Abstract
Gram-negative bacterium-released outer-membrane vesicles (OMVs) and Gram-positive bacterium-released membrane vesicles (MVs) share significant similarities with mammalian cell-derived MVs (eg, microvesicles and exosomes) in terms of structure and their biological activities. Recent studies have revealed that bacterial OMVs/MVs could (1) interact with immune cells to regulate inflammatory responses, (2) transport virulence factors (eg, enzymes, DNA and small RNAs) to host cells and result in cell injury, (3) enhance barrier function by stimulating the expression of tight junction proteins in intestinal epithelial cells, (4) upregulate the expression of endothelial cell adhesion molecules, and (5) serve as natural nanocarriers for immunogenic antigens, enzyme support and drug delivery. In addition, OMVs/MVs can enter the systemic circulation and induce a variety of immunological and metabolic responses. This review highlights the recent advances in the understanding of OMV/MV biogenesis and their compositional remodeling. In addition, interactions between OMVs/MVs and various types of mammalian cells (ie, immune cells, epithelial cells, and endothelial cells) and their pathological/preventive effects on infectious/inflammatory diseases are summarized. Finally, methods for engineering OMVs/MVs and their therapeutic potential are discussed.
Keywords: bacterium-released membrane vesicles, microvesicles, inflammation, vaccine, infectious diseases
Introduction
Shedding of membrane vesicles (MVs) has been recognized as a universal mechanism for intercellular communication and is evolutionarily conserved across eukaryotes, bacteria and archaea1,2,3,4. These extracellular MVs are nanosized, spherical, bilayered proteolipids harboring specific subsets of bioactive proteins, lipids, nucleic acids, and metabolites. Notably, diverse names have been employed to indicate different species-derived extracellular vesicles1,2,3,4. For example, MVs for those of archaea and Gram-positive bacterial origin1,2,3; outer-membrane vesicles (OMVs) for those of Gram-negative bacterial origin1,2,3; and exosomes/microvesicles for those of mammalian cell origin4. Over the past several decades, tremendous efforts have been aimed at elucidating the versatile roles of mammalian cell-released MVs (ie, microvesicles and exosomes) in processes of human disease and injury repai4,5,6,7,8,9,10,11. However, bacterium-released MVs, especially their interactions with mammalian cells and their eventual impact on human health and disease, have received less attention. The first observation of bacterial MVs was reported by Chatterjee and Das in 1967 and revealed that the cell wall of Vibrio cholerae was bulged out and appeared to be pinched off the bacterial surface12. Since then, Gram-negative bacterial OMV-related studies have made steady progress, and the number of publications has increased rapidly in the last 5 years. Furthermore, direct evidence of MV formation in Gram-positive bacteria (eg, Staphylococcus aureus, Bacillus subtilis, Bacillus anthracis, and Streptomyces coelicolor) has also recently been obtained using transmission electron microscopic and proteomic analyses13,14,15,16. Most studies have focused on the analysis of OMVs produced by Gram-negative bacteria, including both pathogenic and non-pathogenic bacteria, whereas the knowledge of MVs secreted from Gram-positive bacteria is limited. Nonetheless, current evidence suggests that both OMVs and MVs can transport their cargo to recipient cells of the same species, other bacterial species, or mammalian cells 17,18,19,20,21. Accordingly, bacterial OMVs/MVs have been shown to play diverse and critical roles in nutrient uptake, antimicrobial defense, horizontal gene transfer, biofilm nucleation, and the trafficking of microbial products such as virulence factors and toxins during infection19,20,21,22,23,24,25. The similarities and differences between MVs and OMVs are briefly listed in Table 1. In this review, we highlight recent findings on 1) the biogenesis and characterization of Gram-negative bacterium-released OMVs and Gram-positive bacterium-released MVs; 2) interactions between OMVs/MVs and mammalian cells; and 3) the pathological/beneficial effects of OMVs/MVs on infectious diseases. Finally, we discuss how to engineer these bacterial MVs as therapeutic agents for the treatment of human diseases.
Table 1. The similarities and differences between MVs and OMVs.
| Membrane vesicles (MVs) | Outer membrane vesicles (OMVs) | |
|---|---|---|
| Origin | Cytoplasmic membrane of Gram positive bacteria | Outer membrane of Gram negative bacteria |
| Size | ∼20–150 nm in diameter | ∼10–300 nm in diameter |
| Components | • Various fatty acids and phospholipids; • Cytoplasmic proteins; • Membrane-associated virulence proteins/enzymes; • lipoteichoic acid (LTA); • Peptidoglycan • DNAs • sRNAs | • Outer membrane proteins; • Periplasmic proteins and virulence factors; • Less cytoplasmic and inner-membrane proteins; • LPSs and phospholipids; • Peptidoglycan • DNAs • sRNAs |
Biogenesis of bacterial MVs
It has been recognized that all Gram-negative bacteria can naturally produce OMVs, which are typically ∼10–300 nm in diameter (Figure 1A)25. Gram-positive bacteria lack an outer membrane, but some strains (eg, S. aureus, B. anthracis, Listeria monocytogenes, Bacillus subtilis, and Clostridium perfringens) also generate MVs (∼20–150 nm in diameter) that are derived from the cytoplasmic membrane (Figure 1B)13. Nonetheless, how Gram-positive bacteria generate MVs remains unclear. Most of the knowledge presented here regarding the biogenesis of bacterial MVs has been collected from studies of Gram-negative bacteria, as summarized below.
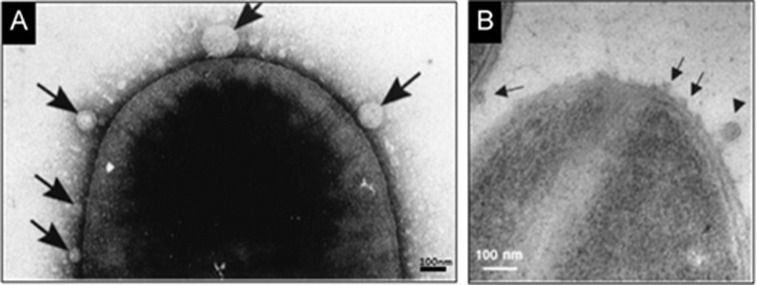
Figure 1.
Transmission electron micrograph of bacteria reveals: (A) OMVs released by Gram-negative bacteria, Serratia marcescens UG-159 (arrows) (This image is adapted from Ref22, Li Z, et al. J Bacteriol 1998); (B) MVs released by Gram-positive bacteria S. aureus (arrows) (This image is adapted from Ref79, Lee EY, et al. Proteomics, 2009).
The envelope structure of Gram-negative bacterium
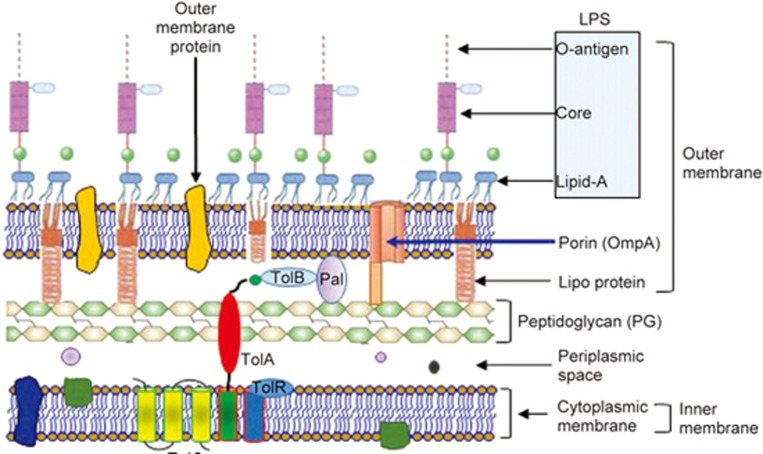
It is becoming clear that multiple mechanisms are involved in the production of OMVs in Gram-negative bacteria26. To easily understand the process of OMV budding and detachment, it is necessary to comprehend the unique architecture of the Gram-negative envelope (Figure 2). The cell wall of Gram-negative bacteria consists of the outer membrane, the cytoplasmic (inner) membrane, and the periplasmic space in between, which contains a layer of peptidoglycan (PG)27. The outer membrane is composed of an exterior leaflet of lipopolysaccharide (LPS: O-antigen, core and lipid A) and an interior leaflet of phospholipids (Figure 2). The inner cytoplasmic membrane consists of a typical phospholipid bilayer that serves as an electrochemical barrier. The periplasm is an oxidative environment that promotes protein folding but does not contain nucleotide sources of energy, such as ATP or GTP28. The net-like PG layer within the periplasm maintains the bacterial shape and provides protection from osmotic changes and shear stress. Envelope proteins are either soluble (periplasmic proteins), membrane associated, integral or anchored into the leaflet of either membrane via covalently attached lipid appendages (lipoproteins). There are multiple crosslinks within the envelope that are important for maintaining the stability of the bacterial wall29,30,31,32,33. These crosslinks include (Figure 2): 1) the covalent crosslinking of Braun's lipoprotein (Lpp) in the outer membrane with the PG sacculus29,30,31; 2) the non-covalent interactions between the PG and outer-membrane protein A (OmpA), which is an outer-membrane porin32,33; and 3) the non-covalent interactions between the PG and the Tol–Pal (peptidoglycan-associated lipoprotein) complex, which spans the envelope from the outer membrane across the periplasm to the cytoplasmic membrane32,33. In general, OMV biogenesis relies on the dissociation of the outer membrane from the underlying PG in areas devoid or depleted of attachments and subsequent fission without compromising envelope integrity34.
Figure 2.
The architecture of the Gram-negative cell envelope. The envelope consists of an outer membrane (OM), an inner membrane (IM), and a periplasm between the two membranes, which contains a thin layer of peptidoglycan (PG).
The models proposed for OMV biogenesis
Depletion/reduction of envelope crosslinks
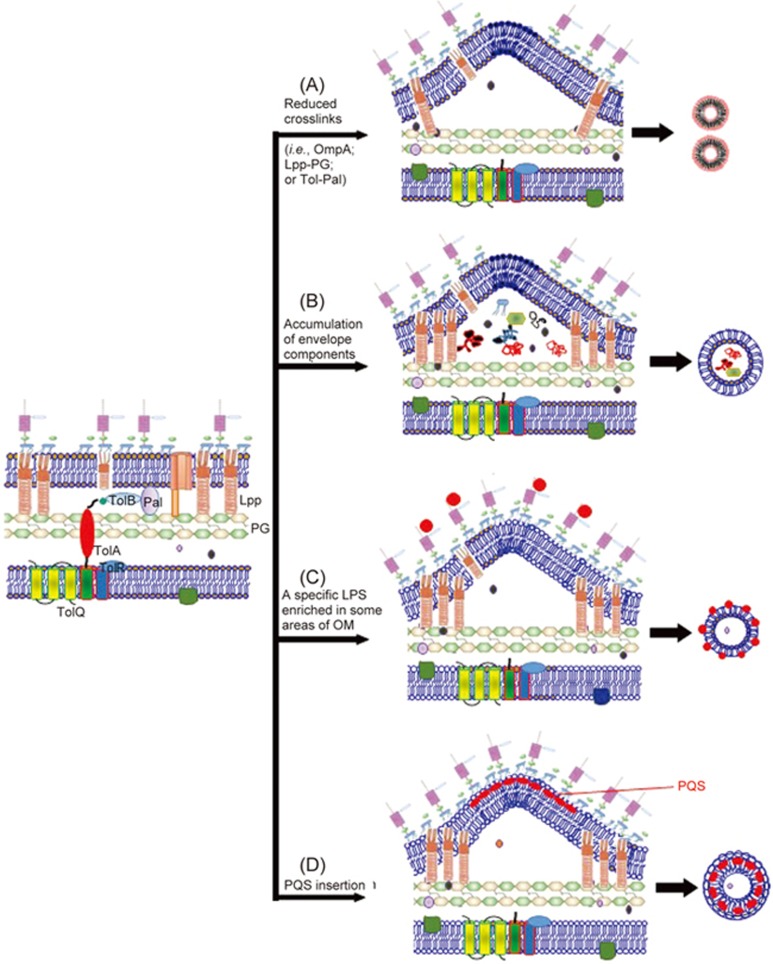
It is well known that crosslinks bridge the outer membrane to the PG in most Gram-negative bacteria (eg, E. coli). For example, OmpA is an outer-membrane porin that contains a periplasmic binding site for diaminopimelic acid (DAP), a PG component35. The Tol–Pal complex is a cell-division component that aids in invagination of the outer membrane and in membrane stability and interacts with PG32,36,37. Lpp is an extremely abundant outer-membrane lipoprotein, one-third of which is covalently crosslinked to PG. Lpp is evenly distributed throughout the entire cell wall, whereas Pal is preferentially located at the cell poles36,37. Accordingly, bacteria lacking OmpA (eg, E. coli, Salmonella spp and Acinetobacter baumannii mutants) display increased OMV production, which is plausibly ascribed to decreased crosslinking between PG and the outer membrane38,39,40,41. Similarly, disruption of the Tol–Pal links causes membrane instability and increased OM shedding32,36,37. Mutations have further shown that vesiculation levels are dependent upon the proteins that crosslink the OM to the cell wall32,36,37. Nonetheless, while either a temporary decrease in overall crosslink abundance or a localized displacement of crosslinks is thought to increase OMV biogenesis, a complete lack of Lpp-PG crosslinks can cause membrane instability, leading to cellular leakage32,39,41. However, in some cases, a partial reduction in the number of Lpp-PG crosslinks could increase OMV production42. For instance, the amount of Lpp crosslinked to PG in the hyper-vesiculating nlpI E. coli mutant was ∼40% lower than that in wild-type E. coli42. NlpI is an outer-membrane lipoprotein that was recently discovered to control the activity of Spr, a PG endopeptidase that cleaves peptide crosslinks in PG42,43,44. Therefore, mutation of nlpI in E. coli prevents the formation of proper crosslinks between PG and Lpp and eventually leads to increased OMV production. Recent studies have also shown that the generation of OMVs in Neisseria meningitidis was affected by PG architecture, as OMVs from this bacterium were found to contain lower levels of the three lytic transglycosylases MltA, MltB and Slt45. Together, these observations support a model in which OMVs bud off at sites with locally decreased levels of crosslinks between the outer membrane and PG and with locally reduced PG hydrolase activity (Figure 3A).
