Abstract
Zebrafish is a widely used animal model in biomedical sciences and toxicology. Although evidence for the presence of phases I and II xenobiotic defense mechanisms in zebrafish exists on the transcriptional and enzyme activity level, little is known about the protein expression of xenobiotic metabolizing enzymes. Given the important role of glutathione S-transferases (GSTs) in phase II biotransformation, we analyzed cytosolic GST proteins in zebrafish early life stages and different organs of adult male and female fish, using a targeted proteomics approach. The established multiple reaction monitoring-based assays enable the measurement of the relative abundance of specific GST isoenzymes and GST classes in zebrafish through a combination of proteotypic peptides and peptides shared within the same class. GSTs of the classes alpha, mu, pi and rho are expressed in zebrafish embryo as early as 4 h postfertilization (hpf). The majority of GST enzymes are present at 72 hpf followed by a continuous increase in expression thereafter. In adult zebrafish, GST expression is organ dependent, with most of the GST classes showing the highest expression in the liver. The expression of a wide range of cytosolic GST isoenzymes and classes in zebrafish early life stages and adulthood supports the use of zebrafish as a model organism in chemical-related investigations.
Keywords: GST, biotransformation, mass spectrometry, multiple reaction monitoring, targeted proteomics
Assessing the risk posed by chemicals to human and environmental health requires appropriate models to investigate the chemicals’ biological activity and toxicity. Especially vertebrate models which are suitable for mechanistic investigations and medium- to high-throughput approaches are needed in order to comply with the requirements imposed by 21st century toxicology (Krewski et al., 2010). Zebrafish (Danio rerio) is such a model. It shares a high degree of homology with other vertebrates, including humans, and zebrafish early life stages attract attention owing to their small size and transparency (Dahm and Geisler, 2006). Yet, despite its popularity and increasing use in biomedical research as well as human and environmental toxicology, knowledge gaps still exist concerning the capacity of zebrafish to biotransform and detoxify chemicals, particularly at early life stages.
The biotransformation potential of zebrafish has been the focus of previous studies, which provided transcriptional evidence for enzymes involved in phases I and II metabolism, including cytochrome P450 (Cyp450), uridine 5′-diphospho-glucuronosyltransferase and glutathione S-transferases (GSTs), already in early stages of the development (Christen and Fent, 2014; Glisic et al., 2016; Goldstone et al., 2010; Timme-Laragy et al., 2013). Additionally, Otte et al. (2017) mapped intrinsic activities of representative enzymes involved in xenobiotic metabolism. The study demonstrated that selected phases I and II enzymes, such as Cyp450 and GSTs, are already active in early developmental stages.
GSTs are an enzyme family that plays a major role in phase II biotransformation processes by catalyzing the conjugation reaction of the tripeptide glutathione (GSH) with electrophilic substrates (Sheehan et al., 2001). This reaction typically results in the formation of more hydrophilic and readily excretable products. Accordingly, GST activity is considered a critical contributor to detoxification and clearance of various intracellular metabolites, but also natural toxins and xenobiotic compounds, including drugs, and their reactive intermediates (Hayes et al., 2005; Sau et al., 2010).
The GST family consists of 3 major groups: membrane-associated, mitochondrial, and cytosolic proteins (Glisic et al., 2015; Hayes et al., 2005). Membrane-associated GSTs belong to the microsomal GST or membrane-associated proteins in eicosanoid and GSH metabolism class, and are involved in the biosynthesis of leukotrienes and prostanoids (Glisic et al, 2015; Jakobsson et al., 1999). Mitochondrial GSTs form the kappa-class (Thomson et al., 2004). Cytosolic GSTs are subdivided into several classes based on enzyme sequence similarities (Glisic et al., 2015; Hayes et al., 2005; Sheehan et al., 2001). These classes are alpha, zeta, theta, mu, pi and omega. In mammalian species, a further cytosolic class, sigma, is present, whereas the cytosolic class rho is specific to teleosts and cephalo-chordates (Glisic et al., 2015). The diversification of cytosolic GSTs into multiple classes provides a broad substrate specificity for the inactivation of potentially harmful endogenous and exogenous compounds, including xenobiotics (Glisic et al., 2015). Increasing our knowledge about cytosolic GSTs therefore will help to better understand the xenobiotic defense mechanisms and their contribution to sensitivity differences among species or life stages.
To date, zebrafish GST studies have focused on 2 levels: GST enzymatic activity (Best et al., 2002; Notch et al., 2011; Otte et al., 2017; Pavagadhi et al., 2012; Wiegand et al., 2000) and mRNA expression (Abunnaja et al., 2017; Glisic et al., 2015, 2016; Timme-Laragy et al., 2013). GST enzymatic activity was detected within the first 4 h of zebrafish development (Notch et al., 2011; Otte et al., 2017; Wiegand et al., 2000) as well as in all examined organs of adult zebrafish (Pavagadhi et al., 2012). Members of all cytosolic GST classes were detectable on the mRNA level during zebrafish development (Glisic et al., 2016; Timme-Laragy et al., 2013). In adult zebrafish, GST mRNA expression levels were found to be tissue- and sex-dependent (Glisic et al., 2015).
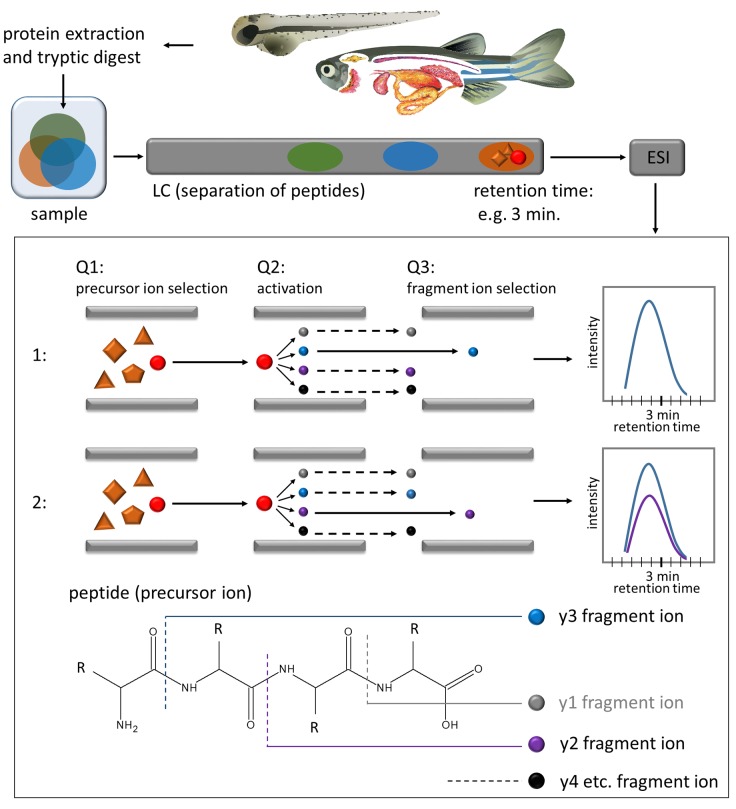
Although enzyme activity and mRNA abundance studies give a valuable overview of the GST family in zebrafish, they have certain limits. Nonspecific substrates used for activity measurements, such as 1-chloro-2, 4-dinitrobenzene (CDNB), do not allow to differentiate between GST isoenzymes (Glisic et al., 2015; Habig et al., 1974). Although the detection of selected isoenzymes is possible via mRNA analysis, the correlation with the protein data is usually poor (Li et al., 2014; Schwanhausser et al., 2011). Proteins are the biomolecules carrying out biotransformation reactions, yet studies mapping GST expression on the protein level are missing in zebrafish. It is, however, possible to analyze enzymes on the protein level within complex biological samples using mass spectrometry-based targeted proteomics (Picotti and Aebersold, 2012). With the multiple reaction monitoring (MRM) technique (Figure 1), multiple pairs of peptide precursor and fragment ions can be monitored over a chromatographic run, allowing the analysis of several proteins within 1 measurement (Bereman et al., 2012; Lange et al., 2008; Picotti and Aebersold, 2012; Surinova et al., 2013).
