Abstract
Cell-derived exosomes (EXs) can modulate target cell differentiation via microRNAs (miRs) that they carried. Previous studies have shown that miR126 is highly expressed in hematopoietic stem cells (HSCs) and plays a role in hematopoiesis via modulating the Notch pathway that participates in progenitors' cell fate decisions. In this study we investigated whether HSC-derived EXs (HSC-EXs) could affect the differentiation of mouse embryonic stem cells (ESCs) into HSCs. We prepared HSC-EXscon, HSC-EXssc and HSC-EXsmiR126 from control HSCs and the HSCs transfected with scramble control or miR126 mimics, respectively. HSC-EXs were isolated by ultracentrifugation and analyzed using nanoparticle tracking analysis. We incubated the collected EXs with mouse ESCs over a 10-d differentiation induction period, during which HSC-EXs and a Notch pathway activator (Jagged1, 100 ng/mL) were added to the cultures every 3 d. After the 10-d differentiation period, the expression levels of miR126, SSEA1, CD117, Sca1, Notch1 and Hes1 in ESCs were assessed. The generated HSCs were validated by flow cytometry using antibodies against HSC markers (CD117, CD34 and Sca1). Our results revealed that: (1) transfection with miR126 mimics significantly increased miR126 levels in HSC-EXsmiR126. (2) HSC-EX co-culture promoted mouse ESCs differentiation into HSCs with the most prominent effect found in the HSC-EXsmiR126 co-culture. (3) HSC differentiation was verified by reduced SSEA1 expression and increased CD117 and Sca1 expression. (4) All the effects caused by HSC-EXs were accompanied by significant reduction of Notch1 and Hes1 expression, thus inhibition of the Notch1/Hes1 pathway, whereas activation of Notch by Jagged1 abolished the effects of HSC-EXsmiR126. In conclusion, HSC-EXs promote hematopoietic differentiation of mouse ESCs in vitro by inhibiting the miR126/Notch1 pathway.
Keywords: hematopoietic stem cells, embryonic stem cells, differentiation, exosomes, miR126, Notch, Jagged1
Introduction
Hematopoietic disease affects millions of people worldwide and can be treated by transplantation of hematopoietic stem cells (HSCs). However, the difficulty in collecting a sufficient number of HSCs from bone marrow and the lack of an efficient in vitro approach for HSC expansion have impeded the clinical use of HSC-based therapy1. Embryonic stem cells (ESCs) derived from the blastocyst of the early embryo could provide a source of HSCs2.
Exosomes (EXs) are extracellular vesicles secreted by various types of cells3,4,5. Accumulating studies have demonstrated that extracellular vesicles such as EXs are important intercellular mediators and can alter the functions of target cells by conveying information from their cargoes, such as microRNAs (miRs)5,6,7. For example, Xin et al reported that EXs from mesenchymal stem cells can promote neural plasticity by transferring miR133b to target cells in an in vivo stroke model7. The extracellular vesicles derived from endothelial progenitor cells can protect endothelial cells against hypoxia/reoxygenation injury by modulating miR126 expression5. Recently, our group found that endothelial progenitor cell-derived vesicles from healthy controls protected the functions of endothelial progenitor cells from diabetic patients via miR126 carried within the vesicles8. Interestingly, Notch signaling pathway proteins, such as the ligand Delta-like 4, can be packaged into endothelial cell-derived EXs and transferred to other endothelial cells to inhibit Notch signaling in the recipient cells. In addition, EXs can alter recipient cell differentiation. It has been shown that EXs derived from neuronal progenitor cells can promote neuronal differentiation from mesenchymal stem cells9. Ismail et al reported that macrophage-derived extracellular vesicles promoted naïve differentiation into macrophage10. However, whether HSC-derived EXs (HSC-EXs) can promote hematopoietic differentiation from ESCs remains unknown.
miRs are considered as master regulators of cellular processes, including cell differentiation. Previous studies have demonstrated that several miRs, such as miR142, miR181a, are predominately expressed in hematopoietic cells11,12. Among these miRs, miR126 is highly expressed and functional within the HSC compartments13. It has been shown that miR126 is progressively downregulated during the early stage of hematopoietic commitment14, indicating its role in HSC differentiation. Overexpression of miR126 in ESCs resulted in a reduction in the number of erythroid colonies, suggesting an inhibitory role of miR126 in erythropoiesis11. Nevertheless, only a few studies have investigated the role of miR126 in HSC differentiation by ESCs.
Recently, Huang et al demonstrated that overexpressing miR126 could enhance mesenchymal stem cells angiogenesis by upregulating the Notch ligand, Delta-like 415, suggesting that miR126 could affect the Notch signaling pathway in stem cells. It has been well documented that Notch signal transduction is based ligand (Dll1, Dll3, Dll4, Jagged1, and Jagged2) binding to the appropriate receptor (Notch1 to 4)16. The Notch signaling pathway governs fundamental processes such as cellular differentiation, proliferation and development17. For example, activation of the Notch pathway by Jagged 1 can promote ESCs differentiation into neural cells18. Previous studies have shown that Notch signaling plays an important role in progenitors' cell fate decisions19,20 and in the development of the hematopoietic system21,22,23. For example, Notch1 activation could result in the differentiation of bone marrow cells into T cells19, and Jagged1 (a Notch ligand) can enhance the differentiation of mesenchymal stem cells into cardiomyocytes20. Activation of Notch1 signaling inhibits the differentiation of Sca+lin− bone marrow cells (a population of HSCs), thus impeding the cells from exiting the stem cell stage into the progenitor cell stage21 and indicating that HSC expansion can be achieved via Notch pathway activation. Since less information is available regarding to the role of Notch in hematopoietic generation from ESCs, we investigated whether the Notch signaling pathway was involved in HSC-EXs effects on hematopoietic differentiation in ESCs.
