) was amplified from genomic DNA using primers specific for TelAP1 open reading frame (ORF) with attB1 and attB2 sites. The polymerase chain reaction (PCR) product was inserted into the intermediate vector pDONR207 by a BP clonase reaction (Invitrogen). This plasmid was used in an LR clonase (Invitrogen) recombination reaction with the pTrypRNAiGate vector to generate the final RNAi construct. For transfections of trypanosomes the plasmid was linearized with NotI. Knockdown was induced with 1 μg/ml tetracycline.
SM TelAP1 OE, 29-13 TelAP1 OE
To generate TelAP1-overexpressing bloodstream and procyclic trypanosomes, the TelAP1 ORF was amplified from genomic DNA and cloned into pLew100v5b1d_Phleo using HindIII and BamHI sites. The resulting plasmid was linearized using NotI for integration in the ribosomal spacer locus in MITat 1.2 SM and 29–13 cells. Expression was induced with 1 μg/ml tetracycline.
BSF ΔTelAP1, PCF ΔTelAP1
PyrFEKO vectors (gift from H.S. Kim) containing either puromycin N-acetyl-transferase (PUR) or hygromycin phosphotransferase (HYG) ORFs fused with Herpes simplex virus thymidine kinase (TK) were used to construct the TelAP1 deletion mutant (38). As the fusion genes PUR-TK and HYG-TK are flanked by loxP sites they can be removed by transient expression of Cre recombinase. To make the knockout vectors, 300 bp of 5′ and 3′ TelAP1 UTRs for targeting were amplified by PCR from genomic DNA and cloned into PvuII–HindIII and SbfI–BamHI sites of pyrFEKO vectors, respectively. The resulting plasmids were linearized with PvuII and SbfI for transfection into MITat 1.2 SM and 427 procyclic cells. Resistance markers were removed in BSF ΔTelAP1 cells by transient transfection with pLew100cre-EP1 (gift from H.S. Kim) and selection with 50 μg/ml Ganciclovir (Sigma, G2536).
SM TbTRF Ty1/-, PCF TbTRF Ty1/-
A PCR-based method was used to delete the first allele of TbTRF. The phleomycin-resistance cassette was amplified from the pLew82 plasmid (33) with primers containing homologous sequences to the TbTRF UTRs for recombination. After deletion of the first allele the second allele was in situ tagged with a Ty1 epitope at the C-terminus by a PCR-based method using the pMOTag2T vector as template as described in (39).
2T1 TRF RNAi
The TRF RNAi cell line was generated using the pGL2084 vector and 2T1 cells as background (40). The resulting plasmid was digested with AscI. Knockdown was induced with 1 μg/ml tetracycline.
ΔTelAP1-Rluc(pro)-Fluc(tel), SM-Rluc(pro)-Fluc(tel)
Dual luciferase reporter cell lines were generated by transfection of BSF ΔTelAP1 or SM cells with BstApI-linearized pCJ25A and SacI/KpnI-linearized pFG14n (41,42). To select for cells that integrated the constructs in the active ES 50 μg/ml of blasticidin and 30 μg/ml hygromycin were used.
ΔTelAP1-Rluc(pro)-Fluc(tel) rescue
For the rescue of TelAP1 the pLew100_TelAP1 construct as used for ectopic TelAP1 overexpression was transfected in ΔTelAP1 dual reporter cells.
Telomere pull-down
Telomere pull-downs were done as previously described (43). In brief, chemically synthesized oligonucleotides with either TTAGGG or TGTGAG repeats were annealed with their complementary oligonucleotides. The dsDNA was phosphorylated with 100 units PNK (Thermo) for 2 h at 37°C and ligated overnight with 20 units T4 ligase (Thermo) at room temperature (RT). The mixture was cleaned by chloroform phenol extraction and the purified DNA was incubated with biotin-dATP (Jena Biosciences) and 60 units Klenow fragment (Thermo) at 37°C overnight. The DNA was re-buffered using a G50 Spin column (GE Healthcare) according to the manufacturer’s instructions. Around 25 μg biotinylated DNA was immobilized on Streptavidin Dynabeads MyOne C1 (Thermo) and incubated with trypanosome PCF whole cell lysate obtained by lysis in modified RIPA buffer [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 0.1% Sodium Deoxycholate, Protease inhibitor cocktail (Roche)]. The binding reaction was performed in protein binding buffer (PBB) (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.5% Igepal CA-630) in the presence of 10 μg sheared salmon sperm DNA (Ambion) at 4°C for 2 h under slight agitation. Unbound proteins were washed with PBB three times and the bound fraction eluted with 1× lithium dodecyl sulfate (LDS) buffer (Thermo).
Co-immunoprecipitation
Per immunoprecipitation (IP), 30 μl of Protein G Sepharose Fast Flow beads (GE Healthcare) were once washed in 1 ml phosphate-buffered saline (PBS) (500 g, 1 min, 4°C) and twice in PBS/1% bovine serum albumin (BSA). Unspecific binding sites were blocked by incubation for 1 h at 4°C with PBS/1%BSA on an orbital mixer. The beads were pelleted by centrifugation (500 g, 1 min, 4°C), supernatant removed, and the antibody added. Beads and antibody were incubated overnight at 4°C under mild agitation on an orbital mixer. Unbound antibody was removed by washing 3× in 1 ml PBS/0.1% BSA.
Per IP, 2 × 108 BSF cells or PCF cells were harvested, washed once with ice-cold TDB (5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2PO4, 2 mM NaH2PO4, 20 mM glucose, pH 7.4) or PBS, respectively, and incubated in 1 ml IP buffer [150 mM NaCl, 0.5% Igepal CA-630, 20 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche)] for 20 min on ice. Cells were lysed by sonication (3 cycles, 30 s on and 30 s off) using a Biorupter (Diagenode). A centrifugation step (10 000 g, 10 min, 4°C) followed to clear the lysate. Protein G sepharose beads coupled to TelAP1 mouse monoclonal IgG or anti-Ty1 mouse monoclonal IgG were first washed in IP buffer and then added to the lysate for overnight incubation at 4°C. The following day the beads were washed 3× with IP buffer for 5 min on ice and harvested by centrifugation (500 g, 1 min, 4°C). Proteins were eluted by incubating the beads in 50 μl sample buffer (NuPAGE®LDS Sample buffer, 100 mM DTT) at 70°C for 10 min. Eluates were analyzed by mass spectrometry (MS).
Mass spectrometry and data analysis
Samples were separated on a 4–12% Novex NuPage gel (Thermo). The in-gel digest was performed according to standard protocol (44). The gel pieces were minced, incubated with 10 mM DTT/0.05 M ammonium bicarbonate pH 8 for 1 h at 56°C and proteins subsequently alkylated with 55 mM iodoacetamide/0.05 M ammonium bicarbonate pH 8 for 30 min in the dark. The proteins were digested with 1 μg trypsin (Promega or Sigma) overnight at 37°C. The tryptic peptides were desalted using a StageTip (45) and stored in the fridge until mass spectrometric measurement.
To analyze the telomere pull-down, the gel was sliced into four fractions per lane and each fraction was measured on an LTQ-Orbitrap XL (Thermo) coupled to an Easy-nLC system (Proxeon). The peptides were eluted in a 105 min non-linear gradient of 2–60% acetonitrile with a Top10 acquisition method using CID fragmentation for MS/MS. For analysis of the protein Co-IPs, a single fraction per lane was measured on a Q Exactive Plus mass spectrometer coupled to an Easy-nLC 1000 (Thermo) with a 75 min non-linear gradient of 2–60% acetonitrile with a Top10 method using HCD fragmentation for MS/MS.
