Abstract
An expansin gene, LeExp2, was isolated from auxin-treated, etiolated tomato (Lycopersicon esculentum cv T5) hypocotyls. LeExp2 mRNA expression was restricted to the growing regions of the tomato hypocotyl and was up-regulated during incubation of hypocotyl segments with auxin. The pattern of expression of LeExp2 was also studied during tomato fruit growth, a developmental process involving rapid cell enlargement. The expression of genes encoding a xyloglucan endotransglycosylase (LeEXT1) and an endo-1,4-β-glucanase (Cel7), which, like LeExp2, are auxin-regulated in etiolated hypocotyls (C. Catalá, J.K.C. Rose, A.B. Bennett [1997] Plant J 12: 417–426), was also studied to examine the potential for synergistic action with expansins. LeExp2 and LeEXT1 genes were coordinately regulated, with their mRNA accumulation peaking during the stages of highest growth, while Cel7 mRNA abundance increased and remained constant during later stages of fruit growth. The expression of LeExp2, LeEXT1, and Cel7 was undetectable or negligible at the onset of and during fruit ripening, which is consistent with a specific role of these genes in regulating cell wall loosening during fruit growth, not in ripening-associated cell wall disassembly.
Plant growth is controlled by numerous hormonal and environmental stimuli that interact to regulate cell division and both the direction and rate of cell expansion. Auxin has been implicated in the control of cell elongation (Cleland, 1995), and auxin-induced elongation has been extensively studied in excised stem and coleoptile segments. There is also recent evidence to suggest that auxin is capable of promoting cell elongation in intact plants (Yang et al., 1993; Romano et al., 1995; Gray et al., 1998). Exogenous auxin is thought to induce rapid elongation in plant tissues through an increase in the mechanical extensibility of the cell wall (Taiz, 1984; Cosgrove, 1993). Several changes in the architecture of the primary wall and in the apoplastic environment have been associated with the action of auxin, including cell wall acidification (Rayle and Cleland, 1992), modification of specific cell wall polymers, and de novo polysaccharide synthesis (Kutschera and Briggs, 1987; Talbott and Ray, 1992). Although still somewhat controversial, the “acid growth” theory proposes that auxin-induced cell wall acidification is an essential component of auxin-induced cell expansion. The reduction in apoplastic pH has been suggested to induce wall loosening through the activation of cell wall-modifying enzymes, which catalyze the breakage of load-bearing bonds and rearrange cell wall polymers (Cleland, 1995). However, the biochemical mechanisms by which cell wall structure is modified in response to auxin remain poorly understood.
The plant primary cell wall is typically described as a complex network of cellulose microfibrils that are interwoven by two classes of matrix polymers, hemicelluloses and pectins, together with other, less-abundant components such as structural proteins (Carpita and Gibeaut, 1993). The major hemicellulose in dicotyledons is xyloglucan, which is thought to coat and tether the cellulose microfibrils together, forming an extensive cellulose-xyloglucan network (Hayashi and Maclachlan, 1984; McCann et al., 1990). Since this network is believed to represent a major constraint to turgor-driven cell expansion, the disassembly of critical load-bearing linkages or associations, such as xyloglucan molecules that cross-link adjacent cellulose microfibrils, may be an essential feature of the wall loosening required for cell expansion. The observation that one of the early effects of auxin-induced growth is xyloglucan depolymerization and solubilization (Labavitch and Ray, 1974; Talbott and Ray, 1992) supports this model.
Candidate enzymes responsible for the reorganization of the cellulose-xyloglucan framework include endo-β-1,4-glucanases (EGases) and xyloglucan endotransglycosylases (XETs). Plant EGases catalyze the endo-hydrolysis of β-1,4 linked glucan chains (Brummell et al., 1994), while XETs catalyze the endo-cleavage of xyloglucan polymers and transfer of the newly generated reducing ends to other xyloglucans (Fry et al., 1992; Nishitani and Tominaga, 1992). An increase in the expression of the EGase and XET genes in response to auxin has been described in several plant species (Xu et al., 1995; Wu et al., 1996; Catalá et al., 1997). Another group of plant proteins, expansins, have been identified as wall-loosening factors, although they have been reported to possess no glycanolytic activity (McQueen-Mason et al., 1992). Unlike EGases and XETs, expansins promote the long-term extension of isolated cell walls in vitro, and have been proposed to catalyze the ability of cell walls to extend at an acidic pH (Cosgrove, 1998).
The mechanism of expansin action is still unknown, but it has been suggested that expansins disrupt hydrogen bonds between cellulose microfibrils and associated hemicelluloses (McQueen-Mason and Cosgrove, 1995). By disrupting the noncovalent associations between xyloglucan and cellulose, expansins could both facilitate the movement of microfibrils relative to each other and alter the accessibility of xyloglucan to cell wall enzymes such as EGases and XETs. Expansin gene families have been identified in several plant species (Shcherban et al., 1995; Link and Cosgrove, 1998), and the expression of some expansin genes has been correlated with cell growth (Cho and Kende, 1997; Orford and Timmis, 1998). However, the complexity of the expansin gene families and the tissue-specific patterns of expression of some of their members suggest that different isoforms may have distinct functions in plant growth and development (Rose et al., 1997; Reinhardt et al., 1998). Most expansins and their homologs identified to date are classified as α-expansins, while β-expansins comprise a divergent class of proteins that contain highly conserved residues with α-expansins, despite only 25% overall amino acid identity (Cosgrove et al., 1997).