Figure 1.
Overview of experimental workflow and schematic representation of the MRM technique. The proteins are extracted from tissue, tryptically digested into peptides, separated with liquid chromatography (LC), ionized via electrospray ionization (ESI) and transferred into the triple-quadrupole mass spectrometer (Q1–Q3). In Q1—the precursor ion (peptide) is selected, in Q2 the peptide is fragmented into fragment ions (y1, y2. y3, etc.), in Q3 the selection of the fragment ion takes place. Subsequently, the intensity of the fragment ion is measured over time. Within 1 measurement, several transitions (peptide and fragment ion) can be monitored.
We performed MRM-based targeted analyses to investigate how cytosolic GST proteins evolve in zebrafish during early life stages and how they are expressed in organs of adult male and female fish. This involved the development of MRM assays for a panel of proteotypic peptides and peptides shared within the same class, enabling the analysis of the relative abundance of specific GST isoenzymes and GST classes in zebrafish.
MATERIALS AND METHODS
Zebrafish maintenance and sampling
Wild-type zebrafish with mixed genetic background from WiK (Max Planck Institute for Developmental Biology, Tübingen, Germany), OBI (Helmholtz Center for Environmental Research established from OBI Baumarkt, Leipzig, Germany) and Qualipet (petshop, Wallisellen, Switzerland) strains were maintained and bred in our facility according to recommended procedures (Nüsslein-Volhard and Dahm, 2002). Fish were reared in a flow-through system filled with a 1:2 mixture of reconstituted water (294.0 mg/l CaCl2·2H2O, 123.2 mg/l MgSO4·7H2O, 64.7 mg/l NaHCO3 and 5.7 mg/l KCl; ISO 15088: 2007(E); 2007) and tap water. Water temperature ranged from 26°C to 28°C; light/dark cycle was 14/10 h. Zebrafish were fed with live food (Artemia nauplia) and dry vitamin flakes (TetraMin, USA) twice daily. Zebrafish eggs were obtained from group crosses. Eggs were collected 1 h after the light in the facility was switched on, washed with reconstituted water and raised in Petri dishes in an incubator at 28°C, 14/10 h light/dark cycle. Zebrafish samples were collected at 4, 8, 24, 48, 72, 96, 120, and 168 h postfertilization (hpf) in order to cover a range of life stages, starting from early embryo and continuing until the end of the transition phase from nonfeeding to larvae capable of independent feeding. Zebrafish embryos and larvae were washed in ice-cold phosphate-buffered saline and snap frozen in liquid nitrogen (60 embryos/sample) in Eppendorf LoBind microcentrifuge tubes (Sigma-Aldrich, USA). The embryos collected at 24 and 48 hpf were dechorionated with forceps prior to sampling, in order to remove chorion proteins. The organs (liver, intestine, gills, brain, gonads, and kidney) were obtained from adult animals aged approximately 1.5 years. For organ collection, adult fish were euthanized with tricaine methanesulfonate (MS222) and dissected. Organs from 4 fish of the same sex were pooled for 1 replicate. The organs were snap frozen in liquid nitrogen in Eppendorf LoBind microcentrifuge tubes (Sigma-Aldrich, USA). Samples were stored at −80°C until further processing. All procedures were in accordance with the animal protection guidelines and approved by the Cantonal Veterinary Office Zurich, Switzerland.
Protein extraction and preparation of tryptic digests
Protein extraction and trypsin digestion were performed as reported previously (Groh et al., 2013) with some modifications. Briefly, samples were taken up in 600 µl ice-cold lysis buffer immediately upon thawing (9 M urea, 2 M thiourea, 0.1 M Tris–HCl, 4% CHAPS, 100 mM DTT, 1× Protease Inhibitor Cocktail, pH 8.5) and homogenized with the soft tissue homogenizing kit (Bertin Instruments, France) using a FastPrep-24 Homogenizer (MP Biomedicals, USA). The sample lysate was centrifuged at 14, 000 × g for 15 min at 4°C and the supernatant was aliquoted in 150 µl amounts into fresh Eppendorf LoBind microcentrifuge tubes (Sigma-Aldrich, USA). Proteins were precipitated from the supernatant using the methanol/chloroform method. The protein pellet was isolated, air-dried for 4 min, wetted with 5 µl NaOH (0.2 M) and redissolved in 25 µl re-solubilization buffer (9 M urea, 2 M thiourea, 50 mM Tris HCl, pH 8). Aliquots of the same sample were recombined and protein concentration determined by the Bradford method. Subsequently, proteins within each sample were diluted with resolubilization buffer to a final protein concentration of 2.5 µg/µl. Samples were reduced with tris(2-carboxyethyl)phosphine (5 mM final concentration) for 30 min in the dark at room temperature, alkylated (carbamidomethylated) with iodacetamide (25 mM final concentration) for 30 min in the dark at room temperature and digested with trypsin (trypsin: protein ratio of 1:100, trypsin sequencing grade, Roche, Switzerland) at 37°C for 16 h. Prior to the trypsin digestion, adult organ samples were spiked with the standards apomyoglobin from equine skeletal muscle and the MS Qual/Quant QC Mix (Sigma-Aldrich, USA). The digestion was terminated through addition of formic acid (1% final concentration). The samples were desalted using reversed-phase cartridges (Sep-Pak Vac tC18, Waters, USA), eluted in 800 µl 80% acetonitrile solution with 0.1% (v/v) formic acid, evaporated using a vacuum centrifuge at 30°C, re-dissolved in 100 µl nanopure water with 0.1% formic acid and filtered through Amicon Ultrafree centrifugal filters, 0.45 µm cutoff (Millipore, USA). The samples were then stored at 4°C or immediately measured on the TSQ Vantage (Thermo Scientific, USA).