In this study, we aimed to investigate whether the EXs from HSCs (HSC-EXs) can promote HSC differentiation from ESCs via modulation of the miR126/Notch pathway.
Materials and methods
Culture of ESCs
Mouse ESCs were purchased from Cyagen Biosciences. The ESCs were cultured and maintained on feeder layers of mitomycin C-treated mouse embryonic fibroblasts (MEFs) in standard ESC complete medium containing knockout high-glucose DMEM supplemented with 15% fetal bovine serum (FBS), 1% nonessential amino acids, 1 mmol/L sodium pyruvate, 0.1 mmol/L 2-mercaptoethanol, 2 mmol/L glutamine, and 1000 U/mL leukemia inhibitory factor (LIF). The culture medium was changed daily. ESCs were regularly passaged every 48–72 h at a proportion of 1:10 on mitomycin C-treated MEF layers after dissociation using Tryple Express (Life Technologies). ESCs were cultured to form embryonic bodies (EBs) for induction of HSC differentiation24. In brief, ESCs were dissociated and cultured with ESC complete medium without leukemia inhibitory factor (LIF) on sterile bacteriological grade petri dish for 4 d. The resultant EBs were used for the differentiation study.
Preparation and collection of EXs released from HSCs
To overexpress miR126, HSCs (obtained from ATCC) were transfected with miR126 mimics or a miR126 scramble control (sc, 1 nmol/L, Thermo Fisher Scientific) for 24 h using Lipofectamine 2000 according to the manufacturer's instructions8. After the 24 h transfection, the cell culture medium was collected for HSC-EXs isolation. In brief, cell culture medium was collected and centrifuged at 300×g for 15 min, followed by 2000×g for 30 min to remove cells and cell debris. Then, the cell-free culture medium was centrifuged at 20 000×g for 70 min, followed by ultracentrifugation at 170 000×g for 6 h to pellet the EXs25. The pelleted HSC-EXscon, HSC-EXsmiR126, and HSC-EXssc were resuspended in phosphate-buffered saline (PBS) that had been passed through a 20 nm-filtered (Whatman, Pittsburgh, PA, USA), aliquoted for nanoparticle tracking analysis (NTA) and co-culture experiments.
Nanoparticle tracking analysis of EXs
The NanoSight NS300 with a 405-nm laser instrument (Malvern Instruments, UK) was used to detect EXs as previously reported25. Briefly, to determine particle size and concentration, the collected HSC-EXs were resuspended with 700 μL of filtered PBS and analyzed using the NTA light-scatter mode. To detect CD63 expression in the EXs, the collected EXs were incubated with CD63-conjugated microbeads (10 μL; Miltenyi Biotec) in a 100-μL reaction volume for 2 h. A magnet (DynaMag-2 magnet; Life Technologies) was then applied to separate the CD63+ EXs from the total EX suspension. After an overnight magnet separation, the CD63+ EXs were resuspended in 100 μL of filtered PBS and incubated with rabbit anti-goat IgG conjugated with Q-dot 655 (1:350; Life Technologies) for 90 min at RT. All samples were analyzed using the NTA fluorescence-scatter mode. Three videos of typically 30 s duration were taken with a frame rate of 30 frames per second. The Data were analyzed using NTA 3.0 software (Malvern Instruments) on a frame-by-frame basis.
Differentiation of HSCs from ESCs
The formed EBs were collected using a cut tip and trypsinized with TrypLE Express (Life Technologies). The EBs were then centrifuged and resuspended with HSC induction medium (knockout high-glucose DMEM supplemented with 15% FBS, 1% nonessential amino acids, 1 mmol/L sodium pyruvate, 0.1 mmol/L 2-mercaptoethanol and 2 mmol/L glutamine). The cells were then counted and reseeded onto 0.1% gelatin-coated plates (4×105 ESCs/well). To determinie the effects of HSC-EXs on HSC differentiation, the ESCs were treated with culture medium only (control) or 40 ng/mL of HSC-EXscon, HSC-EXssc or HSC-EXsmiR126 with or without soluble Jagged1 (100 ng/mL; R&D systems, Minneapolis, MN, USA) for a 10-d differentiation period. HSC-EXs and Jagged1 were added to the culture medium every three days9. At the end of the experiment, the cells were collected for flow cytometry, RT-PCR and Western blot analyses. The miR126 level in the ESCs was determined after 24 h co-culture and on d 10 after differentiation.