Cell expansion also accompanies fruit growth, accounting for increases in cell volume of 10-fold or more (Coombe, 1976), and is likely to involve synthesis and restructuring of the primary wall. It has been suggested that, like growing vegetative tissues, auxin promotes cell expansion in fruit by causing an increase in cell wall extensibility (Gillaspy et al., 1993). However, relatively little information exists about the basis and mechanisms of wall loosening during fruit growth. In contrast, cell wall changes during fruit ripening are well documented and have been associated with an increase in gene expression of specific EGases (Lashbrook et al., 1994), XETs (Arrowsmith and de Silva, 1995; Schröder et al., 1998), and ripening-specific expansins (Rose et al., 1997). Although activities of enzymes degrading β-1,4-glucans or xyloglucans and XET activity are generally high during early fruit growth (Bonghi et al., 1998; Faik et al., 1998), there is little information about the expression of specific members of the XET, EGase, and expansin gene families during fruit growth and their relationship to the expression of related genes during fruit ripening.
We have previously shown that auxin regulates mRNA abundance of an EGase (Cel7) and an XET (LeEXT1) during auxin-induced elongation of tomato (Lycopersicon esculentum cv T5) hypocotyl segments (Catalá et al., 1997). In this paper, we characterize the auxin regulation of a new tomato expansin gene (LeExp2; GenBank accession no. AF096776) and describe the expression of LeExp2, Cel7, and LeEXT1 during fruit development. These auxin-induced genes encode cell wall-modifying enzymes that may act synergistically in restructuring the cellulose-xyloglucan network to allow cell expansion both in elongating hypocotyls and during rapid fruit growth.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum cv T5) seeds were sown on moist vermiculite, and etiolated seedlings were grown in the dark for 7 d at 25°C. The growth rates of different regions of the hypocotyl were measured using paint marks applied at 5-mm intervals along the hypocotyls of 6-d-old seedlings. The distances between the marks were measured in 15 seedlings after a 12-h growth period in the dark.
Fruit and vegetative tissues were harvested from greenhouse-grown tomato plants. Young expanding fruit were staged as I, II, or III, corresponding to fruit diameters of 0.5 to 1 cm, 2 to 3 cm, and 4 to 6 cm, respectively, and ripening fruit were assigned a developmental stage based on color, as described in Lashbrook et al. (1994). Pericarp tissue was isolated from fruit harvested at different developmental stages and frozen immediately in liquid nitrogen. To characterize the rate of tomato fruit growth, flowers were tagged at anthesis and fruit diameters measured over a period of 50 d. A regression curve was established from data obtained with 15 fruit (r2 = 0.96).
Hormone Treatments
Etiolated tomato hypocotyl sections (6 mm) were cut directly below the apical hook and incubated in 25 mm potassium phosphate buffer (pH 6.0), 2% (w/v) Suc for 2 to 3 h (Kelly and Bradford, 1986). Buffer was replaced with fresh buffer or buffer containing the indicated concentration of 2,4-dichlorophenoxyacetic acid (2,4-D), indole-3-acetic acid (IAA), naphthylacetic acid (NAA), or brassinolide, and segments were incubated at 25°C in the dark with gentle agitation.
PCR Amplification and cDNA Library Screening
Degenerate PCR primers were designed from conserved amino acids identified in an alignment of deduced amino acid sequences from nine expansins (Shcherban et al., 1995) and were used to amplify an expansin cDNA fragment from auxin-treated tomato hypocotyl RNA. The 5′ end primer [G(GC)(N) CA(TC) GC(N) AC(N) TT(CT) TA(CT) GG(N) G] corresponded to amino acids 6 to 11 of the consensus sequence and the 3′end primer [(TC) TGCCA(AG) TT(TC) TG(N) CCCCA(AG) TT] to amino acids 182 to 188 (n = A, T, C, or G). cDNA synthesis and PCR amplification were as in Rose et al. (1996), with an annealing temperature of 50°C.
The resulting 540-bp cDNA fragment was cloned into the PCR II vector (Invitrogen, Carlsbad, CA), and the DNA sequence was determined using an automated sequencer (model 377, Perkin-Elmer/Applied Biosystems, Foster City, CA) and dye terminator chemistry with AmpliTaq DNA polymerase (Taq FS, Perkin-Elmer/Applied Biosystems). The PCR fragment, corresponding to amino acids 28 to 207 of the full-length clone, was used to screen a tomato hypocotyl cDNA library in the pARC7 vector (O'Neill et al., 1990). Twenty-four positive clones were identified and six inserts were subcloned from the library vector into the SalI /NotI sites of the pBluescript II SK+ plasmid (Stratagene, La Jolla, CA) and sequenced as for the PCR product. The longest clone was designated LeExp2.