The MS spectra were processed with MaxQuant (version 1.5.2.8) (46) using LFQ quantitation (47) with preset setting except match between runs was activated. For the search, a T. brucei TREU 927 protein database (ver8.1; 11 567 entries) downloaded from www.tritrypdb.org was used. The data of the protein groups file was filtered for contaminant reverse hits and used to generate the volcano plot by calculating median and a P-value (Welch t-test) for each protein group. The data was visualized using the ggplot2 package of R. For an estimation of protein stoichiometry, the average LFQ protein intensities of the control pull-downs (quadruplicates) were subtracted from the LFQ protein intensities from the immunoprecipitated samples (quadruplicates) and normalized to the average LFQ intensity of the bait protein.
Recombinant protein expression
The coding sequence of TbTRF and TelAP1 were amplified from reverse transcribed trypanosome mRNA and cloned into the pCoofy expression system (48). The sequence of both genes was verified by Sanger sequencing. The pCoofy4 constructs (N-terminal His6-MBP) were used to transform BL21(DE3) pRare T1 cells. After induction of protein expression the cell pellet was lysed with Avestin and the soluble fraction subjected to affinity purification using a 1 ml MBPTrapHP column (GE Healthcare), and as second step His-Select Ni Affinity Gel (Sigma). Protein identity and purity was monitored by LC-ESI/MS on a micrOTOF instrument (Bruker).
Telomere pull-down with recombinant proteins
The biotinylated (TTAGGG)n and (TGTGAG)n baits were prepared as previously described (43). HisMBP-TelAP1 was cleaved with HRV-3C protease (Protein Production Core Facility, IMB Mainz) in a 1:200 dilution overnight at 4°C. 5 μg of either HRV-3C protease cleaved TelAP1 or HisMBP-TbTRF as well as a combination of both proteins were incubated with 500 μg Dynabeads MyOne Streptavidin T1 (Life Technologies) coupled with either telomeric or control DNA for 1.5 h at 4°C in PBB buffer (150 mM NaCl, 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 0.5% Igepal CA630) under rotation. After three washing steps with PBB buffer, bound proteins were eluted in 1× NuPAGE LDS buffer (Life Technologies) supplemented with 100 mM DTT by boiling at 70°C for 10 min. Input samples were prepared with 20% of the protein concentration used for the pull-down and boiled directly in 1× LDS/DTT. The separation of the samples was done on a 4–12% NuPAGE Novex Bis-Tris precast gel (Life Technologies) in 1× MOPS at 180 V for 70 min. The gel was afterward stained with Coomassie Blue solution (1.25 g Coomassie Blue G-250, 45% EtOH, 45% DI H2O, 10% acetic acid) and destained in H2O. The picture was taken with a ChemiDoc XRS+ system (Bio-Rad) running with the Image Lab software.
Western blot analysis and antibodies
Protein extracts of 5 × 106 cells were separated on 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene difluoride (PVDF) membranes. For blocking, membranes were incubated in 5% milk powder in PBS at 4°C overnight. Primary antibodies were applied in PBS/1% milk/0.1% Tween 20 solution for 1 h at RT. After three washes with PBS/0.1% Tween 20, IRDye 800CW- and 680LT-coupled secondary antibodies (LI-COR Bioscience) were used to detect primary antibodies. Secondary antibodies were incubated in PBS/1% milk/0.1% Tween 20/0.02% SDS solution for 1 h at RT. Blots were analyzed using a LI-COR Odyssey Imager.
The polyclonal anti-TbH3 rabbit antibody was described in (49). The monoclonal anti-TbPFR mouse antibody L13D6 and the anti-Ty1 mouse antibody BB2 were gifts from K. Gull (University of Oxford) and described in (50). The anti-VSG221 CRD-depleted rabbit antibody was obtained from L. Figueiredo and is described in (42).
Monoclonal antibodies specific for TelAP1 (anti-TelAP1 13D9) and TbTRF (anti-TbTRF 6F5) were raised by immunizing rats with recombinant HisMBP-TelAP1 and HisMBP-TbTRF proteins expressed from bacteria, respectively. In addition, the HisMBP-TelAP1 fusion protein was used to immunize mice to generate the anti-TelAP1 2E6 antibody. The antibody production was carried out at the Helmholtz Centre in Munich by E. Kremmer.
Immunofluorescence
1 × 107 BSF cells were harvested (1500 g, 10 min, RT) and resuspended in 1 ml HMI-9 and fixed in 2% formaldehyde for 5 min at RT. The fixed cells were washed three times with PBS and resuspended in 500 μl PBS. A total of 100 μl of cells were added to poly-L-lysine-coated slides and allowed to settle for 20 min at RT. Attached trypanosomes were then permeabilized with 0.2% Igepal CA-630 in PBS for 5 min at RT. After washing twice with PBS cells were blocked with 1% BSA in PBS for 1 h at 37°C. TelAP1 mouse monoclonal IgG and TbTRF rat monoclonal IgG were used to detect TelAP1 and TbTRF, respectively. The primary antibodies were applied for 1 h at RT. After three washes with PBS, Alexa488- and Alexa594-conjugated secondary antibodies (Life Technologies) and Hoechst to stain DNA were applied for 45 min at RT. After three washing steps with PBS, cells were mounted in Vectashield (Vecta Laboratories Inc.), and images were captured by using an IMIC microscope (TILL Photonics, Gräfelfing, Germany). Deconvolution was carried out using the Huygens Essential software 4.1 (Scientific Volume Imaging).
Differentiation of monomorphic BSF into PCF
BSF cells were grown to ∼1.5 × 106 cells/ml in HMI-9 at 37°C. 2.5 × 107 cells were harvested by centrifugation (1500 g, 10 min, RT) and resuspended in 5 ml Differentiating Trypanosome Medium (DTM) containing 6 mM cis-aconitate (51). Trypanosomes were then cultured at 27°C with 5% CO2. Further cell dilutions were carried out using SDM-79 medium supplemented with 10% heat-inactivated FCS.
Dual-luciferase assay
To measure Firefly luciferase (Fluc) and Renilla luciferase (Rluc) activity the Dual-Luciferase Reporter assay kit (Promega) was used according to the manufacturer’s instructions. Per assay 1 × 106 cells were harvested (1500 g, 10 min, 4°C) and washed once in ice-cold PBS. The cell pellet was then lysed in 100 μl 1× Passive Lysis Buffer (PLB) (Promega). A total of 10 μl of the lysate was added to 45 μl of the substrate for the Firefly luciferase LARII (Luciferase Assay Substrate dissolved in Luciferase Assay Buffer II) (Promega) on a 96-well plate. The luciferase activity was measured using the Tecan reader. To quench the Firefly luminescence 45 μl of Stop&Glo Reagent was added and the Rluc activity was measured.
RESULTS
Identification of novel telomere-binding proteins in T. brucei
African trypanosomes diverged early from the main eukaryotic lineage (52). Therefore, it is not surprising that sequence-based searches for shelterin components in T. brucei have only resulted in the identification of a TRF homolog (6). Other components of the telomeric complex in trypanosomes, e.g. TbRAP1 and TbTIF2, were found by yeast two-hybrid screens or Co-IP experiments (7,8).