Selection of peptides and MRM method development
Protein reference sequences for the cytosolic GSTs were retrieved from NCBI (Supplementary Table 1), imported into Skyline (MacLean et al., 2010) as FASTA files and in silico digested with trypsin. Only fully tryptic peptides in a range of 6–21 amino acids were selected for further processing. Proteotypic peptides (peptides that uniquely identify the protein of interest) and peptides that cover conserved domains (characteristic for 2 or more isoenzymes of the same class, see Figure 2) were selected by running a protein-protein BLAST search against the non-redundant protein sequences from D. rerio (taxid: 7955). As suggested previously (Lange et al., 2008), peptides containing the RP/KP motive, 2 neighboring amino acids (K and/or R) at either cleavage site, and amino acids that are prone to chemical modifications (eg, alkylation, deamination, and oxidation) were avoided whenever possible. In total, 86 peptides representing isoenzymes of the cytosolic GSTs were chosen and commercially synthesized as small scale, unpurified peptides with carbamidomethyl-modified cysteine and C-terminal lysine or arginine (SpikeTides, JPT Peptide Technologies, Germany, see Supplementary Table 2). Their sequence and position within the protein are shown in Supplementary Table 2 and the chromatograms can be downloaded from the Dryad Digital Repository. The synthetic peptides (approximately 54 nmol) were re-suspended in 150 µl aqueous solution containing 20% acetonitrile and 1% formic acid under gentle agitation (30 min, room temperature), aliquoted in volumes of 35 µl and kept at −80°C for long-term storage. For measurements, a pool of all synthetic peptides was generated (10 µl each), evaporated under vacuum at 30°C, redissolved in nanopure water with 0.1% formic acid and stored at 4°C until use.
Figure 2.
Schematic amino acid sequence alignment of GST classes for the visualization of proteotypic peptides (no frame), shared peptides that cover a sequence present in some isoenzymes (dashed frame) and shared peptides that cover a sequence present in all isoenzymes of the respective enzyme class (solid frame). Peptides which were detected only in organs of adult zebrafish are shown with a gray background. All others were also found at certain stages of zebrafish early development.
The MRM methods were developed with Skyline. The charge state of the precursor ions was set to +2 for peptides <15 amino acids and +2 and +3 for peptides >15 amino acids. Only singly charged fragment ions with m/z > precursor were considered. The collision energy for each peptide was calculated for the Thermo TSQ Vantage using Skyline. Cysteine was set to be carbamidomethyl-modified and peptides containing methionine were synthesized and monitored in the oxidized and reduced form.
The synthetic standard mixture in nanopure water with 0.1% formic acid was run on a TSQ Vantage to validate the MRM methods and to obtain peptide retention time (RT) information. The data was processed using Skyline. The 2–3 strongest MRM transitions of detected synthetic peptides were then selected for the analysis of the endogenous peptides.
LC and MS settings
Samples were injected onto a Poroshell 120 EC—C18 (2.7 µm particle size, 2.1 × 100 mm column—Agilent, USA) and separated at a flow rate of 150 µl/min using a 38 min linear gradient from 100% solvent A (1% methanol in water, 0.2% formic acid) to 100% solvent B (98.8% methanol, 0.2% formic acid), followed by a washing step (4 min with 100% solvent B) and a re-equilibration step (8 min with 100% solvent A). The measurements were performed on a TSQ Vantage operating in MRM mode with a scan width of 0.3 m/z and a dwell time of 30 ms. In order to measure all desired transitions with an optimal cycle time, the number of peptides monitored simultaneously was limited to a maximum of 60 per segment. With this, the resulting cycle time was <1.7 s in all segments. The declustering potential was set to zero, the collision cell entrance and exit potential to 10 and 12, respectively. Figure 1 shows the overview of experimental workflow and schematic representation of the MRM technique.
Data analysis
The profiles from endogenous peptides were digitally transformed (Savitzky-Golay smoothing) and evaluated based on the following criteria: RT deviation from the synthetic standard should be ≤1 min, overlapping peak profiles of the transitions, intensity ratio of the monitored transitions and the peak shape comparable to the corresponding synthetic standard, signal intensity >1000 arbitrary units and the signal-to-noise ratio >10. The most sensitive criteria were the overlapping peak profiles and corresponding intensity ratio. All peptides passing these criteria were selected for further analysis. Their sequence and location within the protein sequence is provided in Supplementary Tables 3 and 4, respectively. The chromatograms can be downloaded from the Dryad Digital Repository.
To account for variations in the efficiency of sample digestion and peptide recovery after desalting, as well as for variations between injections, the signal intensities of the target peptides were normalized. For the samples from early life stages (embryos and larvae), an intense and stably expressed housekeeping protein, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was used for normalization. However, within the organs of adult male and female fish, the expression levels of all tested housekeeping proteins (GAPDH, beta-actin and 40S ribosomal protein S18) were organ dependent and therefore could not be used for normalization across different tissues. For this reason, the signal intensities of the target peptides within the adult organs were normalized to a standard protein, apomyoglobin from equine skeletal muscle (Sigma-Aldrich, USA), spiked before digestion. Normalized peak areas of peptides detected in zebrafish embryo and organs of adult zebrafish are summarized in Supplementary Tables 5 and 6, respectively.
RESULTS
We established detection methods for a set of peptides in order to analyze GST classes and GST isoenzymes in zebrafish (GST). Out of 86 peptides preselected according to the criteria described in “Materials and Methods”, 34 peptides could be detected in zebrafish samples, covering all classes of cytosolic GSTs. Eight of those peptides were detected only in tissues of adult zebrafish and with low intensity (Supplementary Table 2). The observed peptides were of 2 types: proteotypic and shared peptides. Proteotypic peptides cover nonconserved protein regions and allow the differentiation of isoenzymes with high-sequence homology. In this way, we were able to unequivocally identify GST isoenzymes belonging to the GST classes alpha (Gsta1, Gsta2), theta (Gstt1a, Gstt1b), mu (Gstm3), pi (Gstp1), rho (Gstr), and omega (Gsto1 and Gsto2) (Figure 2, Supplementary Table 2). The shared peptides cover conserved sequences of 2 or 3 isoenzymes and enable the monitoring of GST class-specific protein expression. We were able to detect shared peptides for the GST classes alpha, zeta, mu, and pi (Figure 2, Supplementary Table 2). GST isoenzymes that share the peptide sequence are indicated through the isoenzyme number separated by comma (eg, peptide shared between isoenzymes Gsta1 and Gsta2 would be indicated as Gsta1, 2).