Flow cytometry analysis of the generated HSCs
After the 10-d differentiation period, the cells were washed with PBS, digested with 0.25% trypsin-0.02% EDTA, centrifuged and resuspended with 100 μL PBS. Then, each sample was incubated with PE-conjugated CD117, FITC-conjugated Sca1, or efluor 660-conjugated CD34 for 30 min in the dark. FITC-conjugated IgG, PE-conjugated IgG and efluor 660-conjugated IgG were used as isotype controls. After incubation, all samples were analyzed by flow cytometry (FACS canto flow cytometer)26, and 50 000 events were collected for data analysis. A gate (P1) was set on the FSC-A vs SSC-A plot for the collected cell population. A second gate (P2) was set on the CD117-conjugated PE-A vs SSC-A for the analysis of CD117+ cells. HSCs were defined as CD117+CD34+Sca1+ cells. The percentage of HSCs was calculated as CD117+CD34+Sca1+ cells/total cells×100%.
Isolating HSCs from the differentiating ESCs
To determine whether the generated HSCs were functional, we used magnetic activated cell sorting (MACS) to isolate the generated HSCs with anti-CD117 microbeads (Miltenyi Biotec) according to the manufacturer's instructions. In brief, after the 10-d differentiation period, ESCs from different groups were harvested and incubated with anti-CD117 microbeads antibody (10 μL anti-CD117 microbeads per 107 total cells) in a 100 μL reaction volume for 20 min in the refrigerator. Then, the CD117+ cells in each group were separated using a magnet separator (DynaMag-2 magnet; Life Technologies) as previously reported27. The isolated CD117+ cells were used for the colony-forming unit (CFU) assay described below.
Hematopoietic colony-forming unit assay
The colony-forming unit (CFU) assay is a commonly used in vitro functional assays for HSCs28. The CFU assay for the isolated CD117+ HSCs was performed using MethoCult™ medium (Stem Cell Technologies) according to the manufacture's instructions. Briefly, 1×104 dissociated single CD117+ cells were mixed with methylcellulose medium supplemented with stem cell factor (100 ng/mL), granulocyte-macrophage-CSF (100 ng/mL), IL-3 (10 ng/mL), IL-6 (10 ng/mL), granulocyte colony-stimulating factor (100 ng/mL), and erythropoietin (4 units/mL) in a 35 mm petri dish and cultured for 12 d. The number of colonies formed was counted under an inverted microscope (EVOS, 10× magnification).
Quantitative RT-PCR analysis
Total RNA was extracted from cells using Tri-reagent (Sigma). The cDNA was generated by reverse transcription with a cDNA synthesis kit (Qiagen) in a 20 μL volume according to manufacturer's instructions. The qRT-PCR analysis was performed by using SYBR green II qPCR Premix (Takara, Japan) in 96-well PCR plates on an ABI PRISM 7500 real-time PCR system (Bio-tek). The PCR conditions were 95 °C for 10 min and 50 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. U6 was used as the internal control. The relative expression of each gene was calculated using the 2−ΔΔCT method29.
Western blot analysis
After the 10-d differentiation period, cell proteins were extracted with cell lysis buffer (Thermo Fisher Scientific) supplemented with a protease inhibitor tablet (Roche). Protein lysates were electrophoresed with SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% non-fat milk for 1 h and incubated with primary antibodies against notch intracellular domain (NCID, 1:500; Abcam), Hes1 (1:100; Santa Cruz) or β-actin (1:4000; Sigma) at 4 °C overnight. After washing with T-PBS, the membranes were incubated with horseradish-peroxidase-conjugated IgG (Jackson Immuno Research Lab) for 1 h at RT. Blots were developed with enhanced chemiluminescence developing solutions and quantified using ImageJ software.
Statistical analysis
Each experimental measurement was repeated four times. The data are expressed as the mean±SEM. Multiple comparisons were analyzed by one- or two-way ANOVA followed by an LSD post-hoc test. SPSS 17.0 statistical software was used. For all measurements, P<0.05 was considered significant.
Results
MiR126 mimics increased miR126 levels in HSCs and their released EXs
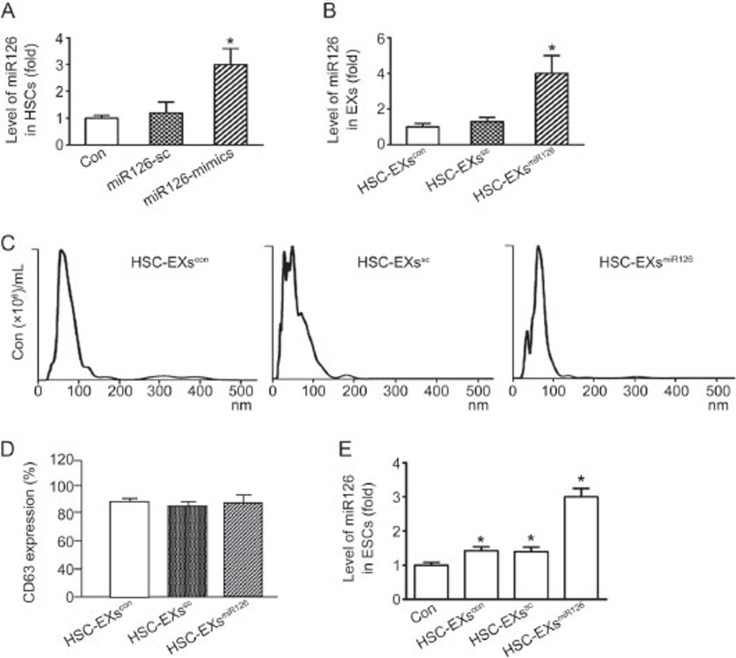
After 24-h transfection with sc or miR126-mimics, HSCs were collected for real time RT-PCR analysis. As shown in Figure 1A, the miR126 level was significantly increased in HSCs transfected with miR126-mimics when compared to that in HSCs transfected with control or sc. These results suggested the successful miR126 transfection.
