Abstract
During mild replication stress provoked by low dose aphidicolin (APH) treatment, the key Fanconi anemia protein FANCD2 accumulates on common fragile sites, observed as sister foci, and protects genome stability. To gain further insights into FANCD2 function and its regulatory mechanisms, we examined the genome-wide chromatin localization of FANCD2 in this setting by ChIP-seq analysis. We found that FANCD2 mostly accumulates in the central regions of a set of large transcribed genes that were extensively overlapped with known CFS. Consistent with previous studies, we found that this FANCD2 retention is R-loop-dependent. However, FANCD2 monoubiquitination and RPA foci formation were still induced in cells depleted of R-loops. Interestingly, we detected increased Proximal Ligation Assay dots between FANCD2 and R-loops following APH treatment, which was suppressed by transcriptional inhibition. Collectively, our data suggested that R-loops are required to retain FANCD2 in chromatin at the middle intronic region of large genes, while the replication stress-induced upstream events leading to the FA pathway activation are not triggered by R-loops.
INTRODUCTION
Fanconi anemia (FA) is a rare hereditary disorder characterized by progressive bone marrow failure, congenital anomalies, cancer predisposition, and chromosome instability. FA proteins, which are encoded by at least 22 genes (FANCA-W), together play a key role in the DNA interstrand cross link (ICL) repair pathway (1–4). ICLs can arise endogenously by aldehydes produced as metabolic byproducts, and this may contribute to genome instability, leading to impaired function of hematopoietic stem cells and subsequent leukemogenesis (5). Eight of the FA proteins and other associated proteins form the FA core E3 ligase complex, which monoubiquitinates the FANCD2-FANCI (D2-I) complex during the DNA damage response (DDR). This monoubiquitination event, which is triggered by ATR-ATRIP phosphorylation of FANCI (6,7), leads to relocalization of the D2-I complex to damaged chromatin. The monoubiquitinated D2-I proteins assemble foci at the DNA damage sites in chromatin to facilitate DNA repair, serving to protect the genome. As a master regulator of ICL repair, FANCD2 protein coordinates ICL incision (‘unhooking’ by SLX4-XPF or FAN1 nuclease), which leads to DNA double-strand break generation and repair by homologous recombination (HR) (1,3). In addition, FANCD2 and FA pathway components including BRCA2 and RAD51 also function in the protection of stalled replication forks by preventing Mre11-mediated fork degradation (8–10).
Treatment with low-dose aphidicolin (APH), a DNA polymerase α/δ/ϵ inhibitor, induces mild replication stress, which is accompanied by ATR activation but the absence of downstream Chk1 kinase activation (11); this mild replication stress results in breaks and gaps in a set of chromosomal locations, termed common fragile sites (CFSs; e.g. FRA3B, FRA16D, etc.) (12–14) Although CFSs were originally identified in lymphocytes from normal individuals, it has more recently been shown that the genomic deletions and rearrangements in cancer often coincide with CFSs, suggesting that CFS instability is caused by oncogene-induced replication stress (during oncogenesis) (12–14). The presence of oncogenes or tumor suppressors inside or near CFS loci is also notable (15,16). CFS-containing genes span huge regions with a relative paucity of replication origins. Therefore, during replication stress, in which forks are stalled or moving slower, they must progress from flanking regions and onward through CFSs to complete replication (13). These huge genes, if actively transcribed, need to continue transcription during S phase because of the extent of the gene and the slower speed of RNA polymerase II (Pol II) progression. These properties lead to inevitable collisions of transcription and replication machineries that generate incompletely replicated CFS regions, exacerbating replication stress and genome instability (17). The CFS fragility is expressed when SLX4-MUS81-EME1 mediates incision of left-over replication intermediates in early M phase, which in turn leads to mitotic DNA synthesis (MiDAS) in a manner dependent on RAD52, POLD3, and RecQ5, but not on FANCD2 or RAD51 (18–20). If MiDAS does not occur, the under-replicated CFS regions form ultra-fine chromosomal bridges (UFB) (between sister chromatids) coated by BLM or PICH helicase during M phase (21,22). UFBs are converted to 53BP1 bodies in the next G1 phase, or result in chromosomal instability, which can be recognized as micronuclei.
Interestingly, FANCD2 accumulates at CFSs during replication stress caused by APH treatment, forming sister foci at the roots of ultrafine bridges (UFB) in early M phase (21). FANCD2 contributes to genome stability at the CFSs by suppressing CFS expression (23); however, recruitment of MUS81-EME1 nuclease, which cleaves the replication intermediate or stalled fork at the CFS, is independent of the FA pathway (24). Recently, it was reported that FANCD2 is required for efficient genome replication, in particular that of CFSs (25), by resolving R-loops caused by transcription-replication conflicts (25–27). R-loops, which consist of DNA:RNA hybrids and displaced single-stranded DNA, are physiologically relevant in the genome and associate with immunoglobulin class switching, replication of mitochondrial DNA as well as transcriptional promoters or terminators (28). Removal of DNA:RNA hybrids alleviates replication perturbation caused by defects in FANCD2 (25–27). FANCD2 supports replication in a manner dependent on its monoubiquitination by the FA core complex (26,27), or independent of the other upstream FA proteins (25). In any case, untimely formation of R-loops is a major threat to genome instability (29), possibly in a manner dependent on specific histone modification (30). Nonetheless, mechanistically how FANCD2 contributes to CFS stability and replication stress response in terms of R-loop formation remains unclear.
In this study, we performed chromatin immunoprecipitation (ChIP)-sequencing (ChIP-seq) analysis, and we provide a genome-wide landscape of replication stress response involving FANCD2 in U2OS cells. Our results revealed that FANCD2 accumulation mostly occurs in the central portion of large transcribed genes during mild replication stress. This accumulation appeared to be dependent on R-loop formation and therefore likely depended on transcription-replication collision. In addition, we also found that the upstream events leading to the FA pathway activation are not triggered by R-loops.
MATERIALS AND METHODS
Cell culture and transfection
U2OS cells were cultured in DMEM (Nacalai Tesque, high glucose) supplemented with 10% fetal calf serum (Gibco). For plasmid transfections, Lipofectamine 3000 (Invitrogen) was used according to the manufacturer's instructions. To generate replication stress, cells were treated with APH at 0.4 μM for 24 h, unless stated otherwise.
Antibodies and reagents
The following antibodies were obtained from commercial sources: anti-DNA:RNA (S9.6, Kerafast); anti-DDDDK-tag (MBL); anti-FLAG (M2, Sigma-Aldrich); anti-FLAG M2 magnetic beads (Sigma); normal mouse IgG (Santa Cruz); anti-FANCA (Bethyl); anti-FANCD2 (Novus); anti-PCNA (PC10, Santa Cruz); anti-γH2AX (JBW301, Millipore); anti-RPA (9H8, Abcam); anti-a-tubulin (T5168, Sigma). Aphidicolin (Wako), mitomycin C (MMC) (Kyowa Hakko Kirin), or cordycepin (Wako) were used at the indicated concentrations. Primers and siRNA oligos are summarized in Supplemental Table S1.
Plasmids
The cDNA encoding human RNaseHI (without mitochondrial localization signal) was amplified from HeLa cell cDNA and cloned into pEGFP-C1. The Hybrid binding (HBD) domain of RNaseHI was then amplified using this plasmid and subcloned into pENTR entry vector (Invitrogen). The inserts in these entry vectors were transferred to the expression vectors or lentiviral constructs by LR Clonase II (Invitrogen). pcDNA3.1-Cre was generated by cloning Cre cDNA from pBS185 (Invitrogen) into pcDNA3.1 (Invitrogen).