Figure 3.
Proposed models for the biogenesis of outer membrane vesicles (OMVs). (A) The linkage between the outer membrane and the underneath peptidoglycan layer is disrupted. (B) A physical force induced by accumulation of misfiled or overexpressed envelope proteins pushes out outer membrane vesicles. (C) The accumulation of LPS molecules with atypical structures or charges leads to the curvature of outer membrane. (D) Local curvature of bacterial outer membrane is stimulated by extracellular signals (ie, PQS).
Accumulation of misfolded proteins or envelope components in the periplasmic space
Schwechheimer et al46 recently investigated several bacterial mutants and discovered that increased OMV production is not solely dependent on Lpp crosslinking; rather, it can also occur via a mechanism that is independent of the total levels of PG-bound Lpp. These hypervesiculating bacteria exhibited accumulation of misfolded proteins or envelope proteins, PG fragments and/or aberrant LPS in the periplasmic space46. Such accumulations can be triggered by, for example, defects in cell wall remodeling or temperature stress47,48,49. Following the accumulation of these contents in some regions of the periplasmic space, the outer membrane then bulges outward and buds off, effectively removing the undesirable envelope components from the cell47,48,49 (Figure 3B).
Enrichment of the outer membrane with lipids and lipid-binding molecules
Recent studies have shown that Pseudomonas syringae OMV lipids differ from the lipids of the OM of the bacterium50. These findings have led to a model in which membrane curvature is induced by the accumulation of LPS molecules with atypical structures or charges (Figure 3C). LPS is the major constituent of the outer leaflet of the OM of most Gram-negative bacteria. The LPS molecules themselves are not homogeneous, as the length and content of the polysaccharide chain varies among the different molecules. It has been proposed that subsets of these molecules may gather in patches along the OM, inducing higher degrees of membrane curvature at particular locations, either due to charge repulsion51 or their molecular shape52. In addition, the Pseudomonas quinolone signal (PQS) of Pseudomonas aeruginosa can enhance anionic repulsions between lipopolysaccharide molecules, resulting in membrane blebbing by sequestering divalent cations, which are important in forming stabilizing salt bridges between the negatively charged B-band lipopolysaccharide molecules53,54,55. Recently, it was proposed that the PQS induces OMV formation through a mechanism of asymmetric expansion of the outer leaflet of the OM53,54,55 (Figure 3D). Although the PQS-based model is one of the best studied so far, whether it is utilized by other strains of Gram-negative bacteria to produce OMVs remains unclear.
Phospholipid accumulation in the outer leaflet of the OM causes OMV biogenesis
Recently, Roier et al56 reported a new model for OMV biogenesis involving the VacJ/Yrb ABC (ATP-binding cassette) transport system, a proposed phospholipid (PL) transporter. They observed that disruptions within the VacJ/Yrb ABC (ATP-binding cassette) transport system increased OMV production in the human pathogens H. influenzae and V. cholerae without compromising OM integrity. Similarly, mutations in homologues of E. coli also increased vesiculation. Using lipidome analyses, they further found that OMVs from VacJ/Yrb-defective mutants in H. influenzae were enriched in phospholipids and certain fatty acids. Given that PL transporters are essential for maintaining the lipid asymmetry in the OM, the asymmetric expansion of phospholipids in the outer leaflet would initiate an outward bulging of the OM, leading to the generation of OMVs. These findings suggest a new general mechanism of OMV biogenesis based on phospholipid accumulation in the outer leaflet of the outer membrane. Importantly, this mechanism is highly conserved among Gram-negative bacteria and can account for OMV formation under all growth conditions56. Thus, this model of OMV biogenesis may be applicable to a broad range of Gram-negative bacteria and might have important pathophysiological roles in vivo.
Other models for OMV biogenesis
Recently, two additional novel models of OMV biogenesis have been reported. The first model, proposed by Elhenawy et al57, shows that lipid A deacylation could impose shape modifications that result in the curvature of the outer membrane and subsequent OMV formation. They utilized Salmonella enterica serovar Typhimurium as a model organism and tested the effect of lipid A remodeling on OMV biogenesis. They observed that expression of the lipid A deacylase PagL resulted in increased vesiculation without inducing an envelope stress response. Mass spectrometry analysis further revealed profound differences in the patterns of lipid A expression in the OM and OMVs, with deacylated lipid A forms accumulating exclusively in OMVs. These findings suggest a novel mechanism for OMV biogenesis that involves outer membrane remodeling through lipid A modification.
The second model of OMV biogenesis was proposed by Turnbull et al58, who showed that the explosive cell lysis of Pseudomonas aeruginosa produces shattered membrane fragments that rapidly form MVs. They identified that a prophage endolysin encoded within the R- and F-pyocin gene cluster is essential for explosive cell lysis. Endolysin-deficient mutants are defective in MV production. These findings reveal that explosive cell lysis, mediated through the activity of a cryptic prophage endolysin, acts as a mechanism for the production of bacterial OMVs.
Growth conditions and genetic factors that affect OMV biogenesis
The formation of OMVs was initially thought to be a physical process associated with the routine wear and tear of the OM of the cell52. However, recent discoveries show that OMV generation is an active resource-depleting process, which suggests that OMVs are not by-products of simple material shearing of the membrane but that energy expenditure is likely required for the sorting of biomolecules into OMVs25. While the exact signaling pathway of OMV biogenesis is not currently known, many factors have been identified to affect the generation of OMVs, as described below.
Growth conditions and environmental stress
Numerous studies have indicated that the production of OMVs changes with different growth conditions. For example, high growth temperatures can increase the number of OMVs produced. McBroom & Kuehn47 found that temperature stress in E. coli leads to accumulation of misfolded proteins that ultimately are removed by being packaged in OMVs. Although they did not discuss it, the increased fluidity of the OM at higher temperatures may also have partially contributed to the increased generation of OMVs. In addition, it has been reported that oxygen stress can increase vesiculation in P. aeruginosa and Neisseria meningitidis59,60; DNA-damaging antibiotics can stimulate OMV production in P. aeruginosa via the SOS response61; and medium composition can influence vesiculation in Lysobacter sp. XL162. In the case of OMVs of Pseudomonas putida DOT-T1E, chemical stresses such as toxic concentrations of long-chain alcohols and EDTA (which chelates ions needed for the normal growth of bacterial cells), as well as physical stresses such as osmotic pressure and heat shock, caused the bacterium to release OMVs63,64. Notably, while OMV production is increased in bacteria in response to stress conditions, OMVs are also released even when cells grow without external stress65. Indeed, exponentially growing bacteria produce more OMVs, and similar findings were observed in growing bacteria at division septa, possibly because of the increased peptidoglycan turnover that occurs during cell division65,66. In addition, pathogenic species of bacteria generally release more OMVs than their non-pathogenic counterparts, and it is likely that OMV secretion has been adapted by pathogens to enhance their virulence67,68. For example, a P. aeruginosa strain isolated from a cystic fibrosis patient was found to release 3- to 4-fold more OMVs than the laboratory-adapted strain PAO168. Finally, nutrient availability can also affect OMV biogenesis, and the specific effects are dependent on the species of bacteria69,70. OMV biogenesis is upregulated in Lysobacter in response to low nutrient conditions, whereas for Pseudomonas fragi, upregulation of OMV biogenesis occurs in response to nutrient-rich meat media69,70. Considering these observations, it can be concluded that OMV production occurs in normal growth conditions but is also affected by multiple stress conditions.
Genetic factors
It is well recognized that both OMV production and OMV cargo loading are gene-regulated and do not result from random occurrences or cell death26,51,71. For example, the lipids of OMVs from P. aeruginosa are mainly composed of a highly charged form of LPS known as B-band LPS, whereas this molecule is only a minor component of the bacterial OM51. The enrichment of B-band LPS in OMVs rules out random membrane shedding or lysis, which would be expected to produce vesicles that are identical to the cell envelope in composition. Indeed, using an E. coli transposon mutant screen, McBroom et al34 identified a number of gene mutations that could upregulate OMV production. Two of the genes they identified encode putative cell-envelope-localized proteins (ypjA and nlpA). Recently, Kulp et al72 also assessed the genetic basis of OMV production by measuring vesiculation levels in the gene-deletion strains of the E. coli Keio library. They observed that nearly 150 new genes affected the process of vesiculation, suggesting that OMV production can be stimulated or inhibited through a gene-modulated pathway.
Furthermore, it has been observed that the OMVs of S. enterica contain higher levels of PagC and OmpX proteins73. Accordingly, overexpression of PagC and OmpX genes in bacteria could accelerate the generation of OMVs73. Similarly, the E. coli inner-membrane protein NlpA was found to regulate the biogenesis of OMVs74. By contrast, it was previously shown that lipoproteins are specifically depleted or decreased in OMVs of E. coli32. Accordingly, disruptions in the expression of lipoprotein genes (eg, OmpA and the Tol–PAL complex) in E. coli have been shown to increase OMV production32. It is important to note here that Pseudomonas aeruginosa possesses fewer lipoproteins than E. coli75, which may explain why P. aeruginosa usually produces significantly higher levels of OMVs than E. coli76. Along this line, some OM proteins such as Omps (proteins related to Tol–Pal systems and YbgF) are suspected to have a role in the bulging of the bacterial OM, which marks the onset of OMV production 77. Together, these observations provide multiple lines of compelling evidence showing that OMV biogenesis could be regulated at the genetic level.
Composition of bacterial MVs
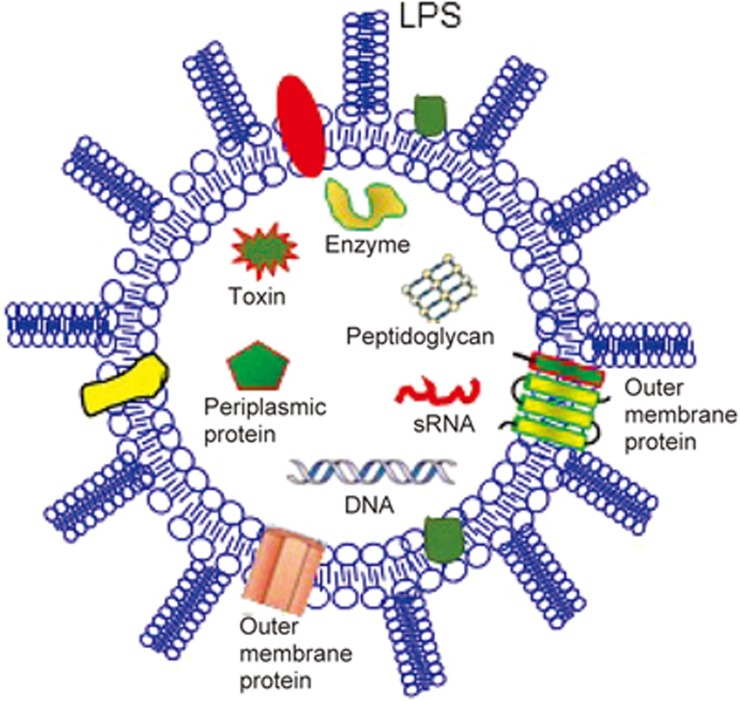
Structural studies of OMVs/MVs have invariably shown that they are spherical and enclosed by a membrane bilayer and have a strain-specific characteristic size distribution13,14,15. The contents of OMVs/MVs from various species of bacteria have been characterized using protein mass spectrometry (MS), lipid and SDS-PAGE profiles, and RNA sequencing25,26,27,78,79,80,81,82,83,84. It has been shown that OMVs contain outer-membrane proteins, lipopolysaccharide, periplasmic components including peptidoglycan and virulence factors, and inner-membrane and cytoplasmic proteins as well as DNA and RNA78,79,80,81,82,83,84 (Figure 4), whereas MVs contain various fatty acids and phospholipids, cytoplasmic proteins, membrane-associated virulence proteins/enzymes, lipoteichoic acid (LTA), peptidoglycan, DNA and sRNAs. These OMV/MV contents can be found in the EVpedia public database (http://evpedia.info).
