Abstract
Genomic binding of transcription factors, like the glucocorticoid receptor (GR), is linked to the regulation of genes. However, as we show here, GR binding is a poor predictor of GR-dependent gene regulation even when taking the 3D organization of the genome into account. To connect GR binding sites to the regulation of genes in the endogenous genomic context, we turned to genome editing. By deleting GR binding sites, individually or in combination, we uncovered how cooperative interactions between binding sites contribute to the regulation of genes. Specifically, for the GR target gene GILZ, we show that the simultaneous presence of a cluster of GR binding sites is required for the activity of an individual enhancer and that the GR-dependent regulation of GILZ depends on multiple GR-bound enhancers. Further, by deleting GR binding sites that are shared between different cell types, we show how cell type-specific genome organization and enhancer-blocking can result in cell type-specific wiring of promoter–enhancer contacts. This rewiring allows an individual GR binding site shared between different cell types to direct the expression of distinct transcripts and thereby contributes to the cell type-specific consequences of glucocorticoid signaling.
INTRODUCTION
Transcription factors (TFs) play a pivotal role in specifying which genes are expressed in a given cell. The regulation of gene expression requires the binding of these TFs to cis-regulatory elements. Such cis-regulatory elements can either act promoter-proximal or control the expression of genes that can be located up to several Mb away (1,2). TFs can bind to tens of thousands of genomic binding sites (3), yet they seem to regulate a much smaller number of genes (4–6). Consequently, it is mostly unclear which TF-bound regions are responsible for the regulation of individual genes and what discriminates productive TF binding events that are linked to the regulation of genes from apparently non-productive interactions of TFs with the genome. For distal TF-bound elements, productive binding is more likely when the 3D organization of the genome brings such regions in proximity to a promoter as was shown using genome-wide chromosome conformation capture-based approaches such as Hi-C (7). The CCCTC-binding factor (CTCF) plays a key role in the organization of chromatin architecture and the formation of chromatin loops to facilitate long-range interactions (8,9). Moreover, CTCF is required for the enhancer-blocking activity of insulator elements (10,11), which can prevent cis-regulatory elements from acting on nearby promoters and thus influence whether a TF binding event contributes to the regulation of a nearby gene or not. In addition to the 3D organization of the genome, the combinatorial nature of regulatory elements might explain why only certain TF binding events result in the regulation of genes (12). For example, a productive TF binding event might require the concerted recruitment of the right combination of TFs to a response element. Accordingly, reporter assays have shown examples where individual binding sites for a TF do not activate transcription and that synergistic interactions between either multiple binding sites for the same TF or between binding sites for different TFs are required for activity (13,14). Regulation of some genes might even require the concerted action of multiple response elements located at distinct genomic loci that can either act additively or synergistically (15,16). However, combinatorial regulation by multiple TF binding sites has only been investigated for a limited number of endogenous genomic loci. Additionally, most studies suffer from a limited resolution because TF binding sites were deleted in their genomic context as part of bigger genomic fragments.
Enhancer usage is not hard-wired but dynamic and can change depending on cell type (17,18), differentiation state (19) and environmental conditions (20). For example, the glucocorticoid receptor (GR), a hormone-activated TF, regulates distinct sets of genes depending on the cell type (21,22). Those cell type-specific transcriptional consequences of glucocorticoid signaling can be partially explained by differential binding patterns of GR in different cell types (23,24). Another mechanism to generate cell type-specific consequences of TF binding could be via cell type-specific synergy at GR-bound regulatory elements harboring binding sites for GR and for other TFs (13). Finally, cell type-specific gene regulation could arise from changes in the regulatory activity of GBSs shared between cell types. To date, however, the role of TF binding sites that are shared between cell types on cell type-specific gene expression programs has not been investigated in the genomic context.
Here, we set out to study the relation between GR binding and the regulation of genes in the genomic context using two distinct approaches. First, we analyzed the genome-wide connection between GR binding sites and the GR-dependent regulation of genes computationally. Importantly, only few studies have determined which fraction of the observed TF-dependent regulation can be explained by TF binding rather than assessing statistically if TF binding and the regulation of genes are connected. Our approach uncovered that GR binding and the regulation of genes are associated and that this connection becomes tighter when the cell type-specific 3D organization of the genome in the nucleus is taken into account. However, GR binding is a poor predictor of gene regulation with the majority of genes not changing their expression upon GR binding. Therefore, as a second approach, we exploited genome-editing tools (25,26) to study how individual GR binding sites contribute to the regulation of genes. By deleting a number of GR binding sites individually or in combination in two different cell types, we show that cooperative interactions between multiple GR binding sites are needed for GR-dependent gene regulation. Furthermore, we uncover how cell type-specific wiring of promoter–enhancer contacts contributes to the cell type-specific consequences of glucocorticoid signaling.
MATERIALS AND METHODS
Cell lines, plasmids, transient transfections, genome editing using CRISPR/Cas9, luciferase assays, FISH and RNA preparation and analysis
A549 (ATCC CCL-185), U2OS and U2OS-GR18 cells stably expressing rat GRα (27) were cultured in DMEM supplemented with 5% FBS. Transient transfections, genome editing, immunoblotting, cloning, RNA isolation and analysis, FISH and luciferase assays are described in detail in the Supplemental Methods.
dCas9-SAM activation of endogenous target genes
The ability of dCas9-SAM to activate endogenous target genes was tested by transfecting 600 ng each of dCas9-VP64, MS2-p65-HSF1 activator complex, the modified gRNA including two MS2 stem loops and a GFP expression construct by nucleofection (Lonza) according to the manufacturer's instructions. Twenty four hours post-transfection, GFP-positive cells were isolated by FACS sorting, RNA was isolated and analyzed by qPCR.
ChIP, ChIP-seq and 4C-seq
ChIP assays targeting GR were performed as described (28). ChIP assays targeting CTCF (polyclonal CTCF-antibody from Active Motif, Cat. No. 61311) were essentially done as described (28) except that a modified RIPA wash buffer (50 mM HEPES–KOH, 1 mM EDTA, 1% NP40, 0.7% Na-deoxycholat, 500 mM LiCl, pH 7.5) was used. For ChIP assays targeting H3K27ac, 1 μg of Diagenode ChIP-seq grade rabbit polyclonal antibody (H3K27Ac; pAb-196-050) were used per ChIP. 4C experiments were performed as previously described (29), using 5 × 106 dexamethasone-treated cells (1 μM, 90 min). For more details see Supplemental Methods.
Computational analysis
Correlating genomic GR binding with gene regulation
RNA-seq data after 0 and 4 h of treatment with 100 nM dexamethasone was downloaded from GEO (GEO:GSE91305, GEO:GSE91243). Differentially expressed genes were determined with DESeq2 (30). ChIP-seq data for GR binding after 1 h of dexamethasone treatment and H3K27ac after 4 h of dexamethasone treatment were obtained as fastq from (GEO:GSE91285, GEO:GSE91357 and GEO:GSE91347, GEO:GSE91282), mapped with Bowtie2 (31) to reference genome GRCh38, filtered and duplicate reads were removed. Peak calling for GR was performed with MACS2 (32). Chromatin interactions and chromatin domains from Hi-C after 4 h dexamethasone treatment were of taken from (GEO:GSE92804). For more details see Supplemental Methods.
ChIP-seq and 4C-seq analysis
ChIP-seq data were mapped with Bowtie2 (31) to reference genome hg19, filtered and duplicate reads were removed. For 4C-seq, primer sequences were extended to the next 3′ restriction site and clipped from short reads allowing up to three mismatches for the identification of primers. Clipped short reads were mapped in single-end mode to reference genome hg19 using BWA-MEM v0.7.12 (33,34) and sorted by name afterward. The reference genome was digested virtually according to the first cutter CviQI to obtain restriction fragments. Reads were counted per fragment to obtain interaction profiles. For more details, see Supplemental Methods.
DNA sequence motif analysis
To identify CTCF binding motif-matches at CTCF-ChIP peaks (Figure 7C and Supplementary Figure S7), we analyzed regions of interest using the Transcription Factor Affinity Prediction (TRAP) webtool (35). For more details, see Supplemental Methods.