The full deduced amino acid sequence of LeExp2 was aligned with the corresponding sequences of α-expansins, including the signal peptides, present in the GenBank database using CLUSTAL with substitution matrix BLOSUM30 (Gap opening penalty of 10 and gap extension penalty of 0.05). A phylogenetic tree was constructed using PHYLIP software (Felsenstein, 1989) with the parsimony method PROTPARS using the neighbor-joining method and bootstrap analysis with 1,000 replicates. The GenBank accession numbers are: Arabidopsis AtExp1, U30476, AtExp2, U30481, AtExp5, U30478, and AtExp6, U30480; rape BnExp1, AJ000885; cotton GhExp1, AF043284; cucumber CsExp1, U30482; CsExp2, U30460; pea PsExp1, X85187; Phleum pollen allergen Phlp1, X78813; pine PtExp4, U64892; rice OsExp1, Y07782; OsExp2, U30477; OsExp3, U30479; OsExp4, U85246; tobacco NtExp2, AF049351; NtExp3, AF049352; NtExp4, AF049353; NtExp5, AF049354; tomato LeExp1, U82123; LeExp2, AF096776; LeExp3, AF059487; LeExp4, AF059488; LeExp5, AF059489; LeExp18, AJ004997.
DNA and RNA Gel-Blot Analysis
Genomic DNA was isolated from tomato leaves as described in Murray and Thompson (1980). Ten micrograms of genomic DNA was digested with a range of restriction enzymes, fractionated by electrophoresis on 0.8% (w/v) agarose gels, and transferred to positively charged nylon membrane (Boehringer Mannheim/Roche, Basel, Switzerland), as in Sambrook et al. (1989). A 652-bp SacI/StuI fragment, including the first 43 bp of the 3′ untranslated region, was used as the LeExp2 probe. Cel7 and LeEXT1 probes were prepared as described in Catalá et al. (1997). A 200-bp tomato actin cDNA fragment was used as a loading control for a gene expressed constitutively throughout fruit development. Probes were radiolabeled by random hexamer priming using [α-32P]dATP (3,000 Ci mmol−1, Dupont, Wilmington, DE) and Klenow DNA polymerase (New England Biolabs, Beverly, MA). Hybridizations were performed at 42°C in 50% (w/v) formamide, 6× SSPE, 0.5% (v/v) SDS, 5× Denhardt's solution, and 100 mg mL−1 sonicated salmon sperm DNA. Membranes were washed three times in 5× SSC, 1% (w/v) SDS at 42°C for 15 min, followed by three washes in 0.2× SSC, 0.5% (w/v) SDS at 65°C for 20 min (8°C below Tm).
Total RNA was isolated from etiolated tomato hypocotyls using a kit (RNeasy Plant Total RNA kit, Qiagen, Valencia, CA) according to the manufacturer's instructions. Total RNA (15 μg per lane) was subjected to electrophoresis on 1.2% (w/v) agarose, 10% (v/v) formaldehyde gels, visualized with ethidium bromide to test for equal loading, and transferred to Hybond-N membrane (Amersham-Pharmacia Biotech, Uppsala, Sweden) as in Sambrook et al. (1989). RNA was extracted from frozen pericarp and vegetative tissues by the method of Wan and Wilkins (1994). Poly(A)+ RNA was isolated with the Oligotex mRNA kit (Qiagen), and 1 μg per lane was subjected to electrophoresis as described above. RNA blots were hybridized and washed at a stringency equivalent to that of the DNA-blot analysis described above.
RESULTS
Cloning and Phylogenetic Analysis of LeExp2
To identify expansins expressed in response to auxin, RNA was isolated from auxin-treated tomato hypocotyls and used for reverse transcriptase-PCR reactions, together with degenerate oligonucleotide primers based on deduced amino acid domains conserved between expansins. The sequence of an amplified 540-bp cDNA fragment corresponded to a new tomato expansin, and was designated LeExp2. Subsequent screening of a tomato hypocotyl cDNA library identified a full-length LeExp2 clone (1,147 bp) encoding a predicted polypeptide of 247 amino acids, including a signal peptide of 20 amino acids, as predicted using SignalP (Henrik et al., 1997). An expansin gene isolated from tomato leaves was previously described as LeExp2 (Reinhardt et al., 1998); however, the authors did not report the nucleotide or amino acid sequence. We have subsequently confirmed that the LeExp2 DNA sequences are identical (S. McQueen-Mason, personal communication).