Generation of lentivirus and transduction
To produce a U2OS cell line expressing GFP-HBD under tetracycline-controlled transcriptional activation, cDNA in the entry vector pENTR was transferred to a puromycin resistant derivative (a gift of Dr. Makoto Nakanishi) of lentiviral plasmid CSIV-TRE-RfA-UbC-KT (a gift from Dr Hiroyuki Miyoshi) (1) using LR Clonase II (Invitrogen). U2OS cells were infected with the respective lentivirus and selected with puromycin (2 μg/ml). Cells were induced by treatment with 2 μg/ml doxycycline for 24 h. GFP expression was confirmed with a FACSCalibur flow cytometer (BectonDickinson). To produce a U2OS cell line expressing FLAG-FANCI, cDNAs were transferred to lentiviral plasmid CSII-CMV-MCS-IRES-Bsd (a gift from Dr Hiroyuki Miyoshi) rendered Gateway system-compatible (Invitrogen). Following lentivirus infection, Blasticidin S (5 μg/ml) resistant populations were selected and expanded.
siRNA transfections
All siRNA duplexes used in this study were purchased from Invitrogen or Sigma. Transfection and co-transfection was carried out using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cells were harvested 72 h after transfection. The individual siRNA duplexes used were: siFANCD2 (5′CAGAGUUUGCUUCACUCUCUATT-3′) (Invitrogen) (4) siFANCA (5′AAGGGUCAAGAGGGAAAAAUA-3′) (Invitrogen) (5); Luciferase Control (Ctrl) (5′UCGAAGUAUUCCGCGUACGTT-3′) (Invitrogen) (8).
Construction of TALEN and FANCD2-3xFLAG knock-in vectors
The TAL effector nuclease (TALEN) pair targeting the termination codon of the FANCD2 gene was created using the original Golden Gate TALEN and TAL Effector Kit obtained from Addgene (a gift from the Voytas lab) with some modifications (31). To create the FANCD2–3xFLAG knock-in vector, a human ∼1kb genomic fragment containing the FANCD2 termination codon (in exon 44) was amplified by PCR using primers KD12–106 and KD12–109, and cloned into the NotI/SalI sites of pBS plasmid. Using this plasmid as a template, inverse PCR was carried out using primers KD12–107 and KD12–108, resulting in replacement of the termination codon with an EcoRI site. The loxP-mPGK-puro-loxP cassette in pBS derived from OCT4-eGFP-PGK-puro (Addgene #31937, a gift from Rudolf Jaenisch) was fused with 3xFLAG sequence amplified from TAL2255 (Addgene #36698, a kind gift from Keith Joung). This cassette was inserted into the EcoRI site of the knock-in vector, resulting in an in-frame fusion of the coding sequence with 3xFLAG tag in the FANCD2 exon 44 sequence.
Generation of U2OS cells expressing FANCD2-3xFLAG
U2OS (one million) cells were transfected with 12 μg of targeting vector and TALEN plasmids (4 μg each, precipitated and dissolved in PBS) using a Neon electroporator (ThermoFisher) (1200 V, 10 ms, 4 pulses). After overnight culture in 10 cm dishes, cells were seeded into 96-well plates (∼50 cells in 0.1 ml medium per well). The next day, selection was initiated by adding 0.1 ml of complete medium containing puromycin (1 μg/ml). Two weeks later, single colonies were picked up and expanded. Clones with correct knock-in events were identified with genomic PCR using KOD-FX polymerase (TOYOBO). Finally, the puromycin cassette was removed by transient transfection of a Cre expression plasmid (pcDNA3.1-Cre) followed by subcloning. The cells were further verified by genomic PCR and western blotting.
Anti-FLAG Chromatin immunoprecipitation (ChIP) for next generation sequencing
Cells (4 × 107 cells) were treated as indicated (0.4 μM APH or DMSO for 24 h) and fixed for 10 min with 1% paraformaldehyde until quenching with the addition of 1/10 volume of 1.25 M Glycine. Then cells were washed twice with PBS and resuspended in 1 ml of SDS lysis buffer (50 mM Tris–HCl pH 8.0, 10mM EDTA, 20% SDS, protease inhibitor cocktail (Roche), 2 mM PMSF). Lysates were briefly sonicated using a Bioruptor (Cosmo Bio) and centrifuged at 14 000 rpm for 10 min. After part of the supernatant was saved for input, it was diluted with nine volumes of ChIP dilution buffer (50 mM Tris–HCl pH 8.0, 167 mM NaCl, 1.1% Triton X-100, 0.11% NaDeoxycholate (DOC), protease inhibitor cocktail, 2 mM PMSF) and then incubated with Dynabeads protein G (Life Technologies) with anti-FLAG M2 Ab (Sigma), or normal mouse IgG (Santa Cruz) for 20 h at 4°C. Beads were washed once with 1× RIPA buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.1% NaDOC) with 150 mM NaCl, followed by one wash with 1× RIPA buffer with 500 mM NaCl, once with the LiCl wash buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP40, 0.5% NaDOC), and twice with 1× TE (10 mM Tris–HCl pH8.0, 1 mM EDTA), and bound complexes were eluted with 1.4 ml of elution buffer (10 mM Tris–HCl pH 8.0, 5 mM EDTA, 300 mM NaCl, 0.5% SDS). Input samples and eluates were incubated at 65°C for 6 h, treated with 4 mg/ml RNase A at 37°C for 30 min, followed by 0.1 mg/ml Protease K at 55°C for 2.5 h. DNA was recovered using the QIAquick PCR Purification Kit according to manufacturer's protocol (Qiagen).
Next generation sequencing (NGS)
Immunoprecipitated DNA and input DNA were sequenced by Illumina GAIIx (one sample per lane). 75 bp paired-end runs were performed with a TruSeq SBS Kit v5 (Illumina). For the sequencing, DNA libraries were prepared using the NEBNext ChIP-Seq library prep reagent set for Illumina (NEB) and the NEBNext multiplex oligos for Illumina (NEB). Total RNA was separated from U2OS-D2-FLAG cells with or without APH treatment using an RNAeasy kit (Qiagen) and treated with a Ribo-Zero rRNA Removal kit (Illumina). RNA sequencing was done by Macrogen Japan (Kyoto, Japan).
NGS data analysis
For ChIP-seq data, adapter sequences were trimmed using cutadapt (32). Reads with many (>80%) low-quality (score < 20) bases were discarded. Low quality (score < 20) bases were trimmed from both ends of the reads. Reads shorter than 20nt were discarded. The filtered reads were aligned to the reference genome hg19 using Bowtie (33). Peak calling was performed using MACS (34). The detected peaks were annotated using SnpEff (35). For RNA-seq data, the quality control was performed with ShortRead package (36) in R/Bioconductor (https://www.R-project.org/) (37). Low quality (score < 20) bases were trimmed from both ends of the reads. Trimmed reads shorter than 40nt or containing many (>20%) low-quality (score < 20) bases were discarded. Read pairs were retained for the subsequent analysis only if both ends passed. The filtered reads were aligned to the reference genome hg19 using TopHat2 with –no-novel-juncs option (38). The aligned reads were filtered using SAMtools with -f 3 option (39), which means each read was required to be mapped in a proper pair. The mapped reads were counted for each known gene using HTSeq (40).
ChIP-qPCR analysis
For FLAG (FANCD2–3xFLAG), PCNA or γH2AX ChIP, Dynabeads Protein G (Life Technologies) were incubated with anti-FLAG (M2), anti-PCNA (PC10) or anti-γH2AX (JBW301) antibody, respectively. Samples were processed as in ChIP for NGS described above. The plasmid encoding GFP-RNaseHI was transiently transfected into U2OS-D2-FLAG cells using Lifofectamine 3000. GFP-Trap-M (ChromoTek) was used for GFP-HBD immunoprecipitation. Primers used for qPCR were: NRG3 5′ primers (KD15–215 and 216), peak primers (KD15–282 and 283), 3′ primers (KD15–221 and 222); WWOX (KD15–375 and 376); CDH13 (KD15–247 and 248); LRP1B (KD15–385 and 386). Reactions were run using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). All qPCR was performed using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The dilution factor was adjusted and the percentage of the input signal was calculated.