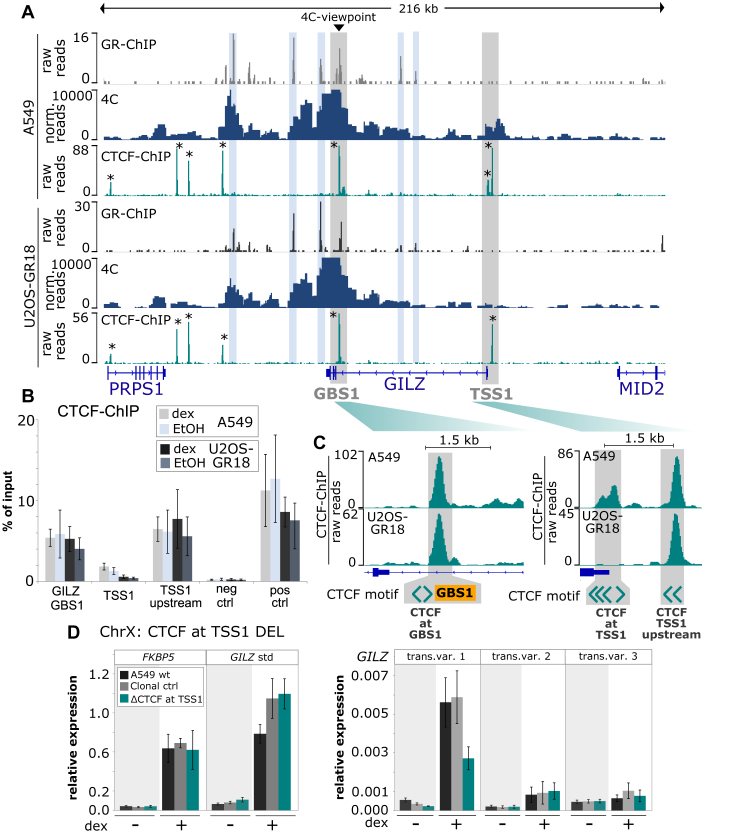
Figure 7.
Cell type-specific 4C-seq and CTCF binding profiles. (A) GR ChIP-seq, 4C-seq and CTCF ChIP-seq for a 216 kb region encompassing the GILZ gene are shown for (top) A549 and (bottom) U2OS-GR18 cells treated with dexamethasone. The GILZ GBS1 viewpoint for the 4C experiment and the promoter region of transcript variant 1 (TSS1) are highlighted in gray, GR-bound regions in blue and CTCF-bound regions with an *. (B) ChIP-qPCR of CTCF-binding at GILZ GBS1 and around GILZ TSS1 in wild type U2OS-GR18 and A549 cells. Average percentage of input immunoprecipitated ± SEM (n = 3) are shown for cells treated with vehicle (EtOH) and for cells treated for 90 minutes with 1 μM dex. (C) Zoom-in and schematic representation of CTCF binding, GBS1 and the location and orientation of CTCF motif-matches at the GILZ GBS1 and TSS1 regions. (D) Relative mRNA expression levels in A549 cells as determined by qPCR for FKBP5 and GILZ transcript variants as indicated for wt A549, for the clonal cell line with deleted CTCF motifs at the promoter region of GILZ transcript variant 1 or for clonal lines unedited at the GILZ locus. Averages ± SEM are shown for three independent experiments in cells treated with vehicle and for cells treated overnight with 1 μM dex.
RESULTS
Target gene prediction based on genome-wide GR binding benefits from integrating information regarding the 3D organization of the genome
To study the global connection between GR binding and GR-dependent gene regulation, we combined data from genome-wide GR binding experiments (Chromatin Immunoprecipitation followed by sequencing (ChIP-seq)) with RNA-seq data regarding gene expression changes upon GR-activation in A549 cells (3). Similar to the Jin et al. study (7) we restricted our analysis to GR peaks with high H3K27ac levels in hormone-treated cells (active GR peaks). We grouped genes by the distance between the transcription start site (TSS) and the nearest active GR peak. As expected, we find that genes with GR peaks are more likely to be regulated by GR than genes that do not harbor a GR peak, especially when the GR ChIP-seq peaks are close the TSS (Figure 1A). However, regardless of the distance between the GR peak and the TSS of a gene, the majority of genes that have a GR peak are not regulated by GR. Consequently, GR binding is a poor predictor of GR-dependent gene regulation and additional information is needed to discriminate productive GR binding events that result in the regulation of associated genes from non-productive binding events that do not result in obvious changes in gene expression. Part, but likely not all, of the disconnect between GR binding and regulation might be explained by false-positive GR ChIP-seq peaks and by genes that are regulated at other time points than the one examined (4 h) and are thus incorrectly classified as non-regulated. Furthermore, assigning enhancers to target genes is complicated by the fact that they can either regulate the expression of the closest gene, but also of other genes that are located further away on the linear genome (2,36,37).
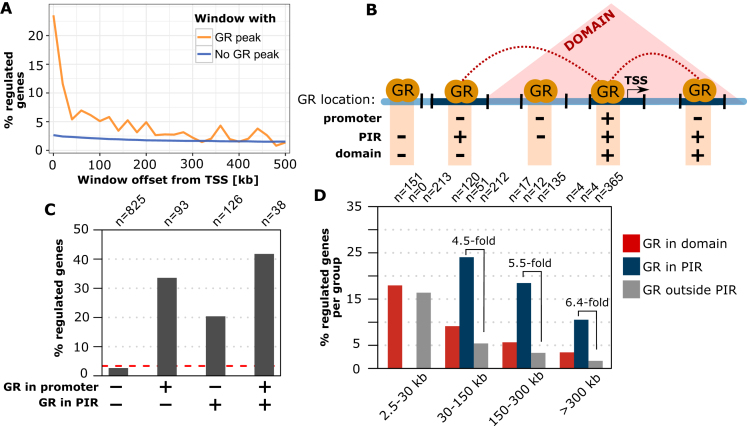
Figure 1.
Linking GR binding to the GR-dependent regulation of genes. (A) Percentage of genes regulated by GR in A549 cells (absolute log2 fold change (|log2FC|) upon dexamethasone treatment >0.5 and adjusted P-value (P-adj) < 0.01) versus distance between the TSS and the nearest active GR peak (orange curve). Genes were grouped in distance intervals of 20 kb. Genes without an active GR peak between the TSS and the end of the distance interval are shown as a blue curve. (B) Cartoon depicting the different regions that were used to test if the link between GR binding and gene regulation benefits from incorporating information regarding the 3D organization of the genome. Promoter regions: TSS of genes ±5 kb; PIR: promoter interacting region with increased contact frequencies with promoter regions based on Hi-C data (GEO: GSE92804); Domain: contact domains obtained from Hi-C. (C) Percentage of genes regulated by GR (|log2FC| > 0.5 and P-adj < 0.01) for different GR binding configurations: (i) without an active GR peak in the promoter region and all promoter interacting regions. The dashed red line represents the percentage of all genes that is regulated (ii) active GR peak in the promoter region only, (iii) active GR peak in PIR only and (iv) active GR peak in the promoter region and at least one PIR. (D) Percentage of genes regulated by GR (|log2FC| > 0.5 and P-adj < 0.01) for genes grouped by the location of the nearest active GR peak: (i) in a PIR (blue bars), (ii) in the same chromatin domain (red bars), and (iii) outside of PIRs (gray bars).
The link between GR binding and regulation is especially weak for distal peaks located at large distances from the TSS of genes. One likely explanation for this is that many of these remote peaks lack the physical proximity to the promoter, which can be provided by chromatin looping. We therefore hypothesized that integrating Hi-C data, regarding the 3D organization of the genome in the nucleus of A549 cells (3), might help to explain why only certain GR binding events result in the regulation of genes. To test our hypothesis, we first grouped TSSs based on whether they contain a GR peak in either (i) the promoter region, (ii) in a distal promoter interacting region (PIR) looping to the promoter region, (iii) both promoter region and PIR or (iv) GR peak in neither promoter region nor distal PIR (Figure 1B). Consistent with our previous analysis, we found that genes with peaks in either their promoter region or in PIRs are more likely to be regulated by GR than genes that do not harbor a GR peak (Figure 1C). Interestingly, a high percentage (>40%) of genes was regulated when a GR peak is found in both promoter region and PIR. This implies that GR-dependent gene regulation is more likely when GR binds at multiple distinct regulatory regions. To test if long range-interactions based on Hi-C are a better predictor of gene regulation than assigning GR peaks to genes based on proximity, we compared GR peaks that fall within distal PIRs with their counterparts that are located at the same distance interval but do not display long-range interactions with the promoter. As a third group, we also analyzed GR peaks that map to the same contact domain (regions with increased contact frequencies, Figure 1B) in A549 cells as specified by another study (3). This analysis showed that incorporating data regarding the 3D organization of the genome strengthens the association between GR binding and gene regulation (Figure 1D). A tighter link between GR binding and regulation was found especially with PIRs, but also when contact domains were used for the analysis and was most evident for genes with GR peaks located >30 kb from the TSS (Figure 1D). Notably, the A549 cells we used here to study the correlation between GR-binding and GR-dependent gene regulation are derived from a cancer cell line with infinite life span. However, similar results were also obtained using primary human fibroblast cells (IMR90) for the computational analysis (Supplementary Figure S1).
Taken together, we find that (i) GR-binding and the regulation of genes are connected, (ii) that GR-dependent gene regulation is more likely when GR binds at multiple distinct regulatory regions and (iii) that the link between binding and regulation benefits from integrating information regarding the 3D organization of the genome in the nucleus, especially for binding sites located at distances >30 kb from the TSS of genes. Nonetheless, our data support findings by others (38) that TF binding is a poor predictor of gene regulation and consequently, experimental approaches are needed to causatively link GR binding to the regulation of genes.