A phylogenetic tree generated from the alignment of the deduced amino acid sequences of LeExp2 and other expansin genes identified different subgroups of expansins (Fig. 1). Four groups were indicated and, although group I is supported with a low bootstrap value, the same groups have been identified in another phylogenetic analysis based on nucleotide sequences of expansins (Link and Cosgrove, 1998). LeExp2 aligned within a sublineage of group I containing expansins from tobacco (NtExp5, 89% amino acid identity), cucumber (CuExp1, 78% identity), and pine (PtExp4, 76% identity). This group also contained expansins from rice (OsExp4) and cotton (GhExp1), whose expression is correlated with elongation growth (Cho and Kende, 1997; Orford and Timmis, 1998). Five other tomato expansins aligned within different phylogenetic groups and showed 49% to 71% identity with LeExp2. Group IV contained three expansin homologs: LeExp4, expressed in young tomato fruit (Brummell et al., 1999) and LeExp1 and LeExp18, which are expressed during fruit ripening and leaf primordia formation, respectively (Rose et al., 1997; Reinhardt et al., 1998). The more divergent tomato expansin LeExp3 aligns outside the four principal groups.
Figure 1.
Phylogenetic tree of the alignment of LeExp2 deduced amino acid sequence with other α-expansins. Protein sequences were aligned using CLUSTAL, and a phylogenetic tree was constructed using PHYLIP software with the PROTPARS program. Numbers above the branches indicate the bootstrap values. A Phleum pollen allergen (Phlp1), identified as a β-expansin, was used as an outgroup. Full details and accession numbers are given in “Materials and Methods.”
Genomic Analysis of LeExp2
A cDNA fragment corresponding to part of the LeExp2 coding region was used as a probe for genomic DNA gel-blot analysis (Fig. 2). The probe hybridized strongly to a single genomic fragment, indicating that LeExp2 is a single-copy gene and that the probe is gene specific. Additional faint bands were present in some lanes and may represent other distantly related genes. The same LeExp2 probe was used subsequently at the same relative stringency to probe the RNA gel blots.
Figure 2.
Genomic DNA analysis of LeExp2. Genomic DNA (10 μg per lane) was digested with the indicated restriction enzymes, gel blot hybridized with the LeExp2 cDNA probe, and washed with 0.2× SSC, 0.5% (w/v) SDS at 65°C (8°C below the Tm).
LeExp2 mRNA Accumulates in the Apical Elongation Zone of Tomato Etiolated Hypocotyls
LeExp2 expression was examined in tomato vegetative tissues at the level of mRNA abundance using the same cDNA probe as above. RNA gel-blot analysis using poly(A+) RNA from leaf, stem, hypocotyl, and root revealed a band of 1.2 kb (Fig. 3a). The strongest hybridization signal was detected in hypocotyls, while expression was low in roots. A steep gradient in growth rate from the apical to basal regions of etiolated seedlings has been observed in several plant species, with the maximal rate in the elongation zone immediately below the apical hook (Shinkle et al., 1992; Gendreau et al., 1997). An analysis of LeExp2 mRNA abundance in different regions of etiolated hypocotyls showed high levels in the apical elongating zone (elongation rate, 0.73 ± 0.15 mm h−1), while levels decreased toward region B (elongation rate, 0.36 ± 0.07 mm h−1) and C (elongation rate, 0.14 ± 0.08 mm h−1) (Fig. 3b). LeExp2 was not detected in zone D adjacent to the root, which exhibited negligible elongation.
Figure 3.
Analysis of LeExp2 mRNA abundance in tomato vegetative tissues. a, Poly(A+) RNA-blot analysis of LeExp2 expression in vegetative tissues. Each lane contained 1 μg of poly(A+) RNA from leaf, stem, hypocotyl, or root tissues. b, Total RNA gel-blot analysis of LeExp2 expression along the tomato etiolated hypocotyl. Each lane contained 15 μg of total RNA isolated from consecutive 1-cm regions of the hypocotyl. Ethidium bromide staining of the gel is shown below the blot as a loading control. The growth rates of regions A through D were measured in 15 seedlings over a 12-h period, as described in “Materials and Methods.” RNA gel blots were hybridized and washed at the same stringency as in Figure 2.
LeExp2 Expression Is Auxin Regulated in Tomato Etiolated Hypocotyl Segments
The effect of auxin on LeExp2 mRNA abundance was examined using previously described conditions (Kelly and Bradford, 1986; Catalá et al., 1997). Etiolated hypocotyl segments were pre-incubated for 2 h in buffer to deplete endogenous auxin and then incubated in buffer alone, or in buffer plus the synthetic auxin 2,4-D. In this system it has been shown that the growth of tomato hypocotyl segments is auxin-stimulated over a 12-h incubation period (Kelly and Bradford, 1986; Catalá et al., 1997). We extended the analysis with treatments up to 24 h to be able to detect a prolonged or delayed accumulation of transcripts, such as has been observed for some auxin-induced genes (Sitbon and Perrot-Rechenmann, 1997), including endo-1,4-β-glucanases (Verma et al., 1975; Catalá et al., 1997). Segments were incubated for 24 h in a range of 2,4-D concentrations (Fig. 4a) or in 5 μm 2,4-D over a time course of up to 24 h (Fig. 4b).
Figure 4.