GST Classes and Isoenzymes Show Distinct Expression Patterns Throughout Zebrafish Development
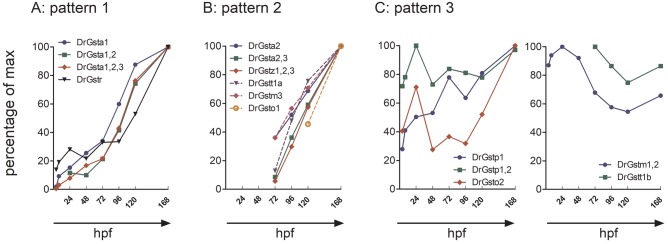
By comparing the time-resolved abundance of cytosolic GSTs, we identified 3 distinct expression patterns: (1) early detection followed by continuous increase throughout the development (Figure 3A), (2) first occurrence after 72 hpf, followed by a continuous increase (Figure 3B), and (3) variable trend with no consistent change (Figure 3C). The expression pattern of shared peptides generally was similar to the expression of 1 dominating isoenzyme. For instance, the expression patterns of the shared peptides Gsta1, 2, 3 and Gsta1, 2 were similar to those exhibited by Gsta1 whereas Gsta2, 3 showed a pattern comparable to Gsta2 (Figure 3).
Figure 3.
GST expression patterns observed during zebrafish early development. A, Early detection followed by continuous increase throughout the development (pattern 1). B, First occurrence after 72 hpf, followed by an increase (pattern 2), and (C), variable trend with no consistent change (pattern 3). Median of the normalized peak area is shown as percentage of the maximal value.
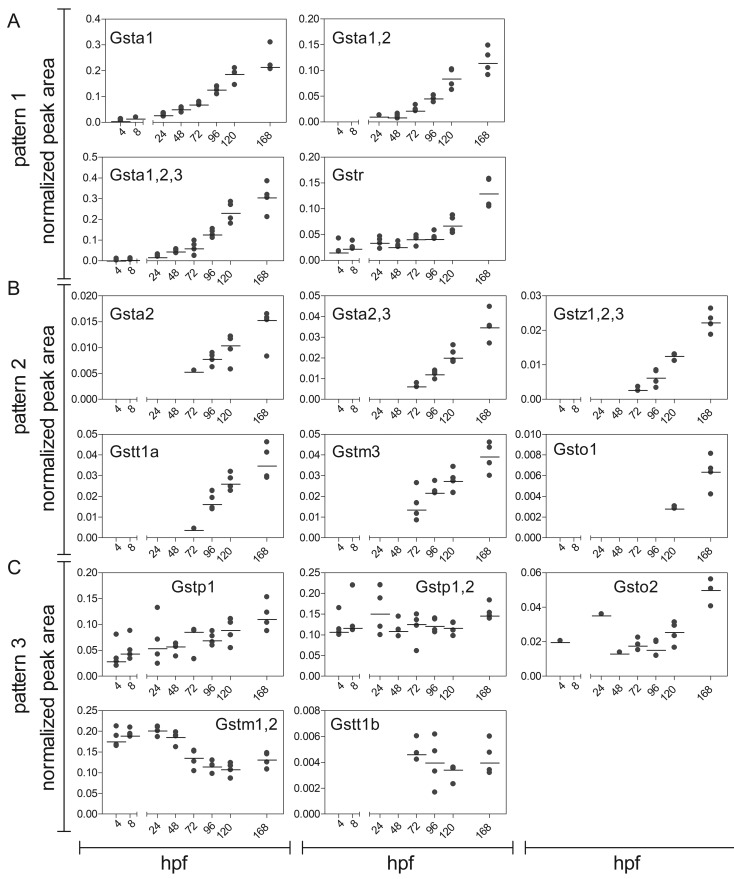
Members of the class alpha and Gstr followed the expression pattern (1) (Figure 4A). Gsta1, Gsta1, 2, 3, and Gstr were detected as early as 4 hpf and their expression level increased throughout the development. In case of Gsta1, Gsta1, 2, and Gsta1, 2, 3 we observed a strong increase in the expression already after 24 hpf, whereas the difference between 120 and 168 hpf was less distinct. For Gstr, the expression increased slowly but continuously until 96 hpf, and only after 96 hpf a sharp increase in the expression level was observed.
Figure 4.
GST expression in zebrafish embryos and larvae. The graphs are sorted in accordance to the expression patterns described in Figure 3. Data are shown as peak area normalized to the housekeeping protein GAPDH at 4, 8, 24, 48, 72, 96, 120, and 168 hpf. Each replicate (a sample of 60 pooled embryos) is shown in addition to the median (black line). For visualization, the normalized peak area of peptides belonging to the same enzyme (in case of proteotypic peptides) or several isoenzymes from the same class (in case of shared peptides) were cumulated. The number and characteristics of the cumulated peptides are summarized in Supplementary Tables 2 and 5.
The majority of GSTs (Gsta2, Gsta2, 3, Gstz1, 2, 3, Gstt1a, Gstm3, and Gsto1) could be assigned to the expression pattern (2) (Figure 4B). Those enzymes were first detected after hatching (72 hpf) and their expression subsequently increased.
In some cases, the GST expression showed no consistent or strong change with age (Figure 4C). Gstp1 and Gstp1, 2 were detected already at 4 hpf in all replicates. Whereas Gstp1 expression increased slightly over time, Gstp1, 2 showed a steady expression throughout the development. Gsto2 was detected at 4, 24, and 48 hpf in 1 replicate each; at 72 and 96 hpf the expression level stayed constant and increased only after 120 hpf.
The expression level of Gstm1, 2 appeared to be highly dynamic. It was detected at 4 hpf, decreased at 72 hpf and stayed at a constant level thereafter. However, this dynamic trend could only be observed for 1 peptide that covered a conserved sequence of Gstm1 and 2. The second shared peptide was observed only after hatching (72 hpf) and expression continuously increased until 168 hpf (Figure 4 C, Supplementary Table 5). Gstt1b was first observed after hatching and was expressed at a constant level until 168 hpf.
Altogether, representatives of 4 classes (alpha, mu, pi, and rho) could be detected as early as 4 hpf in more than 1 replicate. Representatives of the remaining classes (zeta, theta, and omega) were expressed after hatching (72 hpf). In general, the expression level of most GST classes increased throughout the zebrafish development.