Individual GR binding sequences contribute to the regulation of GR target genes
To experimentally test the contribution of the smallest regulatory units of gene regulation, individual GR binding sequences (GBSs), we used the CRISPR/Cas9-system (25) to delete selected GBSs in their endogenous genomic context. We chose a GBS located 1.5 kb upstream of the GR target gene GILZ (glucocorticoid induced leucine zipper, alias TSC22D3) and one GBS 1.5 kb upstream of the target gene DUSP1 (dual specificity phosphatase 1). These two genes play a role in mediating the immune-suppressive and anti-inflammatory actions of glucocorticoids (39,40). Candidate GBSs were chosen for several reasons. First, both GBSs map to GR-bound regions and are located near the TSS of GR target genes in U2OS-GR18 cells (Figure 2A–C), a U2OS osteosarcoma cell line stably expressing rat GRα (Supplementary Figure S5A) (27,41,42). Such TSS-proximal peaks are more likely to influence the expression of nearby genes than distal peaks (Figure 1A). Second, the selected GBSs contain a protospacer adjacent motif (PAM) necessary to direct the Cas9 nuclease to a strategic position within the core GR binding sequence (Figure 2D and E). Due to the positioning of the PAM most small and large deletions as well as insertions at the DNA cleavage site, which is located 3 bp upstream of the PAM (43), disrupt the spacing of the GR half-sites which is essential for GR binding (Supplementary Figure S2A). The importance of this became evident when we tried to disrupt GBSs at other loci where the PAM was directly flanking the GBS. For these GBSs, the typically short CRISPR/Cas9-induced insertions or deletions failed to disrupt the targeted GBS (data not shown). Third, studies by others have shown that disruption of the DUSP1 and GILZ GBSs in luciferase reporters containing the GR ChIP-seq region in which they are embedded, results in a marked reduction of GR-dependent transcriptional activation (41,42).
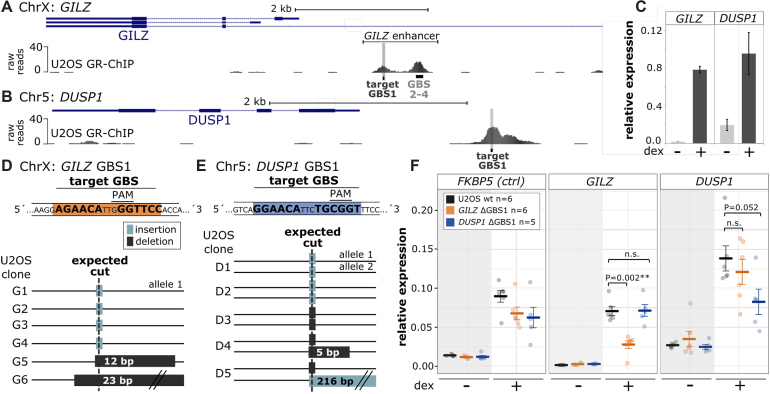
Figure 2.
Deletion of individual genomic GR binding sequences blunts the GR-dependent transcriptional regulation of nearby genes. (A, B) GR ChIP-seq tag density for dexamethasone (dex)-treated U2OS-GR18 cells stably expressing GR for (A) a promoter region of the GR target gene GILZ (hg18, ChrX:106 844 000–106 852 000) and (B) the GR target gene DUSP1 (hg18, Chr5:172 128 000–171 134 000). The GBS targeted for deletion is highlighted in gray for each locus. (C) Relative mRNA expression of the GILZ and DUSP1 genes upon overnight treatment with 1 μM dex was determined by qPCR. Averages ± standard error of mean (SEM) are shown (n = 3). (D, E) Top: Target GBS (highlighted in orange) and the position of the PAM for both loci. Bottom: Genotyping results of successfully edited single cell-derived clonal cell lines containing CRISPR/Cas9-induced insertions (light gray) or deletions (dark gray) at the locus as indicated. (F) Relative mRNA expression levels as determined by qPCR for the FKBP5, GILZ and DUSP1 genes are shown for wt U2OS-GR18, for clonal lines with deleted GILZ GBS1 (n = 6) and for clonal lines with deleted DUSP1 GBS1 (n = 5). Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight. Here and elsewhere: Statistical tests we done using an unpaired two-sided Mann–Whitney U test. n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
Using gRNAs targeting the DUSP1 GBS1 or the GILZ GBS1, we generated single-cell-derived clonal lines in U2OS-GR18 cells stably expressing GR (27) containing insertions or deletions for both the GILZ GBS1 and the DUSP1 GBS1, respectively (Figure 2D, E and Supplementary Figure S8). These insertions and deletions effectively delete the canonical GBS match consisting of two half-sites separated by a 3 bp spacer, by disrupting the 3 bp spacing between the half-sites. For each clonal line, we ascertained that the induced mutations disrupted the GBS motif-match based on visual inspection of the sequence and by calculating how the mutation influences the motif score (Supplementary Figure S2B). When we targeted the GILZ GBS1, genotyping resulted in the detection of a single allelic variant, whereas genotyping of the edited DUSP1 GBS1 clones resulted in the detection of up to two differentially edited alleles (Figure 2D, E and Supplementary Figure S8). We were surprised to find a single allelic GILZ GBS1 variant, given that it is located on the X-chromosome and U2OS cells are derived from a female. However, DNA Fluorescence in situ hybridization (FISH) experiments provided a trivial explanation, by showing that only a single allele is present for the GILZ locus in U2OS-GR18 cells (Supplementary Figure S3A). Taken together, using the CRISPR/Cas9-system we efficiently generated clonal cell lines containing disrupted GBSs upstream of the GILZ and DUSP1 genes.
Next, we tested whether the GBS deletions influence GR-dependent transcriptional regulation of the nearby gene. To ascertain that the effects we observed were a consequence of deletion of the binding site and not due to clonal variation, the results we show are an average of multiple clonal lines (6 for the GILZ GBS1 deletion and 5 for the DUSP1 GBS1 deletion). For the DUSP1 gene, deletion of the GBS1 resulted in a small reduction (39%) of the hormone-induced expression level when compared to parental U2OS-GR18 cells (Figure 2F). It was similarly lower compared to clonal lines where the GILZ GBS1 was deleted, which served as an additional control to test if the effect of disrupting the GBS was specific for the nearby gene (Figure 2F). In addition, deletion of the GBS did not influence basal DUSP1 expression nor did it selectively influence the GR-dependent regulation of FKBP5, a GR target gene located on another chromosome (Figure 2F). Notably, the DUSP1 gene is still activated in response to hormone treatment when the GBS is deleted. This indicates that additional motifs and or other enhancers also contribute to the GR-dependent regulation of the gene. Similarly, we find that deletion of the GILZ GBS1 resulted in a partial reduction (62%) of the hormone-induced GILZ levels when compared to either the parental U2OS-GR18 cells or to the clonal lines where the DUSP1 GBS1 was deleted (Figure 2F). Together, we efficiently disrupted individual GBSs located near two GR-regulated genes and found that the deletion resulted in a partial loss of GR-dependent regulation indicating that (i) these individual GBSs are functional and (ii) that multiple binding sites contribute to the GR-dependent regulation of both the GILZ and the DUSP1 genes.
A cluster of GBSs regulates GILZ gene expression as an interdependent functional unit
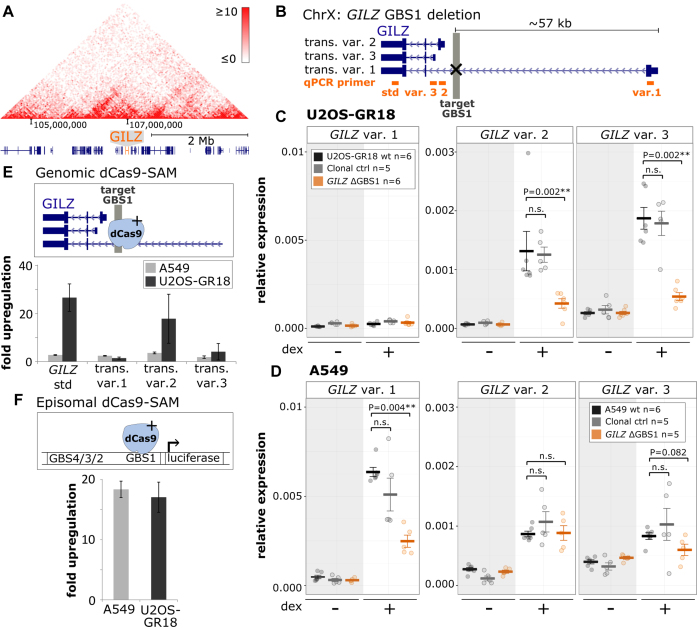
For both the GILZ and the DUSP1 gene, deletion of the GBS1 resulted in a partial loss of GR-dependent transcriptional regulation indicating that additional GR-bound loci contribute to the observed regulation. The GBSs we deleted are part of enhancers that contain additional GBSs (41,42). Specifically, for the GILZ gene, ChIP-seq and combined sequence motif searches identified the presence of three additional GBSs (GILZ GBS2–4), which cluster approximately 500 bp downstream of GBS1 (Figure 2A and Supplementary Figure S4A) (41). Furthermore, luciferase reporter assays have shown that deletion of GILZ GBS2–4 results in reduced GR-dependent transcriptional regulation (41). To investigate the role of GBS2–4 in the GR-dependent regulation of the GILZ gene, we generated single-cell-derived clonal U2OS-GR18 cell lines in which we deleted the region containing GBS2–4 (Supplementary Figure S9). Similar to what we observed when we deleted GBS1, deletion of GBS2–4 resulted in a ∼50% reduction of the hormone-induced GILZ levels when compared to either the parental U2OS-GR18 cells or to the clonal lines where the DUSP1 GBS1 was deleted (Figure 3A). The effect was specific, given that deletion of GBS2–4 did not affect GR-dependent regulation of the FKBP5 gene (Figure 3A).