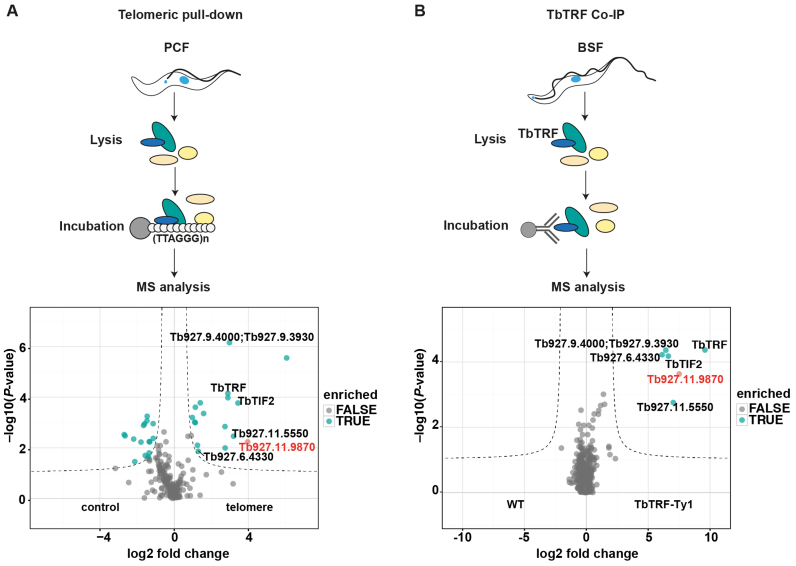
In order to identify novel proteins that associate with telomeres in T. brucei, we used two independent biochemical approaches (Figure 1). First, a pull-down assay with telomeric repeat oligonucleotides to isolate proteins that bind directly or indirectly to telomeric DNA (43). Second, we used the telomeric protein TbTRF in an IP experiment and quantified co-purifying proteins by MS. The telomeric DNA pull-down was performed in quadruplicates with whole cell lysates of PCF cells incubated either with TTAGGG-repeat oligonucleotides or a shuffled control sequence (TGTGAG). The bound proteins were eluted and analyzed by label-free quantitative proteomics. From this approach we identified among 210 proteins 17 candidates that were statistically significantly (P < 0.01) enriched in the telomeric DNA pull-downs (Figure 1A).
Figure 1.
Label-free interactomics identifies novel telomere-binding proteins in Trypanosoma brucei. (A) Experiment design and volcano plot showing how telomeric DNA-binding proteins were identified. Quadruplicates of procyclic cell extracts were incubated either with telomeric TTAGGG-repeat or control TGTGAG-repeat oligonucleotides. Seventeen proteins were significantly enriched with telomeric DNA compared to control oligonucleotides. (B) Experiment design of the TbTRF Co-IP and resulting volcano plot showing TbTRF-Ty1 interacting proteins. In both plots only the overlapping candidates are shown (complete datasets are summarized in Supplementary Tables S1 and 2). Six telomeric proteins were identified in both the experiments: TbTIF2, TbTRF, Tb927.6.4330, Tb927.11.5550, Tb927.9.4000/3930 and Tb927.11.9870. The candidate Tb927.11.9870, which was selected for detailed analyses, is highlighted in red.
The TbTRF IP-MS experiment was performed in quadruplicates with an anti-Ty1 antibody using BSF parasites that constitutively expressed a Ty1-tagged TbTRF from the endogenous locus. Extracts of WT cells without the tagged TbTRF were immunoprecipitated in parallel with the Ty1-antibody as controls. The bound proteins were eluted and analyzed by MS (Figure 1B). A total of 636 proteins were co-purified with TbTRF but only five proteins were significantly enriched (P < 0.01) with TbTRF: TbTIF2, Tb927.6.4330 (hypothetical protein, shown to associate with telomeres (53)), Tb927.9.4000/3930 (hypothetical proteins), Tb927.11.5550 (DNA Polymerase theta) and Tb927.11.9870 (hypothetical protein). Tb927.6.4330 was described recently in a genetic screen for defects in telomere-exclusive gene expression (53). All of these five proteins were also identified in our telomere pull-down. The whole dataset of the telomeric pull-down and the TbTRF Co-IP is summarized in Supplementary Tables S1 and 2. We selected the highly enriched uncharacterized protein Tb927.11.9870 for further analysis.
Tb927.11.9870 is a component of the telomere complex in trypanosomes
The hypothetical protein Tb927.11.9870 was named telomere-associated protein 1 (TelAP1) as it was identified in both of our independent biochemical searches for telomere-associated proteins. TelAP1 has a predicted molecular weight of 45 kDa. Bioinformatic analyses of the amino acid sequence revealed neither annotated domains nor any homology to known telomere-binding proteins. The nuclear protein (54) has a potential nuclear localization signal that starts at amino acid position 282 (HTRKRARNA).
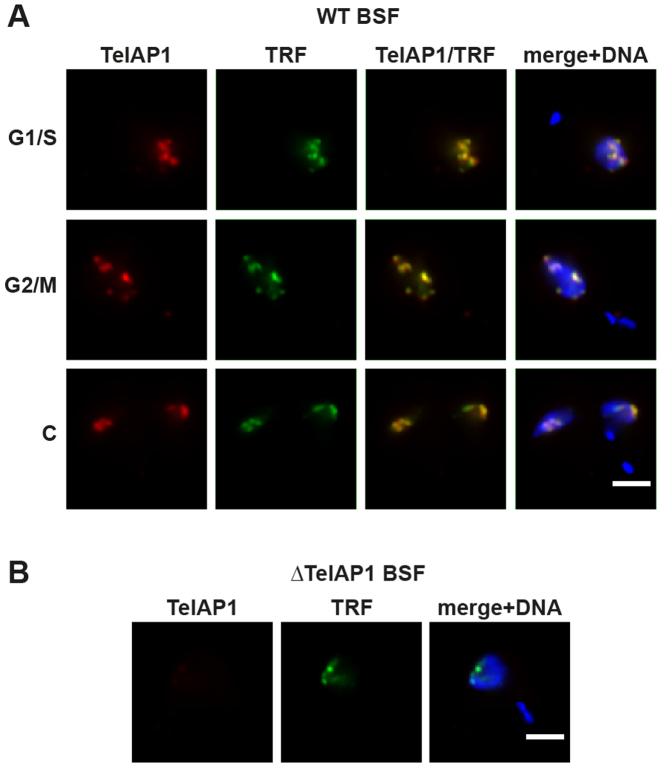
To have a tool for the characterization of endogenous TelAP1, we raised monoclonal antibodies against recombinant TelAP1 full-length protein in mouse and rat (Supplementary Figure S1A and B), as well as a rat anti-TbTRF antibody as a marker for telomeres (6) (Supplementary Figure S1C), and validated all antibodies by western blotting. Immunofluorescence analysis showed that TelAP1 is a nuclear protein and co-localized with TbTRF throughout the cell cycle in BSF cells, suggesting that it is a component of the telomere protein complex in trypanosomes (Figure 2A). In an additional verification of their specificity, the anti-TelAP1 antibody did not label TelAP1 knockout cells (Figure 2B). Similar results were obtained in PCF cells (Supplementary Figure S2).
Figure 2.
TelAP1 co-localizes with TbTRF in the nucleus of BSF cells. (A) Indirect immunofluorescence analysis (IFA) of BSF cells in different cell cycle stages as indicated using monoclonal antibodies specific for TelAP1 (red) and TbTRF (green). (B) Indirect IFA of ΔTelAP1 BSF cells confirmed the specificity of the TelAP1 signal. DNA was stained with Hoechst (blue). Scale bar 2 μm. S/G1 (synthesis/Gap1 phase), G2/M (Gap2 phase/mitosis), C (cytokinesis).
