Abstract
Background
Major hurdles for survival after lung transplantation are rejections and infectious complications. Adequate methods for monitoring immune suppression status are lacking. Here, we evaluated quantification of torque teno virus (TTV) and Epstein-Barr virus (EBV) as biomarkers for defining the net state of immunosuppression in lung-transplanted patients.
Methods
This prospective single-center study included 98 patients followed for 2 years after transplantation. Bacterial infections, fungal infections, viral respiratory infections (VRTI), cytomegalovirus (CMV) viremia, and acute rejections, as well as TTV and EBV levels, were monitored.
Results
The levels of torque teno virus DNA increased rapidly after transplantation, likely due to immunosuppressive treatment. A modest increase in levels of Epstein-Barr virus DNA was also observed after transplantation. There were no associations between either TTV or EBV and infectious events or acute rejection, respectively, during follow-up. When Tacrolimus was the main immunosuppressive treatment, TTV DNA levels were significantly elevated 6–24 months after transplantation as compared with Cyclosporine treatment.
Conclusions
Although replication of TTV, but not EBV, appears to reflect the functionality of the immune system, depending on the type of immunosuppressive treatment, quantification of TTV or EBV as biomarkers has limited potential for defining the net state of immune suppression.
Keywords: biomarker, Epstein-Barr virus, immunosuppression, infection, lung-transplantation, rejection, torque teno virus
For patients with end-stage lung disease, limited life expectancy, and otherwise acceptable physical condition, lung transplantation may be a life-saving therapeutic option. The 5-year survival rate is only approximately 50% in most centers, and 2 major limiting factors for the outcome of lung transplantation are the frequent occurrence of infectious complications and rejection of the transplanted organ [1]. Standard immunosuppression, to prevent graft rejection, consists of a combination of 3 immunosuppressive drugs that subvert B and T cells [2–4]. Currently, there is a lack of an easy-to-use marker that accurately predicts the net effect of the combined immunosuppression. Such a biomarker would be an important tool in the management of patients with compromised immune function to identify over- or underimmunosuppression and evaluate the risk of infectious complications and acute rejections.
Viruses are often only viewed as pathogenic entities, but there are several examples of viruses that are frequently found in healthy individuals without causing obvious disease [5, 6]. Torque teno virus (TTV) is a human DNA virus that causes asymptomatic viremia with a high prevalence in the general population [7–13]. Cell-mediated immunity is considered important in controlling TTV infection [14–17]. Accordingly, serum levels of TTV-DNA increase dramatically in patients who have undergone solid organ transplantation, presumably as a result of immunosuppression [16, 18, 19].
Epstein-Barr virus (EBV) is another DNA virus, commonly acquired early in life, with a high prevalence in US adults [20], that establishes latent infection in memory B cells [21, 22]. The levels of EBV often increase in blood after immunosuppressive treatment, and EBV can cause post-transplant lymphoproliferative disorder (PTLD) when the immune system is repressed. Hence, both TTV and EBV have previously been suggested as surrogate markers of the net state of immunosuppression [16, 23, 24].
The aim of the present study was to evaluate levels of TTV and EBV in relation to the frequency of infectious events and acute rejections over time in a prospective manner in a single-center cohort of lung-transplanted patients.
METHODS
Study Design
All patients over the age of 18 years who received a single or double lung transplant, who survived the initial postoperative intensive care period and remained in Gothenburg during the follow-up period, between January 1, 2009, and April 3, 2012, at the Sahlgrenska University Hospital Transplant Centre, Gothenburg, Sweden, were asked to participate in this prospective single-center cohort study.
All included subjects underwent follow-up (FU) visits at 1, 2, 3, 4.5, 6, 9, 12, 18, and 24 months after transplantation. At every FU visit, whole blood, serum, and nasopharyngeal samples (NPH) were collected. Protocol bronchoscopies were performed at 1, 3, and 12 months post-transplantation and when indicated based on clinical presentation. Bronchoalveolar lavage (BAL) samples were collected at every bronchoscopy. All BAL samples underwent direct microscopy for early detection of Aspergillus hyphae and were cultured for bacteria and fungi and tested for Legionella pneumophila, Pneumocystis jirovecii, and human cytomegalovirus (CMV) using real-time polymerase chain reaction (PCR). BAL samples from patients with cystic fibrosis were routinely cultured for Mycobacterium tuberculosis and atypical Mycobacteria. Bacterial and fungal culture testing from blood, urine, and sputum was performed upon suspicion of infection. NPH and BAL samples from every FU visit, as well as additional NPH samples obtained upon suspicion of airway infection between scheduled FU visits, were analyzed by multiplex real-time PCR for detection of airway pathogens. Serum samples obtained at pretransplant evaluation were used for TTV-DNA and EBV-DNA quantification. For quantification of TTV and EBV during FU, serum and whole blood samples were used, respectively.