Figure 1.
Analysis of miR126 expression in HSCs, HSC-EXs and ESCs co-cultured with various types of HSC-EXs. (A) miR126-mimic transfection increased the level of miR126 expression in HSCs. (B) HSC-EXs released from HSCs transfected with miR126 mimics carried a high level of miR126. (C) Representative image showing the size distribution of HSC-EXs analyzed by NTA. (D) CD63 expression in the HSC-EXs. (E) miR126 levels in the ESCs after 24 h co-culture. *P<0.05 vs con. Data are expressed as the mean±SEM. n=4.
Using real time RT-PCR, we confirmed that the HSC-EXsmiR126 had a higher level of miR126 expression than HSC-EXscon and HSC-EXssc did (Figure 1B), which suggested successful enrichment of miR126 expression in the HSC-EXs. Meanwhile, the sizes of the HSC-EXs collected from the culture medium were assessed by nanoparticle tracking analysis using the NS300 instrument. As shown in Figure 1C, the sizes of the HSC-EXscon, HSC-EXsmiR126, and HSC-EXssc ranged from 30–120 nm in diameter, which was consistent with previous reports by others7,9 and us25. In addition, we characterized the HSC-EXs using the EX specific marker CD63. Our results showed that over 85% of EXs expressed CD63 (Figure 1D).
HSC-EXsmiR126 co-culture increased miR126 expression in ESCs
After 24-h co-culture with HSC-EXsmiR126, we determined the miR126 levels in ESCs. As shown in Figure 1E, the miR126 levels in ESCs were significantly elevated following HSC-EXscon and HSC-EXssc co-culture compared with the control. The expression of miR126 was further increased by HSC-EXsmiR126 co-culture.
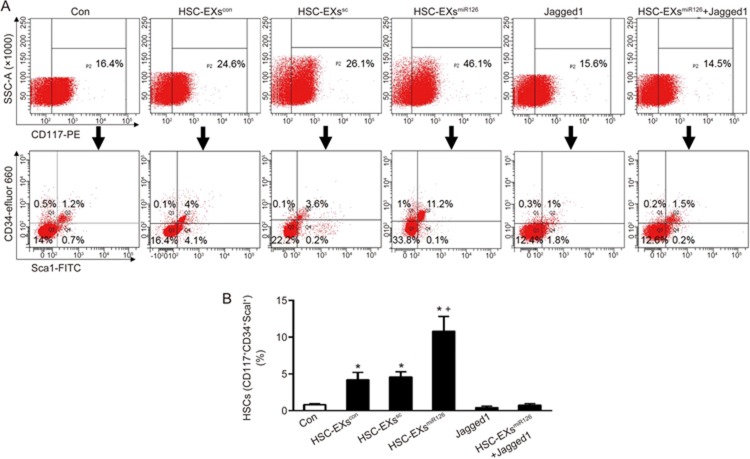
HSC-EXsmiR126 promoted hematopoietic differentiation of ESCs via the miR126/Notch1 pathway to a greater extent than HSC-EXscon and HSC-EXssc did
ESCs were induced for HSC differentiation in the presence of various HSC-EXs. The differentiation rate was analyzed by flow cytometry using the hematopoietic specific markers CD34, CD117 and Sca1. As shown in Figure 2, the percentage of HSCs (defined as CD34+CD117+Sca1+ cells) was higher in the HSC-EXscon treatment group than in the control. We also found that HSC-EXsmiR126 treatment further remarkably increased ESC hematopoietic differentiation (versus HSC-EXssc). The Notch1 ligand Jagged1 did not significantly change the ESC differentiation rate (versus control), although, it significantly abolished the effect of HSC-EXsmiR126 on promoting hematopoietic differentiation.
Figure 2.
HSC-EXsmiR126 co-culture remarkably promoted the differentiation of ESCs into HSCs. (A) Representative flow cytometry plots showing the percentage of HSCs after 10-d differentiation. (B) Summarized data showing that HSC-EXscon and HSC-EXssc treatment promoted the differentiation of ESCs into HSCs, with the greatest differentiation promoted by HSC-EXmiR126. This effect was attenuated by Jagged1. *P<0.05 vs con. +P<0.05 vs HSC-EXscon, or HSC-EXssc, or Jagged1, or HSC-EXsmiR126+Jagged1. Data are expressed as the mean±SEM. n=4.