GST Classes and Isoenzymes Show Distinct Expression Patterns in Organs of Adult Zebrafish
The majority of cytosolic GSTs were expressed in all examined organs of both male and female adult zebrafish (liver, intestine, gill, gonads, brain, and kidney). However, Gstt1a was not observed in the brain, and Gstz1, 2 was not detected in the intestine (Figure 5).
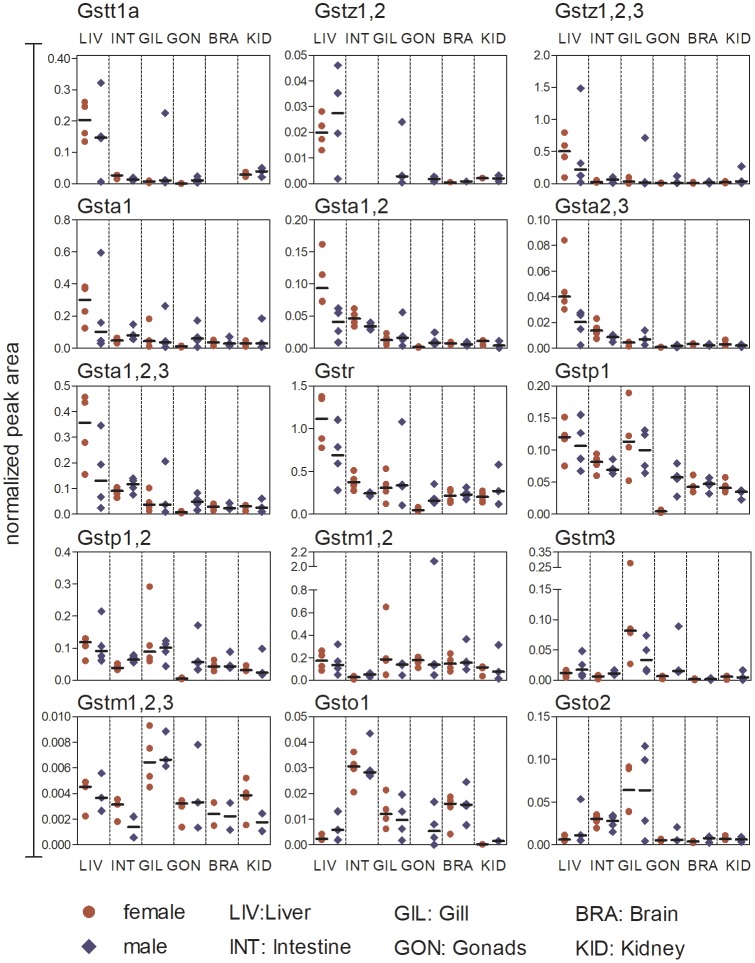
Figure 5.
GST expression in different organs (liver, intestine, gill, gonads, brain, and kidney) of adult female and male zebrafish. Data are shown as peak area normalized to the spiked synthetic standard apomyoglobin. Each replicate (a sample of organs pooled from 4 fish) is shown in addition to the median (black line). For visualization, the normalized peak area of peptides belonging to the same enzyme (in case of proteotypic peptides) or several isoenzymes from the same class (in case of shared peptides) were cumulated. The number and characteristics of the cumulated peptides is summarized in Supplementary Tables 2 and 6.
Differences in the class- and isoenzyme-specific expression level among organs of adult zebrafish were distinguishable despite the partly high variability of GST expression in replicates. All representatives of the alpha-class (except Gsta2), as well as Gstt1a, Gstz1, 2, Gstz1, 2, 3, and Gstr, were predominantly expressed in the liver. The members of the pi-class showed a higher expression in liver and gill compared with other organs. The highest expression of Gstm3, Gstm1, 2, 3, and Gsto2 was observed in the gills, whereby the expression of Gsto2 was also elevated in the intestine. Gsto1 was predominantly expressed in the intestine. In contrast, the expression of Gstm1, 2 in the intestine was below the median of other organs (Figure 5).
The signal intensity of Gsta2 and Gstt1b (Supplementary Table 6) was low, and in some organs the proteins could be observed in only 1 replicate. Therefore, it was not possible to estimate the organ-specific expression of these enzymes.
We did not observe strong sex-dependent differences in the GST expression within most organs. Only female gonads showed a generally low expression of all measured GSTs, with the exception of Gstm1, 2. The isoenzyme Gsto1 and the shared peptide Gstz1, 2 were even below the limit of detection in female gonads.
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.5s32v
DISCUSSION
We present the protein expression of cytosolic GSTs during zebrafish development and in organs of adult zebrafish. With the use of targeted proteomics, we are now able to distinguish selected GST isoenzymes despite their high-sequence identity. By monitoring conserved protein regions, we can also analyze the cumulative expression of enzymes belonging to the same GST class.
Some cytosolic GST family members are involved in the inactivation of endogenous compounds and cell signaling processes (Adler et al., 1999; Cho et al., 2001; Singhal et al., 2015); their expression might be relevant for embryogenesis and post hatching development. Considering that GSTs have a broad and overlapping substrate specificity (Glisic et al., 2015; Mannervik and Danielson, 1988), GST family members with a putative role in endogenous processes may also accept xenobiotics as substrates and be of significance for the protection of the embryo against natural toxins and xenobiotics. Using recombinant enzymes, a previous study demonstrated that cytosolic GST classes catalyze the GSH conjugation with model substrates CDNB and monochlorobimane (MCB), although at different turnover rates (Glisic et al., 2015). The class pi enzymes (Gstp1, Gstp2) and Gstt1a showed the highest enzyme efficiencies towards CDNB and MCB, respectively. However, to conclude on the role each isoenzyme plays in xenobiotic defense processes, not only the enzyme efficiency, but also its expression level at the respective life stage needs to be considered. Therefore, mapping of GST expression during embryogenesis as well as in tissues of adults is necessary in order to ascribe GST isoforms to the biotransformation of xenobiotics.
GST Expression in Early Life Stages of Zebrafish Reflects Important Developmental Events
Prior to the activation of the embryonic genome, the embryo completely relies on the maternally deposited gene products. In zebrafish, maternal mRNA drives cellular processes within the first 3 h after fertilization (Tadros and Lipshitz, 2009). Maternal deposition of GST mRNA transcripts and the presence of all cytosolic GST classes within the first 4 hpf have been demonstrated in previous studies (Glisic et al., 2016; Timme-Laragy et al., 2013). Our study on the protein level confirms the expression of GST classes alpha, mu, pi, and rho as early as 4 hpf. GST enzymatic activity was also detected within the first 4 h of zebrafish development (Notch et al., 2011; Otte et al., 2017; Wiegand et al., 2000). These findings indicate that GST enzymes are not only expressed but also active and capable of catalyzing GSH conjugation reactions with xenobiotic compounds already within the first hours of embryogenesis. Apart from GSH conjugation reactions, GSTs have been shown to interact non-catalytically with different ligands such as kinases (Townsend and Tew, 2003; Sheehan et al., 2001) and perform isomerase and peroxidase reactions (Hurst et al., 1998; Johansson and Mannervik, 2001) which may be of importance for early developmental processes.