Immunosuppression and Antimicrobial Prophylaxis
Induction therapy consisted of rabbit antithymocyte globulin, which was given for 1–3 consecutive days together with methylprednisolone intravenously. Post-transplantation immunosuppression included prednisone 0.3 mg/kg/d and mycophenolate mofetil (MMF) 2 g/d. The patients then received either oral Cyclosporine (CSA; 1–2 mg/kg), adjusted to maintain a serum level of 300–350 ng/mL, or Tacrolimus (TAC; 0.075 mg/kg) given orally divided in 2 doses daily, adjusted to maintain a serum level of 14–16 ng/mL. The choice between CSA and TAC was made based on clinical presentation. Patients previously treated with TAC and patients with cystic fibrosis were given TAC. There was also a preference for TAC if the recipient was younger. The dosage of immunosuppression was gradually lowered during FU. Further changes in immunosuppressive therapy were based on clinical presentation. Four patients, who either underwent retransplantation or had previous immunosuppressive treatment due to autoimmune conditions, were given nonstandard immunosuppression therapy based on previous exposure to immunosuppressive drugs.
All patients received a combination of 80 mg of sulfametoxazol and 400 mg of trimetoprim thrice weekly to prevent Pneumocystis jirovecii infection. Patients seropositive for CMV at the pretransplant evaluation were given valganciclovir at a dose of 900 mg daily for 3 months. Patients seronegative for CMV pretransplant while the organ donor tested positive (ie, CMV mismatches) were given the same treatment for 6 months.
Definition of Infectious Events and Acute Rejections
Bacterial infection was defined as a positive bacterial culture in conjunction with symptoms of clinical infection or, in the absence of positive culture, symptoms consistent with bacterial infection that was treated with antibiotics. Fungal infection was defined as significant presence of fungi in culture from a sterile location in conjunction with symptoms of infection according to the European Organization for Research and Treatment of Cancer (EORTC) criteria [25]. VRTI or Mycoplasma pneumoniae infection was defined as detection of a pathogen by multiplex real-time PCR in NPH or BAL. CMV viremia was defined as elevated levels of CMV-DNA that prompted antiviral therapy. CMV-seronegative recipients were considered to have viremia if CMV-DNA was detected, whereas recipients seropositive for CMV prior to transplantation where considered to have CMV viremia only if the level was above 3.0 log10 (1000) copies/mL. To distinguish milder infections from more severe infectious events, a subgroup of events requiring initial intravenous antimicrobial (antibacterial, antiviral, or antifungal) therapy were defined as severe. Acute rejection was defined as either a lung biopsy showing rejection of ISHLT grade 1A or higher [26] or, in the absence of a biopsy, physical (increased need for oxygen) and radiological findings (progressive infiltrate without signs of infection) consistent with acute rejection followed by a prompt response to high-dose (1 g/d for 3 consecutive days) corticosteroid therapy (methylprednisolone).
DNA Extraction and Quantification of CMV-, EBV-, and TTV-DNA
Nucleic acid isolation was performed with a MagNA Pure LC total nucleic acid or DNA isolation kit for serum (TTV) or whole blood (EBV and CMV) using a standardized protocol, according to the instructions (Roche Diagnostics, Mannheim, Germany). Input/output volume was set to 200/100 µL, and the sample was eluted in extraction buffer. The TTV-DNA levels were determined using a 7300 real-time PCR system (Applied Biosystems, Foster City, CA). Each PCR reaction contained 2 µL of extracted total nucleic acid, 10 µL of 2X Universal Master Mix (Applied Biosystems, Foster City, CA), 0.5 µL and 20 µM of forward and reverse primer, respectively, 0.3 µL and 20 µM of BHQ hydrolysis probe, and 2 µL of RNase-free H2O. The reaction conditions were 50°C for 2 minutes and 95°C for 10 minutes prior to 45 cycles at 95°C for 15 seconds and 60°C for 60 seconds. The assay range was determined by serial dilution of plasmids with an insert of a synthesized sequence matching the TTV PCR product, and quantification was obtained from a plot of Ct values. CMV- and EBV-DNA levels were determined using a method described previously with minor modifications [27]. All primer and probe sequences are listed in Supplementary Table 1. For EBV and CMV quantification, a control sample consisting of unrelated Phocine herpesvirus 1 (PhHV-1) was included prior to nucleic acid extraction.
Digital Droplet PCR for Quantification of TTV-DNA
TTV load was quantified on the QX200 Droplet Digital PCR system using the ddPCR supermix for probes (No dUTP; Bio-Rad, Hercules, CA). Primer and probe concentrations in the 20-µL final reaction were 300 and 200 nM, respectively. The PCR conditions were 50°C for 2 minutes followed by 95°C for 10 minutes, then 30 seconds at 95°C and 1 minute at 58°C for a total of 40 cycles, and finally a step at 98°C for 10 minutes. After overnight incubation at 4°C, the droplet fluorescence signal was determined. Data analysis was done using Bio-Rad Quanta Soft analysis software.
Multiplex Real-Time PCR for Detection of Respiratory Pathogens
Isolated DNA and RNA were analyzed using a multiplex real-time PCR system designed to detect adenovirus, bocavirus, Chlamydophila pneumoniae, human coronavirus (NL63, HKU1, OC43 and 229E), human enterovirus, human metapneumovirus, human rhinovirus, influenza A virus, influenza B virus, Mycoplasma pneumoniae, parainfluenzavirus (1, 2, and 3), and respiratory syncytial virus. The PCR settings and a list of primers and probes have been described previously [28].