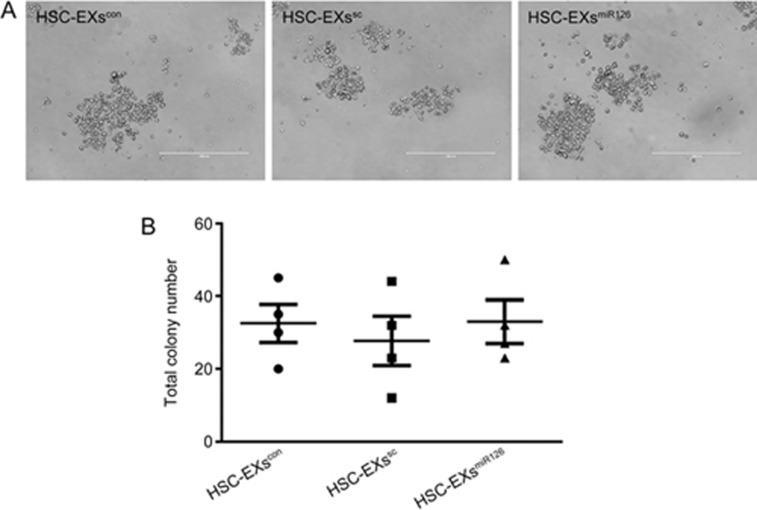
The generated HSCs had self-renewal capability
After the 10-d differentiation period, the generated HSCs were isolated by MACS with anti-CD117 microbeads to analyze their self-renewal ability. The CFU assay showed that the generated HSCs were able to form colony units after 12-d culture in methylcellulose medium (Figure 3).
Figure 3.
Isolated CD117+ cells had colony-forming abilities. (A) Representative images showing the colonies in different groups. (B) Summarized data showing the total number of colonies produced by the isolated CD117+ cells on methylcellulose medium. Each square represents the total colonies from one plate.
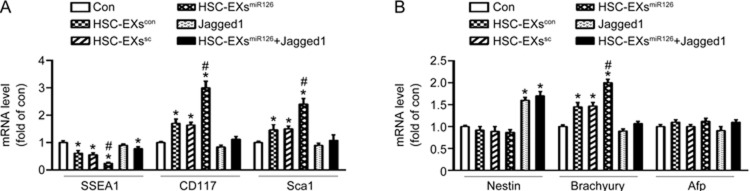
HSC-EXsmiR126 modulated the expression of pluripotent/hematopoietic and mesoderm commitment genes through the miR126/Notch1 pathway to a greater extent than HSC-EXscon and HSC-EXssc did
We analyzed the expression of the pluripotency-related gene SSEA1 and hematopoietic lineage-related genes CD117 and Sca1 in the differentiating ESCs. Our data (Figure 4A) showed that the mRNA level of SSEA1 was decreased, whereas the mRNA levels of CD117 and Sca1 were increased in the differentiating ESCs treated with HSC-EXscon, HSC-EXssc or HSC-EXsmiR126. The HSC-EXsmiR126 exhibited stronger effects than HSC-EXscon or HSC-EXssc did. Activation of Notch1 pathway with Jagged1 alone did not change the mRNA levels of SSEA1, CD117 or Sca1 (versus control), whereas, it significantly abolished the effects of HSC-EXsmiR126 on the mRNA levels of SSEA1, CD117 and Sca1.
Figure 4.
RT-PCR analysis of specific lineage-related genes from ESCs after 10-d differentiation. (A) The mRNA level of the pluripotent gene SSEA1 was decreased, whereas the mRNA levels of the HSC genes CD117 and Sca1 were increased in the ESCs treated with HSC-EXscon and HSC-EXssc that underwent 10-d differentiation. HSC-EXsmiR126 enhanced HSC differentiation which was abolished by Jagged1. (B) The mRNA levels of markers in three germ layers, Nes (ectoderm), Bry (mesoderm) and Afp (endoderm). HSC-EXmiR126 increased mRNA level of Bry, which was inhibited by Jagged1. *P<0.05 vs Con. #P<0.05 vs HSC-EXscon, or HSC-EXssc, or Jagged1, or HSC-EXsmiR126+Jagged1. Data are expressed as the mean±SEM. n=4.
We also measured the expression of the germ-layer early commitment genes Nes (Nestin, for ectoderm), Bry (Brachyury, for mesoderm) and Afp (α-feto protein, for endoderm) in various HSC differentiation groups. We found that HSC-EXscon or HSC-EXssc treatment alone significantly increased the mRNA expression of Bry, which was further increased by HSC-EXsmiR126 treatment. Jagged1 blocked the increase in Bry mRNA expression elicited by HSC-EXsmiR126 and upregulated the mRNA levels of Nes in the differentiating ESCs. No significant changes in Afp expression were observed, suggesting that HSC-EXs could not affect endoderm differentiation of ESCs in our cell culture system (Figure 4B).
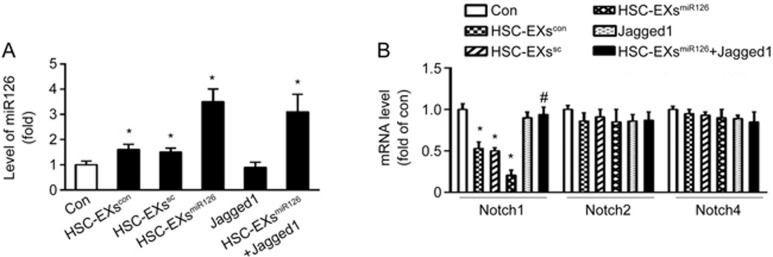
HSC-EXsmiR126 up-regulated miR126 expression and down-regulated Notch1 mRNA levels in ESCs on d 10
As shown in Figure 5A, HSC-EXscon and HSC-EXssc increased the miR126 levels in the differentiating ESCs. HSC-EXsmiR126 treatment remarkably increased the expression of miR126, which was not altered by Jagged1. These data suggested that HSC-EXs can modulate the expression of miR126 in differentiating ESCs.