During zebrafish embryogenesis, the heart forms as the first organ. The zebrafish heart tube starts to beat by 22 hpf and after 24 hpf blood circulation begins (Stainier et al., 1993). At this stage, enzymes of the classes alpha, mu, pi, and rho are expressed, but only Gsta1 shows an increase in the expression. Interestingly, Brox et al. (2016) reported a potential GST biotransformation product of clofibric acid at 28 hpf, indicating that GSTs expressed by the first day of zebrafish development are already active.
The majority of cytosolic GSTs were first observed after 72 hpf followed by a slight increase in expression level. In agreement with this, cytosolic GST enzyme activity experiences an increase after 72 hpf (Otte et al., 2017; Wiegand et al., 2000). Within the first 72 h, the major organ patterning has been completed. (Kimmel et al., 1995). Thus, during the major organ development, only 4 GST classes (alpha, mu, pi, and rho) catalyze the endogenous reactions and protect the embryo from environmental stressors. After 72 hpf, organs, such as liver and intestine, enter the growth phase and continue to develop into fully vascularized and functional organs (Field et al., 2003; Kimmel et al., 1995; Ng et al., 2005). The development of liver and intestine thus clearly correlates with an increase in the expression of most GST isoenzymes and classes.
The 72 hpf also marks the end of the hatching period (42–72 hpf) and the transition of the zebrafish embryo to the free swimming stage (eleutheroembryo) (Kimmel et al., 1995). Timme-Laragy et al., (2013) shows that hatching is associated with changes in the balance of reduced and oxidized GSH (GSH and GSSG, respectively) resulting in a more negative, ie, more reducing, redox potential. It is thus possible that the increase in posthatch GSH levels are linked to the increase in expression levels of some GST family members. It is conceivable that the interplay of GST expression and change in redox state of the organism is important in the protection of the organism from an increase in aerobic metabolism as the embryo enters a period of dynamic growth with an increase in cell proliferation.
One enzyme class, the pi-class, is constantly expressed throughout early zebrafish development. However, we were only able to detect isoenzyme Gstp1 as well as shared peptides for the class pi enzymes. Although Glisic et al., (2015) showed that Gstp2 biotransforms CDNB with the highest enzyme efficiency compared with other analyzed GSTs, its constitutive expression is low during zebrafish development. In contrast, Gstp1, the enzyme with the second highest efficiency for CDNB (Glisic et al., 2015), is present in all analyzed zebrafish life stages and might be dominating the activity assays.
Finally, Gstm1, 2 showed a highly variable expression pattern. However, as this dynamic trend could only be observed for 1 peptide, we cannot distinguish if the observed pattern reflects changes in the enzyme expression or if it is caused by changes in posttranslational modifications over time.
GST Expression in Adult Organs Is Organ-Specific and Variable Among Individuals
Knowledge regarding the organ-specific expression pattern of cytosolic GSTs is of importance, as the lack of individual GST classes can result in a predisposition of specific organs to damage by electrophilic compounds. Additionally, the expression level of specific GSTs in barrier tissues and principal organs of biotransformation provides information regarding the involvement of specific GST enzymes in xenobiotic defense mechanisms. Our study shows a constitutive expression of GST enzymes in all examined organs of zebrafish, indicating that GSH-conjugation is a xenobiotic defense mechanism functioning in all tissues. Nonetheless, some organs show a higher expression of selected GST family members in relation to others. Liver—the presumed main organ responsible for biotransformation—shows the highest protein expression of the alpha-class, zeta-class, Gstt1a and Gstr. Gills, barrier tissues directly exposed to chemicals at the water interface, show an elevated expression of Gstm3, Gstm1, 2, 3, and Gsto2. Overall, these expression patterns are comparable to those reported on the mRNA level by Glisic et al., (2015), with 1 exception: class alpha GSTs were present at low mRNA levels in the liver.
Although in some GST classes, such as in class alpha and zeta, the isoenzymes show consistent trends in tissue expression, this is not true for all GSTs. Gsto1 is predominately expressed in intestine and brain whereas Gsto2 expression is highest in the gills. A comparable expression pattern for Gsto1 was observed on the mRNA level (Glisic et al., 2015). Furthermore, Gstp1 is expressed in all analyzed organs and follows pattern (3) during embryo development, while Gstp2 was not detected. The expression pattern of Gstp1 is potentially indicative of constitutive expression of this isoenzyme. Consistent with this observation, mRNA analysis identified gstp1 as the predominant isoenzyme of the pi-class and its constitutive gene expression was demonstrated to be high in all zebrafish tissues (Glisic et al., 2015).
The expression of many GST classes is low in female gonads, which can be explained by the structure of the organ. A large proportion of the zebrafish ovarian proteome consists of vitellogenins, precursor proteins of egg yolk (Groh et al., 2011). Hence, other proteins are underrepresented in relation to egg yolk and thus, due to limited sensitivity, appear to be expressed at low levels with respect to other organs. In contrast to the sex-dependent mRNA expression of most GST enzymes (Glisic et al., 2015), no further gender differences are apparent within our dataset.
The variability in GST expression is more pronounced in sample replicates of adult zebrafish as compared with the expression data obtained from early life stages. For a better representation of the biological diversity within zebrafish, we chose a wild-type mix over an inbred strain. The wild-type mix chosen for its genetic diversity might explain the higher variability in the adults when compared with the embryos, also because of the limited pool involved (pool of 4 adults vs pool of 60 embryos).
GSTs Expressed in Zebrafish Show Similarities to Humans
Being able to study the expression of GST classes and isoenzymes is of great value because some of their polymorphic variations may be informative about an individual’s susceptibility to develop diseases, such as cancer. In addition, information about GST expression can help to predict a patient’s ability to respond to certain drug treatments (Hollman et al., 2016; McIlwain et al., 2006). Among all members of the GST family, 4 cytosolic classes (GST class alpha, theta, mu, and pi) have been in the center of attention for the last decade, due to their role in antioxidation processes and detoxification of therapeutic drugs, carcinogens as well as environmental pollutants (Hollman et al., 2016). Therefore, it is important to compare the known expression of human GSTs with that determined in zebrafish.