Immunohistochemistry and in situ proximal ligation assay (PLA)
Cells were cultured on 15-mm coverslips and indicated plasmids were transfected. Cells on coverslips were washed with PBS twice and fixed with PBS containing 3% paraformaldehyde, 2% sucrose, 0.5% Triton-X-100, chilled on ice for 30 min, and then permeabilized with 0.5% Triton X-100/PBS for 5 min. After blocking with 2% BSA/PBS, cells were stained with the indicated primary antibody diluted in 2% BSA/PBS for 1 h at RT. The secondary antibodies used were Alexa Fluor 488-conjugated anti-mouse IgG or Alexa Fluor 594-conjugated anti-rabbit IgG (Molecular Probes). PLA was performed with reagents from DuoLink Biosciences in accordance with the manufacturer's instructions. Images were captured using a BZ-9000 fluorescence microscope (Keyence). Quantification of the PLA signal dots and FANCD2 or RPA foci was determined using Hybrid cell count software (Keyence).
Purification of chicken FANCD2 and FANCI protein
Chicken FANCD2 and FANCI proteins were purified by the same method as previously described (41). The D2-I complex was prepared by mixing FANCD2 and FANCI proteins at an equimolar ratio and then incubating at 37°C for 10 min.
DNA:RNA hybrid binding assay
49-mer single-stranded oligonucleotides with the sequences shown in the Supplementary Table S1 were synthesized and annealed to prepare DNA:RNA or DNA:DNA duplexes. All of the oligonucleotides were purified by HPLC, and the RNA and DNA concentrations are expressed in moles of nucleotides. The synthetic DNA:RNA hybrid (5 μM) and double stranded (ds) DNA (5 μM) were mixed with 0.05–0.20 μM of the D2-I complex in 10 μl of reaction buffer, containing 20 mM Tris–HCl pH 8.0, 70 mM NaCl, 2% glycerol, 0.3 mM MgCl2, 5 mM DTT and 10 μg/ml BSA. The samples were incubated at 37°C for 10 min, and were then analyzed by 3.5% PAGE in TBE (18 mM Tris-borate and 0.4 mM EDTA) buffer. RNAs and DNAs were visualized by SYBR Gold (Invitrogen) staining.
D-loop or R-loop binding and competition assay
80-mer and 90-mer single-stranded oligonucleotides with the sequences shown in the Supplementary Table S1 were synthesized and annealed to prepare D-loop (oligos A/B/C) or R-loop (oligos A/B/D) as described with slight modifications (42). All of the oligonucleotides were purified by HPLC, and the RNA and DNA concentrations are expressed in moles of nucleotides. The synthetic D-loop (5 μM in nucleotides) and R-loop (5 μM in nucleotides) were mixed with FANCD2 (0.1, 0.2, 0.4, 0.6 μM), FANCI (0.1, 0.2, 0.4, 0.6 μM), the D2-I complex (0.1, 0.2, 0.3, 0.4 μM) in 10 μl of reaction buffer, containing 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 2% glycerol, 0.1 mM MgCl2, 5 mM DTT and 10 μg/ml BSA. The samples were incubated at 37°C for 10 min, and were then analyzed by 3.5% PAGE in TBE (18 mM Tris-borate and 0.4 mM EDTA) buffer. RNAs and DNAs were visualized by SYBR Gold (Invitrogen) staining. For competition assay, Cy5-labeled oligonucleotide A was used to prepare labeled D-loops or R-loop. The labelled D-loop (2.5 μM in nucleotides) or R-loop (2.5 μM in nucleotides), the D2-I complex (0.2 μM) and unlabeled competitor D-loop (2.5, 5.0, 12.5, and 25 μM in nucleotides) or R-loop (2.5, 5.0, 12.5, and 25 μM in nucleotides) were incubated at 37°C for 10 min, in 10 μl of reaction buffer, containing 20 mM Tris–HCl pH 8.0, 70 mM NaCl, 2% glycerol, 0.3 mM MgCl2, 5 mM DTT and 10 μg/ml BSA. The samples were analyzed by 3.5% PAGE in TBE buffer, and the bands were detected by Amersham Typhoon (GE Healthcare).
Statistical analysis
Prism (GraphPad Software) was used to perform unpaired two-tailed Student's t-tests and calculation of the correlation coefficient.
Accession number
The ChIP-seq data and RNA-seq data in this study have been deposited in the NCBI Gene Expression Omnibus with accession numbers GSE104464 and GSE104465, respectively.
RESULTS
ChIP-sequencing analysis in engineered U2OS cells expressing FANCD2-3xFLAG from the endogenous locus upon APH replication stress
To precisely examine the genome-wide distribution of FANCD2 protein in chromatin upon replication stress, we set out to create a derivative of human cell line U2OS, which incorporated a 3xFLAG epitope tag into the FANCD2 termination codon in-frame using a TALEN-assisted knock-in strategy (Supplementary Figure S1A). The puromycin resistance gene cassette was removed by Cre-loxP excision. Obtained cell clones were verified with genome PCR (Supplementary Figure S1B). Western blotting using anti-FANCD2 and anti-FLAG antibodies showed all of the expressed FANCD2 alleles were correctly knocked-in with the 3xFLAG tag (Supplementary Figure S1C).
In one of the clones (termed U2OS-D2-FLAG), we performed anti-FLAG ChIP-seq analysis with or without 24 hr treatment with low-dose (0.4 μM) aphidicolin (APH) as explained in Materials and Methods. The U2OS-D2-FLAG cells responded to this treatment by inducing FANCD2 monoubiquitination (Supplementary Figure S1C) and formation of FANCD2/FANCI colocalizing foci (Supplementary Figure S1D), which was dependent on FANCA as expected (Supplementary Figure S1E). The fraction of cells with S-phase DNA content was increased, indicating that replication fork progression was perturbed in this condition (Supplementary Figure S1F).
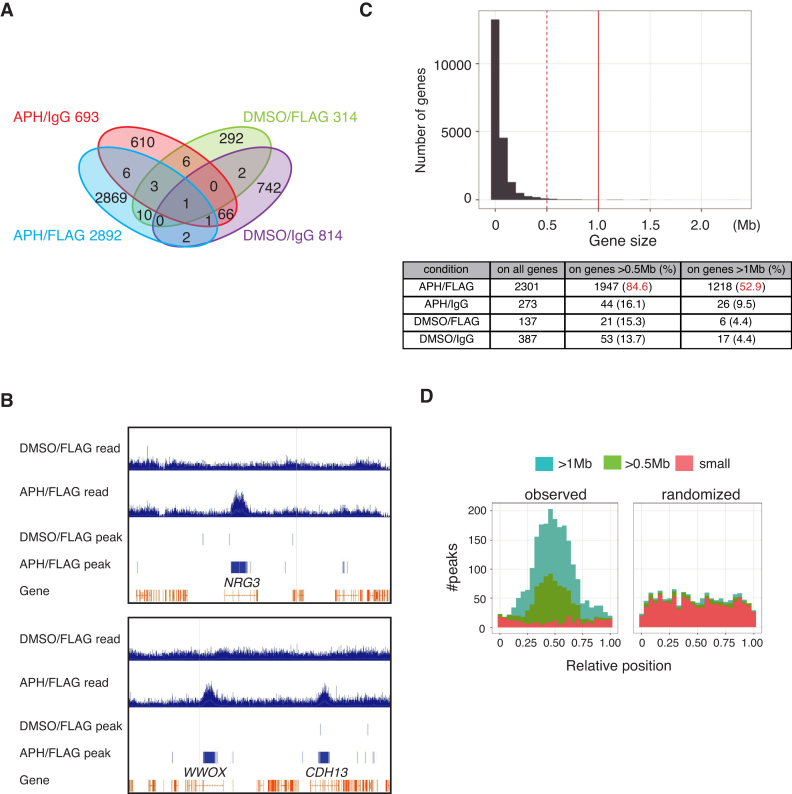
In anti-FLAG ChIP-seq experiments from cells with APH treatment, we observed a much greater number of FANCD2-FLAG (APH/FLAG) peaks compared to the DMSO-treated sample precipitated with anti-FLAG (DMSO/FLAG) or negative control ChIP-seq results (APH- or DMSO-treated sample precipitated with non-specific IgG (APH/IgG or DMSO/IgG) (Figure 1A). The peaks were defined by the accumulation of pair-ends sequencing reads using MACS software. The overlaps in locations between APH/FLAG peaks and negative control ChIP-seq peaks were limited, indicating highly specific enrichment of APH-induced FANCD2 binding sites in our experiments. In addition, DMSO/FLAG peaks were not extensively shared by APH/FLAG peaks. Thus FANCD2 is recruited to chromatin upon replication stress as reflected by foci formation.