Statistics
Comparisons on group level for numerical variables were performed using the Mann-Whitney U test. Pearson’s correlation and linear regression were used to evaluate the relationship between the real-time PCR and ddPCR measurements. Cox regression with TTV, EBV, and both as time-dependent covariates was used to analyze the relationship between the 2 biomarkers and the outcomes VRTI, fungal infection, bacterial infection, CMV viremia, or any infectious event. Each patient was treated as a cluster to handle multiple infections occurring in a single individual. Univariable logistic regression was used in each of 4 periods, q1 (1–3 months), q2 (3–6), q3 (6–12), and q4 (12–24), with the outcomes VRTI, fungal infection, bacterial infection, CMV viremia infection, or acute rejection and the predictors age, treatment, CMV mismatch, log TTV (beginning of period), or log EBV (beginning of the period). A P value <.05 was considered significant. R software, version 3.3.1 (R core team, 2016, R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org/), was used for all statistical analyses except for Pearson’s correlation analysis, which was performed using GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA).
Ethical Consideration
The study was approved by the Regional Ethical Review Board in Gothenburg (DNR: 791-08), and all subjects provided written and oral consent to participate.
RESULTS
Study Design
In total, 98 lung transplant recipients were included in the study, of which 79 (80%) were still alive 24 months after transplantation. Detailed patient description is outlined in Table 1. Blood-, BAL-, and nasopharyngeal samples (NPH) were collected longitudinally at follow-up (FU) visits until 24 months post–lung transplantation (LTx; n = 837), including pretransplant samples. The total nucleic acid content was isolated from serum or whole blood samples and analyzed for TTV-, EBV-, and CMV-DNA load by real-time PCR. The total nucleic acid content was isolated from the NPH and BAL samples and analyzed for respiratory pathogens using multiplex real-time PCR.
Table 1.
Patient Characteristics at Baseline
| General Description | Median (range) n | % |
|---|---|---|
| Age, y | 56 (18–73) | |
| No. of patients | 98 | |
| Female sex | 60 | 61 |
| Double lung recipientsa | 67 | 68 |
| CMV serology positive pre-LTx | 84 | 86 |
| CMV mismatchb | 9 | 9 |
| EBV serology positive pre-LTx | 95 | 97 |
| EBV mismatch | 9 | 9 |
| Dominant CNI | ||
| Cyclosporine | 79 | 80 |
| Tacrolimus | 19 | 20 |
| Underlying disease | ||
| Pulmonary fibrosis | 30 | 31 |
| COPD | 32 | 33 |
| Alfa-1 antitrypsin deficiency | 10 | 10 |
| Cystic fibrosis | 6 | 6 |
| Retransplantation | 6 | 6 |
| Pulmonary artery hypertension | 4 | 4 |
| RA bronchiolitis | 2 | 2 |
| Sarcoidosis | 2 | 2 |
| Sclerodermia | 2 | 2 |
| Bronchiectasies | 2 | 2 |
| Alveolitis | 1 | 1 |
| Lymphangioleiomyomatosis | 1 | 1 |
Abbreviations: CMV, human cytomegalovirus; CNI, calcineurin inhibitor; COPD, chronic obstructive pulmonary disease; EBV, Epstein-Barr virus; LTx, lung transplantation; RA bronchiolitis, rheumatoid arthritis bronchiolitis.
aAll other received single lung transplantations.
bDonor being positive for antibodies against CMV/EBV and recipient being negative prior to transplantation.