Figure 4.
Composition of outer membrane vesicles (OMVs). Generally, OMVs contain outer membrane components (ie, OM-anchored lipoproteins, OM phospholipids and LPS), periplasmic proteins (ie, autolytic enzymes), peptidoglycan, and virulence factors (ie, toxins in pathogenic bacterium-derived OMVs), DNAs and small non-coding RNAs (sRNAs).
Proteins
The protein content of OMVs/MVs has been studied thoroughly by many groups using different bacterial strains78,79,80,81,82,83,84. The proteins identified in these Gram-negative OMVs include OM structural proteins, porins, ion channels, transporters for different molecules, periplasmic and cytoplasmic enzymes, and proteins related to stress responses84. Over the last decade, mass spectrometry-based proteomic studies have identified more than 3500 proteins that are encased in OMVs (http://evpedia.info). It is important to note that 1) not all outer-membrane proteins from a given strain are sorted into OMVs85, although membrane proteins make up a significant portion of the OMV proteome; 2) cytoplasmic and inner-membrane proteins identified in OMVs are not contaminants from lysed cells in the cultures but might have resulted from an active sorting mechanism84; 3) enzymes found in OMVs include proteases, peptidases, nucleases and β-lactamases85,86,87,88; and 4) protein profiles are different in the OMVs harvested from different strains of bacteria and from the same strain at different growth phases or in response to different stresses 89. OMVs may also contain toxins and other virulence factors25. The presence of virulence factors and toxins is usually observed in the OMVs produced by different pathogenic bacteria78.
More recently, researchers have characterized the protein compositions of MVs from several Gram-positive bacteria, such as S. aureus, B. anthracis, Listeria monocytogenes, Bacillus subtilis, and Clostridium perfringens13,14,15,54,79,90. The first high-throughput proteomic analysis was performed by Lee et al on S. aureus ATCC 14458 extracellular vesicles79. They identified a total of 90 proteins in S. aureus-derived MVs. Importantly, S. aureus-derived MVs are especially enriched with transporter proteins (eg, the protein translocase SecD/SecF and hydroxamate siderophores), antibiotic resistance-related proteins [β-lactamase, membrane-associated global regulator (MsrR), and penicillin-binding proteins (PBP1, PBP2, and PBP3)], diverse virulence proteins (toxins, adhesins, proteolysin, coagulase, and other related enzymes), and many cytoplasmic proteins, such as ribosomal proteins, DNA polymerases, tRNA synthetases, and several metabolic enzymes. Furthermore, Jiang et al90 recently identified a total of 431 proteins in the MVs released from Gram-positive C. perfringens. They found the most abundant proteins in C. perfringens-derived MVs were membrane (24%) and cytoplasm (18%) proteins, but they found no alpha-toxin or NetB toxin in these MVs. Additional MV proteomic studies on other Gram-positive bacteria have revealed that there are some differences in the specific protein compositions of these vesicles. Together, these results indicate that the makeup of the OMV/MV proteome may help to shed light on the pathophysiological functions of OMVs/MVs.
Phospholipids
In an early study, the OMVs of E. coli were identified to have a similar phospholipid profile to the OM91. Various studies have subsequently been carried out to compare the phospholipids from the OM and OMVs and have confirmed that OM phospholipids are present in the membranes of OMVs50,78,80,92. However, there is evidence that OMVs contain some lipids that are not detected in the OM93. Tashiro et al 92 recently observed that the phospholipid composition of the OM and OMVs from P. aeruginosa was different. In the OM, phosphatidylethanolamine was abundant, while phosphatidylglycerol was found in OMVs92. Furthermore, they found that the relative amount of saturated fatty acyl chains was higher in OMVs than in the OM92, making the OMVs more rigid than the OM. In a later study, Tashiro et al94 proposed that the release of OMVs may make the bacterial OM more fluid and that P. aeruginosa may shed OMVs to maintain the optimum fluidity for the prevailing growth conditions. Recently, several lipidomic analyses revealed that Gram-positive bacterial MVs contain various fatty acids21,95,96. For example, both B. anthracis and S. pneumoniae MVs are enriched in short-chain saturated fatty acids (C12–C16), such as myristic and palmitic acid, compared with bacterial cell membranes95,96. Using a matrix-assisted laser desorption ionization (MALDI)-based mass spectrometry approach, Resch et al21 recently identified that Group A streptococcus (GAS)-derived MVs contain more than 85 individual lipid species corresponding to glycoglycerolipids and both anionic and cationic phospholipids (PLs). They further observed that anionic phosphatidylglycerols (PGs) were significantly enriched in GAS MVs. In contrast, cardiolipin (CL) comprised only a minor fraction of MV lipids relative to the GAS membrane. Differential enrichment of MV lipid species relative to the membrane provides further evidence for the existence of a regulated mechanism contributing to MV biogenesis, as opposed to nonspecific membrane shedding.
LPSs
It is well known that LPSs in the outer membrane are a characteristic feature of the Gram-negative cell envelope. Therefore, it is obvious that bacterium-released OMVs contain LPS molecules; however, not all LPSs of the parent cell appear in OMVs 50,51,52,97. For example, OMVs from P. aeruginosa are mainly composed of the negatively charged B-band of LPS instead of the more neutral A-band in the outer membrane51. Haurat et al52 further showed that in the case of the dental pathogen P. gingivalis, deacetylated lipid A was accumulated in OMVs. By contrast, no differences were observed in the lipid A profiles of the outer membrane and OMVs in Bacteroides fragilis97. Collectively, these observations suggest that LPSs are selectively sorted into OMVs.
Nucleic acids (DNA and RNA)
It has been demonstrated that MVs/OMVs contain DNA components, including both plasmid DNA and chromosomal DNA 98,99,100. Although DNA fragments from lysed cells could potentially bind to the positively charged surface of OMVs, treatment of OMVs with DNase proved that some DNA fragments were trapped in the lumen of the OMVs100. A detailed study of the presence of nuclease-resistant DNA in MVs and OMVs from Gram-positive and Gram-negative bacteria, respectively, was carried out by Dorward & Garon (1990)101. They observed that DNA in the form of circular plasmids, linear plasmids and chromosomal fragments is likely to be present in Gram-negative OMVs but not in Gram-positive MVs. However, Jiang et al90 recently showed that Gram-positive C. perfringens-derived MVs did contain partial chromosomal DNA fragments encoding the 16S rRNA, plc and pfoA genes. At present, the mechanisms by which DNA is sorted into MVs/OMVs are not clear. It has been suggested that MVs/OMVs could pick up free DNA fragments present in their environment and that DNA can enter through the periplasm during MV/OMV biogenesis100. Notably, DNA has not been found in the OMVs of many other bacteria strains102. For example, three different strains of Porphyromonas gingivalis were found to produce OMVs that did not contain any DNAs102.
Recently, using an RNA deep sequencing (RNA-Seq) approach, several groups have identified that both MVs and OMVs contain a significant amount of RNAs21,83,103. For example, Sjöström et al103 detected more than 200 short RNA (sRNA) sequences in the OMVs released by V. cholerae strain A1552. Interestingly, the majority of the most abundant RNA sequences identified in V. cholerae OMVs correspond to non-coding regions. Similarly, Koeppen et al83 identified 481,480 unique sRNA sequences in OMVs purified from the supernatants of three planktonic cultures of P. aeruginosa strain PA14. They observed that the median sequence length of the sRNAs was 24 nucleotides, with a minimum of 15 and a maximum of 45 nucleotides. The 1733 most abundant sRNA sequences in OMVs (ie, at least 64 sequence reads) mapped to 68 loci in the P. aeruginosa genome. These authors further used bioinformatic approaches and found that the ten most abundant sRNAs in OMVs were likely to form stable secondary structures and that these sRNAs could potentially interact with human mRNAs.
In the case of Gram-positive MVs, Resch et al21 recently characterized RNA species present in MVs released by Streptococcus pyogenes, also known as group A streptococcus (GAS), using RNA deep-sequencing. They found that a total of 207 RNA species were differentially abundant in MVs relative to bacteria. Of these, 120 RNAs were more abundant in MVs, including the zinc-responsive adhesive competence repressor adcR, anaerobic ribonucleoside triphosphate reductase nrdG, exonuclease-helicase subunit A rexA, and type 1 restriction endonuclease hsdR genes; and 87 RNAs were less abundant in the MV fraction, including the streptolysin S gene sagA and the glucosamine-6-phosphate isomerase gene nagB. These findings indicate that Gram-positive bacterial MVs are enriched with intragenic RNAs of the originating bacteria. Nonetheless, whether non-coding shorter RNAs are included in these MVs remains unclear.
Other molecules
MVs/OMVs also contain a variety of ions, quorum sensing signaling molecules and metabolites104,105. However, the appearance of these molecules in MVs/OMVs has not been studied extensively to date. Certainly, many other types of bioactive molecules will be identified in MVs/OMVs in the future.
Roles of bacterial MVs in bacterial survival
Many different functions have been demonstrated for MVs/OMVs, making them important players in the physiology of Gram-positive and -negative bacteria13,15,16,17,18,19,20,21,22,23,24,25,26. The prevalent idea is that MVs/OMVs are indispensable for the survival of these bacteria22. The contributions of MVs, mostly OMVs, to bacterial survival have been extensively investigated over the past several decades13,15,16,17,18,19,20,21,22,23,24,25,26. Some of the prominent functions are discussed below.
Defense and resistance
The outer membrane of Gram-negative bacterial cells is the first line of defense against external stress conditions. In response to unfavorable conditions, bacteria have developed mechanisms of generating MVs to quickly remove surface-attacking agents and internal misfolded or toxic materials22. For example, upon treatment of bacteria with lytic bacteriophages, vesiculation of the bacterial outer membrane increases, generating an abundance of decoy membranes in the environment, which could help the bacteria avoid infection by phages 106,107. OMVs can also bind or absorb some antibacterial molecules (eg, complement and antibiotics) and, consequently, inactivate these molecules or transport them away from bacteria80,107,108,109,110. For example, in E. coli, the addition of OMVs or the use of a hypervesiculating mutant increased immediate resistance to the antimicrobial peptides polymyxin B and colistin107. Similarly, OMVs from P. syringae reduced the levels of colistin and melittin (another antimicrobial peptide) in solution by sequestering these compounds80. Moreover, MVs/OMVs can also carry enzymes that mediate antibiotic protection. For instance, β-lactamase can be packaged into MVs released from Gram-positive bacteria, and co-incubation of such MVs with β-lactam-sensitive species improves resistance to these antibiotics108,110. Recently, OMVs from amoxicillin-resistant M. catarrhalis (Gram-negative) were found to carry active β-lactamase and to protect amoxicillin-sensitive M. catarrhalis from antibiotic-induced killing86. Furthermore, it has been demonstrated that the OMVs produced by one bacterial species are able to protect other bacterial species from antibiotic stress, emphasizing the ecological role of OMVs in microbial niches53,86,88. For example, OMVs from amoxicillin-resistant M. catarrhalis have also been shown to improve the amoxicillin resistance of non-typeable H. influenzae and S. pneumoniae, two bacteria that typically co-infect the respiratory tract86.