Figure 1.
Genome-wide ChIP sequencing analysis of FANCD2–3xFLAG. (A) A Venn diagram showing anti-FLAG ChIP sequencing results from U2OS-D2-FLAG cells. Cells were treated with APH 0.4 μM for 24 h or mock-treated with DMSO, and were subjected to ChIP using anti-FLAG or non-specific IgG. Numerals indicate FANCD2–3xFLAG peak numbers defined in Materials and Methods. (B) Anti-FLAG ChIP-seq reads (blue histogram) and peak (blue bars) profiles on the NRG3 gene region or the WWOX and CDH13 gene region from cells with APH or DMSO treatment. Exons and introns of each gene are indicated in orange. (C) Histogram showing size of each gene (X-axis) versus number of genes (Y-axis). All of the human genes in GRCh37/hg19 are analyzed. The table below shows the number and percentage of ChIP-seq peaks on genes >0.5 Mb or >1.0 Mb among all genes. (D) The distribution of anti-FLAG ChIP-seq peaks from cells with APH stimulation are shown. The transcribed region of each gene was normalized to [0, 1]. Genes were classified by size: >1 Mb (blue), 0.5–1 Mb (green) and <0.5 Mb (red). The observed peaks were enriched near the center of large genes (left, >0.5 Mb) more frequently than expected by random chance (right). The dispersion and kurtosis of the observed distribution were smaller and larger, respectively, than any of 1000 randomized distributions (P< 0.001; only one of them is shown here).
FANCD2 binds to middle intronic regions of a set of large transcribed genes during mild replication stress
Although it has been reported that replication stress-induced FANCD2 foci coincide with common fragile sites (CFSs) such as FRA3B or FRA16D (21), it was a bit surprising for us that there were so many known CFS among the top hits by the number of FANCD2 APH/FLAG peaks per gene/region (Table 1). In general, FANCD2 peaks were largely accumulated in gene bodies of the very large genes (Figure 1B and C). Specifically, out of 2892 APH/FLAG FANCD2 peaks, 2301 peaks (79.6%) were within genes. The rest of the peaks were in intergenic regions, but most of them were not clustered (therefore they were not on the top-hits list). Furthermore, 84.6% or 52.9% of the APH/FLAG peaks on genes were on extremely large genes spanning >0.5 Mb or >1 Mb, respectively (Figure 1C). Even though these genes are huge, of all the genic regions (20 418 genes in total occupy 1.22 Giga base), only 19% or 7% of them were found to be associated with 282 genes spanning >0.5 Mb or by 61 genes spanning >1 Mb, respectively. Thus, their huge size could not explain the preferential distribution of APH/FLAG peaks on them. Interestingly, these FANCD2 binding sites were enriched near the center of the genic region of large genes (Figure 1B and D).
Table 1. 30 top hits genes/fragile sites in FANCD2-FLAG ChIP-seq analysis after APH treatment.
| Top hits order | Chromosome localization | Gene | CFS | Number of reads | Percent in total reads | Gene size (Kb) | CFS size (Mb) |
|---|---|---|---|---|---|---|---|
| 1 | 10q23 | NRG3* | - | 8608 | 10.0 | 1111 | - |
| 2 | 16q23 | WWOX | FRA16D | 7929 | 9.2 | 1113 | 1.8 |
| 3 | 16q23 | CDH13* | - | 4704 | 5.5 | 1169 | - |
| 4 | 9p21 | LINGO2* | FRA9C | 3832 | 4.4 | 1264 | 13.3 |
| 5 | 7p22 | SDK1* | FRA7B | 3395 | 3.9 | 967 | 7.3 |
| 6 | 11q14 | DLG2* | FRA11F | 3302 | 3.8 | 2172 | 2.2 |
| 7 | 3p14 | FHIT* | FRA3B | 3082 | 3.6 | 1502 | 4.1 |
| 8 | 3q11 | EPHA6* | - | 3074 | 3.6 | 934 | - |
| 9 | 3p26 | CNTN4* | - | 2906 | 3.4 | 959 | - |
| 10 | 12p12 | SOX5* | - | 2241 | 2.6 | 1030 | - |
| 11 | 10q11 | PRKG1 | FRA10C | 2091 | 2.4 | 1307 | 17.7 |
| 12 | 21p11 | miR-3648 | - | 2002 | 2.3 | 0.18 | - |
| 13 | 7q31 | IMMP2L* | FRA7K | 1937 | 2.2 | 899 | 0.2 |
| 14 | 21p11 | miR-3687 | - | 1929 | 2.2 | 0.061 | - |
| 15 | 5p14 | CDH12* | - | 1800 | 2.1 | 1102 | - |
| 16 | 1p31 | NEGR1* | FRA1L | 1392 | 1.6 | 879 | 23.6 |
| 17 | 5p15 | CTNND2* | - | 1072 | 1.2 | 932 | - |
| 18 | 8q24.3 | TRAPPC9* | FRA8D | 1001 | 1.2 | 726 | 6.5 |
| 19 | 20p12 | MACROD2* | - | 891 | 1.0 | 2057 | - |
| 20 | 4p15 | - | FRA4D | 860 | 1.0 | - | 24.5 |
| 21 | 1p36 | - | FRA1A | 797 | 0.9 | - | 28.0 |
| 22 | 11q13 | - | FRA11H | 765 | 0.9 | - | 13.7 |
| 23 | 7q11 | MAGI2* | - | 636 | 0.7 | 1436 | - |
| 24 | 7q11 | AUTS2* | FRA7J | 616 | 0.7 | 1194 | 17.6 |
| 25 | 13q33 | FAM155A | - | 497 | 0.6 | 698 | - |
| 26 | 4q35 | FRG2 | - | 401 | 0.5 | 2.89 | - |
| 27 | 9p32 | ASTN2* | - | 380 | 0.4 | 989 | - |
| 28 | 22q13 | - | FRA22A | 346 | 0.4 | - | 13.7 |
| 29 | Xq22 | DIAPH2 | - | 339 | 0.4 | 915 | - |
| 30 | 2q22 | THSD7B | - | 322 | 0.4 | 686 | - |
*Genes related with autism, neurodevelopmental or psychiatric disorders (https://www.omim.org). Numbers of the ChIP-seq reads in a gene or in a region consisting of combined gene and CFS, which are defined as described in Bignell et al. (51), are shown. When there were two or more genes overlapping with a CFS, the names and size of the genes were omitted.
It should be noted that the top-hits list also included large non-CFS genes (Table 1). This seems to be consistent with a previous notion that all large genes if actively transcribed may be fragile (43). For example, the Neuregulin 3 (NRG3) gene spans ∼1.1 Mb and is similarly as huge as CFS-associated genes, albeit the protein coding sequence is small (the longest spliced transcript is ∼2.1 kb). NRG3 accumulated the highest number of FANCD2 peaks as a single gene (Table 1). NRG3 protein is the ligand for ErbB4 receptor tyrosine kinase, and it is expressed mostly in neural cells, but not in lymphocytes. We speculate that NRG3 might be regulated/operated in neural cells (and we find it is expressed in U2OS cells, see below) in a manner similar to conventional CFS genes that have mostly been described in lymphocytes. Thus, it is likely that NRG3 is ‘fragile’ in neurons as well as in U2OS cells. This gene is known to correspond to a susceptibility locus for schizophrenia and autism (44).