Auxin regulation of LeExp2 mRNA levels in etiolated hypocotyl segments. a, Effect of auxin concentration on LeExp2 accumulation. Apical segments were pre-incubated for 2 h in buffer and then transferred to fresh buffer plus the indicated auxin concentration and incubated for 24 h. NI, Non-incubated; BR, brassinolide. b, Time course of LeExp2 mRNA accumulation. Apical segments were pre-incubated for 2 h in buffer (0-h time point) and then for the indicated times in buffer alone (control) or buffer plus 5 μm 2,4-D. c, Effect of other auxins on LeExp2 mRNA accumulation. Apical segments were pre-incubated for 2 h in buffer (0-h time point) and then for 24 h in buffer alone (control) or buffer plus the indicated concentration of 2,4-D, αNAA, or IAA. After incubation, RNA was isolated and total RNA-blot analysis (15 μg per lane) was performed. Ethidium bromide staining of the gel is shown below the blots as a loading control. RNA gel blots were hybridized and washed at the same stringency as in Figure 2.
Although LeExp2 is expressed at high levels in the apical regions of growing hypocotyls, it was not detected in hypocotyl segments following incubation for 24 h in buffer alone (Fig. 4a). However, mRNA levels were substantially higher upon incubation with 1 μm 2,4-D. LeExp2 mRNA accumulation was induced to a similar extent with 5 μm 2,4-D, and 10 μm 2,4-D resulted in slightly higher levels. Treatment of segments for 24 h with 0.1 to 1.0 μm of the growth-inducing hormone brassinolide did not increase LeExp2 mRNA levels (Fig. 4a). Analysis of changes in LeExp2 expression with time showed that mRNA levels in the control segments declined over the 6 h following the addition of fresh buffer, and became undetectable after 12 h (Fig. 4b). Segments incubated in the presence of 2,4-D accumulated substantially higher levels of LeExp2 mRNA after 2 h, and by 6 h, LeExp2 mRNA abundance had been restored to similar or greater levels than those at the beginning of the incubation period. Although LeExp2 mRNA abundance also decreased during incubation of the segments with auxin, the decrease was delayed and transcript levels were substantially higher than in the corresponding control segments after 24 h.
The effect of the natural auxin IAA and the auxin analog αNAA on the steady-state levels of LeExp2 mRNA was also examined. Hormone concentrations shown to be optimal in stimulating hypocotyl segment elongation were used (Kelly and Bradford, 1986; Catalá et al., 1997). A higher concentration of IAA was also used to compensate for the instability of this compound. As observed with 2,4-D, incubation of segments for 24 h with IAA or αNAA also elevated the levels of LeExp2 mRNA over the control incubated in buffer alone (Fig. 4c).
LeExp2 and Auxin-Regulated XET and EGase Genes Are Expressed during Tomato Fruit Growth
The expression of LeExp2 during tomato fruit development was examined, together with the expression of genes encoding an XET (LeEXT1) and an EGase (Cel7), which, like LeExp2, are auxin regulated in etiolated hypocotyls (Catalá et al., 1997).
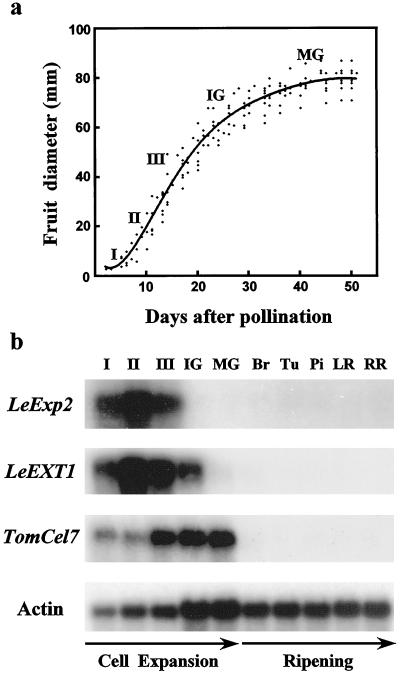
Tomato fruit growth followed a sigmoidal curve (Fig. 5a). After fruit set and cell division (beginning of stage I), a growth phase, characterized by a rapid increase in fruit diameter (stages II and III), was followed by a period of continued growth but at a reduced rate (end of stage III, immature green stage). The growth rate continued to decline until the end of fruit expansion (mature green stage).
Figure 5.
Expression of expansin (LeExp2), XET (LeEXT1), and EGase (Cel7) genes during tomato fruit development. Flowers were tagged at anthesis and fruit diameter measured over a period of 50 d. A regression curve was established from data obtained with 15 fruit. Poly(A+) RNA-blot analysis of LeExp2, LeEXT1, and Cel7 expression was done during tomato fruit development. Northern blots (1 μg per lane) were hybridized successively with the LeExp2, LeEXT1, Cel7, and actin cDNA probes. (I, 0.5- to 1.0-cm diameter fruit; II, 2- to 3-cm diameter fruit; III, 4- to 6-cm diameter fruit; IG, immature green; MG, mature green; Br, breaker; Tu, turning; Pi, pink; LR, light red; RR, red-ripe). RNA gel blots were hybridized and washed at the same stringency as in Figure 2.