Nutrient acquisition
Numerous studies have shown that OMVs are able to promote nutrient acquisition in bacterial communities and thereby enhance bacterial survival22,45,53,105,111. This is mainly ascribed to the preferential packaging of diverse enzymes (eg, glycosidases, proteases, alkaline phosphatase and enolase), metal ion-binding proteins/molecules, and surface receptors in OMVs45,53,105,111. For example, the signal molecule 2-heptyl-3-hydroxy-4-quinolone (also known as Pseudomonas quinolone signal, PQS) is highly enriched in P. aeruginosa OMVs and binds iron, which is essential for bacterial viability and often limiting in biological environments53,111. PQS-Fe-OMVs could then be absorbed into the OM by fusion, or OMVs could release PQS-bound iron in the vicinity of the cell and thereby enable the bacteria to scavenge iron, leading to enhanced survival. Notably, the OMVs from N. meningitidis are also enriched in the zinc acquisition proteins ZnuA and ZnuD45, suggesting that the role of OMVs in metal acquisition is not limited to iron.
In addition, OMVs themselves could serve as nutrient sources for bacteria. For example, OMVs from the marine cyanobacterium Prochlorococcus have been shown to support growth of the heterotrophs Alteromonas and Halomonas as the sole carbon source105. Similarly, OMV-associated DNA and proteins may serve as a source of nitrogen and phosphorous for bacterial growth.
Facilitation of biofilm formation
Biofilms are surface-adhering structures made from a matrix consisting of exopolysaccharides, DNA, proteins and many other molecules, in which bacterial cells remain embedded112. In general, bacteria form biofilms as a response to stress112. The number of OMVs released by bacteria increases during stress, suggesting a relationship between OMVs and biofilm formation63,112,113. Indeed, it has been reported that the addition of OMVs to an H. pylori culture stimulated biofilm production114. Interestingly, exogenous addition of DNA to bacteria can also stimulate biofilm production115. Considering that OMVs carry DNA, it is therefore possible that OMVs trigger biofilm formation through surface-associated DNA. However, the detailed mechanism underlying the role of OMVs in biofilm formation remains to be investigated.
Killing of competing microbial cells
As discussed above, OMVs can help protect bacteria from antibiotics and other stress conditions. In addition to this function, OMVs produced from one bacterium can also kill other competing microbes in the vicinity. For example, Li et al previously showed that the OMVs of various Gram-negative strains could kill many Gram-positive and Gram-negative bacteria22. Their study investigated the effectiveness of OMVs in killing target bacteria with different peptidoglycan chemotypes. They found that killing was most effective when the target possessed a peptidoglycan that was similar to the OMV donor. If the peptidoglycan hydrolases present in the OMVs are the same as those of the target strain, then they cannot cleave the peptidoglycan layer. Along this line, Vasilyeva et al 62 recently found that Lysobacter sp. XL1 secreted bacteriolytic enzymes in OMVs. This activity of OMVs shows that they might be capable of distinguishing between self and non-self cells in a mixed community. At present, while the concept of using OMVs as a new antimicrobial agent has been explored, whether they can be a commercially feasible option remains to be tested in the future.
In summary, bacteria tend to produce more MVs/OMVs as a survival mechanism under stress conditions. In fact, hyper-vesiculating mutants have been shown to better adapt to stress conditions compared with wild-type strains25,107. During temperature stress, OMVs can quickly remove misfolded proteins64. Upon antibiotic exposure, OMVs can sequester107 or degrade108 the antibiotics. When stress is induced by antibodies or bacteriophages, OMVs can act as decoy targets, protecting cells by titration of the antibodies or phages107. The production of OMVs is also found to increase with nutrient stress, which shows that OMVs have a role in bacterial nutrition, as discussed above. Finally, OMVs are able to facilitate biofilm formation and kill competing microbial cells.
Versatile effects of bacterial MVs on mammalian cells
Beneficial effects of OMVs on epithelial cells
The integrity of the epithelial barrier is critical in maintaining homeostasis in the body, and its dysfunction is linked to inflammatory, allergic and metabolic diseases116,117. Interactions between the gut microbiota and the intestinal epithelium are crucial for the integrity of this barrier. Therefore, alterations in the microbiota composition or aberrant responses to luminal bacteria or dietary components can result in increased intestinal permeability, which may lead to the development of the aforementioned pathologies. In this context, many studies have been conducted to investigate the therapeutic potential of certain commensal and probiotic strains to ameliorate inflammatory bowel diseases in clinical trials (see reviews elsewhere118,119) or in animal models of colitis120,121,122,123,124,125. In mouse colitis models, beneficial bacteria reduce inflammatory cytokines, normalize gut permeability, and reinforce the epithelial barrier124. Recently, several studies have indicated that these effects may be mediated, at least in part, by bacterial-secreted OMVs124,125,126. For example, Alvarez et al126 reported that OMVs and soluble factors released by probiotic E. coli Nissle 1917 and commensal ECOR63 were able to enhance barrier function by upregulating ZO-1 and claudin-14 and downregulating claudin-2 in intestinal epithelial cells.
Pathogenic effects of OMVs on epithelial cells
By contrast, MVs/OMVs released from pathogenic bacteria in the gut could interact with epithelial cells and ultimately cause inflammation127,128,129. H. pylori is an example of a pathogen that resides in the mucosa and colonizes the gut of approximately 50% of the human population130. In most patients, this infection is asymptomatic, but in approximately 20% of patients, the infection leads to chronic gastritis as well as an increased risk of developing peptic ulcer disease, gastric lymphoma, or gastric carcinoma130. Surprisingly, H. pylori has been found to be a non-invasive pathogen. However, LPS-enriched OMVs secreted from H. pylori bind to and invade gastric epithelial cells partly through clathrin-mediated endocytosis131. By adhering to cells via the adhesins BabA and SabA and delivering the vacuolating cytotoxin VacA, H. pylori-derived OMVs could trigger inflammation through the persistent delivery of antigens such as proteases and urease to the gastric mucosa132,133.
Enterotoxigenic E. coli, a major cause of diarrhea and infant mortality in third world countries, has also been analyzed for OMV binding to epithelial cells134,135,136. The heat-labile enterotoxin LT is secreted by the outer membrane of the bacteria as well as OMVs136. LT-containing vesicles and OMVs lacking the toxin were compared for their capacity to invade adrenal and intestinal epithelial cells. It was clearly shown that only LT-carrying vesicles could invade cells in a time-, temperature-, and receptor-dependent manner136. The vesicles attached to the cells at 4°C, and the LT was internalized at 37°C136. This was mediated through cholesterol-rich lipid rafts, and OMVs sometimes co-localized with the endocytosis protein caveolin. Kesty et al136 concluded that the OMVs are targeted transport vesicles of the enterotoxin.
Furthermore, Cronobacter sakazakii, a causative agent of infant meningitis and enterocolitis, produces OMVs that are internalized by intestinal epithelial cells, inducing cell proliferation and IL-8 secretion and facilitating bacterial persistence137. Moreover, bacterial RNA contained within OMVs produced by E. coli, P. aeruginosa, P. gingivalis, and V. cholerae has immuno-stimulatory capabilities in epithelial cells81,83,138,139.
Additionally, OMVs have the potential to bind to epithelial cells in the respiratory tract as well as in the mucosa, anatomical sites where bacteria interact with their host140,141. For instance, Legionella pneumophila could secrete OMVs that attach to lung alveolar epithelial cells, stimulating the secretion of inflammatory cytokines (ie, IL-6, IL-7, IL-8, and IL-13 as well as GM-CSF, IFN-γ, and MCP-1)140. The OMVs of M. catarrhalis, a respiratory pathogen that commonly causes otitis media in children, also bind to epithelial cells141. M. catarrhalis OMVs bind to TLR2 in epithelial cell lipid rafts, which are subdomains of the membrane with distinct protein and lipid compositions and enriched in cholesterol and sphingolipids141. The OMVs are subsequently internalized, causing a pro-inflammatory response and resulting in increased IL-8 secretion and ICAM-1 expression. Moreover, OMVs can be observed as bacterial tools that interact with and regulate the inflammatory response of epithelial cells at sites that are distant from the parent bacteria141.
Versatile effects of OMVs on immune cells
In addition to interacting with the host's epithelial cells, bacterium-released OMVs can directly affect various immune cell populations, including neutrophils, macrophages and dendritic cells (DCs). Some effects of OMVs on immune cells are discussed below.
Effects of OMVs on neutrophils.
OMVs regulate neutrophil function in different ways that are dependent on their composition and bacterial origin. Lapinet et al142 showed that stimulation of human neutrophils with N. meningitides-derived OMVs caused secretion of TNF and IL-1β and upregulation of CXCL8, CCL3 and CCL4 expression. These responses were further enhanced by IFNγ, suggesting that N. meningitidis OMV-triggered immune responses can be potentiated during a chronic inflammatory state in which IFNγ is present142. By contrast, Davis et al143 observed that OMVs from non-pathogenic E. coli encase cytotoxic necrotizing factor type 1 (CNF1), a bacterial toxin that impairs the phagocytic and chemotactic abilities of neutrophils. Interestingly, OMVs from N. meningitidis and Histophilus somni can induce the formation of neutrophil extracellular traps (NETs), which contain DNA, antimicrobial peptides and histones that form extracellular fibers to trap and kill extracellular pathogens144,145,146.
Effects of OMVs on macrophages.
Recent studies have elucidated the mechanisms by which OMVs can induce macrophage-mediated diseases. Legionella pneumophila (L. pneumophila) is an intracellular, Gram-negative pathogen that causes a severe form of pneumonia147. In humans, it infects alveolar macrophages, where it blocks lysosomal degradation and forms a specialized replication vacuole147. Recently, Jung et al148 demonstrated that incubating differentiated THP-1 macrophages with L. pneumophila OMVs led to a TLR2-dependent classical activation of the macrophages and the release of pro-inflammatory cytokines in a dose-dependent manner. Furthermore, treatment of THP-1 cells with OMVs prior to infection reduced L. pneumophila replication in the THP-1 cells. However, with prolonged infection times, the OMV pre-treated macrophages became more permissive for bacterial replication than untreated cells. They further identified that L. pneumophila OMVs were initially potent pro-inflammatory stimulators of macrophages, acting via TLR2, IRAK-1, and NF-κB, while at later time points, OMVs facilitated L. pneumophila replication by miR-146a-dependent IRAK-1 suppression. Therefore, OMVs might promote the spread of L. pneumophila in the host.
Furthermore, when examining lung tissue from patients infected with L. pneumophila, Jager et al149 observed that OMVs were distributed within the cytoplasm of infected macrophages, suggesting that OMVs directly interact with macrophages in vivo. Indeed, fluorophore labeling was used to confirm the direct binding and internalization of L. pneumophila OMVs into human macrophages, where they consequently stimulated TNF-α production148. Similarly, incubation of monocytes and macrophages with N. meningitidis OMVs has been shown to induce the production of CCL2, CCL3, CCL5 (also known as RANTES), CXCL8, IL-1β, IL-6, IL-10, IL-12p40, IL-12p70 and TNF150. Notably, OMVs activate macrophages to induce adaptive immune responses, as N. meningitidis OMVs upregulate the expression of HLA-DR, the co-stimulatory molecules CD80 and CD86, and intercellular adhesion molecule 1 (ICAM1) by macrophages150.
In addition to having pro-inflammatory effects, OMVs are able to elicit anti-inflammatory effects that benefit the pathogen of origin or that promote secondary bacterial infections. For example, Winter et al151 recently reported that H. pylori OMVs could induce the production of the immunosuppressive cytokine IL-10 by human peripheral blood mononuclear cells to suppress inflammation and facilitate bacterial survival. Likewise, P. gingivalis OMVs could inhibit CD14 expression in macrophages, rendering these cells unresponsive to TLR4 signaling triggered by secondary E. coli LPS stimulation152. Pollak et al153 also showed that B. abortus OMVs attenuated TLR2, TLR4 and TLR5 responses, dampened IFNγ-induced MHC class II expression and promoted the internalization of B. abortus bacteria by THP-1 cells. Collectively, these studies indicate that OMVs play an important role by interacting with macrophages/monocytes and act both as pro- and anti-inflammatory mediators, possibly depending on the specific strain and on environmental conditions.
Effects of OMVs on dendritic cells (DCs)
OMVs have been shown to be internalized into dendritic cells (DCs) and to program DCs to induce the differentiation of IL-10-producing CD4+Foxp3+ Treg cells in a PSA-dependent manner125. Alaniz et al154 reported that OMVs from Salmonella spp. induced the expression of CD86 and MHC class II molecules on DCs and the production of TNF and IL-12 and promoted the development of protective B cell and T cell responses. Similarly, Durand et al155 showed that N. meningitidis OMVs induced DC maturation, characterized by the upregulation of MHC class II and co-stimulatory molecules and the production of pro-inflammatory cytokines and chemokines. Mechanistically, N. meningitidis OMVs are able to facilitate their delivery to and internalization by DCs by binding to the host factor bactericidal permeability-increasing protein (BPI)156. BPI is a host immune factor that binds and neutralizes endotoxin in addition to promoting bacterial clearance157. Taken together, these observations reveal that OMVs affect DCs by inducing their maturation and facilitating antigen presentation and by limiting their cytokine responses to either OMVs or secondary bacterial antigens.