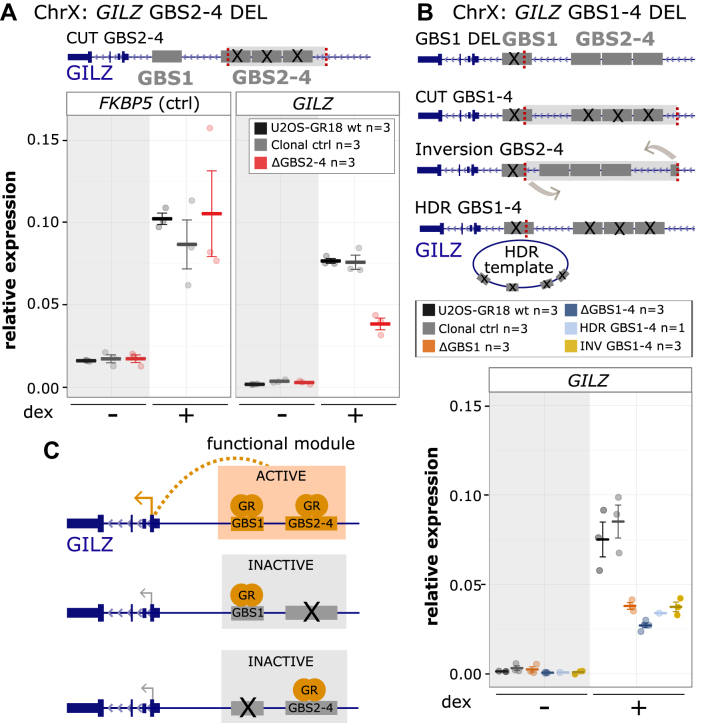
Figure 3.
Cooperative interactions between multiple GR binding sequences of the GILZ GBS1–4 enhancer. (A) Top: Schematics of the GILZ GBS1–4 enhancer locus with GBSs 1–4 highlighted in gray and the location of the gRNAs used to cut out GBS 2–4 in red. Bottom: Relative mRNA expression levels as determined by qPCR for the FKBP5 and GILZ genes are shown for wt U2OS-GR18 (n = 3), for clonal lines lacking a region containing GILZ GBSs 2–4 (n = 3) and unedited clonal lines (n = 3) as additional control. Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight. (B) Top: Schematics of the GILZ GBS1–4 enhancer and the various clonal lines that were generated to assess the role of the GBSs found at this locus, targets for gRNAs highlighted in red. Clonal cell lines with GBS1–4 deletion were either generated by cutting with two gRNAs or by using a homology directed repair (HDR) template to mutate each of the four GBSs by HDR. Bottom: Relative GILZ expression levels as determined by qPCR for clonal lines with GBS1 deletion (n = 3), cut GBS1–4 deletion (n = 3), inversion GBS1–4 (n = 3) and HDR GBS1–4 deletion (n = 1). Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight. (C) Cartoon depicting how the activity of the GILZ GBS1–4 enhancer depends on simultaneous GR binding at both GBS1 and GBSs2–4.
To unravel how multiple GBSs cooperate within the GILZ GBS1–4 enhancer, we generated clonal cell lines in which both GBS1 and GBS2–4 were deleted simultaneously. First, using a pair of gRNAs to remove a ∼600 bp genomic segment containing all four GBSs (CUT GBS1–4 DEL) (Figure 3B and Supplementary Figure S10). Second, because cutting removes a large DNA fragment potentially containing additional GBSs and other regulatory elements, we also introduced point mutations in each of the four GBSs at positions critical for GR-binding (HDR GBS1–4 DEL) (Figure 3B and Supplementary Figure S10) using a homology directed repair (HDR) template. As expected, deletion of the region harboring GBS1–4 resulted in a reduction of the hormone-induced GILZ levels (Figure 3B). However, unexpectedly this effect was comparable to the reduction observed when only GBS1 or GBS2–4 were deleted. Similarly, the effect of mutating all 4 GBSs by HDR was comparable to the effect when only GBS1 or GBS2–4 were deleted (Figure 3A, B). For some clonal cell lines, we obtained an inversion of the targeted genomic fragment, which placed GBS2–4 closer to the TSS of the GILZ gene and simultaneously deleted GBS1 (Figure 3B and Supplementary Figure S9). Again, the effect of the inversion on GR-dependent regulation of the GILZ gene was comparable to the effect of deleting GBS1 alone, arguing that placing GBS2–4 into closer proximity to the TSS of GILZ does not rescue the effect of GBS1 deletion. Here and elsewhere, we also see some fluctuation between groups of clonal lines in basal GILZ gene expression levels. However, in contrast to the effects on the hormone-induced levels, these effects do not consistently correlate with deletions at the GILZ GBS1–4 enhancer and might reflect difficulties to quantify these transcripts by qPCR due to low expression levels in the absence of hormone treatment. Together, our findings indicate that GBS1 and GBS2–4 act interdependently to generate a functional regulatory module that is only capable of contributing to the regulation of the nearby gene when all of the GBSs are present (Figure 3C).
One possible explanation for the cooperative interaction between GBSs at the GILZ GBS1–4 enhancer is that protein:protein interactions between TFs can stabilize their interaction with the genome (44,45). To test if deletion of an individual GBS influences GR occupancy at the edited enhancer, we analyzed GR binding by ChIP-qPCR in a representative clonal cell line in which we deleted the GILZ GBS1. As predicted, deletion of GBS1 resulted in a clear reduction of GR binding at the deleted GBS itself (Figure 4B). In contrast, GR binding at nearby GBSs 2–4 was essentially unaffected by the deletion of GILZ GBS1 (Figure 4B). Next, we examined if deletion of an individual GBS influences GR binding at distal GR-bound loci at the GILZ locus. Similar to our observations directly at the edited GILZ locus, we found that the deletion of GILZ GBS1 did not influence GR binding at other selected peaks in the region (Figure 4C).
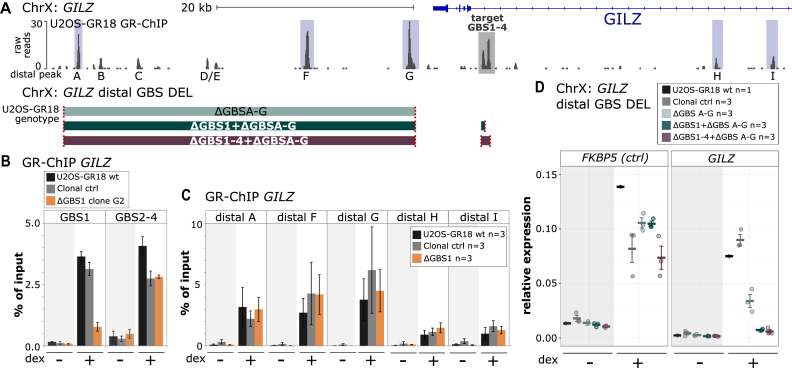
Figure 4.
Effect of GBS1 deletion on GR binding at the GILZ region. (A) Top: GR ChIP-seq tag density for dexamethasone (dex)-treated U2OS-GR18 cells stably expressing GR for the genomic region surrounding the GR target gene GILZ. The GILZ GBS1–4 enhancer is highlighted in gray. The main GR-ChIP peaks in the region selected for quantitative analysis by ChIP-qPCR are highlighted in blue. Bottom: Schematics of the generated clonal cell lines with deletions of a downstream region harboring several distal GR-peaks either alone or in combination with deletions at the GILZ GBS1–4 enhancer. The colored blocks correspond to the genomic regions (see panel a) targeted for deletion. The targets of the gRNAs are indicated in red. (B, C) GR occupancy levels for clonal lines as indicated was analyzed by chromatin immunoprecipitation followed by qPCR for cells treated with either dex (1 μM, 1.5 h) or ethanol as vehicle control. Average percentage of input precipitated ± SEM from three independent experiments for wild type cells, an unedited clonal control cell line and GBS1 deletion clone (G2) are shown. (D) Relative mRNA expression levels as determined by qPCR for the FKBP5 and GILZ genes are shown for wt U2OS-GR18 and clonal lines as indicated. Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight.