LeExp2 mRNA levels were high in young expanding fruit, exhibiting a peak at stage II, corresponding to the greatest rate of fruit expansion, but decreased dramatically at the immature green stage (Fig. 5b). LeExp2 mRNA was undetectable during ripening. The XET gene, LeEXT1, showed a similar pattern of expression to LeExp2, with the highest mRNA levels at stages II and III, prior to a decrease at the immature green stage. Expression was barely detectable in mature green fruit that had reached full expansion and was not observed at the onset of or during ripening (Fig. 5b). The EGase gene Cel7 was also expressed during fruit expansion, but showed a different pattern from LeExp2 and LeEXT1. Levels of Cel7 mRNA increased at stage III of fruit expansion and remained high in the immature and mature green fruit. Cel7 was not detected at the breaker stage, which marks the onset of autocatalytic ethylene production and fruit ripening, or in later ripening stages (Fig. 5b).
DISCUSSION
We have shown that a new member of the tomato expansin gene family, LeExp2, is regulated by auxin, and its expression pattern is consistent with a role in plant cell expansion. LeExp2 mRNA accumulated primarily in elongating regions of the tomato hypocotyl and was absent in mature, non-expanding basal regions (Fig. 3b). The LeExp2 mRNA decayed to undetectable levels during a 24-h incubation of apical hypocotyl sections in buffer alone, which may indicate a low stability of the transcript. However, the addition of auxin resulted in the maintenance of higher LeExp2 mRNA levels during the incubation period (Fig. 4b). This increase in LeExp2 mRNA levels may be controlled at the level of gene transcription and/or mRNA stability. Regulation of gene expression by auxin has been most extensively studied for members of the SAUR (small auxin-up-regulated genes) and Aux/IAA gene families. These rapidly induced, auxin-responsive mRNAs are under transcriptional regulation involving regulatory sequences in the promoter region (Abel and Theologis, 1996). As with LeExp2, these auxin-responsive mRNAs also decline to undetectable levels when organ sections are excised and incubated in the absence of auxin for up to 24 h.
Relatively short half-lives have been measured for some of these transcripts; however, auxin does not significantly alter the stability of certain SAUR and Aux/IAA mRNAs (Koshiba et al., 1995; Gil and Green, 1996). Transient accumulation of LeExp2 transcript in the presence of auxin (Fig. 4b) is consistent with a short mRNA half-life; however, whether LeExp2 mRNA levels are regulated through gene transcription and/or post-transcriptional events remains to be determined. Treatment of segments with the hormone brassinolide, which also promotes tissue elongation (Zurek et al., 1994), caused no increase in LeExp2 mRNA levels.
It has been demonstrated that brassinosteroid treatment of tomato hypocotyl segments does not induce members of the tomato SAUR gene family or other auxin-inducible genes, and it has been suggested that the molecular mechanism(s) of brassinosteroid-promoted elongation is likely to differ from that of auxin-induced elongation (Clouse et al., 1992; Zurek et al., 1994). Other expansin gene(s) may be involved in brassinosteroid-induced elongation. This suggests that the maintenance of LeExp2 mRNA steady-state levels is part of an auxin-regulated pathway and could act as an important mechanism for sustained auxin-induced growth. We also examined the expression of LeExp2 during tomato fruit development, and showed that it is selectively expressed during a period of rapid and substantial cell expansion but not during fruit ripening (Fig. 5).
Phylogenetic analysis of expansin sequences has revealed the existence of at least three subfamilies of expansin genes (Fig. 1) that contain characteristic conserved amino acid sequences (Link and Cosgrove, 1998). This suggests that each expansin subclass comprises members with similar specific physiological roles, which may be reflected in common patterns of expression and hormonal regulation. The LeExp2 sequence is highly homologous to the tobacco expansin NtExp5 and is closely related to a biochemically characterized cucumber expansin (CsExp1) that promotes extension in isolated cell walls of cucumber hypocotyls (McQueen-Mason et al., 1992). LeExp2 and CsExp1 belong to a sublineage of group I. This group also contains an expansin gene expressed specifically during elongation of cotton fibers (Orford and Timmis, 1998) and a rice expansin (OsExp4) that is up-regulated by gibberellic acid, a treatment that promotes rapid internode elongation (Cho and Kende, 1997). Therefore, this group includes expansins involved in facilitating rapid cell expansion and regulated by growth-promoting hormones.
Tomato expansins comprise a gene family with individual members showing distinct and overlapping expression during different phases of fruit development (Brummell et al., 1999). LeExp4, like LeExp2, is expressed during rapid fruit expansion; however, these genes align within divergent phylogenetic groups: I and IV, respectively (Fig. 1). It is likely that divergent tomato expansins are expressed in specific fruit cell types, are regulated differentially by environmental and hormonal stimuli, or act on different cell wall components. Furthermore, the tissue-specific expression of other tomato expansins from group IV, LeExp18 and LeExp1, suggests their involvement in distinct processes involving differentiation (such as primordium initiation; Reinhardt et al., 1998) and and morphogenesis (such as wall disassembly during fruit ripening; Rose et al., 1997). It is interesting that the tomato ripening-related expansin LeExp1 has a diametrically opposite pattern of expression to LeExp2 during fruit development and is regulated by ethylene, a hormone that coordinates many ripening-regulated pathways (Rose et al., 1997). More detailed analyses of tissue-specific expression and hormonal regulation of individual members of the expansin gene family may help elucidate the significance of evolutionary divergence among expansins that has given rise to the different phylogenetic groups.