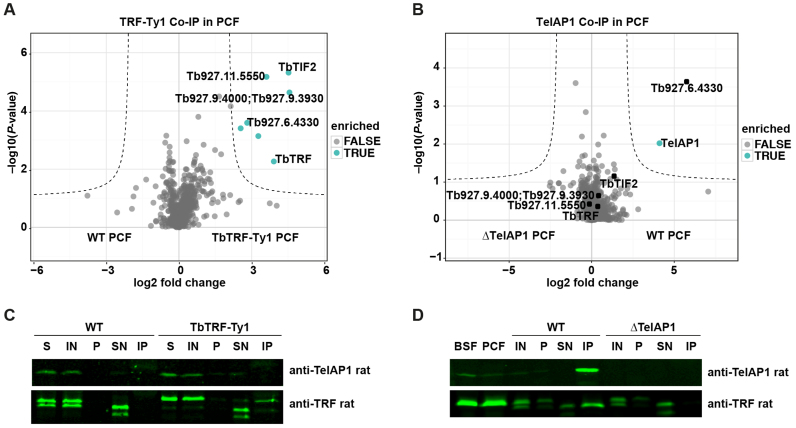
Additionally, we performed IP-MS using the TelAP1-specific mouse antibody and whole cell lysates of BSF cells with a TelAP1 knockout cell line (ΔTelAP1) serving as control. Eight proteins were significantly enriched (P < 0.01) in the TelAP1 Co-IP including the bona fide telomere-binding proteins TbTRF, TbTIF2, TbRAP1 and Tb927.6.4330 (Figure 3A). The whole dataset of the TelAP1 Co-IP is summarized in Supplementary Table S3. The interaction of TelAP1 with TbTRF was further corroborated by IP and western blot (Figure 3B). Additionally, we tested if TelAP1 can bind directly to telomeric repeats (Supplementary Figure S3). An in vitro pull-down assay with immobilized TTAGGG-containing oligonucleotides and purified recombinant TelAP1 revealed that TelAP1 does not bind directly to telomeric DNA. Overall, our data demonstrate that TelAP1 is a component of the telomere complex interacting with TbTRF, TbTIF2, TbRAP1 and Tb927.6.4330 and co-localizes with TbTRF in T. brucei BSF and PCF cells.
Figure 3.
TelAP1 is a component of the telomere complex in Trypanosoma brucei. (A) Volcano plot showing interaction partners of TelAP1. Co-IP was performed in four independent experiments with WT and ΔTelAP1 cells. Precipitates were analyzed by MS. The x-axis shows the log2 fold change of detected proteins between WT and ΔTelAP1 cells. The y-axis represents the P-value. (B) Western blot analysis of TelAP1 Co-IP confirmed interaction of TelAP1 and TbTRF. Twenty-fold more of the pellet and IP samples were loaded compared to IN and SN samples. About 13% of TbTRF input was co-precipitated with TelAP1. W (whole cell lysate), IN (input), P (pellet), SN (supernatant), IP (immunoprecipitate).
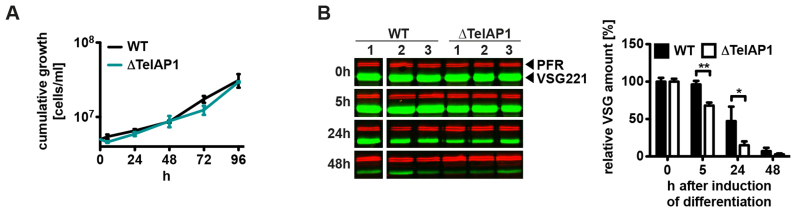
TelAP1 is not essential for procyclic and bloodstream form growth
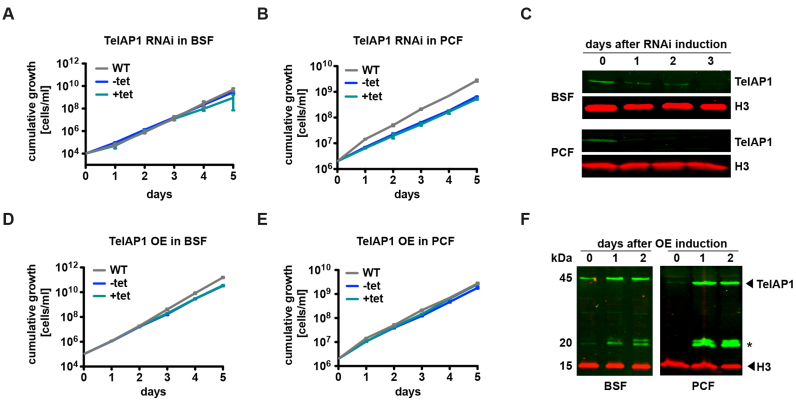
We next tested whether TelAP1 is essential for cell growth. Inducible TelAP1 RNAi knockdown cell lines were generated in both life cycle stages (Figure 4A and B). RNAi-mediated downregulation of TelAP1 expression was confirmed by western blot using the rat monoclonal anti-TelAP1 antibody (Figure 4C). TelAP1 protein was undetectable in BSF cells 3 days after RNAi induction, while in PCF cells it was not detectable already after day 1. TelAP1 depletion did not affect the viability of BSF nor of PCF cells. As the TelAP1 knockdown results suggested that the protein is not essential for cell viability, we generated knockout mutants (ΔTelAP1) in both life cycle stages (Supplementary Figure S4). Replacement of both TelAP1 alleles with drug selection markers was validated by PCR amplification of the TelAP1 locus from genomic DNA from each cell line (Supplementary Figure S4A–D). Growth was mildly slowed in the BSF null mutant compared to WT, with population doubling times of 7.0 and 6.4 h, respectively (Supplementary Figure S4E). TelAP1 deletion in PCF cells showed no effect on cell viability (Supplementary Figure S4F). Next, we analyzed the impact of inducible TelAP1 overexpression on cell viability in BSF and PCF cells (Figure 4D and E). Increase of TelAP1 protein expression was confirmed by western blot using the rat TelAP1-specific antibody. Ectopic TelAP1 overexpression led to 2- to 8-fold increase of TelAP1 in BSF cells compared to WT levels, and to 20- to 40-fold increase of TelAP1 in PCF cells (Figure 4F, quantification not shown). Overexpression of TelAP1 had no effect on the growth rates of either BSF or PCF cells. We concluded that the viability of BSF and PCF cells is independent of TelAP1 protein expression levels. This is in stark contrast to previously characterized telomeric proteins that are all essential for cell growth and indispensable for telomere integrity and genome stability. This suggests that TelAP1 function might be uncoupled from the function of TbTRF, TbTIF2 and TbRAP1.
Figure 4.
Cell viability of both life cycle stages is independent of TelAP1 protein expression levels. (A) Cumulative growth of WT, non-induced (−tet) and induced (+tet) TelAP1 RNAi BSF cell lines. (B) Cumulative growth of TelAP1-depleted PCF cells. (C) Western blot analysis confirmed TelAP1 depletion in BSF and PCF cells. Histone H3 was used as loading control. (D) Cumulative growth of BSF and (E) PCF cells after induction of ectopic TelAP1 overexpression. (F) Western blot analysis of TelAP1 overexpression. Two additional bands (asterisk) are detectable after overexpression. Histone H3 was used as loading control. All growth curves represent the cumulative mean cell number ± SD of three biological replicates of one clone.
TelAP1 protein expression is upregulated in bloodstream form trypanosomes
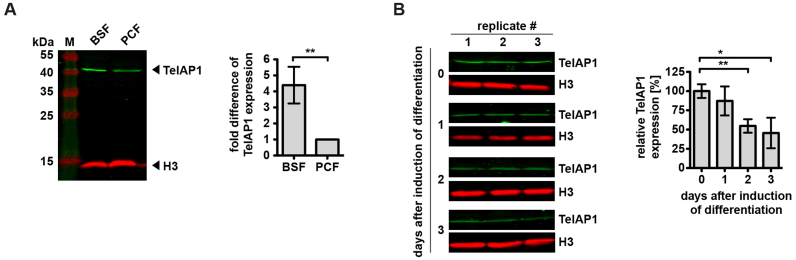
To elucidate the function of TelAP1, we first wanted to validate stage-specific protein expression pattern, which was described recently. A 1.68-fold (55) and a 2.48-fold (56) higher TelAP1 expression was detected in BSF parasites in MS-based comparative proteome studies. To verify this, we compared TelAP1 expression levels between BSF and PCF cells by quantitative western blot (Figure 5A). For western blot analysis whole cell lysates of BSF and PCF were analyzed using the TelAP1-specific antibody. Quantification of the TelAP1 signal confirmed a 4.39-fold upregulation of TelAP1 in BSF cells compared to PCF cells (Figure 5A).