Kinetics of TTV- and EBV-DNA Levels
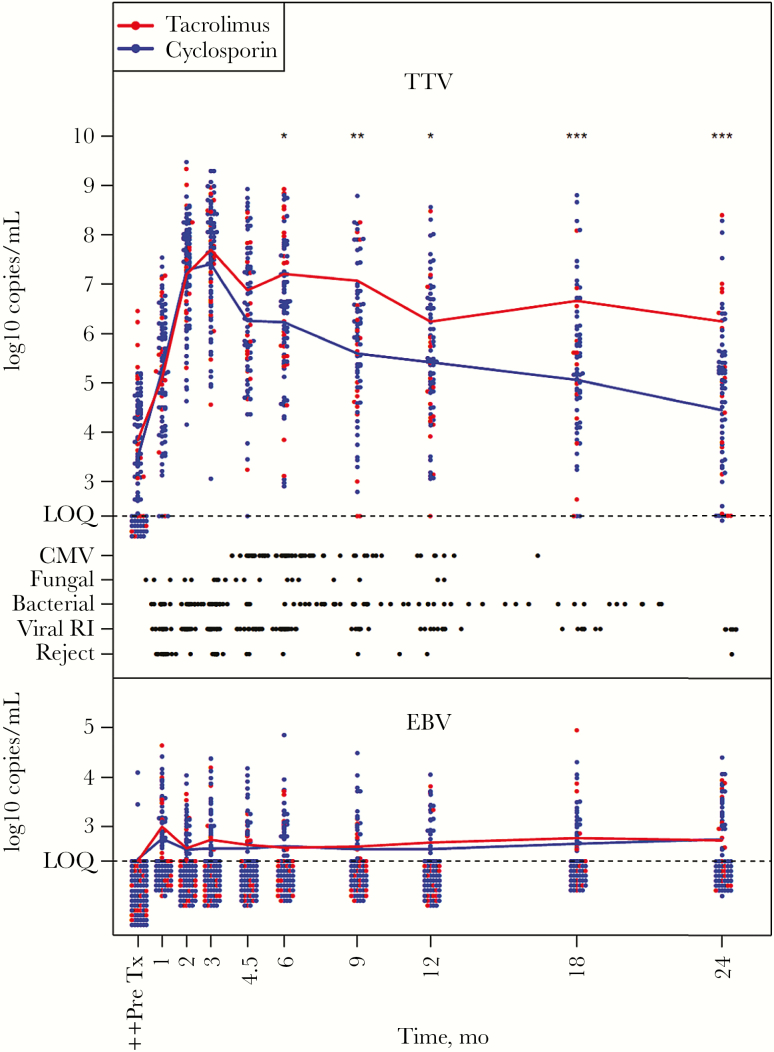
A majority of the patients had detectable TTV-DNA levels before transplantation (pre-LTx), and the levels subsequently increased until a peak was reached at 3 months post-LTx, after which the levels gradually decreased (Figure 1). The mean TTV-DNA levels for the respective time periods were 6.7 log10 1–3 months post-LTx, 6.4 log10 3–6 months post-LTx, 5.8 log10 6–12 months post-LTx, and 5.2 log10 12–24 months post-LTx, respectively. Conversely, a majority of the patients had no detectable EBV-DNA prior to transplantation as determined in serum. At 1 month post-LTx, a peak in mean EBV-DNA levels was observed, which decreased already 2 months post-LTx and thereafter remained at a relatively constant level, determined in whole blood (Figure 1). The mean EBV-DNA levels for the respective time periods were 2.7 log10 1–3 months post-LTx, 2.6 log10 3–6 months post-LTx, 2.6 log10 6–12 months post-LTx, and 2.7 log10 12–24 months post-LTx, respectively. Overall, fewer patients had detectable levels of EBV-DNA compared with TTV-DNA (Figure 1). Although the individual TTV-DNA levels varied, a majority of the patients had detectable levels across the entire FU period. In comparison, only a minority of the patients had detectable levels of EBV-DNA over a longer consecutive period, and a large fraction had EBV-DNA levels below the level of quantification (LOQ) at any given time point during FU (Figure 1). Three patients developed suspected PTLD. No case was biopsy-verified, and all were reversed upon modification of immunosuppression. Detailed individual TTV- and EBV-DNA level kinetics for each patient, including information regarding all infectious events and acute rejections, are displayed in Supplementary Figure 1.
Figure 1.
Kinetics of torque teno virus (TTV)– and Epstein-Barr virus (EBV)–DNA levels, in serum and whole blood respectively, before and during the follow-up period after lung transplantation. TTV-DNA levels in serum starting pre–lung transplantation (LTx) were determined by real-time polymerase chain reaction (PCR). Individual TTV levels are indicated by dots. EBV-DNA levels in whole blood post-LTx were determined by real-time PCR, and individual EBV levels are indicated by dots. Patients receiving either Tacrolimus treatment (n = 19) or Cyclosporine treatment (n = 79) are indicated by red and blue dots, respectively. The mean levels of TTV and EBV in Tacrolimus-treated patients are indicated by red lines in their respective graphs. The mean levels of TTV and EBV in Cyclosporine-treated patients are indicated by blue lines in their respective graphs. Infectious events and acute rejections are indicated by black dots. Statistical calculations were done using Mann-Whitney U (P values are indicated by *<.05, **<.01, and ***<.001). ++The levels of EBV-DNA pre-LTx were determined in serum samples. Abbreviations: CMV, cytomegalovirus, reject: acute rejection of which all but 3 were biopsy-verified; LOQ, level of quantification; viral RI, viral respiratory tract infection.
Type of Immunosuppressant Influences TTV- but not EBV-DNA Levels
Comparison of TTV- and EBV-DNA levels in lung transplant recipients who received either Tacrolimus- or Cyclosporine-based therapy revealed that Cyclosporine-treated patients had significantly lower TTV-DNA levels in serum at month 6 post-LTx and onwards, compared with the Tacrolimus-treated patients (Figure 1). There was no significant difference in EBV-DNA levels in whole blood between Tacrolimus- and Cyclosporine-treated patients at any time point during FU (Figure 1).
Infectious Events and Acute Rejections
The FU was divided into 4 periods, denoted q1 through q4, defined as 1–3 (q1), 3–6 (q2), 6–12 (q3), and 12–24 (q4) months after transplantation. The periodic intervals were chosen to distinguish the initial phase of intensive immunosuppression from the later periods when immunosuppressive therapy was gradually lowered. Viral infections (VRTI) were more common during the first 6 months, while bacterial and fungal infections were evenly distributed throughout the period and CMV viremia occurred at a higher frequency 3–12 months after LTx after discontinuation of prophylactic ganciclovir treatment (Table 2). Events of acute rejections were most common during the first postoperative months and then decreased over time (Figure 1). All but 3 episodes of acute rejections were biopsy-verified.