Effects of OMVs on other types of mammalian cells
Endothelial cells and OMVs
In an early study, Shoberg and Thomas158 showed that Borrelia burgdorferi produces OMVs that contain some of the outer-surface proteins of the bacterium (eg, OspA and OspB). They further observed that such OMVs can bind and enter into cultured human umbilical vein endothelial cells (HUVECs) in a dose-dependent, OMV component-dependent and passage number-dependent manner. Consistent with this, several studies later reported that P. gingivalis-derived OMVs were capable of stimulating the expression of E-selectin and ICAM-1 and suppressing capillary tube formation by cultured HUVECs39,159,160,161. Recently, Kim et al162 found that OMVs derived from intestinal E. coliupregulate functional ICAM-1, E-selectin, or VCAM-1 expression on the surface of human microvascular endothelial cells via the activation of NF-κB. When administered intraperitoneally to mice, these OMVs induce neutrophil aggregation in the lung endothelium in an ICAM1- and TLR4-dependent manner, suggesting that OMVs are used by pathogens to sequester leukocytes in the endothelium162. Likewise, Dr. Sullivan's group163,164demonstrated that E. coli OMVs induce the production of IL-6, tissue factor, thrombomodulin, and the adhesion molecules P-selectin and E-selectin by human endothelial cells, which leads to the recruitment of pro-inflammatory leukocytes, platelet aggregation and coagulation. A recent study by Jia et al165 further showed that P. gingivalis OMVs suppressed endothelial nitric oxide synthase (eNOS) in both cultured HUVECs and the mouse aortic endothelium.
By contrast, OMVs have also been observed to limit inflammatory responses at the endothelium; for example, the addition of P. gingivalis OMVs to IFNγ-stimulated endothelial cells inhibited the upregulation of MHC class II molecules on the cell surface, thus modulating antigen presentation and favoring survival of the pathogen166. Hence, similar to the dual effects that are discussed above in epithelial cells and immune cells, OMVs have both immunostimulatory and immunomodulatory effects on endothelial cells.
Platelets and OMVs
It has been reported that P. gingivalis OMVs are potent inducers of mouse platelet aggregation in vitro167. Similarly, N. meningitidis OMVs contribute to thrombosis via increased platelet-platelet and platelet-leukocyte aggregation168.
Fibroblasts, osteoblasts and synovial cells
Friedrich et al169 recently showed that OMVs released by T. forsythia not only interact with human macrophages but also stimulate human periodontal ligament fibroblasts to release pro-inflammatory mediators that are potentially relevant to the development of periodontitis (i.e., TNF-α, IL-6, IL-8, and MCP-1). Consistent with this, multiple groups observed that P. gingivalis OMVs could be internalized into human oral keratinocytes and gingival fibroblasts139,160,170. In addition, osteoblasts and synovial cells can internalize K. kingae OMVs, leading to the production of GM-CSF and IL-6, both of which are implicated in pathogen-induced bone and tissue inflammation and destruction171.
Pathological roles of bacterial MVs in infectious/inflammatory diseases
As discussed above, MVs/OMVs can be internalized into various types of host cells and subsequently induce the activation of inflammatory pathways. Furthermore, MVs/OMVs can be detected in the blood and cerebrospinal fluid during clinically severe bacterial infections. It has been proposed that MVs/OMVs in the blood and body fluids may significantly contribute to inflammation and mortality in human diseases172,173,174.
Gram-negative bacterium-derived OMVs in infectious/inflammatory diseases
Currently, it is recognized that OMVs released by Gram-negative bacteria can evoke pathological conditions in vivo. For example, OMVs from pathogenic bacteria contribute to the hypercoagulable response in sepsis by influencing the inflammatory and coagulation cascades163,164. In a mouse model, intraperitoneal injection of OMVs derived from intestinal E. coli induced a sepsis-like syndrome associated with systemic induction of TNF-α and IL-6 and with significant lethality173. Similarly, continuous infusion of E. coli OMVs into rats elicited physiological, biochemical, and histological changes that resembled clinical sepsis symptoms174. Indeed, LPS-carrying OMVs were found to contribute to fatal endotoxin levels observed in the serum of a patient 175.
In addition, some Gram-negative bacterial OMVs can mediate pulmonary inflammation. For instance, OMVs derived from M. catarrhalis and Klebsiella pneumoniae have been shown to induce neutrophil and lymphocyte infiltration into the lungs of mice141,176. Moreover, Park et al173 recently showed that intranasal administration of P. aeruginosa OMVs into mice caused pulmonary inflammation without any live bacteria in vivo. Thus, it is speculated that bacterial OMVs may be causative agents in the pathogenesis of specific infectious diseases. However, little is known about the dissemination and distribution of these vesicles in vivo, although this information could provide critical clues into their functions in living organisms. Recently, for the first time, Jang et al177 examined the in vivo distribution of E. coli OMVs after intraperitoneal injection in mice. They observed that the OMVs spread throughout the body and accumulated in the liver, lungs, spleen, and kidney within 3 h after administration. They also performed functional analyses on the consequences of OMV administration by investigating systemic and lung inflammation177 and observed (1) increased levels of TNF-α and IL-6 in serum and bronchoalveolar lavage fluid as an early systemic inflammatory response, (2) fewer leukocytes and platelets in the blood, (3) decreased body temperature, and (4) increased ICAM-1 levels in the lung. These observations suggested that bacterial OMVs could act as effective mediators of long distance communication in vivo.
Gram-positive bacterium-derived MVs in infectious/inflammatory diseases
While direct evidence of MV formation in Gram-positive bacteria has just emerged, several recent studies have also suggested that these Gram-positive bacterial MVs may significantly contribute to pathophysiological conditions during bacterial infection178,179. For example, Hong et al178 observed that the in vitro application of S. aureus EVs increased the production of pro-inflammatory mediators (IL-6, thymic stromal lymphopoietin, macrophage inflammatory protein-1α, and eotaxin) in dermal fibroblasts. The in vivo application of S. aureus EVs into tape-stripped mouse skin caused severe atopic dermatitis-like skin inflammation and epidermal thickening by increasing the infiltration of eosinophils and mast cells. S. aureus MVs were also observed to induce both Th1 and Th17 neutrophilic pulmonary inflammation and facilitated airway hypersensitivity to inhaled allergens. More interestingly, Kim et al179 observed the presence of S. aureus MVs in house dust and demonstrated their role as important causative agents of pulmonary inflammatory diseases.
Other Gram-positive bacteria, including Bacillus spp., B. anthracis, S. coelicolor, L. monocytogenes, S. pneumoniae, C. perfringens, and S. mutans, also produce MVs13. However, their possible pathological roles in vivo have not been investigated. It is clear that these MVs could also contribute significantly to disease processes during bacterial infection.
Preventive and therapeutic effects of bacterial MVs in human diseases
OMVs are naturally released exocytic vesicles of Gram-negative bacteria that function in a number of processes, including biofilm formation, virulence protein secretion, and cell-to-cell communication26. Importantly, because of their distinct immunological and structural features, including nanometer-scale vesicle structure, self-adjuvant effectiveness, potential for genetic modification, and the ability to present exogenous proteins and carry immune stimulators, OMVs have received increased attention as emerging and feasible vaccine carriers180. Numerous studies have shown that OMV-derived vaccines could induce protective immunity against infections with pathogenic bacteria, such as N. meningitides, A. baumannii, P. gingivalis, S. Typhimurium, H. pylori, and V. cholerae, in animal models (see reviews elsewhere180,181,182). The following sections highlight recent findings on OMV-derived vaccines and therapeutic agents for human diseases.
Neisseria meningitides-derived OMV vaccine
For several serogroups of N. meningitidis (A, C, W, X), effective conjugate vaccines consisting of a capsular polysaccharide coupled to a carrier protein are on the market. However, for serogroup B, this approach is not feasible because this polysaccharide exhibits homology to molecular structures in the human brain, generating the risk of autoimmunity183. Therefore, the development of an N. meningitidis serogroup B (MenB) vaccine has focused on other approaches, and MenB-derived OMVs, as an effective vaccine, have been extensively studied over the last several decades183. Thus far, OMV vaccines have been used successfully to combat outbreaks of meningitis in Cuba, Norway and New Zealand, where monovalent detergent-extracted OMVs were prepared from a manufacturing strain matching the circulating strains causing the epidemic182. In the Netherlands, a multivalent immuno-dominant OM protein porin A (PorA)-based OMV vaccine has been developed based on genetically engineered strains in which multiple porA genes were inserted184. Recently, trivalent native OMVs derived from three genetically modified N. meningitidis serogroup B strains have been evaluated immunologically in mice, rabbits and human infants185. This study suggests that the cross-reactive antibodies generated by one or more OMV antigens may provide further protection against serogroup B meningococcal disease caused by heterologous strains.
Acinetobacter baumannii-derived OMV vaccine
Acinetobacter baumannii, a non-fermenting, Gram-negative, aerobic coccobacillus, is currently one of the major causes of hospital-acquired infections and is usually associated with pneumonia and sepsis in intensive care unit patients186. Notably, A. baumannii has been shown to secrete OMVs into the blood circulation, through which it sheds virulence factors (i.e., AbOmpA protein) and delivers them to mammalian cells, inducing cell death187. In recent years, epidemics caused by multi- or pan-drug-resistant strains of A. baumannii have been widely investigated and reported186. Thus, there is an urgent need to develop innovative approaches to prevent and treat such multi- or pan-drug-resistant infections. To this end, several groups recently identified various vaccine candidates [eg, inactivated whole cell, outer-membrane complexes , single outer-membrane proteins (AbOmpA, Bap, and Ata)] that have an effect on controlling A. baumannii infections (see reviews elsewhere188). McConnell et al189 also indicated that vaccination using OMVs could be considered as a viable strategy for preventing A. baumannii infection. However, OMVs contain a wide range of LPSs that may induce uncontrolled inflammatory responses. Therefore, reducing LPS endotoxicity is an important goal for achieving an optimized immune response. Recently, it was identified that heterologous expression of the 3-O-deacylase PagL could remove the 3-O-linked acyl chain from the disaccharide backbone of lipid A190. This suggests that OMVs purified from a bacterium that expresses PagL could be used as an optimized vaccine candidate against A. baumannii infection. Indeed, Badmasti et al191 evaluated the immunological effects of OMVs containing the signal peptides Bap(1-487aa) and AbOmpA(8–346aa) and PagL as well as their combinations [PagL-OMVs/Bap(1-487aa) and AbOmpA(8-346aa)/Bap(1-487aa)] in a C57BL/6 murine model of disseminated sepsis. They observed that although the AbOmpA and Bap peptides were excellent vaccine targets, they did not provide full protection by themselves against A. baumannii infection. By contrast, PagL-OMVs/Bap(1-487aa)-immunized mice displayed robust humoral/cellular immune responses, resulting in full protection against A. baumannii ATCC 19606 and MDR AB-44 strains. Together, these findings may provide a new strategy for developing an A. baumannii vaccine.
Other bacterium-derived OMV vaccines
Vibrio cholerae-derived OMV vaccine
The Camilli laboratory192,193,194 has recently developed a V. cholerae OMV vaccine delivered intranasally, which represents an alternative vaccine approach to the currently licensed whole-cell-killed (WCK) and live attenuated vaccines, both delivered orally. Schild's group195 also reported that immunization with genetically detoxified V. cholerae OMVs could elicit a protective immune response. They tested the potential of LPS-modified and toxin-negative OMVs isolated from V. cholerae and enterotoxigenic E. coli (ETEC) as a combined OMV vaccine candidate. They found that immunization with V. cholerae or ETEC OMVs induced a species-specific immune response, whereas the combination of both OMV species resulted in a high-titer, protective immune response against both pathogens195.
Porphyromonas gingivalis-derived OMV vaccine
Periodontitis is the most prevalent infectious disease related to oral and systemic health, and a novel prophylaxis to prevent the disease is therefore highly desirable196. Recently, the Senpuku laboratory reported that intranasal immunization using a combination of P. gingivalis OMVs and a TLR3 agonist, Poly (I:C), efficiently elicited a P. gingivalis-specific antibody response196. They observed that intranasal vaccination of OMVs in mice elicited the production of P. gingivalis-specific antibodies in blood and saliva in a dose-dependent manner, which was dramatically enhanced by the addition of the TLR3 agonist Poly (I:C). Serum samples from these immunized mice revealed significant inhibition of the gingipain proteolytic activity of not only the vaccine strain but also heterologous strains.