Since the deletion of the GILZ GBS1–4 enhancer resulted in only a partial loss of GR-dependent regulation, we next analyzed the contribution of a region downstream of GILZ containing multiple GR ChIP-seq peaks (Figure 4A). Specifically, we used a pair of gRNAs to remove a ∼38 kb region containing peaks A-G in wild type U2OS-GR18 cells (Figure 4A and Supplementary Figure S11) or in cells where either GBS1 or the entire GILZ GBS1–4 enhancer was deleted (Figure 4A and Supplementary Figures S11, S12). In wild type, we found that deletion of the downstream region resulted in a 55% reduction of the hormone-induced expression level when compared to parental U2OS-GR18 cells (Figure 4D). When combined with either deletion of GBS1 or of the entire GILZ GBS1–4 enhancer, GR-dependent regulation was virtually lost (>90% reduction) indicating that the combined action of the proximal enhancer and distal GR binding sites in the downstream region are responsible for the GR-dependent regulation of the GILZ gene (Figure 4D).
Transcriptional consequences of disrupting GBSs vary between cell types
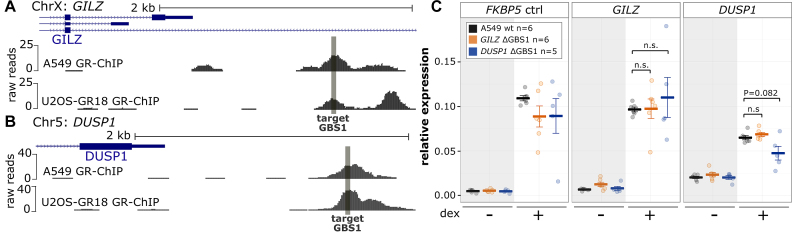
The genes regulated by GR show little overlap between cell types (22), which could be due to cell type-specific binding of GR (23,24). In addition, it could result from the same binding sites regulating the activity of distinct promoters depending on the cellular context. To test if the consequences of disrupting the GILZ and DUSP1 GBSs on GR-dependent gene regulation are conserved across different cell types, we also deleted them in A549 cells. Notably, the origin of the U2OS-GR18 and the A549 cell lines differs: The U2OS-GR18 cell line is derived from a human osteosarcoma, whereas the A549 cell line is derived from a human lung adenocarcinoma. In contrast, both cell lines show a similar level of GR expression (Supplementary Figure S5a). As previously shown (41,42), the targeted loci are bound by GR in both A549 and U2OS-GR18 cells (Figure 5A, B). Furthermore, GR regulates the expression of the GILZ and DUSP1 genes in both cell lines (Figures 2C, 5C and Supplementary Figure S5B). For each GBS, we generated five single-cell-derived clonal cell lines with deleted GBSs for each allele (Supplementary Figures S5C, D and S13) and analyzed the effect of the deletion on GR-dependent transcriptional regulation in A549 cells. Regarding DUSP1, similar to our observation in U2OS-GR18 cells, we found that deletion of the GBS1 did not influence basal DUSP1 expression levels but resulted in a small (27%) and partial reduction of hormone-induced expression of the DUSP1 gene when compared both to the parental A549 cells and to clonal lines for which the GILZ GBS1 was deleted (Figure 5C). Arguing that the effect is specific for the DUSP1 gene, no clear reduction of hormone-induced expression is observed for two other GR target genes, FKBP5 and GILZ (Figure 5C). Thus, for both A549 and U2OS-GR18 cells, GR-dependent activation of the DUSP1 gene is reduced, but not lost, when we delete the promoter–proximal GBS1.
Figure 5.
Effect of GILZ and DUSP1 GBS1 deletion in A549 cells. (A) GR ChIP-seq tag density for dex-treated (top) A549 cells and U2OS-GR18 cells (bottom) for a promoter region of the GR target gene GILZ. (B) Same as for (A) except that for the DUSP1 gene. (C) Relative mRNA expression levels as determined by qPCR for the FKBP5, GILZ and DUSP1 genes are shown for wt A549 (n = 6), clonal A549 lines with deleted GILZ GBS1 (n = 5) and for clonal lines with deleted DUSP1 GBS1 (n = 5). Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight. Unpaired two-sided Mann–Whitney U test: n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
To our surprise and in contrast to what we found in U2OS-GR18 cells, deletion of the GILZ GBS1 in A549 cells did not diminish the hormone-dependent activation of the GILZ gene (Figure 5C). However, enhancers can regulate the activity of distal promoters and the Hi-C contact matrix of A549 cells indicates that the GILZ GBS1 locus is part of a topologically associated domain, which encompasses several genes including transcript variants of the GILZ gene with alternative promoters (Figure 6A). Therefore, we first tested if the GILZ GBS1 deletion influences the GR-dependent regulation of MID2 and PRPS1, two genes flanking the GILZ gene that are regulated by GR in U2OS-GR18 cells. However, disruption of the GILZ GBS1–4 enhancer did not influence the GR-dependent regulation of either MID2 or PRPS1 (Supplementary Figure S4B), indicating that the GILZ GBS1–4 enhancer does not contribute to their GR-dependent regulation. For A549, neither MID2 nor PRPS1 showed a clear change in expression in response to hormone treatment (Supplementary Figure S5E). In addition, we analyzed three individual GILZ transcript variants, using variant-specific primer pairs, each binding in the first variant-specific exon (Figure 6B). Of note, up until now we had analyzed the effects on the GILZ gene using primers that according to RefSeq gene annotation (46) target the 3′ UTR shared by all transcript variants (std qPCR primer, Figure 6B). The TSSs of transcript variants 2 and 3 are located 1.5 and 2.5 kb downstream from GBS1 respectively, whereas the distance to the TSS of transcript variant 1 is ∼57 kb. Mirroring what we found before using primers that target the 3′ UTR shared by all annotated GILZ transcript variants, GILZ GBS1 deletion resulted in reduced hormone-induced levels of transcript variants 2 and 3 in U2OS-GR18 cells (Figure 6C) whereas deletion in A549 cells had no effect on these variants (Figure 6D). In contrast, for transcript variant 1 we observed a robust hormone-dependent regulation in A549 cells, which was markedly reduced (by 61%) upon GBS1 deletion and little to no regulation in U2OS-GR18 cells. Furthermore, the fact that we fail to see an effect of the deletion of GILZ GBS1 when we use primers targeting the supposedly transcript-invariant 3′ UTR argues that the GILZ transcripts in A549 cells differ from the RefSeq annotation.
Figure 6.
Cell type- and GILZ isoform-specific consequences of TF binding at the GILZ GBS1–4 enhancer. (A) Hi-C-derived intra-chromosomal contact frequencies for a region encompassing the GILZ gene at chromosome X for A549 cells (data from (47)). (B) Schematics of the GILZ locus, the deleted GBS1 (highlighted in gray) and three individual GILZ transcript variants that were analyzed using transcript variant-specific qPCR primers (target of primers highlighted in orange). (C) Relative mRNA expression levels in U2OS-GR18 cells as determined by qPCR for GILZ transcript variants as shown for wt U2OS-GR18, for clonal lines with deleted GILZ GBS1 (n = 6) or for clonal lines unedited at the GILZ locus. Circles indicate values for each individual clonal line. Horizontal lines and error bars: Averages ± SEM for cells treated with vehicle or 1 μM dex overnight. Unpaired two-sided Mann-Whitney U test: n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001. (D) Same as for (C) except that data is shown for A549 cells. (E) Comparison between U2OS-GR18 and A549 cells of the transcriptional induction of endogenous GILZ transcript variants by dCas9 synergistic activation mediator (dCAS9-SAM). Average fold induction by targeted recruitment to the GILZ GBS1 relative to gRNAs targeting other loci ± SEM (n = 3) is shown. (F) Comparison between U2OS-GR18 and A549 cells of the transcriptional induction by dCas9-SAM of a transiently transfected luciferase reporter harboring the GILZ promoter region, including GBS1–4 and the endogenous GILZ transcript variant 2 promoter. Average fold induction by targeted recruitment to GILZ GBS1 relative to a gRNA that targets dCas9-SAM to another chromosome ± SEM (n = 3) is shown.
Together, our results indicate that GR binding to the GILZ GBS1 influences the transcriptional activity of distinct promoters depending on the cell type. For U2OS-GR18 cells, the GILZ GBS1 acts on two proximal promoters whereas in A549 no effect on these proximal promoters was observed. Instead, for A549 cells we identified a functional long-range promoter–enhancer interaction between GILZ GBS1 and the TSS of GILZ transcript variant 1.