Table 2.
Number of Infectious Events and Acute Rejections During Each Period, q1 (1–3 Months Post-LTx), q2 (3–6 Months Post-LTx), q3 (6–12 Months Post-LTx), and q4 (12–24 Months Post-LTx)
| Type of Event | q1 | q2 | q3 | q4 |
|---|---|---|---|---|
| Fungal infections | 7 (4) | 8 (4) | 7 (1) | 2 (1) |
| Bacterial infections | 17 (13) | 17 (9) | 26 (16) | 18 (8) |
| Viral RI | 62 | 44 | 28 | 21 |
| CMV viremia | 0 | 30 (4) | 26 (1) | 5 |
| Acute rejections | 13 | 8 | 3 | 0 |
| Deaths | 2 | 4 | 5 | 8 |
| Patients at risk | 98 | 96 | 92 | 87 |
Number of severe infectious events that prompted intravenous treatment are denoted within parentheses.
Abbreviations: CMV, human cytomegalovirus; LTx, lung transplantation; RI, respiratory infections.
The frequency of infectious events (Figure 1 and Table 2) was compared with the mean TTV-DNA or EBV-DNA levels during each period (q1–q4) and the during the total FU period, respectively. No statistically significant association was found. With logistic regression, log TTV-DNA or log EBV-DNA levels in the beginning of the period did not predict any infectious event in any of the periods (Supplementary Table 2).
Next, we performed a Cox regression analysis to establish if TTV- or EBV-DNA levels were associated with either viral respiratory tract infections, bacterial infections, fungal infections, CMV viremia, or any infectious event. TTV and EBV levels were added as time-varying covariates to provide the most accurate statistical model possible. No significant associations were found in a univariable analysis (Table 3) or when both were used as predictors (data not shown). A separate Cox regression analysis, including only patients with severe infectious events who required intravenous treatment, was performed to establish the relation between TTV- or EBV-DNA levels and bacterial infection, fungal infection, CMV viremia, or a combination of the 3 (Supplementary Table 3). No significant associations were found using this criterion.
Table 3.
Cox Regression in the Time Frame 3–24 Months With TTV and EBV as Time-Dependent Covariates Was Used to Analyze the Relationship With Respective Outcome Variables
| TTV | EBV | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |||
| VRTI | 1.09 | 0.92 | 1.29 | .32 | 1.20 | 0.82 | 1.75 | .37 |
| Bact | 0.94 | 0.73 | 1.20 | .62 | 1.62 | 0.99 | 2.66 | .06 |
| Fungi | 0.97 | 0.48 | 1.92 | .92 | 1.77 | 0.78 | 4.02 | .17 |
| CMV | 0.91 | 0.76 | 1.10 | .34 | 1.02 | 0.62 | 1.67 | .95 |
| IE | 0.98 | 0.87 | 1.11 | .77 | 1.11 | 0.90 | 1.46 | .43 |
Abbreviations: Bact, bacterial infection; CI, confidence interval; CMV, human cytomegalovirus viremia; EBV, Epstein-Barr virus; Fungi, fungal infection; HR, hazard ratio; IE, infectious event of any type; TTV, torque teno virus; VRTI, viral respiratory tract infections.
When comparing other factors relevant to infections and acute rejection, using logistic regression, we found that age, CMV mismatch, and dominant immunosuppressive treatment did not predict outcome in any of the 4 periods denoted q1 through q4 (Supplementary Table 4).
Most acute rejection events (ARs) occurred within the first 6 months post-LTx (Figure 1). Logistic regression with initial TTV-DNA or EBV-DNA for each period, respectively, did not predict whether an acute rejection event would occur during that period (data not shown).
Absolute Quantification of TTV-DNA in Serum
Previous studies [15, 24] have described higher mean TTV-DNA levels in lung transplant patients than reported here. In order to verify the real-time PCR results, TTV-DNA levels of patients with mean TTV-DNA levels above 8 log10 in period q3 were assessed by digital droplet PCR (ddPCR). The TTV-DNA level was underestimated by more than 1 log10 by real-time PCR in 2 samples, and overall the levels obtained by real-time PCR correlated well with the results obtained by ddPCR (Pearson r = 0.67, P = .009) (Supplementary Table 5).
DISCUSSION
TTV and EBV has been proposed as biomarkers reflecting the functionality of the immune system after LTx. In this prospective study, evaluating serial samples for TTV and EBV and carefully recording infectious and rejection events during an FU period of 24 months, we found no associations between the biomarkers and the risk of complications.