Figure 5.
TelAP1 expression is stage-specifically regulated. (A) Quantitative western blot analysis of TelAP1 expression in BSF and PCF cells revealed a 4-fold upregulation in BSF cells. A representative blot of four independent experiments and their quantification is shown. TelAP1 signal intensity was normalized to histone H3 protein. PCF expression level was set to 1. Error bars represent the standard deviation of four replicates. (B) Western blot analysis and its quantification of TelAP1 expression during differentiation from BSF to PCF. Three independent experiments were analyzed. TelAP1 signal intensity was normalized to Histone H3 protein. Error bars represent the standard deviation of three replicates. Statistical significance was determined by an unpaired t-test *P < 0.05, **P < 0.01.
In addition, TelAP1 expression levels were monitored during the differentiation process from BSF to PCF cells (Figure 5B), as TelAP1 was not detected in the differentiation proteome (57). The quantification of TelAP1 signals revealed that TelAP1 downregulation starts early during the differentiation process. The protein expression level is decreased about 50% already 48 h post induction of differentiation.
Telomere protein complex composition changes in procyclic parasites
Given that TelAP1 is stage-specifically regulated, we asked whether the composition of the telomere complexes might be developmentally regulated as well. To address this question, we investigated the interactions of TelAP1 and TbTRF in PCF parasites by Co-IP coupled to MS (Figure 6). While TelAP1 is most likely an indirect interaction partner of TbTRF in the BSF stage, no interaction of TelAP1 and TbTRF was observed by MS in either the TbTRF or the TelAP1 Co-IPs in PCF cells. Previously identified TelAP1 interactions in the BSF stage are shifted toward the origin of the plot (Figure 6B). Only Tb927.6.4330 was co-purified efficiently with TelAP1 in PCF parasites. In addition, no co-enrichment of TelAP1 and TbTRF was detected by western blot analysis of the TbTRF Co-IP (Figure 6C). In contrast to the MS data (Figure 6A and B) and the western blot analysis of TbTRF Co-IP, analysis of TelAP1 Co-IP by western blot showed a weak interaction of TbTRF with TelAP1 (Figure 6D). The protein interactions of TbTRF and TelAP1 in different stages described above are summarized in a graph showing relative LFQ intensities of enriched proteins normalized to TbTRF and TelAP1 (Supplementary Figure S5).
Figure 6.
TbTRF and TelAP1 Co-IPs in procyclic cells. Co-IPs were performed in four biological replicates and enriched proteins were analyzed by MS. (A) Volcano plot representing TbTRF interactions in PCF cells. Six proteins were significantly enriched with TbTRF-Ty1 including four proteins, which were also found in the BSF TbTRF-Ty1 Co-IP: TbTIF2, Tb927.11.5550, Tb927.9.4000/3930, Tb927.6.4330. The whole dataset is summarized in Supplementary Table S4. (B) Volcano plot showing TelAP1 interacting proteins in PCF cells. Besides TelAP1 the telomere-associated candidate Tb927.6.4330 was enriched. (C) Western blot analysis of TbTRF-Ty1 Co-IP with anti-TelAP1 and anti-TbTRF antibodies. TbTRF was proteolytically degraded during the IP experiment as additional shorter bands appeared, which were detected by the monoclonal TbTRF antibody. Nevertheless, Ty1 epitope-tagged TbTRF was precipitated. TbTRF-Ty1 migrates slower in the SDS-PAGE than the WT TbTRF. TelAP1 could not be detected in the eluate. Twenty-fold more of the IP sample was loaded compared to IN and SN samples. (D) Western blot analysis of TelAP1 Co-IP with anti-TbTRF antibody confirmed interaction of TelAP1 with TbTRF in PCF stage. Again, TbTRF showed signs of proteolytic degradation after cell lysis. A smaller TbTRF fragment was co-purified with TelAP1. BSF and PCF whole cell lysates served as control and showed only one TbTRF product. Twenty-fold more of the pellet and IP sample were loaded compared to IN and SN samples. About 5% of TbTRF input was co-purified with TelAP1. S (starting material after lysis), IN (input), P (pellet), SN (supernatant), IP (immunoprecipitate).
To exclude that the reduced expression of TelAP1 in PCF stage influences the recovery of interacting proteins after TelAP1 Co-IP, we ectopically overexpressed TelAP1 in PCF to a protein expression level comparable with BSF parasites (Supplementary Figure S6A) and repeated the TelAP1 Co-IP. However, TelAP1 overexpression in PCF cells did not lead to an efficient recovery of the BSF telomere complex (Supplementary Figure S6B). Only TbTIF2 of the BSF telomere complex could be significantly co-enriched additionally to Tb927.6.4330 suggesting that assembly of theTelAP1-containing protein complex is regulated by other mechanisms than protein expression.
In summary, Co-IP data suggest that the protein–protein interactions of the telomere complex changes in the PCF stage and that two distinguishable complexes might form in BSF. According to our Co-IP results in PCF stage Tb927.6.4330 seems to be the strongest interaction partner of TelAP1 leading to the hypothesis that this interaction might form a TbTRF-independent complex in PCF parasites. However, TelAP1 is still associated with telomeres in PCF parasites as TelAP1 was found by the telomeric pull-down assay using PCF cells (Figure 1A). Furthermore, two different telomeric complexes could be isolated in BSF. One that consists of TbTRF, TelAP1, Tb927.6.4330, TbTIF2, Tb927.11.5550 and Tb927.9.3930 by TbTRF Co-IP. Another one that consist of TbTRF, TelAP1, Tb927.6.4330, TbTIF2 and additionally TbRAP1 and Tb927.10.2520 (PPL2, (58)) by TelAP1 Co-IP.
Deletion of TelAP1 promotes VSG silencing during differentiation
Although the function of TelAP1 seems to be uncoupled from known telomere complex components, we still expected a role in VSG regulation as this has been shown for all currently known telomeric proteins in trypanosomes. Hence, we analyzed ES silencing of WT and ΔTelAP1 BSF cells during differentiation to PCF cells. The analysis of VSG protein expression provides indirect information about the status of ES transcriptional activity during the differentiation process. Although other mechanisms like mRNA (59) and protein stability (60) influence VSG protein levels during differentiation, the decrease of ES promoter activity also has a direct influence on VSG gene transcription (28). To analyze ES transcriptional activity during early differentiation, WT and ΔTelAP1 BSF cells were differentiated and whole cell lysates prepared after 5, 24 and 48 h (Figure 7B). VSG expression during differentiation was monitored by quantitative western blot using anti-VSG221 and anti-paraflagellar rod (PFR) antibodies. The VSG221 protein amount was normalized against PFR protein levels. The quantification of the relative VSG221 protein amount showed a faster VSG downregulation in ΔTelAP1 BSF cells. The highest difference of VSG expression between WT and ΔTelAP1 cells was observed 24 h after differentiation induction. ΔTelAP1 cells express 3-fold less VSG protein compared to WT cells. This result indicates that TelAP1 influences VSG expression early during differentiation to PCF cells. Notably, this does not affect growth or cell viability during developmental transition from BSF to PCF stage (Figure 7A).
Figure 7.
VSG silencing during differentiation from BSF to PCF is faster in ΔTelAP1 cells. (A) Cumulative growth of WT and ΔTelAP1 cells during differentiation. The graph shows the cumulative mean cell number and standard deviation (n = 3). (B) Western blot and its quantitative analysis of VSG221 expression in WT and ΔTelAP1 cells during differentiation. The experiment was performed in triplicates. VSG221 levels were normalized to PFR protein expression. Time point 0 h was set as 100%. Error bars represent the standard deviation of three biological replicates. Statistical significance was determined by an unpaired t-test *P < 0.05, **P < 0.01.