In tomato, XETs and EGases, like expansins, comprise gene families with divergent members showing differential developmental or hormonal regulation (Rose and Bennett, 1999). In common with LeExp2, the tomato genes LeEXT1 and Cel7, encoding an XET and an EGase, respectively, are expressed during auxin-induced elongation of hypocotyl segments (Catalá et al., 1997) and could participate in the cell wall restructuring necessary for sustained fruit expansion. We have shown that, like LeExp2, LeEXT1 and Cel7 are selectively expressed during tomato fruit growth (Fig. 5), with negligible mRNA levels present during fruit ripening. The mRNA levels of both LeExp2 and LeEXT1 peaked during the stages of higher rates of fruit growth, while Cel7 expression increased and remained high during the later stages of fruit expansion, when the growth rate is lower. This trend was similar to the time dependence of mRNA accumulation observed for these genes during the incubation of hypocotyl segments with auxin (Fig. 4b; Catalá et al., 1997). In hypocotyl segments, LeExp2 and LeEXT1 transcript levels declined after the period of maximal elongation, while Cel7 mRNA accumulation, which lagged behind that of LeEXT1, remained high after segment elongation had ceased.
Disassembly of the cellulose-xyloglucan network by the potentially coordinated action of expansins and enzymes such as XETs and EGases has been suggested to mediate reversible wall loosening (Rose and Bennett, 1999). Several models may be proposed whereby the products of the LeExp2 and LeEXT1 genes have a synergistic or complementary action to allow controlled restructuring of the cellulose-xyloglucan matrix during cell expansion. The action of expansin may disrupt the association between hemicellulose and the cellulose microfibrils, allowing transient wall loosening, while at the same time XETs catalyze incorporation of newly synthesized xyloglucan into the expanding wall, maintaining structural integrity and tensile strength. The kinetics of the accumulation of Cel7 mRNA are consistent with Cel7 acting as the rate of cell expansion decreases, perhaps hydrolyzing free ends of newly incorporated xyloglucans. Extensive trimming of the non-cellulose-bound domains of xyloglucans was reported in studies of rapidly growing rose cell-suspension cultures (Thompson and Fry, 1997). The significance of such modifications is unclear; however, it should be noted that hydrolysis of xyloglucan chains might generate substantial populations of xyloglucan oligosaccharides that can act as biologically active growth regulators (Creelman and Mullett, 1997).
The patterns of expression of LeExp2, LeEXT1, and Cel7 during the period of rapid fruit growth suggest that they are part of a developmental and/or hormonal signal transduction network controlling cell expansion in fruit. LeExp2, LeEXT1, and Cel7 are regulated by auxin in etiolated hypocotyl segments, and since auxin levels peak coincident with the initiation of cell expansion during tomato fruit development (Hocher et al., 1992; Buta and Spaulding, 1994), it is likely that these genes are also up-regulated by auxin during tomato fruit growth. Down-regulation of LeExp2, LeEXT1, and Cel7 expression is coincident with the cessation of fruit cell expansion and the initiation of ripening. In contrast, ripening-related EGase, XET, and expansin genes have been identified and in some cases shown to be ethylene up-regulated (Lashbrook et al., 1994; Arrowsmith and de Silva, 1995; Rose et al., 1997). This suggests that during fruit development the structure of the cell wall is modified by the actions of different subsets of these classes of enzymes whose gene expression is regulated by different hormonal signals such as auxin and ethylene, coordinating fruit growth and ripening, respectively.