We found that TTV-DNA levels increased rapidly; 1 month after LTx, the levels were already markedly elevated, presumably due to ablation of the functional effector cells of the immune system that normally control the TTV infection [14, 15]. At 3 months post-LTx, the mean TTV-DNA level peaked and then gradually declined, reflecting the gradual moderation of immunosuppressive therapy. Interestingly, the TTV-DNA level was lower in patients who received Cyclosporine treatment, possibly mirroring a different immune modulatory mechanism for Cyclosporine compared with Tacrolimus. Görzer et al. [16] previously suggested that this might be due to Cyclosporine being less efficient as an immunosuppressant. However, we found no association between type of immunosuppressive regimen and acute rejection or other events that would indicate a difference in net immunosuppressive effect. Recently, TTV-DNA has been associated with chronic lung allograft dysfunction after lung transplantation [29], but in our study this was not included as an outcome.
It is possible that the observed higher mean TTV-DNA levels in other studies could be explained by differences in induction therapy and distribution of respective immunosuppressive regimen. For example, in the study by Görzer et al. [16], the type of induction therapy was not defined and only 26% of the patients were treated with Cyclosporine. There is some evidence that TTV replication is dependent on the type of induction therapy that appears to reflect changes in lymphocyte concentration; however, the duration is brief, after which the levels of TTV recover [30]. In the present study, TTV viral load was determined in serum samples, whereas plasma was used in previous studies [16, 31]. However, to our knowledge, there are no apparent reasons for variations in TTV-DNA load, depending on whether plasma or serum is being used for analysis.
To compare results regarding TTV levels between different transplantation centers, it is vital to reliably quantify the concentrations of TTV-DNA in a standardized manner. Here it was shown that the real-time PCR method was adequate as compared with the ddPCR method, which is considered a more reliable method for accurate quantification [32]. Nonetheless, real-time PCR is prone to interlaboratory differences, and utilization of ddPCR for quantification has been suggested to circumvent this problem, facilitating direct comparison of results between centers [33, 34]. Digital droplet PCR depends on partitioning of each master mix followed by end point PCR. Quantitation is determined using Poisson statistics to generate a result that is not dependent on a relation to a standard curve and should therefore exhibit lower variability between laboratories. However, the ddPCR assays still depend on amplification of a specific target sequence, and depending on the design of the primer and probe, the targeted sequence can vary. Recent work has shown that viral DNA in plasma samples is to a large extent free and not associated with viral particles and is also subjected to various degrees of degradation, influencing the results, depending on the amplicon length determined by the PCR-assay design [35]. Thus, standardization of the entire assay including primer and probe design, use of international standards, and assessment of the commutability of reference material is needed before a direct comparison of interlaboratory results can be made with absolute confidence, even with the use of ddPCR [36, 37].
It remains to be clarified in which cell types TTV replication occurs, but CD4+ T cells and CD8 + 57+ T cells appear to be important for controlling the infection, whereas EBV resides within the B-cell pool and is controlled by CD4+ and CD8+ T cells [14, 15, 17, 21, 22, 38–41]. In this work, we show that the choice of calcineurin inhibitor affects the TTV- but not the EBV-DNA levels, possibly reflecting separate mechanisms for viral replication rather than merely ablation of the T-cell pool. As the choice of immunosuppressive regimen was made at the discretion of the treating physician, these findings should be interpreted with caution but warrant further investigation.
In this study, infectious events were defined as symptoms prompting antimicrobial therapy also in cases without a positive culture (bacterial and fungal infections) or simply detection of a potential pathogen by PCR (VRTI, CMV). Attempting to focus on more severe infectious events, subgroup analysis of events requiring intravenous therapy was made. Also, this analysis failed to reveal any significant association between TTV- or EBV-DNA levels and infectious events. However, no proper validated scoring system for grading of the severity of infectious complications was used in this study, and we cannot exclude that our definitions cause underestimation of severe complications. Further studies using proper grading of events are needed.
EBV has previously been suggested as a surrogate marker for immunosuppression as an association between elevated EBV-DNA levels and reduced incidence of acute rejection of lung transplants was observed [23]. In addition, EBV-DNA was safely used as a trigger for reduction of immunosuppression late after lung transplantation and was found to be associated with a shorter time to development of infectious complications or solid tumors, but not PTLD [24, 42]. We found no association between EBV-DNA and acute rejection at any time during FU. A direct comparison could, however, be hampered by the fact that EBV-DNA was analyzed in this study, and EBV-encoded RNA in an earlier study [23].
In the present study, mean EBV-DNA levels did not increase as markedly as TTV-DNA levels after transplantation, even though EBV-specific T cells most likely are subverted by immunosuppressive therapy [43]. The seroprevalence pre-LTx and number of patients positive for EBV-DNA at any time during FU were comparable to previous studies [24, 42, 44]. One possible caveat when determining EBV DNA load is the use of valgangciclovir for preventing reactivation or primary infection of CMV. All patients received 3 months of valgangciclovir treatment initially after LTx; there are a few studies showing that valgangciclovir treatment reduces EBV load, which may have affected the results in this study [45, 46].