Transcription is a prerequisite for the FANCD2 accumulation upon replication stress
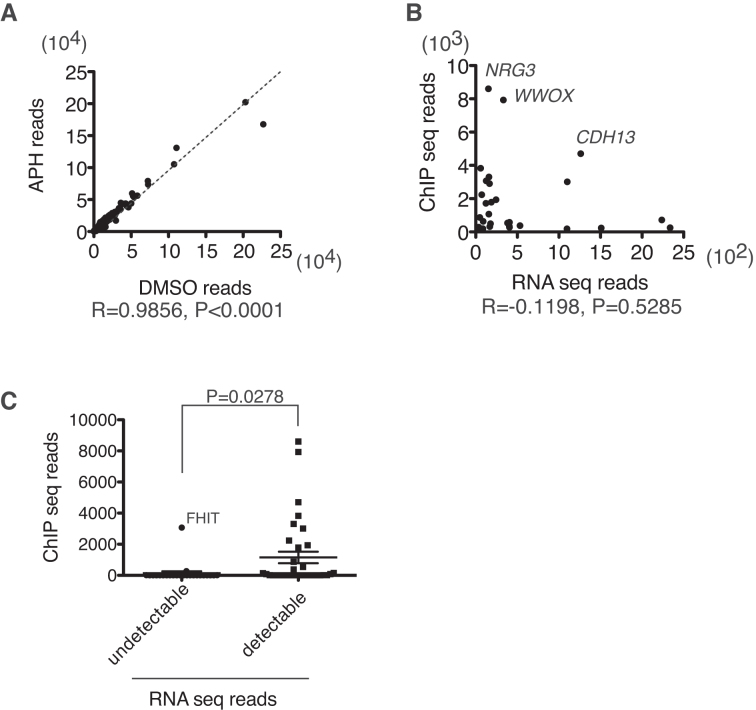
Our ChIP-seq experiments indicated that FANCD2 accumulates mostly on gene bodies upon replication stress. Therefore, we hypothesized that transcription might be a prerequisite for the FANCD2 accumulation upon replication stress. To detect transcripts at steady state levels, we subjected total RNA isolated from U2OS-D2-FLAG cells before and after APH treatment to next generation sequencing (RNA-seq). The levels of transcripts were not significantly altered after APH treatment in most of the genes (Figure 2A). There was no significant correlation between the FANCD2 ChIP-seq peak number and the transcript levels (reads number) among the 30 top hits (Figure 2B). However, it was obvious that large genes without detectable RNA-seq reads did not accumulate high levels of FANCD2 reads except for FHIT (Figure 2C).
Figure 2.
Correlation between the FANCD2-FLAG ChIP-seq read number and the transcript levels. (A) The RNA-seq read number before and after APH (0.4 μM for 24 h) was plotted per gene. The correlation coefficient was calculated with Prism software. (B) FANCD2-FLAG ChIP-seq read number and RNA-seq read number after APH treatment were plotted for the 30 genes with the highest D2 ChIP-seq reads. The correlation coefficient was calculated with Prism software. (C) Comparison of FANCD2-FLAG ChIP-seq read number between genes among the 60 largest genes with undetectable or detectable transcript levels. Means ± SEM are shown (P-value, unpaired, two-tailed Student's t test). We were able to detect FHIT transcripts in U2OS cells by RT-PCR (not shown), suggesting that FHIT was indeed transcribed, albeit at relatively low levels.
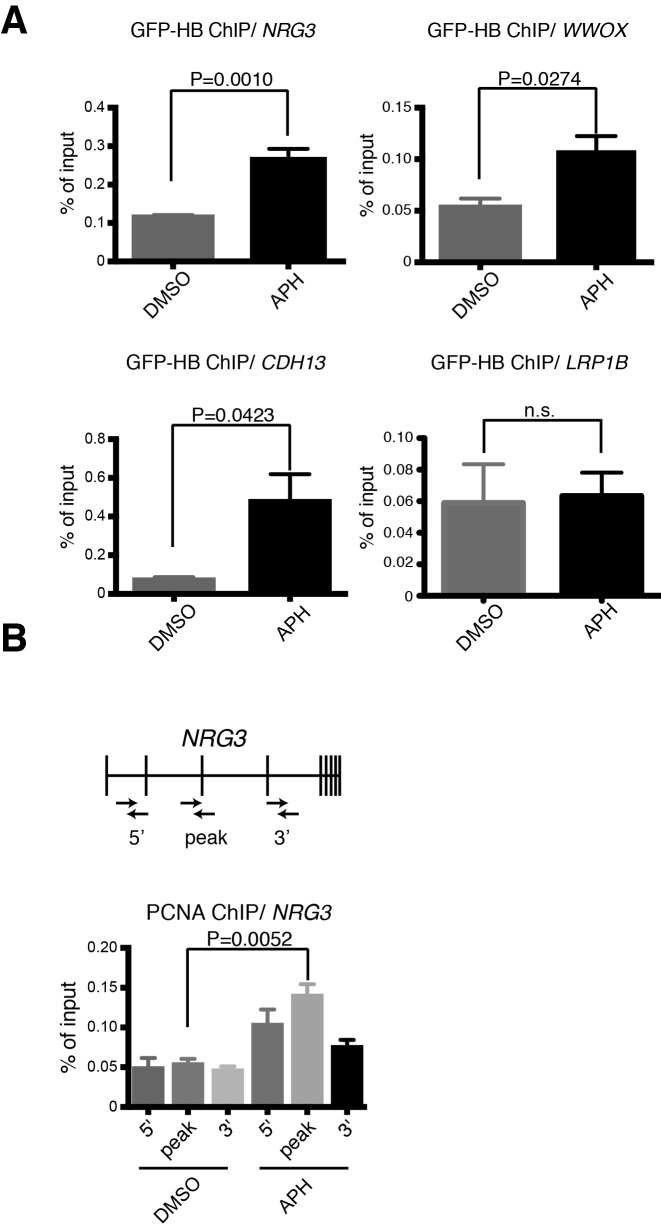
Previous studies indicated that transcription of huge genes takes more time than a single cell cycle, leading to the transcription-replication collision with R-loop formation (17). R-loops consist of DNA:RNA hybrids and displaced ssDNA. To detect R-loops at specific genomic loci, we utilized expression of the DNA:RNA hybrid-binding domain (HBD) from human RNase HI fused with enhanced green fluorescent protein (EGFP) (GFP-HBD) (Supplementary Figure S2A). This method has been previously verified for detecting R-loops (45). In U2OS-D2-FLAG cells transduced with lentivirus carrying DOX-inducible GFP-HBD, high levels of GFP expression were observed following DOX addition (Supplementary Figure S2B). In this cell line, R-loop accumulation was detected at the NRG3, CDH13 and WWOX (Figure 3A) loci by ChIP-qPCR analysis using a GFP-trap antibody. We were also able to see increased anti-PCNA ChIP signals on the NRG3 gene, which appeared to be highest in the central part of the gene (Figure 3B). This suggests that there were slowed or stalled replication forks on NRG3 upon APH stress.
Figure 3.
Detection of DNA:RNA hybrids resulting from transcription-replication collision during mild replication stress. (A) GFP-HBD ChIP-qPCR analysis of the NRG3 (with the peak primers), WWOX, and CDH13 genes in U2OS cells stably expressing GFP-HBD after APH treatment. LRP1B gene was used as a negative control since it is a huge gene (∼2 Mb) without FANCD2 accumulation. Percentage of input is plotted (n = 3). Means and SEM are shown (unpaired, two-tailed Student's t test). Position of the NRG3 amplicon is shown in (B). (B) Anti-PCNA ChIP-qPCR analysis of NRG3 in U2OS cells treated with APH. Percentage of input is plotted (n = 3). Means ± SEM are shown (unpaired, two-tailed Student's t test). Positions of the amplicons are shown.