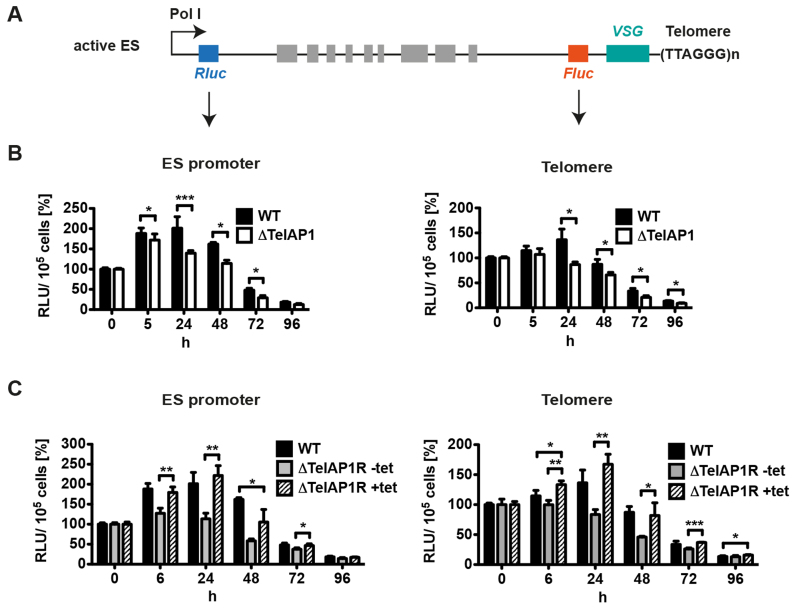
It has been described previously that either ES activity or the untranslated regions (UTRs) of the VSG mRNA can influence VSG protein expression levels (28,59) during differentiation and in different life cycle stages (61,62). To distinguish whether the faster VSG silencing in ΔTelAP1 cells is regulated transcriptionally or translationally, we set up a reporter assay. The active ES of WT and ΔTelAP1 cells was doubly marked with a Renilla luciferase (Rluc) reporter gene downstream of the RNA polymerase I promoter and a Firefly luciferase (Fluc) reporter gene upstream of the VSG221 gene (Figure 8A). The Rluc reporter gene expression is regulated by tubulin UTRs and the Fluc reporter gene by an actin 3′UTR and an aldolase 5′UTR. These UTRs do not influence mRNA stability or translation during differentiation (63,64). To guarantee that the reporter gene constructs integrate in the active ES, antibiotic concentrations for selection were increased about 10-fold compared to standard culture conditions as shown previously (42). These extremely high antibiotic concentrations prohibit survival of cells with construct integration into the inactive ES because the transcriptional activity of these sites is very low compared to an active ES (65). Using two different luciferase reporter genes enabled us to monitor simultaneously ES silencing kinetics during differentiation at the ES promoter and at the telomere. WT and ΔTelAP1 reporter cells were differentiated and the luciferase activity measured at different time points post induction (Figure 8B). WT and ΔTelAP1 reporter cells start with comparable absolute values (data not shown) indicating that TelAP1 deletion has no effect on the transcriptional activity of the active ES in BSF cells. In the first 24 h WT cells showed an increase of luciferase activity at both regions, downstream of the Pol I promoter and adjacent to the telomere. The increase was stronger at the ES promoter. Forty-eight hours post-induction the luciferase activity dropped at both regions. The decrease of Fluc activity was stronger compared to the Rluc activity, indicating that ES silencing happens earlier at the telomere. The decrease of both luciferase activities started earlier in ΔTelAP1 reporter cells compared to WT cells. The most significant difference (P < 0.001) is observed at 24 h upon differentiation induction (1.5-fold difference). These data strongly suggest that ES silencing kinetics are faster in ΔTelAP1 reporter cells compared to WT reporter cells and thus confirm the influence of TelAP1 on ES transcriptional activity early during differentiation.
Figure 8.
Dual luciferase reporter reveals faster silencing kinetics during differentiation in ΔTelAP1 cells. (A) Illustration of the dual luciferase reporter. The active ES of WT and ΔTelAP1 cells was doubly marked with an Rluc reporter gene downstream of the ES promoter and an Fluc reporter gene upstream of VSG gene. Graphic is not to scale. (B) Analysis of luciferase activity at the ES promoter (Pol I) and at the telomere during differentiation of WT and ΔTelAP1 reporter cell lines. Differentiation of reporter cell lines (n = 3) was induced and luciferase activity was measured at the time points indicated. Luciferase activity is shown as relative light units (RLU) and standard deviation. Time point 0 h was set as 100%. (C) Reintroduction of TelAP1 in ΔTelAP1 reporter cell line slows down ES silencing kinetics during differentiation. Analysis of luciferase activity at the ES promoter and at the telomere during differentiation of ΔTelAP1R reporter cells with and without tetracycline induction. One clone was analyzed in triplicates. WT values of the experiment shown in B were included into the graphs for better comparison. Reintroduction of TelAP1 leads to ES silencing kinetics similar to WT cells. Statistical significance was determined by an unpaired t-test *P < 0.05, **P < 0.01, ***P < 0.001.
To confirm that the observed effect on the ES transcriptional activity during differentiation was TelAP1-specific, the protein was reintroduced into ΔTelAP1 reporter cells (ΔTelAP1R) (Figure 8C). The ectopic expression resulted in a 2-fold higher level compared to WT cells after full induction of the system (Supplementary Figure S7A). Twenty-four hours after induction ΔTelAP1R reporter cells were differentiated to PCF cells and the luciferase activity was compared with uninduced cells. The luciferase assays revealed higher values in induced ΔTelAP1R reporter cells at the ES promoter and at the telomere compared to uninduced ΔTelAP1R cells. When luciferase activities of the rescue cell line were compared to WT, induced ΔTelAP1R reporter cells showed identical kinetics of ES silencing during differentiation indicating that only deletion of TelAP1 was responsible for the transcriptional effect observed in the ΔTelAP1 mutants during differentiation.
Our data strongly indicate that TelAP1 is a stage-specific telomere-associated protein and a regulator of ES silencing during early differentiation events in T. brucei. This is the first evidence that a telomere-associated protein influences transcriptional activity of the ES during developmental differentiation of African trypanosomes. Furthermore, TelAP1 function illustrates that developmental ES silencing is a fine-tuned process, which involves stage-specific changes in telomere complex composition.
DISCUSSION
Telomeres are essential structural components that mediate transcriptional control of VSG genes in T. brucei. Compared to mammals and yeast, where telomeres are well studied, less is known about the composition of telomeres in trypanosomes. Previous studies described three homologs of the mammalian shelterin complex components in trypanosomes (TbTRF, TbTIF2 and TbRAP1) and demonstrated a link between telomere biology and antigenic variation (6–8,20–22). However, how these proteins influence the transcriptional control of VSG genes exactly remains unknown. In addition, less is known about telomeres and their role in developmental differentiation. To understand the contribution of telomere-binding proteins to antigenic variation and developmental silencing it is therefore essential to identify all telomere complex components.