In human embryogenesis, cytosolic classes, alpha, mu and pi are already expressed (Raijmakers et al., 2001). Similar to human data, GST classes alpha, mu, and pi were expressed very early during zebrafish embryogenesis, indicating that those classes may be most critical for the protection and functioning of cells during early phases of vertebrate development. In addition to the human orthologues, zebrafish embryos express a GST enzyme designated to the class rho. Class rho is an evolutionarily distinct member of the GST family that is present in teleosts and cephalo chordate (Glisic et al., 2015). It is assumed to play a role in microcystein toxicity and shows reactivity towards some model substrates (Glisic et al., 2015; Hao et al., 2009; Liang et al., 2007).
In human adult liver, GSTA1 is the dominant form. In addition, classes mu and theta are reported to be expressed in the liver of healthy adults (Coles and Kadlubar, 2003; Mainwaring et al., 1996; Rowe et al., 1997). Similarly, class alpha and theta in zebrafish have a pronounced expression in the liver, indicating conserved roles of these GST-classes in fish and mammals.
GSTP1 is expressed in most adult human tissues including digestive, urinary and respiratory organs (Schnekenburger et al., 2014). Similarly, class pi was expressed in all zebrafish organs with an elevated level in the liver, intestine and gill.
All cytosolic GST classes are present on the protein level during zebrafish development and in organs of adults. The early expression of GSTs during zebrafish embryogenesis and the similarities to humans support the use of zebrafish as model in research applications that depend on functional biotransformation pathways. The targeted proteomics methods developed within this study allow to determine specific isoenzymes of the GST classes, thereby opening new avenues for understanding the role of GSTs in endogenous processes and upon exposure to chemicals.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
We thank M Zimmermann and P Reichlin from the Department of Environmental Toxicology (Eawag, Switzerland) for the assistance in maintaining the zebrafish facility.
FUNDING
This work was supported by a discretionary fund of the Swiss Federal Institute of Aquatic Science and Technology (Eawag).
REFERENCES
- Abunnaja M. S., Kurogi K., Mohammed Y. I., Sakakibara Y., Suiko M., Hassoun E. A., Liu M. C. (2017). Identification and characterization of the zebrafish glutathione S-transferase Pi-1. J. Biochem. Mol. Toxicol. 31, doi: 10.1002/jbt.21948, 10.1002/jbt.21948. [DOI] [PubMed] [Google Scholar]
- Adler V., Yin Z., Fuchs S. Y., Benezra M., Rosario L., Tew K. D., Pincus M. R., Sardana M., Henderson C. J., Wolf C. R. et al. , (1999). Regulation of JNK signaling by GSTp. Embo J. 18, 1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereman M. S., MacLean B., Tomazela D. M., Liebler D. C., MacCoss M. J. (2012). The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics 12, 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best J. H., Pflugmacher S., Wiegand C., Eddy F. B., Metcalf J. S., Codd G. A. (2002). Effects of enteric bacterial and cyanobacterial lipopolysaccharides, and of microcystin-LR, on glutathione S-transferase activities in zebra fish (Danio rerio). Aquat. Toxicol. 60, 223–231. [DOI] [PubMed] [Google Scholar]
- Brox S., Seiwert B., Haase N., Kuster E., Reemtsma T. (2016). Metabolism of clofibric acid in zebrafish embryos (Danio rerio) as determined by liquid chromatography-high resolution-mass spectrometry. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 185–186, 20–28. [DOI] [PubMed] [Google Scholar]
- Cho S. G., Lee Y. H., Park H. S., Ryoo K., Kang K. W., Park J., Eom S. J., Kim M. J., Chang T. S., Choi S. Y. et al. , (2001). Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 276, 12749–12755. [DOI] [PubMed] [Google Scholar]
- Christen V., Fent K. (2014). Tissue-, sex- and development-specific transcription profiles of eight UDP-glucuronosyltransferase genes in zebrafish (Danio rerio) and their regulation by activator of aryl hydrocarbon receptor. Aquat. Toxicol. 150, 93–102.http://dx.doi.org/10.1016/j.aquatox.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Coles B. F., Kadlubar F. F. (2003). Detoxification of electrophilic compounds by glutathione S-transferase catalysis: Determinants of individual response to chemical carcinogens and chemotherapeutic drugs? (Reprinted from Thiol Metabolism and Redox Regulation of Cellular Functions). Biofactors 17, 115–130.http://dx.doi.org/10.1002/biof.5520170112 [DOI] [PubMed] [Google Scholar]
- Dahm R., Geisler R. (2006). Learning from small fry: The zebrafish as a genetic model organism for aquaculture fish species. Mar. Biotechnol. 8, 329–345.http://dx.doi.org/10.1007/s10126-006-5139-0 [DOI] [PubMed] [Google Scholar]
- Field H. A., Ober E. A., Roeser T., Stainier D. Y. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279–290. [DOI] [PubMed] [Google Scholar]
- Glisic B., Hrubik J., Fa S., Dopudj N., Kovacevic R., Andric N. (2016). Transcriptional profiles of glutathione-S-Transferase isoforms, Cyp, and AOE genes in atrazine-exposed zebrafish embryos. Environ. Toxicol. 31, 233–244. [DOI] [PubMed] [Google Scholar]
- Glisic B., Mihaljevic I., Popovic M., Zaja R., Loncar J., Fent K., Kovacevic R., Smital T. (2015). Characterization of glutathione-S-transferases in zebrafish (Danio rerio). Aquat. Toxicol. 158, 50–62. [DOI] [PubMed] [Google Scholar]
- Goldstone J. V., McArthur A. G., Kubota A., Zanette J., Parente T., Jonsson M. E., Nelson D. R., Stegeman J. J. (2010). Identification and developmental expression of the full complement of cytochrome P450 genes in Zebrafish. Bmc Genomics 11, 643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh K. J., Nesatyy V. J., Segner H., Eggen R. I. L., Suter M. J. F. (2011). Global proteomics analysis of testis and ovary in adult zebrafish (Danio rerio). Fish Physiol. Biochem. 37, 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh K. J., Schonenberger R., Eggen R. I. L., Segner H., Suter M. J. F. (2013). Analysis of protein expression in zebrafish during gonad differentiation by targeted proteomics. Gen. Comp. Endocr. 193, 210–220. [DOI] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. (1974). Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139. [PubMed] [Google Scholar]
- Hao L., Xie P., Fu J., Li G. Y., Xiong Q., Li H. Y. (2009). The effect of cyanobacterial crude extract on the transcription of GST mu, GST kappa and GST rho in different organs of goldfish (Carassius auratus) (vol 90, pg 1, 2008). Aquatic Toxicology 911, 99–99. [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Flanagan J. U., Jowsey I. R. (2005). Glutathione transferases. Annu. Rev. Pharmacol. 45, 51–88.http://dx.doi.org/10.1146/annurev.pharmtox.45.120403.095857 [DOI] [PubMed] [Google Scholar]