Transcription inhibition and GFP-RNaseHI expression abrogate replication stress-induced FANCD2 foci as well as FANCD2 accumulation at the NRG3 gene
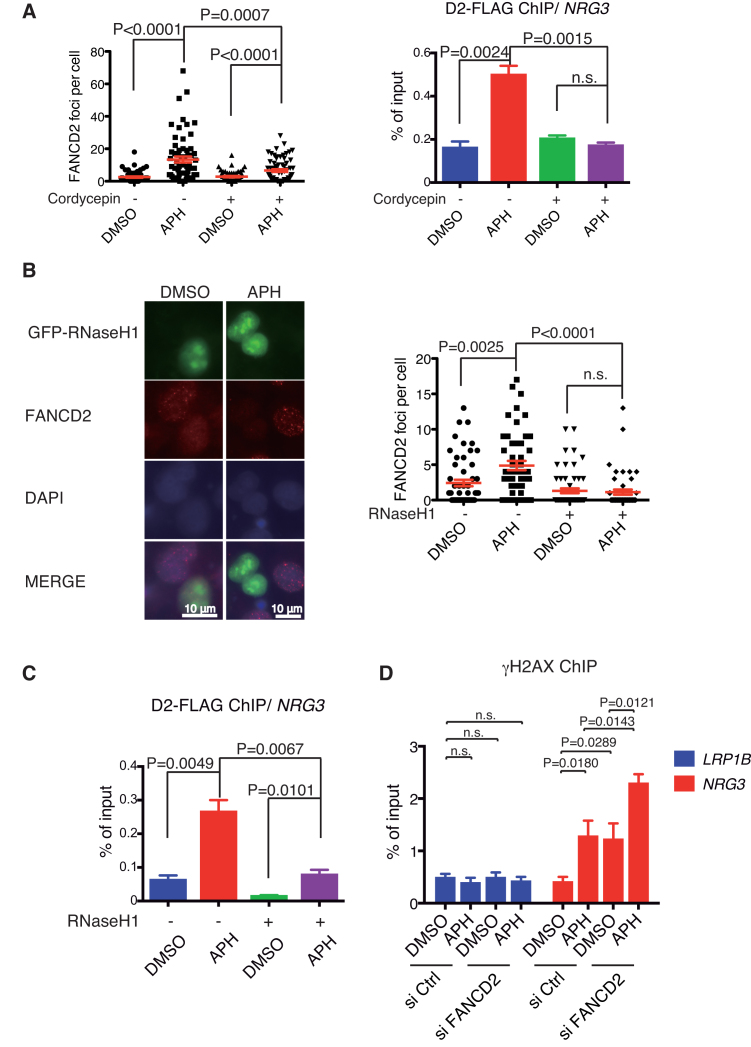
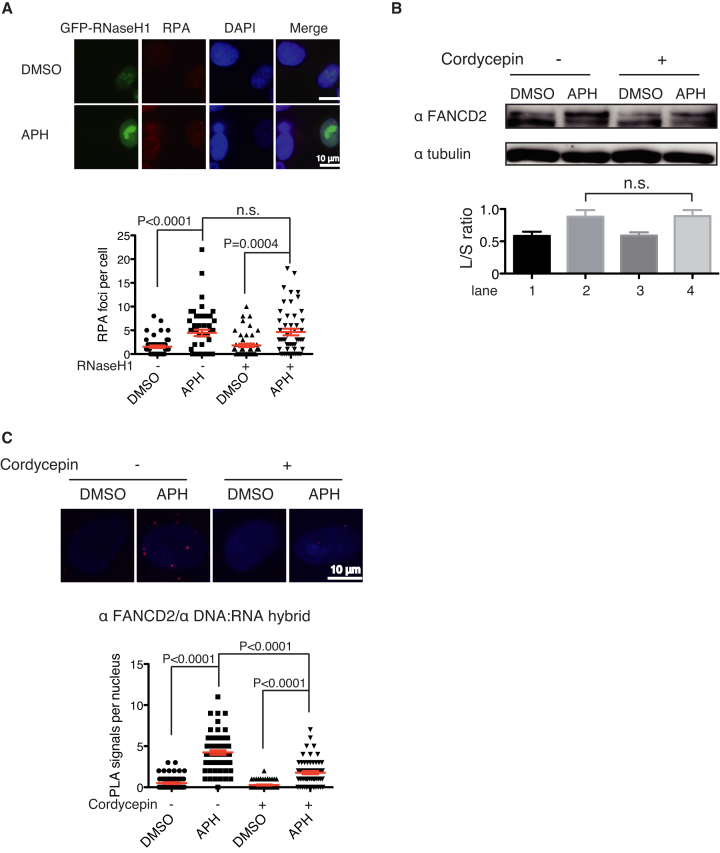
It has previously shown that the FANCD2 accumulation at chromatin might be promoted by R-loop formation during replication stress (27). Thus we treated cells with cordycepin, an adenosine derivative and a transcription inhibitor, which is incorporated into RNA and terminates RNA synthesis, and thus prevents R-loop formation. Indeed, treatment with cordycepin for 3 h abrogated FANCD2 foci in U2OS cells (Figure 4A, left panel). We confirmed by FLAG-ChIP qPCR that FANCD2 accumulation at the NRG3 gene was also significantly decreased by cordycepin treatment (Figure 4A, right panel). We also tried to use RNaseHI as another experimental approach to eliminate R-loops in the cell. As we could not isolate U2OS cells with stable as well as inducible expression of GFP-RNaseHI, we switched to transient expression of a transfected GFP-RNaseHI expression construct. Consistent with the previous study (27), FANCD2 foci formation was mostly abrogated in GFP expressing cells compared to non-transfected cells (Figure 4B). Moreover, FANCD2 accumulation at the NRG3 gene following APH treatment was also significantly decreased, as shown by anti-FLAG ChIP-qPCR in GFP-RNaseHI transfected cells (Figure 4C). Taken together, these results indicated that R-loops formed during replication stress play a critical role in recruitment and accumulation of FANCD2 at gene bodies of transcribed large genes. In cells depleted of FANCD2, higher levels of DNA damage were detected with or without APH at the NRG3 gene by γH2AX ChIP-qPCR (Figure 4D), suggesting critical roles of FANCD2 in handling replication stress.
Figure 4.
Transcription inhibition and GFP-RNaseHI expression abrogate R-loop and FANCD2 foci following APH treatment. (A) Cordycepin treatment reduced APH-induced FANCD2 foci levels, and anti-FLAG ChIP-qPCR signals at the NRG3 gene, evidenced using the NRG3 peak primers depicted in Figure 2B. Results of the immunofluorescence analysis of nuclei (n > 50) (left panel) and ChIP-qPCR analysis (n = 3) (right panel) are shown. Cells were treated with APH for 24 h and/or the transcription inhibitor cordycepin (50 μM) for 3 h. Means ± SEM are shown. P values were calculated using unpaired, two-tailed Student's t test. (B) GFP-RNaseH1 overexpression abrogated FANCD2 foci in U2OS cells (n = 50) treated with APH. P-values were calculated by unpaired, two-tailed Student's t test. (C) D2-FLAG ChIP-qPCR analysis of NRG3, using the peak primers, in GFP-RNaseHI overexpressed U2OS-D2-FLAG cells treated with APH. Percentage of input is plotted (n = 3). Means ± SEM are shown. P-values were calculated by unpaired, two-tailed Student's t test. (D) Anti-γH2AX ChIP-qPCR analysis of LRP1B and NRG3 in FANCD2-depleted U2OS cells after APH treatment. Percentage of input is plotted (n = 6). Means and SEM are shown (unpaired, two-tailed Student's t test). LRP1B gene was used as a negative control. The NRG3 qPCR was done using the peak primers.
R-loops are dispensable for FANCD2 monoubiquitination upon replication stress
To establish whether upstream events leading to FA pathway activation are also dependent on R-loops, we examined RPA foci formation in cells expressing GFP-RNaseHI. RPA foci were induced by the APH treatment, and their levels appeared to be only slightly decreased by RNaseHI, yet this was a statistically insignificant difference (Figure 5A), suggesting that the extent of ssDNA exposed during APH-induced replication stress was mostly independent of R-loops. The mild decrease could be explained by the presence of RPA on the displaced ssDNA in R-loops (46). The RPA foci should recruit ATRIP-ATR kinase via binding of RPA and ATRIP, leading to ATR activation (11), FANCI phosphorylation, and subsequent FANCD2 monoubiquitination (1,6,7,47). This seems consistent with our observation that cordycepin treatment did not decrease APH-induced FANCD2 monoubiquitination levels (Figure 5B). Taken together, our results suggest that R-loop formation is not essential for the FA pathway activation but is required for FANCD2 foci formation, and raised an interesting possibility that R-loops contribute to the retention of FANCD2 in chromatin when FANCD2 is monoubiquitinated. We also subjected U2OS cells to PLA using anti-FANCD2 and anti-DNA:RNA (S9.6) antibody. We detected increased levels of PLA dots following APH treatment, and these levels were reduced by cordycepin treatment (Figure 5C), indicating spatial proximity between R-loops and FANCD2 in chromatin.