In conclusion, TTV- but not EBV-DNA load appears to reflect the function of the immune system after lung transplantation, depending on the type of immunosuppressive treatment. However, we found no association between either TTV- or EBV-DNA load and infectious events or acute rejections, which suggests a limited clinical applicability as biomarkers predicting short-term outcomes related to the net state of immunosuppression. Further studies are warranted to define the effect of immunomodulation on TTV and EBV replication in relation to various immunosuppressive regimens.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This project was supported by funding from Stiftelsen Professor Lars-Erik Gelins Minnesfond, FoU Laboratoriemedicin, Sahlgrenska University Hospital, the Swedish Heart-Lung Foundation, the Region Västra Götaland Research funds (VGFOUREG-228341, VGFOUREG-82811), Regional ALF funds (ALFGBG-217671, ALFGBG-439391), and Region Halland.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chambers DC, Yusen RD, Cherikh WS et al. ; International Society for Heart and Lung Transplantation The Registry of the International Society for Heart and Lung Transplantation: Thirty-fourth Adult Lung And Heart-Lung Transplantation Report-2017; focus theme: allograft ischemic time. J Heart Lung Transplant 2017; 36:1047–59. [DOI] [PubMed] [Google Scholar]
- 2. Allison AC, Eugui EM. Purine metabolism and immunosuppressive effects of mycophenolate mofetil (MMF). Clin Transplant 1996; 10:77–84. [PubMed] [Google Scholar]
- 3. Tedesco D, Haragsim L. Cyclosporine: a review. J Transplant 2012; 2012:230386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomson AW, Bonham CA, Zeevi A. Mode of action of tacrolimus (FK506): molecular and cellular mechanisms. Ther Drug Monit 1995; 17:584–91. [DOI] [PubMed] [Google Scholar]
- 5. Norder H, Karlsson M, Mellgren Å et al. . Diagnostic performance of five assays for anti-hepatitis E virus IgG and IgM in a large cohort study. J Clin Microbiol 2016; 54:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis 1983; 147:676–84. [DOI] [PubMed] [Google Scholar]
- 7. Gallian P, Berland Y, Olmer M et al. . TT virus infection in French hemodialysis patients: study of prevalence and risk factors. J Clin Microbiol 1999; 37:2538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spandole S, Cimponeriu D, Berca LM, Mihăescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol 2015; 160:893–908. [DOI] [PubMed] [Google Scholar]
- 9. Nishizawa T, Okamoto H, Konishi K et al. . A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun 1997; 241:92–7. [DOI] [PubMed] [Google Scholar]
- 10. Biagini P. Classification of TTV and related viruses (anelloviruses). Curr Top Microbiol Immunol 2009; 331:21–33. [DOI] [PubMed] [Google Scholar]
- 11. Biagini P, Uch R, Belhouchet M et al. . Circular genomes related to anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequence-independent single primer amplification approach. J Gen Virol 2007; 88:2696–701. [DOI] [PubMed] [Google Scholar]
- 12. Okamoto H. History of discoveries and pathogenicity of TT viruses. Curr Top Microbiol Immunol 2009; 331:1–20. [DOI] [PubMed] [Google Scholar]
- 13. Focosi D, Antonelli G, Pistello M, Maggi F. Torquetenovirus: the human virome from bench to bedside. Clin Microbiol Infect 2016; 22:589–93. [DOI] [PubMed] [Google Scholar]
- 14. Moen EM, Sagedal S, Bjøro K et al. . Effect of immune modulation on TT virus (TTV) and TTV-like-mini-virus (TLMV) viremia. J Med Virol 2003; 70:177–82. [DOI] [PubMed] [Google Scholar]
- 15. Maggi F, Focosi D, Albani M et al. . Role of hematopoietic cells in the maintenance of chronic human torquetenovirus plasma viremia. J Virol 2010; 84:6891–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Görzer I, Haloschan M, Jaksch P et al. . Plasma DNA levels of torque teno virus and immunosuppression after lung transplantation. J Heart Lung Transplant 2014; 33:320–3. [DOI] [PubMed] [Google Scholar]
- 17. Christensen JK, Eugen-Olsen J, SŁrensen M et al. . Prevalence and prognostic significance of infection with TT virus in patients infected with human immunodeficiency virus. J Infect Dis 2000; 181:1796–9. [DOI] [PubMed] [Google Scholar]
- 18. De Vlaminck I, Khush KK, Strehl C et al. . Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 2013; 155:1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Béland K, Dore-Nguyen M, Gagné MJ et al. . Torque teno virus in children who underwent orthotopic liver transplantation: new insights about a common pathogen. J Infect Dis 2014; 209:247–54. [DOI] [PubMed] [Google Scholar]
- 20. Balfour HH Jr, Sifakis F, Sliman JA et al. . Age-specific prevalence of Epstein-Barr virus infection among individuals aged 6-19 years in the United States and factors affecting its acquisition. J Infect Dis 2013; 208:1286–93. [DOI] [PubMed] [Google Scholar]
- 21. Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity 1998; 9:395–404. [DOI] [PubMed] [Google Scholar]