Figure 5.
HSC-EXs increased miR126 levels and decreased mRNA levels of Notch1 in ESCs on d 10. (A) HSC-EXs increased the level of miR126 in the differentiating ESCs. (B) The summarized data showing that the expressions of Notch1, Notch2 and Notch4 in the cells that underwent 10-d differentiation. *P<0.05 vs con. #P<0.05 vs HSC-EXscon, HSC-EXssc, HSC-EXsmiR126, or Jagged1. Data are expressed as the mean±SEM. n=4.
To explore whether the Notch signaling pathway was involved in this process, we analyzed the mRNA expression levels of Notch receptors 1, 2 and 4 on the differentiating ESCs. We found that the HSC-EXscon and HSC-EXssc significantly decreased the mRNA levels of Notch1. HSC-EXsmiR126 decreased Notch1 levels to the greatest extent, and this effect was blocked by Jagged1. The expression levels of Notch2 and Notch4 were not different among groups (Figure 5B). These findings suggested that HSC-EXsmiR126 promoted HSC differentiation by inhibiting the Notch1 pathway.
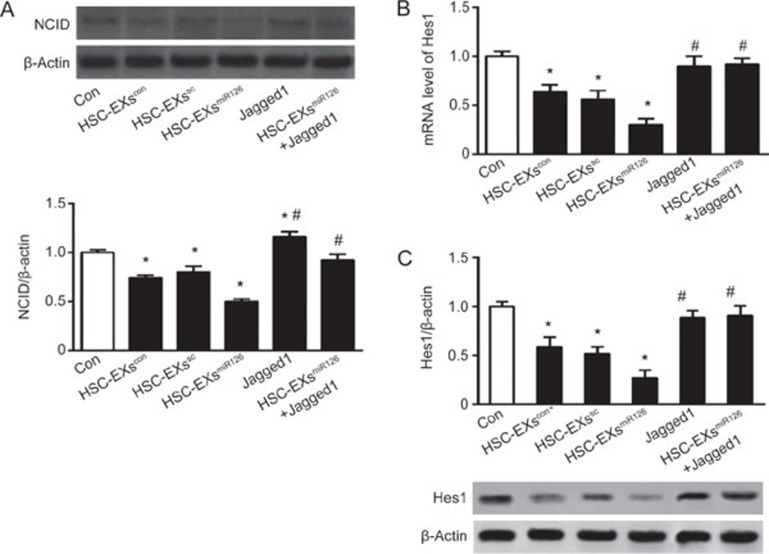
Enrichment of miR126 in HSC-EXs promoted the downregulation of NCID/Hes1 in ESCs on d 10
As shown in Figure 6A, NCID expression was significantly decreased in the differentiating ESCs treated with HSC-EXscon or HSC-EXssc as compared with that of the control group in, which NCID expression was further downregulated by HSC-EXsmiR126. Jagged1 alone increased NCID expression and reversed the downregulation of NCID induced by HSC-EXsmiR126. These data suggest that HSC-EXsmiR126 can inhibit Notch signaling in the differentiating ESCs.
Figure 6.
HSC-EXs down-regulated NCID expression and the mRNA and protein levels of Hes1 in ESCs on d 10. (A) Top: representative bands of NCID and β-actin. Bottom: summarized data showing the expression of NCID in the differentiating cells; (B, C) The summarized data showing the mRNA and protein levels of Hes1 in the differentiating ESCs. *P<0.05 vs Con. #P<0.05 vs HSC-EXscon, HSC-EXssc, or HSC-EXsmiR126. Data are expressed as the mean±SEM. n=4.
Hes1 is a major downstream gene of the Notch1 signaling pathway. Compare with the HSC-EXscon and HSC-EXssc, HSC-EXsmiR126 decreased Hes1 mRNA to a greater extent in the differentiating cells. Jagged 1 inhibited this effect (vs HSC-EXsmiR126, P<0.05). Similarly, Western blot analysis demonstrated that the expression of Hes1 was down-regulated by HSC-EXscon and HSC-EXssc. The HSC-EXsmiR126 further decreased Hes1 expression in the differentiating ESCs, and this effect was blocked by Jagged1 (Figure 6B, 6C). The data further confirmed that HSC-EXs can modulate the miR126/Notch/Hes1 pathway.
Discussion
The major findings of the present study are that HSC-EXs can promote HSC differentiation from ESCs and that overexpression of miR126 can increase the efficiency of HSC-EXs-mediated differentiation. In addition, our data suggested that miR126-mediated inhibition of the Notch1/Hes1 pathway is the underlying mechanism for this process.
EXs, which are small vesicles derived from intracellular multivesicular bodies of various cell types3,30, are enriched in bioactive molecules such as proteins, mRNAs and miRs from the cells of origin. EXs are released constitutively or in response to different stimuli and impact the genetic content and function of recipient cells by delivering bioactive molecules4,6,7. In this study, we found that HSC-EXs co-culture promoted the differentiation of ESCs into CD117+CD34+Sca1+ HSCs, indicating the important role of EXs in hematopoietic differentiation. This finding is supported by previous studies that showed the participation of EXs in neuronal9 or macrophage differentiation10, which represents a novel feedback mechanism from the differentiated cells.