We performed a DNA pull-down assay with telomeric DNA as bait and found 17 interaction partners including the already known direct and indirect telomeric proteins TbTRF (6) and TbTIF2 (8), respectively. The recently identified telomere-associated factor Tb927.6.4330 was also found by our approach (53). The identification of previously known telomere-binding proteins validates that our screening method is suitable to identify new telomere-binding proteins and their interaction partner. However, TbRAP1 was not found by our method. Like TbTIF2, TbRAP1 does not bind telomeric DNA directly (7). It associates with telomeres through its interaction with TbTRF. Co-IP experiments carried out by Yang et al. demonstrated a weak interaction between TbRAP1 and TbTRF (7). Only 3–14% of endogenously FLAG-HA-HA-tagged TbRAP1 were co-precipitated with TbTRF. Yang et al. also performed chromatin immunoprecipitation experiments to validate TbRAP1 as a telomeric protein. Using a TbRAP1-specific antibody and formaldehyde cross-linked material they revealed an association of TbRAP1 with telomeric DNA. Samples in which the proteins were not cross-linked to DNA showed no significant enrichment of telomeric DNA with TbRAP1, indicating a weak interaction of TbRAP1 with TbTRF. This might be the reason why TbRAP1 was not detected in our approach. Nevertheless, the TbTRF Co-IP partially verified the telomeric pull-down assay results because all five TbTRF-interacting proteins were also enriched, indicating that these five candidates are bona fide telomere-associated proteins.
PrimPol-like protein 2 (PPL2) (58) and TbRAP1 were identified in the TelAP1 Co-IP, but not in the TbTRF Co-IP, which might suggest that a separate TelAP1-PPL2/TbRAP1 complex is present in BSF cells. Tb927.6.4330 might also be a component of this complex because the TelAP1 Co-IP in PCF cells indicates direct interaction of TelAP1 with Tb927.6.4330. The second and larger telomere complex might be composed of TbTRF, TbRAP1, TbTIF2, Tb927.11.5550, Tb927.6.4330, Tb927.9.4000/3930 and TelAP1 as all these proteins were found in the TbTRF and TbTelAP1 BSF Co-IPs. Further hints that different complexes exist in trypanosomes are provided by comparison of TbTRF and TelAP1 Co-IPs in PCF cells. These data show that a TbTRF complex and a TelAP1 complex are present at telomeres. However, in PCF cells, TelAP1 interacts only with Tb927.6.4330 and is missing in the TbTRF complex as shown by TRF Co-IP.
Strikingly, the work on TelAP1 revealed that the telomere complex composition is stage-specifically regulated. The TelAP1 data are to our knowledge the first evidence for dynamic changes of telomere complexes during differentiation from BSF to PCF cells. The analyses of the interaction partners of TelAP1 and TbTRF in BSF and PCF cells lead us not only to the conclusion that different telomere complexes might be present at telomeres but also that the composition of the telomere complexes may differ between the life cycle stages of the parasite. In the PCF, TelAP1 interacts only with Tb927.6.4330 and is missing in the TbTRF complex. The interaction of TbTRF with TbTIF2, Tb927.94000/3930, Tb927.6.4330, Tb927.115550 does not change in PCF. It could be that TelAP1 dissociates from the telomere complex containing TbTRF and is degraded during differentiation to PCF as our data indicate that TelAP1 might be a part of TbTRF complex and additionally assembles an own complex in the BSF with PPL2. Expression levels of TbTRF and TbRAP1 only change mildly during differentiation compared to TelAP1. According to a recently published differentiation proteome (55), TbRAP1 is 1.26-fold upregulated in PCF cells and TbTRF shows a 1.23-fold upregulation in the BSF stage. No information is available for TbTIF2 in this report. To our knowledge TelAP1 is the first telomere-associated protein, which is highly stage-specifically regulated in trypanosomes.
What might be the reason for different telomere complexes in the BSF and PCF cells? It has been described already that chromatin structure and nuclear architecture are different between BSF and PCF cells, indicating that chromatin restructuring takes place during developmental transition (66). Several reports support the concept of developmentally regulated telomere structure in trypanosomes (67). First, telomeric DNA modifications differ between BSF and PCF parasites. A modified nucleotide β-D-glucosyl-hydroxymethyluracil (base J) replaces a part of the thymidines in the telomeric DNA of BSF cells but not of PCF cells (68,69). Although Jehi et al. have reported that TbTRF binding to telomeric DNA is not affected by base J (21), it is still unknown if base J influences the binding affinity of other telomere-binding proteins. Second, telomeric silencing of reporter constructs differs between the BSF and PCF stage. In BSF cells transcription from three different promoters inserted in an inactive ES near telomeres was silenced (70). In PCF only the ES promoter kept its repressed status indicating a developmentally regulated silencing effect of telomeres. Finally, the chromatin accessibility of inactive ESs in the BSF alters upon the differentiation to PCF stage (27). There, the ES chromatin becomes inaccessible suggesting that chromatin remodeling is developmentally regulated as well. We hypothesize that this is facilitated by distinguishable composition or regulation of telomeric complexes in BSF and PCF stages.
In this study, we focused on the characterization of TelAP1 as this protein was found in the telomeric pull-down assay as well as in the BSF TbTRF Co-IP. Interestingly, functional analyses revealed that TelAP1 is not essential for cell viability in BSF and PCF cells. This might be another hint that the function of TelAP1 is uncoupled from the function of the known telomere-binding proteins TbTRF, TbTIF2 and TbRAP1, which are all essential for cell viability (6,7). The presence of a TbTRF-independent TelAP1 complex supports this hypothesis. Furthermore, we used quantitative proteomics to analyze changes of VSG expression pattern in ΔTelAP1 cells. We did not detect derepression of silent VSG genes or enhanced switching rates (data not shown). This is in contrast to the phenotype in Tb927.6.4330-depleted cells in which derepression of a silent ES VSG could be observed (53). This hints to a function of TelAP1, which is distinguishable from Tb927.6.4330-mediated processes. However, since TelAP1 is not able to bind telomeric repeats directly, it might need Tb927.6.4330 to be recruited to telomeres where it transiently antagonizes differentiation-dependent silencing of the ES.