LITERATURE CITED
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith DA, de Silva J. Characterisation of two tomato fruit-expressed cDNAs encoding xyloglucan endo-transglycosylases. Plant Mol Biol. 1995;28:391–403. doi: 10.1007/BF00020389. [DOI] [PubMed] [Google Scholar]
- Bonghi C, Ferrarese L, Ruperti B, Tonutti P, Ramina A. Endo-β-1,4-glucanases are involved in peach fruit growth and ripening, and regulated by ethylene. Physiol Plant. 1998;102:346–352. [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999;39:161–169. doi: 10.1023/a:1006130018931. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Lashbrook CC, Bennett AB. Plant endo-1,4-β-glucanases: structure, properties and physiological function. Am Chem Soc Symp Ser. 1994;566:100–129. [Google Scholar]
- Buta JG, Spaulding DW. Changes in indole-3-acetic acid and abscisic acid levels during tomato (Lycopersicon esculentum Mill.) fruit development and ripening. J Plant Growth Reg. 1994;13:163–163. [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. Auxin regulation and spatial localization of an endo-1,4-β-d-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J. 1997;12:417–426. doi: 10.1046/j.1365-313x.1997.12020417.x. [DOI] [PubMed] [Google Scholar]
- Cho H-T, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland RE. Auxin and cell elongation. In: Davies PJ, editor. Plant Hormones. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 214–227. [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME. Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol. 1992;100:1377–1383. doi: 10.1104/pp.100.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe B. The development of fleshy fruits. Annu Rev Plant Physiol. 1976;27:507–528. [Google Scholar]
- Cosgrove DJ. Wall extensibility: its nature, measurement, and relationship to plant cell growth. New Phytol. 1993;124:1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Oligosaccharins, brassinolides and jasmonates: nontraditional regulators of plant growth, development and gene expression. Plant Cell. 1997;9:1211–1223. doi: 10.1105/tpc.9.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Desveaux D, Maclachlan G. Enzymic activities responsible for xyloglucan depolymerization in extracts of developing tomato fruit. Phytochemistry. 1998;49:365–376. [Google Scholar]
- Felsenstein J. PHYLIP phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Green PJ. Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 1996;15:1678–1686. [PMC free article] [PubMed] [Google Scholar]
- Gillaspy GH, Ben-David H, Gruissem W. Fruits: a developmental perspective. Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Maclachlan G. Pea xyloglucan and cellulose: I. Macromolecular organization. Plant Physiol. 1984;75:596–604. doi: 10.1104/pp.75.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrik N, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Hocher V, Sotta B, Maldiney R, Bonnet M, Miginac E. Changes in indole-3-acetic acid levels during tomato (Lycopersicon esculentum Mill.) seed development. Plant Cell Rep. 1992;11:253–256. doi: 10.1007/BF00235076. [DOI] [PubMed] [Google Scholar]
- Kelly M, Bradford K J. Insensitivity of the diageotropica tomato mutant to auxin. Plant Physiol. 1986;82:713–717. doi: 10.1104/pp.82.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong LM, Theologis A. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum) J Mol Biol. 1995;235:396–413. doi: 10.1006/jmbi.1995.0562. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Briggs WR. Rapid auxin-induced stimulation of cell wall synthesis in pea internodes. Proc Natl Acad Sci USA. 1987;84:2747–2751. doi: 10.1073/pnas.84.9.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labavitch JM, Ray PM. Turnover of cell wall polysaccharides in elongating pea stem segments. Plant Physiol. 1974;53:669–673. doi: 10.1104/pp.53.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and α-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Wells B, Roberts K. Direct visualization of cross-links in the primary cell wall. J Cell Sci. 1990;96:323–334. [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Expansin mode of action on cell walls. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyses transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- O'Neill SD, Tong Y, Sporlein B, Forkmann G, Yoder JI. Molecular genetic analysis of chalcone synthase in Lycopersicon esculentum and an anthocyanin-deficient mutant. Mol Gen Genet. 1990;224:279–288. doi: 10.1007/BF00271562. [DOI] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN. Specific expression of an expansin gene during elongation of cotton fibers. Biochim Biophys Acta. 1998;1398:342–346. doi: 10.1016/s0167-4781(98)00065-7. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Robson PRH, Smith H, Estelle M, Klee HJ. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6–1 hypocotyl elongation and axr1 auxin resistant mutants. Plant Mol Biol. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999;4:176–183. doi: 10.1016/s1360-1385(99)01405-3. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Brummell DA, Bennett AB. Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in nasturtium. Plant Physiol. 1996;110:493–499. doi: 10.1104/pp.110.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schröder R, Atkinson RG, Langenkämper G, Redgwell RJ. Biochemical and molecular characterisation of xyloglucan endotransglycosylase from ripe kiwifruit. Planta. 1998;204:242–251. doi: 10.1007/s004250050253. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason S, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkle JR, Sooudi SK, Jones RL. Adaptation to dim-red light leads to a nongradient pattern of stem elongation in Cucumis seedlings. Plant Physiol. 1992;99:808–811. doi: 10.1104/pp.99.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon F, Perrot-Rechenmann CP. Expression of auxin-regulated genes. Physiol Plant. 1997;100:443–455. [Google Scholar]
- Taiz L. Plant cell expansion: regulation of cell wall mechanical properties. Annu Rev Plant Physiol. 1984;35:585–657. [Google Scholar]
- Talbott LD, Ray PM. Changes in molecular size of previously deposited and newly synthesized pea cell wall matrix polysaccharides. Plant Physiol. 1992;98:369–379. doi: 10.1104/pp.98.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JE, Fry SC. Trimming and solubilization of xyloglucan after deposition in the walls of cultured rose cells. J Exp Bot. 1997;48:297–305. [Google Scholar]
- Verma DPS, Maclachlan GA, Byrne H, Ewings D. Regulation and in vitro translation of messenger ribonucleic acid for cellulase from auxin-treated pea epicotyls. J Biol Chem. 1975;250:1019–1026. [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton. Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Wu S-C, Blumer JM, Darvill AG, Albersheim P. Characterization of an endo-β-1,4-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiol. 1996;110:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Law DM, Davies PJ. Magnitude and kinetics of stem elongation induced by exogenous indole-3-acetic acid in intact light-grown pea seedlings. Plant Physiol. 1993;102:717–724. doi: 10.1104/pp.102.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD. Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol. 1994;104:505–513. doi: 10.1104/pp.104.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]