It is well known that miRs are important regulators of hematopoiesis, and miR126 is highly expressed in HSC compartments and downregulated during hematopoietic commitment14. Our previous study demonstrated that EX-carried miR126 can be delivered to endothelial cells4. Nevertheless, whether miR126 participates in the hematopoietic differentiation of ESCs elicited by HSC-EXs remains elusive. We determined that miR126 expression was higher in the HSC-EXscon co-cultured cells. To further explore the role of miR126 in the differentiation of ESCs in HSCs, we overexpressed miR126 in HSCs to obtain EXs with high levels of miR126 (HSC-EXsmiR126). Interestingly, our data revealed that HSC-EXsmiR126 remarkably increased the ESC hematopoietic differentiation rate, indicating that miR126 is the major underlying mechanism responsible for the effect of HSC-EXs on ESC differentiation. However, whether miR126 alone can affect hematopoietic differentiation of ESCs requires further study. Meanwhile, knock down of miR126 using inhibitors could be used to further confirm the role of miR126 in ESC hematopoietic differentiation. Whether other sources of EXs such as endothelial cell-derived EXs and fibroblast cell derived EXs could offer such a regulatory effect requires more investigation. In the present study, to test whether the generated HSCs were functional, we isolated the generated HSCs using MACS with anti-CD117 conjugated microbeads and performed a colony formation unit (CFU) assay in vitro. The generated HSCs were capable of forming colony units on methylcellulose medium. Meanwhile, we found that there was no difference in colony unit formation among the HSCs generated from ESCs treated with HSC-EXscon, HSC-EXssc, or HSC-EXsmiR126. These findings suggested that miR126 can promote HSC differentiation from ESCs but does not affect the function of the generated HSCs.
Previous studies have revealed that activation of Notch signaling in ESCs leads to preferential neuroectoderm fate determination, whereas inactivation of this pathway facilitates the mesodermal fate31,32,33. In this study, we found that ESCs co-cultured with HSC-EXscon or HSC-EXssc promoted differentiation into the Brachyury-positive mesodermal lineage, and the HSC-EXsmiR126 enhanced this effect. These data are line with the observations made using the hematopoietic specific markers CD117 and Sca1. Here, we found that Jagged1 blocked the effect of HSC-EXsmiR126 and promoted the expression of Nestin (ectoderm lineage) in the differentiating ESCs, which is in agreement with a previous study showing that overexpression of Jagged1 can promote the differentiation of ESCs under neural differentiation induction18. Collectively, these data indicated that HSC-EXs can regulate fate choice in ESCs through the miR126/Notch signaling pathway.
Cellular receptor expression varies among different cell types, which elicits distinct responses during differentiation process. Jagged1-mediated Notch signaling results in enhanced Hes5 expression and diminished Hey1 expression, leading to the differentiation of ESCs toward a neural lineage, whereas, Delta-like 4 reverses the expression pattern of these genes and results in mesodermal commitment of ESCs18. In this study, we found that HSC-EXsmiR126 co-culture down-regulated Notch1 mRNA levels, which led to the differentiation of ESCs towards HSCs. Our findings are supported by a previous study indicating that ectopic expression of Notch intracellular domain suppressed hematopoietic differentiation of FLK+ mesodermal progenitors34.
It is well-known that the Notch signaling involves the Hes and Hey family of transcription co-repressors downstream. Hes1 is a direct target gene of Notch and is highly expressed in ESCs35,36. Our results showed that expression of Hes1 in the differentiating cells was significantly decreased by HSC-EXscon and HSC-EXssc and more so by HSC-EXsmiR126. These findings suggested that enrichment of miR126 can enhance the inhibition of HSC-EXs on Notch signaling in the differentiating ESCs. To directly elucidate the relationship between HSC-EXsmiR126 and the Notch pathway, we added Jagged1, a Notch ligand, to activate the Notch pathway in the differentiating ESCs. As expected, Jagged1 activated Notch signal as evidenced by the increased level of NCID expression. Moreover, it reversed the downregulation of Hes1 induced by HSC-EXsmiR126, which supported the observation that HSC-EXsmiR126 can inhibit the Notch pathway. Taken together, these findings suggest that HSC-EXs can promote HSC differentiation through the miR126-modulated Notch1/Hes1 pathway.
Conclusions
In conclusion, HSC-EXs accelerate HSC differentiation from ESCs by modulating the miR126/Notch1 signal pathway, providing a novel approach to increase the production of HSCs for clinical use.
Author contribution
Yanfang CHEN, Bin ZHAO, Ji BIHL, Yi YANG and Ri-ling CHEN designed the experiments; Feng-ling LIAO, Lin TAN and Xiao-tang MA performed the experiments; Feng-ling LIAO, Lin TAN, Xiao-tang MA, Ri-ling CHEN, Jin-ju WANG, Yanfang CHEN and Ji BIHL analyzed the data; Feng-ling LIAO, Lin TAN, Hua LIU, Jin-ju WANG, Bin ZHAO, Yi YANG, Ri-ling CHEN and Yanfang CHEN wrote the paper. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No 81400360); the Science and Technology Planning Project of Zhanjiang (No [2012] 172); and the Competitive Project of Zhanjiang (2014A01022).