Cell type-specific differences in the genomic context prevent GILZ GBS1 from acting on proximal promoters
To understand the mechanism responsible for the cell type-specific effect of deletion of GILZ GBS1 on the proximal promoters, we first tested if it can be recapitulated using an episomal luciferase reporter. This luciferase reporter contains the endogenous promoter of GILZ transcript variant 2 and ∼2 kb upstream regulatory sequence including GBS1–4 (genomic sequence for A549 and U2OS-GR18 cells is identical for this region). However, for both U2OS-GR18 and A549 cells, we saw a robust hormone-dependent activation of the reporter, which was markedly reduced when we deleted GBS1 (Supplementary Figure S6A). This suggests that the lack of effect of GBS1 deletion on the proximal transcript variants in A549 cells is not a consequence of missing cofactors, but rather indicates that the cell type-specific effect of GBS1 deletion requires its endogenous genomic context.
One explanation for the lack of effect of deletion of the GILZ GBS1 in A549 cells on the nearby transcript variants could be that the region harbors multiple GR-bound loci that might serve redundant functions. To assess the individual regulatory capacity of the GBS1 locus in its genomic context, we made use of a nucleolytically inactive Cas9-VP64 fusion that can activate gene expression with the help of a modified gRNA containing two MS2 aptamers to recruit multiple additional transcriptional activator domains (dCas9-SAM) (26). Targeting of dCas9-SAM to the GBS1 in U2OS-GR18 cells (in the absence of hormone treatment) resulted in a robust activation of the GILZ gene, specifically of variant 2 (Figure 6E). In addition, we observed a more modest activation of GILZ variant 3. In contrast, targeting dCas9-SAM to the GBS1 in A549 cells resulted in a much weaker activation of the GILZ transcripts, specifically for variant 2 and using the primers that target multiple GILZ variants (26.7-fold activation for U2OS-GR18, 2.8-fold for A549; Figure 6E). The weaker activation of the proximal transcript variants in A549 cells is specific for the GBS1 locus given that targeting the promoter of transcript variant 1 with dCas9-SAM resulted in a comparable activation of GILZ transcript variant 1 in both cell lines (Supplementary Figure S6B). Interestingly, when we direct dCas9-SAM to GBS1 of the luciferase reporter containing the endogenous promoter of GILZ variant 2, we observed comparable levels of activation for both A549 and U2OS-GR18 cells (Figure 6F), confirming that the genomic context is required to recapitulate the cell type-specific effects of placing an activator at GBS1. These findings suggests that the lack of effect of GILZ GBS1 deletion on the proximal transcript variants in A549 cells might not be a consequence of functional redundancy where other GR-bound regions can compensate for the absence of GR binding at GBS1 in A549 cells. Rather, it suggests that the genomic context in A549 cells blocks the ability of transcriptional activators, like GR and dCas9-SAM, bound at GBS1 to activate the proximal GILZ promoters.
Cell type-specific regulation coincides with differential long-range interactions and CTCF binding
Chromatin loops can bring enhancers in close proximity to distal promoters to regulate their activity (1) and the Hi-C contact matrix for A549 cells is indicative of high contact frequencies between the GILZ GBS1 locus and its surrounding region (Figure 6A). To determine if cell type-specific 3D organization of the genome can explain the cell type-specific effect of deletion of GILZ GBS1 on GILZ transcript variants, we performed circular chromosome conformation capture (4C) experiments (29) in U2OS-GR18 and A549 cells. Using GILZ GBS1 as a viewpoint, we found similar 4C-profiles for A549 and U2OS-GR18 cells, suggesting that the majority of DNA-looping interactions for this locus are not cell type-specific (Figure 7A). The TSS of GILZ variant 1 however, consistent with the cell-type specific functional interplay we observed, shows an increased relative interaction frequency with the GILZ GBS1 for A549 cells when compared to U2OS-GR18 cells (Figure 7A, highlighted as gray-shaded area). Thus, a plausible explanation for the observed A549-specific transcriptional effect of deletion of the GILZ1 GBS on variant 1 could be A549-specific looping interactions, which functionally connect GBS1 with the promoter of GILZ transcript variant 1. Furthermore, in both cell types we find several distal GR-bound regions with increased 4C contact frequencies (Figure 7A, highlighted as blue-shaded areas). Of note, these distal GR-bound regions map to the locus downstream of the GILZ gene that contributes to the GR-dependent regulation of the GILZ gene in U2OS-GR18 cells (GBS A-G, Figure 4A, D).
CTCF plays a key role in coordinating the 3D organization of the genome with the majority of looping contacts coinciding with CTCF-bound motifs that are oriented in a convergent manner (8,9,47,48). To study the possible role of CTCF in coordinating the cell type-specific organization of the GILZ locus, we assayed genome-wide CTCF binding in both A549 and U2OS-GR18 cells by ChIP. For A549 cells, we found 7 clear CTCF peaks (Figure 7A marked with an *) in a 200 kb window encompassing the GILZ gene and six of these seven peaks were also bound by CTCF in U2OS-GR18 cells. The A549-specific CTCF-bound region, which is bound both in the presence and absence of hormone (Figure 7B), is located directly at the TSS of GILZ variant 1 (Figure 7A–C). At the other end of the putative loop, CTCF binds at the GILZ GBS1 in both A549 and U2OS-GR18 cells (Figure 7A–C). The A549-specific CTCF peak at TSS1 harbors multiple consensus CTCF binding sequences. Three of these are in a reverse orientation relative to the CTCF peak at the GILZ GBS1, which harbors two CTCF binding sequences including one in the forward orientation (Figure 7C and Supplementary Figure S7A). Since nearly all DNA looping (>90%) coincides with CTCF sites that are organized in a convergent orientation (47), we assayed the contribution of the three CTCF binding sequences in the reverse orientation at TSS1 using a pair of gRNAs to remove a 129 bp region in A549 cells (Figure 7D and Supplementary Figure S7A, B). Notably, the interpretation of the effect of this deletion on the GR-dependent regulation of GILZ transcript variant 1 is complicated by fact that it overlaps with the 5′ UTR of GILZ transcript variant 1. Consequently, the deletion might influence gene regulation directly independent of effects on long-range contacts. Nonetheless, the observed ∼2-fold reduction in GR-dependent regulation of GILZ transcript variant 1 upon deletion of this region (Figure 7D) is consistent with a role of these CTCF sites in mediating long-range contacts and facilitating the A549-specific regulation of GILZ transcript variant 1.
DISCUSSION
Genome-wide approaches, for example to map TF binding sites, have uncovered a myriad of putative cis-regulatory elements in the genome (3,49). However, linking such putative cis-regulatory elements to the regulation of genes remains a major challenge. Here, we studied the connection between TF binding at cis-regulatory elements and gene regulation using GR, a hormone-activated TF, as a model. Consistent with previous studies of other signaling pathways (7,50,51) we found that GR binding and gene regulation are connected and that GR binding sites that loop to the promoters of genes are more likely to coincide with changes in gene expression than binding sites that do not show such interactions (Figure 1). To identify additional inputs that discriminate productive GR binding events from those that do not result in changes in gene expression, we disrupted GR binding sites in their endogenous genomic context. The mutation of GILZ and DUSP1 GBS1 disrupts the 3bp spacing between the half-sites while leaving the half-sites themselves intact. However, for the GILZ gene, our GR binding and functional analysis indicate that the remaining half-sites are neither bound by GR nor do they contribute to GR-dependent regulation, indicating that proper spacing of the half-sites is essential for GBS1 activity at the GILZ GBS1–4 enhancer. Furthermore, this approach uncovered several operating principles that help explain why only certain GR binding events result in changes in gene expression. First, we found that GR-dependent regulation by the promoter-proximal GILZ GBS1–4 enhancer requires the presence of a cluster of GR binding sites. When one of the binding sites in the cluster is disrupted, GR can still bind to the remaining binding sites, but the GILZ GBS1–4 enhancer no longer contributes to GR-dependent regulation of the GILZ gene (Figure 3C). Consistent with the notion that only certain GR-binding events are productive, recent studies reported that only a subset of GR-bound regions show regulatory potential in reporter gene assays (52,53). Second, we found indications that cell type-specific enhancer-blocking can prevent cis-regulatory elements from interacting with nearby promoters, whereas cell type-specific 3D genome organization might direct the activity of an individual enhancer to distinct promoters. Third, our results corroborate findings by others (18,54) that multiple enhancers are needed for the robust regulation of genes. Specifically, we found that deletion of the GILZ GBS1–4 enhancer alone only results in a partial loss of the GR-dependent regulation whereas additional deletion of a region downstream of the gene harboring multiple GR peaks resulted in an almost complete loss of GR-dependent regulation of the GILZ gene (Figure 4D). Similarly, genomic deletion of individual cis-regulatory elements was repeatedly reported to result in only marginal effects on gene expression (17,55,56). Nevertheless, this does not imply the absence of regulatory potential of enhancers. Rather, the small effects of disrupting individual enhancers could reflect that multiple enhancers act in an additive manner or that certain genes are not direct TF targets. Alternatively, functional redundancy between enhancers (57) can mask the contribution of individual enhancers to the regulation of individual genes or enhancers might show their regulatory potential only during a specific cell-stage, in a specific cell type or under certain environmental conditions (17–20,58,59).