Figure 5.
Replication stress-induced RPA foci and FANCD2 monoubiqitination were not significantly affected by RNaseH1 or cordycepin treatment. (A) RPA foci levels in GFP-RNaseHI overexpressing U2OS cells treated with APH. Means ± SEM from 50 scored nuclei are shown. P-values were calculated by unpaired, two-tailed Student's t test. (B) APH-induced FANCD2 monoubiquitination was not decreased by cordycepin treatment. (C) The PLA results between anti-FANCD2 and S9.6 in U2OS cells treated with APH (24 h) and/or cordycepin (3 h). Means ± SEM scored from more than 50 nuclei are shown. P-values were calculated by unpaired, two-tailed Student's t test.
These results prompted us to test the possibility that FANCD2 (and FANCI) could bind with DNA:RNA hybrids or R-loops by an in vitro binding assay using purified recombinant chicken FANCD2, FANCI, and the D2-I complex, which was prepared by mixing and pre-incubating the two proteins (Supplementary Figure S3A) (41,48). Binding of short DNA:RNA hybrids or dsDNA was detected with purified chicken FANCD2, FANCI and the D2-I complex (Supplementary Figure S3B). We also prepared D-loop or R-loop in vitro, using synthesized ssDNA and ssRNA which were annealed by heating followed by gradual cooling. Gel shift assay and competition assay revealed clear binding of R-loop with purified chicken FANCD2 and the D2-I complex or FANCI (Supplementary Figure S3C and S3D), with comparable affinity compared to D-loop having the same nucleotide sequence.
DISCUSSION
To shed light on the function and regulation of the key FA factor FANCD2, we looked into the genome-wide distribution of FANCD2 following mild replication stress, and found that the protein binds to chromatin in a set of very large genes. The FANCD2 binding pattern in chromatin formed bell-shaped curves with their peaks at the middle region of the genes (mostly in introns). This is in sharp contrast to the situation reported in a recent study, where the presence of introns at a given genomic locus decreases the propensity of R-loop formation by recruiting protein factors that may sterically hinder hybridization of nascent RNA with genomic DNA (49). We speculate that large introns in these genes may increase the probability of R-loop formation since co-transcriptional splicing out of introns may take longer, allowing primary transcripts to anneal with the template DNA. The FANCD2 binding appeared to be dependent on R-loop formation likely caused by transcription-replication collision. This may suggest that the collision mainly occurs at the central part of these large genes. Further research is needed to clarify how this occurs. Furthermore, our data provide a novel insight into the activation mechanisms of the FA pathway (summarized in Figure 6). Since MMC-induced FANCD2 foci formation is also affected by RNaseHI expression (27), similar mechanisms likely operate during ICL repair.
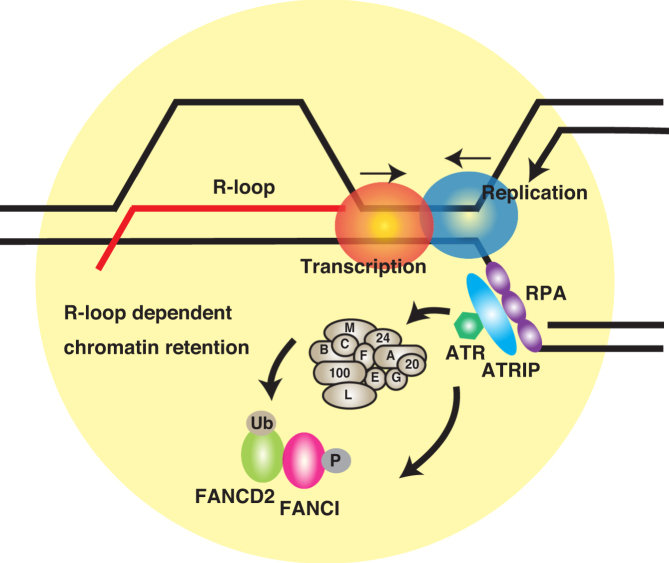
Figure 6.
Our proposed model for the role of R-loops in FA pathway activation. Upon mild replication stress, the polymerase–helicase uncoupling or stalled replication forks create extended regions of ssDNA and transcription-replication collision causes accumulation of R-loops that may further exacerbate replication stress. ssDNA recruits RPA, leading to ATR/ATRIP kinase activation. In turn, ATR phosphorylates FANCI and triggers D2-I complex monoubiquitination, and the ubiquitinated proteins may be retained in a manner dependent on R-loops accumulated near or at the stalled forks.
R-loops support FANCD2 chromatin binding but are not critical for its monoubiquitination
We addressed how R-loops participate in FA pathway activation by employing RNaseHI overexpression or the transcriptional inhibitor cordycepin. In both cases, we found that FANCD2 foci formation induced by replication stress was abrogated. Similar observations in the absence of APH treatment have been previously described (26,27). Though we were not able to provide FANCD2 monoubiquitination data in cells transfected with an RNaseHI expression plasmid because of cellular toxicity in this condition, cordycepin treatment revealed that FANCD2 monoubiquitination can proceed unaffected during replication stress in the lower levels of R-loops. The APH treatment stalls or slows down replication forks, leading to exposed ssDNA with RPA accumulation (and hence ATR activation), which seems mostly independent of R-loops. This is reflected by the intact RPA foci and FANCD2 monoubiquitination in cells depleted of R-loops. It seemed an interesting possibility that R-loops might function as a landing platform for FANCD2 and FANCI once they are monoubiquitinated, while it is clear that D2-I complex can efficiently bind with dsDNA (Supplementary Figure S3B). Thus we compared FANCD2 or the D2-I complex binding with D-loop versus R-loop in vitro. The data failed to show higher affinity of FANCD2 with R-loops than D-loops. Future research will be intended to clarify how R-loops contribute to chromatin retention of FANCD2 during replication stress.
Operational mechanisms specific to the huge genes might contribute to how FANCD2 accumulates during replication stress
The CFS genes are generally large and are implicated in neuronal development, synapse function, and neural cell adhesion. They are often implicated in neuronal disorders like autism or psychiatric disorders. We found that FANCD2 during mild replication stress binds to introns of a set of transcribed large genes that include CFSs, and its accumulation is mostly restricted to the central intronic portion of these genes. Although it is at the moment impossible to provide a sufficient explanation for why FANCD2 has a particular propensity to bind in this region of the large genes, we provide evidence that these FANCD2 peaks are dependent on R-loop formation. Previous studies indicated that transcription complexes may collide with replication forks in large transcribed genes, since transcription in these genes takes more time than a single cell cycle (17), causing R-loops and CFS expression. It is highly interesting to note that the cumulative pattern of APH-induced genomic gain or loss in CFSs in previous reports (12,43) strikingly resembles the bell-shaped curve of FANCD2 peaks.
Although U2OS is derived from an osteosarcoma, these huge genes are generally expressed in neurons, and they are involved in neuronal function. These genes are also transcribed in U2OS but their expression levels (read numbers in RNA-seq) did not correlate with FANCD2 peak number. Nonetheless, the transcription itself appeared to be required for large genes to accumulate FANCD2. We suggest there should be specific regulatory/operational mechanisms for these huge neuronal genes, making them particularly sensitive to APH treatment. Elucidating these mechanisms is an important task that will eventually lead to full understanding of how these genes evolved, why such huge size is required for neurons despite the apparent disadvantage of genome instability, and how FANCD2 accumulation is triggered at the middle part of these genes. It is interesting to note that a recent study finds increased levels of double-strand breaks in neuronal progenitor cells upon replication stress, suggesting potential roles by which DNA damage may impact neuronal development (50). The list of affected genes in their study partially overlaps with ours (50).