Burkholderia pseudomallei-derived OMV vaccine
Burkholderia pseudomallei (Bps) is the causative agent of melioidosis and is endemic in regions of northern Australia and Southeast Asia197. Bps is naturally resistant to multiple antibiotics, and there is currently no approved vaccine against the organism197. Recently, the Morici laboratory has shown that immunization with natural Bps-derived OMVs provides significant protection against lethal aerosol and systemic infections in BALB/c mice197. They further evaluated the safety and immunogenicity of escalating doses of the OMV vaccine in rhesus macaques and observed the generation of humoral immune responses with no associated toxicity or reactogenicity in Bps OMV-immunized animals198. Nonetheless, future studies are warranted to evaluate the protective efficacy of the OMV vaccine in a nonhuman primate model of melioidosis.
Engineered OMVs as a universal vaccine
The following points are well appreciated: 1) heterologous proteins expressed in E. coli cells can be distributed in the periplasmic space and packed into shedding OMVs25; 2) expressed heterologous proteins can anchor to the surface of E. coli cells and are also presented on the shedding OMVs199; 3) technologically, fusion with an intrinsic outer-membrane protein (eg, a pore-forming hemolytic protein, ClyA) can bring exogenous proteins to the surface of OMVs200; and 4) exogenous proteins can also be presented in the inner lumen of OMVs using an appropriate leader protein25. Thus, OMVs could be engineered by genetic manipulation to present exogenous proteins of interest. Such engineered OMVs can serve as an antigen-delivery platform for the efficient induction of specific antibody responses. Huang et al201 recently utilized recombinant gene technology to overexpress a previously identified immunogenic outer-membrane protein of A. baumannii, Omp22, in the non-pathogenic E. coli DH5α strain. They observed that E. coli DH5α-derived OMVs (Omp22-OMVs) could display a complete Omp22 protein on their surfaces without any alterations in their OMV morphology. Immunization with Omp22-OMVs induced high titers of Omp22-specific antibodies. In a murine sepsis model, Omp22-OMV immunization significantly protected mice from lethal challenge with a clinically isolated A. baumannii strain, as evidenced by 1) the increased survival rate of the mice; 2) the reduced bacterial burdens in the lungs, spleen, liver, kidney, and blood; and 3) the suppressed serum levels of inflammatory cytokines. These results clearly indicate that engineered OMVs could display a whole heterologous protein on the surface and effectively trigger specific antibody responses and that OMVs may therefore be utilized as a new and feasible antigen delivery tool for a vaccine to induce protective immunity.
However, there are several concerns associated with the commercialization of OMVs as vaccines in humans: 1) although most of the OMVs considered safe for use in vaccine or drug delivery are LPS-free, increasing evidence suggests that other components of the OMVs besides LPS can also provoke systemic inflammatory response syndrome, leading to lethality in mice; and 2) bacteria release relatively low quantities of OMVs, indicating that their cost-effective mass production may become inconvenient. Regarding these concerns, Kim et al202 recently developed bacterial protoplast-derived nanosized vesicles (PDNVs), which are depleted of outer-membrane components, as a safe and effective adjuvant-free vaccine delivery system. By fabricating protoplasts of genetically engineered antigen-expressing E. coli strains, antigen-loaded PDNVs can be easily produced with a significantly higher yield, and, importantly, these PDNVs are less toxic than E. coli OMVs. Moreover, they observed that immunization with antigen-loaded PDNVs induced potent antigen-specific humoral and cellular immune responses, leading to effective protection against both Gram-negative and Gram-positive bacterial sepsis in murine models. Given that the preventive effect of a vaccine is specific to the specific antigen, the use of PDNVs loaded with a single or multiple bacterial antigens may enhance future vaccination efficacy against most virulent bacterial infections. Hence, these nanosized bacterial protoplast-derived vesicles may represent a versatile system that could accommodate either viral or bacterial antigens for future applications as a novel universal vaccine to prevent various infectious diseases. Nonetheless, further studies are required to assess the overall safety of PDNV-based vaccination and to elucidate the key components and detailed mechanism underlying the induction of antigen-specific protective immune responses by PDNVs.
Conclusions
Although bacteria-released OMVs have been studied for almost 4 decades, only in recent years have we witnessed the great advances in our understanding of the mechanisms by which OMVs contribute to bacterial-triggered inflammation and pathology and of their ability to modulate the immune system by suppressing inflammation and facilitating the survival of their parent bacterium in the host1,2,3. The recent identification of MVs produced by some Gram-positive bacteria, which have immuno-pathological and potentially immuno-modulatory roles similar to those of OMVs, indicates that there is still much to learn about the involvement of bacterial MVs in human diseases133. Notably, the observation of OMVs present in indoor dust may suggest their possible contributions to airway disease179, which should be further explored. In addition, there is increased interest in exploring the possible contributions of the host microbiota to the development of immunity to gastrointestinal diseases181. Given the beneficial contribution of commensal OMVs to gastrointestinal integrity, future studies are needed to better understand the potential protective mechanisms of commensal OMVs against pathogens and how to genetically engineer probiotic E. coli OMVs.
Moreover, there has been limited use of OMV-based technologies to deliver antigens and protective proteins to specific organs/tissues. Recently, a mutant E. coli strain that exhibits reduced endotoxicity toward human cells was engineered to generate OMVs displaying a human epidermal growth factor receptor 2 (HER2)-specific affibody in the membrane as a targeting ligand. These OMVs delivered anti-tumorigenic cargo that resulted in cell death and tumor regression in mice203. This study has expanded our potential use of OMVs beyond vaccine carriers and implicates bioengineered OMVs as new cell-specific drug-delivery vehicles for treating various cancers. Finally, while several OMV vaccines have been clinically applied to combat some infectious diseases, concerns such as serum resistance still remain180,181,182. With the technological advances in our ability to modify the composition of OMVs, it is expected that OMVs will have broad therapeutic and preventive applications for human health.
Author contribution
You-jiang YU wrote the manuscript; Xiao-hong WANG edited the references; and Guo-Chang FAN wrote and edited the manuscript.
Acknowledgments
The study in Dr Fan's laboratory was supported by an American Heart Association (AHA) Established Investigator Award (17EIA33400063) and National Institutes of Health (NIH) grants HL-087861 and GM-112930 (Dr Guo-Chang FAN).
References
- Haurat MF, Elhenawy W, Feldman MF. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol Chem 2015; 396: 95–109. [DOI] [PubMed] [Google Scholar]
- Avila-Calderón ED, Araiza-Villanueva MG, Cancino-Diaz JC, López-Villegas EO, Sriranganathan N, Boyle SM, et al. Roles of bacterial membrane vesicles. Arch Microbiol 2015; 197: 1–10. [DOI] [PubMed] [Google Scholar]
- Gould SB, Garg SG, Martin WF. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol 2016; 24: 525–34. [DOI] [PubMed] [Google Scholar]
- Ailawadi S, Wang X, Gu H, Fan GC. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim Biophys Acta 2015; 1852: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Azuma E, Muramatsu M, Hamashima T, Ishii Y, Sasahara M. Significance of extracellular vesicles: pathobiological roles in disease. Cell Struct Funct 2016; 41: 137–43. [DOI] [PubMed] [Google Scholar]
- Withrow J, Murphy C, Liu Y, Hunter M, Fulzele S, Hamrick MW. Extracellular vesicles in the pathogenesis of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther 2016; 18: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrochers LM, Antonyak MA, Cerione RA. Extracellular vesicles: satellites of information transfer in cancer and stem cell biology. Dev Cell 2016; 37: 301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang W, Liu G, Cai W, Millard RW, Wang Y, et al. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J Mol Cell Cardiol 2014; 74: 139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gu H, Qin D, Yang L, Huang W, Essandoh K, et al. Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis. Sci Rep 2015; 5: 13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essandoh K, Yang L, Wang X, Huang W, Qin D, Hao J, et al. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim Biophys Acta 2015; 1852: 2362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gu H, Huang W, Peng J, Li Y, Yang L, et al. Hsp20-mediated activation of exosome biogenesis in cardiomyocytes improves cardiac function and angiogenesis in diabetic mice. Diabetes 2016; 65: 3111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol 1967; 49: 1–11. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee J, Park J, Gho YS. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol 2015; 40: 97–104. [DOI] [PubMed] [Google Scholar]
- Kim GH, Choi CW, Park EC, Lee SY, Kim SI. Isolation and proteomic characterization of bacterial extracellular membrane vesicles. Curr Protein Pept Sci 2014; 15: 719–31. [DOI] [PubMed] [Google Scholar]
- Pathirana RD, Kaparakis-Liaskos M. Bacterial membrane vesicles: Biogenesis, immune regulation and pathogenesis. Cell Microbiol 2016; 18: 1518–24. [DOI] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Functional advantages conferred by extracellular prokaryotic membrane vesicles. J Mol Microbiol Biotechnol 2013; 23: 131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald KL, Beveridge TJ. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can J Microbiol 2002; 48: 810–20. [DOI] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 2016; 165: 1106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman J, Auer M. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ Microbiol 2013; 15: 347–54. [DOI] [PubMed] [Google Scholar]
- Jurkoshek KS, Wang Y, Athman JJ, Barton MR, Wearsch PA. Interspecies communication between pathogens and immune cells via bacterial membrane vesicles.. Front Cell Dev Biol 2016; 4: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, et al. A Two-Component Regulatory System Impacts Extracellular Membrane-Derived Vesicle Production in Group A Streptococcus. MBio 2016; 7: e00207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 1998; 180: 5478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald KL, Beveridge TJ. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can J Microbiol 2002; 48: 810–20. [DOI] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, et al. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 2011; 6: e27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 2010; 64: 163–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 2015; 13: 605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2010; 2: a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojer K, Riemer J. Balancing oxidative protein folding: the influences of reducing pathways on disulfide bond formation. Biochim Biophys Acta 2014; 1844: 1383–90. [DOI] [PubMed] [Google Scholar]
- Braun V, Wolff H. Attachment of lipoprotein to murein (peptidoglycan) of Escherichia coli in the presence and absence of penicillin FL 1060. J Bacteriol 1975; 123: 888–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 1975; 415: 335–77. [DOI] [PubMed] [Google Scholar]
- Huang YX, Ching G, Inouye M. Comparison of the lipoprotein gene among the Enterobacteriaceae. DNA sequence of Morganella morganii lipoprotein gene and its expression in Escherichia coli. J Biol Chem 1983; 258: 8139–45. [PubMed] [Google Scholar]
- Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 2002; 184: 754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. The function of OmpA in Escherichia coli. Biochem Biophys Res Commun 2002; 292: 396–401. [DOI] [PubMed] [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 2006; 188: 5385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gramnegative bacterial outer membrane. FASEB J 2011; 26: 219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 2007; 63: 1008–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The caulobacter Tol–Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol 2010; 192: 4847–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, Kim SI, et al. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol 2012; 50: 155–60. [DOI] [PubMed] [Google Scholar]
- Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. Biogenesis of bacterial membrane vesicles. Mol Microbiol 2009; 72: 1395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol 2008; 70: 100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol 1978; 136: 280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Rodriguez DL, Kuehn MJ. NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiol Open 2015; 4: 375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara M, Wu HC, Sankaran K, Rick PD. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J Bacteriol 1999; 181: 4318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 2012; 86: 1036–51. [DOI] [PubMed] [Google Scholar]