The analysis of the effect of deletion of multiple GBSs within the GILZ GBS1–4 enhancer uncovered that these GBSs act in a highly interdependent manner. To our knowledge, this is the first demonstration that the functioning of an enhancer element critically depends on the simultaneous presence of multiple TF binding sites in the endogenous genomic context. This finding is in contrast to findings for the super enhancer WAP, for which the multiple STAT5 binding sites present at this locus act in an additive manner (16). Theoretically, the observed interdependency of GBSs at the GILZ GBS1–4 enhancer might come from cooperative binding of multiple different TFs, including GR, as was shown for so-called transcriptional ‘hotspots’ (60–62). Interestingly however, deletion of one of the GBSs in the cluster appeared not to influence binding of GR to other GBSs nearby or at other GR-bound regions in the region as detected by ChIP-qPCR (Figure 4B, C). Although these findings argue that the functional cooperativity we observe is not a consequence of an inability of GR to bind to nearby binding sites when GBS1 is deleted, it is unclear if ChIP experiments have the sensitivity to detect subtle changes in occupancy or binding kinetics. Alternative explanations for the observed interdependency include synergetic recruitment of co-activators by multiple TFs as was shown for the IFNβ enhanceosome (63), or of the basal RNA polymerase machinery (64,65). Notably, we do not assume that the presence of multiple GR binding sites is a general operating principle that defines all productive GR-binding events for several reasons. First, a single GR binding site is sufficient to facilitate GR-dependent gene regulation of a reporter gene either episomally or when genomically integrated (52,53). Furthermore, unpublished studies from our lab indicate that addition of a single GR binding site at the promoter can be sufficient to make a normally unresponsive gene a GR target. Nonetheless, our findings demonstrate that GR binding alone, which is still observed when one of the GBSs is missing at the GILZ GBS1–4 enhancer, is insufficient to create a productive GR-binding event thus explaining why only a fraction of GR binding events, namely those for which the right combination of TFs bind, result in changes in gene expression.
GR-responsive genes show little overlap between cell types (21,22), which likely explains the striking differences in the physiological consequences of glucocorticoid signaling for different tissues which range from inducing gluconeogenesis in the liver, to suppression of inflammation when acting on various cell types of the hematopoietic lineage. The cell type-specific transcriptional consequences of glucocorticoid signaling could be due to the limited overlap of GR-bound loci which has been documented for cell lines (23,24), although it is unclear if the same applies for healthy cells. In addition, subtle differences between cell types in the binding pattern of GR (e.g. differences between cell types in the relative ChIP-seq signal at the GBS1 and GBS2–4, Figure 5A) and in the repertoire of TFs that bind at a locus could contribute to cell type-specific transcriptional responses to glucocorticoid signaling. Our data suggests an additional explanation, namely that specificity can arise from a rewiring of promoter–enhancer contacts. Similar to observations made at the β-globin locus, where the promoter-distal locus control region can influence the activity of different transcripts during erythroid differentiation (1), we find that a GR binding site shared between cell types can contribute to the regulation of cell type-specific gene expression (Figure 8). In contrast to the locus control region of the β-globin locus, here we show that the regulatory activity of an individual promoter-proximal TF binding site motif can be rewired depending on the cell type. In the case of GILZ GBS1, our data suggests that the activity of a promoter-proximal enhancer may be rewired to either a nearby or a distal promoter via a combination of cell type-specific genome organization and enhancer-blocking. Furthermore, to our knowledge we are the first to show that the same enhancer can contribute to the regulation of different promoters of the same gene rather than promoters of different genes. Cell type-specific long-range interactions might allow an enhancer shared between cell types to contribute to the GR-dependent regulation of distinct GILZ transcript variants with potentially different functions. In the case of GILZ, the longer GILZ transcript variant 1 codes for a protein, L-GILZ, which interacts with p53, whereas the proteins encoded by the shorter transcript variants 2 and 3 do not. Consequently, the observed cell-type specific expression of GILZ transcript variant 1 could result in cell-type-specific effects of GR activation on cell proliferation (66).
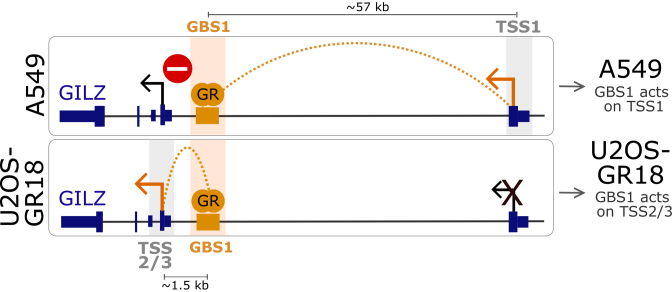
Figure 8.
Model depicting how cell type-specific long-range interactions and enhancer-blocking can direct the activity of an enhancer shared between cell types to distinct transcripts. Top: An A549-specific long-range DNA-looping interaction between GBS1 and TSS1 enables GR-dependent regulation of GILZ transcript variant 1. Simultaneously, enhancer-blocking insulates GILZ transcript variants 2 and 3 from the regulatory activity of GBS1. Bottom: In U2OS-GR18 cells the absence of both looping and enhancer-blocking facilitates GR-dependent regulation of the nearby transcript variants 2 and 3.
Our finding that targeting the transcriptional activator dCas9-SAM to the GBS1 recapitulates the cell type-specific effect of GBS1 deletion suggests that cell type-specific enhancer-blocking and looping can determine if an enhancer shared between cell types can act on a transcript or not. Interestingly, CTCF plays a role in both enhancer-blocking and in organizing long-range chromatin interactions (67–69). These two functions of CTCF may be connected which could also explain why the orientation of the CTCF binding site is important for both enhancer-blocking activity (70,71) and for long-range chromatin interactions (8). Thus, one hypothesis explaining the cell type-specific promoter–enhancer wiring at the GILZ GBS1–4 enhancer could be that the A549-specific CTCF binding at the TSS1 facilitates both the A549-specific long-range interaction with the GILZ GBS1–4 enhancer and that this looping might simultaneously block the ability of the enhancer to act on the nearby TSSs (Figure 8). However, arguing against this hypothesis, we find that although deletion of convergently oriented CTCF binding sequences results in a reduced activation of GILZ transcript variant 1, this does not coincide with increased activation of variants 2 and 3 as could be expected when deletion of the CTCF binding sites would perturb enhancer blocking (Figure 7D). Furthermore, the GR-dependent regulation of GILZ transcript variant 1 is reduced and not completely lost upon deletion of the CTCF binding sites (Figure 7D), indicating that this A549-specific CTCF-bound region only partially explains the regulation of GILZ transcript variant 1 by distal GR binding sites. Notably, the 4C profile shows that several distal GR peaks map to regions with increased contact frequencies but without CTCF binding (Figure 7A) thus corroborating observations by others (72) that many long-range chromatin interactions appear CTCF-independent and might rely on other proteins for loop formation. What those proteins are and the underlying mechanisms responsible for the observed cell type-specific enhancer blocking and looping will be the subject of future studies.
The staggering number of potential cis-regulatory elements in the genome—depending on the cell type up to several tens of thousands of genomic regions are bound by GR—complicates a systematic analysis of their function. The analysis is complicated even further by the combinatorial nature of gene regulation. Computational approaches provide an alternative path to link cis-regulatory elements to the regulation of genes. The predictive power of computational approaches improves when additional layers of information, e.g. the chromatin state of regulatory elements and information regarding the 3D organization of the genome are added. Yet, although these models work quite well at meta-level, their limited accuracy restricts their potential to predict activity for individual cis-regulatory elements. Here, we show that the functional analysis of a small number of cis-regulatory elements can help uncover several operational principles of active regulatory elements. These gained mechanistic insights can be used to refine computational approaches and might eventually lead to more accurate predictions and a better mechanistic understanding of how individual cis-regulatory elements control the expression of genes.
DATA AVAILABILITY
ChIP-seq and 4C-seq data are deposited in the ArrayExpress repository. Access number and login details as specified below:
E-MTAB-5350: 4C-seq (U2OS-GR18/A549)
E-MTAB-5352: CTCF ChIP-seq (U2OS-GR18/A549)
E-MTAB-5355: H3K27ac ChIP-seq (IMR90)
Supplementary Material
ACKNOWLEDGEMENTS
We thank Edda Einfeldt for excellent technical support, Martin Franke and Ivana Jerkovic for help setting up the 4C-seq and CTCF ChIP assays respectively.