References
- Gori JL, Butler JM, Chan YY, Chandrasekaran D, Poulos MG, Ginsberg M, et al. Vascular niche promotes hematopoietic multipotent progenitor formation from pluripotent stem cells. J Clin Invest 2015; 125: 1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman S, Daley G. Derivation of hematopoietic stem cells from murine embryonic stem cells. J Vis Exp 2007; (2): 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuda T, Kosaka N, Takeshita F, Ochiya T. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics 2013; 13: 1637–53. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen S, Ma X, Cheng C, Xiao X, Chen J, et al. Effects of endothelial progenitor cell-derived microvesicles on hypoxia/reoxygenation-induced endothelial dysfunction and apoptosis. Oxid Med Cell Longev 2013; 2013: 572729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanueli C, Shearn AI, Angelini GD, Sahoo S. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascul Pharmacol 2015; 71: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res 2010; 107: 1047–57. [DOI] [PubMed] [Google Scholar]
- Xin H, Li Y, Liu Z, Wang X, Shang X, Cui Y, et al. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells 2013; 31: 2737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Yang Y, Zhong Y, Ammar HM, Zhang P, Guo R, et al. The effects of microvesicles on endothelial progenitor cells are compromised in type 2 diabetic patients via downregulation of the miR-126/VEGFR2 pathway. Am J Physiol Endocrinol Metab 2016; 310: E828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda YS, Xu Q. Neuronal differentiation of human mesenchymal stem cells using exosomes derived from differentiating neuronal cells. PLoS One 2015; 10: e0135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 2013; 121: 984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Gschweng E, Van HB, Cheng D, Mikkola HK, Witte ON. Regulated expression of microRNAs-126/126* inhibits erythropoiesis from human embryonic stem cells. Blood 2011; 117: 2157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83–6. [DOI] [PubMed] [Google Scholar]
- Nikolic I, Plate KH, Schmidt MH. EGFL7 meets miRNA-126: an angiogenesis alliance. J Angiogenes Res 2010; 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 2012; 11: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Zhu X, Hu XQ, Fang ZF, Tang L, Lu XL, et al. Mesenchymal stem cells modified with miR-126 release angiogenic factors and activate Notch ligand Delta-like-4, enhancing ischemic angiogenesis and cell survival. Int J Mol Med 2013; 31: 484–92. [DOI] [PubMed] [Google Scholar]
- Dikic I, Schmidt MH. Notch: Implications of endogenous inhibitors for therapy. Bioessays 2010; 32: 481–7. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006; 7: 678–89. [DOI] [PubMed] [Google Scholar]
- Ramasamy SK, Lenka N. Notch exhibits ligand bias and maneuvers stage-specific steering of neural differentiation in embryonic stem cells. Mol Cell Biol 2010; 30: 1946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 1999; 11: 299–308. [DOI] [PubMed] [Google Scholar]
- Li H, Yu B, Zhang Y, Pan Z, Xu W, Li H. Jagged1 protein enhances the differentiation of mesenchymal stem cells into cardiomyocytes. Biochem Biophys Res Commun 2006; 341: 320–5. [DOI] [PubMed] [Google Scholar]
- Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood 2002; 99: 2369–78. [DOI] [PubMed] [Google Scholar]
- Ohishi K, Varnum-Finney B, Bernstein ID. Delta-1 enhances marrow and thymus repopulating ability of human CD34+CD38− cord blood cells. J Clin Invest 2002; 110: 1165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Ye Q, Moore MA. A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic progenitor cells. Blood 2000; 95: 1616–25. [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008; 132: 661–80. [DOI] [PubMed] [Google Scholar]
- Wang J, Guo R, Yang Y, Jacobs B, Chen S, Iwuchukwu I, et al. The novel methods for analysis of exosomes released from endothelial cells and endothelial progenitor cells. Stem Cells Int 2016; 2016: 2639728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A 2009; 75: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhong Y, Ma X, Xiao X, Cheng C, Chen Y, et al. Analyses of endothelial cells and endothelial progenitor cells released microvesicles by using microbead and Q-dot based nanoparticle tracking analysis. Sci Rep 2016; 6: 24679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood 2006; 108: 2095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Liu H, Zheng C, Zhao Y, Liao X, Wang Y, et al. Microvesicles derived from inflammation-challenged endothelial cells modulate vascular smooth muscle cell functions. Front Physiol 2016; 7: 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008; 10: 619–24. [DOI] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol 2006; 4: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T, Fraser ST, Ogawa M, Nishikawa S, Oka C, Bornkamm GW, et al. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc Natl Acad Sci U S A 2003; 100: 4018–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res 2006; 98: 1471–8. [DOI] [PubMed] [Google Scholar]
- Huang C, Jackson M, Samuel K, Taylor AH, Lowell S, Forrester LM. Haematopoietic differentiation is inhibited when Notch activity is enhanced in FLK1+ mesoderm progenitors. Stem Cell Res 2013; 11: 1273–87. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature 1995; 377: 355–8. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Mizuno H, Imayoshi I, Furusawa C, Shirahige K, Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev 2009; 23: 1870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]