- 22. Hurley EA, Thorley-Lawson DA. B cell activation and the establishment of Epstein-Barr virus latency. J Exp Med 1988; 168:2059–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahya VN, Douglas LP, Andreadis C et al. . Association between elevated whole blood Epstein-Barr virus (EBV)-encoded RNA EBV polymerase chain reaction and reduced incidence of acute lung allograft rejection. J Heart Lung Transplant 2007; 26:839–44. [DOI] [PubMed] [Google Scholar]
- 24. Bakker NA, Verschuuren EA, Erasmus ME et al. . Epstein-Barr virus-DNA load monitoring late after lung transplantation: a surrogate marker of the degree of immunosuppression and a safe guide to reduce immunosuppression. Transplantation 2007; 83:433–8. [DOI] [PubMed] [Google Scholar]
- 25. De Pauw B, Walsh TJ, Donnelly JP et al. ; European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group; National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 2008; 46:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stewart S, Fishbein MC, Snell GI et al. . Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant 2007; 26:1229–42. [DOI] [PubMed] [Google Scholar]
- 27. Kullberg-Lindh C, Olofsson S, Brune M, Lindh M. Comparison of serum and whole blood levels of cytomegalovirus and Epstein-Barr virus DNA. Transpl Infect Dis 2008; 10:308–15. [DOI] [PubMed] [Google Scholar]
- 28. Andersson ME, Olofsson S, Lindh M. Comparison of the FilmArray assay and in-house real-time PCR for detection of respiratory infection. Scand J Infect Dis 2014; 46:897–901. [DOI] [PubMed] [Google Scholar]
- 29. Görzer I, Jaksch P, Strassl R et al. . Association between plasma torque teno virus level and chronic lung allograft dysfunction after lung transplantation. J Heart Lung Transplant 2017; 36:366–8. [DOI] [PubMed] [Google Scholar]
- 30. Focosi D, Macera L, Boggi U et al. . Short-term kinetics of torque teno virus viraemia after induction immunosuppression confirm T lymphocytes as the main replication-competent cells. J Gen Virol 2015; 96:115–7. [DOI] [PubMed] [Google Scholar]
- 31. Görzer I, Jaksch P, Kundi M et al. . Pre-transplant plasma torque teno virus load and increase dynamics after lung transplantation. PLoS One 2015; 10:e0122975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicot F, Cazabat M, Lhomme S et al. . Quantification of HEV RNA by droplet digital PCR. Viruses 2016; 8:doi: 10.3390/v8080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson GL, Bibby DF, Wong S et al. . A MIQE-compliant real-time PCR assay for Aspergillus detection. PLoS One 2012; 7:e40022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hindson CM, Chevillet JR, Briggs HA et al. . Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013; 10:1003–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tong Y, Pang XL, Mabilangan C, Preiksaitis JK. Determination of the biological form of human cytomegalovirus DNA in the plasma of solid-organ transplant recipients. J Infect Dis 2017; 215:1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang L, Sun Y, Buelow D et al. . Quantitative assessment of commutability for clinical viral load testing using a digital PCR-based reference standard. J Clin Microbiol 2016; 54:1616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preiksaitis JK, Hayden RT, Tong Y et al. . Are we there yet? Impact of the first international standard for cytomegalovirus DNA on the harmonization of results reported on plasma samples. Clin Infect Dis 2016; 63:583–9. [DOI] [PubMed] [Google Scholar]
- 38. Subklewe M, Paludan C, Tsang ML et al. . Dendritic cells cross-present latency gene products from Epstein-Barr virus-transformed B cells and expand tumor-reactive CD8(+) killer T cells. J Exp Med 2001; 193:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Münz C, Bickham KL, Subklewe M et al. . Human CD4(+) T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J Exp Med 2000; 191:1649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Focosi D, Bestagno M, Burrone O, Petrini M. CD57+ T lymphocytes and functional immune deficiency. J Leukoc Biol 2010; 87:107–16. [DOI] [PubMed] [Google Scholar]
- 41. Maggi F, Fabrizio M, Ricci V et al. . Changes in CD8 + 57+ T lymphocyte expansions after autologous hematopoietic stem cell transplantation correlate with changes in torquetenovirus viremia. Transplantation 2008; 85:1867–8. [DOI] [PubMed] [Google Scholar]
- 42. San-Juan R, De Dios B, Navarro D et al. . Epstein-Barr virus DNAemia is an early surrogate marker of the net state of immunosuppresion in solid organ transplant recipients. Transplantation 2013; 95:688–93. [DOI] [PubMed] [Google Scholar]
- 43. Rickinson AB, Moss DJ. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu Rev Immunol 1997; 15:405–31. [DOI] [PubMed] [Google Scholar]
- 44. Engelmann I, Welte T, Fühner T et al. . Detection of Epstein-Barr virus DNA in peripheral blood is associated with the development of bronchiolitis obliterans syndrome after lung transplantation. J Clin Virol 2009; 45:47–53. [DOI] [PubMed] [Google Scholar]
- 45. Hierro L, Díez-Dorado R, Díaz C et al. . Efficacy and safety of valganciclovir in liver-transplanted children infected with Epstein-Barr virus. Liver Transpl 2008; 14:1185–93. [DOI] [PubMed] [Google Scholar]
- 46. Yager JE, Magaret AS, Kuntz SR et al. . Valganciclovir for the suppression of Epstein-Barr virus replication. J Infect Dis 2017; 216:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.