- Lappann M, Otto A, Becher D, Vogel U. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J Bacteriol 2013; 195: 4425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kulp A, Kuehn MJ. Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 2014; 14: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 2007; 63: 545–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KJ, Castelli ME, Vescovi EG, Feldman MF. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol 2012; 194: 3241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R, Lo M, Khoo C, Zhang H, Boysen RI, Picardeau M, et al. Precipitation of iron on the surface of Leptospira interrogans is associated with mutation of the stress response metalloprotease HtpX. Appl Environ Microbiol 2013; 79: 4653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Jagannadham MV. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta 2013; 1834: 231–9. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 1995; 177: 3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 2011; 286: 1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 2005; 437: 422–5. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Ichikawa S, Nakajima-Kambe T, Uchiyama H, Nomura N. Pseudomonas quinolone signal affects membrane vesicle production in not only Gram-negative but also Gram-positive bacteria. Microbes Environ 2010; 25: 120–5. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 2008; 69: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 2016; 7: 10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. LPS remodeling triggers formation of outer membrane vesicles in salmonella. MBio 2016; 7: e00940–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 2016; 7: 11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W, Lunsdorf H, Zeng AP. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology 2003; 149: 2789–95. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd B, Zomer G, van den Ijssel J, van Keulen L, Eppink MH, van der Ley P, et al. Cysteine depletion causes oxidative stress and triggers outer membrane vesicle release by Neisseria meningitidis; implications for vaccine development. PLoS One 2013; 8: e54314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, et al. Vesiculation from Pseudomonas aeruginosa under SOS. Sci World J 2012; 2012: 402919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS. Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J 2008; 275: 3827–35. [DOI] [PubMed] [Google Scholar]
- Baumgarten T, Sperling S, Seifert J, von Bergen M, Steiniger F, Wick LY, et al. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl Environ Microbiol 2012; 78: 6217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten T, Vazquez J, Bastisch C, Veron W, Feuilloley MG, Nietzsche S, et al. Alkanols and chlorophenols cause different physiological adaptive responses on the level of cell surface properties and membrane vesicle formation in Pseudomonas putida DOT-T1E. Appl Microbiol Biotechnol 2012; 93: 837–45. [DOI] [PubMed] [Google Scholar]
- Mug-Opstelten D, Witholt B. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli. Biochim Biophys Acta 1978; 508: 287–95. [DOI] [PubMed] [Google Scholar]
- Katsui N, Tsuchido T, Hiramatsu R, Fujikawa S, Takano M, Shibasaki I. Heat-induced blebbing and vesiculation of the outer membrane of Escherichia coli. J Bacteriol 1982; 151: 1523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem 2000; 275: 12489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman SJ, Kuehn MJ. Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol 2009; 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SS, Naidu YM, Pestka JJ. Ultrastructural localization of an extracellular protease in Pseudomonas fragi by using the peroxidase-antiperoxidase reaction. Appl Environ Microbiol 1985; 50:1038–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilyeva NV, Tsfasman IM, Kudryakova IV, Suzina NE, Shishkova NA, Kulaev IS, et al. The role of membrane vesicles in secretion of Lysobacter sp. bacteriolytic enzymes. J Mol Microbiol Biotechnol 2013; 23: 142–51. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Sullivan CJ, Kuehn MJ. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 2013; 52: 3031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp AJ, Sun B, Ai T, Manning AJ, Orench-Rivera N, Schmid AK, et al. Genome-Wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 2015; 10: e0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Takaya A, Ohya M, Mizunoe Y, Takade A, Yoshida S, et al. Biogenesis of Salmonella enterica serovar typhimurium membrane vesicles provoked by induction of PagC. J Bacteriol 2010; 192: 5645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodero MD, Pilonieta MC, Munson GP. Repression of the inner membrane lipoprotein NlpA by Rns in enterotoxigenic Escherichia coli. J Bacteriol 2007; 189: 1627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HH, Heilmann HD, Preusser HJ. State of the rigid-layer in celll walls of some gram-negative Bacteria. Arch Mikrobiol 1972; 83: 332–46. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 2006; 61: 839–46. [DOI] [PubMed] [Google Scholar]
- Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubès R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 1998; 180: 4872–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 2014; 160: 2109–21. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, et al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009; 9: 5425–36. [DOI] [PubMed] [Google Scholar]
- Kulkarni HM. Swamy ChV, Jagannadham MV. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res 2014; 13: 1345–58. [DOI] [PubMed] [Google Scholar]
- Sjöström AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Sci Rep 2015; 5: 15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altindis E, Fu Y, Mekalanos JJ. Proteomic analysis of s outer membrane vesicles. Proc Natl Acad Sci U S A 2014; 111: E1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, et al. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 2016; 12: e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim OY, Gho YS. Proteomic profiling of Gram-negative bacterial outer membrane vesicles: current perspectives. Proteomics Clin Appl 2016; 10: 897–909. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DS, Kim KP, Gho YS. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev 2008; 27:535–55. [DOI] [PubMed] [Google Scholar]
- Schaar V, Nordström T, Mörgelin M, Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother 2011; 55: 3845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar V, Paulsson M, Mörgelin M, Riesbeck K. Outer membrane vesicles shield Moraxella catarrhalis β-lactamase from neutralization by serum IgG. J Antimicrob Chemother 2013; 68: 593–600. [DOI] [PubMed] [Google Scholar]
- Schaar V, Uddbäck I, Nordström T, Riesbeck K. Group A streptococci are protected from amoxicillin-mediated killing by vesicles containing β-lactamase derived from Haemophilus influenzae. J Antimicrob Chemother 2014; 69: 117–20. [DOI] [PubMed] [Google Scholar]
- Berlanda Scorza F, Colucci AM, Maggiore L, Sanzone S, Rossi O, Ferlenghi I, et al. High yield production process for Shigella outer membrane particles. PLoS One 2012; 7: e35616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Kong Q, Roland KL. Curtiss R 3rd. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 2014; 304: 431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta 1976; 455: 889–99. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima-Kambe T, et al. Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa. Biosci Biotechnol Biochem 2011; 75: 605–7. [DOI] [PubMed] [Google Scholar]
- Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog 2002; 32: 1–13. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Uchiyama H, Nomura N. Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ Microbiol 2012; 14: 1349–62. [DOI] [PubMed] [Google Scholar]
- Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 2010; 107: 19002–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martin-Pena R, Gonzalez-Reyes JA, Jimenez-Munguia I, et al. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumonia. J Proteomics 2014; 106: 46–60 [DOI] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. MBio 2014; 5: e00909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, Mawhinney EL. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl Environ Microbiol 2005; 71: 4248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 1999; 65: 1843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004; 150: 2161–9. [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl Environ Microbiol 1990; 56: 1960–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett 1998; 163: 223–8. [DOI] [PubMed] [Google Scholar]
- Sjöström AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Sci Rep 2015; 5: 15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. MBio 2012; 3: e00297–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. Bacterial vesicles in marine ecosystems. Science 2014; 343: 183–6. [DOI] [PubMed] [Google Scholar]
- Loeb MR, Kilner J. Release of a special fraction of the outer membrane from both growing and phage T4-infected Escherichia coli B. Biochim Biophys Acta 1978; 514: 117–27. [DOI] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 2011; 11: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. Chromosomal betalactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 2000; 45: 9–13. [DOI] [PubMed] [Google Scholar]
- Tan TT, Morgelin M, Forsgren A, Riesbeck K. Haemophilus influenzae survival during complementmediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 2007; 195: 1661–70. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, et al. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother 2013; 57: 2589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst 2008; 4: 882–8. [DOI] [PubMed] [Google Scholar]
- Wang W, Chanda W, Zhong M. The relationship between biofilm and outer membrane vesicles: a novel therapy overview. FEMS Microbiol Lett 2015; 362: e117. [DOI] [PubMed] [Google Scholar]
- Fulsundar S, Harms K, Flaten G E, Johnsen PJ, Chopade BA Nielsen KM. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol 2014; 80: 3469–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, et al. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol 2009; 9: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science 2002; 295: 148. [DOI] [PubMed] [Google Scholar]
- Hering NA, Fromm M, Schulzke JD. Determinants of colonic barrier function in inflammatory bowel disease and potential therapeutics. J Physiol 2012; 590: 1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Miwa H. Gastrointestinal mucosal barrier function and diseases. J Gastroenterol 2016; 51: 768–78. [DOI] [PubMed] [Google Scholar]
- Chibbar R, Dieleman LA. Probiotics in the management of ulcerative colitis. J Clin Gastroenterol 2015; 49: S50–S55. [DOI] [PubMed] [Google Scholar]
- Wasilewski A, Zielinska M, Storr M, Fichna J. Beneficial effects of probiotics, prebiotics, synbiotics and psychobiotics in inflammatory bowel disease. Inflamm Bowel Dis 2015; 21: 1674–82. [DOI] [PubMed] [Google Scholar]
- Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol 2008; 295: G1025–G1034. [DOI] [PubMed] [Google Scholar]
- Arribas B, Rodríguez-Cabezas ME, Camuesco D, Comalada M, Bailón E, Utrilla P, et al. A probiotic strain of Escherichia coli, Nissle 1917, given orally exerts local and systemic anti-inflammatory effects in lipopolysaccharide-induced sepsis in mice. Br J Pharmacol 2009; 157: 1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Chain F, Miquel S, Lu J, Gratadoux JJ, Sokol H, et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis 2014; 20: 417–30. [DOI] [PubMed] [Google Scholar]
- Souza ÉL, Elian SD, Paula LM, Garcia CC, Vieira AT, Teixeira MM, et al. Escherichia coli strain Nissle 1917 ameliorates experimental colitis by modulating intestinal permeability, the inflammatory response and clinical signs in a faecal transplantation model. J Med Microbiol 2016; 65: 201–10. [DOI] [PubMed] [Google Scholar]
- Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One 2013; 8: e76520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012; 12: 509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez CS, Badia J, Bosch M, Giménez R, Baldomà L. Outer membrane vesicles and soluble factors released by probiotic Escherichia coli Nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front Microbiol 2016; 7: 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S, Hampton MB, Keenan JI. Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun 2003; 71: 5670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala G, Torres L, Espinosa M, Fierros-Zarate G, Maldonado V, Meléndez-Zajgla J. External membrane vesicles from Helicobacter pylori induce apoptosis in gastric epithelial cells. FEMS Microbiol Lett 2006; 260: 178–85. [DOI] [PubMed] [Google Scholar]
- Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun 2010; 78: 5054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Neddermann M, Maubach G, Naumann M. Pathogenesis of Helicobacter pylori infection. Helicobacter 2016; 21: 19–25. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Nygård Skalman L, Obi I, Lundmark R, Arnqvist A. Uptake of Helicobacter pylori vesicles is facilitated by clathrin-dependent and clathrin-independent endocytic pathways. MBio 2014; 5: e00979–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson A, Vallstrom A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S, et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol Microbiol 2010; 77: 1539–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun 2010; 78: 5054–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp JM, Witholt B. K88-mediated binding of Escherichia coli outer membrane fragments to porcine intestinal epithelial cell brush borders. Infect Immun 1981; 31:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkan H, Kuehn MJ. Context-dependent activation kinetics elicited by soluble versus outer membrane vesicle-associated heat-labile enterotoxin. Infect Immun 2011; 79: 3760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J 2004; 23: 4538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzahrani H, Winter J, Boocock D, De Girolamo L, Forsythe SJ. Characterization of outer membrane vesicles from a neonatal meningitic strain of Cronobacter sakazakii. FEMS Microbiol Lett 2015; 362: e085. [DOI] [PubMed] [Google Scholar]