- Hollman A. L., Tchounwou P. B., Huang H. C. (2016). The association between gene-environment interactions and diseases involving the human GST superfamily with SNP variants. Int. J. Environ. Res. Public Health 13, 379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst R., Bao Y., Jemth P., Mannervik B., Williamson G. (1998). Phospholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases. Biochem J. 332, 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson P. J., Morgenstern R., Mancini J., Ford-Hutchinson A., Persson B. (2008). Common structural features of MAPEG - A widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein Sci. 8, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. S., Mannervik B. (2001). Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J. Biol. Chem. 276, 33061–33065.http://dx.doi.org/10.1074/jbc.M104539200 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of Embryonic-Development of the Zebrafish. Dev. Dynam. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Krewski D., Acosta D. Jr, Andersen M., Anderson H., Bailar J. C. 3rd, Boekelheide K., Brent R., Charnley G., Cheung V. G., Green S. Jr et al. , (2010). Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 13, 51–138. 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. F., Li G. G., He S., Huang Y. (2007). Transcriptional responses of alpha- and rho-class glutathione S-Transferase genes in the liver of three freshwater fishes intraperitoneally injected with Microcystin-LR: Relationship of inducible expression and tolerance. J Biochem Mol Toxic 215, 289–298. [DOI] [PubMed] [Google Scholar]
- Lange V., Picotti P., Domon B., Aebersold R. (2008). Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Bickel P. J., Biggin M. D. (2014). System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2, e270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainwaring G. W., Williams S. M., Foster J. R., Tugwood J., Green T. (1996). The distribution of theta-class glutathione S-transferases in the liver and lung of mouse, rat and human. Biochem J. 318, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik B., Helena Danielson U., Ketterer B. (1988). Glutathione transferases–structure and catalytic activity. CRC Crit. Rev. Biochem. 23, 283–337.http://dx.doi.org/10.3109/10409238809088226 [DOI] [PubMed] [Google Scholar]
- McIlwain C. C., Townsend D. M., Tew K. D. (2006). Glutathione S-transferase polymorphisms: Cancer incidence and therapy. Oncogene 25 25, 1639–1648.http://dx.doi.org/10.1038/sj.onc.1209373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. N., de Jong-Curtain T. A., Mawdsley D. J., White S. J., Shin J., Appel B., Dong P. D., Stainier D. Y., Heath J. K. (2005). Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol. 286, 114–135. [DOI] [PubMed] [Google Scholar]
- Notch E. G., Miniutti D. M., Berry J. P., Mayer G. D. (2011). Cyanobacterial LPS potentiates cadmium toxicity in zebrafish (Danio rerio) embryos. Environ. Toxicol. 26, 498–505. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Dahm R. (2002). Zebrafish: A Practical Approach. New York: Oxford University Press. [Google Scholar]
- Otte J. C., Schultz B., Fruth D., Fabian E., van Ravenzwaay B., Hidding B., Salinas E. R. (2017). Intrinsic xenobiotic metabolizing enzyme activities in early life stages of zebrafish (Danio rerio). Toxicol. Sci. 159, 86–93. [DOI] [PubMed] [Google Scholar]
- Pavagadhi S., Gong Z., Hande M. P., Dionysiou D. D., de la Cruz A. A., Balasubramanian R. (2012). Biochemical response of diverse organs in adult Danio rerio (zebrafish) exposed to sub-lethal concentrations of microcystin-LR and microcystin-RR: A balneation study. Aquat. Toxicol. 109, 1–10. [DOI] [PubMed] [Google Scholar]
- Picotti P., Aebersold R. (2012). Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566.http://dx.doi.org/10.1038/nmeth.2015 [DOI] [PubMed] [Google Scholar]
- Raijmakers M. T., Steegers E. A., Peters W. H. (2001). Glutathione S-transferases and thiol concentrations in embryonic and early fetal tissues. Hum. Reprod. 16, 2445–2450. [DOI] [PubMed] [Google Scholar]
- Rowe J. D., Nieves E., Listowsky I. (1997). Subunit diversity and tissue distribution of human glutathione S-transferases: Interpretations based on electrospray ionization MS and peptide sequence-specific antisera. Biochem J. 325, 481–486.http://dx.doi.org/10.1042/bj3250481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sau A., Tregno F. P., Valentino F., Federici G., Caccuri A. M. (2010). Glutathione transferases and development of new principles to overcome drug resistance. Arch. Biochem. Biophys. 500, 116–122. [DOI] [PubMed] [Google Scholar]
- Schnekenburger M., Karius T., Diederich M. (2014). Regulation of epigenetic traits of the glutathione S-transferase P1 gene: From detoxification toward cancer prevention and diagnosis. Front. Pharmacol. 5, ARTN 170 10.3389/fphar.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Sheehan D., Meade G., Foley V. M., Dowd C. A. (2001). Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16.http://dx.doi.org/10.1042/bj3600001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal S. S., Singh S. P., Singhal P., Horne D., Singhal J., Awasthi S. (2015). Antioxidant role of glutathione S-transferases: 4-Hydroxynonenal, a key molecule in stress-mediated signaling. Toxicol. Appl. Pharmacol. 289, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D. Y., Lee R. K., Fishman M. C. (1993). Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development 119, 31–40. [DOI] [PubMed] [Google Scholar]
- Surinova S., Huttenhain R., Chang C. Y., Espona L., Vitek O., Aebersold R. (2013). Automated selected reaction monitoring data analysis workflow for large-scale targeted proteomic studies. Nat. Protoc. 8, 1602–1619.http://dx.doi.org/10.1038/nprot.2013.091 [DOI] [PubMed] [Google Scholar]
- Townsend D. M., Tew K. D. (2003). The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 2247, 7369–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: A play in two acts. Development 136, 3033–3042. 10.1242/dev.033183.http://dx.doi.org/10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Thomson R. E., Bigley A. L., Foster J. R., Jowsey I. R., Elcombe C. R., Orton T. C., Hayes J. D. (2004). Tissue-specific expression and subcellular distribution of murine glutathione S-transferase class kappa. J. Histochem. Cytochem. 52, 653–662. [DOI] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Goldstone J. V., Imhoff B. R., Stegeman J. J., Hahn M. E., Hansen J. M. (2013). Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med. 65, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand C., Pflugmacher S., Oberemm A., Steinberg C. (2000). Activity development of selected detoxication enzymes during the ontogenesis of the zebrafish (Danio rerio). Int. Rev. Hydrobiol. 85, 413–422. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.