DATA AVAILABILITY
The ChIP-seq data and RNA-seq data in this study have been deposited in the NCBI Gene Expression Omnibus with accession numbers GSE104464 and GSE104465, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr James Hejna (Kyoto University) for critical reading of the manuscript; Drs Patrick Calsou, Daniel Voytas, Rudolf Jaenisch, Hiroyuki Miyoshi, Makoto Nakanishi and Keith Joung for reagents; Drs Yasutoshi Agata (Shiga University of Medical Science), Naoyuki Kataoka (The University of Tokyo), Daisuke Kaida (University of Toyama) for advice in ChIP procedure; Drs Mari Miyaji, Kimiko Tsutsui and Ken Tsutsui (Okayama University Medical School), or Nozomi Sugimoto, and Masatoshi Fujita (Kyushu University) for discussions; technical and secretarial help from Chinatsu Ohki, Akiko Watanabe, Seiko Arai, Ayumi Katayama, and Fan Peng. The Radiation Biology Center, Kyoto University, is a Joint Usage Research Center supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. A part of the authors' work has been performed in collaborative research projects in the Joint Usage Research Center. The authors declare no competing financial interests.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
JSPS KAKENHI [JP23114010 to M.T., JP26550026 to M.T., JP15H01738 to M.T., JP26281021 to M.I., JP25116002 to H.K., JP17H01408 to H.K.]; Ministry of Health, Labour and Welfare (to M.T.); Uehara Memorial Foundation (to M.T.); Waseda University (to H.K.). Funding for open access charge: Uehara Memorial Foundation (to M.T.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Ishiai M., Sato K., Tomida J., Kitao H., Kurumizaka H., Takata M.. Activation of the FA patihway mediated by phosphorylation and ubiquitination. Mutat. Res. 2017; 803–805:89–95. [DOI] [PubMed] [Google Scholar]
- 2. Ceccaldi R., Sarangi P., D’Andrea A.D.. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell. Biol. 2016; 17:337–349. [DOI] [PubMed] [Google Scholar]
- 3. Kottemann M.C., Smogorzewska A.. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013; 493:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walden H., Deans A.J.. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu. Rev. Biophys. 2014; 43:257–278. [DOI] [PubMed] [Google Scholar]
- 5. Garaycoechea J.I., Patel K.J.. Why does the bone marrow fail in Fanconi anemia. Blood. 2014; 123:26–34. [DOI] [PubMed] [Google Scholar]
- 6. Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T. et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008; 15:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shigechi T., Tomida J., Sato K., Kobayashi M., Eykelenboom J.K., Pessina F., Zhang Y., Uchida E., Ishiai M., Lowndes N.F. et al. ATR-ATRIP kinase complex triggers activation of the Fanconi anemia DNA repair pathway. Cancer Res. 2012; 72:1149–1156. [DOI] [PubMed] [Google Scholar]
- 8. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato K., Shimomuki M., Katsuki Y., Takahashi D., Kobayashi W., Ishiai M., Miyoshi H., Takata M., Kurumizaka H.. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5′-DNA end. Nucleic. Acids. Res. 2016; 44:10758–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koundrioukoff S., Carignon S., Técher H., Letessier A., Brison O., Debatisse M.. Stepwise activation of the ATR signaling pathway upon Increasing replication stress impacts fragile site integrity. PLoS Genet. 2013; 9:e1003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glover T.W., Wilson T.E., Arlt M.F.. Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer. 2017; 17:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O.. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012; 28:22–32. [DOI] [PubMed] [Google Scholar]
- 14. Minocherhomji S., Hickson I.D.. Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol. 2014; 24:321–327. [DOI] [PubMed] [Google Scholar]
- 15. Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B.. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002; 1:89–97. [DOI] [PubMed] [Google Scholar]
- 16. Sarni D., Kerem B.. The complex nature of fragile site plasticity and its importance in cancer. Curr. Opin. Cell Biol. 2016; 40:131–136. [DOI] [PubMed] [Google Scholar]
- 17. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 18. Bhowmick R., Minocherhomji S., Hickson I.D.. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell. 2016; 64:1117–1126. [DOI] [PubMed] [Google Scholar]
- 19. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D.. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 20. Di Marco S., Hasanova Z., Kanagaraj R., Chappidi N., Altmannova V., Menon S., Sedlackova H., Langhoff J., Surendranath K., Hühn D. et al. RECQ5 helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Mol. Cell. 2017; 66:658–671. [DOI] [PubMed] [Google Scholar]
- 21. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D.. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature. 2009; 11:753–760. [DOI] [PubMed] [Google Scholar]
- 22. Chan K.L., North P.S., Hickson I.D.. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007; 26:3397–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howlett N.G., Taniguchi T., Durkin S.G., D’Andrea A.D., Glover T.W.. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005; 14:693–701. [DOI] [PubMed] [Google Scholar]
- 24. Naim V., Wilhelm T., Debatisse M., Rosselli F.. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 2013; 15:1008–1015. [DOI] [PubMed] [Google Scholar]
- 25. Madireddy A., Kosiyatrakul S.T., Boisvert R.A., Herrera-Moyano E., García-Rubio M.L., Gerhardt J., Vuono E.A., Owen N., Yan Z., Olson S. et al. FANCD2 facilitates replication through common fragile sites. Mol. Cell. 2016; 64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez-Martinez D., Liang C.-C., Cohn M.A., Gibbons R.J., Deans A.J., Niedzwiedz W.. The fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell. 2015; 60:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. García-Rubio M.L., Pérez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A.. The fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steyaert S., Rajpurkar A., Ginno P.A., Xu X.. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016; 63:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 30. García-Pichardo D., Cañas J.C., García-Rubio M.L., Gómez-González B., Rondón A.G., Aguilera A.. Histone mutants separate R loop formation from genome instability induction. Mol. Cell. 2017; 66:597–609. [DOI] [PubMed] [Google Scholar]
- 31. Sakuma T., Hosoi S., Woltjen K., Suzuki K.-I., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y. et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013; 18:315–326. [DOI] [PubMed] [Google Scholar]
- 32. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–13. [Google Scholar]
- 33. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R.. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009; 25:2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sato K., Toda K., Ishiai M., Takata M., Kurumizaka H.. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic. Acids. Res. 2012; 40:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao W., Steinfeld J.B., Liang F., Chen X., Maranon D.G., Jian Ma C., Kwon Y., Rao T., Wang W., Sheng C. et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature. 2017; 550:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilson T.E., Arlt M.F., Park S.H., Rajendran S., Paulsen M., Ljungman M., Glover T.W.. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015; 25:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kao W.-T., Wang Y., Kleinman J.E., Lipska B.K., Hyde T.M., Weinberger D.R., Law A.J.. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2015; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 46. Nguyen H.D., Yadav T., Giri S., Saez B., Graubert T.A., Zou L.. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 2017; 65:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomida J., Itaya A., Shigechi T., Unno J., Uchida E., Ikura M., Masuda Y., Matsuda S., Adachi J., Kobayashi M. et al. A novel interplay between the Fanconi anemia core complex and ATR-ATRIP kinase during DNA cross-link repair. Nucleic. Acids. Res. 2013; 41:6930–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., Tachiwana H., Takizawa Y., Kagawa W., Kitao H., Dohmae N. et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012; 31:3524–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bonnet A., Grosso A.R., Elkaoutari A., Coleno E., Presle A., Sridhara S.C., Janbon G., Géli V., de Almeida S.F., Palancade B.. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol. Cell. 2017; 67:608–621. [DOI] [PubMed] [Google Scholar]
- 50. Wei P.-C., Chang A.N., Kao J., Du Z., Meyers R.M., Alt F.W., Schwer B.. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. 2016; 164:644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C. et al. Signatures of mutation and selection in the cancer genome. Nature. 2010; 463:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq data and RNA-seq data in this study have been deposited in the NCBI Gene Expression Omnibus with accession numbers GSE104464 and GSE104465, respectively.