Here, we provide the first direct evidence for a telomere-binding protein playing a role in the regulation of developmental silencing of the VSG ES. Our characterization of TelAP1 revealed two novel aspects of telomere biology of trypanosomes. First, the telomere complex composition is dynamic and changes during developmental transition of BSF to PCF cells. Second, telomere-associated proteins regulate the kinetics of developmental ES silencing. Our current hypothesis is that TelAP1 transiently maintains an open chromatin status or an active ES promoter early during differentiation to coordinate transcriptional silencing of the ES with other cellular processes or environmental cues.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elisabeth Kremmer for providing antibodies. We thank the Biochemistry Core Facility at the MPI of Biochemistry for help with recombinant protein expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Rhineland Palatinate Forschungsschwerpunkt GeneRED; Deutsche Forschungsgemeinschaft (DFG) [Bu2996/1]. Funding for open access charge: DFG [Bu2996/1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Blackburn E.H. Structure and function of telomeres. Nature. 1991; 350:569–573. [DOI] [PubMed] [Google Scholar]
- 2. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 3. Hockemeyer D., Collins K.. Control of telomerase action at human telomeres. Nat. Struct. Mol. Biol. 2015; 22:848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kappei D., Butter F., Benda C., Scheibe M., Draskovic I., Stevense M., Novo C.L., Basquin C., Araki M., Araki K. et al. HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J. 2013; 32:1681–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armstrong C.A., Tomita K.. Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol. 2017; 7:160338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li B., Espinal A., Cross G.A.. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol. Cell. Biol. 2005; 25:5011–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B.. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009; 137:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jehi S.E., Wu F., Li B.. Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res. 2014; 24:870–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crabbe L., Cesare A.J., Kasuboski J.M., Fitzpatrick J.A., Karlseder J.. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012; 2:1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taddei A., Gasser S.M.. Structure and function in the budding yeast nucleus. Genetics. 2012; 192:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tham W.H., Zakian V.A.. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002; 21:512–521. [DOI] [PubMed] [Google Scholar]
- 12. Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007; 8:299–309. [DOI] [PubMed] [Google Scholar]
- 13. Moretti P., Shore D.. Multiple interactions in sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 2001; 21:8082–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008; 452:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merrick C.J., Duraisingh M.T.. Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 2006; 62:612–620. [DOI] [PubMed] [Google Scholar]
- 16. Horn D. Antigenic variation in African trypanosomes. Mol. Biochem. Parasitol. 2014; 195:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I. et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008; 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Navarro M., Gull K.. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001; 414:759–763. [DOI] [PubMed] [Google Scholar]
- 19. Horn D., McCulloch R.. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 2010; 13:700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jehi S.E., Nanavaty V., Li B.. Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One. 2016; 11:e0156746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jehi S.E., Li X., Sandhu R., Ye F., Benmerzouga I., Zhang M., Zhao Y., Li B.. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res. 2014; 42:12899–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pandya U.M., Sandhu R., Li B.. Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res. 2013; 41:7673–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alsford S., Kawahara T., Isamah C., Horn D.. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol. Microbiol. 2007; 63:724–736. [DOI] [PubMed] [Google Scholar]
- 24. Roditi I., Schwarz H., Pearson T.W., Beecroft R.P., Liu M.K., Williams R.O., Overath P.. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol. 1989; 108:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amiguet-Vercher A., Perez-Morga D., Pays A., Poelvoorde P., Van Xong H., Tebabi P., Vanhamme L., Pays E.. Loss of the mono-allelic control of the VSG expression sites during the development of Trypanosoma brucei in the bloodstream. Mol. Microbiol. 2004; 51:1577–1588. [DOI] [PubMed] [Google Scholar]
- 26. Schlimme W., Burri M., Bender K., Betschart B., Hecker H.. Trypanosoma brucei brucei: differences in the nuclear chromatin of bloodstream forms and procyclic culture forms. Parasitology. 1993; 107:237–247. [DOI] [PubMed] [Google Scholar]
- 27. Navarro M., Cross G.A.M., Wirtz E.. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 1999; 18:2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landeira D., Navarro M.. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J. Cell Biol. 2007; 176:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Batram C., Jones N.G., Janzen C.J., Markert S.M., Engstler M.. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife. 2014; 3:e02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aresta-Branco F., Pimenta S., Figueiredo L.M.. A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei. Nucleic Acids Res. 2016; 44:3131–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schulz D., Mugnier M.R., Paulsen E.M., Kim H.S., Chung C.W.W., Tough D.F., Rioja I., Prinjha R.K., Papavasiliou F.N., Debler E.W.. Bromodomain proteins contribute to maintenance of bloodstream form stage identity in the African trypanosome. PLoS Biol. 2015; 13:e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirumi H., Hirumi K.. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989; 75:985–989. [PubMed] [Google Scholar]
- 33. Wirtz E., Leal S., Ochatt C., Cross G.A.M.. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 34. Alsford S., Horn D.. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 2008; 161:76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brun R., Schonenberger M.. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979; 36:289–292. [PubMed] [Google Scholar]
- 36. Burkard G., Fragoso C.M., Roditi I.. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007; 153:220–223. [DOI] [PubMed] [Google Scholar]
- 37. Kalidas S., Li Q., Phillips M.A.. A Gateway(R) compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 2011; 178:51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim S., Kim C.H.. Evaluation of the Cre-loxP system for construction of auxotrophic strains in Kluyveromyces marxianus. Yeast. 2013; 30:133–133. [Google Scholar]
- 39. Oberholzer M., Morand S., Kunz S., Seebeck T.. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 2006; 145:117–120. [DOI] [PubMed] [Google Scholar]
- 40. Jones N.G., Thomas E.B., Brown E., Dickens N.J., Hammarton T.C., Mottram J.C.. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS Pathog. 2014; 10:e1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janzen C.J., van Deursen F., Shi H., Cross G.A., Matthews K.R., Ullu E.. Expression site silencing and life-cycle progression appear normal in Argonaute1-deficient Trypanosoma brucei. Mol. Biochem. Parasitol. 2006; 149:102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Figueiredo L.M., Janzen C.J., Cross G.A.. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008; 6:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Casas-Vila N., Scheibe M., Freiwald A., Kappei D., Butter F.. Identification of TTAGGG-binding proteins in Neurospora crassa, a fungus with vertebrate-like telomere repeats. BMC Genomics. 2015; 16:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 45. Rappsilber J., Mann M., Ishihama Y.. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007; 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- 46. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 47. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scholz J., Besir H., Strasser C., Suppmann S.. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 2013; 13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gassen A., Brechtefeld D., Schandry N., Arteaga-Salas J.M., Israel L., Imhof A., Janzen C.J.. DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosoma brucei. Nucleic Acids Res. 2012; 40:10302–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bastin P., Bagherzadeh A., Matthews K.R., Gull K.. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996; 77:235–239. [DOI] [PubMed] [Google Scholar]
- 51. Overath P., Czichos J., Haas C.. The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur. J. Biochem. 1986; 160:175–182. [DOI] [PubMed] [Google Scholar]
- 52. Adl S.M., Simpson A.G., Lane C.E., Lukes J., Bass D., Bowser S.S., Brown M.W., Burki F., Dunthorn M., Hampl V. et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012; 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Glover L., Hutchinson S., Alsford S., Horn D.. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goos C., Dejung M., Janzen C.J., Butter F., Kramer S.. The nuclear proteome of Trypanosoma brucei. PLoS One. 2017; 12:e0181884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Butter F., Bucerius F., Michel M., Cicova Z., Mann M., Janzen C.J.. Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite's host adaptation machinery. Mol. Cell. Proteomics. 2013; 12:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Urbaniak M.D., Guther M.L., Ferguson M.A.. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One. 2012; 7:e36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dejung M., Subota I., Bucerius F., Dindar G., Freiwald A., Engstler M., Boshart M., Butter F., Janzen C.J.. Quantitative proteomics uncovers novel factors involved in developmental differentiation of Trypanosoma brucei. PLoS Pathog. 2016; 12:e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rudd S.G., Glover L., Jozwiakowski S.K., Horn D., Doherty A.J.. PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol. Cell. 2013; 52:554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berberof M., Vanhamme L., Tebabi P., Pays A., Jefferies D., Welburn S., Pays E.. The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. EMBO J. 1995; 14:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ziegelbauer K., Stahl B., Karas M., Stierhof Y.D., Overath P.. Proteolytic release of cell surface proteins during differentiation of Trypanosoma brucei. Biochemistry. 1993; 32:3737–3742. [DOI] [PubMed] [Google Scholar]
- 61. Siegel T.N., Hekstra D.R., Wang X., Dewell S., Cross G.A.. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010; 38:4946–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clayton C. The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog. 2013; 9:e1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garcia-Salcedo J.A., Perez-Morga D., Gijon P., Dilbeck V., Pays E., Nolan D.P.. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004; 23:780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hug M., Carruthers V.B., Hartmann C., Sherman D.S., Cross G.A.M., Clayton C.. A possible role for the 3′-untranslated region in developmental regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 1993; 61:87–96. [DOI] [PubMed] [Google Scholar]
- 65. Kassem A., Pays E., Vanhamme L.. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8943–8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Navarro M., Penate X., Landeira D.. Nuclear architecture underlying gene expression in Trypanosoma brucei. Trends Microbiol. 2007; 15:263–270. [DOI] [PubMed] [Google Scholar]
- 67. Alsford S., duBois K., Horn D., Field M.C.. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev. Mol. Med. 2012; 14:e13. [DOI] [PubMed] [Google Scholar]
- 68. van Leeuwen F., Wijsman E.R., Kuyl-Yeheskiely E., van der Marel G.A., van Boom J.H., Borst P.. The telomeric GGGTAA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996; 24:2476–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gommers-Ampt J., Lutgerink J., Borst P.. A novel DNA nucleotide in Trypanosoma brucei only present in the mammalian phase of the life-cycle. Nucleic Acids Res. 1991; 19:1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Horn D., Cross G.A.M.. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995; 83:555–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.