Author contributions: V.T., M.C.R., L.V.G, N.L., P.D. and S.H.M. performed and conceived experiments and analyzed the data. R.S. and K.S. conducted the computational analyses. V.T., R.S., H.C., M.V. and S.H.M. designed and supervised the study and wrote the manuscript with input from all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W.. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002; 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 2. Amano T., Sagai T., Tanabe H., Mizushina Y., Nakazawa H., Shiroishi T.. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009; 16:47–57. [DOI] [PubMed] [Google Scholar]
- 3. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sandmann T., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E.. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006; 10:797–807. [DOI] [PubMed] [Google Scholar]
- 5. Gitter A., Siegfried Z., Klutstein M., Fornes O., Oliva B., Simon I., Bar-Joseph Z.. Backup in gene regulatory networks explains differences between binding and knockout results. Mol. Syst. Biol. 2009; 5:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cusanovich D.A., Pavlovic B., Pritchard J.K., Gilad Y.. The functional consequences of variation in transcription factor binding. PLoS Genet. 2014; 10:e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B.. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 9. Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015; 162:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell A.C., West A.G., Felsenfeld G.. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999; 98:387–396. [DOI] [PubMed] [Google Scholar]
- 11. Ghirlando R., Felsenfeld G.. CTCF: making the right connections. Genes Dev. 2016; 30:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 13. Strahle U., Schmid W., Schutz G.. Synergistic action of the glucocorticoid receptor with transcription factors. EMBO J. 1988; 7:3389–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bienz M., Pelham H.R.. Heat shock regulatory elements function as an inducible enhancer in the Xenopus hsp70 gene and when linked to a heterologous promoter. Cell. 1986; 45:753–760. [DOI] [PubMed] [Google Scholar]
- 15. Hay D., Hughes J.R., Babbs C., Davies J.O., Graham B.J., Hanssen L.L., Kassouf M.T., Oudelaar A.M., Sharpe J.A., Suciu M.C. et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016; 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L.. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016; 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.-C. et al. Dynamic control of enhancer repertoires trives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perry M.W., Boettiger A.N., Levine M.. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13570–13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunipace L., Saunders A., Ashe H.L., Stathopoulos A.. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev. Cell. 2013; 26:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frankel N., Davis G.K., Vargas D., Wang S., Payre F., Stern D.L.. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010; 466:490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love M.I., Huska M.R., Jurk M., Schopflin R., Starick S.R., Schwahn K., Cooper S.B., Yamamoto K.R., Thomas-Chollier M., Vingron M. et al. Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation. Nucleic Acids Res. 2016; 45:1805–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. John S., Johnson T.A., Sung M.H., Biddie S.C., Trump S., Koch-Paiz C.A., Davis S.R., Walker R., Meltzer P.S., Hager G.L.. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology. 2009; 150:1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gertz J., Savic D., Varley K.E., Partridge E.C., Safi A., Jain P., Cooper G.M., Reddy T.E., Crawford G.E., Myers R.M.. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell. 2013; 52:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science (New York, N.Y.). 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rogatsky I., Trowbridge J.M., Garabedian M.J.. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 1997; 17:3181–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R.. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science (New York, N.Y.). 2009; 324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van de Werken H.J., de Vree P.J., Splinter E., Holwerda S.J., Klous P., de Wit E., de Laat W.. 4C technology: protocols and data analysis. Methods Enzymol. 2012; 513:89–112. [DOI] [PubMed] [Google Scholar]
- 30. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heng L. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics. 2013; 1303:https://arxiv.org/abs/1303.3997. [Google Scholar]
- 35. Manke T., Heinig M., Vingron M.. Quantifying the effect of sequence variation on regulatory interactions. Hum. Mut. 2010; 31:477–483. [DOI] [PubMed] [Google Scholar]
- 36. Levings P.P., Bungert J.. The human beta-globin locus control region. Eur. J. Biochem. 2002; 269:1589–1599. [DOI] [PubMed] [Google Scholar]
- 37. Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A.. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015; 33:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B.. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Owens D.M., Keyse S.M.. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007; 26:3203–3213. [DOI] [PubMed] [Google Scholar]
- 40. Ayroldi E., Riccardi C.. Glucocorticoid-induced leucine zipper (GILZ): a new important mediator of glucocorticoid action. FASEB J. 2009; 23:3649–3658. [DOI] [PubMed] [Google Scholar]
- 41. Wang J.C., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C., Yamamoto K.R.. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15603–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shipp L.E., Lee J.V., Yu C.Y., Pufall M., Zhang P., Scott D.K., Wang J.C.. Transcriptional regulation of human dual specificity protein phosphatase 1 (DUSP1) gene by glucocorticoids. PLoS One. 2010; 5:e13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.). 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mirny L.A. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y.S., Carey M., Ptashne M., Green M.R.. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990; 345:359–361. [DOI] [PubMed] [Google Scholar]
- 46. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014; 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuin J., Dixon J.R., van der Reijden M.I.J.A., Ye Z., Kolovos P., Brouwer R.W.W., van de Corput M.P.C., van de Werken H.J.G., Knoch T.A., van IJcken W.F.J. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson D.S., Mortazavi A., Myers R.M., Wold B.. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, N.Y.). 2007; 316:1497–1502. [DOI] [PubMed] [Google Scholar]
- 50. Cheng C., Alexander R., Min R., Leng J., Yip K.Y., Rozowsky J., Yan K.K., Dong X., Djebali S., Ruan Y. et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012; 22:1658–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ouyang Z., Zhou Q., Wong W.H.. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21521–21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vockley C.M., D’Ippolito A.M., McDowell I.C., Majoros W.H., Safi A., Song L., Crawford G.E., Reddy T.E.. Direct GR binding sites potentiate clusters of TF binding across the human genome. Cell. 2016; 166:1269–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schone S., Jurk M., Helabad M.B., Dror I., Lebars I., Kieffer B., Imhof P., Rohs R., Vingron M., Thomas-Chollier M. et al. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nat. Commun. 2016; 7:12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chatterjee S., Kapoor A., Akiyama J.A., Auer D.R., Lee D., Gabriel S., Berrios C., Pennacchio L.A., Chakravarti A.. Enhancer variants synergistically drive dysfunction of a gene regulatory network In Hirschsprung disease. Cell. 2016; 167:355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sanjana N.E., Wright J., Zheng K., Shalem O., Fontanillas P., Joung J., Cheng C., Regev A., Zhang F.. High-resolution interrogation of functional elements in the noncoding genome. Science (New York, N.Y.). 2016; 353:1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R.. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016; 34:192–198. [DOI] [PubMed] [Google Scholar]
- 57. Cannavo E., Khoueiry P., Garfield D.A., Geeleher P., Zichner T., Gustafson E.H., Ciglar L., Korbel J.O., Furlong E.E.. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol.: CB. 2016; 26:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O’Meara M.M., Bigelow H., Flibotte S., Etchberger J.F., Moerman D.G., Hobert O.. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics. 2009; 181:1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crocker J., Abe N., Rinaldi L., McGregor A.P., Frankel N., Wang S., Alsawadi A., Valenti P., Plaza S., Payre F. et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015; 160:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siersbaek R., Nielsen R., John S., Sung M.H., Baek S., Loft A., Hager G.L., Mandrup S.. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011; 30:1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moorman C., Sun L.V., Wang J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X.J., White K.P., Bussemaker H.J. et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siersbaek R., Baek S., Rabiee A., Nielsen R., Traynor S., Clark N., Sandelin A., Jensen O.N., Sung M.H., Hager G.L. et al. Molecular architecture of transcription factor hotspots in early adipogenesis. Cell Rep. 2014; 7:1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Merika M., Williams A.J., Chen G., Collins T., Thanos D.. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell. 1998; 1:277–287. [DOI] [PubMed] [Google Scholar]
- 64. Tjian R., Maniatis T.. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994; 77:5–8. [DOI] [PubMed] [Google Scholar]
- 65. Ptashne M., Gann A.. Transcriptional activation by recruitment. Nature. 1997; 386:569–577. [DOI] [PubMed] [Google Scholar]
- 66. Ayroldi E., Petrillo M.G., Bastianelli A., Marchetti M.C., Ronchetti S., Nocentini G., Ricciotti L., Cannarile L., Riccardi C.. L-GILZ binds p53 and MDM2 and suppresses tumor growth through p53 activation in human cancer cells. Cell Death Differ. 2015; 22:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ong C.T., Corces V.G.. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ali T., Renkawitz R., Bartkuhn M.. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016; 37:17–26. [DOI] [PubMed] [Google Scholar]
- 69. Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S.. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bell A.C., Felsenfeld G.. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000; 405:482–485. [DOI] [PubMed] [Google Scholar]
- 71. Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M.. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000; 405:486–489. [DOI] [PubMed] [Google Scholar]
- 72. Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015; 163:1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and 4C-seq data are deposited in the ArrayExpress repository. Access number and login details as specified below:
E-MTAB-5350: 4C-seq (U2OS-GR18/A549)
E-MTAB-5352: CTCF ChIP-seq (U2OS-GR18/A549)
E-MTAB-5355: H3K27ac ChIP-seq (IMR90)