- Fritz JV, Heintz-Buschart A, Ghosal A, Wampach L, Etheridge A, Galas D, et al. Sources and functions of extracellular small RNAs in human circulation. Annu Rev Nutr 2016; 36: 301–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MH, Chen CH, Goodwin JS, Wang BY, Xie H. Functional advantages of porphyromonas gingivalis vesicles. PLoS One 2015; 10: e0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka F, Wai SN, Kusch H, Engelmann S, Hecker M, Schmeck B, et al. Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun 2008; 76: 1825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar V, de Vries SP, Perez Vidakovics ML, Bootsma HJ, Larsson L, Hermans PW, et al. Multicomponent Moraxella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell Microbiol 2011; 13: 432–49. [DOI] [PubMed] [Google Scholar]
- Lapinet JA, Scapini P, Calzetti F, Pérez O, Cassatella MA. Gene expression and production of tumor necrosis factor alpha, interleukin-1beta (IL-1beta), IL-8, macrophage inflammatory protein 1alpha (MIP-1alpha), MIP-1beta, and gamma interferon-inducible protein 10 by human neutrophils stimulated with group B meningococcal outer membrane vesicles. Infect Immun 2000; 68: 6917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Carvalho HM, Rasmussen SB, O'Brien AD. Cytotoxic necrotizing factor type 1 delivered by outer membrane vesicles of uropathogenic Escherichia coli attenuates polymorphonuclear leukocyte antimicrobial activity and chemotaxis. Infect Immun 2006; 74: 4401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science 2004; 303: 1532–5. [DOI] [PubMed] [Google Scholar]
- Lappann M, Danhof S, Guenther F, Olivares-Florez S, Mordhorst IL, Vogel U. In vitro resistance mechanisms of Neisseria meningitidis against neutrophil extracellular traps. Mol Microbiol 2013; 89: 433–49. [DOI] [PubMed] [Google Scholar]
- Hellenbrand KM, Forsythe KM, Rivera-Rivas JJ, Czuprynski CJ, Aulik NA. Histophilus somni causes extracellular trap formation by bovine neutrophils and macrophages. Microb Pathog 2013; 54: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Luo ZQ. Cell biology of infection by Legionella pneumophila. Microbes Infect 2013; 15: 157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung AL, Stoiber C, Herkt CE, Schulz C, Bertrams W, Schmeck B. Legionella pneumophila-derived outer membrane vesicles promote bacterial replication in macrophages. PLoS Pathog 2016; 12: e1005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger J, Marwitz S, Tiefenau J, Rasch J, Shevchuk O, Kugler C, et al. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun 2014; 82: 275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano R, Franzoso S, Cecchini P, Cartocci E, Oriente F, Aricò B, et al. The membrane expression of Neisseria meningitidis adhesin A (NadA) increases the proimmune effects of MenB OMVs on human macrophages, compared with NadA-OMVs, without further stimulating their proinflammatory activity on circulating monocytes. J Leukoc Biol 2009; 86: 143–53. [DOI] [PubMed] [Google Scholar]
- Winter J, Letley D, Rhead J, Atherton J, Robinson K. Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect Immun 2014; 82:1372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Yoshioka M, Chandad F, Grenier D. Loss of lipopolysaccharide receptor CD14 from the surface of human macrophage-like cells mediated by Porphyromonas gingivalis outer membrane vesicles. Microb Pathog 2004; 36: 319–25. [DOI] [PubMed] [Google Scholar]
- Pollak CN, Delpino MV, Fossati CA, Baldi PC. Outer membrane vesicles from Brucella abortus promote bacterial internalization by human monocytes and modulate their innate immune response. PLoS One 2012; 7: e50214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaniz RC, Deatherage BL, Lara JC, Cookson BT. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J Immunol 2007; 179: 7692–701. [DOI] [PubMed] [Google Scholar]
- Durand V, Mackenzie J, de Leon J, Mesa C, Quesniaux V, Montoya M, et al. Role of lipopolysaccharide in the induction of type I interferon-dependent cross-priming and IL-10 production in mice by meningococcal outer membrane vesicles. Vaccine 2009; 27: 1912–22. [DOI] [PubMed] [Google Scholar]
- Schultz H, Hume J, Zhang DS, Gioannini TL, Weiss JP. A novel role for the bactericidal/permeability increasing protein in interactions of gram-negative bacterial outer membrane blebs with dendritic cells. J Immunol 2007; 179: 2477–84. [DOI] [PubMed] [Google Scholar]
- Weiss J, Elsbach P, Shu C, Castillo J, Grinna L, Horwitz A, et al. Human bactericidal/permeability-increasing protein and a recombinant NH2-terminal fragment cause killing of serum-resistant gram-negative bacteria in whole blood and inhibit tumor necrosis factor release induced by the bacteria. J Clin Invest 1992; 90: 1122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoberg RJ, Thomas DD. Specific adherence of Borrelia burgdorferi extracellular vesicles to human endothelial cells in culture. Infect Immun 1993; 61: 3892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisatjaluk R, Doyle RJ, Justus DE. Outer membrane vesicles of Porphyromonas gingivalis inhibit IFN-gamma-mediated MHC class II expression by human vascular endothelial cells. Microb Pathog 1999; 27: 81–91. [DOI] [PubMed] [Google Scholar]
- Bartruff JB, Yukna RA, Layman DL. Outer membrane vesicles from Porphyromonas gingivalis affect the growth and function of cultured human gingival fibroblasts and umbilical vein endothelial cells. J Periodontol 2005; 76: 972–9. [DOI] [PubMed] [Google Scholar]
- Ho MH, Guo ZM, Chunga J, Goodwin JS, Xie H. Characterization of innate immune responses of human endothelial cells induced by Porphyromonas gingivalis and their derived outer membrane vesicles. Front Cell Infect Microbiol. 2016; 6: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yoon YJ, Lee J, Choi EJ, Yi N, Park KS, et al. Outer membrane vesicles derived from Escherichia coli up-regulate expression of endothelial cell adhesion molecules in vitro and in vivo. PLoS One 2013; 8: e59276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soult MC, Lonergan NE, Shah B, Kim WK, Britt LD, Sullivan CJ. Outer membrane vesicles from pathogenic bacteria initiate an inflammatory response in human endothelial cells. J Surg Res 2013; 184: 458–66. [DOI] [PubMed] [Google Scholar]
- Soult MC, Dobrydneva Y, Wahab KH, Britt LD, Sullivan CJ. Outer membrane vesicles alter inflammation and coagulation mediators. J Surg Res 2014; 192: 134–42. [DOI] [PubMed] [Google Scholar]
- Jia Y, Guo B, Yang W, Zhao Q, Jia W, Wu Y. Rho kinase mediates Porphyromonas gingivalis outer membrane vesicle-induced suppression of endothelial nitric oxide synthase through ERK1/2 and p38 MAPK. Arch Oral Biol 2015; 60: 488–95. [DOI] [PubMed] [Google Scholar]
- Srisatjaluk R, Kotwal GJ, Hunt LA, Justus DE. Modulation of gamma interferon-induced major histocompatibility complex class II gene expression by Porphyromonas gingivalis membrane vesicles. Infect Immun 2002; 70: 1185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Novak EK, Sojar HT, Swank RT, Kuramitsu HK, Genco RJ. Porphyromonas gingivalis platelet aggregation activity: outer membrane vesicles are potent activators of murine platelets. Oral Microbiol Immuno 2000; 15: 393–6. [Google Scholar]
- Mirlashari MR, Hagberg IA, Lyberg T. Platelet-platelet and platelet-leukocyte interactions induced by outer membrane vesicles from N. meningitidis. Platelets 2002; 13: 91–9. [DOI] [PubMed] [Google Scholar]
- Friedrich V, Gruber C, Nimeth I, Pabinger S, Sekot G, Posch G, et al. Outer membrane vesicles of Tannerella forsythia: biogenesis, composition, and virulence. Mol Oral Microbiol 2015; 30: 451–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thay B, Damm A, Kufer TA, Wai SN, Oscarsson J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-κB activation. Infect Immun 2014; 82: 4034–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Wei R, Kachlany SC, Kazi M, Balashova NV. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microb Pathog 2011; 51: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Takashiba S, Kosono S, Yoshida M, Watanabe H, Ohnishi M, et al. Effect of Porphyromonas gingivalis outer membrane vesicles on gingipain-mediated detachment of cultured oral epithelial cells and immune responses. Microbes Infect 2014; 16: 6–16. [DOI] [PubMed] [Google Scholar]
- Park KS, Choi KH, Kim YS, Hong BS, Kim OY, Kim JH, et al. Outer membrane vesicles derived from Escherichia coli induce systemic inflammatory response syndrome. PLoS One 2010; 5: e11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B, Sullivan CJ, Lonergan NE, Stanley S, Soult MC, Britt LD. Circulating bacterial membrane vesicles cause sepsis in rats. Shock 2012; 37: 621–8. [DOI] [PubMed] [Google Scholar]
- Namork E, Brandtzaeg P. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 2002; 360: 1741. [DOI] [PubMed] [Google Scholar]
- Lee JC, Lee EJ, Lee JH, Jun SH, Choi CW, Kim SI, et al. Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol Lett 2012; 331: 17–24. [DOI] [PubMed] [Google Scholar]
- Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J, et al. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small 2015; 11: 456–61. [DOI] [PubMed] [Google Scholar]
- Hong SW, Choi EB, Min TK, Kim JH, Kim MH, Jeon SG, et al. An important role of α-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One 2014; 9: e100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Hong SW, Choi EB, Lee WH, Kim YS, Jeon SG, et al. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy 2012; 67: 1271–81. [DOI] [PubMed] [Google Scholar]
- Collins BS. Gram-negative outer membrane vesicles in vaccine development. Discov Med 2011; 12: 7–15. [PubMed] [Google Scholar]
- Unal CM, Schaar V, Riesbeck K. Bacterial outer membrane vesicles in disease and preventive medicine. Semin Immunopathol 2011; 33: 395–408. [DOI] [PubMed] [Google Scholar]
- van der Pol L, Stork M, van der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnol J 2015; 10: 1689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton SI. The global evolution of meningococcal epidemiology following the introduction of meningococcal vaccines. J Adolesc Health 2016; 59: S3–S11. [DOI] [PubMed] [Google Scholar]
- Bousema JC, Ruitenberg J. Need for optimisation of immunisation strategies targeting invasive meningococcal disease in the Netherlands. Int J Health Policy Manag 2015; 4: 757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wen Z, Lin J, Xu H, Herbert P, Wang XM, et al. Improving the immunogenicity of a trivalent Neisseria meningitidis native outer membrane vesicle vaccine by genetic modification. Vaccine 2016; 34: 4250–6. [DOI] [PubMed] [Google Scholar]
- Zarrilli R, Casillo R, Di Popolo A, Tripodi MF, Bagattini M, Cuccurullo S, et al. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin Microbiol Infect 2007; 13: 481–9. [DOI] [PubMed] [Google Scholar]
- Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, et al. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 2011; 6: e17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Quintanilla M, Pulido MR, McConnell MJ. First steps towards a vaccine against Acinetobacter baumannii. Curr Pharm Biotechnol 2013; 14: 897–902. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Rumbo C, Bou G, Pachón J. Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine 2011; 29: 5705–10. [DOI] [PubMed] [Google Scholar]
- Asensio CJ, Gaillard ME, Moreno G, Bottero D, Zurita E, Rumbo M, et al. Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine 2011; 29: 1649–56. [DOI] [PubMed] [Google Scholar]
- Badmasti F, Ajdary S, Bouzari S, Fooladi AA, Shahcheraghi F, Siadat SD. Immunological evaluation of OMV(PagL)+Bap(1-487aa) and AbOmpA(8-346aa)+Bap(1-487aa) as vaccine candidates against Acinetobacter baumannii sepsis infection. Mol Immunol 2015; 67: 552–8. [DOI] [PubMed] [Google Scholar]
- Bishop AL, Schild S, Patimalla B, Klein B, Camilli A. Mucosal immunization with Vibrio cholerae outer membrane vesicles provides maternal protection mediated by antilipopolysaccharide antibodies that inhibit bacterial motility. Infect Immun 2010; 78: 4402–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Tarique AA, Patimalla B, Calderwood SB, Qadri F, Camilli A. Immunization of mice with vibrio cholerae outer-membrane vesicles protects against hyperinfectious challenge and blocks transmission. J Infect Dis 2012; 205: 412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lazinski DW, Camilli A. Immunity provided by an outer membrane vesicle cholera vaccine is due to O-antigen-specific antibodies inhibiting bacterial motility. Infect Immun 2016; 85: e00626–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner DR, Lichtenegger S, Temel P, Zingl FG, Ratzberger D, Roier S, et al. A combined vaccine approach against Vibrio cholerae and ETEC based on outer membrane vesicles. Front Microbiol 2015; 6: e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Hasegawa H, Dongying B, Ohnishi M, Senpuku H. Assessment of outer membrane vesicles of periodontopathic bacterium Porphyromonas gingivalis as possible mucosal immunogen. Vaccine 2016; 34: 4626–34. [DOI] [PubMed] [Google Scholar]
- Nieves W, Petersen H, Judy BM, Blumentritt CA, Russell-Lodrigue K, Roy CJ, et al. A Burkholderia pseudomallei outer membrane vesicle vaccine provides protection against lethal sepsis. Clin Vaccine Immunol 2014; 21: 747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen H, Nieves W, Russell-Lodrigue K, Roy CJ, Morici LA. Evaluation of a Burkholderia pseudomallei outer membrane vesicle vaccine in nonhuman primates. Procedia Vaccinol 2014; 8: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, et al. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci U S A 2010; 107: 3099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Doody AM, Chen DJ, Cremona GH, Shuler ML, Putnam D, et al. Engineered bacterial outer membrane vesicles with enhanced functionality. J Mol Biol 2008; 380: 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Yao Y, Wang S, Xia Y, Yang X, Long Q, et al. Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii. Sci Rep 2016; 6: e20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OY, Choi SJ, Jang SC, Park KS, Kim SR, Choi JP, et al. Bacterial protoplast-derived nanovesicles as vaccine delivery system against bacterial infection. Nano Lett 2015; 15: 266–74. [DOI] [PubMed] [Google Scholar]
- Gujrati V, Kim S, Kim SH, Min JJ, Choy HE, Kim SC, et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 2014; 8: 1525–37. [DOI] [PubMed] [Google Scholar]